This work examines how protein arginine methylation of Rpc31, a subunit of RNA Pol III, promotes negative regulation of tRNA biogenesis in the context of cellular stress.
Abstract
Protein arginine methylation is an important means by which protein function can be regulated. In the budding yeast, this modification is catalyzed by the major protein arginine methyltransferase Hmt1. Here, we provide evidence that the Hmt1-mediated methylation of Rpc31, a subunit of RNA polymerase III, plays context-dependent roles in tRNA gene transcription: under conditions optimal for growth, it positively regulates tRNA gene transcription, and in the setting of stress, it promotes robust transcriptional repression. In the context of stress, methylation of Rpc31 allows for its optimal interaction with RNA polymerase III global repressor Maf1. Interestingly, mammalian Hmt1 homologue is able to methylate one of Rpc31’s human homologue, RPC32β, but not its paralogue, RPC32α. Our data led us to propose an efficient model whereby protein arginine methylation facilitates metabolic economy and coordinates protein-synthetic capacity.
Introduction
In eukaryotes, RNA polymerase III (Pol III) transcribes small, untranslated RNAs such as 5S rRNAs and tRNAs, highly abundant molecules that comprise ∼15% of total cellular RNA by weight and are requisite elements for protein synthesis (reviewed in Geiduschek and Tocchini-Valentini (1988), Turowski and Tollervey (2016), Lesniewska and Boguta (2017)). The Pol III transcription apparatus is highly conserved among eukaryotes. In the budding yeast Saccharomyces cerevisiae, the holoenzyme is composed of 17 subunits, 12 of which are either shared with, or have homologs among the other RNA polymerases. Of the remaining five subunits, Rpc82, Rpc34, and Rpc31 form a Pol III-specific trimeric subcomplex that contributes to transcription initiation (Werner et al, 1992; Thuillier et al, 1995). Pol III is recruited to tRNA genes by two general transcription factors, TFIIIB and TFIIIC. The latter is a complex that recognizes sequence-specific promoter elements and guides the concerted binding of three TFIIIB subunits (TBP, Brf1, and Bdp1) to the transcription start site. The resulting TFIIIB/C complex recruits Pol III and contributes to the formation of an open promoter complex (reviewed in Graczyk et al (2018)).
Pol III-mediated transcription is robust under optimal conditions. However, under nonfavorable growth conditions or after exposure to other forms of stress, Pol III transcription is repressed by the negative regulator Maf1 (Upadhya et al, 2002). In the latter context, Maf1 is dephosphorylated and then translocates from the cytoplasm to the nucleus, where it binds to and regulates the activity of Pol III (reviewed in Boguta (2013), Willis and Moir (2018)). Cryo-EM–based analysis of the structure of a Pol III–Maf1 complex revealed that Maf1 binding leads to rearrangement of the Rpc82/34/31 subcomplex (Vannini et al, 2010) and that this inhibits the interaction between Rpc34 and TFIIIB subunit Brf1 and thus prevents recruitment of Pol III to promoters.
The Pol III machinery has been shown to undergo several types of post-translational modifications and these have been implicated in its regulation (reviewed in Chymkowitch and Enserink (2018)). Recently, a comprehensive proteomics analysis revealed that human RPC4 (yeast Rpc53) and RPC7 (yeast Rpc31) can undergo protein arginine methylation (Geoghegan et al, 2015). This modification is catalyzed by members of the protein arginine methyltransferase (PRMT) family of enzymes, which are divided into four subtypes based on the type of methylarginine formed (reviewed in Bedford and Richard (2005), Morales et al (2016), Blanc and Richard (2017)). PRMT1 is the most conserved of the type I PRMTs and it catalyzes the formation of monomethylarginine and asymmetric dimethylarginine. In yeast, Hmt1 (also termed Rmt1) is the homolog of mammalian PRMT1, and it is the only known type I PRMT in the budding yeast (Gary et al, 1996; Henry & Silver, 1996).
