Abstract
Luteolin (3,4,5,7-tetrahydroxyflavone) is a natural flavonoid that has been found to exhibit anticancer properties in certain types of cancers. In the present study, the role of luteolin and its underlying mechanisms were explored in colorectal cancer (CRC) cells. First, the effects of luteolin on CRC cells proliferation, migration and invasion were examined by CCK-8, wound healing and Transwell assays, respectively. It was demonstrated that luteolin had no effects on CRC cells proliferation while inhibited cells migration and invasion both in vitro and in vivo. Then, expression of pleiotrophin (PTN) and miR-384 was detected in cells and CRC tissues by qPCR. Luteolin was found to upregulate miR-384 and downregulate PTN expressions both in CRC cells and tissues. miR-384 inhibition and PTN overexpression partially reversed the inhibition of HT-29 cells migration and invasion induced by luteolin. Target analysis revealed that miR-384 directly regulates PTN expression. The correlation analysis between PTN expression and clinical characteristics revealed that PTN expression was positively related to cancer progression. The present study demonstrated that luteolin exerts anticancer effects against CRC cells by modulating PTN via miR-384 expression suggested that PTN may serve as a promising candidate for therapeutic applications in CRC treatment.
Keywords: luteolin, colorectal cancer, miR-384, PTN, invasion, metastasis
Introduction
Luteolin (3,4,5,7-tetrahydroxyflavone) is a natural flavonoid abundant in various fruits and vegetables. Luteolin has been reported to display a wide range of bioactive properties, such as anti-virus (1,2), hepatoprotective (3), anti-inflammatory (4) and anti-diabetic effects (5). Recently, the antitumor effects of luteolin have been extensively investigated in breast and gastric cancer (6), melanoma (7), esophageal carcinoma (8), glioblastoma (9), hepatocellular carcinoma (10), placental choriocarcinoma (11) and lung adenocarcinoma (12). The role of luteolin in colorectal cancer (CRC) has also been investigated. Luteolin has been found to inhibit CRC cells proliferation, induce cells apoptosis and cell cycle perturbation (13–15). Liu et al also reported that luteolin was able to inhibit CRC cells epithelial to mesenchymal transition by suppressing CREB1 expression (16). Luteolin was demonstrated to play the protective roles in 1,2-dimethylhydrazine and azoxymethane (AOM)-induced experimental colon carcinogenesis (17,18). Moreover, luteolin increased the sensitivity of CRC cells to chemotherapy (19,20).
MicroRNAs (miRNAs) are a category of small, non-coding, single-stranded RNAs of 18–25 nucleotides in length which bind to the 3′-untranslated regions (3′-UTRs) of their target mRNAs. miRNAs regulate the expressions of target genes through post-transcriptional silencing or induction of degradation (21,22). Recent studies have revealed critical functions of miRNAs in tumor biological processes, including cell proliferation, differentiation, metastasis and chemosensitivity (23,24). By regulating a variety of target genes in cancer cells, miRNAs act as oncogenes or tumor suppressors.
Several studies have reported the involvement of miRNAs in the effects induced by luteolin. In prostate cancer cells, luteolin treatment inhibited prostate cancer cells proliferation and induced apoptosis through downregulation of miR-301 by triggering DEDD2 expression (25). Zhou et al found that luteolin upregulated miR-34 expression in gastric cancer cells and upregulation of miR-34 enhanced the susceptibility of cells to luteolin (6). Luteolin also inhibited the tumorigenesis and induced the apoptosis of non-small cell lung cancer cells by upregulation of miR-34a-5p by targeting MDM4 (26). Thus, we hypothesized that miRNAs play significant roles during luteolin treatment in CRC cells and this has not yet been investigated.
In the present study, it was observed that luteolin inhibited CRC cells migration and invasion both in vitro and in vivo while had no effects on cells proliferation. miR-384 expression was markedly upregulated and inhibition of miR-384 partly reversed the inhibitory effects induced by luteolin. However, the expression of pleiotrophin (PTN) was found to be downregulated after luteolin treatment and was confirmed to be a direct target of miR-384. By analyzing CRC tissues, we found that miR-384 expression was downregulated while PTN expression was upregulated compared with the paired-matched adjacent tissues. The correlation analysis between PTN expression and clinical characteristics revealed that PTN expression is positively related to the cancer progression. Hence, the present study demonstrated that luteolin exerts anticancer effects against CRC cells by modulating PTN via miR-384 expression suggesting that PTN may serve as a promising candidate for therapeutic applications in CRC treatment.
Materials and methods
Cells and reagents
HT-29, SW480, SW620 and LoVo human CRC cell lines were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). The normal human mucosal epithelial cell line NCM460 was purchased from the National Institute of Cells (Shanghai, China). All cells were maintained in RPMI-1640 medium (HyClone; GE Healthcare, Chicago, IL, USA) supplemented with 10% fetal bovine serum (FBS; Gibco™, Thermo Fisher Scientific, Inc., Waltham, MA, USA) and 1% penicillin-streptomycin in a humidified incubator containing 5% CO2 at 37°C. Luteolin was purchased from Sigma-Aldrich/Merck (Shanghai, China) and dissolved in dimethyl sulfoxide (DMSO).
CRC tissues and relative patient clinical information
We collected a total of 50 cases of CRC and matched normal tissues from patients who underwent surgery at Hangzhou Hospital of Traditional Chinese Medicine from May 2017 to May 2018. All postoperative pathology was determined to be colorectal adenocarcinoma. The following patient characteristics were recorded: age, sex, histologic differentiation, TNM stage and serum levels of carcinoembryonic antigen (CEA) before surgery and all the data are listed in Table I. Approval of the present study was obtained from the Ethics Committee of Hangzhou Hospital of Traditional Chinese Medicine and informed consent was acquired from all the patients.
Table I.
