Abstract
Breast cancer (BC) has a complex etiology and pathogenesis, and is the most common malignant tumor type in females, in USA in 2018, yet its relevant molecular mechanisms remain largely unknown. The collagen type V α-1 chain (COL5A1) gene is differentially expressed in renal and ovarian cancer. Using bioinformatics methods, COL5A1 was determined to also be a significant gene in BC, but its association with BC has not been sufficiently reported. COL5A1 microarray and relevant clinical data were collected from the Gene Expression Omnibus, The Cancer Genome Atlas and other databases to summarize COL5A1 expression in BC and its subtypes at the mRNA and protein levels. All associated information was comprehensively analyzed by various software. The clinical significance of the mutation was obtained via the cBioPortal. Furthermore, Gene Ontology functional annotation and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment were also performed to investigate the mechanism of COL5A1 in BC. Immunohistochemistry was also conducted to detect and confirm COL5A1 expression. It was determined that COL5A1 was highly expressed in BC tissues, compared with normal tissues at the mRNA level [standard mean difference, 0.84; 95% confidence interval (CI), 0.60-1.07; P=0.108]. The area under the summary receiver operator characteristic curve for COL5A1 was 0.87 (95% CI, 0.84-0.90). COL5A1 expression was altered in 32/817 (4%) sequenced samples. KEGG analysis confirmed the most notable pathways, including focal adhesion, extracellular matrix-receptor interaction and regulation of the actin cytoskeleton. Immunohistochemical detection was used to verify the expression of COL5A1 in 136 selected cases of invasive BC tissues and 55 cases of adjacent normal tissues, while the rate of high expression of COL5A1 in BC was up to 90.4%. These results indicated that COL5A1 is highly expressed at the mRNA and protein levels in BC, and the prognosis of patients with BC with high COL5A1 expression may be reduced; therefore, COL5A1 may be used independently or combined with other detection factors in BC diagnosis.
Keywords: breast cancer, collagen typeV α-1 chain, prognosis, molecular mechanism, immunohistochemistry
Introduction
According to the latest 2018 cancer statistics for USA, breast cancer (BC) is the most common malignant tumor type in females, accounting for 30% of estimated new diagnoses (1). Simultaneously, the mortality rate of females with malignant breast tumors is second only to those with lung cancer in USA in 2011–2015 (1,2). Additionally, in economically developed countries and regions, BC is prevalent. According to the latest reports in USA, there are ~250,000 new cases annually, and the mortality toll is estimated at 411,000 annually in 2017 (3,4). However, as economies develop further and living standards improve, the incidence of BC increases year after year (5). Globally, ~1.7 million females are diagnosed with BC annually, resulting in >500,000 mortalities annually, which makes BC the most common malignant tumor type affecting the lives and health of females, not only in USA, but also globally in 2012 (5). At present, BC treatment primarily includes surgery, chemotherapy, endocrine therapy, radiotherapeutics and biotherapy (6,7). Nevertheless, the incidence and mortality of BC remain high, and BC still affects the quality of life of females (8). At present, scholars have reached a consensus that BC, similar to the majority of cancer types, is considered a specific gene-associated disease, which means that the pathogenic process of BC involves a multi-factor, multi-stage and multi-step complex effect (9). With research deepening on the level of gene molecules and screening for differentially-expressed genes (DEGs) associated with BC, research on the functional annotation and participating signaling pathways of DEGs helps to improve understanding regarding BC pathogenesis. These in-depth studies notably assist in predicting the recurrence, metastasis and prognosis of BC, which also provides a theoretical basis for individualized precision treatment. The urgent requirement for the early diagnosis and treatment of BC, as well as for improving the prognosis and reducing the mortality rate, has led researchers to shift focus to the molecular levels of BC, aiming to determine its pathogenic mechanisms and to provide an evidence-based foundation for targeted therapy.
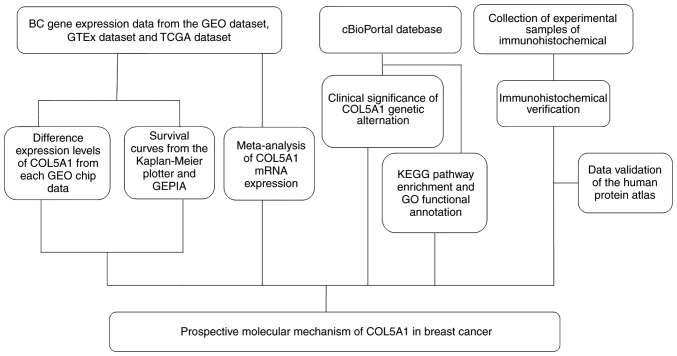
The present study attempts to screen and analyze BC-relevant gene expression profiles in the Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) databases by computational biology. Following consulting a large body of literature on the subject and using computational biology statistical calculations, COL5A1 was determined to be a crucial gene in the oncogenesis of BC (10,11). However, coverage of the clinical value of COL5A1 remains insufficient (12). In the present study, computational biology-associated methods are used to mine the expression levels of associated genes in the GEO database and analyze their prognostic value with numerous methods. To support the calculation of the biological data results, immunohistochemical methods are used to verify whether the expression level of COL5A1 in BC tissue is increased, compared with normal breast tissue. Furthermore, the association between the COL5A1 gene and the clinicopathological features and prognosis of BC were determined, as well as the clinical significance of COL5A1 alterations (Fig. 1).
Figure 1.
Flow chart for the research. BC, breast cancer; GEO, Gene Expression Omnibus; TCGA, The Cancer Genome Atlas; GTEx, The Genotype Tissue Expression; GEPIA, Gene Expression Profiling Interactive Analysis; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; COL5A1, collagen type V α-1 chain.
Methods and materials
mRNA and protein expression levels of COL5A1
Clinical data collection of COL5A1 mRNA expression
The GEO database (https://www.ncbi.nlm.nih.gov/geo/) is the largest and most comprehensive public gene expression database available today (13). The BC gene probe expression matrix and probe annotation file were mined from the GEO dataset, and probes corresponding to the COL5A1 gene were located based on the probe annotation file. According to the data requirements for BC, the present study screened the chips in the GEO database. The key word was set as ‘breast cancer’, and the chip filter conditions were as follows: i) Human breast tissue; ii) comparative RNA expression data chip with BC tissue and paracancer or normal breast tissue; iii) invasive BC sample; and iv) patients without adjuvant therapies, including radiotherapy and chemotherapy. The exclusion conditions were as follows: i) Cell line; ii) methylation; iii) samples that have been treated with adjuvant therapies; and iv) cancer samples without comparison of paracancer or normal tissue. Considering that the specific classification of BC is associated with its prognosis, all selected GEO chips underwent secondary screening. Invasive BC is divided into the invasive non-specific and invasive specific types, in which non-specific BC is subdivided into invasive ductal carcinoma (IDC) and invasive lobular carcinoma (ILC) (8). The criteria aforementioned currently applies to the classification of each BC case. The lack of subdivisions for invasive specific BC may be due to fewer clinical cases, and, if necessary, the present study uniformly classified it as invasive specific type. If there was no specific classification in the GEO chip, the sample was included as an invasive BC type. A corresponding histogram was then created using GraphPad Prism 7.0 (GraphPad Software, Inc., La Jolla, CA, USA) to depict the difference in COL5A1 expression levels in invasive BC and normal tissues from each piece of GEO chip data.
The Kaplan-Meier plotter (http://kmplot.com/analysis/index.php?p=service) (14,15) is an online drawing tool used to evaluate the influence of 54,685 genes on survival by mining information from 10,471 samples collected from 5,243 patients with BC, along with detailed survival data originating in the European Genome-phenome Archive, TCGA and the GEO (Affymetrix microarray only) databases. Additionally, the clinical values of specific genes were analyzed by the Kaplan-Meier plotter with survival curves. Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/) (16–18) collected 9,736 tumor samples from the TCGA and Genotype-Tissue Expression databases and constituted a web tool for analyzing the RNA sequencing expression data. In the research, the expression level of COL5A1 and the prognostic value material were obtained via GEPIA and Kaplan-Meier plotter.
Prediction of clinical significance of COL5A1 genetic alteration
The cBioPortal (http:/cbioportal.org/) (19,20), which provides data from 20 types of cancer studies including >5,000 tumor samples, was utilized to investigate the clinical significance of COL5A1 genetic alteration in BC, including detailed information regarding the mutation of COL5A1 in multiple samples, including genetic sites, mutation types and amino acid changes. The Kaplan-Meier survival estimate of COL5A1 in patients with BC was also analyzed using cBioPortal.
Potential mechanism of COL5A1 in BC
The co-expressed genes associated with COL5A1 were obtained to investigate the possible mechanism of COL5A1 in BC. The relative genes were processed with KEGG pathway enrichment and GO functional annotation through WebGestalt (http://www.webgestalt.org/option.php), which is a tool to perform functional enrichment analysis (21,22). The KEGG enrichment analysis was performed with COL5A1-associated genes using the ggplot2 package in R (23). For further investigation, the biological process of cellular component organization, which had the most accumulation and alteration rate of co-expressed genes associated with COL5A1, was selected to analyze and discuss the potential mechanism of BC.
Meta-analysis of COL5A1 mRNA expression
To conduct a holistic evaluation of COL5A1 expression levels, Stata 12.0 (StataCorp LP, College Station, TX, USA) was used for the meta-analysis of continuous variables. Additionally, if the meta-analysis was revealed to be heterogeneous, sources of heterogeneity were further identified through sensitivity analysis. After removing the chips that may cause heterogeneity in the meta-analysis, the remaining chips were re-analyzed to confirm that heterogeneity had been excluded. Heterogeneity indicated that the results of GEO data were inconsistent, which may result in bias in the results of meta-analysis. Provided that the heterogeneity value of the results was <50%, the results of the meta-analysis were considered credible. Finally, if the standard mean difference (SMD) was <0, and 95% confidence interval (CI) that did not cross the 0-point coordinate line (0.60-1.07), then the target gene was differentially expressed in BC tissues relative to the normal control group. Furthermore, Deeks' test was conducted to identify any sources of publication bias.
Additionally, the expression of COL5A1 was imported into IBM SPSS 22.0 (IBM Corp., Armonk, NY, USA) to calculate the number of true positive (TP), true negative (TN), false positive (FP) and false negative (FN) cases. In the calculation process, cancer tissue was defined as the control group for the gene highly expressed in BC tissue. GraphPad Prism 7.0 was used to plot the Receiver Operating Characteristic (ROC) curves to show the potential diagnostic value of the target gene in each chip. Finally, a summary ROC curve (sROC) was plotted using Stata 12.0 to further evaluate the potential diagnostic value of the gene of interest as a whole. The aforementioned steps were repeated for all sub-types of BC to further study the association of COL5A1 these subtypes. Rversion 3.4.1 was used to statistically analyze the relationship between CLO5A1 expression levels and clinical parameters, molecular typing in BC cases. Ggplot2 presenting as a drawing supplement package that can be applied to R software and was used to graphically display differential genes in potential pathway analysis.
Immunohistochemical verification
Source of experimental material
The study protocol was approved by The Ethical Committee of First Affiliated Hospital of Guangxi Medical University (Nanning, China). A total of 136 cases were included in the present study, all of which were female, aged 29–79 years old, with a median age of 47 years old. The tissue collection location was the breast cancer and adjacent tissue. All tissue samples were collected from the Department of Pathology of the First Affiliated Hospital of Guangxi Medical University between January and December 2012.