Our previous study revealed that Hmt1 associates with most tRNA genes in vivo and the tRNA abundance in Hmt1 loss-of-function mutants are lower than in WT (HMT1) counterparts (Milliman et al, 2012). However, the molecular mechanism by which Hmt1 influences tRNA abundance remained unclear. In this study, we demonstrate that the degree of association of Hmt1 with tRNA genes correlates with the transcriptional activities of the latter. We also show that Hmt1 methylates Rpc31 in vitro, that this ability is conserved in one of the two human homologs of Rpc31, and that under optimal growth conditions, Rpc31 methylation promotes biogenesis of precursor tRNAs (pre-tRNAs), but in the setting of stress, it represses the biogenesis of these pre-tRNAs. Our finding further suggests that the observed difference in outcomes is due to Rpc31 methylation affecting with the interaction between Pol III and its repressor, Maf1.
Results
Hmt1 occupancy at tRNA genes correlates with RNA Pol III transcriptional activity
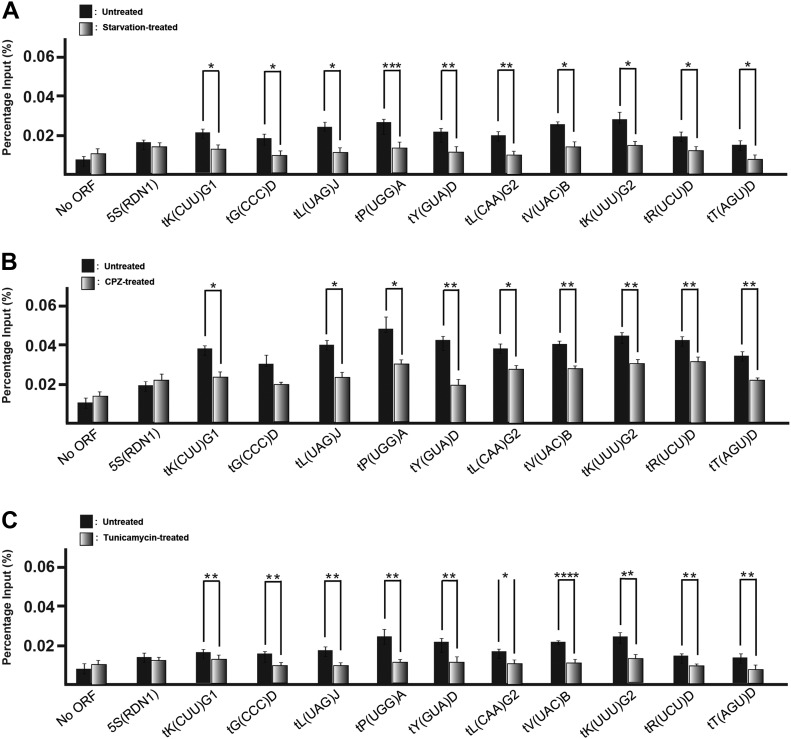
In vivo, Hmt1 is associated with many tRNA genes, and in both hmt1Δ and Hmt1-G68R (a catalytically inactive mutant of Hmt1) cells grown in rich medium, tRNA abundance is higher than that in their HMT1 counterparts (Milliman et al, 2012). These observations led us to investigate the relationship between the transcription of tRNA genes and the association of Hmt1 with these loci. The transcriptional activity of tRNA genes is high in rich medium and low in the contexts of nutrient deprivation (Clarke et al, 1996), treatment with the antifungal compound chlorpromazine (CPZ) (Upadhya et al, 2002), and treatment with tunicamycin (Li et al, 2000). We subjected exponentially growing cultures of yeast cells expressing Myc-tagged Hmt1 to each of these conditions and used chromatin immunoprecipitation (ChIP) to measure the extent to which Hmt1 associated with tRNA genes before and after treatment (Fig 1). This analysis revealed reductions in the association of Hmt1 with these tRNA genes upon nutrient deprivation (Fig 1A), treatment with CPZ (Fig 1B), or treatment with tunicamycin (Fig 1C). In particular, the reduction in association after nutrient deprivation is similar to the reduction in occupancy seen for the RNA Pol III machinery under the same conditions (Roberts et al, 2003). We note that Hmt1 occupancy in untreated samples across these treatments had varied level of occupancy and this is likely attributed to technical differences in the immunoprecipitation efficiency for each immunoprecipitation, as the background signals seen with the negative control genes were also slightly higher as well. Nevertheless, the trend we observed for each of the three different treatments reflects a decreased association of Hmt1 with tRNA genes that correlates with the levels of transcription of the latter.