Clinicopathological characteristics of PTN expression in the human CRC cases.
| Expression of PTN | |||
|---|---|---|---|
| Characteristics | Low (n=25) | High (n=25) | P-value |
| Age (years) | 0.7761 | ||
| >60 | 15 | 13 | |
| ≤60 | 10 | 12 | |
| Sex | 0.3961 | ||
| Male | 14 | 10 | |
| Female | 11 | 15 | |
| Differentiation | 0.1963 | ||
| Well | 15 | 14 | |
| Moderate | 10 | 8 | |
| Poor | 0 | 3 | |
| T factor | 0.0477a | ||
| 1+2 | 16 | 9 | |
| 3+4 | 9 | 16 | |
| N factor | 0.0449a | ||
| 0 | 18 | 11 | |
| 1+2 | 7 | 14 | |
| M factor | 0.0414a | ||
| 0 | 19 | 12 | |
| 1 | 6 | 13 | |
| CEA (ng/ml) | 0.3686 | ||
| >5 | 13 | 9 | |
| ≤5 | 12 | 16 | |
P<0.05, significant results. PTN, pleiotrophin; CRC, colorectal cancer; CEA, carcinoembryonic antigen.
Cell viability assay
Cell proliferation ability was detected by Cell Counting Kit-8 (CCK-8) assay kit (Beyotime Institute of Biotechnology, Shanghai, China). In brief, CRC cells were plated in a 96-well plate at the initial density of 1,000 cells/well and cultured in medium without or with luteolin at concentrations of 0, 10, 50 and 100 µM for 24, 48 and 72 h. CCK-8 solutions were added into the cell cultures 2 h before harvesting. The optical density at 450 nm was measured using an ultra-microplate reader (EMax; Molecular Devices, Sunnyvale, CA, USA).
In vivo nude mouse study
Thirty-nine BALB/c nude mice (17–20 g) were utilized to evaluate the effects of luteolin on tumor growth and tumor metastasis in vivo. Five- to six-week-old female BABL/c nude mice were purchased from Shanghai Slac Animal (Shanghai, China) and maintained in the animal facility at the Zhejiang University (Hangzhou, China). Mice were provided with water and food ad libitum and kept under standard conditions (temperature 24±2°C, humidity, 50–70 %, 12-h light/dark cycle). The study received ethical approval from the Animal Care and Use Committee of Zhejiang University, and experiments and animal care were performed according to the approved protocols.
For the tumor growth assay, HT-29 cells (1×106) were subcutaneously injected into the right flanks of the nude mice. When the volumes of xenograft tumors reached an average of 100 mm3, the mice were randomly divided into two groups: the control group and the luteolin group with 10 mice in each group (20 mice in total). Mice received luteolin were intragastrically administered with luteolin at 100 mg/kg every 2 days for 30 days. The control group mice received the same volume of phosphate-buffered saline (PBS). Tumor measurements were performed with a caliper by measuring the largest diameter and its perpendicular length every 3 days. According to the rules of our ethics committee, humane endpoints set for the tumor experiments were as follows: the tumor weight was >10% of the original body weight or the average tumor diameter was >20 mm in mice. Thus, when the tumor almost reached 20 mm, the experiment was terminated. The mice were sacrificed by cervical dislocation, and tumors were harvested and weighed on day 30. Tumor volume was calculated using the following formula: V = ½ × (length × width2). To note, we repeated this experiment for the same setting for three times (8 mice for the first experiment, and (6 mice for the other two experiments).
For liver metastasis assay, mice (n=19) were anesthetized with 75 mg/kg pentobarbital by intraperitoneal injection. Mice were fixed and then a small left abdominal flank incision (1 cm) was made and the spleen was exteriorized for the intrasplenic injection. HT-29 cells (5×106) suspended in 100 µl ice-cold PBS were injected into the spleen with a 27-gauge needle. A sterile cotton was held over the site of injection for 30 sec to prevent tumor cell leakage and bleeding. The injected spleen was returned to the abdomen and the wound was sutured with 5-0 black silk. Mice receiving luteolin were intragastrically administered with luteolin at 100 mg/kg every 2 days for 30 days. The control mice received the equal volume of PBS. After 3 weeks, the mice were sacrificed by cervical dislocation, spleen and liver were removed and observed. The liver metastatic tumors on the surface were calculated and H&E staining was performed. H&E staining was performed at Servicebio Co., Ltd. (Wuhan, China). To note, we also repeated this experiment three times (6 mice for the first two experiments and 7 mice for the third trial).
Wound healing assay
HT-29 cells were seeded in 24-well plate until reaching 80% confluency in the complete medium. The monolayer was wounded using a sterile 200-µl micropipette tip. The cells were washed with PBS three times to rinse off the detached cells. Cells were incubated for 48 h in 2% FBS medium containing the indicated concentrations of luteolin. Images of the wound morphology were acquired under a light microscopy at ×100 magnification.
Transwell migration and invasion assays
Cell migration and invasion potential were assessed using 8-µm Transwell chambers (Corning Costar, Corning, NY, USA). Briefly, 8×104 CRC cells suspended in 200 µl serum-free medium treated with luteolin or not were seeded in the upper chambers with the lower chamber filled with 600 µl of medium supplemented with 20% FBS. For the invasion assay, the upper surfaces of the membranes were coated with 50 µl Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) 6 h before cells were seeded. Cells were allowed to migrate for 36 h before being fixed with ice-cold methanol and stained with 0.1% crystal violet for 30 min. Cells that passed through the membrane were photographed and counted with an Olympus CKX41 light microscope (Olympus, Shanghai, China; magnification, ×100).
Western blotting
Cells or CRC tissues for protein extraction were lysed with RIPA buffer (Beyotime Institute of Biotechnology). Protein concentration was determined by BCA assay (Cwbiotech, Beijing, China). Total proteins (30 µg) were separated by 10% SDS-PAGE gels and transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were incubated with primary antibodies against human MMP-16 (cat. no. E-AB-32127), MMP-2 (cat. no. 10373-2-AP), MMP-3 (cat. no. 17873-1-AP), MMP-9 (cat. no. 10375-2-AP), PTN (cat. no. 27117-1-AP) (all diluted 1:1,000) and GAPDH (cat. no. 60004-1-Ig) (dilution 1:5,000) at 4°C overnight. Subsequently, the membranes were incubated with the peroxidase (HRP)-conjugated goat anti-rabbit (dilution 1:5,000; cat. no. SA00001-2) or goat anti-mouse (dilution 1:5,000; cat. no. SA00001-1) secondary antibody at room temperature for 2 h. MMP-16 was purchased from Elabscience (Shanghai, China). All the other antibodies were purchased from Proteintech Group Inc. (Chicago, IL, USA). The protein bands were detected with FluorChem E System (ProteinSimple, Santa Clara, CA, USA).