Archived sections of patients with invasive ductal breast carcinoma who underwent surgical resection and pathological confirmation from January 2012 to December 2012 were selected as experimental material. The selection criteria for the experimental samples were as follows: i) Female patients; ii) patients diagnosed with invasive breast carcinoma by the World Health Organization's 2012 fourth edition of the Breast Tumor Histological Classification (24); iii) patients with well-preserved clinical pathology data via estrogen (ER), progesterone (PR) and oncogenes [human epidermal growth factor receptor-2 (HER-2), P53 and Ki-67] immunohistochemical staining; iv) patients who had not received adjuvant therapy, including radiotherapy, chemotherapy and endocrine therapy prior to the operation; v) patients for whom the recorded breast carcinoma was their first-discovered primary tumor; and vi) patients who received standardized treatments following operation. Exclusion criteria were as follows: i) Male patients; ii) patients without complete clinical records or complete ER, PR, HER-2, P53 and Ki-67 immunohistochemical staining or no clinical staging; and iii) patients whose BC tissues in the archived paraffin specimens were too small to be re-cut (25–27).
Determination principles of immunohistochemistry in experimental samples
Human bladder collagen tissue was used as a positive control for COL5A1, and PBS was used as a negative control replacing the primary antibody. Each of the specimens was strictly prepared according to the manufacturer's protocols of a mouse and rabbit specific HRP/DAB (ABC) Detection immunohistochemistry kit (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and each stain was provided with a positive control and a blank control.
The fixative solution of tissue specimens was 10% neutral formalin solution, the amount of fixative solution was 5–10 times of the volume of the specimen, and the fixed time of the specimens was 12–24 h at room temperature 15–28 °C. The specimens were embedded in Spurr resin to form a paraffin block. Immunohistochemistry was performed on 4 µm sections of the tissue paraffin block. They were deparaffinized in xylene at 37°C and rinsed in medicinal-graded ethanol (100, 95, 85, 75 and 50%). Subsequently, 3% H2O2 prepared fresh from distilled water was used to inactivate endogenous peroxidase, which lasted for 15 min at room temperature and was repeatedly soaked in PBS solution. Antibody reaction used rabbit anti-human concentrated polyclonal COL5A1 antibody (dilution, 1:1,000; cat. no. HPA030769; Sigma-Aldrich; Merck KGaA) in 37°C for 1 h, and then thoroughly flushed by PBS solution. After washed, slides were incubated with fast enzyme-labeled goat anti-mouse/rabbit IgG polymer (cat. no. KIT-5030; Fuzhou Maixin Biotech Co., Ltd., Fuzhou, China) as secondary antibody in 37°C for 30 min. Finally, a freshly prepared DAB colorant (cat. no. DAB-1031; Fuzhou Maixin Biotech Co., Ltd.) was used for dyeing. The color developing effect was observed with a light microscope with an OLYMPUS VANOX micrographic system (magnification, ×100 and ×200).
There were two experienced pathologists who read each film by a double-blind method. If the results of the two pathologists' interpretations had been inconsistent, a third pathologist would have been asked to review the film. Positive controls were required to exhibit positive results for each test, and negative controls were required to exhibit negative results.
The results of the immunohistochemical staining were mostly consistent with the relevant literature (28). Immunohistochemical results of sections were evaluated according to the following established criteria. The evaluation standards were as follows: i) Results were scored according to color intensity (I) (light yellow was one point; yellow was two points; and yellow-brown was three points); ii) results were digitized (1–100%) in accordance with the percentages (P) that positive cells occupied; and iii) the two numbers were multiplied to obtain Q, which meant that Q=PxI. When the score was Q=0, it was considered to be negative; when the score was 0<Q≤120, it was considered to be weakly positive; when the score was 120<Q≤210, it was considered to be moderately positive; and when the score was 210<Q≤300, it was considered to be strongly positive. This experiment defined negative and weakly positive as low-level expression levels, and moderately positive and strongly positive as high-level expression levels.
Prognosis and patient follow-up
The return visits of the patients were assessed with the hospital management information system. For those who did not return in the course of the follow-up period, the patients or their family members were provided relevant follow-up information by telephone. The survival time of all patients was counted from the day of surgery to March 15, 2018. In the event that the patient succumbed, or was lost to follow-up, the date of mortality or loss to follow-up was considered the end date.
Data analysis
The experimental data were statistically analyzed and plotted using SPSS 22.0, R version 3.4.1 (https://www.r-project.org/), GraphPad Prism 7.0 and Stata 12.0 software. All results are presented as mean ± standard deviation. SPSS 22.0 was applied to calculate the χ2 of the COL5A1 protein expression levels, as well as the association between the clinicopathological features and the expression of antibodies in BC. The correlation analysis of antibodies and clinicopathological parameters was conducted using Spearman's rank correlation test. The Kaplan-Meier plotter was also used to perform survival analysis to demonstrate the association between COL5A1 and BC prognosis. P<0.05 was considered to indicate a statistically significant difference. The independent sample t-test was used to compare the expression levels of COL5A1 between two continuous variables. Student's t-test was used to compare the expression levels between the two paired variables. One-way ANOVA analysis of variance was used to compare more than two different groups. Multiple comparisons between the groups was performed using the Student-Newman-Keuls method. All expression data were log2 (TPM+1) transformed for differential analysis.
Protein validation of COL5A1 expression
The immunohistochemical results were verified via the Human Protein Atlas (HPA; http://www.proteinatlas.org/) (29,30). The website aims at mapping the distribution of human proteins in cells, tissues and organs using integration technologies. To ensure the staining results were sufficiently representative, all tissues examined were derived from 144 different individuals and 216 tumor tissues, and immunohistochemical techniques were used to detect the distribution and expression of COL5A1 in 48 normal human tissues, 20 tumor tissues, 47 cell lines and 12 blood cells (24–26).
Results
Association of COL5A1 mRNA expression level and clinical significance
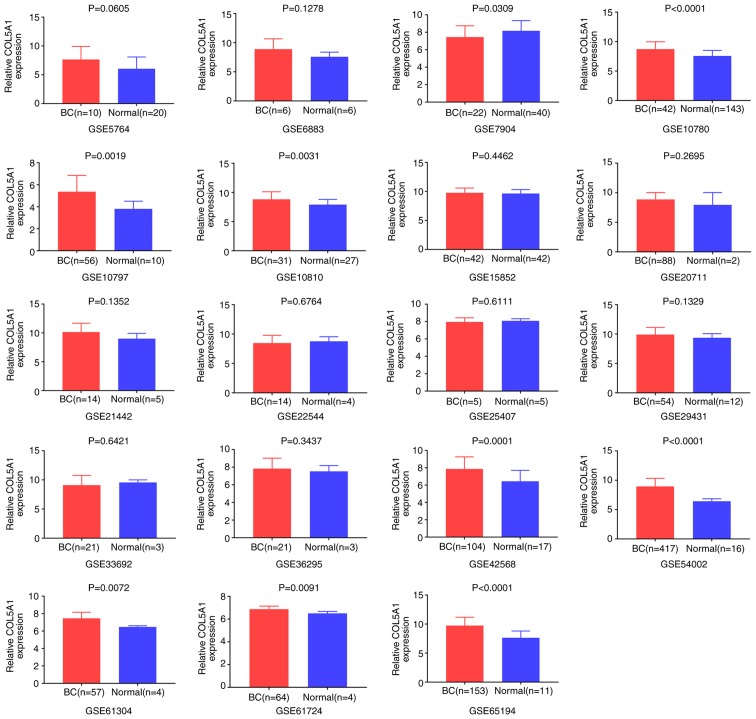
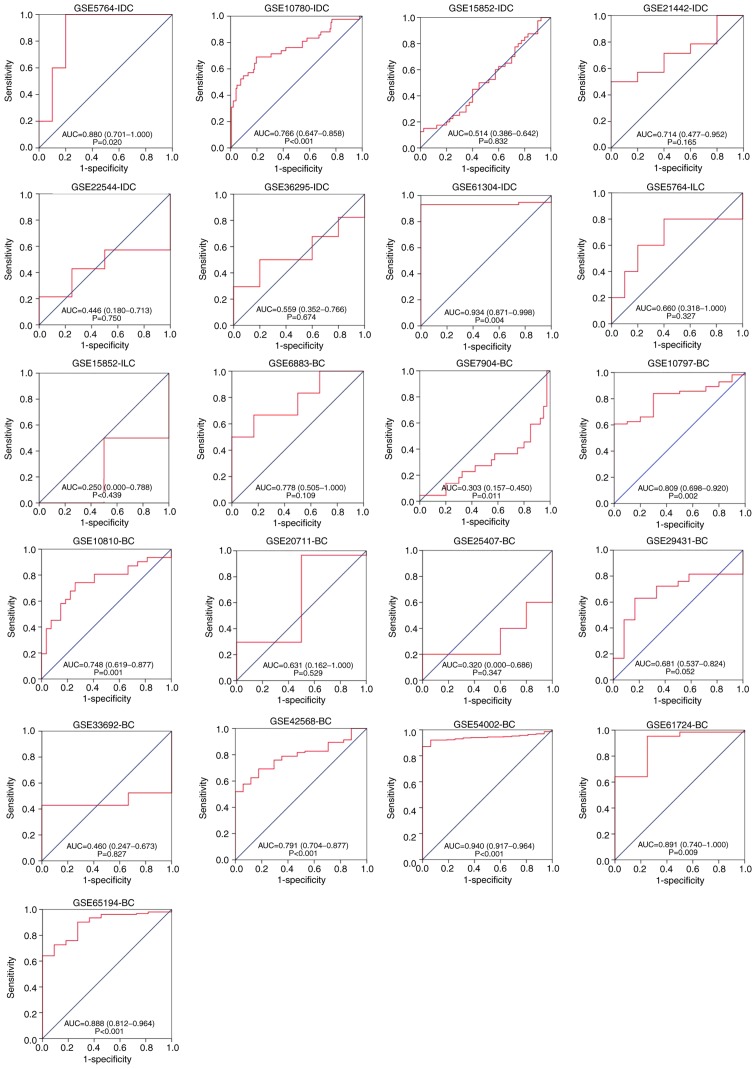
Overall, 20 GEO chip data were included after a preliminary screening in the present study. The chip GSE61723, without COL5A1 expression data, was excluded. According to the gene chip probe annotation file, the target gene, COL5A1, corresponded with the probes 203325_s_at, 212488_at and 212489_at, and the investigation of COL5A1 the expression value was based on the arithmetic mean value of the three probes above. According to the established principles, seven microarrays containing IDC were screened, including GSE5764, GSE10780, GSE15852, GSE21442, GSE22544, GSE36295 and GSE61304. The microarrays containing ILC were GSE5764 and GSE15852. Due to the lack of adequate data for invasive specific BC, the expression analysis could not be performed. Following integrating the data, GraphPad Prism 7.0 was used to pool the box charts, which directly demonstrated the difference in the expression levels of COL5A1 between BC and normal tissues. In the overall data analysis of 19 GEO chips, the results revealed that eight selected COL5A1 chips exhibited significantly increased expression in carcinoma tissues, compared with normal tissues (P<0.05; Fig. 2).
Figure 2.
Expression of COL5A1 in cancerous and non-cancerous tissues based on 19 Gene Expression Omnibus chips. P<0.05 is considered statistically significant. COL5A1, collagen type V α-1 chain; BC, breast cancer.