Figure 1. Hmt1 occupancy at tRNA genes is decreased under stress conditions.
(A–C) Hmt1 occupancy across tRNA genes was measured in yeast cells before and after nutrient deprivation (A), treatment with CPZ (B), or treatment with tunicamycin (C). qPCR results for products of ChIP are displayed in bar graphs. Percentage of input is calculated as ΔCT, with error bars representing the SEM of three biological samples (n = 3). P-value as calculated by t test: *<0.05; **<0.01; and ***<0.001.
Hmt1 methylates the Pol III subunit Rpc31 in vitro
The RNA hybridization data for Hmt1-G68R hinted that a substrate of this methyltransferase may be crucial for Pol III transcription (Milliman et al, 2012). In hmt1Δ cells, the loss of methylation of such a substrate is likely responsible for the observed changes in tRNA levels. Examination of the amino acid sequences of all Pol III subunits for the presence of RGG tripeptides or RG repeats, which are common methylation motifs in substrates of Hmt1, identified Rpc31 as a candidate (Fig 2A). Notably, this protein had previously been suggested as a putative Hmt1 substrate based on another in silico analysis (Frankel & Clarke, 1999).
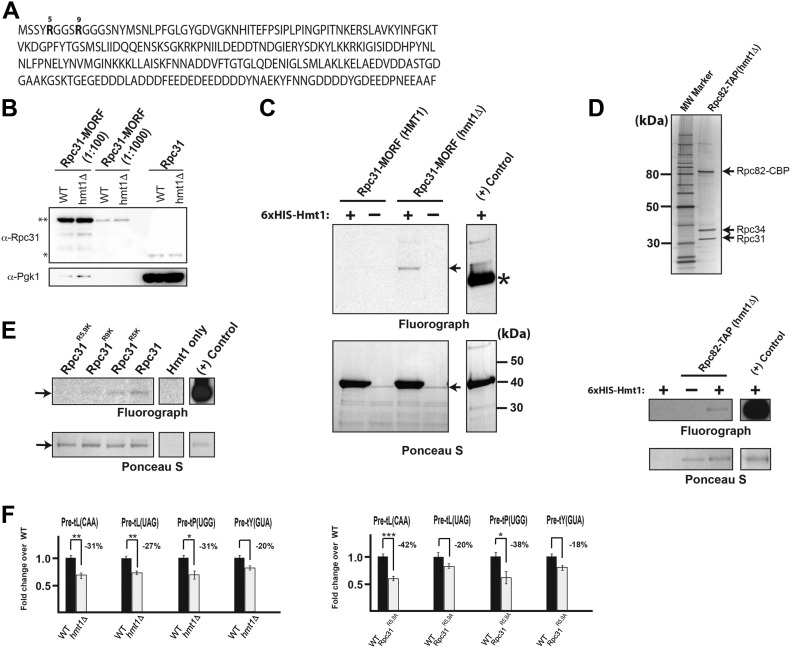
Figure 2. Under optimal growth conditions, Hmt1 methylates Rpc31 and loss of this modification adversely affects biogenesis of pre-tRNAs.
