Abstract
Cytokeratin 18 (CK18), one of the major components of intermediate filaments (IF) in simple epithelial cells, undergoes caspase-mediated cleavage upon epithelial cell necrosis and apoptosis. CK18 has been used as a biomarker of several cancers and has been reported to be dysregulated in cervical cancers. The effects of dysregulated expression of CK18 at a molecular level are, however, unclear. In the present study, the function of CK18 in HeLa cells, a cell line derived from a cervical cancer cells, was investigated using shRNA knockdown. Reduced levels of CK18 led to a significant decrease in cell apoptosis, compared with control cells. Notably, RNA-seq analysis of the transcriptomes of HeLa cells, with or without CK18 knockdown, revealed that genes in the NF-κB pathway, and certain apoptosis pathways, were under global transcriptional and alternative splicing regulation. Quantitative RT-PCR confirmed the CK18-regulated transcription of apoptotic genes FAS and FADD, as well as immune genes CXCL2 and CD79B, in addition to alternative splicing of FAS and CTNNB1. Western blot analysis further revealed that CK18 knockdown led to reduced expression of CASP8. In conclusion, the present study indicated that CK18 played a role in apoptosis, which may be mediated via a feed-back regulation loop and may involve regulation of transcription and alternative splicing of a number of genes in apoptotic pathways.
Keywords: CK18, RNA-seq, apoptosis, transcriptional regulation, alternative splicing
Introduction
Cervical cancer is one of the most common malignant tumors among women worldwide, with more than 85% of cervical cancer-related deaths occurring in developing countries (1). Although the development of diagnosis techniques has improved the detection of cervical cancer and the use of vaccines can effectively prevent the disease, the overall survival rate of cervical cancer patients at five years remains only ~50% since most patients are diagnosed when the cancer is at an advanced stage (2). It is, therefore, crucially important to develop effective novel therapeutic strategies to improve the survival rate of patients.
Keratins, also known as cytokeratins, are the intermediate filament (IF)-forming proteins of epithelial cells. The primary role of keratins is to protect epithelial cells from both mechanical and non-mechanical stressors (3). Keratins have also been reported to have many roles in cancer, including as diagnostic markers (4–7) and prognostic markers in epithelial tumors (8–10), as well as roles in tumorigenesis (11,12) and drug responsiveness (13–16).
Keratins 8, 18 and 19 (CK8, CK18 and CK19), which are the most abundant keratins in simple epithelial cells, are extensively used as the diagnostic markers (17,18). CK18, also known as KRT18, and its caspase-cleaved fragment can be released into the circulation and are indicative of epithelial cell necrosis and apoptosis, respectively. Caspase-cleaved CK18 (M30) and full length CK18 (M65) are assessed using tissue polypeptide antigen (TPA) and tissue polypeptide specific antigen (TPS) (19). The M30 and M65 assays may provide important prognostic and predictive biomarkers in many malignancies (14,18,20–23).
Notably, several studies have revealed that CK18 is not only a biomarker but also a regulator in many diseases, including cancer (24,25). CK18 knockdown can decrease cell migration and increase chemosensitivity in non-small cell lung cancer (15), decrease cell migration in renal carcinoma (RCC5) cells (26), and increase cytokine-induced apoptosis in HeLa cells (27), suggesting that CK18 gene positively regulates tumorigenesis. Conversely, CK18 has been reported to suppress tumor aggressiveness and paclitaxel-resistance in paclitaxel-resistant prostate cancer (DU145-TxR) cells (16), as well as to induce cell adhesion and regression of malignancy in breast cancer (28). However, it remains unclear how CK18 exerts these biological functions.
Notably, CK18 has been reported to physically interact with LRP16, also known as O-Acetyl-ADP-Ribose Deacetylase MACROD1, thereby sequestering LRP16 in the cytoplasm and thus inhibiting the proliferation of ERα-positive breast tumor cells (29). It could be possible that CK18 regulates gene expression via protein-protein interaction. CK18 has also been revealed to be associated with mRNAs and could, therefore, be an RNA binding protein (RBP) (30). RBPs are known to coordinate RNA processing and post-transcriptional gene regulation (31). It is, however, unclear whether CK18 is able to affect post-transcriptional gene regulation in the context of exerting biological functions.
In the present study, we firstly aimed to elucidate the function of CK18 in HeLa cells, which are derived from cervical cancer cells. It was revealed that knockdown of CK18 led to significantly reduced apoptosis. RNA-seq analysis of the effect of CK18 on the transcription and alternative splicing of the HeLa transcriptome revealed that CK18 regulated the expression and alternative splicing of a number of genes involved in apoptosis, including FAS, FADD and CASP8. Additionally, CK18 was revealed to regulate the transcription and alternative splicing of many genes enriched in immunity and cancer-related pathways. Our study revealed a novel apoptotic function of CK18, which was linked to a feedforward regulation of apoptosis genes FAS and CASP8, and to CK18-regulated transcriptomes favoring apoptosis at both the transcription and alternative splicing levels. These findings elucidated the dysregulated expression of CK18 in cancers and supported an important role for CK18 in tumorigenesis.
Materials and methods
Cell culture and transfections
HeLa (immortalized human cervical cancer) cells were purchased from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, HepG2 (immortalized human liver cancer) cells were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). The cell lines were identified by short terminal repeat (STR) genotyping, which revealed a correspondence of >80% of the markers tested (32). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS).
To knockdown CK18, different shRNAs against human CK18 (shRNA1: 5′-GATGACACCAATATCACACGA-3′; shRNA2: 5′-CTTCATGAAGAAGAACCACGA-3′; shRNA3: 5′-CCTGCTGAACATCAAGGTCAA-3′) were designed, and scrambled shRNA (Scr-shRNA) that targeted a non-specific sequence (5′-ACTGGACCAGGCAGCAGCGTCAGAAGACT-3′) was used as the control. These shRNAs were transfected into HeLa cells using Lipofectamine 2000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's protocol. Transfected cells were harvested after 48 h for analysis.
Assesment of knockdown by shRNA
Total RNA was isolated from cells using TRIzol reagent (Ambion; Thermo Fisher Scientific, Inc) and cDNA synthesis was carried out using standard procedures. qPCR was performed on the Bio-Rad S1000 Thermal Cycler, using Bestar SYBR-Green RT-PCR Master Mix (DBI Bioscience, Shanghai, China). The PCR conditions are consisted of denaturing at 95°C for 10 min, 40 cycles of denaturing at 95°C for 15 sec, annealing and extension at 60°C for 1 min. The primers of CK18 used for quantitative real-time PCR (qPCR) were: Forward, AAAGGCCTACAAGCCCAGAT and reverse, CACTGTGGTGCTCTCCTCAA. Gene expression levels were calculated using the 2−ΔΔCq method (33) and CT values were normalized using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as an internal standard. The primers of GAPDH were: Forward, GGTCGGAGTCAACGGATTTG and reverse, GGAAGATGGTGATGGGATTTC.