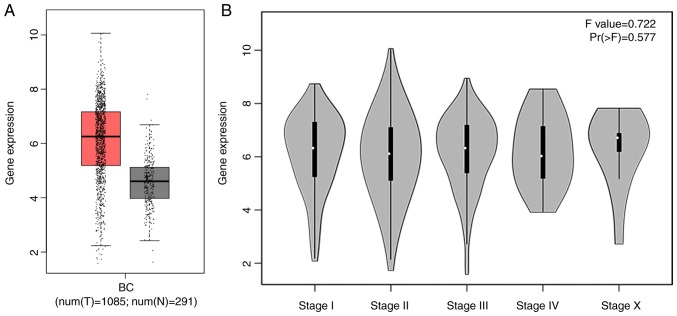
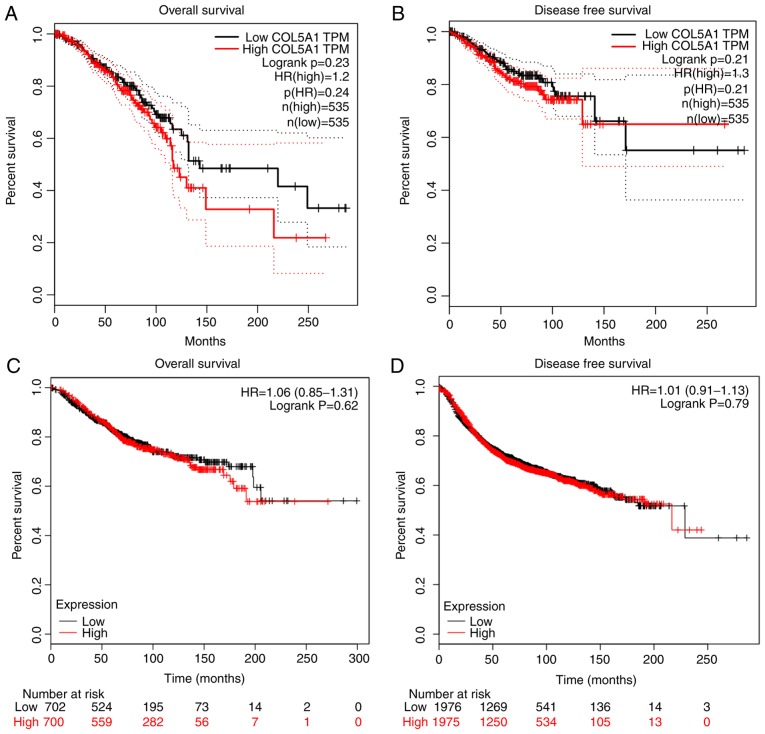
Additionally, the box chart of the overall COL5A1 expression level indicated that the gene expression was significantly increased in cancerous BC tissues, compared with non-cancerous BC tissues (Fig. 3A). The clinical BC stage made no significant difference on COL5A1 expression (Fig. 3B). The survival prognoses of COL5A1 in BC, plotted by GEPIA and the Kaplan-Meier plotter, demonstrated that there was no significant difference in the overall survival (OS) and disease-free survival (DFS) time (Fig. 4). Nevertheless, a difference was observed in the curves between low and high expression levels. To further investigate the association between clinical parameters and BC, ER negative (P=4.3×10−5) and PR negative (P=0.00037) were determined to be significant with DFS in BC, as well as histological grade III (P=0.0024) (Fig. 5), while there was no direct statistical evidence to reveal the association between ER, PR and HER-2, and OS.
Figure 3.
The Cancer Genome Atlas data mining provided by Gene Expression Profiling Interactive Analysis. (A) The expression level of COL5A1 between cancerous (1,085 cases) and non-cancerous (291 cases) tissue. Red represents cancerous tissue, grey represents non-cancerous tissue and black spots represent individual cases. (B) Arithmetic mean expression level of COL5A1 in different clinical stages of patients with BC tissues. The expression data are first log2(TPM+1) transformed for differential analysis and the log2FC is defined as median(Tumor)-median(Normal)BC, breast cancer; COL5A1, collagen type V α-1 chain; N, non-cancerous tissue; T, cancerous tissue; TPM, transcripts per million.
Figure 4.
Survival curves of patients with BC based on COL5A1 levels based on GEPIA and Kaplan-Meier plotter. Red lines represent the survival time of patients with cancer with high COL5A1 expression levels and black lines represent the survival time of patients with cancer with low COL5A1 expression levels. (A) Overall survival of patients with BC based on COL5A1 levels provided by Kaplan-Meier plotter. (B) Disease-free survival of patients with BC based on COL5A1 levels provided by Kaplan-Meier plotter; (C) Overall survival of patients with BC based on COL5A1 levels provided by GEPIA; (D) Disease-free survival of patients with BC based on COL5A1 levels provided by GEPIA. GEPIA, Gene Expression Profiling Interactive Analysis; BC, breast cancer; COL5A1, collagen type V α-1 chain; HR, hazard ratio; TPM, transcripts per million.
Figure 5.
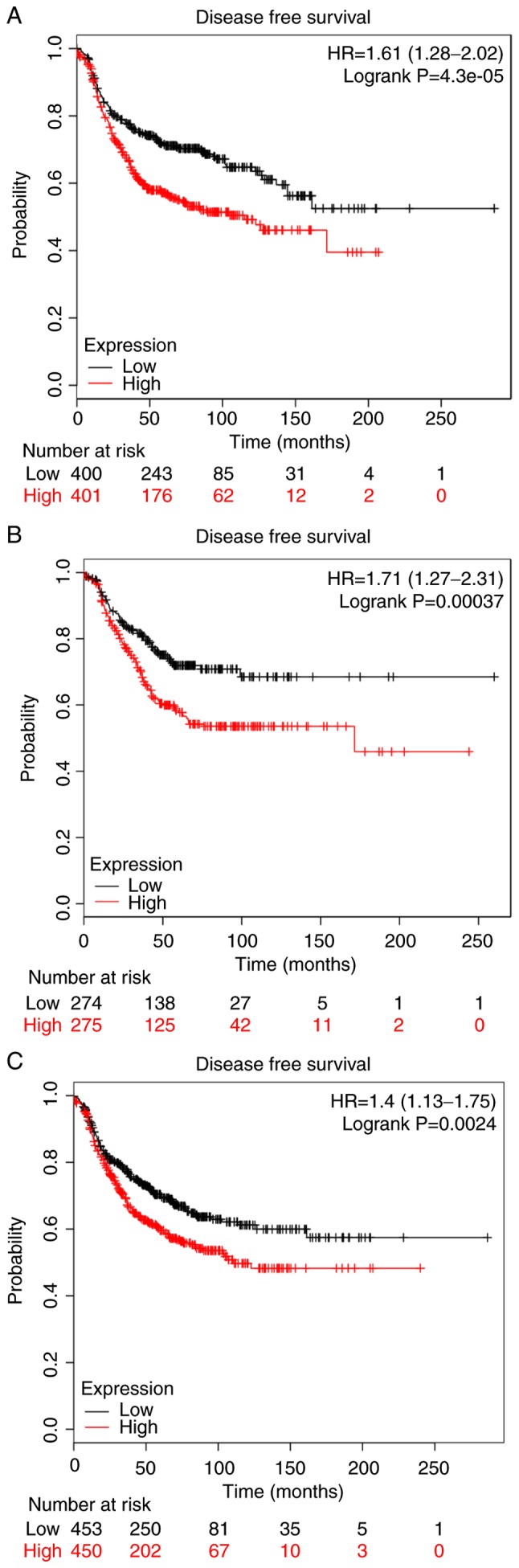
Disease-free survival of patients with BC based on COL5A1 levels and clinical parameters in breast cancer based on Kaplan-Meier plotter. Red lines represent the survival time of patients with cancer with high COL5A1 expression levels and black lines represent the survival time of patients with cancer with low COL5A1 expression levels. (A) ER negative, (B) PR negative and (C) histological grade III. HR, hazard ratio; COL5A1, collagen type V α-1 chain.
Clinical significance of COL5A1 genetic alteration
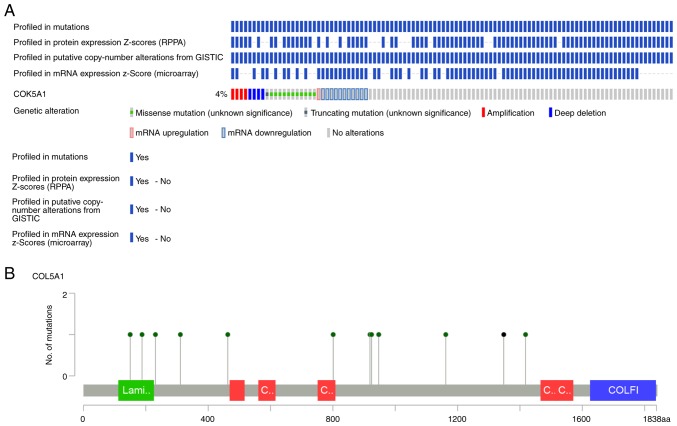
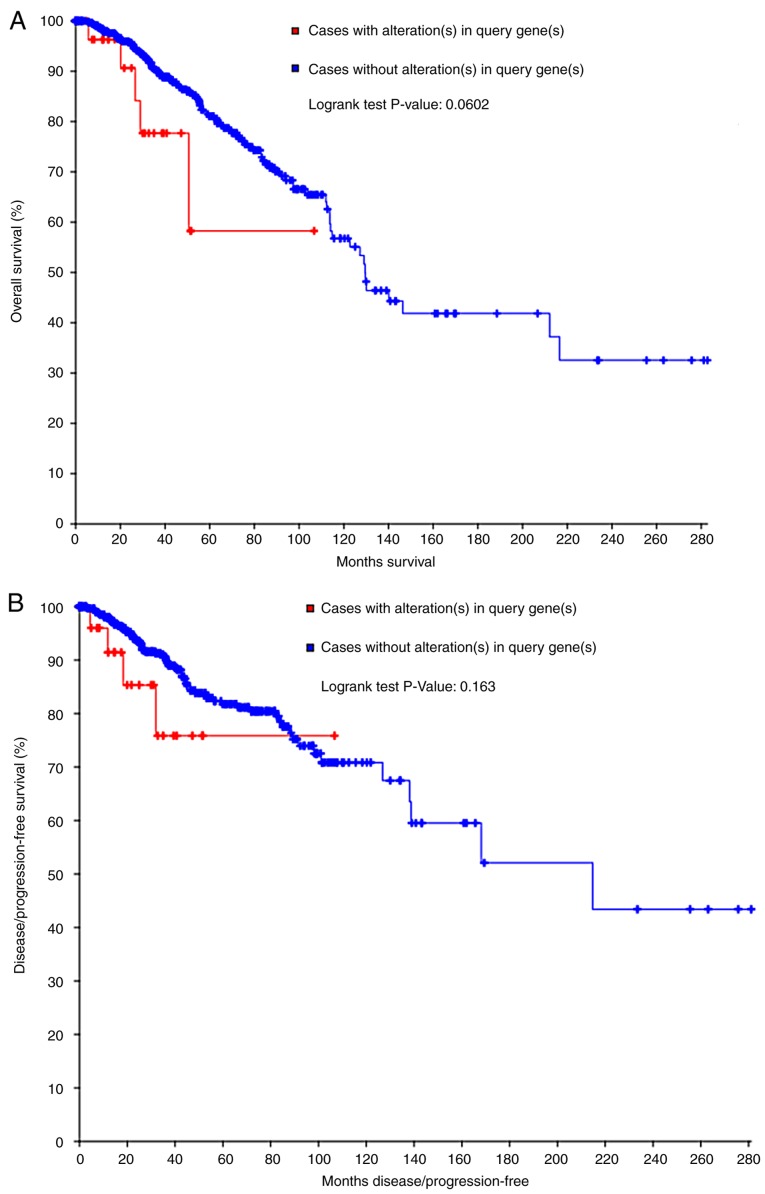
Mining the OncoPrint data demonstrated that COL5A1 altered in 32/817 (4%) sequenced samples within four cases of amplification, 11 cases of missense mutation, four cases of deep deletion, one case of mRNA upregulation and 11 cases of mRNA downregulation (Fig. 6A). Summarizing and analyzing the data revealed the somatic mutation frequency of COL5A1 to be 1.5%, and missense mutations were most prominent in BC (Fig. 6B). From the survival curves, neither the OS nor DFS estimations demonstrated statistical differences. However, a separating tendency was observed in Fig. 7, which indicated that patients without COL5A1 alterations had improved prognoses, compared with those with COL5A1 alterations.
Figure 6.
Genetic alteration data of COL5A1 in BC based on OncoPrint. (A) The OncoPrint schematic revealed that the COL5A1 gene altered in 32/817 (4%) sequenced samples with 4 cases of amplification, 11 cases of missense mutation, 4 cases of deep deletion, 1 case of mRNA upregulation, 1 case of truncating mutation and 11 cases of mRNA downregulation. (B) The mutated locations of COL5A1 based on OncoPrint: Somatic mutation frequency of COL5A1 was 1.5% and missense mutations were most prominent in BC. BC, breast cancer; COL5A1, collagen type V α-1 chain.