(A) The amino acid sequence of yeast Rpc31 with methylated arginines at positions 5 and 9 denoted in bold lettering. (B) Immunoblotting showing the relative levels of Rpc31-MORF to endogenous Rpc31 was analyzed using lysates made from WT and hmt1Δ cells. Double asterisks denote MORF-tagged Rpc31 and a single asterisk denotes endogenous Rpc31. The level of Pgk1 is used as a loading control for the relative total protein levels loaded. (C) In vitro methylation of Rpc31 from the yeast MORF collection, after purification from WT and hmt1Δ cells, using recombinant Hmt1 and [methyl-3H]-SAM. The full protein complement in each reaction was resolved on a 4–12% SDS–PAGE; methylation was visualized by fluorography (arrow) and protein levels by Ponceau S staining. Recombinant GST-tagged Rps2 served as a positive control (highlighted by asterisk). (D) Biochemical purification of TAP-tagged Rpc82 from hmt1Δ cells. TAP was performed from hmt1Δ cells expressing TAP-tagged Rpc82 (top panel). The purified proteins were resolved on a 4–12% SDS–PAGE and the gel was silver stained to determine the protein composition (bottom panel). The TAP-purified Rpc82 and associated proteins were subjected to an in vitro methylation assay using recombinant Hmt1 and [methyl-3H]-SAM. The full protein complement in each reaction was resolved on a 4–12% gel by SDS–PAGE. Methylation of Rpc31 was visualized by fluorography and protein levels by Ponceau S staining. Recombinant GST-tagged Rps2 served as a control. (E) In vitro methylation of GST-tagged WT Rpc31, Rpc31R5K, Rpc31R9K, and Rpc31R5,9K, after purification from Escherichia coli, using recombinant Hmt1 and [methyl-3H]-SAM. Visualization of methylation and protein as in Part (B). Recombinant GST-tagged Rps2 served as a positive control. (F) Fold change in levels of pre-tRNAs in hmt1Δ or Rpc31R5,9A versus WT cells under optimal growth condition in either SC + glucose (for WT versus hmt1Δ cells) or YPD (for WT versus Rpc31R5,9A cells), as assessed by hybridization of probes to intronic regions. Bars show abundance of pre-tRNAs tL(CAA), tL(UAG), tP(UGG), and tY(GUA) in hmt1Δ (left panel) or Rpc31R5,9A (right panel) relative to those in WT cells. In each case, the signal was normalized based on the levels of U4 snRNA. Error bars represent the SEM of three biological replicates (n = 3). P-value as calculated by t test: *<0.05 and **<0.01.
To test whether Rpc31 can be methylated by Hmt1, we used the yeast moveable ORF (MORF) expression plasmid collection (Gelperin et al, 2005) to overexpress and purify C-terminal epitope-tagged Rpc31 in the presence of endogenous Rpc31 expression. The rationale for choosing this approach is because C-terminal tagging of endogenous Rpc31 has proven to be detrimental for cell growth and N-terminal tagging of Rpc31 is likely to interfere with the predicted methylation motif located close to the N-terminal end of the protein. The level of MORF-tagged Rpc31 is much more in abundance than endogenous Rpc31, based on our immunoblot analysis (Fig 2B). Using C-terminal epitope-tagged Rpc31 purified from HMT1 and hmt1Δ cells, we set up an in vitro methylation assay, and our results revealed a signal corresponding to methylation of Rpc31 in only the Rpc31 purified from hmt1Δ cells (Fig 2C, compare position of asterisk on the fluorograph in the HMT1 and hmt1Δ lanes). This is consistent with Rpc31 purified from hmt1Δ cells being in the hypomethylated form and thus serving as a good substrate for in vitro methylation but Rpc31 purified from HMT1 cells being at least partially (and potentially fully) methylated and thus a poorer substrate for methylation in vitro.
To demonstrate the physiological relevance of Rpc31 methylation, we carried out a tandem-affinity purification (TAP) of TAP-tagged Rpc82 that was expressed in hmt1Δ cells. Our purification results revealed Rpc34 and Rpc31 that were co-purified with TAP-tagged Rpc82 in corresponding stoichiometry (Fig 2D, top). When we subjected the entire TAP-purified fractions to an in vitro methylation assay, a signal that corresponds to the co-purified Rpc31 was detected (Fig 2D, bottom). Thus, endogenous Rpc31 can be methylated when present as a complex along with Rpc34 and Rpc82, which resembles a more physiological relevant condition that exists in a cell. Taken together, our data collectively demonstrated that Rpc31 is an in vitro substrate of Hmt1 and is likely to be methylated by Hmt1 under physiological conditions.