MTT assay
An MTT assay was used to assess cell proliferation. Briefly, HeLa cells were cultured in 96-well plates and transfected with the vector using Lipofectamine 2000, according to the manufacturer's protocol. The cells were then incubated at 37°C for 48 h. Subsequently, MTT solution (5 mg/ml, 0.025 ml) was added to each well, and the cells were incubated for another 4 h. After centrifugation at 4,000 rpm for 15 min, the supernatant was removed from each well. The colored formazan crystals produced from MTT in each well were dissolved in DMSO (0.15 ml) and the optical density (OD) values were measured at 490 nm.
Flow cytometric analysis of cell apoptosis
HeLa cells (5×104) were seeded in 24-well culture plates. Once the cells reached 70% confluence, the cells were were transfected with the vector using Lipofectamine 2000, according to the manufacturer's protocol. The cells were then incubated at 37°C for 48 h and viable cells were harvested and washed twice with phosphate-buffered saline (PBS). Viable cells were double-stained with fluorescein isothiocyanate (FITC)-conjugated Annexin V and 7-amino actinomycin D (7-AAD) (4A Biotech Co., Ltd. Beijing, China).
The percentage of cell apoptosis was defined as the sum of the right lower quadrant and upper quadrant.
Library preparation and sequencing
The RNA was treated with RQ1 DNase (Promega Corp., Madison, WI, USA) to remove DNA. The quality and quantity of the purified RNA were assessed by measuring the absorbance at 260 nm/280 nm (A260/A280) using SmartSpec Plus spectrophotometer (BioRad Laboratories, Inc., Hercules, CA, USA). The integrity of each RNA sample was further verified by 1.5% agarose gel electrophoresis.
For each sample, a total of 1 µg RNA was used for RNA-seq library preparation. PolyA mRNAs were purified and concentrated with oligo (dT)-conjugated magnetic beads (Invitrogen; Thermo Fisher Scientific, Inc.) before directional RNA-seq library preparation. The fragmented RNAs were end repaired and an adaptor sequence was ligated at the 5′end. Reverse transcription was then performed using an RT primer harboring a 3′adaptor sequence and randomized hexamer. The cDNAs were purified, amplified, and stored at −80°C until they were used for sequencing.
For high-throughput sequencing, the libraries were prepared following the manufacturer's instructions. An Illumina HiSeq 4000 sequencing system (ABLife, Inc., Wuhan, China) was used to collect data from 151-bp pair-end sequencing.
Clean and alignment of RNA-Seq raw data
Raw reads were filtered to remove the adaptors, PolyN reads, and low-quality bases using FASTX-Toolkit (version 0.0.13). Short reads, less than 16 nt, were also discarded. Clean reads were then aligned to the GRCh38 genome using TopHat2 software (34), with four mismatches. Uniquely mapped reads were used to calculate the reads number and FPKM value (fragments per kilobase of transcript per million fragments mapped) for each gene.
Analysis of differentially expressed genes (DEGs)
EdgeR software (35) was used to measure FPKM values and to analyze the differential expression of genes using RNA-Seq data, in order to identify DEGs. To determine whether a gene was differentially expressed, the results were analyzed based on the fold change (fold change ≥2 or ≤0.5) and a false discovery rate (FDR<0.05).
We also analyzed the two replicates separately (named as simple pair). In detail, the DEGs were identified between shCK18-1st vs. Ctrl-1st and shCK18-2nd vs. Ctrl-2nd, respectively. We then obtained the overlapping genes from the DEGs of each replicate. The upregulated and downregulated DEGs shared by these two simple pairs were then overlapped with those identified by edgeR as biological replicates.
Gene Ontology (GO) and enriched KEGG pathway analyses were carried out using the KOBAS 2.0 server (36) to predict the gene function and calculate the frequency distribution of functional categories. The hypergeometric test and Benjamini-Hochberg FDR controlling procedure were used to define the enrichment of each pathway (corrected P-value <0.05).
Alternative splicing analysis
The alternative splicing events (ASEs) and regulated alternative splicing events (RASEs) between the samples were defined and quantified by using the ABLas pipeline, as previously described (37). In brief, eight types of ASEs were identified, based on the splice junction reads. The eight possible types of ASE included Cassette exon (CassetteExon), Exon skipping (ES), Mutual exclusive exon skipping (MXE), A5SS, A3SS, the MXE combined with alternative 5′promoter (5pMXE) combined with alternative polyadenylation site (3pMXE) and intron retention.
Having identified the ASEs in each RNA-seq sample, Fisher's exact test was selected to determine statistical significance, using the alternative reads and model reads of the samples as input data. The altered ratio of alternatively spliced reads and constitutively spliced reads between compared samples was defined as the RASE ratio. The P-value <0.05 and RASE ratio >0.2 were set as the thresholds for detection of RASEs.
Real time qPCR validation of DEGs and ASEs
To determine the validity of the RNA-seq data, qPCR was performed for selected DEGs and normalized with the housekeeping gene GAPDH. Primers are designed in exon regions, and sequences are presented in Table I. The same RNA samples for RNA-seq and RNA samples isolated from CK18 knockdown in HepG2 (using the same shRNAs) were used for qPCR. The PCR conditions consisted of denaturing at 95°C for 10 min, 40 cycles of denaturing at 95°C for 15 sec, and annealing and extension at 60°C for 1 min. PCR amplifications were performed in triplicate for each sample.
Table I.
Primer sequences used in q-PCR experiments.
| DEGsa | Forward | Reverse | |
|---|---|---|---|
| FAS | AAGCGGTTTACGAGTGACTTGG | AGCATGGTTGTTGAGCAATCCT | |
| FADD | TCTACCTCCGAAGCGTCCTGAT | AGGTGGTCTGTGGCTCACTCA | |
| CD79B | GGGCTGGAGACAAATGGCAG | TGAAGTGGTCTGTAGGTGAGCA | |
| CXCL2 | CTTGGATTCCTCAGCCTCTAT | GGTTTGCAGATATTCTCTAGTC | |
| RASGsb | Model forward | AS forward | Model/AS reverse |
| CTNNB1 | CATCCTTTAGCTGTATTGTC | TTTATACAGCCTGTATTGTC | AACAAGCAAGGCTAGGGTTTGA |
| FAS | AAAGAGGAAGGATCCAGATC | AAAGAGGAAGTGAAGAGAAA | AGTTGGAGATTCATGAGAACCT |
Indicates CK18-regulated genes that were differentially expressed genes in RNA-seq analysis and were subjected to qPCR validation. The primer sequences are listed.
Indicates genes that contained CK18-regulated alternative splicing events identified in RNA-seq and were subjected to qPCR validation. The primer sequences are listed. DEGs, differentially expressed genes; RASGs, alternative splicing genes; CK18, cytokeratin 18.
Concurrently, a qPCR assay was used to analyze ASEs. The primers for detecting the pre-mRNA splicing are presented in Table I. To detect one of the alternative isoforms, one primer was designed in the alternative exon and an opposing primer was designed in a constitutive exon. Other alternative isoforms were detected using a boundary-spanning primer for the sequence encompassing the exon-exon junction, with the opposing primer in a constitutive exon.
