Abstract
The present study aimed to investigate the clinical significance and prospective molecular mechanism of cystatin (CST) genes in patients with hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC). The role of CST genes in the molecular mechanism of HCC was revealed through bioinformatics analysis. The clinical significance of CST genes was investigated using GSE14520-derived data from patients with HBV-related HCC. Gene set enrichment analysis (GSEA) was used to identify pathways in which the CST genes were enriched, as well as the association between these pathways and HCC. The expression levels of CST1, CST2, CST5, CSTA and CSTB genes were higher in HCC tissue compared with in normal tissue; conversely, CST3 and CST7 were reduced in HCC tissue. Subsequent receiver operating characteristic analysis of the CST genes demonstrated that CST7 and CSTB genes may function as potential diagnostic markers for HCC. Furthermore, the expression levels of CST6 and CST7 were strongly associated with recurrence-free survival and overall survival of patients with HBV-related HCC. GSEA of the CST genes revealed that CST7 was significantly enriched in tumor evasion and tolerogenicity, cancer progenitors, liver cancer late recurrence, liver cancer progression and several liver cancer subclasses. In addition, CST genes demonstrated homology in terms of protein structure and were revealed to be strongly co-expressed. The present findings suggested that CST7 and CSTB genes may serve as potential prognostic and diagnostic biomarkers for HCC.
Keywords: clinical significance, molecular mechanism, cystatin, hepatitis B virus, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) was reported to be the sixth most common cancer and the fourth most common cause of malignancy-associated mortality worldwide in 2018. Each year, ~841,000 new cases of HCC are diagnosed and 782,000 deaths occur due to HCC worldwide (1). Notably, ~50% of newly diagnosed HCC cases and HCC-related deaths are thought to occur in China, with ~466,100 newly diagnosed patients and ~422,100 deaths occurring in China in 2015 (2). Primary liver cancer includes several pathological types, of which HCC is the predominant form that accounts for 75–85 % of all cases, with an incidence of 6.20 cases per 100,000 (1,3). Compared with other types of cancer, HCC in China carried the worst prognosis between 2003 and 2005, with a 1-year survival rate of <50% and a 5-year-survival rate of only 10.1% (4,5). Hepatitis B virus (HBV) infection is the primary cause of the high incidence of HCC in China (6). Given the poor prognosis of this disease, early HCC detection and treatment is of the utmost importance (7). Recent advances in genetic research have promoted a comprehensive understanding of the role of genetic mutations in HCC, allowing for the identification of diagnostic and prognostic HCC biomarkers (8).
Cysteine proteases are critical in promoting the progression of various types of tumor (9). There are eight subfamilies in the cystatin (CST) family group (Family 1, Family 2, Family 3, HRG, Fetuins, CRP, Spp24 and CRES) (10). Cysteine proteases are inhibited by CSTs (11) and are concentrated in the leading edge of tumor cells, where they dissolve extracellular matrix (ECM) proteins to promote invasion (12,13), thus enhancing tumor progression. Several types of CST have been discovered to possess significantly distinct expression profiles in HCC compared with their expression in healthy tissues. CST3, CSTA and CSTB are significantly highly expressed in HCC tissue compared with in adjacent healthy tissue, and the expression levels of CSTA and CSTB are strongly associated with node metastasis for HCC (14,15). Additionally, it has been reported that CST3 and CSTB may function as serum markers for HCC (15,16). Therefore, further investigations into the role of CST genes in HCC are warranted. The present study aimed to uncover the prognostic and diagnostic values of Family 1 CSTs (CSTA and CSTB) and Family 2 CSTs (CST1, CST2, CST3, CST4, CST5, CST6, CST7 and CST8) in patients with HCC using freely available data derived from public genomic databases.
Materials and methods
Bioinformatics analysis of CST genes
Database for Annotation, Visualization and Integrated Discovery (DAVID, version 6.8; david.ncifcrf.gov/home.jsp) (17,18) was accessed on December 17, 2018 for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation, Gene Ontology (GO) functional annotation and enrichment analysis of CST genes. An enrichment P<0.05 was considered to indicate a statistically significant difference. Gene-gene interactions of CST genes were constructed using GeneMANIA (www.genemania.org, accessed December 17, 2018) (19,20), whereas protein-protein interactions of CST genes were constructed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; string-db.org, accessed December 17, 2018) (21,22).
Data source
The GSE14520 dataset (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14520, accessed December 17, 2018), which comprises clinical data of patients with HBV-related HCC as well as their CST gene expression profiles, was extracted from the Gene Expression Omnibus database (23–25). Due to multiple probe sets in GSE14520, the expression value of each gene was regarded as the average value corresponding to the same gene and was normalized using the limma package of the R platform (version 3.5.1.; www.r-project.org).
Analysis of gene association and assessment of diagnostic value
Correlations between the CST genes were analyzed using Pearson's correlation coefficient and were depicted using the corrplot function of the R platform (version 3.5.1.; www.r-project.org); P<0.05 as considered to indicate a statistically significant difference. Differential expression of the CST genes between healthy liver tissues and HCC tumor tissues were statistically analyzed using Student's t-test in SPSS software (version 22.0; IBM Corp.); P<0.05 as considered to indicate a statistically significant difference. Receiver operating characteristic (ROC) curve analysis was used to assess the diagnostic value of CST genes in predicting HCC (26,27).
Survival analysis
Based on the median value of gene expression, patients were grouped into either the low or high gene expression group. Each CST gene was analyzed for survival using Kaplan-Meier analysis with log-rank test, and a Cox proportional hazards regression model was conducted to analyze the association of CST genes with clinical parameters that were strongly associated with OS (P<0.05). The CST genes associated with survival of patients with HCC (adjusted P<0.05) were analyzed in combination to explore their joint effects on survival analysis using Kaplan-Meier analysis and log-rank test, and Cox proportional hazards regression model. Nomograms based on biological and clinical variables were used to construct a statistical prognostic model of overall survival (OS) for HCC in accordance with survival analysis results and the Cox proportional hazards regression model (28). Data processing and plot generation were conducted in R platform (version 3.5.1.; www.r-project.org) with rms package. A scale that was marked on both ends of the line corresponding to each variable represented the value range of the variable, and the length of the line segment reflected the contribution of this factor to the outcome event.
Gene set enrichment analysis (GSEA)
The biological pathways targeted by CST genes were further explored with GSEA (accessed December 17, 2018) (29) using data derived from the Molecular Signatures Database of c2 (c2.all.v6.1 symbols) and c5 (c5.all.v6.1 symbols) (30). GSEA-derived gene enrichment sets that attained a false discovery rate (FDR) of <0.25 and P<0.05 were determined to confer statistical significance.
Statistical analysis
Statistical data processing was conducted using SPSS (version 22.0; IBM Corp.) and R (version 3.5.1.; www.r-project.org). The relative risk of patients with HCC based on CST gene expression was expressed in terms of 95% confidence intervals (CIs) and hazard ratios (HRs). Univariate survival analysis of the CST genes and clinical parameters was performed using Kaplan-Meier analysis with log-rank test. CST genes and patient clinical parameters that were strongly correlated with OS (P<0.05) were further subjected to a multivariate Cox proportional hazards regression model. Pearson's correlation coefficient was used to assess the relationship between co-expressed CST genes. P<0.05 was considered to indicate a statistically significant difference. FDR control of GSEA was achieved using the Benjamini-Hochberg procedure and adjusted for multiple testing (31–33).