RNA isolation and quantitative real-time PCR (qPCR)
Total RNA was extracted from the cells or the surgically resected CRC and adjacent non-tumor tissues using TRIzol reagent (Thermo Fisher Scientific, Inc.). Then, 1 µg of total RNA was reverse transcribed using the HiFiScript cDNA Synthesis kit or miRNA cDNA Synthesis kit (Cwbiotech) according to the manufacturer's instructions. The PTN expression levels were determined with UltraSYBR Mixture (Cwbiotech) and the miR-384 levels were determined with miRNA qPCR Assay Kit (Cwbiotech) on CFX96 Touch Real-Time PCR Detection system (Bio-Rad Laboratories, Hercules, CA, USA). The miR-384 and U6 primers were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China), and the PTN and β-actin primers were synthesized by Thermo Fisher Scientific, Inc. The primers were as follows: miR-384 forward, 5′-TGTTAAATCAGGAATTTTAA-3′ and reverse, 5′-TGTTACAGGCATTATGAA-3′; U6 forward, 5′-CTCGTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; PTN forward, 5′-ACCAGTGAGTCATCCGTCCA-3′ and reverse, 5′-TGCAAATTTTCGACGCTGCT-3′; β-actin forward, 5′-GTATCCTGACCCTGAAGTACC-3′ and reverse, 5′-TGAAGGTCTCAAACATGATCT-3′. The thermocycling conditions were as follows: initial denaturation at 95°C for 10 min, followed by 40 cycles of 15 sec at 95°C and 1 min at 60°C. Relative fold-change in expression of miR-384 and PTN was normalized to U6 and β-actin expression, respectively. The relative expression was analyzed using the 2−∆∆Cq (27) method.
Plasmid, miR-384 mimic, miR-384 inhibitor and transfection
miR-384 mimic, miR-384 inhibitor and its relative control miRNAs were synthesized by Shanghai GenePharma Co., Ltd. The sequences are as follows (28): miR-384 mimic: 5′-AUUCCUAGAAAUUGUUCAUA-3′, miR-384 inhibitor: 5′-UAUGAACAAUUUCUAGGAAU-3′, miR-384 mimic NC: 5′-UUCUCCGAACGUGUCACGU-3′, miR-384 inhibitor NC: 5′-CAGUACUUUUGUGUAGUACAA-3′. Full length clone DNA of human pleiotrophin was obtained from Sino Biological Inc. (Shanghai, China). Transfections were performed using Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions.
Mapping of prediction algorithm data
Target genes for miR-384 were predicted using miRDB (http://www.mirdb.org) and TargetScan v.5.1 (http://www.targetscan.org/). From the TargetScan, we found that PTN ranked the top 2 genes that was the targets of miR-384. We then confirmed it in the miRDR database and found that PTN ranked 86. Thus, we chose PTN as one candidate of miR-384 target genes.
Luciferase reporter assay
Wild-type (WT) or the mutated type (MUT) of PTN-3′-UTR containing miR-384 binding sites was synthesized by Shanghai GenePharma Co., Ltd., and cloned downstream of the luciferase reporter gene. HT-29 cells were seeded into 24-well plates (5×104 cells/well) one day before and then co-transfected with the WT PTN-3′-UTR or the MUT PTN-3′-UTR vectors (500 ng) and the miR-384 mimics or negative controls with Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, Inc.). After 48 h of transfection, cells were harvested, and the luciferase activity was detected with the Dual-Luciferase Reporter Assay system (Promega Corporation, Madison, WI, USA).
Oncomine database
Oncomine database (http://www.oncomine.org) was selected to search the PTN expression levels between the CRC and normal groups.
Statistical analysis
Data are expressed as means ± SEM. Statistical significance was determined by Student's t-test or one-way analysis of variance (ANOVA) followed by Dunnett's test. The relationship between the PTN expression and the CRC clinical features was analyzed with the Chi-square test. P-value <0.05 was considered as indicative of statistical significance.
Results
Luteolin inhibits CRC cells migration and invasion both in vitro and in vivo
In order to explore the functions of luteolin in CRC, we first detected HT-29 and SW480 cells proliferation after luteolin treatment by CCK-8 assay. The results showed that luteolin had no effects on CRC cells proliferation (Fig. 1A). We then examined cells migration and invasion by Transwell and wound healing assays. As shown in Fig. 1B, we observed that luteolin-treated HT-29 and SW620 cells displayed significantly reduced cells migration and invasion. Consistently, luteolin treatment induced a slower closing of scratch wounds in the HT-29 cells (Fig. 1C), indicating that luteolin inhibited cells migratory ability. We also detected the expressions of several matrix metalloproteinases (MMPs), which are strongly related with tumor migration and invasion and overexpressed in human CRCs. As shown in Fig. 1D, luteolin obviously downregulated MMP-2, MMP-3, MM-9 and MMP-16 expressions.
Figure 1.
Luteolin inhibits CRC cells migration and invasion in vitro and in vivo. (A) HT-29 and SW480 cells were treated with luteolin at the concentrations of 10, 50 and 100 µM. CCK-8 assay was performed after culturing for 24, 48 and 72 h. (B) Migration and invasion capacity of HT-29 and SW480 cells after treated with luteolin were determined and representative images were taken. Original magnification, ×100. (C) Monolayers of HT-29 cells were wounded by a tip and treated with the indicated concentrations of luteolin. The representative images of the wound healing were taken at 0, 6, 12 and 24 h after the scratch was created. Original magnification, ×100. (D) Expressions of MMP-2, MMP-3, MMP-9 and MMP-16 in HT-29 and SW480 cells were detected by western blotting after stimulation with luteolin for 24 h. Results were obtained from 3 independent experiments and are expressed as the means ± SEM. Statistical significance was determined by one-way analysis of variance (ANOVA) followed by Dunnett's test. *P<0.05, **P<0.01, ***P<0.001. CRC, colorectal cancer; CCK-8, Cell Counting Kit-8; DMSO, dimethyl sulfoxide.