Figure 7.
The survival curves for collagen type V α-1 chain alterations. (A) Overall survival and (B) disease-free survival.
Mechanism of COL5A1 in BC
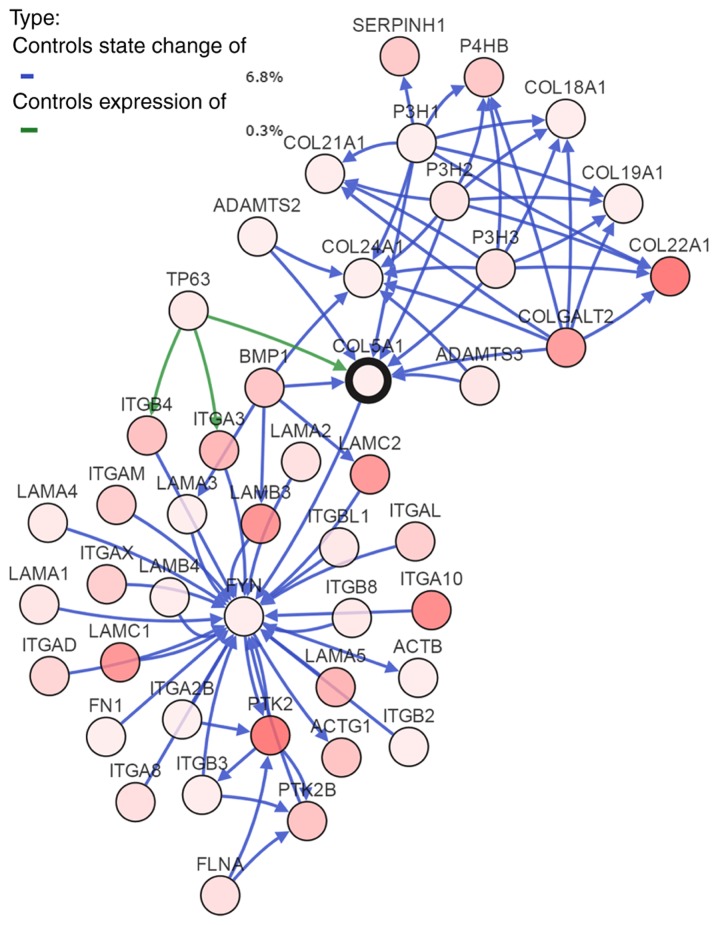
To investigate the mechanism of COL5A1 in BC, the most frequently altered neighbor genes were obtained from cBioPortal. A total of 50 associated genes were displayed as a gene interaction network (Fig. 8). Furthermore, GO functional annotation and KEGG pathway enrichment were performed on these genes. Subsequently, following setting the filter to 5, 6 and 7%, the thresholds of total alteration frequencies, genes with alteration frequencies below these thresholds were filtered out. Results of the GO functional annotation were obtained, as depicted in Fig. 9.
Figure 8.
The gene interaction network of COL5A1-associated genes. The depth of color in each circle represents the alteration frequency. The darker the color, the higher the frequency of alteration. COL5A1, collagen type Vα-1 chain.
Figure 9.
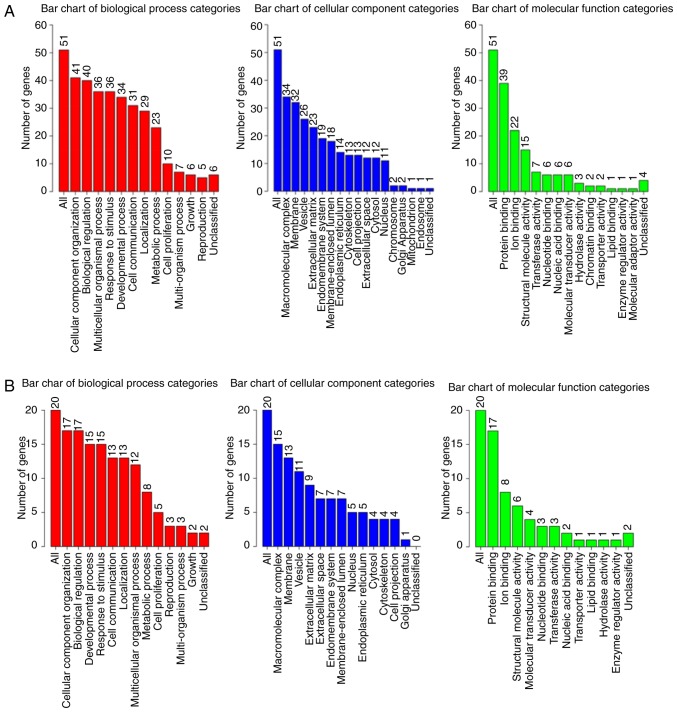
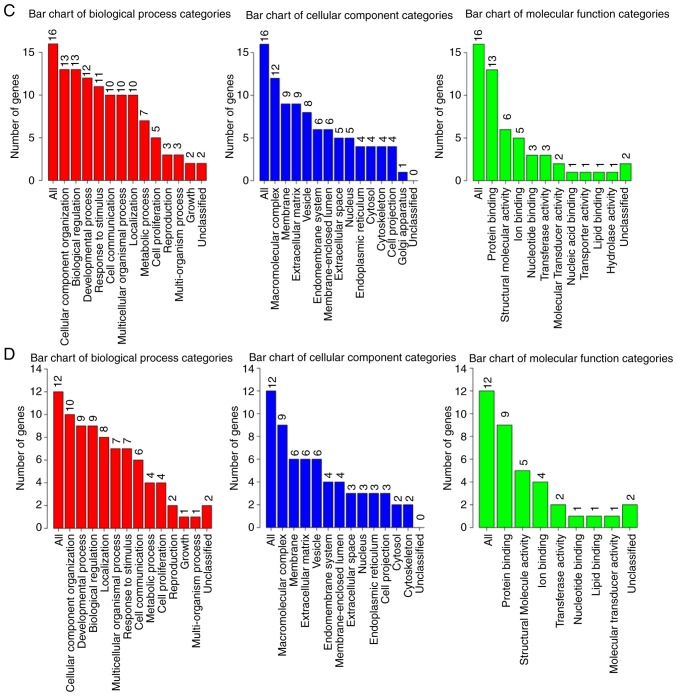
GO analysis bar chart. The columns represent different GO analysis terms. (A) A total of 50 COL5A1-associated genes. (B) COL5A1-associated genes with 5% alteration rate. (C) COL5A1-associated genes with 6% alteration rate. (D) COL5A1-associated genes with 7% alteration rate. GO, Gene Ontology; COL5A1, collagen type V α-1 chain.
GO analysis of the 50 associated genes indicated that the three most significant biological processes included cellular component organization, biological regulation and a multicellular organismal process. Particularly in the cellular component, all three processes involved macromolecular complex, membrane and vesicle; and, in the molecular function, the three most significant remained protein binding, ion binding and structural molecular activity. In the genes of 5, 6 and 7% alteration rates, all three GO analyses demonstrated that the most significant biological process was cellular component organization, along with macromolecular complex in the cellular component and protein binding in the molecular function. The outcomes of alteration rates were consistent with that of COL5A1-associated genes.
Additionally, KEGG analysis confirmed that the most notable DEG-enriched pathways included focal adhesion, extracellular matrix (ECM)-receptor interaction and regulation of the actin cytoskeleton (Fig. 10).
Figure 10.
KEGG pathways of COL5A1-associated genes. The KEGG enrichment analysis was performed with COL5A1-associated genes using the ggplot2 package in R. The color tint indicates the P-value. The size of the circle represents the number of selected genes in the pathway. Rich factor expresses the percentage of the ratio of COL5A1-associated genes in the present study vs. the total genes in the pathway. COL5A1, collagen type V α-1 chain; KEGG, Kyoto Encyclopedia of Genes and Genomes; ECM, extracellular matrix; PI3K, phosphoinositide 3-kinase.
Meta-analysis of COL5A1 mRNA expression
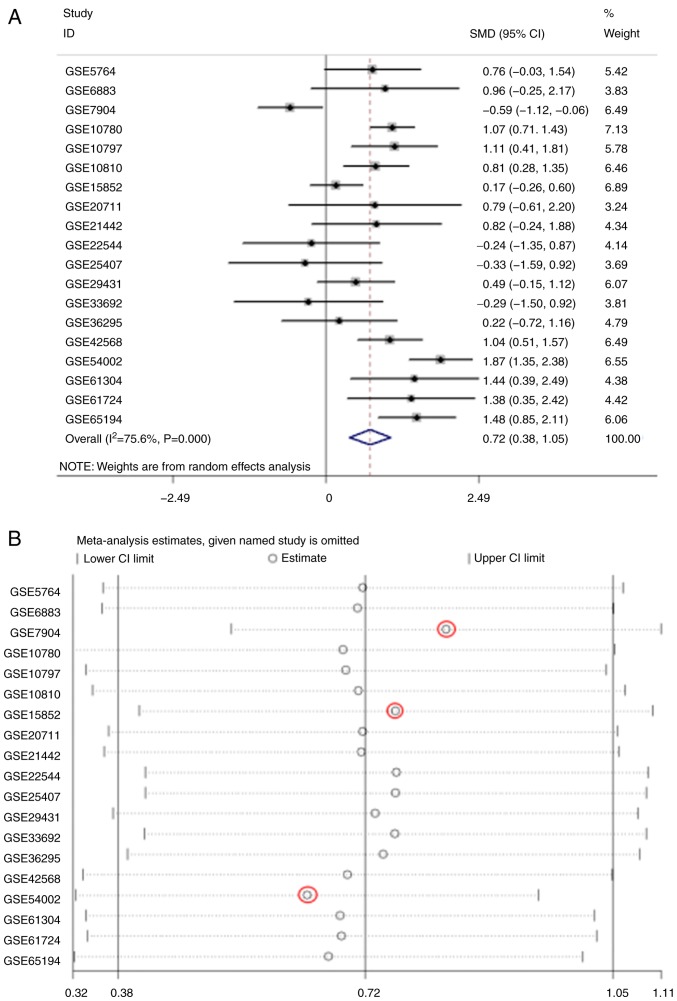
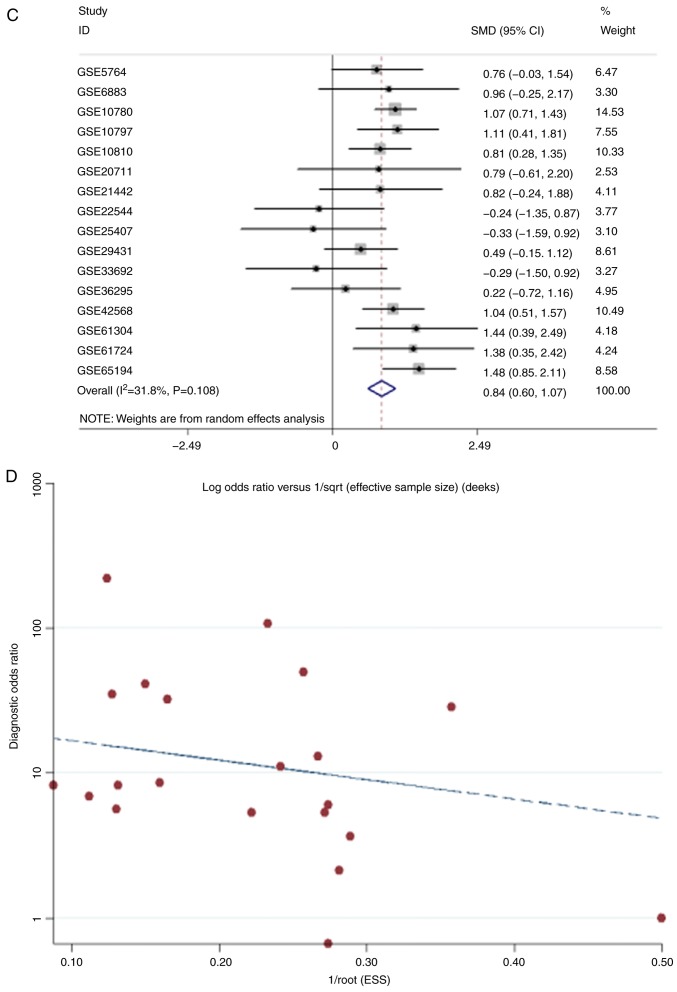
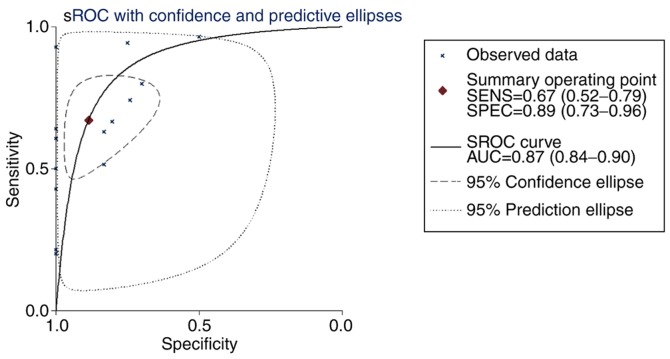
The meta-analysis of COL5A1 expression levels demonstrated high heterogeneity from the research (I2>50%; Fig. 11A). To eliminate heterogeneity and reduce research error, a sensitivity analysis was conducted to further determine the source of heterogeneity. As a result, GSE7904, GSE15852 and GSE54002 were considered the main sources of heterogeneity (Fig. 11B). Following removing these three GEO chips, research heterogeneity was reduced (I2=31.8%; P=0.108; Fig. 11C). Additionally, Deeks' test demonstrated that the bias value of P>|t| was 0.468, also indicating that the study of COL5A1 diagnostic significance had no notable publication bias (Fig. 11D). In conclusion, COL5A1 mRNA expression levels were increased in cancerous tissues, compared with non-cancerous tissues, from the GEO database for BC (SMD, 0.84; 95% CI, 0.60-1.07). The sROC curves were plotted based on TP, FN, TN and FP cases obtained from the GEO datasets. The area under the sROC was 0.87 (95% CI, 0.84-0.90), indicating that COL5A1 has a strong potential diagnostic value for BC (Fig. 12).