Results
Bioinformatics analysis of CST genes
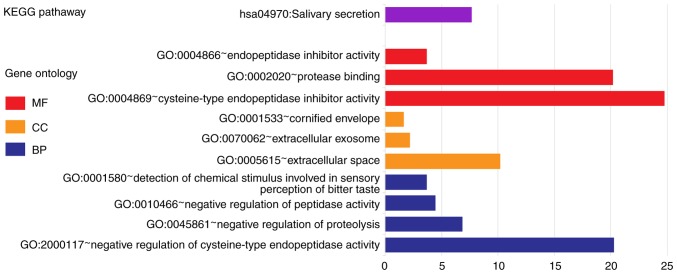
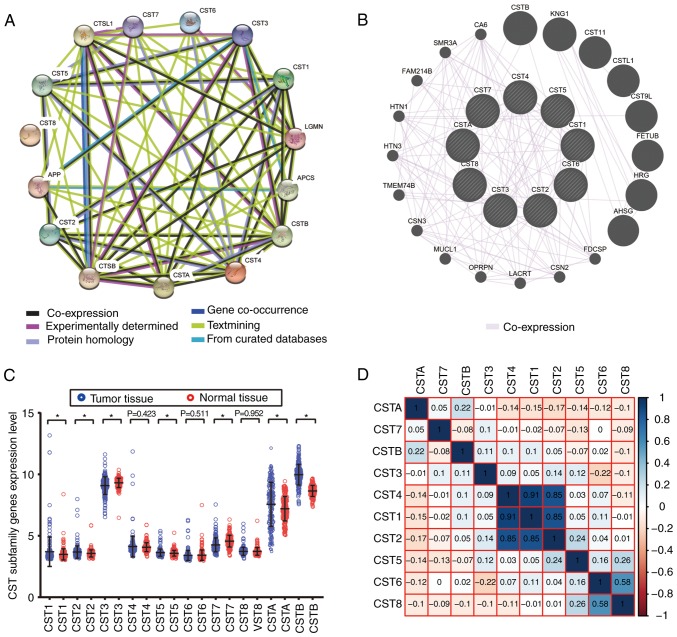
Biological functions (biological processes, cellular components and molecular functions) of CST1, CST2, CST3, CST4, CST5, CST6, CST7, CST8, CSTA and CSTB were subjected to a GO analysis using DAVID. Each of these genes was markedly enriched in ‘extracellular space’, ‘cysteine-type endopeptidase inhibitor activity’ and ‘protease binding’ (Fig. 1). KEGG pathway analysis using DAVID suggested that CST1, CST2, CST3, CST4 and CST5 were involved in ‘salivary secretion’ (Fig. 1). CST1, CST2, CST4, CSTA and CSTB genes and proteins had significant co-expression relationships (Fig. 2B) and strong protein homology (Fig. 2A) with each other.
Figure 1.
KEGG pathway and GO term analysis of cystatin genes. BP, biological process; CC, cellular component; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; MF, molecular function.
Figure 2.
Interaction and correlation analysis of CST genes, and the expression level of CST genes between HCC and normal tissues in the GSE14520 dataset. (A) STRING protein-protein association networks of the CST genes. (B) GeneMANIA gene-gene interaction networks of the CST genes. (C) Expression levels of CST genes between HCC and normal tissues in the GSE14520 dataset. (D) Matrix graphs of Pearson correlation analysis of the CST genes in the GSE14520 dataset. *P<0.05. CST, cystatin; HCC, hepatocellular carcinoma.
Data source
The present study derived its data only from the Affymetrix HT Human Genome U133A Array of GSE14520 in order to avoid a batch effect. The majority of subjects in this cohort had HBV-related HCC, whereas the remainder of non-HBV-related cases and those with no survival data were eliminated. This resulted in data from 204 adjacent healthy liver tissues and 212 HBV-related HCC tumor tissues. Data regarding patient prognosis were available for all patients.
Analysis of gene association and assessment of diagnostic value
CST gene co-expression in HCC neoplastic tissues was analyzed using the Pearson correlation coefficient. CST1, CST2 and CST4 were closely associated with each other in GSE14520 (Fig. 2D). Furthermore, CST1, CST2, CST5, CSTA and CSTB expression levels were markedly increased in HCC tumor tissue in the GSE14520 dataset, whereas CST3 and CST7 expression levels were markedly decreased in HCC tumor tissue (Fig. 2C). There was no significant difference in the expression of CST4, CST6 and CST8 between HCC tumor tissues and healthy liver tissues.
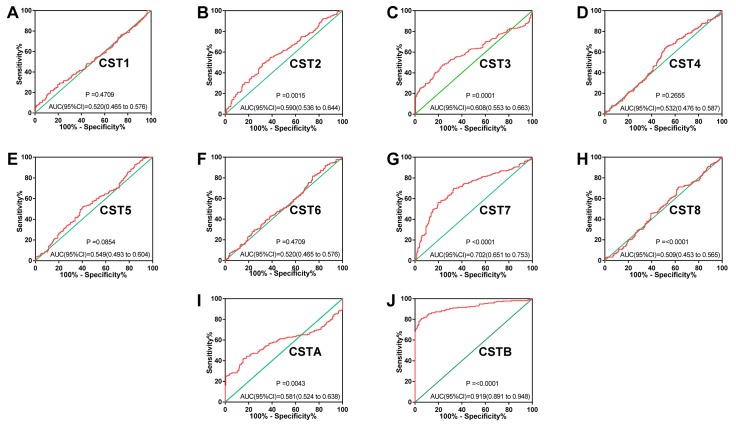
ROC analysis of CST genes revealed that the expression levels of CST7 and CSTB had significant diagnostic values in differentiating between healthy and malignant hepatic tissues. The area under the ROC curves of CST7 and CSTB were 0.702 (95% CI: 0.651-0.753; Fig. 3G) and 0.919 (95% CI: 0.891-0.948; Fig. 3J), respectively. The other CST genes did not exhibit significant diagnostic values.
Figure 3.
ROC curve analysis of the ability of CST genes to discriminate between hepatocellular carcinoma and adjacent healthy tissues in the GSE14520 dataset. ROC curves for (A) CST1, (B) CST2, (C) CST3, (D) CST4, (E) CST5, (F) CST6, (G) CST7, (H) CST8, (I) CSTA and (J) CSTB. AUC, area under the curve; CST, cystatin; ROC, receiver operating characteristic.
Survival analysis
In GSE14520, patients with an advanced Barcelona Clinic Liver Cancer (BCLC) stage (34), larger tumor volume (diameter, >5 cm), higher serum α-fetoprotein (AFP; >300 ng/ml) and cirrhosis were at high risk of death due to HBV-related HCC (Table SI). Cirrhotic patients, males and those with advanced BCLC stages were also more at risk of recurrence of HBV-related HCC (Table SI). No other clinical parameters were revealed to impact recurrence-free survival (RFS) or OS.
