Abstract
Background: Emerging evidences have indicated that long non-coding RNAs (LncRNAs) play vital roles in cancer development and progression. Previous studies have suggested that overexpression of SPRY4 intronic transcript 1 (SPRY4-IT1) predicates poor prognosis and promotes tumor progress in cervical cancer (CC). However, the underlying mechanism of SPRY4-IT1 in CC remains unknown. The aim of the present study is to evaluate the function and mechanism of SPRY4-IT1 in CC.
Methods: SPRY4-IT1 was detected by quantitative PCR. Wound-healing assay and Transwell assay were performed to detect cell migration and invasion, respectively. Western blotting assays were used to analyze the protein expression of E-cadherin, N-cadherin and vimentin. Tumor xenografts experiments were performed to detect the effect of SPRY4-IT1 in vivo. Dual luciferase reporter assay was used to investigate potential molecular mechanism of SPRY4-IT1 in CC cells.
Results: SPRY4-IT1 was up-regulated in CC cell lines. Knockdown of SPRY4-IT1 significantly inhibited CC cells migration and invasion in vitro and in vivo. Moreover, knockdown of SPRY4-IT1 significantly suppressed the epithelial–mesenchymal transition (EMT) of CC by increased E-cadherin expression and decreased the N-cadherin and vimentin expression. Mechanically, SPRY4-IT1 could directly bind to miR-101-3p and effectively act as a competing endogenous RNA (ceRNA) for miR-101-3p to regulate the expression of the target gene ZEB1.
Conclusions: Our findings indicate that the SPYR4-IT1/miR-101-3p/ZEB1 axis contributes to CC migration and invasion, which may provide novel insights into the function of lncRNA-driven tumorigenesis of CC.
Keywords: cervical Cancer, EMT, Long non-coding RNA, metastasis, miR-101-3p, SPRY4-IT1
Introduction
Cervical cancer (CC) is the fourth most commonly diagnosed cancer and the fourth leading cause of cancer-related deaths among females worldwide [1]. With the improvement of diagnostic techniques and therapeutic strategies, the incidence and mortality rates of CC has decreased [2]. However, the overall prognosis of CC patients still remains poor, especially in developing countries [3]. Tumor invasion and metastasis are key processes affecting the efficacy and prognosis of CC [4]. Therefore, exploring ways to inhibit these processes has important implications for improving the prognosis and quality of life of patients with CC.
Epithelial–mesenchymal transition (EMT) defines an orchestrated series of transcriptional and morphological program during which cells lose their typical epithelial characteristics and acquire mesenchymal properties [5]. Recent studies have suggested that EMT may play an important role in epithelial cancer progression and metastasis including cervical carcinoma, which have dramatic phenotypic changes by the loss of epithelial marker proteins such as E-cadherin and the acquisition of mesenchymal marker protein such as vimentin [6,7]. EMT is thought to be critical for the initial transformation from benign to invasive carcinoma, whereas MET (the reverse of EMT) is critical for the later stages of metastasis [8]. Just as a critical EMT event is the down-regulation or silencing of E-cadherin, the re-expression of E-cadherin is proposed to be the important hallmark of MET [9], thereby representing a potently novel strategy for development of new methods to improve therapeutic interventions of CC.
ZEB1 is increasingly recognized as a central regulator of EMT and invasion in solid tumors through transcriptional repression of E-cadherin gene via direct interaction with its E-boxes [10–12]. Increased ZEB1 expression has been reported in CC tissue, indicating that ZEB1 expression may be associated with progression of CC [13]. Importantly, up-regulation of ZEB1 at the post-transcriptional level could be induced by the loss or repression of microRNAs (miRNAs) that selectively target ZEB1 in cancer [5]. MiR-101, one of these potential miRNAs, is negatively regulated in different types of cancers including CC and considered to be a tumor suppressor [14–16]. While miR-101 expression was shown to correlate with ZEB1 signaling in breast cancer cells [16], little is known about the role of miR-101/ZEB1 signaling in regulating the EMT process of CC.
Accumulating evidence has demonstrated that long non-coding RNAs (lncRNAs) play a non-negligible role in tumorigenesis [17], and a new post-transcriptional regulatory mechanism that LncRNAs can function as a natural miRNA sponge has been recently revealed [18]. For example, MALAT1 mediates Rac1 expression by acting as an miR-101b sponge [19]. LncRNA Unigene56159 acts as a sponge for miR-140-5p to modulate ZEB2 expression in hepatocellular carcinoma [20]. SPRY4 intronic transcript 1 (SPRY4-IT1), an lncRNA derived from an intron within SPRY4 gene, has been recently revealed as oncogenic regulatory hubs or tumor suppressors in different cancers. For example, SPRY4-IT1 was down-regulated in gastric cancer and contributed to gastric cancer cells metastasis partly via regulating the EMT process [21]. By contrast, it was reported to promote metastasis of bladder cancer and colorectal cancer by targeting miR-101, and knockdown of its expression inhibited cell growth, invasion and induced cell apoptosis [22,23]. A previous study found that SPRY4-IT1 was up-regulated in CC [24], whereas its function and mechanism of action is not well documented.
In the present study, we explore the biological roles of SPRY4-IT1 on the phenotypes of CC cells both in vitro and in vivo. Furthermore, mechanistic analysis reveals that SPRY4-IT1 functions as a miRNA sponge to positively regulate the expression of ZEB1 through sponging miR-101-3p, thus playing an oncogenic role in CC pathogenesis. Together, our study elucidates the role of LncRNA SPRY4-IT1 as regulations of CC progression, and sheds new light on LncRNA-directed diagnostics and therapeutics in CC.
Materials and methods
Cell culture
The CC cell lines (HeLa and CaSki) were purchased from American Type Culture Collection (ATCC, Manassas VA, U.S.A.). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM)/F12 supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 mg/ml streptomycin (Gibco) and maintained in humidified air at 37°C with 5% CO2 atmosphere.