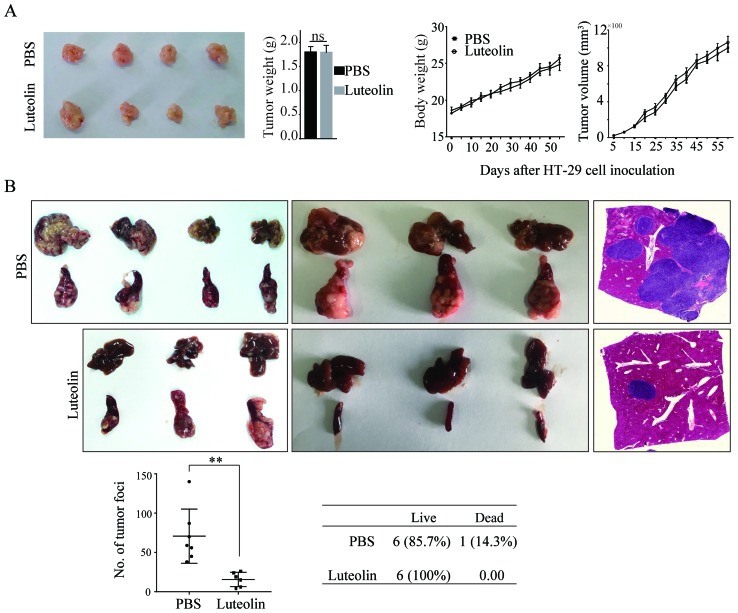
We then performed the in vivo assay to observe the tumor growth and tumor metastasis by injecting HT-29 cells into the nude mice subcutaneously and into the spleen, respectively. In accordance with the in vitro results, intake of luteolin did not impact the tumor growth as evidenced by no differences in tumor volume and tumor weight (Fig. 2A). However, luteolin significantly inhibited HT-29 cells metastasis from the spleen to the liver (Fig. 2B). The number of metastatic nodules in the liver was significantly decreased. Taken together, all these data suggest that luteolin plays a significant role in suppressing the migration and invasion of CRC cells both in vitro and in vivo while has no effects on cells proliferation.
Figure 2.
Luteolin inhibits CRC cells metastasis in vivo. (A) Nude mice were inoculated subcutaneously with HT-29 cells followed by PBS or luteolin treatment. The volume of the tumor and the body weight was monitored. Tumors were photographed at the end of the experiment. (B) Nude mice were intrasplenically injected with HT-29 cells followed by PBS and luteolin treatment. At the end of the experiment, tumor lesions in the liver were photographed and counted. Original magnification, ×4. Results were obtained from 3 independent experiments and are expressed as the means ± SEM. **P<0.01; ns, not significant. CRC, colorectal cancer; PBS, phosphate-buffered saline.
Luteolin inhibits CRC cells migration and invasion by upregulating miR-384
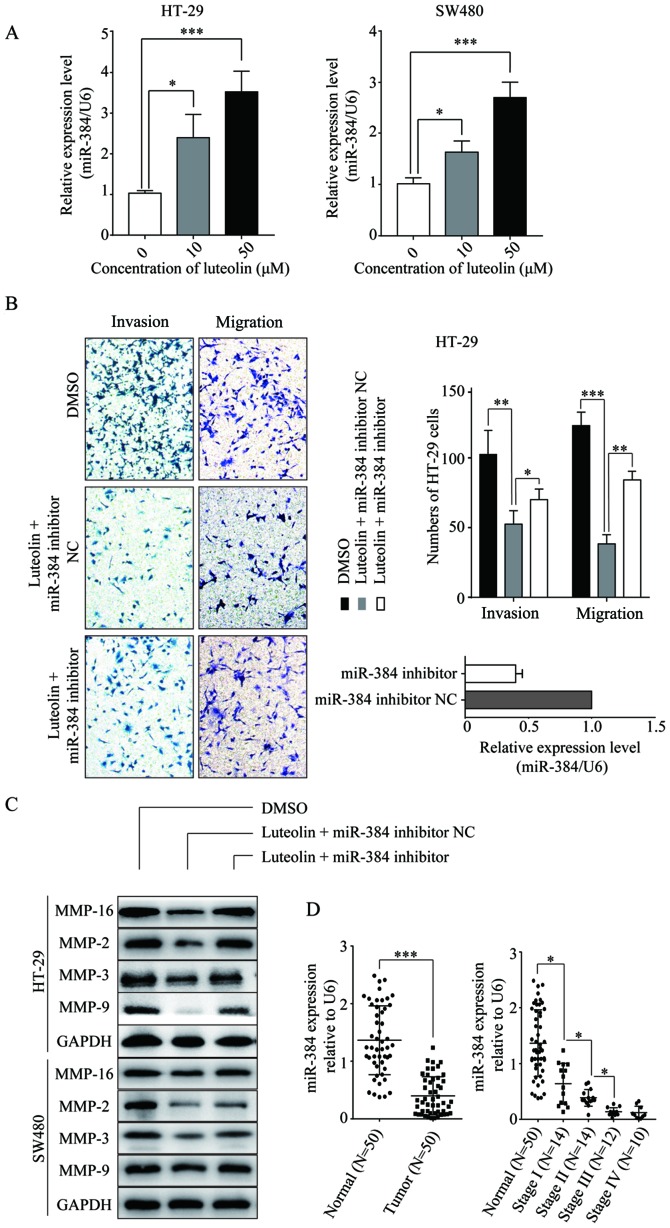
To identify the specific miRNA involved in the luteolin-induced inhibition of CRC cells migration and invasion, the expression level of miR-384 was determined in CRC cells treated with luteolin. It was found that the expression of miR-384 was significantly elevated in the CRC cell lines following treatment with luteolin in a dose-dependent manner compared with no-luteolin stimulation (Fig. 3A). Then, we constructed the miR-384 inhibitor and transfected into the CRC cells before luteolin treatment. As shown in Fig. 3B, the miR-384 inhibitor partially reversed the inhibition of cells migration and invasion induced by luteolin. Simultaneously, the expression levels of MMP-2, MMP-3, MMP-9 and MMP-16 which were decreased by luteolin treatment showed an increase compared to the miR-384 inhibitor NC group (Fig. 3C). All these results imply that miR-384 is involved in the anticancer effects of luteolin. We then detected the miR-384 expression level in CRC tissues. Downregulation of miR-384 expression was noted in the CRC tissues compared with that observed in the non-tumor tissues (P<0.001; Fig. 3D). Moreover, with the progression of the tumor, the expression was decreased significantly. The expression of miR-384 was lowest in patients with TNM stage IV.
Figure 3.