Western blot analysis
CK18 knockdown and control Hela cells were lysed in RIPA buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1.0% deoxycholate, 1% Triton X-100, 1 mM EDTA and 0.1% SDS. Following centrifugation of the homogenate (20,000 × g, 15 min) the supernatants were used for western blotting. Protein concentrations were measured using the BCA protein assay (Beyotime Institute of Biotechnology, Nanjing, China) with bovine serum albumin (BSA) as a standard. Equal amounts (20 µg/lane) of protein samples were loaded on 12% SDS-PAGE gel for separation and then transferred to a polyvinylidene difluoride (PVDF) membrane. The membrane was incubated with blocking solution (1X TBS; 0.05% Tween-20; 5% non-fat milk) at room temperature for 1 h and incubated overnight with primary antibodies raised against CK18 (dilution 1:1,000; cat. no. A0389), CASP8 (dilution 1:1,000; cat. no. A0215) and GAPDH (dilution 1:1,000; cat. no. AC027) (all from ABclonal, Wuhan, China). Immunoreactive proteins were detected using an ECL chemiluminescence system (Clinx Science Instruments Co., Ltd., Shanghai, China) with default settings and GAPDH as the normalized control.
Accession number
RNA-seq data presented in this study have been deposited in the Gene Expression Omnibus of NCBI and are accessible through GEO series accession no. GSE119255.
Statistical analysis
Experimental data are presented as the mean ± standard deviation of at least three experiments. Statistical analyses were performed with SPSS software (version 17.0; SPSS, Inc., Chicago, IL, USA). Significance of differences was evaluated with Student's t-test when only two groups were compared. P<0.05 was considered to indicate a statistically significant difference.
Results
Effect of CK18 knockdown on proliferation and apoptosis of HeLa cells
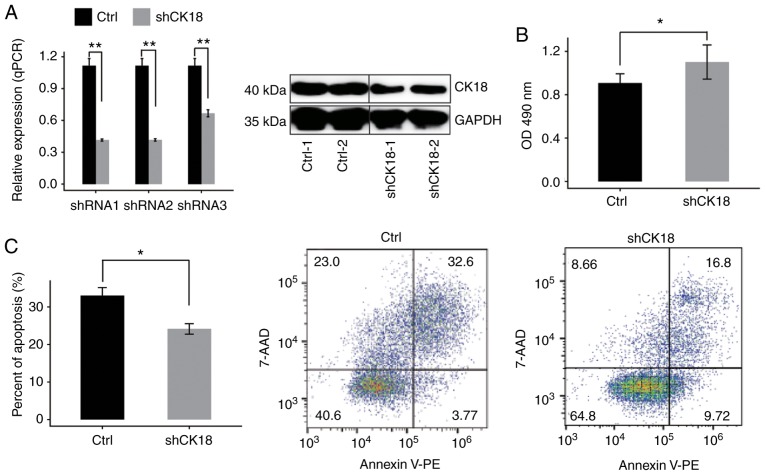
The expression of CK18 was examined in HeLa cells transduced with different shRNAs (shRNA1, shRNA2 and shRNA3) or an empty vector by RT-qPCR. Compared with cells treated with the empty vector mRNA expression was effectively reduced in cells treated with CK18-shRNAs. The effective shRNA (shRNA1) also led to a significant reduction in the protein level (Fig. 1A). Therefore, CK18-shRNA1 was used in subsequent experiments in order to knockdown CK18 expression in HeLa cells. Knockdown of CK18 (CK18 KD) led to a significant increase in cell proliferation and a significant decrease in cell apoptosis (Fig. 1B and C).
Figure 1.
CK18 KD promotes the proliferation and attenuates the apoptosis of HeLa cells. (A) CK18 mRNA expression in HeLa cells after transient transfection with CK18-specific shRNAs or control vector, as determined by qRT-PCR (left); CK18 protein expression was decreased by shRNA1, as revealed by western blotting (right). GAPDH was used as an internal control. The control samples and shCK18 samples were from the same gel in the western blotting experiment, but not side by side. (B) An MTT assay revealed that transfection with the CK18-shRNA1 upregulated cell proliferation in HeLa cells; (C) Plots and quantification of flow cytometric analysis of apoptosis following CK18 KD in HeLa cells. Data are presented as the mean ± standard deviation of three independent experiments. *P<0.05 and **P<0.01. CK18, cytokeratin 18; Knockdown, KD; shCK18, CK18 knockdown by shRNA in HeLa cells; Ctrl, HeLa cells transfected with scrambled plasmid as the control.
RNA-Seq and DEG analysis
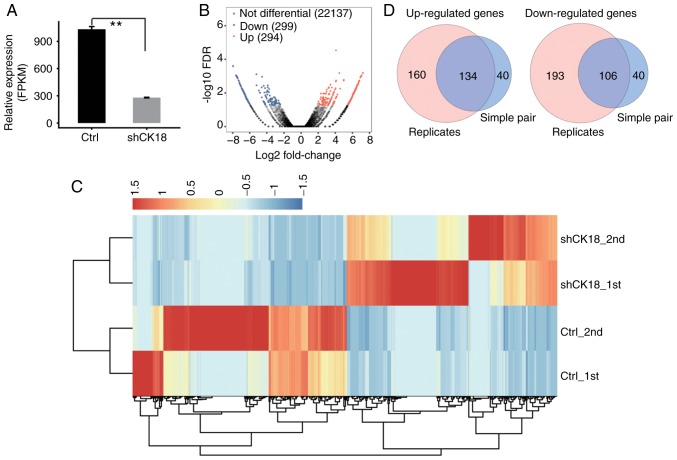
CK18 KD and control cells were used to construct cDNA libraries for sequencing on an Illumina HiSeq 4000 platform. Two biological replicates were used and a total of 76.7±4.9M 150 nucleotide paired-end raw reads per sample were obtained. The raw reads were filtered by removing low-quality reads and reads containing N and adaptor sequences, leaving 73.5±4.7M clean reads for downstream bioinformatics analysis. The clean reads were then mapped onto the human GRCh38 genome using TopHat2: 77.99-81.98% were aligned and ~86.77-95.05% were uniquely mapped (Table II). Quantification of genes and transcripts was reassessed using Cufflinks (38), to compare gene expression patterns across individuals. FPKM values were then calculated. There were 22,677 genes expressed in the RNA-seq data (available upon request). Effective KD of CK18 was further confirmed by in parallel RNA-seq analysis (Fig. 2A).
Table II.
Summary of RNA-seq reads used in the analysis.
| Sample | shCK18_1st | ShCK18_2nd | shCtrl_1st | shCtrl_2nd | |
|---|---|---|---|---|---|
| Raw reads | 75207886 | 83976994 | 73556828 | 73880306 | 76655503±4932995a |
| Clean reads | 72139395 | 80470749 | 70542901 | 70836199 | 73497311±4700455 |
| Paired-end reads | 68071008 | 68521042 | 69757064 | 77558436 | 70976888±4445225 |
| Total mapped | 55639546 | 56175922 | 54402225 | 62167878 | 57096393±3461607 |
| (81.74%b) | (81.98%) | (77.99%) | (80.16%) | ||
| Total uniquely mapped | 52472346 | 53397850 | 51511527 | 53943874 | 52831399±1069182 |
| (94.31%c) | (95.05%) | (94.69%) | (86.77%) | ||
| Splice reads | 28437401 | 30330300 | 29524140 | 29318683 | |
| (54.2%d) | (56.8%) | (57.32%) | (54.35%) |
Mean and standard deviation across the 4 samples.