DNA constructs and cell transfection
Short hairpin RNA (shRNA) against SPRY4-IT1 (sh-SPRY4-IT1: 5′-TGCTTTATCTGTAGGACAT-3′) and negative control shRNA (sh-NC: 5′-GTTCTCCGAACGTGTCACGT-3′) were synthesized by Genema (Shanghai, China). miR-101-3p mimics (sense: 5′-UACAGUACUGUGAUAACUGAA-3′; antisense: 5′-UUCAGUUAUCACAGUACUGUA-3′), miR-101-3p inhibitor (5′-UUCAGUUAUCACAGUACUGUA-3′) and plasmids were purchased from Genema (Shanghai, China). The ZEB1 full-length was cloned into pcDNA3.1 plasmid. And human ZEB1 3′-untranslated region (UTR) fragment and SPRY4-IT1 gene containing putative binding sites for miR-101-3p reporter vector were synthesized by Genechem from RiboBio.
HeLa or CaSki cells (5 × 105) were planted in six-well plates 24 h prior to transfection with shRNAs, miR-101-3p mimics, miR-101-3p inhibitor, and pcDNA3.1-ZEB1 with 60–70% confluence, then transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, U.S.A.) according to the manufacturer’s instructions. The transfected cells were harvested at 48 h after transfection.
RNA extraction, reverse transcription and quantitative PCR
Total RNA was isolated with TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Complementary DNA was synthesized with random primers using a reverse transcription kit PrimeScript RT reagent Kit (Takara Biomedical Technology, Dalian, China) or commercial miRNA reverse transcription PCR kit (RiboBio). Quantitative real-time PCR (qPCR) analysis was carried out using the SYBR Premix Ex Taq kit (Takara Biomedical Technology). The primer set for SPRY4-IT1 was 5′-AATATGCCCAGTGGAGCCAT-3′ (forward) and 5′- GGCCTTGGAATCAGAAAGCA-3′ (reverse). The primer set for miR-101-3p was 5′-GCGCGCATACAGTACTGTGATA-3′ (forward) and 5′- CGGCCCAGTGTTCAGACTAC-3′ (reverse). The primer set for ZEB1 was 5′-TATGAATGCCCAAACTGCAA-3′ (forward) and 5′-TGGTGATGCTGAAAGAGACG-3′ (reverse). The primer set for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was 5′-CCAGGTGGTCTCCTCTGA-3′ (forward) and 5′-GCTGTAGCCAAATCGTTGT-3′ (reverse). The primer set for U6 was 5′-CTCGCTTCGGCAGCACA-3′ (forward) and 5′-AACGCTTCACGAATTTGCGT-3′ (reverse). All data analyses were operated using the StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA, U.S.A.). All reactions were run in triplicate with 7500 real-time PCR System (Applied Biosystems). RNA relative expression was calculated as fold change using the comparative threshold cycle (CT) method (2−ΔΔCT) with GAPDH or U6 serving as the internal control gene.
Western blotting analysis
Cells were collected and lysed using RIPA protein extraction reagent (Beyotime, Beijing, China) supplemented with a protease inhibitor cocktail (Roche, Pleasanton, CA, U.S.A.) and phenylmethylsulfonyl fluoride (Roche). The concentrations of protein samples were detected using a BCA Protein assay kit (Beyotime). Equal amounts of protein extracts were separated by 10% SDS/PAGE gels electrophoresis, then transferred to PVDF membranes. The membranes were blocked for 1 h in Tris-buffered saline (TBS) containing 5% nonfat milk and incubated with primary antibodies at 4°C for 12 h. Antibodies against E-cadherin (#3195), N-cadherin (#5246), and Vimentin (#5741) were purchased from Cell Signaling Technology (Danvers, MA, U.S.A.). GAPDH signal was used as a control. Membranes were then incubated with goat anti-mouse or anti-rabbit secondary antibody (Beyotime, Shanghai, China) and visualized using enhanced chemiluminescence (ECL, Thermo Scientific, Shanghai, China).
Wound-healing assay and cell transwell invasion assay
Cells were seeded on to six-well plates and cultured overnight. Wounds were created by scratching cell layer with a sterile 200-μl plastic pipette tips and washed with culture medium. Cells were further cultured with medium containing 1% FBS in 48 h, images were acquired at different time points by a microscope (Olympus, Tokyo, Japan) at 100× magnification. For the invasion assays, we used a 24-well transwell chamber (Corning: 8 μm) with the upper chamber coated with Matrigel (BD Bioscience). Cells (1 × 105 cells in 100 μl serum-free medium) were seeded in the top chamber, 500 ml medium containing 10% FBS was placed into the lower chamber. After incubation for 24 h, cells on the upper membrane surface were wiped off using a cotton swab and the lower membrane surface was fixed with methanol, stained with 0.1% Crystal Violet, and counted in five random fields at 100× magnification.
Luciferase reporter assays
The wild-type and mutant regions of SPRY4-IT1 containing putative binding site with miR-101-3p were synthesized and constructed into the pGL3-control reporter vector (Promega, Madison, WI, U.S.A.). The introduction of mutations is listed in Figure 3A. The similar strategy was performed to assess the regulation relationship between ZEB1 and miR-101-3p. The wild-type fragment or three mutants of human ZEB1 3′UTR containing putative binding sites for miR-101-3p were synthesized. The introduction of mutations is listed in Figure 4A. HeLa and CaSki cells were harvested 24 h after transfection and the luciferase activity was detected by Dual Luciferase Reporter Assay Kit (Promega) according to the manufacturer’s instructions. Firefly, luciferase activities were normalized to Renilla luciferase activity. All experiments were performed in triplicate.