Luteolin inhibits CRC cells migration and invasion by upregulating miR-384. (A) Total RNA was extracted from HT-29 and SW480 cells treated with luteolin (10 and 50 µM) for 24 h and miR-384 expression was determined by qPCR. (B) HT-29 and SW480 cells were transfected with the miR-384 inhibitor NC or miR-384 inhibitor for 24 h, and then treated with 50 µM luteolin and cell migration and invasion abilities were determined by Transwell assay. (C) HT-29 and SW480 cells were transfected with the miR-384 inhibitor NC or miR-384 inhibitor for 24 h and cells were treated with luteolin for 24 h before protein was extracted for western blotting to determine MMP-2, MMP-3, MMP-9 and MMP-16 expression. (D) Total RNA was extracted from 50 pairs of CRC tissues and the corresponding adjacent normal tissues and qPCR was performed to determine the miR-384 expression. Results are representative of 3 independent experiments and are expressed as mean ± SEM. Statistical significance in (A and B) was determined by one-way analysis of variance (ANOVA) followed by Dunnett's test and statistical significance in D was determined by Student's t-test. *P<0.05, **P<0.01, ***P<0.001. CRC, colorectal cancer; DMSO, dimethyl sulfoxide.
PTN is a direct target of miR-384
miRNAs exert their function by regulating the expression of their target genes. Thus, we took advantage of the prediction algorithms (miRDB and TargetScan v.5.1) to identify the potential targets of miR-384. Among the potential target genes, pleiotrophin (PTN) caught our interest. Thus, we detected the PTN expression after luteolin stimulation. The results showed that PTN expression was downregulated in a dose-dependent manner (Fig. 4A). Overexpression of PTN was able to significantly reverse the luteolin-mediated inhibition of cell migration and invasion as well as MMP expressions (Fig. 4B-D). This finding suggests that luteolin inhibited CRC cells migration and invasion by downregulating PTN expression. We next detected the relationship between PTN and miR-384. The expression of PTN was decreased when cells transfected with miR-384 mimic (Fig. 4E). Then miR-384 mimic or miR-384 mimic NC and PTN were co-transfected into CRC cells and the luciferase activity was detected. Compared with the control, a decrease in relative luciferase activity was observed when the WT PTN 3′-UTR was co-transfected with miR-384. However, the miR-384 mimic did not affect luciferase activity in the mutant construct (Fig. 4F). These data indicate that miR-384 directly modulates PTN expression by binding to its 3′-UTR. We also detected PTN expression after cells transfected with the miR-384 inhibitor followed by treatment with luteolin. Results showed that miR-384-inhibitor rescued the PTN downregulation induced by luteolin (Fig. 4G), confirming that luteolin-induced suppression of PTN expression was mediated by miR-384.
Figure 4.
PTN is a direct target of miR-384. (A) The protein level of PTN in HT-29, and SW480 cells treated with luteolin after 24 h was determined by western blotting. (B) HT-29 and SW480 cells were transfected with the vector or PTN overexpression plasmid for 24 h, and the transfection efficiency was determined by western blotting. (C) Transfected cells were treated with luteolin for 24 h before protein was extracted for western blotting to determine MMP-2, MMP-3, MMP-9 and MMP-16 expression. (D) Transfected cells were treated with luteolin for 24 h and cell migration and invasion abilities were determined by Transwell assay. Original magnification, ×100. (E) HT-29 and SW480 cells were transfected with miR-384 mimic NC or miR-384 mimic for 24 h before proteins were extracted for western blotting to determine PTN expression. (F) Putative Wt and Mut miR-384 binding sites in the 3′-UTR of PTN. Relative luciferase activities were analyzed in HT-29 and SW48T0 cells co-transfected with Wt or Mut reporter plasmids and miR-384 mimic or miR-384 mimic NC. (G) HT-29 and SW480 cells were transfected with miR-384 inhibitor NC or miR-384 inhibitor for 24 h before cells were treated with luteolin for 24 h and then protein was extracted for western blotting to determine PTN expression. Results are representative of 3 independent experiments and are expressed as mean ± SEM. Statistical significance in D was determined by one-way analysis of variance (ANOVA) followed by Dunnett's test and statistical significance in F was determined by Student's t-test. ns, no significant; *P<0.05, **P<0.01, ***P<0.001; ns, not significant. PTN, pleiotrophin; DMSO, dimethyl sulfoxide; Wt, wild-type; Mut, mutant.
Expression of PTN in CRC tissues and its relationship with clinical characteristics
Data from Oncomine database were analyzed to examine the differential expression levels of PTN DNA between CRC and normal tissues. As shown in Fig. 5A, PTN was upregulated in tumor tissues (P<0.001). We also collected 50 cases of human CRC tissues and the paired adjacent normal tissues. The relative expression level of PTN was measured by qPCR (Fig. 5B) and western blotting (Fig. 5C). Results showed that PTN was highly expressed in CRC tissues. Furthermore, as the tumor progressed, the expression level of PTN increased as well. We also measured the PTN expression level in four CRC cell lines (SW480, SW620, LoVo and HT-29) and one normal colon epithelial cell (NCM460). The expression level of PTN was markedly higher in CRC cell lines than that noted in the NCM460 cells (Fig. 5D). Then, we analyzed the expression levels of miR-384 and PTN in tumor tissues, and we found that the Pearson's correlation coefficient was −0.58, demonstrating a negative correlation between miR-384 and PTN (Fig. 5E).
Figure 5.
PTN expression is downregulated in CRC tissues and cell lines. (A) Comparison of DNA expression of PTN in CRC and normal tissues from Oncomine database. (Normal, n=445; CRC, n=302) (B) The PTN mRNA expression in 50 pairs of CRC tumor tissues and the corresponding adjacent normal tissues were determined by qPCR (Normal, n=50; Stage I, n=14; Stage II, n=14; Stage III, n=12; Stage IV, n=10). (C) The PTN protein expression in 30 pairs of CRC tissues (T) and the corresponding adjacent normal tissues (N) were determined by western blotting. (D) The mRNA and protein level of PTN in four CRC cell lines (SW480, SW620, LoVo and HT-29) and one normal human colon epithelium cell line NCM460 were determined by qPCR and western blotting, respectively. Results are representative of 3 independent experiments. (E) Pearson's correlation analysis of the negative correlation between the expression of miR-384 and PTN in colorectal cancer tissues. Results are expressed as mean ± SEM. ***P<0.001. PTN, pleiotrophin; CRC, colorectal cancer.