Percentage of paired-end reads that were mapped to the genome.
Percentage of unique reads mapped out of the total mapped reads.
Percentage of uniquely mapped reads that were mapped to the splice site. CK18, cytokeratin 18.
Figure 2.
Differential gene expression in response to CK18 KD. (A) Quantification of CK18 expression using RNA-seq data, FPKM values were calculated as explained in Materials and methods. (B) Volcano plot of the CK18-regulated genes using edgeR software (filtering criteria, FC ≥2 and FDR<0.05) treating RNA-seq data from two samples in each group as experimental replicates. Red indicates upregulated genes and blue indicates downregulated genes. (C) Heat map of all the 593 differentially expressed genes in the control and CK18 shRNA-treated samples. (D) Overlap of two sets of DEGs. One set was identified as described in B. The other set was identified by treating one CK18 shRNA-treated and one control RNA-seq data as a simple pair using edgeR software; the resulting two pairs of DEGs were then overlapped to obtain the common simple-pair DEGs. CK18, cytokeratin 18; FPKM, fragments per kilobase of transcript per million fragments mapped; DEGs, differentially expressed genes.
Using criteria of an absolute fold change ≥2 and FDR <0.05 with the edge R package (35), 294 upregulated and 299 downregulated genes related with CK18 KD (data not shown) were identified. A volcano plot was constructed to display the significantly expressed genes that were associated with CK18 KD (Fig. 2B). The heatmap demonstrated distinguishable transcription profiles between CK18 KD and the control groups (Fig. 2C).
The two replicate RNA-seq datasets were divided into two groups (case 1 with control 1; case 2 with control 2). Using the criteria of an absolute fold change ≥2 with the EdgeR software package, 174 co-upregulated and 146 co-downregulated genes overlapping in two groups associated with CK18 KD were identified. A Venn-diagram demonstrated the upregulated and downregulated genes between the two different analytical strategies (Fig. 2D).
Functional analysis of CK18-regulated genes
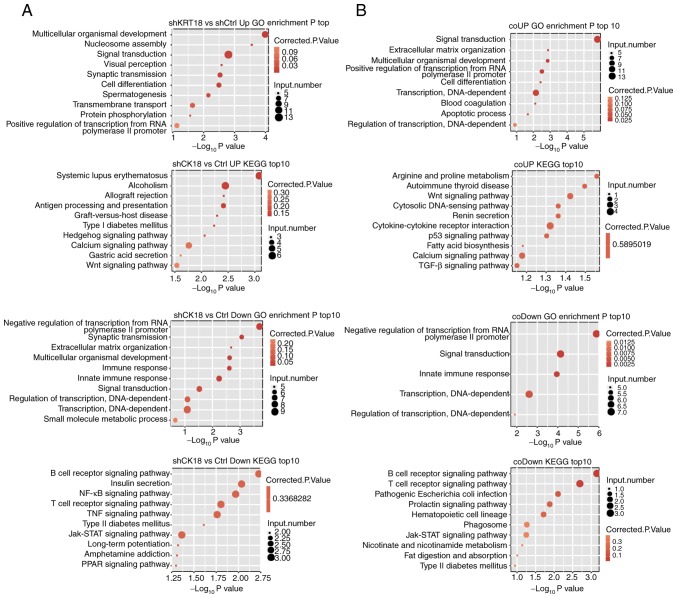
GO enrichment analysis was performed to further explore the biological function of DEGs with two different analytical strategies. All three ontologies of GO analysis, molecular function, cellular component and biological process, were obtained. The top biological process terms of the GO analysis that involved upregulation or downregulation of genes following CK18 KD are presented in Fig. 3A (repetition) and B (simple pair). Most of the terms overlapped; the upregulated genes were mainly associated with processes involved in multicellular organismal development, signal transduction, cell differentiation and the apoptosis process. The downregulated genes, conversely, were mainly associated with negative regulation of transcription from RNA polymerase II promoter, innate immune response, signal transduction, and regulation of transcription, DNA-dependent. Based on the KEGG analysis, the top ten pathways involved in up- and downregulated DEGs were also presented in Fig. 3. Coinciding with the GO analyses, many pathways were associated with the immune response and apoptosis. The B-cell receptor signaling pathway, the T-cell receptor signaling pathway and the Jak-STAT signaling pathway were all significantly enriched in downregulated gene sets.
Figure 3.
Functional enrichment analysis of DEGs. GO and KEGG analysis were performed on DEGs obtained using the two different strategies (Fig. 2D). (A) Top representatives of GO biological process terms and KEGG pathways up- and downregulated by CK18 of the DEGs in CK18-knockdown and control groups (named as replicates). (B) Top representatives of GO biological process terms and KEGG pathways up- and downregulated by CK18 of the DEGs overlapping in the two replicate groups (named as simple pair). DEGs, differentially expressed genes; CK18, cytokeratin 18.
Analysis of CK18-regulated ASEs
Regulation of alternative splicing (AS) of CK18 in the transcriptome sequencing data was also explored. Between 54.2 and 57.32% of the uniquely mapped reads from CK18 KD and control samples were junction reads (Table II). A total of 58.9% of annotated exons (216,356 out of 367,321) were detected. For splicing junctions, 147,540 annotated and 86,961 novel junctions were detected using TopHat2.
ASEs were analyzed using ABLas software to investigate global changes in the occurrence of alternative splicing. We detected 15,786 known ASEs in the model gene that we designated in the reference genome, along with 40,946 novel ASEs, excluding intron retention (Table III).
Table III.
Known or novel splicing events detected from ABLas.
| Known splicing events | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample | 3pMXE | 5pMXE | A3SS | A3SS&ES | A5SS | A5SS&ES | ES | IntronR | MXE | Cassette exon | Totalb | Detected junctionc | ||||||||||||||||||
| shCtrl_1st | 274 | 562 | 2614 | 296 | 3177 | 385 | 2058 | 1013 | 233 | 1463 | 12075 | 134568 | ||||||||||||||||||
| shCtrl_2nd | 181 | 383 | 1729 | 197 | 2103 | 277 | 1359 | 953 | 139 | 923 | 8244 | 114474 | ||||||||||||||||||
| shCK18_1st | 189 | 399 | 1838 | 202 | 2222 | 307 | 1460 | 959 | 135 | 1000 | 8711 | 119159 | ||||||||||||||||||
| shCK18_2nd | 224 | 451 | 2319 | 255 | 2725 | 371 | 1759 | 1009 | 194 | 1199 | 10506 | 128578 | ||||||||||||||||||
| All samplesa | 362 | 733 | 3455 | 384 | 4118 | 515 | 2573 | 1443 | 322 | 1881 | 15786 | 147540 | ||||||||||||||||||
| Novel splicing events | ||||||||||||||||||||||||||||||
| Sample | 3pMXE | 5pMXE | A3SS | A3SS&ES | A5SS | A5SS&ES | ES | IntronR | MXE | Cassette exon | Totalb | Detected junctiond | ||||||||||||||||||
| shCtrl_1st | 612 | 1654 | 3795 | 485 | 5032 | 574 | 1872 | 3782 | 205 | 660 | 18671 | 46290 | ||||||||||||||||||
| shCtrl_2nd | 341 | 972 | 2644 | 321 | 3395 | 390 | 1382 | 3072 | 120 | 401 | 13038 | 22297 | ||||||||||||||||||
| shCK18_1st | 381 | 1061 | 2737 | 360 | 3494 | 407 | 1485 | 3466 | 104 | 396 | 13891 | 24036 | ||||||||||||||||||
| shCK18_2nd | 503 | 1420 | 3305 | 391 | 4101 | 490 | 1642 | 3647 | 139 | 508 | 16146 | 34627 | ||||||||||||||||||
| All samplesa | 1313 | 3282 | 8950 | 1201 | 10843 | 1360 | 4380 | 7750 | 416 | 1451 | 40946 | 86961 | ||||||||||||||||||
Numbers in this line indicate the unique alternative splicing events identified from all four samples. Since many ASEs were detected in more than one sample, the numbers in this line are less than the sum of the four individual numbers.