Figure 11.
The meta-analysis of COL5A1 expression levels between cancerous tissues and non-cancerous tissues in BC based on the GEO database. (A) The forest plot of COL5A1 expression levels based on 19 microarrays. (B) Sensitivity analysis. Red circles represent the chips from sources of heterogeneity. (C) The forest plot of GEO meta-analysis after eliminating three chips considered to be sources of heterogeneity. (D) The publication bias. Red dots represent each GEO chip and the dotted line represents the regression line. SMD, standard mean difference; CI, confidence interval; COL5A1, collagen type V α-1 chain; GEO, Gene Expression Omnibus.
Figure 12.
The sROC curve of COL5A1 based on 16 microarrays. The y-axis represents specificity and the x-axis represents sensitivity. AUC=0.87 (95% CI, 0.84-0.90). sROC, summary receiver operating curve; AUC, area under the curve; SENS, sensitivity; SPEC, specificity.
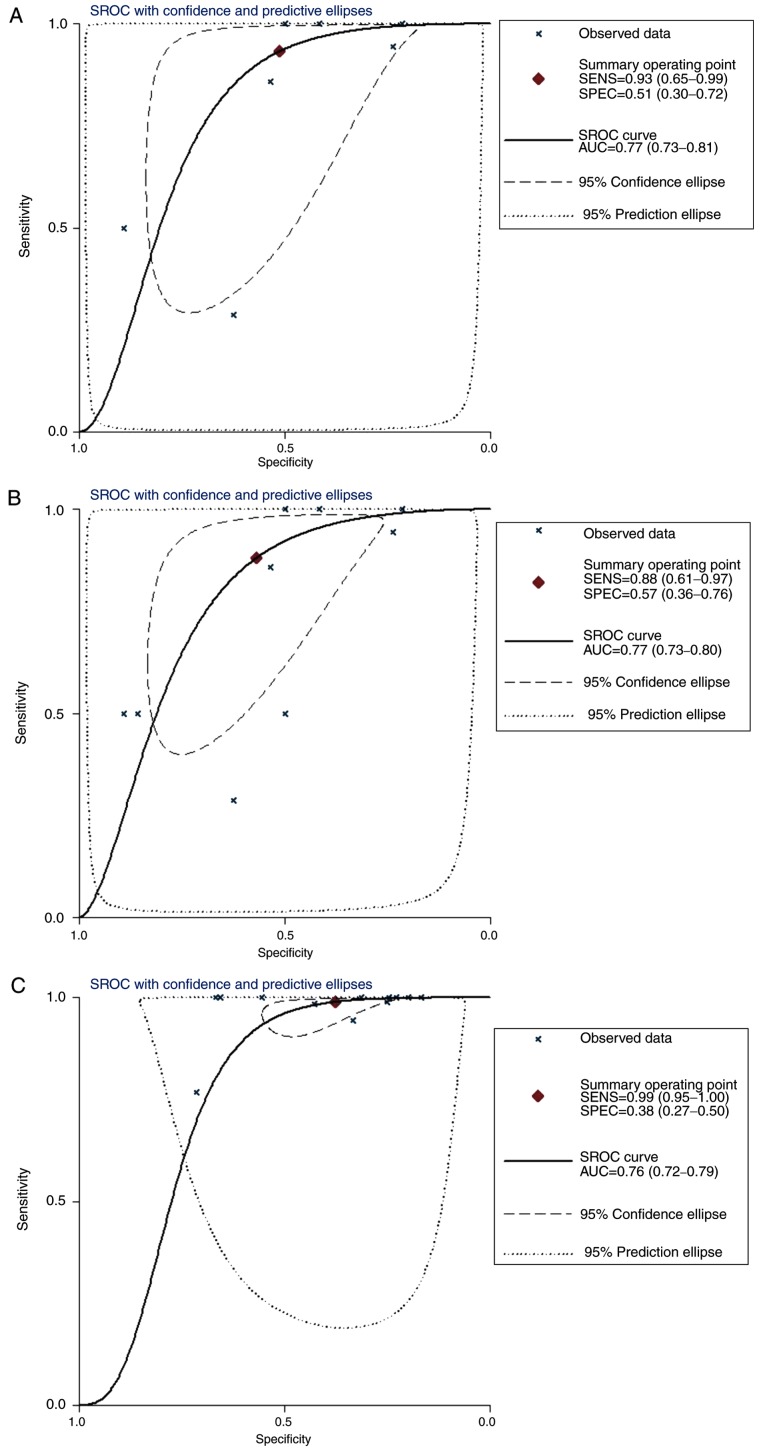
To clarify the significance of COL5A1 in BC subtypes, the subgroup analysis was performed as aforementioned by Stata 12.0 software. All analysis results were displayed in the output window of the Stata 12.0 software. From the outcome of the meta-analysis, high expression levels of COL5A1 were determined to have clinical significance in invasive ductal BC (P=0.004), as well as in un-subdivided invasive BC (P=0.002). The results of the meta-analysis, including ILC, which did not indicate clinical significance (P=0.717), indicated that the expression of COL5A1 in BC was significantly increased, compared with non-cancer tissues (P<0.001) (data not shown). The forest plot for subgroup analysis demonstrated increased heterogeneity in the IDC and BC subgroups. After excluding the three GEO chips aforementioned, the forest plot indicated decreased heterogeneity (P>0.05; Fig. 13). The ROC curves of the chip data for each subtype are depicted in Fig. 14, and the sROC curves for each subtype are depicted in Fig. 15. The AUC was 0.77 (95% CI, 0.73-0.81) in IDC, 0.77 (95% CI, 0.73-0.80) in both IDC and ILC, and 0.76 (95% CI, 0.72-0.79) in un-subdivided invasive BC.
Figure 13.
The subgroup meta-analysis of COL5A1 expression levels between cancerous and non-cancerous tissues in BC based on the Gene Expression Omnibus database. The forest plot of COL5A1 expression levels for (A) IDC, ILC and BC subgroups, and (B) following the removal of GSE7904, GSE15852 and GSE54002. COL5A1, collagen type V α-1 chain; SMD, standard mean difference; CI, confidence interval; BC, breast cancer; IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma.
Figure 14.
The receiver operating curves of collagen type V α-1 chain in BC subtypes between cancerous and non-cancerous BC tissues based on each Gene Expression Omnibus chip. The y-axis represents 1-specificity and the x-axis represents sensitivity. BC, breast cancer; IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma; AUC, area under the curve.
Figure 15.
The sROC curve of collagen type V α-1 chain based on microarrays for BC subgroups. The y-axis represents specificity and the x-axis represents sensitivity. (A) The AUC was 0.77 with a 95% CI of 0.73-0.81 in IDC, (B) 0.77 with a 95% CI of 0.73-0.80 in both IDC and ILC and (C) 0.76 with a 95% CI of 0.72-0.79 in un-subdivided invasive breast cancer. BC, breast cancer; IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma; AUC, area under the curve; sROC, summary receiver operating curve; SENS, sensitivity; SPEC, specificity.
Validation of COL5A1 protein expression levels detected by immunohistochemistry
Selected BC cases and associated clinical data
According to the selection criteria in the present study, 136 cases of BC were considered alongside 55 pairs of normal breast tissue adjacent to cancer. The selected patients were all female, aged 29–79 years old, with a median age of 47 years old. The clinical and pathological features of the 136 BC cases are presented in Table I. During the tracking period, the longest follow-up time was 2,242 days, and the shortest was 162 days. There were 12 cases of death due to BC; 15 cases were lost to follow-up, and the remaining 121 cases were entered as samples for survival analysis.
Table I.
A total of 136 cases of clinical and pathological features of invasive ductal carcinoma.
| Clinicopathological variables | Cases | % |
|---|---|---|
| Age (years) | ||
| ≤50 | 90 | 66.2 |
| >50 | 46 | 33.8 |
| Histological grade (8) | ||
| I | 9 | 6.6 |
| II | 88 | 64.7 |
| III | 39 | 28.7 |
| Pathological stage (pTNM) (8) | ||
| Tumor size (cm) | ||
| ≤2 | 33 | 24.3 |
| 2-5 | 80 | 58.8 |
| >5 | 23 | 16.9 |
| Lymph node metastasis (8) | ||
| 0 | 49 | 36 |
| 1-3 | 43 | 31.6 |
| 4-9 | 26 | 19.2 |
| ≥10 | 18 | 13.2 |
| Distant metastasis (8) | ||
| M0 | 129 | 94.9 |
| M1 | 7 | 5.1 |
| Clinical stage (8) | ||
| I | 17 | 12.5 |
| II | 68 | 50 |
| III | 44 | 32.4 |
| IV | 7 | 5.1 |
| Molecular types (8) | ||
| Luminal A | 16 | 12.5 |
| Luminal B (HER-2 negative) | 63 | 49.1 |
| Luminal B (HER-2 positive) | 15 | 11.7 |
| HER-2 (overexpression) | 16 | 12.5 |
| Triple negative type | 18 | 14.1 |
M, metastasis; HER-2, human epidermal growth factor receptor-2; TNM, Tumor Node Metastasis.
Experimental validation of COL5A1 expression in BC and adjacent tissue
While the IHC method was applied to stain all sections, in 136 cases of BC tissues and adjacent tissues, zero cases were negative, 13 cases were weakly positive, 55 were moderately positive and 68 cases had strongly positive expression levels of COL5A1 (Table II). Additionally, zero cases of BC tissues exhibited negative COL5A1 expression, 25 cases were weakly positive, 25 cases were moderately positive and 21 cases were strongly positive (Figs. 16 and 17). The results are displayed in Table III.