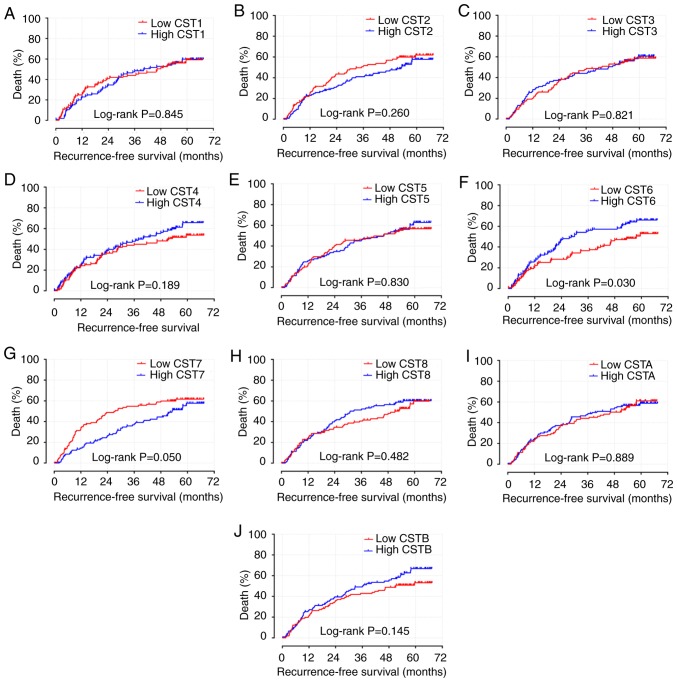
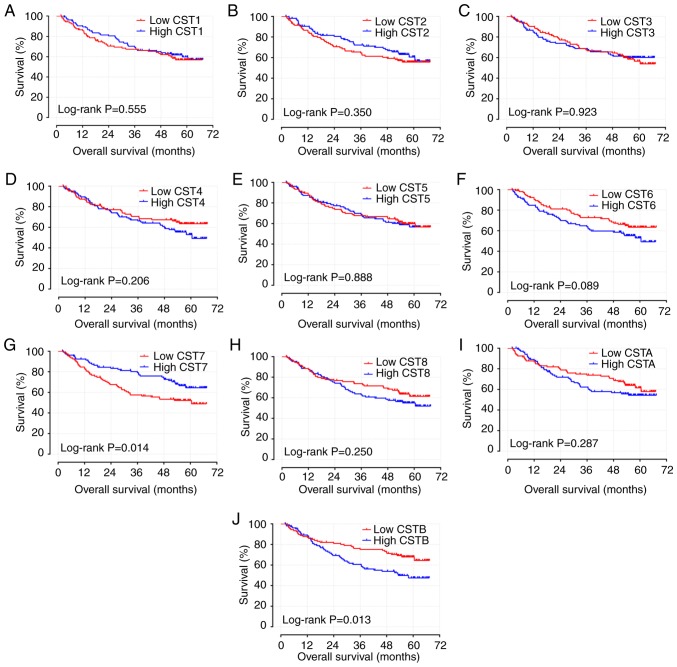
The results of survival analyses of the CST genes are presented in Figs. 4A-J, 5A-J and Table I. Low expression levels of CST6 (adjusted P=0.009; adjusted HR=1.651; 95% CI: 1.136-2.398; Table I; Fig. 4F) and high expression levels of CST7 (adjusted P=0.048; adjusted HR=0.688; 95% CI: 0.475-0.966; Table I; Fig. 4G) were strongly associated with the increased RFS of patients with HBV-related HCC, adjusted for sex, BCLC stage and cirrhosis. Low expression levels of CST6 (adjusted P=0.036; adjusted HR=1.618; 95% CI: 1.033-2.533; Table I; Fig. 5F) and high expression levels of CST7 (adjusted P=0.014; adjusted HR=0.559; 95% CI: 0.351-0.891; Table I; Fig. 5G) were also strongly associated with the increased OS of patients with HBV-related HCC, adjusted for tumor size, AFP, BCLC stage and cirrhosis.
Figure 4.
Kaplan-Meier survival curve analysis of recurrence-free survival for CST genes in hepatitis B virus-related hepatocellular carcinoma in the GSE14520 dataset. RFS curves for (A) CST1, (B) CST2, (C) CST3, (D) CST4, (E) CST5, (F) CST6, (G) CST7, (H) CST8, (I) CSTA and (J) CSTB. CST, cystatin.
Figure 5.
Kaplan-Meier survival curve analysis of overall survival for CST genes in hepatitis B virus-related hepatocellular carcinoma in the GSE14520 dataset. OS curves for (A) CST1, (B) CST2, (C) CST3, (D) CST4, (E) CST5, (F) CST6, (G) CST7, (H) CST8, (I) CSTA and (J) CSTB. CST, cystatin.
Table I.
Prognostic values of CST gene expression in patients with hepatitis B virus-related hepatocellular carcinoma from the GSE14520 dataset.
| RFS | OS | ||||||||||||
| Gene expression | Patients (n=212) | No. of events | MRT (months) | Crude HR (95% CI) | Crude P-value | Adjusted HR (95% CI) | Adjusted P-valuea | No. of events | MST (months) | Crude HR (95% CI) | Crude P-value | Adjusted HR (95% CI) | Adjusted P-valueb |
| CST1 | |||||||||||||
| Low | 106 | 57 | 46 | 1 | 1 | 42 | NA | 1 | 1 | ||||
| High | 106 | 59 | 40 | 0.964 (0.670-1.388) | 0.846 | 0.900 (0.624-1.299) | 0.575 | 40 | NA | 0.878 (0.569-1.354) | 0.555 | 0.931 (0.602-1.44) | 0.748 |
| CST2 | |||||||||||||
| Low | 106 | 62 | 35 | 1 | 1 | 44 | NA | 1 | 1 | ||||
| High | 106 | 54 | 53 | 0.811 (0.563-1.169) | 0.261 | 0.739 (0.511-1.068) | 0.108 | 38 | NA | 0.813 (0.527-1.256) | 0.351 | 0.768 (0.492-1.198) | 0.244 |
| CST3 | |||||||||||||
| Low | 106 | 57 | 43 | 1 | 1 | 42 | NA | 1 | 1 | ||||
| High | 106 | 59 | 46 | 1.043 (0.725-1.501) | 0.821 | 0.940 (0.647-1.365) | 0.746 | 40 | NA | 0.979 (0.635-1.509) | 0.923 | 0.803 (0.513-1.259) | 0.339 |
| CST4 | |||||||||||||
| Low | 106 | 52 | 51 | 1 | 1 | 35 | NA | 1 | 1 | ||||
| High | 106 | 64 | 40 | 1.278 (0.886-1.843) | 0.19 | 1.186 (0.821-1.713) | 0.365 | 47 | 60 | 1.325 (0.855-2.054) | 0.208 | 1.230 (0.791-1.913) | 0.357 |
| CST5 | |||||||||||||
| Low | 106 | 57 | 46 | 1 | 1 | 41 | NA | 1 | 1 | ||||
| High | 106 | 59 | 45 | 1.041 (0.723-1.498) | 0.83 | 1.066 (0.738-1.539 | 0.735 | 41 | NA | 1.032 (0.699-1.591) | 0.888 | 1.074 (0.693-1.664) | 0.75 |
| CST6 | |||||||||||||
| Low | 106 | 55 | 57 | 1 | 1 | 35 | NA | 1 | 1 | ||||
| High | 106 | 66 | 28 | 1.499 (1.037-2.166) | 0.031 | 1.651 (1.136-2.398) | 0.009 | 47 | 60 | 1.459 (0.942-2.260) | 0.091 | 1.618 (1.033-2.533) | 0.036 |
| CST7 | |||||||||||||
| Low | 106 | 63 | 27 | 1 | 1 | 49 | 60 | 1 | 1 | ||||
| High | 106 | 53 | 53 | 0.695 (0.482-1.003) | 0.052 | 0.688 (0.475-0.966) | 0.048 | 33 | 0.579 (0.372-0.901) | 0.016 | 0.559 (0.351-0.891) | 0.014 | |
| CST8 | |||||||||||||
| Low | 106 | 56 | 51 | 1 | 1 | 37 | NA | 1 | 1 | ||||
| High | 106 | 60 | 30 | 1.139 (0.791-1.641) | 0.483 | 1.106 (0.764-1.602) | 0.592 | 45 | NA | 1.290 (0.835-1.994) | 0.251 | 1.382 (0.883-2.164) | 0.157 |
| CSTA | |||||||||||||
| Low | 106 | 59 | 46 | 1 | 1 | 38 | NA | 1 | 1 | ||||
| High | 106 | 57 | 40 | 1.026 (0.713-1.477) | 0.889 | 1.173 (0.811-1.696) | 0.397 | 44 | NA | 1.266 (0.820-1.955) | 0.288 | 1.473 (0.943-2.299) | 0.088 |
| CSTB | |||||||||||||
| Low | 106 | 54 | 51 | 1 | 1 | 34 | NA | 1 | 1 | ||||
| High | 106 | 62 | 36 | 1.311 (0.910-1.889) | 0.147 | 1.164 (0.803-1.698) | 0.423 | 48 | 53 | 1.734 (1.115-2.694) | 0.014 | 1.468 (0.935-2.307) | 0.096 |
Adjusted for sex, cirrhosis and BCLC stage using multivariate Cox proportional hazards regression model
adjusted for tumor size, cirrhosis, BCLC stage and α-fetoprotein using multivariate Cox proportional hazards regression model. BCLC, Barcelona Clinic Liver Cancer; CI, confidence interval; CST cystatin; HR, hazard ratio; MRT, median recurrence time; MST, median survival time; NA, not available; OS, overall survival; RFS, recurrence-free survival.