We next divided the patients into two groups according to the PTN expression level (25 patients in each group). The relationship between PTN expression and the clinical characteristics including age, sex, tumor differentiation, TNM stage and serum CEA level was determined. The results are shown in Table I. The differential expression of PTN was significantly associated with tumor size, N stage and M stage of CRC patients (all P<0.05).
Discussion
Currently, the prognosis or therapy for patients with advanced stage of colorectal cancer (CRC) is poor. Treatment includes chemotherapy, radiotherapy and surgery. Recently, traditional Chinese medicine has attracted much attention in the treatment of cancer due to their extensive tumor-suppressive activity, safety and inexpensiveness. One such reagent is luteolin. In the present study, luteolin was found to inhibit CRC cells migration and invasion both in vitro and in vivo, but it did not affect the proliferation of CRC cells. Further experiments revealed the important role of the miR-384/PTN axis in the luteolin-induced effects.
Many reports have reported that luteolin can induce cell cycle arrest and apoptosis in CRC cells. For example, Krifa et al reported that luteolin induced cytotoxicity in human CRC cell line BE and the IC50 value was 90.13 and 48.32 µM for 24 and 48 h, respectively (13). Kang et al showed that luteolin inhibited HT-29 cells proliferation in a dose-dependent manner from 5 to 100 µg/ml (29). Luteolin also decreased the cell viability of human colon carcinoma cell line Caco-2 (30). In the present study, we found that luteolin had no effects on HT-29 and SW480 cells proliferation. We hypothesized that the CRC cells used were different and thus the sensitivity to luteolin may be varied. Thus, the concentrations of 0–100 µM of luteolin in this study had no effects on cells proliferation.
Pleiotrophin (PTN) is a small heparin-binding cytokine which can be induced during tumorigenesis (31). Levels of PTN have been found to be increased in several cancer cell lines and primary tumors, including pancreatic cancer (32), hepatocellular carcinoma (33), breast (34) and papillary thyroid cancer (35). PTN promotes tumor progression either through direct effects on tumor cells or through stimulation of angiogenesis and remodeling of the tumor microenvironment (36–38). In CRC, Yamakawa et al first reported that the PTN expression level was decreased in CRC compared with those in normal adjacent mucosae (39). Kong et al reported that PTN was highly expressed in CRC tissues and its expression was related to CRC differentiation and TNM staging, and a high level of PTN is a predictor of poor prognosis (40). In this study, by analyzing the data from the Oncomine database and clinical samples, we also found that PTN expression was much higher in CRC tissues than that in the normal tissues and its expression was related to TNM staging while PTN had no relationship with tumor differentiation (Table I). We speculate that the probable reason is that the sample size in the present study was too small. Further research with an increased number of samples is needed.
Growing evidence suggests that miR-384 is a potent tumor-suppressor molecule. The functions of miR-384 have been studied in various types of cancers including papillary thyroid (41), pancreatic (42) and non-small cell lung cancer (43). Astrocyte elevated gene-1 (43,44), PTN (33), piwi-like RNA-mediated gene silencing 4 (PIWIL4) (45), KRAS and CDC42 (28) have been identified as target genes of miR-384. Meanwhile, miR-384 was found to be negatively regulated by long non-coding RNA CRNDE (45,46). In CRC, miR-384 was downregulated and closely related with the progression. Inhibition of miR-384 promoted the invasive and metastatic abilities of CRC cells (28). Sun et al reported that in papillary thyroid cancer cells PTN was found to be a downstream target of miR-384 (41). We also confirmed by luciferase reporter assay that in CRC cells PTN is a direct target of miR-384.
In summary, our results demonstrated the antitumor role of luteolin in CRC and the mechanism of which is partly mediated via the miR-384/PTN axis. Thus, miR-384 or PTN can be developed as a therapeutic target in the treatment of CRC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
YY, CR and SW participated in the conception and design of the study. YY, CR and GZ performed the statistical analysis and were involved in the preparation of the figures. YY and SW reviewed the results and participated in the discussion of the data. YY, CR and SW prepared the manuscript and revised it. All authors read and approved the manuscript and agreed to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Approval of present study was obtained from the Ethics Committee of the Hangzhou Hospital of Traditional Chinese Medicine and informed consent was acquired from all the patients. The animal study received ethical approval from the Animal Care and Use Committee of Zhejiang University, and experiments and animal care were performed according to the approved protocols.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Peng M, Watanabe S, Chan KW, He Q, Zhao Y, Zhang Z, Lai X, Luo D, Vasudevan SG, Li G. Luteolin restricts dengue virus replication through inhibition of the proprotein convertase furin. Antiviral Res. 2017;143:176–185. doi: 10.1016/j.antiviral.2017.03.026. [DOI] [PubMed] [Google Scholar]
- 2.Peng M, Swarbrick CMD, Chan KW, Luo D, Zhang W, Lai X, Li G, Vasudevan SG. Luteolin escape mutants of dengue virus map to prM and NS2B and reveal viral plasticity during maturation. Antiviral Res. 2018;154:87–96. doi: 10.1016/j.antiviral.2018.04.013. [DOI] [PubMed] [Google Scholar]