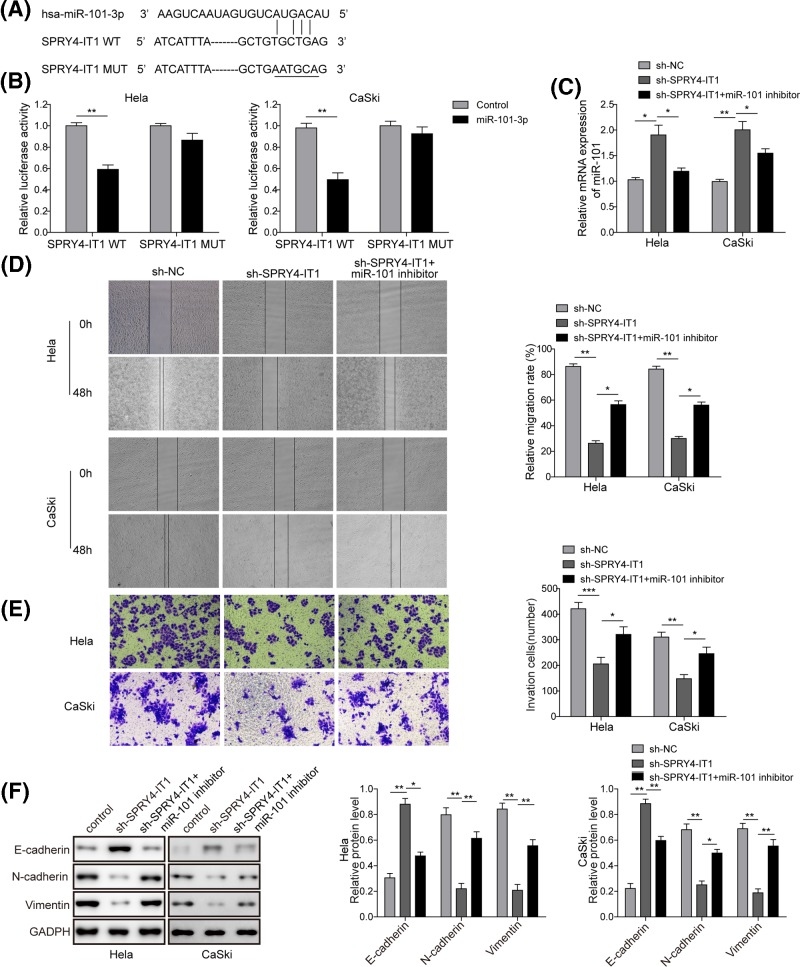
Figure 3. SPRY4-IT1 targets miR-101-3p to regulate the EMT process of CC.
(A) Sequence alignment of miR-101-3p with the putative binding sites within the wild-type or mutant regions of SPRY4-IT1. (B) The luciferase report assay demonstrated that overexpression of miR-101-3p reduced the intensity of fluorescence in HeLa and CaSki cells transfected with wild-type SPRY4-IT1 vector, while it had on significant effect on mutant-type of SPRY4-IT1. (C) qPCR showed that knockdown of miR-101-3p with miR-101-3p inhibitor completely reversed the increased miR-101-3p mRNA expression induced by sh-SPRY4-IT1 in HeLa and CaSki cells. (D) Wound healing assay showed that knockdown of miR-101-3p with miR-101-3p inhibitor partly reversed cell migration induced by sh-SPRY4-IT1 in HeLa and CaSki cells. (E) Transwell assay showed that knockdown of miR-101-3p with miR-101-3p inhibitor partly reversed cell invasion induced by sh-SPRY4-IT1 in HeLa and CaSki cells. (F) Western blotting analysis showed that knockdown of miR-101-3p with miR-101-3p inhibitor partly reversed the up-regulation of E-cadherin and down-regulation of N-cadherin and vimentin induced sh-SPRY4-IT1 in HeLa and CaSki cells. *P<0.05, **P<0.01, ***P<0.001 compared with the sh-SPRY4-IT1 group.
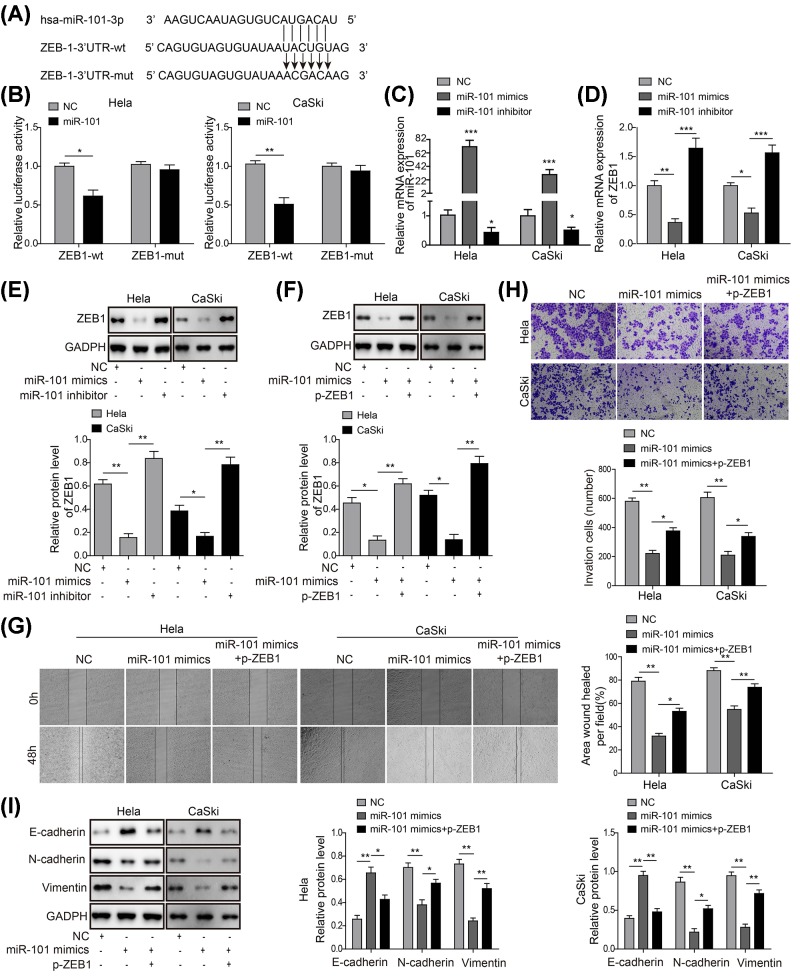
Figure 4. miR-101-3p targets ZEB1 to regulate the EMT process of CC.