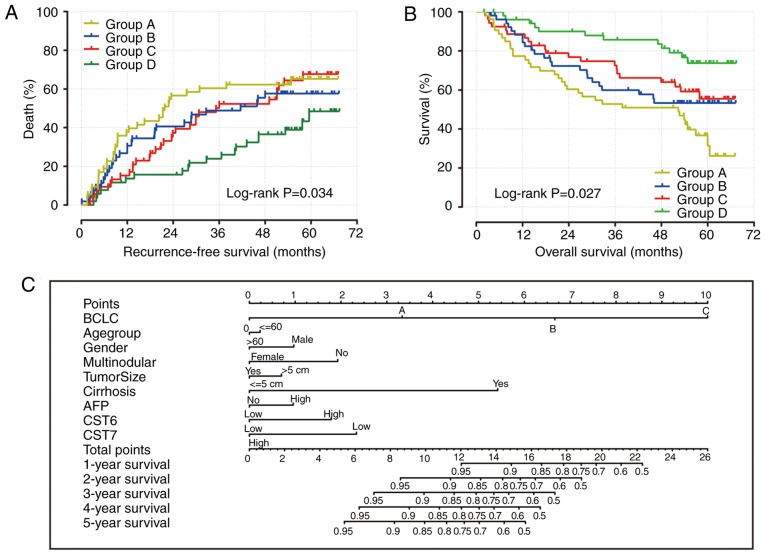
The results of CST gene survival analysis indicated that the expression levels of CST6 and CST7 may be significantly associated with the recurrence and mortality of patients with HBV-related HCC. The combined impact of CST6 and CST7 on OS and RFS of patients with HBV-related HCC was then further analyzed. Patients were divided into four groups according to CST6 and CST7 expression: Group A, high CST6 and low CST7 expression; Group B, low CST6 and low CST7 expression; Group C, high CST6 and high CST7 expression; Group D, low CST6 and high CST7 expression. Patients who had low CST6 expression and high CST7 expression had a decreased risk of recurrence (adjusted P=0.003; adjusted HR=0.431; 95% CI: 0.264-0.754; Table II; Fig. 6A) and mortality (adjusted P=0.001; adjusted HR=0.315; 95% CI: 0.115-0.641; Table III; Fig. 6B) in HBV-related HCC. In addition, the nomogram indicated that both CST6 and CST7 may make a contribution to the prognosis of HCC (Fig. 6C).
Table II.
Joint effects analysis of CST6 and CST7 expression in the recurrence-free survival of patients with hepatocellular carcinoma.
| Group | CST6 | CST7 | Patients | No. of events | MRT (months) | Crude HR (95% CI) | Crude P-value | Adjusted HR (95% CI) | Adjusted P-valuea |
|---|---|---|---|---|---|---|---|---|---|
| A | High | Low | 53 | 34 | 22 | 1 | – | 1 | – |
| B | Low | Low | 53 | 29 | 42 | 0.788 (0.480-1.294) | 0.48 | 0.753 (0.458-1.237) | 0.263 |
| C | High | High | 53 | 32 | 36 | 0.815 (0.503-1.322 | 0.408 | 0.880 (0.541-1.4330) | 0.608 |
| D | Low | High | 53 | 21 | NA | 0.452 (0.262-0.779) | 0.004 | 0.431 (0.264-0.754) | 0.003 |
Adjusted for sex, cirrhosis and Barcelona Clinic Liver Cancer stage in the GSE14520 cohort using multivariate Cox proportional hazards regression model. CI, confidence interval; CST, cystatin; HR, hazard ratio; MRT, median recurrence time; NA, not available.
Figure 6.
Combined effect of CST6 and CST7 on the overall survival and recurrence-free survival of patients, and nomogram for predicting 1-, 2- and 3-year events. (A) Recurrence-free survival curves for the combined effect of CST6 and CST7; (B) overall survival curves for the combined effect of CST6 and CST7. Group A, high CST6 and low CST7 expression; Group B, low CST6 and low CST7 expression; Group C, high CST6 and high CST7 expression; Group D, low CST6 and high CST7 expression. (C) Nomogram for predicting 1-, 2- and 3-year events (death) that combine clinical data with CST6 and CST7 expression. AFP, α-fetoprotein; BCLC, Barcelona Center Liver Cancer; CST, cystatin.
Table III.
Joint effects analysis of CST6 and CST7 expression in the overall survival of patients with hepatocellular carcinoma.
| Group | CST6 | CST7 | Patients | No. of events | MST (months) | Crude HR (95% CI) | Crude P-value | Adjusted HR (95% CI) | Adjusted P-valuea |
|---|---|---|---|---|---|---|---|---|---|
| A | High | Low | 53 | 26 | 61 | 1 | – | 1 | – |
| B | Low | Low | 53 | 23 | NA | 0.849 (0.484-1.488) | 0.567 | 0.785 (0.447-1.739) | 0.399 |
| C | High | High | 53 | 21 | NA | 0.723 (0.407-1.286) | 0.27 | 0.721 (0.396-1.310) | 0.283 |
| D | Low | High | 53 | 12 | NA | 0.367 (0.185-0.728) | 0.004 | 0.315 (0.115-0.641) | 0.001 |
Adjusted for tumor size, cirrhosis, Barcelona Clinic Liver Cancer stage and α-fetoprotein in the GSE14520 cohort using multivariate Cox proportional hazards regression model. CI, confidence interval; CST, cystatin; HR, hazard ratio; MST, median survival time; NA, not available.