- 3.Zhang H, Tan X, Yang D, Lu J, Liu B, Baiyun R, Zhang Z. Dietary luteolin attenuates chronic liver injury induced by mercuric chloride via the Nrf2/NF-κB/P53 signaling pathway in rats. Oncotarget. 2017;8:40982–40993. doi: 10.18632/oncotarget.17334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo YF, Xu NN, Sun W, Zhao Y, Li CY, Guo MY. Luteolin reduces inflammation in Staphylococcus aureus-induced mastitis by inhibiting NF-κB activation and MMPs expression. Oncotarget. 2017;8:28481–28493. doi: 10.18632/oncotarget.16092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zang Y, Igarashi K, Li Y. Anti-diabetic effects of luteolin and luteolin-7-O-glucoside on KK-Ay mice. Biosci Biotechnol Biochem. 2016;80:1580–1586. doi: 10.1080/09168451.2015.1116928. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Y, Ding BZ, Lin YP, Wang HB. MiR-34a, as a suppressor, enhance the susceptibility of gastric cancer cell to luteolin by directly targeting HK1. Gene. 2018;644:56–65. doi: 10.1016/j.gene.2017.10.046. [DOI] [PubMed] [Google Scholar]
- 7.Byun EB, Song HY, Mushtaq S, Kim HM, Kang JA, Yang MS, Sung NY, Jang BS, Byun EH. Gamma-irradiated luteolin inhibits 3-isobutyl-1-methylxanthine-induced melanogenesis through the regulation of CREB/MITF, PI3K/Akt, and ERK pathways in B16BL6 melanoma cells. J Med Food. 2017;20:812–819. doi: 10.1089/jmf.2016.3890. [DOI] [PubMed] [Google Scholar]
- 8.Chen P, Zhang JY, Sha BB, Ma YE, Hu T, Ma YC, Sun H, Shi JX, Dong ZM, Li P. Luteolin inhibits cell proliferation and induces cell apoptosis via down-regulation of mitochondrial membrane potential in esophageal carcinoma cells EC1 and KYSE450. Oncotarget. 2017;8:27471–27480. doi: 10.18632/oncotarget.15832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Q, Wang H, Jia Y, Pan H, Ding H. Luteolin induces apoptosis by ROS/ER stress and mitochondrial dysfunction in gliomablastoma. Cancer Chemother Pharmacol. 2017;79:1031–1041. doi: 10.1007/s00280-017-3299-4. [DOI] [PubMed] [Google Scholar]
- 10.Cao Z, Zhang H, Cai X, Fang W, Chai D, Wen Y, Chen H, Chu F, Zhang Y. Luteolin promotes cell apoptosis by inducing autophagy in hepatocellular carcinoma. Cell Physiol Biochem. 2017;43:1803–1812. doi: 10.1159/000484066. [DOI] [PubMed] [Google Scholar]
- 11.Lim W, Yang C, Bazer FW, Song G. Luteolin inhibits proliferation and induces apoptosis of human placental choriocarcinoma cells by blocking the PI3K/AKT pathway and regulating sterol regulatory element binding protein activity. Biol Reprod. 2016;95:82. doi: 10.1095/biolreprod.116.141556. [DOI] [PubMed] [Google Scholar]
- 12.Shen XF, Teng Y, Sha KH, Wang XY, Yang XL, Guo XJ, Ren LB, Wang XY, Li J, Huang N. Dietary flavonoid luteolin attenuates uropathogenic Escherichia. Coli invasion of the urinary bladder. Biofactors. 2016;42:674–685. doi: 10.1002/biof.1314. [DOI] [PubMed] [Google Scholar]
- 13.Krifa M, Leloup L, Ghedira K, Mousli M, Chekir-Ghedira L. Luteolin induces apoptosis in BE colorectal cancer cells by downregulating calpain, UHRF1, and DNMT1 expressions. Nutr Cancer. 2014;66:1220–1227. doi: 10.1080/01635581.2014.951729. [DOI] [PubMed] [Google Scholar]
- 14.Xavier CP, Lima CF, Preto A, Seruca R, Fernandes-Ferreira M, Pereira-Wilson C. Luteolin, quercetin and ursolic acid are potent inhibitors of proliferation and inducers of apoptosis in both KRAS and BRAF mutated human colorectal cancer cells. Cancer Lett. 2009;281:162–170. doi: 10.1016/j.canlet.2009.02.041. [DOI] [PubMed] [Google Scholar]
- 15.Pandurangan AK, Dharmalingam P, Sadagopan SK, Ramar M, Munusamy A, Ganapasam S. Luteolin induces growth arrest in colon cancer cells through involvement of Wnt/β-catenin/GSK-3β signaling. J Environ Pathol Toxicol Oncol. 2013;32:131–139. doi: 10.1615/JEnvironPatholToxicolOncol.2013007522. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, Lang T, Jin B, Chen F, Zhang Y, Beuerman RW, Zhou L, Zhang Z. Luteolin inhibits colorectal cancer cell epithelial-to-mesenchymal transition by suppressing CREB1 expression revealed by comparative proteomics study. J Proteomics. 2017;161:1–10. doi: 10.1016/j.jprot.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 17.Manju V, Nalini N. Protective role of luteolin in 1,2-dimethylhydrazine induced experimental colon carcinogenesis. Cell Biochem Funct. 2007;25:189–194. doi: 10.1002/cbf.1305. [DOI] [PubMed] [Google Scholar]
- 18.Ashokkumar P, Sudhandiran G. Protective role of luteolin on the status of lipid peroxidation and antioxidant defense against azoxymethane-induced experimental colon carcinogenesis. Biomed Pharmacother. 2008;62:590–597. doi: 10.1016/j.biopha.2008.06.031. [DOI] [PubMed] [Google Scholar]
- 19.Chian S, Li YY, Wang XJ, Tang XW. Luteolin sensitizes two oxaliplatin-resistant colorectal cancer cell lines to chemotherapeutic drugs via inhibition of the Nrf2 pathway. Asian Pac J Cancer Prev. 2014;15:2911–2916. doi: 10.7314/APJCP.2014.15.6.2911. [DOI] [PubMed] [Google Scholar]
- 20.Qu Q, Qu J, Guo Y, Zhou BT, Zhou HH. Luteolin potentiates the sensitivity of colorectal cancer cell lines to oxaliplatin through the PPARγ/OCTN2 pathway. Anticancer Drugs. 2014;25:1016–1027. doi: 10.1097/CAD.0000000000000125. [DOI] [PubMed] [Google Scholar]
- 21.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet. 2016;17:719–732. doi: 10.1038/nrg.2016.134. [DOI] [PubMed] [Google Scholar]
- 23.Eichmuller SB, Osen W, Mandelboim O, Seliger B. Immune modulatory microRNAs involved in tumor attack and tumor immune escape. J Natl Cancer Inst. 2017;109 doi: 10.1093/jnci/djx034. [DOI] [PubMed] [Google Scholar]