Indicates the sum of all different types of alternative splicing events detected in each sample or all samples.
Indicates the known spliced junctions detected in each sample or all samples.
Indicates the novel spliced junctions detected in each sample or all samples. ASEs, alternative splicing events.
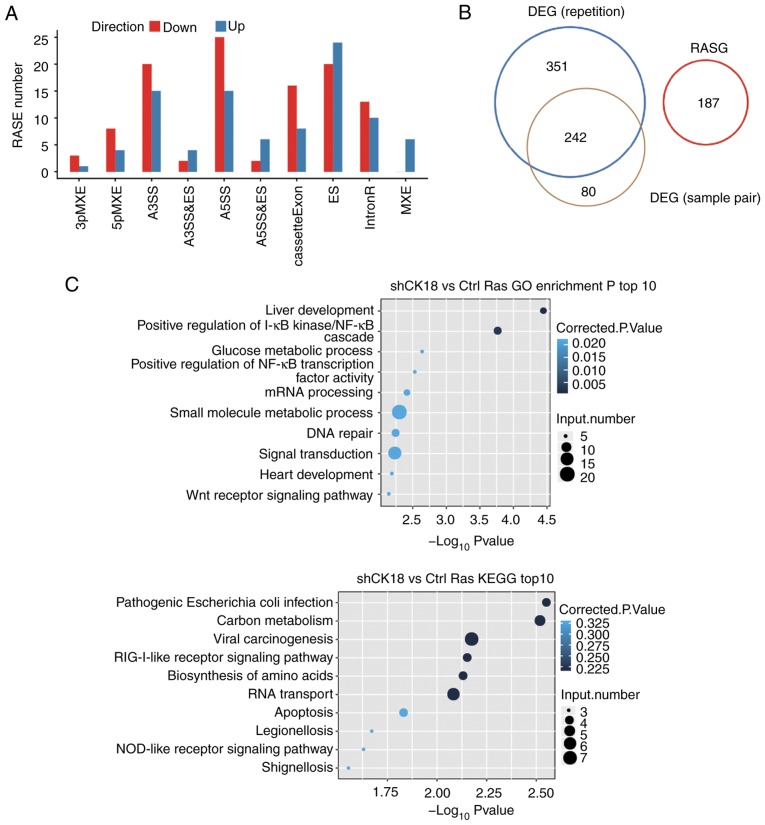
We identified 263 high confidence RASEs by using a stringent cutoff of P≤0.05, along with changed AS ratio ≥0.2. CK18-regulated ASEs, which included 35 alternative 3'splice sites (A3SS), 40 alternative 5'splice sites (A5SS), 44 examples of exon skipping (ES) and 24 cassette exons, 12 mutually exclusive 5′UTRs (5pMXE), four mutually exclusive 3′UTRs (3pMXE), six mutually exclusive exons (MXE), 84 examples of intron retention (IR), eight examples of alternative 5'splice site & exon skipping (A5SS & ES) and six examples of alternative 3'splice site & exon skipping (A3SS & ES), are summarized in Fig. 4A. These results indicated that CK18 KD had a broad influence on splicing. Coupled to the transcription data, there were no significant expression changes in alternative splicing genes (Fig. 4B). The alternative spliced genes that were identified by GO analyzed were, however, enriched in liver development, positive regulation of I-κB kinase/NF-κB cascade, glucose metabolic process, positive regulation of NF-κB transcription factor activity, mRNA processing and small molecule metabolic process. These results were similar to those obtained by the transcriptional GO analysis, further confirming the important regulatory role of CK18 in these biological functions. The detailed results of the GO and KEGG pathway analyses are presented in Fig. 4C. It was observed that alternative splicing of CASP8 and FAS, the known upstream genes regulating the production of CK18, was under the regulation of CK18.
Figure 4.
Identification and functional analysis of CK18-regulated splicing events. (A) Classification of different types of alternative splicing events regulated by CK18 protein. (B) Analysis of the overlap between CK18-regulated genes (DEGs from the two different analyses (Fig. 2D) and RASGs. (C) Top ten GO biological processes and KEGG functional pathways enriched by alternative gene splicing. CK18, cytokeratin 18; DEGs, differentially expressed genes; RASG, alternative splicing genes.
To confirm the important regulatory function of CK18 on both gene expression levels of alternative splicing in HeLa and HepG2 cell lines, we validated certain DEGs and ASEs that were important in immunity and apoptosis by RT-qPCR. The ASEs detected by PCR primer pairs (Table I) were designed to amplify both long splicing isoforms and the short splicing isoforms in the same reaction.
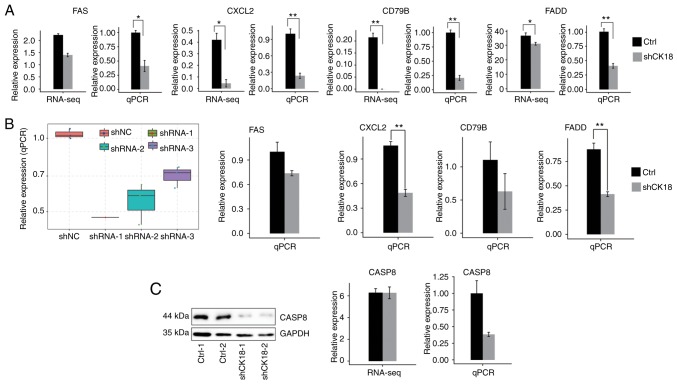
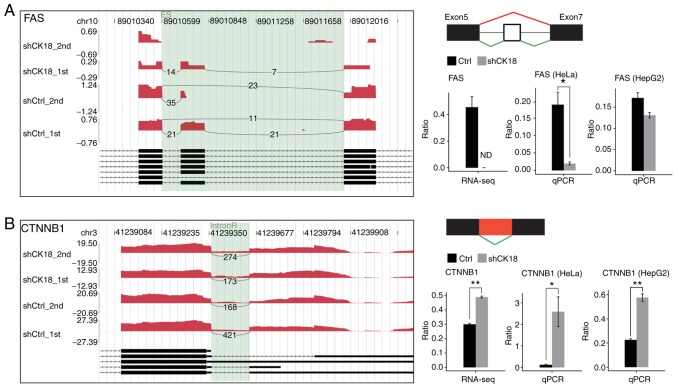
Most of the DEGs associated with immune function and apoptosis that we validated were in agreement with RNA-seq results (Fig. 5A and B). For CASP8 gene expression, however, there was no significant change in RNA-seq data, whereas the qPCR and western blotting results revealed an evident decrease in CK18 KD cells (Fig. 5C). Two important ASEs were located in FAS and CTNNB1, which have been well validated as key genes in apoptotic pathways (Fig. 6).