(A) Sequence alignment of miR-101-3p with the putative binding sites within the wild-type or mutant regions of ZEB1. (B) The luciferase report assay demonstrated that overexpression of miR-101-3p reduced the intensity of fluorescence in HeLa and CaSki cells transfected with wild-type but not mutant-type ZEB1 vector. (C) qPCR showed that overexpression of miR-101-3p with miR-101-3p mimics up-regulated the levels of miR-101-3p mRNA, which was completely reversed by transfection of miR-101-3p inhibitor. (D) qPCR showed that overexpression of miR-101 with miR-101-3p mimics down-regulated the levels of ZEB1 mRNA, which was completely reversed by transfection of miR-101-3p inhibitor. (E) Western blotting analysis showed that miR-101-3p inhibitor completely reversed the suppression of ZEB1 induced by miR-101-3p minics in HeLa and CaSki cells. (F) Western blotting analysis showed that overexpression of ZEB1 by plasmid transfection (p-ZEB1) completely reversed the suppression of ZEB1 induced by miR-101-3p mimics in HeLa and CaSki cells. (G) Wound healing analysis showed that overexpression of ZEB1 by plasmid transfection (p-ZEB1) partly reversed cell migration induced by miR-101-3p mimics in HeLa and CaSki cells. (H) Transwell analysis showed that p-ZEB1 partly reversed cell invasion induced by miR-101-3p mimics in HeLa and CaSki cells. (I) Western blotting analysis showed that p-ZEB1 partly reversed the up-regulation of E-cadherin and down-regulation of N-cadherin and vimentin induced miR-101-3p mimics in HeLa and CaSki cells. *P<0.05, **P<0.01 and ***P<0.001 compared with the negative control (NC) group.
Tumor xenografts experiments in vivo
All BALB/c nude mice (4 weeks old, female) were maintained under pathogen-free conditions and all procedures for the mouse experiments were approved by the Animal Care Committee of Shandong Provincial Hospital Affiliated to Shandong University. For the tumor xenografts experiments, stable HeLa and CaSki cells (5 × 106, 100 μl) transfected with SPRY4-IT1 shRNA or negative control were subcutaneously injected into mice (n=6 per group). Tumor growth was examined every 5 days after injection, and tumor volumes were calculated using the equation: length × width2 × 0.5. At 30 days post injection, mice were killed, and the tumors were excised, photographed and tissue sections were obtained for further study.
Statistical analyses
All statistical analyses were performed with SPSS 20.0 (SPSS, Chicago, U.S.A.). Data were represented as mean ± standard deviation based on at least three repeats. Group difference was assessed using Student’s t test or one-way analysis of variance followed by post hoc Dunnett’s multiple comparisons test. P<0.05 was considered as statistically significant.
Results
Knockdown of SPRY4-IT1 inhibits cell proliferation, cell migration, cell invasion and EMT of CC in vitro
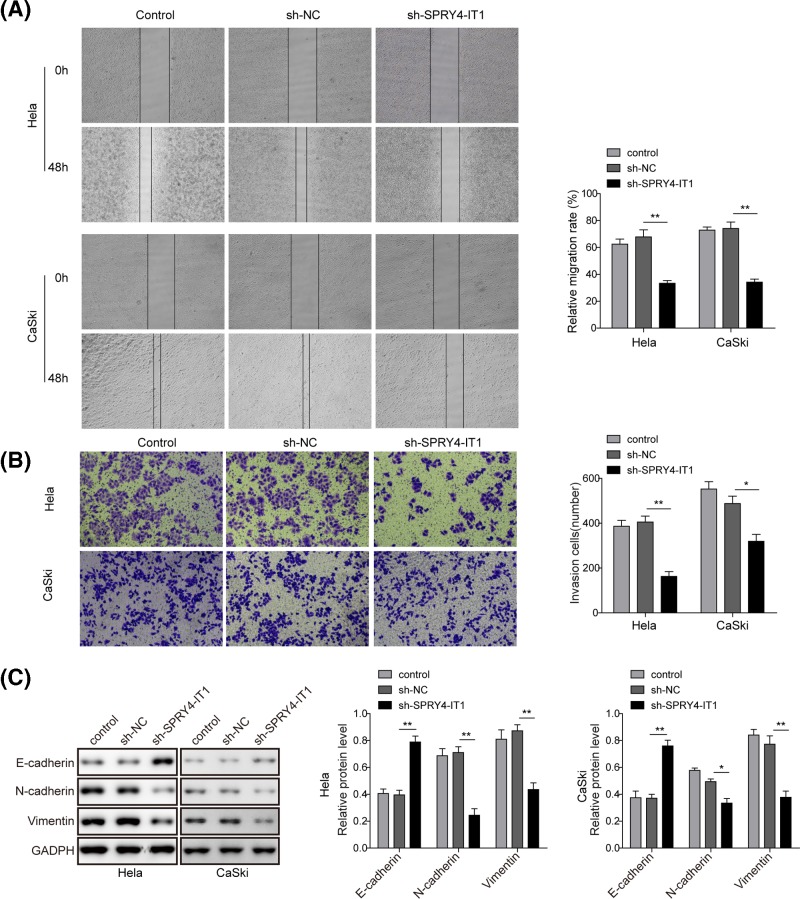
The expression of SPRY4-IT1 was significantly up-regulated in CC cell (P<0.05, Supplementary Figure S1A). Knockdown of SPRY4-IT1 by shRNA inhibited the cell viability of HeLa and CaSki cells (P<0.05, Supplementary Figure S1B,C). Moreover, we further investigated the effect of knockdown of SPRY4-IT1 on cell migration and invasion by an in vitro scratch assay and transwell assay, respectively. Knockdown of SPRY4-IT1 indicated approximately 50% decrease in migration compared with sh-NC treatment in HeLa and CaSki cell lines (Figure 1A). The results of the invasion assay shows that knockdown of SPRY4-IT1 significantly inhibits CC cell invasion in both cell lines (Figure 1B). Finally, we investigated the effects of SPRY4-IT1 knockdown on the protein expression of E-cadherin, N-cadherin and vimentin (Figure 1C). Western blot analysis showed that knockdown of SPRY4-IT1 up-regulated the expression level of the epithelial marker, E-cadherin, and down-regulated the mesenchymal maker, N-cadherin and vimentin in HeLa and CaSki cells. Taken together, the data suggest that down-regulation of SPRY4-IT1 inhibits the migration and invasion of CC cells, which may be possibly via the EMT process.