Table II.
Immunohistochemical staining results of COL5A1 in cancerous and adjacent tissues.
| COL5A1 | |||||
|---|---|---|---|---|---|
| Group | Negative | Weakly positive | Moderately positive | Strongly positive | Sum |
| BRCA tissues | 0 | 13 | 55 | 68 | 136 |
| Adjacent tissues | 0 | 9 | 25 | 21 | 55 |
| Sum | 0 | 22 | 80 | 89 | 191 |
BC, breast cancer; COL5A1, collagen type V α-1 chain.
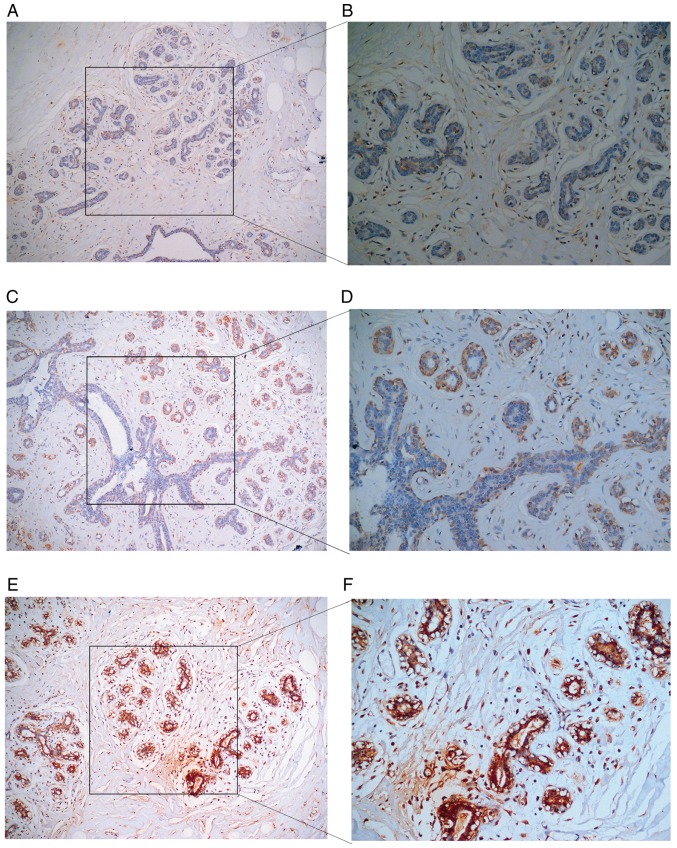
Figure 16.
Expression of collagen type V α-1 chain protein in paraneoplastic tissues. (A) Weakly positive expression in paraneoplastic breast epithelial tissues (original magnification, ×100). (B) Weakly positive expression in paraneoplastic breast epithelial tissues (original magnification, ×200). (C) Moderately positive expression in paracancerous epithelial breast tissues (original magnification, ×100). (D) Moderately positive expression in paracancerous epithelial breast tissues (original magnification, ×200). (E) Strongly positive expression in paraneoplastic epithelial breast tissues (original magnification, ×100). (F) Strongly positive expression in paraneoplastic epithelial breast tissues (original magnification, ×200).
Figure 17.
Expression of collagen type V α-1 chain in breast cancer. (A) Weakly positive expression in breast cancer tissues (original magnification, ×100). (B) Weakly positive expression in breast cancer tissues (original magnification, ×200). (C) Moderately positive expression in breast cancer tissues (original magnification, ×100). (D) Moderately positive expression in breast cancer tissues (original magnification, ×200). (E) Strongly positive expression in breast cancer tissues (original magnification, ×100). (F) Strongly positive expression in breast cancer tissues (original magnification, ×200).
Table III.
COL5A1 expression in BC and adjacent tissues.
| COL5A1 | |||||
|---|---|---|---|---|---|
| Group | Low expression (%) | High expression (%) | Sum | χ2 | P-value |
| BRCA tissues | 13 (9.6) | 123 (90.4) | 136 | 1.779 | 0.182 |
| Adjacent tissues | 9 (16.4) | 46 (83.6) | 55 | ||
| Sum | 22 | 169 | 191 | ||
BC, breast cancer; COL5A1, collagen type V α-1 chain.
The expression of COL5A1 protein in BC tissues demonstrated no statistically significant difference between ER, PR, HER-2, P53 and Ki-67 (P>0.05) (Table IV), nor did it indicate a statistically significant difference in age, histological grade, tumor size, lymph node metastasis, distant metastasis, clinical stage and molecular type (P>0.05; Table V).
Table IV.
Association between COL5A1 and ER, PR, P53, HER-2 and Ki-67 expression in breast cancer.
| COL5A1 | |||||
|---|---|---|---|---|---|
| Factors | Cases | Low expression | High expression | χ2 | P-value |
| ER | 0.247 | 0.619 | |||
| Negative | 45 | 3 | 42 | ||
| Positive | 91 | 10 | 81 | ||
| PR | 0.247 | 0.619 | |||
| Negative | 45 | 3 | 42 | ||
| Positive | 91 | 10 | 81 | ||
| HER-2 (128 cases) | 0.05 | 0.823 | |||
| Negative | 98 | 9 | 89 | ||
| Positive | 30 | 3 | 27 | ||
| P53 | 0.003 | 0.957 | |||
| Negative | 88 | 9 | 79 | ||
| Positive | 48 | 4 | 44 | ||
| Ki-67 | 0.158 | 0.691 | |||
| <14% | 21 | 2 | 19 | ||
| ≥14% | 115 | 11 | 104 | ||
ER, estrogen; PR, progesterone; HER-2, human epidermal growth factor receptor-2; COL5A1, collagen type V α-1 chain.
Table V.
Expression of COL5A1 in breast cancer and its association with clinicopathological features.
| COL5A1 | |||||
|---|---|---|---|---|---|
| Clinicopathological variables | Cases | Low expression | High expression | χ2 | P-value |
| Age (years) | 0.138 | 0.710 | |||
| ≤50 | 90 | 8 | 82 | ||
| >50 | 46 | 5 | 41 | ||
| Histological grade (8) | 0.248 | 0.619 | |||
| I+II | 97 | 8 | 89 | ||
| III | 39 | 5 | 34 | ||
| Tumor size (cm) | 0.946 | 0.623 | |||
| ≤2 | 33 | 4 | 29 | ||
| 2-5 | 80 | 8 | 72 | ||
| >5 | 23 | 2 | 21 | ||
| Lymph node metastasis (8) | 0.013 | 0.911 | |||
| No (N0) | 49 | 5 | 44 | ||
| Yes (N1-N3) | 87 | 8 | 79 | ||
| Distant metastasis (8) | 0.693 | 0.405 | |||
| M0 | 129 | 11 | 118 | ||
| M1 | 7 | 2 | 5 | ||
| Clinical stage (8) | 0.142 | 0.707 | |||
| I+II | 85 | 7 | 78 | ||
| III+IV | 51 | 6 | 45 | ||
| Molecular type (8) (128 cases) | 0.748 | 0.862 | |||
| Luminal A | 16 | 1 | 15 | ||
| Luminal B | 77 | 8 | 70 | ||
| HER-2 (overexpression) | 16 | 2 | 14 | ||
| Triple negative type | 18 | 1 | 17 | ||
COL5A1, collagen type V α-1 chain; N, node; M, metastasis.
Spearman's correlation analysis of COL5A1 expression and clinicopathological parameters
Spearman's correlation analysis between COL5A1 and the clinicopathological parameters demonstrated that the specific gene in BC had no statistical significance in terms of tumor size (r=0.032; P=0.713), lymph node metastasis (r=−0.033; P=0.700), histological grade (r=−0.093; P=0.282), clinical stage (r=−0.027; P=0.757), molecular typing (r=0.023; P=0.791), ER (r=−0.069; P=0.424), PR (r=−0.069; P=0.4246), Ki-67 (r=0.032; P=0.714), P53 (r=0.031; P=0.722) or HER-2 (r=0.061; P=0.493) (data not shown).
Association between COL5A1 protein expression level and prognosis
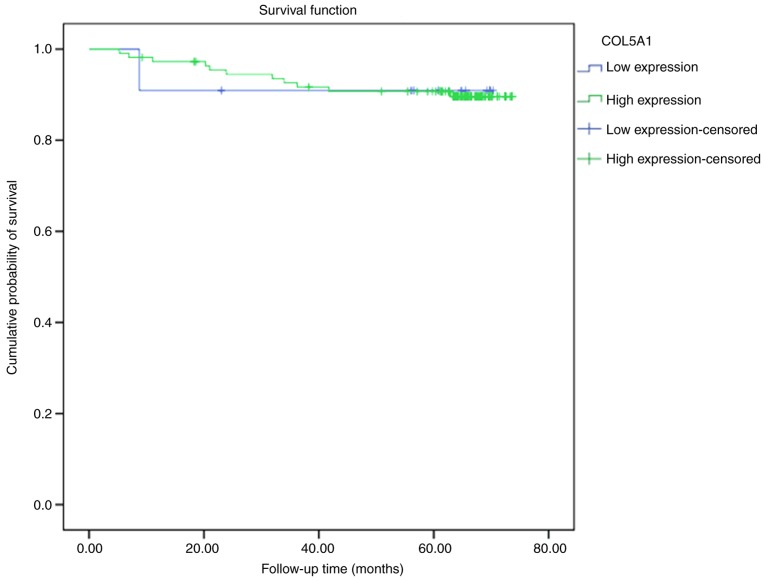
In the 121 studied cases of BC, 11 cases had low expression levels of COL5A1, with one case of mortality, and the mean survival time was 64.72 months (95% CI, 54.27-75.18), and 110 cases were high-expression cases, with 11 mortalities and a mean survival time of 68.89 months (95% CI, 65.05-71.73) (data not shown). A log-rank test demonstrated no statistical difference between the low and high expression levels of the COL5A1 protein in survival curves (P=0.985; Fig. 18).
Figure 18.
Kaplan-Meier survival curves of COL5A1 expression and overall survival. The y-axis represents the cumulative probability of survival and the x-axis represents follow-up time. COL5A1, collagen type V α-1 chain.
HPA Database verification
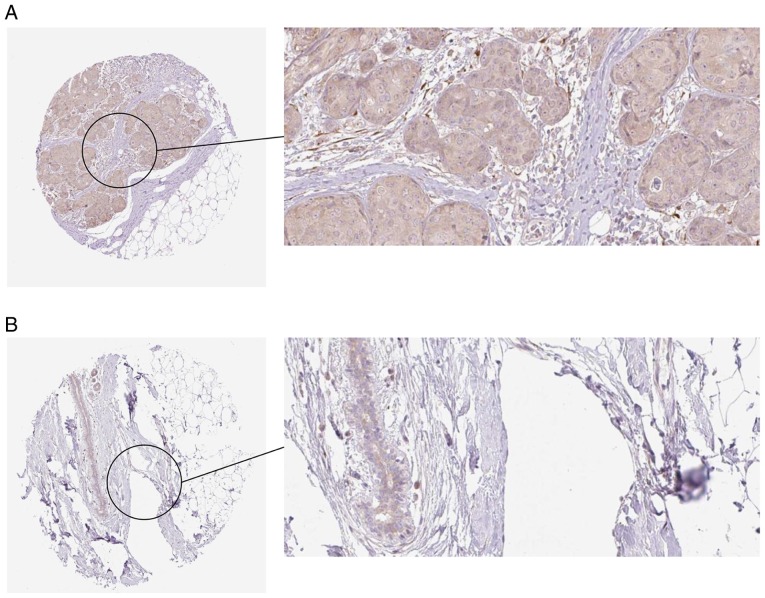
The results from the HPA Database further demonstrated a trend toward a high expression of COL5A1 at the protein level in BC (Fig. 19).
Figure 19.
COL5A1 expression in BC and non-tumor tissues from the Human Protein Atlas database. Immunohistochemical staining revealed that (A) COL5A1 exhibited high expression in BC tissues and (B) low expression in non-tumor breast tissue samples. COL5A1, collagen type V α-1 chain; BC, breast cancer.