Figure 5.
(A-C) Validation of CK18-regulated genes (DEGs) by quantitative PCR and (C) western blotting in (A and C) HeLa cells, and in (B) HepG2 cells. (B) The effectiveness of three different CK18-specific shRNAs (left) and the effect of multiple CK18-specific shRNAs on the expression of four CK18-regulated genes (right) are presented. (C) Validation of the effect of CK18-specific shRNA1 on the protein level (left) and mRNA level (right) of CASP8. Experimental data are presented as the mean ± standard deviation of at least three experiments. *P<0.05 and **P<0.01. CK18, cytokeratin 18; DEGs, differentially expressed genes.
Figure 6.
Validation of CK18-regulated ASEs. (A) Validation of an ASE of FAS in HeLa and HepG2 cells. (B) Validation of ASEs of CTNNB1 in HeLa and HepG2 cells. Left panels reveal the IGV-sashimi plots for alternative splicing changes that occurred in HeLa cells in response to CK18 knockdown. The annotated transcripts for the gene are presented below. Right top panels reveal the schematic diagrams depicting the structures of alternative splicing events, AS1 (shown in red) and AS2 (shown in green); exon sequences are denoted by boxes, and intron sequences by the horizontal lines. Right bottom panels reveal the RNA-seq and RT-qPCR quantification of alternative splicing events. The altered ratio of ASEs in RNA-seq were calculated using the formula: AS1 junction reads/AS1 junction reads+AS2 junction reads; while the altered ratio of AS events in qPCR were calculated using the formula: AS1 transcripts level/AS2 transcripts level. CK18, cytokeratin 18; ASEs, alternative splicing events. *P<0.05 and **P<0.01.
Discussion
Knockdown of the expression level of CK18 by a shRNA in HeLa cells led to a significant reduction of apoptosis, indicating that CK18 is an apoptotic gene in the cancer cells. Through genome-wide transcriptional and ASE analysis of CK18 knockdown and control HeLa cells using RNA-seq, it was revealed that the FAS-mediated apoptosis pathway was regulated by CK18. Additionally, CASP8, which is downstream of the FAS-induced apoptosis pathway, was revealed to be feedback-regulated by its cleavage substrate, CK18. These findings indicated that CK18 had more diverse regulatory functions than have been generally recognized.
We have unbiasedly analyzed DEGs whose expression was regulated by CK18. These DEGs were enriched in multiple GO functional clusters and KEGG pathways. The finding that CK18 regulates the expression of genes enriched in multicellular organismal development and signal transduction was consistent with the reported function of CK18 in maintaining integrity of epithelial cells and organismal development (39,40). CK18-regulated genes were also enriched in immune response-related pathways, which includes the B-cell receptor signaling pathway, the T-cell receptor signaling pathway, the TNF signaling pathway and the Jak-STAT signaling pathway.
In addition to transcriptional regulation, CK18 also regulates alternative splicing of a large number of genes, which are enriched in NF-κB regulatory processes. NF-κB transcription factors are central coordinators of innate and adaptive immune responses. Activated NF-κB regulates the expression of over 300 different genes (41). Activated NF-κB generally inhibits apoptosis by activation of anti-apoptotic genes, although in some cases NF-κB promotes apoptosis (42,43). Fortier et al have reported that PI3K/Akt/NF-κB is activated when both CK8 and CK18 are knocked down in HepG2 and KLE cells, thus increasing cell migration and invasion (44).
CK18 has caspase recognition sites, which can be cleaved during apoptosis caused by tumor pathogenesis or chemotherapy. Fas- and TNF-mediated apoptosis are associated with caspase-mediated CK18 degradation. A previous study revealed a higher incidence of FasL-induced apoptosis in CK18− HeLa cells compared with CK18+ HeLa cells (27). Another study revealed that siRNA-mediated knockdown of K8/18 filament expression enhanced the expression of the apoptotic gene FAS (>70% of cells) in granulosa cell tumor cell line, KGN (45). CK18 mutation has, however, been determined to disrupt FAS-mediated apoptosis in the livers of transgenic mice (46).
In the present study, it was revealed that mRNA levels of FAS and FADD in HeLa cells were positively regulated by CK18, indicating that CK18-modulated apoptosis was regulated by FAS but not TNF. A decline in the ratio of FAS exon 6 skipping was also revealed. Skipping of FAS exon 6 resulted in an mRNA encoding a soluble form of FAS receptor that acted as a decoy to prevent cell apoptosis (47–50). It appears, therefore, that CK18 had dual regulatory effects on FAS-mediated apoptosis, i.e., CK18 positively regulated FAS transcription and also regulated production of the decoy isoform of FAS mRNA.
FAS and FADD have been revealed to recruit and activate CASP8, which plays an essential role in cleavage of the actin cytoskeleton, including cytokeratin (51). In the present study, it was revealed that CK18 regulated the alternative splicing of CASP8. In addition, western blot analysis revealed that cleaved (activated) CASP8 was significantly reduced upon CK18 knockdown. It is, therefore, possible that CK18 exerts feedforward regulation of its own cleavage by positively regulating the expression CASP8.
Full-length CK18 (M65) and the caspase-cleaved fragment (M30) are recognized as useful markers in clinical diagnosis and prognostic evaluation (18) and recently, the important regulatory role of CK18 has also attracted attention. A recent study revealed that CK18 could associate with histone H3, leading to aberrant expression of histone deacetylase in hepatocellular carcinoma (52). The finding that CK18 physically interacts with LRP16, sequestering LRP16 in the cytoplasm and thus inhibiting the proliferation of ERα-positive breast cancer cells (29), suggests that CK18 may affect gene expression via protein-protein interaction. Additionally, CK18 was revealed to be a potential RNA binding protein (30). RNA binding proteins play important roles in post-transcriptional gene regulation. We therefore propose that CK18 may regulate gene expression and alternative splicing through its protein and RNA binding functions. Further studies are required to explore this possibility.
CK18 dysregulation occurs widely in various tumors, although the regulatory mechanism in tumors is largely unclear. The knowledge that CK18 regulates the transcription and alternative splicing of NF-κB genes and the transcription of genes related to cancer cell migration and metastasis should expand our understanding of the biological and regulatory functions of CK18. It should also contribute to an improved understanding of the prognostic value and clinicopathological significance of CK18.
Acknowledgements
We are especially grateful to Hong Wu for the language polishing.
Funding
No funding was received.
Availability of data and materials
RNA-seq data mentioned in this article are publicly available at the NCBI Gene Expression Omnibus (GEO) under the accession no. GSE119255.