Figure 1. Knockdown of SPRY4-IT1 inhibits cell proliferation, cell migration, cell invasion and EMT of CC in vitro.
(A) The migration potential of cells with SPRY4-IT1 knockdown detected by wound healing assay. Comparison of cell motility of HeLa or CasKi cells transfected with sh-SPRY4-IT1 or sh-NC was followed, starting at 0 h and continuing through to 48 h. Quantitation of wound healing was based on triplicate assays with error bars indicating the standard deviation (SD) of the mean. (B) The invasive potential of SPRY4-IT1 knockdown in HeLa or CasKi cells detected by Transwell assay. Representative numbers of invading cells through the Matrigel were counted. (C) Expression of E-cadherin, N-cadherin and vimentin in HeLa or CasKi cells transfected with either sh-SPRY4-IT1 or sh-NC analyzed by Western blot analysis. GADPH was used as a load control. Quantitative analysis of relative band intensity demonstrated the significant increase in E-cadherin and the decrease in N-Cadherin and vimentin protein levels upon SPRY4-IT1 knockdown. Each bar represents triplicate analyses of mean ± SD, where the significant difference from the control group is represented by asterisk (*P<0.05, **P<0.01).
Knockdown of SPRY4-IT1 suppresses CC growth and metastasis in vivo
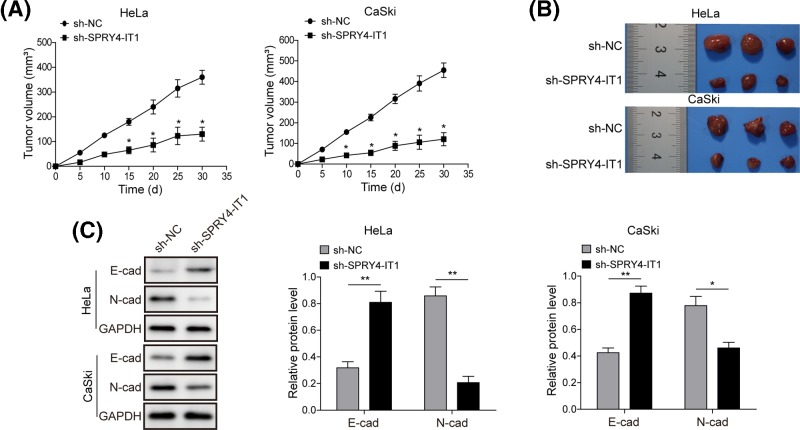
To explore whether SPRY4-IT1 could affect tumorigenesis in vivo, HeLa and CaSki cells transfected with sh-SPRY4-IT1 or sh-NC were used in a nude mice xenograft model. Thirty days after injection, the tumors formed in sh-SPRY4-IT1 group were substantially smaller and lighter (P<0.05) than those in the sh-NC group (Figure 2A,B). Our qPCR analysis also confirmed the low expression of SPRY4-IT4 in tumor tissues of sh-SPRY4-IT4 group (P<0.05). Moreover, Western blot analysis showed that knockdown of SPRY4-IT1 up-regulated E-cadherin expression and down-regulated N-cadherin expression in HeLa and CaSki cells, with the protein levels of E-cadherin was expressed at approximately two-fold higher while the level of N-cadherin was reduced by greater than 50% when compared with the sh-NC group (Figure 2C). Collectively, these data suggested that SPRY4-IT1 contributes to CC metastasis in vivo, which may be partly via affecting EMT process.
Figure 2. Knockdown of SPRY4-IT1 suppresses CC growth and metastasis in vivo.
(A) Tumor volume curve of mouse upon sh-SPRY4-IT1 or sh-NC treatment was analyzed. (B) Tumors collected from mice were exhibited. (C) Tumors developed from sh-SPRY4-IT1 transfected cervical cell lines HeLa and CaSki showed higher E-cadherin protein levels and lower N-cadherin protein levels than tumors developed by negative control cells, determined by Western blot analysis. GADPH was used as a load control. Each bar represents triplicate analyses of mean ± SD, where the significant difference from the sh-NC is represented by *P<0.05 and **P<0.01.
SPRY4-IT1 targets miR-101-3p to regulate the EMT process of CC
To identify the molecular mechanism by which SPRY4-IT1 regulates the metastasis of CC, we first used a bioinformatics approach to predict the putative binding sites between SPRY4-IT1 and miR-101-3p (Figure 3A). To identify the direct binding between SPRY4-IT1 and miR-101-3p, the wild-type and mutant SPRY4-IT1 fragment containing miR-101-3p binding site were synthesized and cloned into downstream of the luciferase reporter gene (pGL3-SPRY4-IT1-WT and pGL3-SPRY4-IT1-Mut) (Figure 3A). After co-trasfection of pGL3-SPRY4-IT1-WT or pGL3-SPRY4-IT1-Mut together with miR-101-3p mimics, luciferase activity was analyzed. The results revealed that the miR-101-3p significantly reduced the luciferase activities in pGL3-SPRY4-IT1-WT-treated HeLa and CaSki cells (40–50% reduction, P<0.01, Figure 3B), while the luciferase activities in pGL3-SPRY4-IT1-Mut-treated cells was not reduced by the expression of miR-101-3p (Figure 3B), suggesting that SPRY4-IT1 is a direct target of miR-101-3p.