Discussion
At present, BC is a cancer seriously affecting women's health globally in 2018 (1,2). Due to its complex pathogenesis, research on its molecular pathogenic factors has become prevalent. With the rapid development of medical technology, increasing numbers of molecular markers associated with BC have been determined and investigated (31,32), continually raising the accuracy of early BC screening. However, due to individual differences, single biomarkers cannot meet the requirements of universal diagnosis, and the markers for multi-gene combined screening are more conducive to diagnosis and evaluation of BC prognosis (33,34). The discovery of markers can also help develop individualized treatment, including molecular targeted therapies. Elucidating the association between gene expression and clinicopathological characteristics can support physicians in selecting appropriate treatments for their patients in clinical practice.
The present study obtained the differential genes associated with BC through screening the GEO and TCGA databases. The genes that appeared more than five times in each database were intersected to gain the significant difference genes. Genes with significant differences were then used to perform, GO and KEGG pathway analyses. The present literature review revealed that COL5A1 has been regarded as a risk biomarker for poor prognoses in ovarian cancer and kidney carcinoma. Considering that COL5A1 may function in BC development, the present study further investigated this target gene.
The COL5A1 gene encodes COL5A1, which is in smaller fibrillar collagen in mammals. Numerous studies on COL5A1 primarily focused on single nucleotide polymorphisms, motor injuries and connective tissue injuries (35–39). COL5A1 is rarely reported in cancer research, which is, instead, primarily predicted by bioinformatics (40,41). However, previous studies indicated that COL5A1 is predicted to have a notable role in BC (42,43). Therefore, the accuracy of this prediction required further experimental verification.
Through a meta-analysis of the BC gene expression profiles from 19 GEO chips, expression levels of COL5A1 were determined to be increased in BC tissues, compared with normal breast tissues, in 14 chips, with nine chips having significantly different expression (P<0.05). Additionally, the expression of COL5A1 was significantly increased in the 1,085 BC cases from TCGA database, compared with their adjacent tissues. A forest plot indicated the significance of COL5A1 expression (SMD, 0.84; 95% CI, 0.60-1.07). In addition, the AUC of COL5A1 was 0.87 (95% CI, 0.84-0.90), indicating that COL5A1 has a strong potential diagnostic value for early BC. In a subgroup analysis, COL5A1 also had diagnostic significance in the IDC subtype (AUC, 0.77; 95% CI, 0.73-0.81). Although the OS and DFS survival curves did not demonstrate statistical significance, a different trend which indicated that elevated COL5A1 mRNA level predicted poor prognosis of BC has been observed. If more cases can be included, the association between COL5A1 and the clinical prognosis of BC may be demonstrated further. Furthermore, research heterogeneity has been reduced following eliminating the three GEO chips considered to be the sources of the heterogeneity (GSE7904, GSE15852 and GSE54002). Therefore, no publication bias existed, and the results were credible. For molecular type, ER and PR negative were determined to be significantly associated with DFS in BC, as well as histological grade III. Therefore, the high expression of COL5A1 mRNA indicated poor prognosis for patients with ER and PR negative. The increased COL5A1 mRNA expression level also indicated that patients may have an increased histological grade type and malignant degree of BC. This indicated that clinicians may predict the prognosis of patients with BC according to their COL5A1 mRNA levels, in order to timely communicate with patients and adjust the treatments.
At the protein level, the HPA database confirmed the high expression of COL5A1 in BC, which was consistent with the in-house immunohistochemistry. There are a number of pathological types of BC, with invasive types accounting for >70% (29,30). The in-house cases were strictly selected to ensure the reliability of experimental results. The present immunohistochemical results confirmed that COL5A1 protein was highly expressed in invasive BC (90.4%), but they also exhibited no statistical significance, compared with normal tissue. The expression level of COL5A1 was not statistically significant with the age, histological grade, tumor size, lymph node metastasis, distant metastasis, clinical stage, molecular type, ER, PR, HER-2, P53 and Ki-67 of patients with BC (P>0.05). However, COL5A1 may be more appropriate as a combined, rather than individual, marker for diagnosing and treating BC. Therefore, it will be necessary for future studies to consider more research samples to verify, and further clarify, these observations. Furthermore, the protein level of COL5A1 in other subtypes of BC will be verified in the planned, follow-up, in-house experiment after sufficient clinical cases are collected.
The present study also determined that the COL5A1 mutation (4% mutation rate) is associated with the prognosis of patients with BC, and patients with BC with COL5A1 mutation may have a reduced prognosis. Investigating the underlying mechanisms, GO analysis indicated that the most important biological process is cellular component organization, as well as macromolecular complexes in molecular function and protein binding in cellular components. The GO pathway for screening genes, based on the alteration rate in line with the aforementioned pathways, indicated the reliability of pathway prediction. According to the KEGG pathway analysis, the 50 most frequently-altered neighboring genes may affect BC by regulating focal adhesion, ECM-receptor interactions and regulation of the actin cytoskeleton. Previous BC studies focused on tumor epithelial cells (44–46), while other factors, including microenvironment, myoepithelial cells and the potential role of stromal cells in tumor progression, have not been well studied. In vivo and in vitro studies demonstrated that cells constituting the microenvironment, including muscle epithelium and endothelial cells, fibroblasts and myofibroblasts, and ECM molecules may regulate the growth and survival of normal breast tissue and the invasion of BC cells (44,47–49). Associated studies confirmed that the ECM-receptor interaction signaling pathway is associated with BC bone metastasis (50), and it is involved in thyroid papillary carcinoma (51), oral squamous cell carcinoma (52) and early lung adenocarcinoma (53). Additionally, research on ovarian cancer research containing 10 genes [adipocyte enhancer-binding protein 1, COL11A1, COL5A1, COL6A2, lysyl oxidase, periostin, snail family transcriptional repressor 2, thrombospondin 2 (THBS2), tissue inhibitor of metalloproteinases 3 (TIMP3) and versican] associated with ovarian cancer predicted a poor prognosis for patients with ovarian cancer and included the joint gene COL5A1 (31,54,55). Subsequently, 10 genes (COL1A1, COL5A1, COL11A1, fibronectin 1, intercellular adhesion molecule 1, integrin subunit αL, integrin subunit αM, integrin subunit β2, THBS2 and TIMP1) were determined to be associated with poor prognosis in renal cell carcinoma, including COL5A1 (56). COL5A1 is featured in all of these studies, and it is the specific gene in the present study that involves the interaction of ECM receptors, all of which indicates that this particular gene serves an essential role in the cancer pathogenesis. Following a comprehensive multi-factor analysis, the present study considered COL5A1 as crucial to BC development.
Notably, a study by Ren et al determined that a high expression of COL5A1 indicated an improved prognosis in patients with BC without lymph node metastasis. Firstly, the study by Ren et al supports the high expression of COL5A1 in IDC of BC (10), which is consistent with the conclusions of the present study. Secondly, due to different sample sources in the two studies, including race, regional environment and lifestyle, may result in different conclusions should be considered. Death factors may also impact lymph node metastasis, which means mortality cases without lymph node metastasis should probably not be considered. The present study evaluated the clinical value of COL5A1 in BC, including OS, DFS, clinical parameters and molecular mechanisms, and supplemented with immunohistochemistry to verify bioinformatics results based on high-throughput data. Additionally, the sample size was large, and the results are highly reliable.
Although COL5A1 has been verified at the mRNA and protein levels, this biomarker is also highly expressed at the transcription level in BC. However, the difference in expression is not reflected at the protein level. Additionally, post-transcriptional modification and post-translational modification, including glycosylation, hydrolytic processing and phosphorylation, may induce protein degradation in vivo during this process, thereby affecting protein expression (57,58). However, in the course of the experiment, unavoidable experimental errors and losses may have occurred in the detection of mRNA and the verification of the immunohistochemistry.
In conclusion, using computational biology and immunohistochemistry, the present study demonstrated that COL5A1 is highly expressed at the mRNA and protein levels in BC. Based on the survival curves, the patients with BC with high COL5A1 expression have a reduced prognosis. Furthermore, COL5A1 may impact the development of BC by regulating pathways, including focal adhesion, ECM-receptor interaction and the regulation of the actin cytoskeleton. Nonetheless, further clinical trials, including more BC cases, are required to verify the results of the present study. It is considered that COL5A1 may be used as an effective single, or combined, indicator for clinical diagnosis and prediction of BC in the future.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- COL5A1
collagen type V α-1 chain
- BC
breast cancer
- IHC
immunohistochemistry
- TCGA
The Cancer Genome Atlas
- GEO
Gene Expression Omnibus
- DEGs
differentially-expressed genes
- GEPIA
Gene Expression Profiling Interactive Analysis
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- ROC
Receiver Operating Characteristic
- sROC
summary ROC curve
- ER
estrogen
- PR
progesterone
Funding
No funding was received.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
MW and QS analyzed and interpreted data, and drafted the manuscript. CHM and JYH performed the majority of the experiments as well as statistical analysis, and supervised the progression of research. JSP, LLP, HPL, YWD, SJF, DT, GC and ZBF participated in sample collection and provided information from the databases. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study protocol was approved by the Ethics Committee of First Affiliated Hospital of Guangxi Medical University. All the patients signed the written informed consents based on the guidelines of the First Affiliated Hospital of Guangxi Medical University prior to participating in the present study. All tissue samples were collected anonymous according to the ethical and legal standards.