GSEA
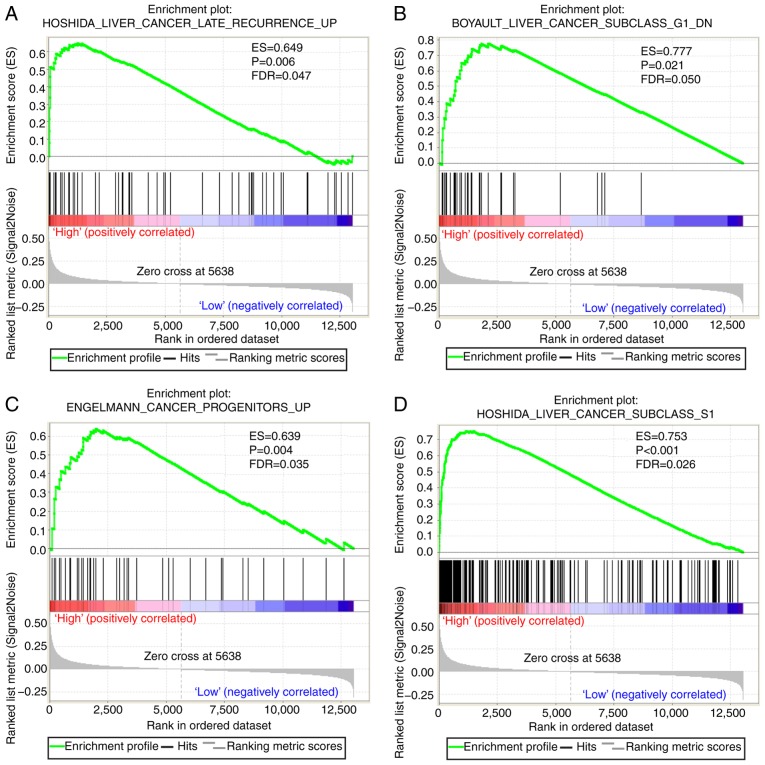
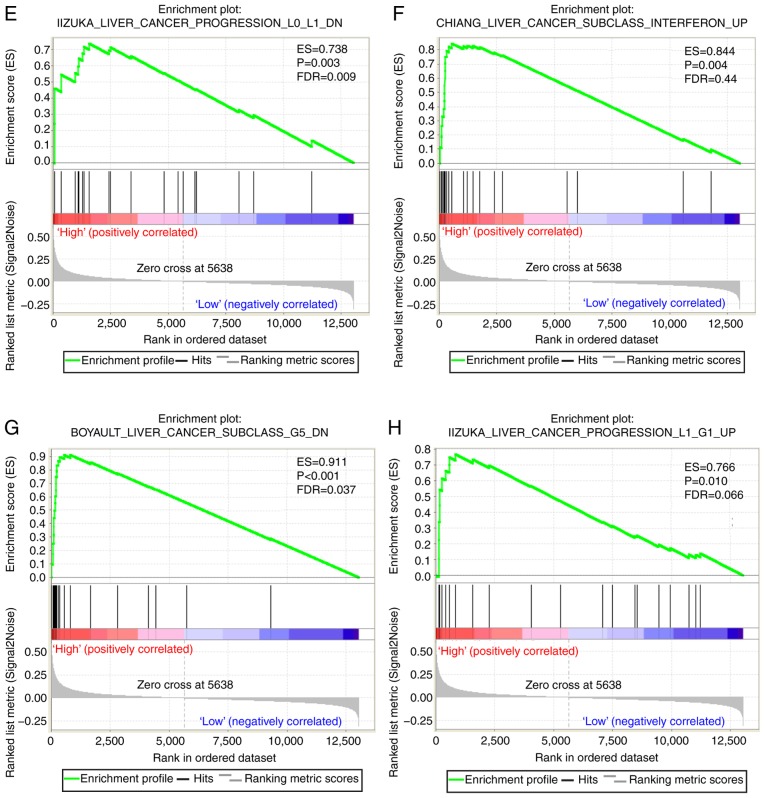
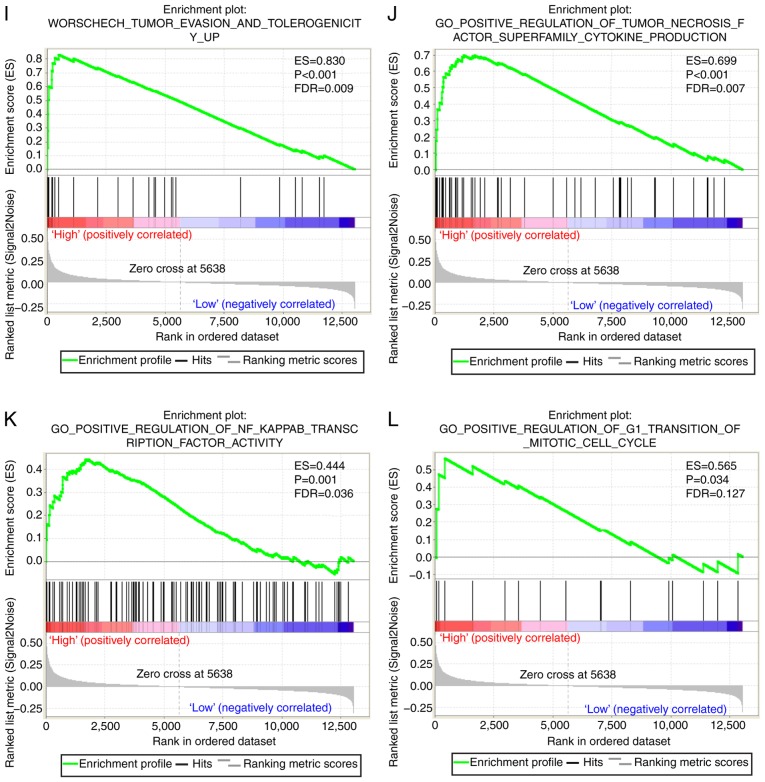
A CST genome-wide RNA sequencing dataset was used for GSEA in order to uncover the potential biological mechanisms of CST6 and CST7 in HCC. The genome expression profile in GSE14520 was categorized based on CST6 and CST7 median gene expression values. GSEA results of the c2 reference gene set are shown in Table SII, in which increased CST7 expression was associated with tumor evasion and tolerogenicity, cancer progenitors, liver cancer late recurrence, liver cancer progression and several liver cancer subclasses (Fig. 7A-I). The enrichment results of c5 are shown in Table SIII; high CST7 expression was revealed to also be involved in positive regulation of the tumor necrosis factor subfamily cytokine production, positive regulation of NF-κB transcription factor and positive regulation of G1-S transition of mitotic cell cycle (Fig. 7J-L). Conversely, the GSEA results of CST6 did not exhibit a significant association between CST6 and biological pathways relevant to HCC.
Figure 7.
GSEA results of CST7 in GSE14520. (A-H) GSEA results of c2-reference gene sets for groups with elevated CST7 expression. GSEA results of CST7 in GSE14520. (I) GSEA results of c2-reference gene sets for groups with elevated CST7 expression; (J-L) GSEA results of c5-reference gene sets for groups with elevated CST7 expression. CST7, cystatin 7; ES, enrichment score; FDR q-val, false discovery rate q-value; FWER p-val, familywise-error rate P-value; GSEA, gene set enrichment analysis; NES, normalized enrichment score; NOM p-val, nominal P-value.