In support of these findings, we observed appproximately two-fold increase in the level of miR-101-3p mRNA in both HeLa and CaSki cells transfected with sh-SPRY4-IT1. Moreover, knockdown of miR-101-3p with miR-101-3p inhibitor reversed this increase induced by sh-SPRY4-IT1 in CC cells (Figure 3C). Meanwhile, knockdown of miR-101-3p partly reversed cell migration and invasion ability inhibited by sh-SPRY4-IT1 (Figure 3D,E). Additionally, we also found that miR-101-3p inhibitor efficiently reversed the up-regulation of E-cadherin protein level and the down-regulation of N-cadherin and vimentin protein levels induced by sh-SPRY4-IT1 in HeLa and CaSki cells (Figure 3F). These data suggested that SPRY4-IT1 modulated the EMT process by targets miR-101-3p.
miR-101-3p targets ZEB1 to regulate the EMT process of CC
To find out if ZEB1 was a direct target of miR-101-3p, 3′UTR fragment containing miR-101-3p-biding site (ZEB1 3′UTR-WT) and its mutant with substitution of six nucleotides (ZEB1 3′UTR-Mut) were synthesized and cloned downstream of the pGL3-control vector (Figure 4A). Dual luciferase reporter assay showed that miR-101-3p reduced the luciferase activities significantly in pGL3-ZEB1 3′UTR-WT-treated HeLa and CaSkis cells, while the luciferase activities in pGL3-ZEB1 3′UTR-Mut-treated cells was not reduced by the expression of miR-101-3p (Figure 4B).
To further verify the mechanism, we determined the levels of miR-101-3p and ZEB1 in HeLa and CaSki cells transfected with either miR-101-3p mimics or miR-101-3p inhibitor. We found that transfection of the miR-101-3p mimics in HeLa and CaSki cells caused an increase in the expression levels of miR-101-3p, while miR-101-3p inhibitor dramatically reduced the miR-101-3p expression (Figure 4C). Importantly, a greater than 50% decrease in the mRNA and protein levels of ZEB1 were observed in cells transfected with the miR-101-3p mimics and miR-101-3p inhibitor led to an opposite effect (Figure 4D,E). Moreover, we found overexpression ZEB1 by plasmid transfection (p-ZEB1) also reversed the suppression of ZEB1 induced by transfection of miR-101-3p mimics (Figure 4F). Overexpression of ZEB1 also partly reversed cell migration and invasion ability induced by miR-101-3p mimics (Figure 4G,H). Additionally, we also found that overexpreesion of ZEB1 efficiently reversed the up-regulation of E-cadherin protein level and the down-regulation of N-cadherin and vimentin protein levels induced by miR-101-3p mimics in HeLa and CaSki cells (Figure 4I). These data suggested that miR-101-3p modulated the EMT process partly through the regulation of ZEB1.
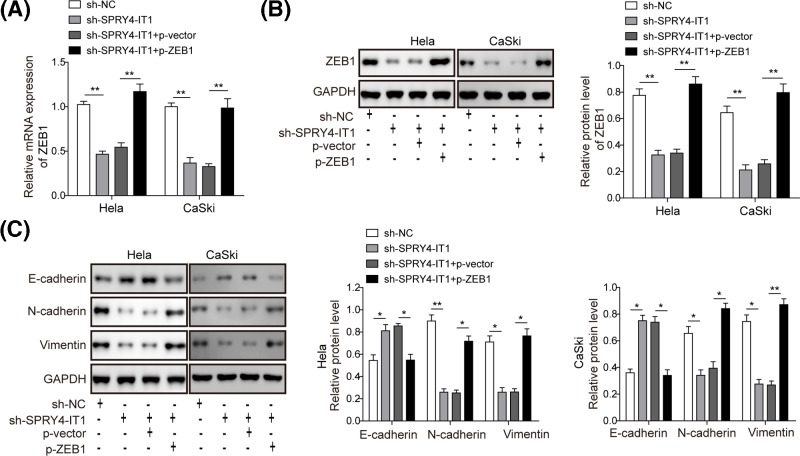
SPRY4-IT1 targets miR-101-3p to regulate ZEB1 expression in the EMT process of CC
Considering the regulatory relationship between SPRY4-IT1/miR-101-3p, and ZEB1/miR-101-3p, we hypothesized that SPRY4-IT1 might regulate the expression levels and functions of ZEB1. To validate this hypothesis, we treated HeLa and CaSki cells with sh-SPRY4-IT1 with or without ZEB1 overexpression vector and determined the expression levels of SPYR4-IT1 and ZEB1. We found that the ZEB1 expression levels were significantly reduced in HeLa and CaSki cells transfected with sh-SPRY4-IT1 compared with sh-NC treatment. But overexpression of ZEB1 by plasmid transfection effectively restored the decreased ZEB1 expression induced by sh-SPRY4-IT1 (Figure 5A). Similarly, the ZEB1 protein levels were significantly reduced in HeLa and CaSki cells transfected with sh-SPRY4-IT1 and overexpression of ZEB1 effectively reversed this decrease, as determined by Western blotting analysis (Figure 5B). Moreover, we found that overexpression of ZEB1 efficiently reversed the up-regulation of E-cadherin protein level and the down-regulation of N-cadherin and vimentin protein levels induced by sh-SPYR4-IT1 in HeLa and CaSki cells (Figure 5C). Taken together, our data suggested that SPRY4-IT1 regulate ZEB1 expression in the EMT process of CC.
Figure 5. SPRY4-IT1 targets miR-101-3P to regulate ZEB1 expression in the EMT process of CC.
(A) qPCR showed that overexpression of ZEB1 by plasmid transfection effectively reversed the reduction in SPRY4-IT1 expression levels induced by sh-SPRY4-IT1. (B) Western blotting analysis showed that overexpression of ZEB1 effectively reversed the reduction in ZEB1 protein levels induced by sh-SPRY4-IT1. (C) Western blotting analysis showed that overexpression of ZEB1 efficiently reversed the up-regulation of E-cadherin protein level and the down-regulation of N-cadherin and vimentin protein levels induced by sh-SPYR4-IT1 in HeLa and CaSki cells. *P<0.05, **P<0.01 compared with the sh-NC group or sh-SPRY4-IT1+p-vector group.
Discussion
Emerging evidence has revealed a key regulatory role of lncRNAs in tumorigenesis, although only a few functional lncRNAs have been identified in CC [4]. Identification of dysregulated lncRNAs will enhance our knowledge of lncRNAs function in the progression and metastasis of CC and could be used as new diagnostic or therapeutic targets. In the present study, we confirmed that shRNA-mediated SPRY4-IT1 knockdown induced significant inhibition of cell migration, and invasion ability in CC both in vitro and in vivo via regulating the EMT process. Mechanistically, we showed that SPRY4-IT1 directly interacted with miR-101-3p at recognized sites. SPYR4-IT1 exerted its oncogenic function on CC in large part due to its role to function as an miR-101-3p sponge, and subsequently initiate ZEB1 signaling pathway. These findings indicate that SPRY4-IT1 could function as an oncogenic regulatory hub and may be useful as a novel prognostic or progression marker for CC.