- 24.Rupaimoole R, Slack FJ. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 25.Han K, Meng W, Zhang JJ, Zhou Y, Wang YL, Su Y, Lin SC, Gan ZH, Sun YN, Min DL. Luteolin inhibited proliferation and induced apoptosis of prostate cancer cells through miR-301. Onco Targets Ther. 2016;9:3085–3094. doi: 10.2147/OTT.S102862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang ZQ, Li MH, Qin YM, Jiang HY, Zhang X, Wu MH. Luteolin inhibits tumorigenesis and induces apoptosis of non-small cell lung cancer cells via regulation of microRNA-34a-5p. Int J Mol Sci. 2018;19:E447. doi: 10.3390/ijms19020447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Wang YX, Chen YR, Liu SS, Ye YP, Jiao HL, Wang SY, Xiao ZY, Wei WT, Qiu JF, Liang L, et al. MiR-384 inhibits human colorectal cancer metastasis by targeting KRAS and CDC42. Oncotarget. 2016;7:84826–84838. doi: 10.18632/oncotarget.12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang KA, Piao MJ, Ryu YS, Hyun YJ, Park JE, Shilnikova K, Zhen AX, Kang HK, Koh YS, Jeong YJ, et al. Luteolin induces apoptotic cell death via antioxidant activity in human colon cancer cells. Int J Oncol. 2017;51:1169–1178. doi: 10.3892/ijo.2017.4091. [DOI] [PubMed] [Google Scholar]
- 30.Abdel Hadi L, Di Vito C, Marfia G, Ferraretto A, Tringali C, Viani P, Riboni L. Sphingosine kinase 2 and ceramide transport as key targets of the natural flavonoid luteolin to induce apoptosis in colon cancer cells. PLoS One. 2015;10:e0143384. doi: 10.1371/journal.pone.0143384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muramatsu T. Midkine and pleiotrophin: Two related proteins involved in development, survival, inflammation and tumorigenesis. J Biochem. 2002;132:359–371. doi: 10.1093/oxfordjournals.jbchem.a003231. [DOI] [PubMed] [Google Scholar]
- 32.Yao J, Zhang LL, Huang XM, Li WY, Gao SG. Pleiotrophin and N-syndecan promote perineural invasion and tumor progression in an orthotopic mouse model of pancreatic cancer. World J Gastroenterol. 2017;23:3907–3914. doi: 10.3748/wjg.v23.i21.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bai PS, Xia N, Sun H, Kong Y. Pleiotrophin, a target of miR-384, promotes proliferation, metastasis and lipogenesis in HBV-related hepatocellular carcinoma. J Cell Mol Med. 2017;21:3023–3043. doi: 10.1111/jcmm.13213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma J, Kong Y, Nan H, Qu S, Fu X, Jiang L, Wang W, Guo H, Zhao S, He J, Nan K. Pleiotrophin as a potential biomarker in breast cancer patients. Clin Chim Acta. 2017;466:6–12. doi: 10.1016/j.cca.2016.12.030. [DOI] [PubMed] [Google Scholar]
- 35.Jee YH, Sadowski SM, Celi FS, Xi L, Raffeld M, Sacks DB, Remaley AT, Wellstein A, Kebebew E, Baron J. Increased pleiotrophin concentrations in papillary thyroid cancer. PLoS One. 2016;11:e0149383. doi: 10.1371/journal.pone.0149383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang Y, Zuka M, Perez-Pinera P, Astudillo A, Mortimer J, Berenson JR, Deuel TF. Secretion of pleiotrophin stimulates breast cancer progression through remodeling of the tumor microenvironment. Proc Natl Acad Sci USA. 2007;104:10888–10893. doi: 10.1073/pnas.0704366104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lampropoulou E, Logoviti I, Koutsioumpa M, Hatziapostolou M, Polytarchou C, Skandalis SS, Hellman U, Fousteris M, Nikolaropoulos S, Choleva E, et al. Cyclin-dependent kinase 5 mediates pleiotrophin-induced endothelial cell migration. Sci Rep. 2018;8:5893. doi: 10.1038/s41598-018-24326-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi Y, Ping YF, Zhou W, He ZC, Chen C, Bian BS, Zhang L, Chen L, Lan X, Zhang XC, et al. Tumour-associated macrophages secrete pleiotrophin to promote PTPRZ1 signalling in glioblastoma stem cells for tumour growth. Nat Commun. 2017;8:15080. doi: 10.1038/ncomms15080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamakawa T, Kurosawa N, Kadomatsu K, Matsui T, Itoh K, Maeda N, Noda M, Muramatsu T. Levels of expression of pleiotrophin and protein tyrosine phosphatase zeta are decreased in human colorectal cancers. Cancer Lett. 1999;135:91–96. doi: 10.1016/S0304-3835(98)00275-4. [DOI] [PubMed] [Google Scholar]
- 40.Kong Y, Bai PS, Nan KJ, Sun H, Chen NZ, Qi XG. Pleiotrophin is a potential colorectal cancer prognostic factor that promotes VEGF expression and induces angiogenesis in colorectal cancer. Int J Colorectal Dis. 2012;27:287–298. doi: 10.1007/s00384-011-1344-z. [DOI] [PubMed] [Google Scholar]
- 41.Sun H, He L, Ma L, Lu T, Wei J, Xie K, Wang X. LncRNA CRNDE promotes cell proliferation, invasion and migration by competitively binding miR-384 in papillary thyroid cancer. Oncotarget. 2017;8:110552–110565. doi: 10.18632/oncotarget.22819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang G, Pan J, Zhang L, Wei Y, Wang C. Long non-coding RNA CRNDE sponges miR-384 to promote proliferation and metastasis of pancreatic cancer cells through upregulating IRS1. Cell Prolif. 2017;50 doi: 10.1111/cpr.12389. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Fan N, Zhang J, Cheng C, Zhang X, Feng J, Kong R. MicroRNA-384 represses the growth and invasion of non-small-cell lung cancer by targeting astrocyte elevated gene-1/Wnt signaling. Biomed Pharmacother. 2017;95:1331–1337. doi: 10.1016/j.biopha.2017.08.143. [DOI] [PubMed] [Google Scholar]
- 44.Song H, Rao Y, Zhang G, Kong X. MicroRNA-384 inhibits the growth and invasion of renal cell carcinoma cells by targeting astrocyte elevated gene 1. Oncol Res. 2018;26:457–466. doi: 10.3727/096504017X15016337254632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng J, Liu X, Wang P, Xue Y, Ma J, Qu C, Liu Y. CRNDE promotes malignant progression of glioma by attenuating miR-384/PIWIL4/STAT3 axis. Mol Ther. 2016;24:1199–1215. doi: 10.1038/mt.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Chen Z, Yu C, Zhan L, Pan Y, Chen L, Sun C. LncRNA CRNDE promotes hepatic carcinoma cell proliferation, migration and invasion by suppressing miR-384. Am J Cancer Res. 2016;6:2299–2309. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.