Patient consent for publication
All patients provided written informed consent before participation and agreed to publication of the present study.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Yang ZJ, Yu Y, Chi JR, Guan M, Zhao Y, Cao XC. The combined pN stage and breast cancer subtypes in breast cancer: A better discriminator of outcome can be used to refine the 8th AJCC staging manual. Breast Cancer. 2018;25:315–324. doi: 10.1007/s12282-018-0833-0. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 4.An N, Shi Y, Ye P, Pan Z, Long X. Association between MGMT promoter methylation and breast cancer: A Meta-analysis. Cell Physiol Biochem. 2017;42:2430–2440. doi: 10.1159/000480196. [DOI] [PubMed] [Google Scholar]
- 5.Casey MC, Sweeney KJ, Brown JA, Kerin MJ. Exploring circulating micro-RNA in the neoadjuvant treatment of breast cancer. Int J Cancer. 2016;139:12–22. doi: 10.1002/ijc.29985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Savci-Heijink CD, Halfwerk H, Koster J, Van de Vijver MJ. Association between gene expression profile of the primary tumor and chemotherapy response of metastatic breast cancer. BMC Cancer. 2017;17:755. doi: 10.1186/s12885-017-3691-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin YH, Hua QF, Zheng JJ, Ma XH, Chen TX, Zhang S, Chen B, Dai Q, Zhang XH. Diagnostic value of ER, PR, FR and HER-2-targeted molecular probes for magnetic resonance imaging in patients with breast cancer. Cell Physiol Biochem. 2018;49:271–281. doi: 10.1159/000492877. [DOI] [PubMed] [Google Scholar]
- 8.Akram M, Iqbal M, Daniyal M, Khan AU. Awareness and current knowledge of breast cancer. Biol Res. 2017;50:33. doi: 10.1186/s40659-017-0140-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Polyak K. Breast cancer: Origins and evolution. J Clin Invest. 2007;117:3155–3163. doi: 10.1172/JCI33295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ren W, Zhang Y, Zhang L, Lin Q, Zhang J, Xu G. Overexpression of collagen type V α1 chain in human breast invasive ductal carcinoma is mediated by TGF-β1. Int J Oncol. 2018 Mar 15; doi: 10.3892/ijo.2018.4317. (Epub ahead of print). doi: 10.3892/ijo.2018.4317. [DOI] [PubMed] [Google Scholar]
- 11.Lee S, Lee J, Sim SH, Lee Y, Moon KC, Lee C, Park WY, Kim NK, Lee SH, Lee H. Comprehensive somatic genome alterations of urachal carcinoma. J Med Genet. 2017;54:572–578. doi: 10.1136/jmedgenet-2016-104390. [DOI] [PubMed] [Google Scholar]
- 12.Chai F, Liang Y, Zhang F, Wang M, Zhong L, Jiang J. Systematically identify key genes in inflammatory and non-inflammatory breast cancer. Gene. 2016;575:600–614. doi: 10.1016/j.gene.2015.09.025. [DOI] [PubMed] [Google Scholar]
- 13.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou GX, Liu P, Yang J, Wen S. Mining expression and prognosis of topoisomerase isoforms in non-small-cell lung cancer by using Oncomine and Kaplan-Meier plotter. PLoS One. 2017;12:e0174515. doi: 10.1371/journal.pone.0174515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 16.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang HL, Chang KK, Mei J, Zhou WJ, Liu LB, Yao L, Meng Y, Wang MY, Ha SY, Lai ZZ, et al. Estrogen restricts the apoptosis of endometrial stromal cells by promoting TSLP secretion. Mol Med Rep. 2018;18:4410–4416. doi: 10.3892/mmr.2018.9428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sas-Korczynska B, Reinfuss M, Mitus JW, Pluta E, Patla A, Walasek T. Radiotherapy alone as a method of treatment for sinonasal mucosal melanoma: A report based on six cases and a review of current opinion. Rep Pract Oncol Radiother. 2018;23:402–406. doi: 10.1016/j.rpor.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. WebGestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017;45:W130–W137. doi: 10.1093/nar/gkx356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai Y, Sun L, Qiang W. A new strategy to uncover the anticancer mechanism of Chinese compound formula by integrating systems pharmacology and bioinformatics. Evid Based Complement Alternat Med. 2018;2018:6707850. doi: 10.1155/2018/6707850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frank GA, Danilova NV, Andreeva I, Nefedova NA. WHO classification of tumors of the breast, 2012. Arkh Patol. 2013;75:53–63. (In Russian) [PubMed] [Google Scholar]
- 24.Gyorffy B, Pongor L, Bottai G, Li X, Budczies J, Szabó A, Hatzis C, Pusztai L, Santarpia L. An integrative bioinformatics approach reveals coding and non-coding gene variants associated with gene expression profiles and outcome in breast cancer molecular subtypes. Br J Cancer. 2018;118:1107–1114. doi: 10.1038/s41416-018-0030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan W, Zhao Y, He J. Anti-breast cancer activity of selected 1,3,5-triazines via modulation of EGFR-TK. Mol Med Rep. 2018;18:4175–4184. doi: 10.3892/mmr.2018.9426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Huang X, Zhang J, Huang H, Zhao L, Yu M, Zhang Y, Wang H. A novel peptide targets CD105 for tumour imaging in vivo. Oncol Rep. 2018;40:2935–2943. doi: 10.3892/or.2018.6643. [DOI] [PubMed] [Google Scholar]
- 27.Noorlag R, van der Groep P, Leusink FK, Leusink FK, van Hooff SR, Frank MH, Willems SM, van Es RJ. Nodal metastasis and survival in oral cancer: Association with protein expression of SLPI, not with LCN2, TACSTD2, or THBS2. Head Neck. 2015;37:1130–1136. doi: 10.1002/hed.23716. [DOI] [PubMed] [Google Scholar]
- 28.Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357:eaan2507. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 29.Thul PJ, Akesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Björk L, Breckels LM, et al. A subcellular map of the human proteome. Science. 2017;356:eaal3321. doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Zhang Y, Huang Q, Li C. Integrated bioinformatics analysis reveals key candidate genes and pathways in breast cancer. Mol Med Rep. 2018;17:8091–8100. doi: 10.3892/mmr.2018.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim GJ, Kim DH, Min KW, Kim YH, Oh YH. Loss of p27kip1 expression is associated with poor prognosis in patients with taxane-treated breast cancer. Pathol Res Pract. 2018;214:565–571. doi: 10.1016/j.prp.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 32.Jimenez-Morales S, Pérez-Amado CJ, Langley E, Hidalgo-Miranda A. Overview of mitochondrial germline variants and mutations in human disease: Focus on breast cancer (Review) Int J Oncol. 2018;53:923–936. doi: 10.3892/ijo.2018.4468. [DOI] [PubMed] [Google Scholar]
- 33.Do SI, Kim HS, Kim K, Lee H, Do IG, Kim DH, Chae SW, Sohn JH. Predictive and prognostic value of sphingosine kinase 1 expression in patients with invasive ductal carcinoma of the breast. Am J Transl Res. 2017;9:5684–5695. [PMC free article] [PubMed] [Google Scholar]
- 34.Kim JH, Jung ES, Kim CH, Youn H, Kim HR. Genetic associations of body composition, flexibility and injury risk with ACE, ACTN3 and COL5A1 polymorphisms in Korean ballerinas. J Exerc Nutrition Biochem. 2014;18:205–214. doi: 10.5717/jenb.2014.18.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim ST, Kim CS, Kim WN, Min SK. The COL5A1 genotype is associated with range of motion. J Exerc Nutrition Biochem. 2015;19:49–53. doi: 10.5717/jenb.2015.15052701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Makoukji J, Makhoul NJ, Khalil M, El-Sitt S, Aldin ES, Jabbour M, Boulos F, Gadaleta E, Sangaralingam A, Chelala C, et al. Gene expression profiling of breast cancer in Lebanese women. Sci Rep. 2016;6:36639. doi: 10.1038/srep36639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu G, Wu K, Sheng Y. Elucidation of the molecular mechanisms of anaplastic thyroid carcinoma by integrated miRNA and mRNA analysis. Oncol Rep. 2016;36:3005–3013. doi: 10.3892/or.2016.5064. [DOI] [PubMed] [Google Scholar]
- 38.An F, Zhang Z, Xia M, Xing L. Subpath analysis of each subtype of head and neck cancer based on the regulatory relationship between miRNAs and biological pathways. Oncol Rep. 2015;34:1745–1754. doi: 10.3892/or.2015.4150. [DOI] [PubMed] [Google Scholar]
- 39.Kumari K, Das B, Adhya AK, Rath AK, Mishra SK. Genome-wide expression analysis reveals six contravened targets of EZH2 associated with breast cancer patient survival. Sci Rep. 2019;9:1974. doi: 10.1038/s41598-019-39122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Di Y, Chen D, Yu W, Yan L. Bladder cancer stage-associated hub genes revealed by WGCNA co-expression network analysis. Hereditas. 2019;156:7. doi: 10.1186/s41065-019-0083-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abrahams Y, Laguette MJ, Prince S, Collins M. Polymorphisms within the COL5A1 3′-UTR that alters mRNA structure and the MIR608 gene are associated with Achilles tendinopathy. Ann Hum Genet. 2013;77:204–214. doi: 10.1111/ahg.12013. [DOI] [PubMed] [Google Scholar]
- 42.Ritelli M, Dordoni C, Venturini M, Chiarelli N, Quinzani S, Traversa M, Zoppi N, Vascellaro A, Wischmeijer A, Manfredini E, et al. Clinical and molecular characterization of 40 patients with classic Ehlers-Danlos syndrome: Identification of 18 COL5A1 and 2 COL5A2 novel mutations. Orphanet J Rare Dis. 2013;8:58. doi: 10.1186/1750-1172-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riches A, Campbell E, Borger E, Powis S. Regulation of exosome release from mammary epithelial and breast cancer cells-a new regulatory pathway. Eur J Cancer. 2014;50:1025–1034. doi: 10.1016/j.ejca.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 44.Rejon C, Al-Masri M, McCaffrey L. Cell polarity proteins in breast cancer progression. J Cell Biochem. 2016;117:2215–2223. doi: 10.1002/jcb.25553. [DOI] [PubMed] [Google Scholar]
- 45.McCuaig R, Wu F, Dunn J, Rao S, Dahlstrom JE. The biological and clinical significance of stromal-epithelial interactions in breast cancer. Pathology. 2017;49:133–140. doi: 10.1016/j.pathol.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 46.Pizon M, Schott DS, Pachmann U, Pachmann K. B7-H3 on circulating epithelial tumor cells correlates with the proliferation marker, Ki-67, and may be associated with the aggressiveness of tumors in breast cancer patients. Int J Oncol. 2018;53:2289–2299. doi: 10.3892/ijo.2018.4551. [DOI] [PubMed] [Google Scholar]
- 47.Luo M, Clouthier SG, Deol Y, Liu S, Nagrath S, Azizi E, Wicha MS. Breast cancer stem cells: Current advances and clinical implications. Methods Mol Biol. 2015;1293:1–49. doi: 10.1007/978-1-4939-2519-3_1. [DOI] [PubMed] [Google Scholar]
- 48.Byler S, Goldgar S, Heerboth S, Leary M, Housman G, Moulton K, Sarkar S. Genetic and epigenetic aspects of breast cancer progression and therapy. Anticancer Res. 2014;34:1071–1077. [PubMed] [Google Scholar]
- 49.Chen X, Pei Z, Peng H, Zheng Z. Exploring the molecular mechanism associated with breast cancer bone metastasis using bioinformatic analysis and microarray genetic interaction network. Medicine (Baltimore) 2018;97:e12032. doi: 10.1097/MD.0000000000012032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao H, Li H. Network-based meta-analysis in the identification of biomarkers for papillary thyroid cancer. Gene. 2018;661:160–168. doi: 10.1016/j.gene.2018.03.096. [DOI] [PubMed] [Google Scholar]
- 51.Li S, Chen X, Liu X, Yu Y, Pan H, Haak R, Schmidt J, Ziebolz D, Schmalz G. Complex integrated analysis of lncRNAs-miRNAs-mRNAs in oral squamous cell carcinoma. Oral Oncol. 2017;73:1–9. doi: 10.1016/j.oraloncology.2017.07.026. [DOI] [PubMed] [Google Scholar]
- 52.Chen M, Liu B, Xiao J, Yang Y, Zhang Y. A novel seven-long non-coding RNA signature predicts survival in early stage lung adenocarcinoma. Oncotarget. 2017;8:14876–14886. doi: 10.18632/oncotarget.14781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Epstein SG, Drucker L, Pomeranz M, Fishman A, Pasmanik-Chor M, Tartakover-Matalon S, Lishner M. First trimester human placenta prevents breast cancer cell attachment to the matrix: The role of extracellular matrix. Mol Carcinog. 2017;56:62–74. doi: 10.1002/mc.22473. [DOI] [PubMed] [Google Scholar]
- 54.Giussani M, Landoni E, Merlino G, Turdo F, Veneroni S, Paolini B, Cappelletti V, Miceli R, Orlandi R, Triulzi T, Tagliabue E. Extracellular matrix proteins as diagnostic markers of breast carcinoma. J Cell Physiol. 2018;233:6280–6290. doi: 10.1002/jcp.26513. [DOI] [PubMed] [Google Scholar]
- 55.Bonnans C, Chou J, Werb Z. Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol. 2014;15:786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boguslawska J, Kedzierska H, Poplawski P, Rybicka B, Tanski Z, Piekielko-Witkowska A. Expression of genes involved in cellular adhesion and extracellular matrix remodeling correlates with poor survival of patients with renal cancer. J Urol. 2016;195:1892–1902. doi: 10.1016/j.juro.2015.11.050. [DOI] [PubMed] [Google Scholar]
- 57.Cheon DJ, Tong Y, Sim MS, Dering J, Berel D, Cui X, Lester J, Beach JA, Tighiouart M, Walts AE, et al. A collagen-remodeling gene signature regulated by TGF-β signaling is associated with metastasis and poor survival in serous ovarian cancer. Clin Cancer Res. 2014;20:711–723. doi: 10.1158/1078-0432.CCR-13-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alqinyah M, Hooks SB. Regulating the regulators: Epigenetic, transcriptional, and post-translational regulation of RGS proteins. Cell Signal. 2018;42:77–87. doi: 10.1016/j.cellsig.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.