Although SPRY4-IT1 can promote migration and invasive phenotype of CC cells, the underlying mechanism is still elusive. Previous study showed that SPRY4-IT1 is decreased in gastric cancer, and the inhibitory effects of SPRY4-IT1 on cell migration and invasion were partly associated with EMT progression [20]. Our data show that knockdown of SPRY4-IT1 indeed reduced the cell motility and invasion in vitro. And in our tumor xenograft models, SPRY4-IT1 knockdown significantly reduced the tumorigenesis ability of HeLa and CaSki cells in vivo. EMT is a key step toward cancer metastasis, a biological process where epithelial cells lose their polarity and undergo transition into a mesenchymal phenotype. Loss of E-cadherin expression is a hallmark of EMT process in epithelial cancer including CC, which has been found to increase cancer cell invasion and metastasis [6]. In our research, we further demonstrated that SPRY4-IT1 promoted CC cell migration and invasion partially via regulating EMT progression, as inhibition of SPRY4-IT1 induced differential enforcement of E-cadherin and reduction in N-cadherin and vimentin both in CC cells and in mice tumor tissues with SPRY4-IT1 knockdown.
One of the mechanisms that lncRNAs function through is to play regulatory roles by acting as target genes for specific miRNAs, then targeting other terminal mRNAs [18]. In bladder cancer, SPRY4-IT1 mediates EZH2 expression by acting as an miR-101b sponge [22]. Similarly, our results confirmed that SPRY4-IT1 could act as an upstream regulator for miR-101-3p and promoted migration and invasion in CC cells. To clarify whether SPRY4-IT1 functions as a sponge for miR-101-3p in CC, bioinformatics was used to analyze the potential miRNA binding sites contained in SPYR4-IT1. Luciferase reporter assay and qPCR showed that SPRY4-IT1 could positively regulate the EMT progression at post-transcriptional level through targeting miR-101-3p in CC.
Emerging evidence has demonstrated that several miRNAs could target the 3ʹ-UTR of the ZEB1, a key transcription factor of EMT, and post-transcriptionally regulate its expression in cancers [5]. Previous study has revealed that miR-101 could directly target ZEB1 to repress ZEB1 expression at post-transcriptional level in breast cancer [16]. While opposite effects of ZEB1 and miR-101-3p in CC whereby ZEB1 functions as an oncogene [12], and miR-101-3p as a tumor suppressor [15] have also been demonstrated, their direct interaction in CC has not yet studied. Our results also demonstrate that miR-101-3p acts as a tumor suppressor in CC. Overexpression of miR-101-3p inhibited cell proliferation and invasion of CC cells in vitro. This may be partially through the regulation of EMT, as overexpression of miR-101-3p also increased the levels of E-cadherin and decreased the levels of N-cadherin and vimentin CC cells. Moreover, the enforced expression of miR-101-3p reduced the ZEB1 expression and miR-101-3p inhibitor, as expected, promoted ZEB1 expression at protein level. Furthermore, ZEB1 activation was responsible for EMT triggered by overexpression of miR-101-3p, as overexpression of ZEB1 restored epithelial phenotype and migration and invasion in CC cells induced by enhanced miR-101-3p expression. Hence, our work verified that ZEB1 is one of the target genes of miR-101-3p, which is involved in cell migration and invasion in CC by regulating EMT. Further exploration showed that SPRY4-IT1 and ZEB1 have shown a positive correlation in CC cells, and knockdown of SPRY4-IT1 decreased ZEB1 at protein levels in vitro. In addition, overexpression of ZEB1 could partially rescue the decreased migration and invasion by SPRY-IT1 knockdown. These results indicate that SPRY4-IT1 could regulate ZEB1 via miR-101-3p.
In summary, we identify that SPRY4-IT1 acts as an oncogene in CC, competitively binds to miR-101-3p and subsequently up-regulates the expression of its target gene ZEB1 to promote the migration and invasion of CC cells. This SPYR4-IT1/miR-101-3p/ZEB1 regulatory network may shed light on tumorigenesis in CC and may be valuable for the development of novel diagnostic and treatment approaches for CC.
Supporting information
Supplementary Figure.
Abbreviations
- CC
cervical cancer
- EMT
epithelial–mesenchymal transition
- FBS
fetal bovine serum
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- LncRNA
long non-coding RNA
- MET
mesenchymal-epithelial transition
- miRNA
microRNA
- qPCR
quantitative real-time PCR
- shRNA
short hairpin RNA
- SPRY4
Sprouty RTK Signaling Antagonist 4
- SPRY4-IT1
SPRY4 intronic transcript 1
- UTR
untranslated region
- ZEB1
Zinc finger E-box-binding homeobox 1
Funding
This work was supported by the Science and Technology Department of Shandong Province [grant number 2014GSF118048].