Discussion
It has been reported that cysteine proteases are involved in the progression of several types of tumor (9). Destruction and remodeling of the ECM is an essential process in tumor progression (35), which can be promoted by cysteine proteases (36), particularly cathepsin B, a representative cysteine protease that serves a key role in tumor cell invasion (37–39). However, biological functions of cysteine proteases are inhibited by CSTs (11). Therefore, CSTs may also be associated with tumor progression. Notably, CSTs have been reported to be associated with the progression of various types of cancer, including bladder cancer (40), breast cancer (41,42), esophageal cancer (43), ovarian cancer (44) and prostate cancer (45). The results of GSEA in the present study indicated that CST7 was significantly enriched in liver cancer progression. Co-expression analysis of the CST genes in GSE14520 using Pearson's correlation coefficient revealed that CST1, CST2 and CST4 are closely associated with each other, verifying the results of GeneMANIA and STRING.
CST7 expression was markedly decreased in HCC tissue samples, whereas CSTB was highly expressed in HCC tumor tissue. ROC analysis indicated that CST7 and CSTB exhibit significant diagnostic values and may serve as potential diagnostic biomarkers. The diagnostic value of CST genes has been reported in previous studies. Having higher expression in HCC compared with in adjacent healthy tissues, the diagnostic value of CSTB for HCC was reported in previous research, and the present investigation provided validation for this (14,16,46). In addition to HCC, the diagnostic value of CSTB has been reported in other tumor types. For example, compared with in normal bladder tissue, CSTB immunohistochemical staining is more intense in bladder cancer tissue (40). CSTB has also been demonstrated to be a diagnostic biomarker of ovarian clear cell carcinoma, due to its high expression in tumor cells based on the results of immunohistochemical analysis, reverse transcription-PCR and western blot analysis (47).
The survival analysis of CST genes indicated that CST6 and CST7 may be strongly associated with the OS and RFS of patients with HBV-related HCC. The combined effects survival analysis indicated that the risks of mortality and recurrence in patients with HBV-related HCC were lowest in those with increased expression of CST7 and attenuated expression of CST6. Therefore, CST7 and CST6 may function as prognostic biomarkers for HCC. The prognostic value of CST genes has been reported across several malignancies. It has been reported that overexpression of CST6 promotes pancreatic cancer growth (48). The function of CST6 was revealed to be similar in the present study, where the survival analysis results indicated that high CST6 expression was associated with poor survival of patients with HCC. However, CST6 has also been reported to act as a human tumor suppressor gene in previous reports (45,49–54). Therefore, these findings indicated that CST6 may exert distinct effects in different types of cancer. The similar role of CST6 in the liver and pancreas may be a result of the liver and pancreas stemming from a common progenitor at the embryo stage (55). The fact that the molecular mechanism of CST6 serves different roles in various types of cancer still requires further exploration. Although no significant association was detected between CSTB and the prognosis of patients with HCC, differences have been reported in the expression of CSTB between tumor tissues and adjacent healthy tissues (16,46,56). In addition, the prognostic value of CSTB has been demonstrated in several other tumors. For example, high CSTB expression is associated with a more favorable prognosis in lung cancer (57). A similar scenario of CSTB functioning as a prognostic biomarker has been reported in gastric cancer, where it restrains tumor development by suppressing proliferation and migration of neoplastic cells (58). The present study did not determine a prognostic value of CST3 in HCC; however, CST3 has been demonstrated to act as a tumor suppresser that restrains tumor cell invasion in previous studies (56,59). A recent study reported that the rate of glomerular filtration of creatinine and CST3 may serve as potential predictors of OS in HCC (60). By reviewing these studies, the different roles of CST genes in numerous types of cancer can be identified. The present results corresponded with the results of previous studies. Although in the same subfamily, the expression levels and biological functions of each CST gene are not the same as those of others, even in different types of cancer. CST genes may also function as oncogenes; however, further studies are needed to validate the present findings.
The GSEA conducted in the present study revealed that CST7 was enriched in tumor evasion and tolerogenicity, cancer progenitors, liver cancer late recurrence, liver cancer progression, several liver cancer subclasses, tumor necrosis factor subfamily cytokine production, regulation of NF-κB transcription factor and regulation of G1-S transition of the mitotic cell cycle. These results suggested that CST7 may be closely associated with liver cancer. However, the association between CST genes and HCC requires further validation in future studies. In addition, although GSEA of CST6 indicated that CST6 was not involved in any pathway or molecular mechanism associated with cancer, the effects of CST6 on different types of cancer have been confirmed by previous studies (45,49–54).
One limitation of the present study was that the sample size was insufficient, which could affect the validity of the results. Secondly, the clinical data obtained from the GSE14520 dataset are not complete, barring the opportunity to carry out a more comprehensive survival analysis using multivariate Cox proportional hazards regression model. In order to better evaluate the association between CST subfamily members and HCC prognosis, likely HCC risk factors, including the presence of a tumor capsule and vascular invasion, Child-Pugh score and alcohol intake, should be taken into consideration. Thirdly, the current investigation only explored the relationship between CST gene mRNA expression and HCC prognosis and did not explore the effects of CST protein levels on HCC prognosis. Finally, further studies are warranted to determine the effects of Family 3 and Family 4 CSTs on HCC.
Although there are several limitations to the present study, to the best of our knowledge, this is the first to discover the clinical significance of CST6 and CST7 in the prognosis of patients with HBV-related HCC. In addition, our result verified the findings of previous reports and suggested that CSTB may act as a diagnostic biomarker for HCC. Furthermore, CST7 was discovered to be enriched in several tumor-related signaling pathways and biological processes, including tumor evasion and tolerogenicity, cancer progenitors, liver cancer late recurrence, liver cancer progression, several liver cancer subclasses, tumor necrosis factor subfamily cytokine production, regulation of NF-κB transcription factor and regulation of G1-S transition of the mitotic cell cycle. The prospective molecular mechanisms underlying the effects of CST7 gene expression on patients with HBV-related HCC were determined using GSEA.
In conclusion, the gene expression levels of CST1, CST2, CST5, CSTA and CSTB were significantly increased in HCC tissue, whereas CST3 and CST7 were overexpressed in normal tissue compared with in HCC tissue. Notably, the present study revealed that CST7 and CSTB may serve as diagnostic markers for HCC, and survival analysis of CST genes indicated that CST6 and CST7 expression levels may be closely associated with the OS and RFS of patients with HCC. However, this investigation requires further validation using a sufficient sample size spread across multiple geographical regions.
Supplementary Material
Acknowledgements
Not applicable.