Author's contributions
YC performed experiments, contributed to analysis and interpretation of the data, and was a major contributor in writing the manuscript. KQ and NH performed experiments, and contributed in drafting and revising the manuscript. ZZ contributed in the data analysis and revision of the manuscript, HX and JZ performed the cell experiments, and contributed in the revision of the manuscript. YZ contributed to the design of the study and the writing and revision of the manuscript. SY contributed to the conception and design of the study, the data analysis and interpretation, and the writing and revision of the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Koh WJ, Greer BE, Abu-Rustum NR, Apte SM, Campos SM, Cho KR, Chu C, Cohn D, Crispens MA, Dorigo O, et al. Cervical Cancer, Version 2.2015. J Natl Compr Canc Netw. 2015;13:395–404. doi: 10.6004/jnccn.2015.0055. [DOI] [PubMed] [Google Scholar]
- 2.Zheng R, Zeng H, Zhang S, Chen T, Chen W. National estimates of cancer prevalence in China, 2011. Cancer Lett. 2016;370:33–38. doi: 10.1016/j.canlet.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Coulombe PA, Omary MB. ‘Hard’ and ‘soft’ principles defining the structure, function and regulation of keratin intermediate filaments. Curr Opin Cell Biol. 2002;14:110–122. doi: 10.1016/S0955-0674(01)00301-5. [DOI] [PubMed] [Google Scholar]
- 4.Greystoke A, Dean E, Saunders MP, Cummings J, Hughes A, Ranson M, Dive C, Renehan AG. Multi-level evidence that circulating CK18 is a biomarker of tumour burden in colorectal cancer. Br J Cancer. 2012;107:1518–1524. doi: 10.1038/bjc.2012.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schneider J. Tumor markers in detection of lung cancer. Adv Clin Chem. 2006;42:1–41. doi: 10.1016/S0065-2423(06)42001-1. [DOI] [PubMed] [Google Scholar]
- 6.Hernandez BY, Frierson HF, Moskaluk CA, Li YJ, Clegg L, Cote TR, McCusker ME, Hankey BF, Edwards BK, Goodman MT. CK20 and CK7 protein expression in colorectal cancer: Demonstration of the utility of a population-based tissue microarray. Hum Pathol. 2005;36:275–281. doi: 10.1016/j.humpath.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 7.Moll R, Divo M, Langbein L. The human keratins: Biology and pathology. Histochem Cell Biol. 2008;129:705–733. doi: 10.1007/s00418-008-0435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahn SK, Moon HG, Ko E, Kim HS, Shin HC, Kim J, You JM, Han W, Noh DY. Preoperative serum tissue polypeptide-specific antigen is a valuable prognostic marker in breast cancer. Int J Cancer. 2013;132:875–881. doi: 10.1002/ijc.27727. [DOI] [PubMed] [Google Scholar]
- 9.Escobar-Hoyos LF, Shah R, Roa-Peña L, Vanner EA, Najafian N, Banach A, Nielsen E, Al-Khalil R, Akalin A, Talmage D, Shroyer KR. Keratin-17 promotes p27KIP1 nuclear export and degradation and offers potential prognostic utility. Cancer Res. 2015;75:3650–3662. doi: 10.1158/0008-5472.CAN-15-0293. [DOI] [PubMed] [Google Scholar]
- 10.Tan HS, Jiang WH, He Y, Wang DS, Wu ZJ, Wu DS, Gao L, Bao Y, Shi JZ, Liu B, et al. KRT8 upregulation promotes tumor metastasis and is predictive of a poor prognosis in clear cell renal cell carcinoma. Oncotarget. 2017;8:76189–76203. doi: 10.18632/oncotarget.19198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung KJ, Padmanaban V, Silvestri V, Schipper K, Cohen JD, Fairchild AN, Gorin MA, Verdone JE, Pienta KJ, Bader JS, Ewald AJ. Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc Natl Acad Sci USA. 2016;113:E854–E863. doi: 10.1073/pnas.1508541113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu J. Doctoral dissertation. Johns Hopkins University; 2017. Nuclear Keratin 17 and its role in the DNA Damage Response. [Google Scholar]
- 13.Saha SK, Choi HY, Kim BW, Dayem AA, Yang GM, Kim KS, Yin YF, Cho SG. KRT19 directly interacts with β-catenin/RAC1 complex to regulate NUMB-dependent NOTCH signaling pathway and breast cancer properties. Oncogene. 2017;36:332–349. doi: 10.1038/onc.2016.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bilici A. Cytokeratin 18 for chemotherapy efficacy in gastric cancer. Transl Gastrointest Cancer. 2015;4:200–206. [Google Scholar]
- 15.Zhang B, Wang J, Liu W, Yin Y, Qian D, Zhang H, Shi B, Li C, Zhu J, Zhang L, et al. Cytokeratin 18 knockdown decreases cell migration and increases chemosensitivity in non-small cell lung cancer. J Cancer Res Clin Oncol. 2016;142:2479–2487. doi: 10.1007/s00432-016-2253-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin B, Zhang M, Zeng Y, Li Y, Zhang C, Getzenberg RH, Song Y. Downregulation of cytokeratin 18 is associated with paclitaxel-resistance and tumor aggressiveness in prostate cancer. Int J Oncol. 2016;48:1730–1736. doi: 10.3892/ijo.2016.3396. [DOI] [PubMed] [Google Scholar]
- 17.Huang YL, Chen J, Yan W, Zang D, Qin Q, Deng AM. Diagnostic accuracy of cytokeratin-19 fragment (CYFRA 21-1) for bladder cancer: A systematic review and meta-analysis. Tumour Biol. 2015;36:3137–3145. doi: 10.1007/s13277-015-3352-z. [DOI] [PubMed] [Google Scholar]
- 18.Nagel M, Schulz J, Maderer A, Goepfert K, Gehrke N, Thomaidis T, Thuss-Patience PC, Al-Batran SE, Hegewisch-Becker S, Grimminger P, et al. Cytokeratin-18 fragments predict treatment response and overall survival in gastric cancer in a randomized controlled trial. Tumor Biol. 2018;40:101042831876400. doi: 10.1177/1010428318764007. [DOI] [PubMed] [Google Scholar]
- 19.Sjöström J, Alfthan H, Joensuu H, Stenman UH, Lundin J, Blomqvist C. Serum tumour markers CA 15-3, TPA, TPS, hCGbeta and TATI in the monitoring of chemotherapy response in metastatic breast cancer. Scand J Clin Lab Invest. 2001;61:431–441. doi: 10.1080/00365510152567068. [DOI] [PubMed] [Google Scholar]
- 20.Yaman E, Coskun U, Sancak B, Buyukberber S, Ozturk B, Benekli M. Serum M30 levels are associated with survival in advanced gastric carcinoma patients. Int Immunopharmacol. 2010;10:719–722. doi: 10.1016/j.intimp.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 21.Demiray M, Ulukaya EE, Arslan M, Gokgoz S, Saraydaroglu O, Ercan I, Evrensel T, Manavoglu O. Response to neoadjuvant chemotherapy in breast cancer could be predictable by measuring a novel serum apoptosis product, caspase-cleaved cytokeratin 18: A prospective pilot study. Cancer Invest. 2006;24:669–676. doi: 10.1080/07357900600981307. [DOI] [PubMed] [Google Scholar]