Hmt1 methylates the Rpc31 arginines at positions 5 and 9
To determine how arginine methylation influences Rpc31 function, it was necessary to identify the residues that are modified by Hmt1. Of the six arginines present in Rpc31, only those in positions 5 and 9 are located within the N-terminal RGG motif (Fig 2A). To test the methylation potential of these two arginines, we mutated them to lysines individually and in combination (Rpc31R5K, Rpc31R9K, or Rpc31R5,9K) and subjected recombinant Rpc31 harboring these substitutions to in vitro methylation (Fig 2E). Our data revealed a reduction in the methylation signal for both Rpc31R5K and Rpc31R9K (Fig 2E, compare the signal from Rpc31 to Rpc31R5K or Rpc31R9K lanes). Moreover, in the Rpc31R5,9K double mutant, this signal was completely abolished (Fig 2E, compare the signal from Rpc31 lane to Rpc31R5,9K lane). Although each substitution had an impact on the total methylation of Rpc31, it was clear that both arginines are methylated.
In both hmt1Δ and Rpc31R5,9A mutants, the biogenesis of pre-tRNAs is impaired when cells are maintained under optimal growth conditions
Our previous conclusions regarding RNA hybridization data for hmt1Δ cells maintained under optimal growth conditions, that is, that tRNA levels increased in this context, were based on normalization to the Pol I transcript 5.8S (Milliman et al, 2012). We have since changed to using levels of the U4 snRNA transcript for normalization, for the following reasons. First, Pol I and III are extensively co-regulated (Li et al, 2000; Briand et al, 2001), making a Pol II transcript a potentially better means of assessing the impact of Hmt1 on the transcriptional activities of Pol III. Second, the U4 snRNA, in particular, has been used in normalizing the results of RNA hybridization studies investigating transcription by Pol III (Sethy-Coraci et al, 1998; Li et al, 2000). Third, the loss of Hmt1 or its catalytic activity does not alter RNA Pol II–mediated gene transcription (Yu et al, 2004).
We repeated our RNA hybridization assay, focusing on tRNAs that were bound by Hmt1 and also contain introns. The presence of an intron on such tRNA allows us to follow the fate of the precursors by using a probe that binds to tRNA introns. This allows us to detect the earliest intermediate resulting from the transcription of these tRNA genes. When we normalized our data using U4, we found the levels of pre-tRNAs to be lower in the hmt1Δ versus WT cells grown in both YPD and synthetic complete (SC) media supplemented with glucose (SC + glu). However, the differences seen in cells grown in YPD were smaller or not as consistent when compared with SC + glu grown cells. When hmt1Δ cells were grown in SC + glu, we consistently see a decrease in the levels of pre-tRNAs to be lower in the hmt1Δ cells across all four pre-tRNAs tested when compared with the WT cells (Fig 2F, left panel). This is in contrast to our previous conclusions, based on normalization of the data to the levels of the 5.8S transcript.
To determine whether methylation of Rpc31 is responsible for this change, we repeated the same experiment using a Rpc31R5,9A mutant grown in YPD. Results from our RNA hybridization data demonstrated a consistent decrease in the levels of pre-tRNAs across the same four pre-tRNAs tested (Fig 2F, right panel). Together, these findings suggest that Hmt1 is required to promote transcription of the tested tRNA genes under optimal growth conditions and that it does so by methylating Rpc31.