Funding
This study was supported in part by the National Natural Science Foundation of China (grant nos. 81560535, 81802874, 81072321, 30760243, 30460143 and 30560133), the Natural Science Foundation of Guangxi Province of China (grant no. 2017JJB140189y), the 2009 Program for New Century Excellent Talents in University (NCET), Guangxi Natural Sciences Foundation (grant no. GuiKeGong 1104003A-7) and the Guangxi Health Ministry Medicine Grant (Key Scientific Research Grant; grant no. Z201018). The present study was also partly supported by the Scientific Research Fund of the Health and Family Planning Commission of Guangxi Zhuang Autonomous Region (grant no. Z2016318), the Key Laboratory of High Incidence Tumor Prevention & Treatment (Guangxi Medical University), Ministry of Education (grant no. GKE2018-01), the Basic Ability Improvement Project for Middle-aged and Young Teachers in Colleges and Universities in Guangxi (grant no. 2018KY0110), the Innovation Project of Guangxi Graduate Education (grant no. JGY2018037) the 2018 Innovation Project of Guangxi Graduate Education (grant no. YCBZ2018036), and the Research Institute of Innovative Think-tank at Guangxi Medical University (the gene-environment interaction in hepatocarcinogenesis in Guangxi HCCs and its translational applications in the HCC prevention). The authors would also like to acknowledge the support provided by the National Key Clinical Specialty Programs (General Surgery & Oncology) and the Key Laboratory of Early Prevention & Treatment for Regional High-Incidence-Tumor (Guangxi Medical University), Ministry of Education, China. This work was also supported in part by the Natural Science Foundation of Guangxi Province of China (grant no. 2018GXNSFBA138013), the Key Laboratory of High-Incidence-Tumor Prevention & Treatment (Guangxi Medical University) and the Guangxi Key R & D Program (grant no. GKEAB18221019).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XZ and TP conceived and designed the study. XW, KH, XL, CY, TY, JL, CH, GZ, HS, WQ, QH, ZL, JH, YG, XY and TP acquired the data and performed data analyses. XZ wrote the manuscript, and TP guided and supervised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declared that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Njei B, Rotman Y, Ditah I, Lim JK. Emerging trends in hepatocellular carcinoma incidence and mortality. Hepatology. 2015;61:191–199. doi: 10.1002/hep.27388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeng H, Zheng R, Guo Y, Zhang S, Zou X, Wang N, Zhang L, Tang J, Chen J, Wei K, et al. Cancer survival in China, 2003-2005: A population-based study. Int J Cancer. 2015;136:1921–1930. doi: 10.1002/ijc.29227. [DOI] [PubMed] [Google Scholar]
- 5.Altekruse SF, McGlynn KA, Reichman ME. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J Clin Oncol. 2009;27:1485–1491. doi: 10.1200/JCO.2008.20.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 7.Volk ML, Marrero JA. Early detection of liver cancer: Diagnosis and management. Curr Gastroenterol Rep. 2008;10:60–66. doi: 10.1007/s11894-008-0010-2. [DOI] [PubMed] [Google Scholar]
- 8.Sawyers CL. The cancer biomarker problem. Nature. 2008;452:548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 9.Gora J, Latajka R. Involvement of cysteine proteases in cancer. Curr Med Chem. 2015;22:944–957. doi: 10.2174/0929867321666141106115624. [DOI] [PubMed] [Google Scholar]
- 10.Ochieng J, Chaudhuri G. Cystatin superfamily. J Health Care Poor Underserved. 2010;21(Suppl 1):S51–S70. doi: 10.1353/hpu.0.0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turk V, Stoka V, Turk D. Cystatins: Biochemical and structural properties, and medical relevance. Front Biosci. 2008;13:5406–5420. doi: 10.2741/3089. [DOI] [PubMed] [Google Scholar]
- 12.Sinha AA, Gleason DF, Deleon OF, Wilson MJ, Sloane BF. Localization of a biotinylated cathepsin B oligonucleotide probe in human prostate including invasive cells and invasive edges by in situ hybridization. Anat Rec. 1993;235:233–240. doi: 10.1002/ar.1092350207. [DOI] [PubMed] [Google Scholar]
- 13.Visscher DW, Sloane BF, Sameni M, Babiarz JW, Jacobson J, Crissman JD. Clinicopathologic significance of cathepsin B immunostaining in transitional neoplasia. Mod Pathol. 1994;7:76–81. [PubMed] [Google Scholar]
- 14.Lin YY, Chen ZW, Lin ZP, Lin LB, Yang XM, Xu LY, Xie Q. Tissue levels of stefin A and stefin B in hepatocellular carcinoma. Anat Rec (Hoboken) 2016;299:428–438. doi: 10.1002/ar.23311. [DOI] [PubMed] [Google Scholar]
- 15.Zinkin NT, Grall F, Bhaskar K, Otu HH, Spentzos D, Kalmowitz B, Wells M, Guerrero M, Asara JM, Libermann TA, Afdhal NH. Serum proteomics and biomarkers in hepatocellular carcinoma and chronic liver disease. Clin Cancer Res. 2008;14:470–477. doi: 10.1158/1078-0432.CCR-07-0586. [DOI] [PubMed] [Google Scholar]
- 16.Lee MJ, Yu GR, Park SH, Cho BH, Ahn JS, Park HJ, Song EY, Kim DG. Identification of cystatin B as a potential serum marker in hepatocellular carcinoma. Clin Cancer Res. 2008;14:1080–1089. doi: 10.1158/1078-0432.CCR-07-1615. [DOI] [PubMed] [Google Scholar]
- 17.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 18.Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID bioinformatics resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res 35 (Web Server issue) 2007:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38 (Web Server issue) 2010:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montojo J, Zuberi K, Rodriguez H, Bader GD, Morris Q. GeneMANIA: Fast gene network construction and function prediction for Cytoscape. F1000Res. 2014;3:153. doi: 10.12688/f1000research.4572.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Research Network. Electronic address, corp-author. wheeler@bcm.edu; Cancer Genome AtlasResearch Network: Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169:1327–1341.e23. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao X, Liu X, Yang C, Wang X, Yu T, Han C, Huang K, Zhu G, Su H, Qin W, et al. Distinct diagnostic and prognostic values of minichromosome maintenance gene expression in patients with hepatocellular carcinoma. J Cancer. 2018;9:2357–2373. doi: 10.7150/jca.25221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roessler S, Long EL, Budhu A, Chen Y, Zhao X, Ji J, Walker R, Jia HL, Ye QH, Qin LX, et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2012;142:957–966e12. doi: 10.1053/j.gastro.2011.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoo ZH, Candlish J, Teare D. What is an ROC curve? Emerg Med J. 2017;34:357–359. doi: 10.1136/emermed-2017-206735. [DOI] [PubMed] [Google Scholar]
- 27.Ma H, Bandos AI, Rockette HE, Gur D. On use of partial area under the ROC curve for evaluation of diagnostic performance. Stat Med. 2013;32:3449–3458. doi: 10.1002/sim.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balachandran VP, Gonen M, Smith JJ, DeMatteo RP. Nomograms in oncology: More than meets the eye. Lancet Oncol. 2015;16:e173–e180. doi: 10.1016/S1470-2045(14)71116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.François O, Martins H, Caye K, Schoville SD. Controlling false discoveries in genome scans for selection. Mol Ecol. 2016;25:454–469. doi: 10.1111/mec.13513. [DOI] [PubMed] [Google Scholar]