Author Contribution
C.-Z.L. and M.-J.F. contributed to the conception and coordination of the study. M.-J.F. and C.-Z.L. designed the study and prepared the manuscript. M.-J.F., Y.-H.Z., P.-J.H., S.Z. and X.-M.S. collected, analyzed the data and interpreted the results. Y.-H.Z. and M.-J.F. collected and processed the samples. All authors read and approved the final manuscript.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J. and Jemal A. (2015) Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2.Lynge E., Rygaard C., Baillet M.V., Dugue P.A., Sander B.B., Bonde J.. et al. (2014) Cervical cancer screening at crossroads. APMIS 122, 667–673 10.1111/apm.12279 [DOI] [PubMed] [Google Scholar]
- 3.Siegel R., Naishadham D. and Jemal A. (2013) Cancer statistics, 2013. CA Cancer J. Clin. 63, 11–30 10.3322/caac.21166 [DOI] [PubMed] [Google Scholar]
- 4.Chi S., Shen L., Hua T., Liu S., Zhuang G., Wang X.. et al. (2017) Prognostic and diagnostic significance of lncRNAs expression in cervical cancer: a systematic review and meta-analysis. Oncotarget 8, 79061–79072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamouille S., Xu J. and Derynck R. (2014) Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196 10.1038/nrm3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee M.Y., Chou C.Y., Tang M.J. and Shen M.R. (2008) Epithelial-mesenchymal transition in cervical cancer: correlation with tumor progression, epidermal growth factor receptor overexpression, and snail up-regulation. Clin. Cancer Res. 14, 4743–4750 10.1158/1078-0432.CCR-08-0234 [DOI] [PubMed] [Google Scholar]
- 7.Thiery J.P., Acloque H., Huang R.Y. and Nieto M.A. (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139, 871–890 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- 8.Yao D., Dai C. and Peng S. (2011) Mechanism of the mesenchymal-epithelial transition and its relationship with metastatic tumor formation. Mol. Cancer Res. 9, 1608–1620 10.1158/1541-7786.MCR-10-0568 [DOI] [PubMed] [Google Scholar]
- 9.Wells A., Yates C. and Shepard C.R. (2008) E-cadherin as an indicator of mesenchymal to epithelial reverting transitions during the metastatic seeding of disseminated carcinomas. Clin. Exp. Metastasis 25, 621–628 10.1007/s10585-008-9167-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmalhofer O., Brabletz S. and Brabletz T. (2009) E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 28, 151–166 10.1007/s10555-008-9179-y [DOI] [PubMed] [Google Scholar]
- 11.Bakiri L., Macho-Maschler S., Custic I., Niemiec J., Guio-Carrion A., Hasenfuss S.C.. et al. (2015) Fra-1/AP-1 induces EMT in mammary epithelial cells by modulating Zeb1/2 and TGFbeta expression. Cell Death Differ. 22, 336–350 10.1038/cdd.2014.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin Y., Tang B., Hu C.J., Xiao Y.F., Xie R., Yong X.. et al. (2016) An hTERT/ZEB1 complex directly regulates E-cadherin to promote epithelial-to-mesenchymal transition (EMT) in colorectal cancer. Oncotarget 7, 351–361 10.18632/oncotarget.5968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ran J., Lin D.L., Wu R.F., Chen Q.H., Huang H.P., Qiu N.X.. et al. (2015) ZEB1 promotes epithelial-mesenchymal transition in cervical cancer metastasis. Fertil. Steril. 103, 1606–1614.e1601-e1602, 10.1016/j.fertnstert.2015.03.016 [DOI] [PubMed] [Google Scholar]
- 14.Shen Q., Bae H.J., Eun J.W., Kim H.S., Park S.J., Shin W.C.. et al. (2014) MiR-101 functions as a tumor suppressor by directly targeting nemo-like kinase in liver cancer. Cancer Lett. 344, 204–211 10.1016/j.canlet.2013.10.030 [DOI] [PubMed] [Google Scholar]
- 15.Liang X., Liu Y., Zeng L., Yu C., Hu Z., Zhou Q.. et al. (2014) miR-101 inhibits the G1-to-S phase transition of cervical cancer cells by targeting Fos. Int. J. Gynecol. Cancer 24, 1165–1172 10.1097/IGC.0000000000000187 [DOI] [PubMed] [Google Scholar]
- 16.Chandra Mangalhara K., Manvati S., Saini S.K., Ponnusamy K., Agarwal G., Abraham S.K.. et al. (2017) ERK2-ZEB1-miR-101-1 axis contributes to epithelial-mesenchymal transition and cell migration in cancer. Cancer Lett. 391, 59–73 10.1016/j.canlet.2017.01.016 [DOI] [PubMed] [Google Scholar]
- 17.Yang G., Lu X. and Yuan L. (2014) LncRNA: a link between RNA and cancer. Biochim. Biophys. Acta 1839, 1097–1109 10.1016/j.bbagrm.2014.08.012 [DOI] [PubMed] [Google Scholar]
- 18.Salmena L., Poliseno L., Tay Y., Kats L. and Pandolfi P.P. (2011) A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu F., Lu Z., Cai J., Huang K., Chen B., Li G.. et al. (2015) MALAT1 functions as a competing endogenous RNA to mediate Rac1 expression by sequestering miR-101b in liver fibrosis. Cell Cycle 14, 3885–3896 10.1080/15384101.2015.1120917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lv J., Fan H.X., Zhao X.P., Lv P., Fan J.Y., Zhang Y.. et al. (2016) Long non-coding RNA Unigene56159 promotes epithelial-mesenchymal transition by acting as a ceRNA of miR-140-5p in hepatocellular carcinoma cells. Cancer Lett. 382, 166–175 10.1016/j.canlet.2016.08.029 [DOI] [PubMed] [Google Scholar]
- 21.Xie M., Nie F.Q., Sun M., Xia R., Liu Y.W., Zhou P.. et al. (2015) Decreased long noncoding RNA SPRY4-IT1 contributing to gastric cancer cell metastasis partly via affecting epithelial-mesenchymal transition. J. Transl. Med. 13, 250 10.1186/s12967-015-0595-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu D., Li Y., Luo G., Xiao X., Tao D., Wu X.. et al. (2017) LncRNA SPRY4-IT1 sponges miR-101-3p to promote proliferation and metastasis of bladder cancer cells through up-regulating EZH2. Cancer Lett. 388, 281–291 10.1016/j.canlet.2016.12.005 [DOI] [PubMed] [Google Scholar]
- 23.Jin J., Chu Z., Ma P., Meng Y. and Yang Y. (2017) Long non-coding RNA SPRY4-IT1 promotes proliferation and invasion by acting as a ceRNA of miR-101-3p in colorectal cancer cells. Tumour Biol. 39, 1010428317716250. 10.1177/1010428317716250 [DOI] [PubMed] [Google Scholar]
- 24.Cao Y., Liu Y., Lu X., Wang Y., Qiao H. and Liu M. (2016) Upregulation of long noncoding RNA SPRY4-IT1 correlates with tumor progression and poor prognosis in cervical cancer. FEBS Open Bio 6, 954–960 10.1002/2211-5463.12102 [DOI] [PMC free article] [PubMed] [Google Scholar]