- 22.Ulukaya E, Yilmaztepe A, Akgoz S, Linder S, Karadag M. The levels of caspase-cleaved cytokeratin 18 are elevated in serum from patients with lung cancer and helpful to predict the survival. Lung Cancer. 2007;56:399–404. doi: 10.1016/j.lungcan.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 23.Ozturk B, Coskun U, Sancak B, Yaman E, Buyukberber S, Benekli M. Elevated serum levels of M30 and M65 in patients with locally advanced head and neck tumors. Int Immunopharmacol. 2009;9:645–648. doi: 10.1016/j.intimp.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 24.Homberg M, Magin TM. Beyond expectations: Novel insights into epidermal keratin function and regulation. Int Rev Cell Mol Biol. 2014;311:265–306. doi: 10.1016/B978-0-12-800179-0.00007-6. [DOI] [PubMed] [Google Scholar]
- 25.Weng YR, Cui Y, Fang JY. Biological functions of cytokeratin 18 in cancer. Mol Cancer Res. 2012;10:485–493. doi: 10.1158/1541-7786.MCR-11-0222. [DOI] [PubMed] [Google Scholar]
- 26.Messai Y, Noman MZ, Derouiche A, Kourda N, Akalay I, Hasmim M, Stasik I, Ben Jilani S, Chebil M, Caignard A, et al. Cytokeratin 18 expression pattern correlates with renal cell carcinoma progression: Relationship with Snail. Int J Oncol. 2010;36:1145–1154. doi: 10.3892/ijo_00000597. [DOI] [PubMed] [Google Scholar]
- 27.Sullivan BT, Cherry JA, Sakamoto H, Henkes LE, Townson DH, Rueda BR. Cytokeratin 18 expression inhibits cytokine-induced death of cervical cancer cells. Int J Gynecol Cancer. 2010;20:1474–1481. doi: 10.1111/IGC.0b013e3181fc3a03. [DOI] [PubMed] [Google Scholar]
- 28.Bühler H, Schaller G. Transfection of keratin 18 gene in human breast cancer cells causes induction of adhesion proteins and dramatic regression of malignancy in vitro and in vivo. Mol Cancer Res. 2005;3:365–371. doi: 10.1158/1541-7786.MCR-04-0117. [DOI] [PubMed] [Google Scholar]
- 29.Meng Y, Wu Z, Yin X, Zhao Y, Chen M, Si Y, Yang J, Fu X, Han W. Keratin 18 attenuates estrogen receptor alpha-mediated signaling by sequestering LRP16 in cytoplasm. BMC Cell Biol. 2009;10:96. doi: 10.1186/1471-2121-10-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 31.Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet. 2014;15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landry JJ, Pyl PT, Rausch T, Zichner T, Tekkedil MM, Stütz AM, Jauch A, Aiyar RS, Pau G, Delhomme N, et al. The genomic and transcriptomic landscape of a HeLa cell line. G3 (Bethesda) 2013;3:1213–1224. doi: 10.1534/g3.113.005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson MD, McCarthy D, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39 (Web Server Issue) 2011:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia H, Chen D, Wu Q, Wu G, Zhou Y, Zhang Y, Zhang L. CELF1 preferentially binds to exon-intron boundary and regulates alternative splicing in HeLa cells. Biochim Biophys Acta Gene Regul Mech. 2017;1860:911–921. doi: 10.1016/j.bbagrm.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 38.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karantza V. Keratins in health and cancer: More than mere epithelial cell markers. Oncogene. 2011;30:127–138. doi: 10.1038/onc.2010.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toivola DM, Boor P, Alam C, Strnad P. Keratins in health and disease. Curr Opin Cell Biol. 2015;32:73–81. doi: 10.1016/j.ceb.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 41.Serasanambati MR, Chilakapati SR. Function of nuclear factor Kappa B (NF-κB) in human diseases-a review. South Indian J Biol Sci. 2016;2:368–387. doi: 10.22205/sijbs/2016/v2/i4/103443. [DOI] [Google Scholar]
- 42.Magné N, Toillon RA, Bottero V, Didelot C, Houtte PV, Gérard JP, Peyron JF. NF-kappaB modulation and ionizing radiation: Mechanisms and future directions for cancer treatment. Cancer Lett. 2006;231:158–168. doi: 10.1016/j.canlet.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 43.Chen X, Kandasamy K, Srivastava RK. Differential roles of RelA (p65) and c-Rel subunits of nuclear factor kappa B in tumor necrosis factor-related apoptosis-inducing ligand signaling. Cancer Res. 2003;63:1059–1066. [PubMed] [Google Scholar]
- 44.Fortier AM, Asselin E, Cadrin M. Keratin 8 and 18 loss in epithelial cancer cells increases collective cell migration and cisplatin sensitivity through claudin1 up-regulation. J Biol Chem. 2013;288:11555–11571. doi: 10.1074/jbc.M112.428920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trisdale SK, Schwab NM, Hou X, Davis JS, Townson DH. Molecular manipulation of keratin 8/18 intermediate filaments: modulators of FAS-mediated death signaling in human ovarian granulosa tumor cells. J Ovarian Res. 2016;9:8. doi: 10.1186/s13048-016-0217-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ku NO, Soetikno RM, Omary MB. Keratin mutation in transgenic mice predisposes to Fas but not TNF-induced apoptosis and massive liver injury. Hepatology. 2003;37:1006–1014. doi: 10.1053/jhep.2003.50181. [DOI] [PubMed] [Google Scholar]
- 47.Cheng J, Zhou T, Liu C, Shapiro JP, Brauer MJ, Kiefer MC, Barr PJ, Mountz JD. Protection from Fas-mediated apoptosis by a soluble form of the Fas molecule. Science. 1994;263:1759–1762. doi: 10.1126/science.7510905. [DOI] [PubMed] [Google Scholar]
- 48.Liu C, Cheng J, Mountz JD. Differential expression of human Fas mRNA species upon peripheral blood mononuclear cell activation. Biochem J. 1995;310:957–963. doi: 10.1042/bj3100957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papoff G, Cascino I, Eramo A, Starace G, Lynch DH, Ruberti G. An N-terminal domain shared by Fas/Apo-1 (CD95) soluble variants prevents cell death in vitro. J Immunol. 1996;156:4622–4630. [PubMed] [Google Scholar]
- 50.Tejedor JR, Papasaikas P, Valcárcel J. Genome-wide identification of Fas/CD95 alternative splicing regulators reveals links with iron homeostasis. Mol Cell. 2015;57:23–38. doi: 10.1016/j.molcel.2014.10.029. [DOI] [PubMed] [Google Scholar]
- 51.Stegh AH, Herrmann H, Lampel S, Weisenberger D, Andrä K, Seper M, Wiche G, Krammer PH, Peter ME. Identification of the cytolinker plectin as a major early in vivo substrate for caspase 8 during CD95- and tumor necrosis factor receptor-mediated apoptosis. Mol Cell Biol. 2000;20:5665–5679. doi: 10.1128/MCB.20.15.5665-5679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lai YC, Cheng CC, Lai YS, Liu YH. Cytokeratin 18-associated histone 3 modulation in hepatocellular carcinoma: A mini review. Cancer Genomics Proteomics. 2017;14:219–223. doi: 10.21873/cgp.20033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNA-seq data mentioned in this article are publicly available at the NCBI Gene Expression Omnibus (GEO) under the accession no. GSE119255.