- 32.Glickman ME, Rao SR, Schultz MR. False discovery rate control is a recommended alternative to Bonferroni-type adjustments in health studies. J Clin Epidemiol. 2014;67:850–857. doi: 10.1016/j.jclinepi.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 33.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 34.Llovet JM, Brú C, Bruix J. Prognosis of hepatocellular carcinoma: The BCLC staging classification. Semin Liver Dis. 1999;19:329–338. doi: 10.1055/s-2007-1007122. [DOI] [PubMed] [Google Scholar]
- 35.Liotta LA, Rao CN, Barsky SH. Tumor invasion and the extracellular matrix. Lab Invest. 1983;49:636–649. [PubMed] [Google Scholar]
- 36.Cudic M, Fields GB. Extracellular proteases as targets for drug development. Curr Protein Pept Sci. 2009;10:297–307. doi: 10.2174/138920309788922207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan S, Sameni M, Sloane BF. Cathepsin B and human tumor progression. Biol Chem. 1998;379:113–123. [PubMed] [Google Scholar]
- 38.Mirković B, Markelc B, Butinar M, Mitrović A, Sosič I, Gobec S, Vasiljeva O, Turk B, Čemažar M, Serša G, Kos J. Nitroxoline impairs tumor progression in vitro and in vivo by regulating cathepsin B activity. Oncotarget. 2015;6:19027–19042. doi: 10.18632/oncotarget.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aggarwal N, Sloane BF. Cathepsin B: Multiple roles in cancer. Proteomics Clin Appl. 2014;8:427–437. doi: 10.1002/prca.201300105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feldman AS, Banyard J, Wu CL, McDougal WS, Zetter BR. Cystatin B as a tissue and urinary biomarker of bladder cancer recurrence and disease progression. Clin Cancer Res. 2009;15:1024–1031. doi: 10.1158/1078-0432.CCR-08-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leto G, Incorvaia L, Flandina C, Ancona C, Fulfaro F, Crescimanno M, Sepporta MV, Badalamenti G. Clinical impact of cystatin C/cathepsin L and follistatin/activin A systems in breast cancer progression: A preliminary report. Cancer Invest. 2016;34:415–423. doi: 10.1080/07357907.2016.1222416. [DOI] [PubMed] [Google Scholar]
- 42.Ai L, Kim WJ, Kim TY, Fields CR, Massoll NA, Robertson KD, Brown KD. Epigenetic silencing of the tumor suppressor cystatin M occurs during breast cancer progression. Cancer Res. 2006;66:7899–7909. doi: 10.1158/0008-5472.CAN-06-0576. [DOI] [PubMed] [Google Scholar]
- 43.Shiba D, Terayama M, Yamada K, Hagiwara T, Oyama C, Tamura-Nakano M, Igari T, Yokoi C, Soma D, Nohara K, et al. Clinicopathological significance of cystatin A expression in progression of esophageal squamous cell carcinoma. Medicine (Baltimore) 2018;97:e0357. doi: 10.1097/MD.0000000000010357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X, Gui L, Zhang Y, Zhang J, Shi J, Xu G. Cystatin B is a progression marker of human epithelial ovarian tumors mediated by the TGF-β signaling pathway. Int J Oncol. 2014;44:1099–1106. doi: 10.3892/ijo.2014.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pulukuri SM, Gorantla B, Knost JA, Rao JS. Frequent loss of cystatin E/M expression implicated in the progression of prostate cancer. Oncogene. 2009;28:2829–2838. doi: 10.1038/onc.2009.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Unić A, Derek L, Duvnjak M, Patrlj L, Rakić M, Kujundžić M, Renjić V, Štoković N, Dinjar P, Jukic A, Grgurević I. Diagnostic specificity and sensitivity of PIVKAII, GP3, CSTB, SCCA1 and HGF for the diagnosis of hepatocellular carcinoma in patients with alcoholic liver cirrhosis. Ann Clin Biochem. 2018;55:355–362. doi: 10.1177/0004563217726808. [DOI] [PubMed] [Google Scholar]
- 47.Takaya A, Peng WX, Ishino K, Kudo M, Yamamoto T, Wada R, Takeshita T, Naito Z. Cystatin B as a potential diagnostic biomarker in ovarian clear cell carcinoma. Int J Oncol. 2015;46:1573–1581. doi: 10.3892/ijo.2015.2858. [DOI] [PubMed] [Google Scholar]
- 48.Li Q, Zheng ZC, Ni CJ, Jin WX, Jin YX, Chen Y, Zhang XH, Chen ED, Cai YF. Correlation of cystatin E/M with clinicopathological features and prognosis in triple-negative breast cancer. Ann Clin Lab Sci. 2018;48:40–44. [PubMed] [Google Scholar]
- 49.Soh H, Venkatesan N, Veena MS, Ravichandran S, Zinabadi A, Basak SK, Parvatiyar K, Srivastava M, Liang LJ, Gjertson DW, et al. Cystatin E/M suppresses tumor cell growth through cytoplasmic retention of NF-κB. Mol Cell Biol. 2016;36:1776–1792. doi: 10.1128/MCB.00878-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shridhar R, Zhang J, Song J, Booth BA, Kevil CG, Sotiropoulou G, Sloane BF, Keppler D. Cystatin M suppresses the malignant phenotype of human MDA-MB-435S cells. Oncogene. 2004;23:2206–2215. doi: 10.1038/sj.onc.1207340. [DOI] [PubMed] [Google Scholar]
- 51.Kioulafa M, Balkouranidou I, Sotiropoulou G, Kaklamanis L, Mavroudis D, Georgoulias V, Lianidou ES. Methylation of cystatin M promoter is associated with unfavorable prognosis in operable breast cancer. Int J Cancer. 2009;125:2887–2892. doi: 10.1002/ijc.24686. [DOI] [PubMed] [Google Scholar]
- 52.Chen X, Cao X, Dong W, Xia M, Luo S, Fan Q, Xie J. Cystatin M expression is reduced in gastric carcinoma and is associated with promoter hypermethylation. Biochem Biophys Res Commun. 2010;391:1070–1074. doi: 10.1016/j.bbrc.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 53.Qiu J, Ai L, Ramachandran C, Yao B, Gopalakrishnan S, Fields CR, Delmas AL, Dyer LM, Melnick SJ, Yachnis AT, et al. Invasion suppressor cystatin E/M (CST6): High-level cell type-specific expression in normal brain and epigenetic silencing in gliomas. Lab Invest. 2008;88:910–925. doi: 10.1038/labinvest.2008.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Veena MS, Lee G, Keppler D, Mendonca MS, Redpath JL, Stanbridge EJ, Wilczynski SP, Srivatsan ES. Inactivation of the cystatin E/M tumor suppressor gene in cervical cancer. Genes Chromosomes Cancer. 2008;47:740–754. doi: 10.1002/gcc.20576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ji NY, Kang YH, Park MY, Lee CI, Kim MK, Kim DG, Kim JW, Song EY. Development of a fluorescent microsphere immunoassay for cystatin B (CSTB) in serum of patients with hepatocellular carcinoma. Clin Chem Lab Med. 2011;49:151–155. doi: 10.1515/CCLM.2011.008. [DOI] [PubMed] [Google Scholar]
- 57.Ma Y, Chen Y, Petersen I. Expression and epigenetic regulation of cystatin B in lung cancer and colorectal cancer. Pathol Res Pract. 2017;213:1568–1574. doi: 10.1016/j.prp.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 58.Zhang J, Shi Z, Huang J, Zou X. CSTB downregulation promotes cell proliferation and migration and suppresses apoptosis in gastric cancer SGC-7901 cell line. Oncol Res. 2016;24:487–494. doi: 10.3727/096504016X14685034103752. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.Coulibaly S, Schwihla H, Abrahamson M, Albini A, Cerni C, Clark JL, Ng KM, Katunuma N, Schlappack O, Glössl J, Mach L. Modulation of invasive properties of murine squamous carcinoma cells by heterologous expression of cathepsin B and cystatin C. Int J Cancer. 1999;83:526–531. doi: 10.1002/(SICI)1097-0215(19991112)83:4<526::AID-IJC15>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 60.Tamai Y, Iwasa M, Kawasaki Y, Yoshizawa N, Ogura S, Sugimoto R, Eguchi A, Yamamoto N, Sugimoto K, Hasegawa H, Takei Y. Ratio between estimated glomerular filtration rates of creatinine and cystatin C predicts overall survival in patients with hepatocellular carcinoma. Hepatol Res. 2019;49:153–163. doi: 10.1111/hepr.13230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.