Investigating repression of tRNA biogenesis under stress in the hmt1Δ and Rpc31R5,9A mutant strains
In the context of stress, Pol III transcription is robustly repressed; this ensures cell survival by promoting metabolic economy (reviewed in Warner (1999)). We examined the extent to which Hmt1 contributes to this process by investigating the ability of the hmt1Δ and Rpc31R5,9A mutants to repress tRNA biogenesis in the context of stress. For the four tRNA species examined in Fig 2F, we carried out RNA hybridizations using total RNA extracted from cells grown in YPD before and after treatment by CPZ. For comparison, we calculated the percentage of pre-tRNA abundance for each strain and treatment and compared these values with the one calculated from untreated WT cells. To ensure that our RNA hybridization results accurately reflects the levels of pre-tRNA examined, we used two internal normalizing controls, U3 and U5, in addition to U4. Both U3 and U5 have been used previously as internal controls for normalization of tRNA abundance (Moir et al, 2006; Arimbasseri et al, 2016). In the four pre-tRNAs examined, we saw that the loss of Hmt1 compromised the cell’s ability to robustly repress pre-tRNA biogenesis in the context of stress (Figs 3A and S1A). This trend is also true for the Rpc31R5,9A mutants where diminished repression of pre-tRNAs was observed in the mutants versus those seen in WT cells (Figs 3B and S1B). Although there are slight variations depending on the use of U3, U4, or U5 as normalizing controls, the overall trend in which tRNA repression is less robust in either hmt1Δ or Rpc31R5,9A mutants remains consistent.
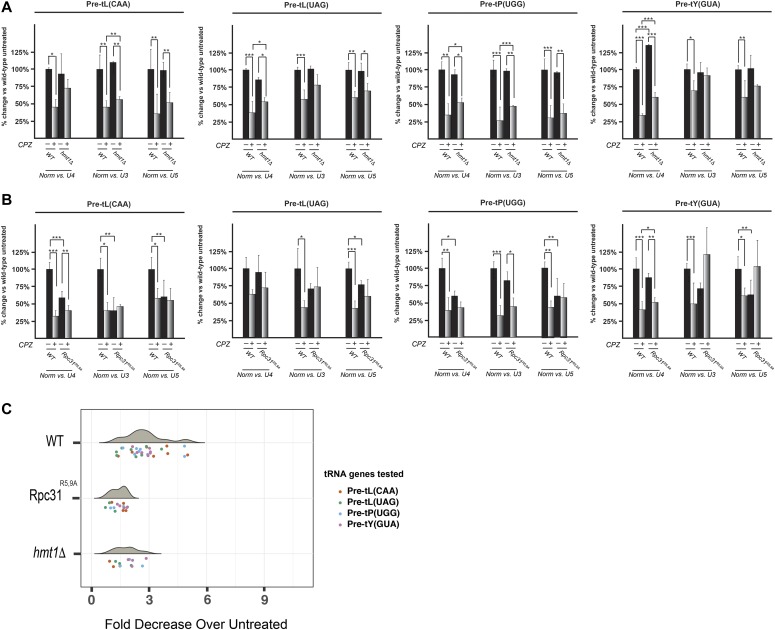
Figure 3. In the context of stress, methylation of Rpc31 is required for robust repression of pre-tRNA biogenesis.
(A, B) RNA hybridization was carried out for four pre-tRNAs in WT versus hmt1Δ cells (A) or WT versus Rpc31R5,9A cells (B) grown in YPD before and after treatment with CPZ. Ratios of signal intensities for each pre-tRNA were individually normalized against three internal controls: U4, U3, and U5. The normalized signals were plotted on a bar graph to compare against signal obtained from the untreated WT cells, which is set to 100%. Error bars represent the SEM of three biological replicates (n = 3). P-value as calculated by t test: *<0.05, **<0.01, and ***<0.001. (C) Fold decrease in expression of four candidate pre-tRNAs in WT, hmt1Δ, or Rpc31R5,9A cells after treatment with CPZ, as assessed by hybridization of probes to intronic regions. Signal was normalized to levels of the U4 snRNA. Analysis of variance (ANOVA) revealed significant variation among the strains(P-value = 1.4 × 10−6). Post hoc Tukey’s Honest significant differences method revealed a significant difference between WT and hmt1Δ (after adjustment for the multiple comparisons, the adjusted P-value is 0.0014), WT and Rpc31R5,9A (adjusted P-value is 2.8 × 10−6). n = at least three per pre-tRNA.
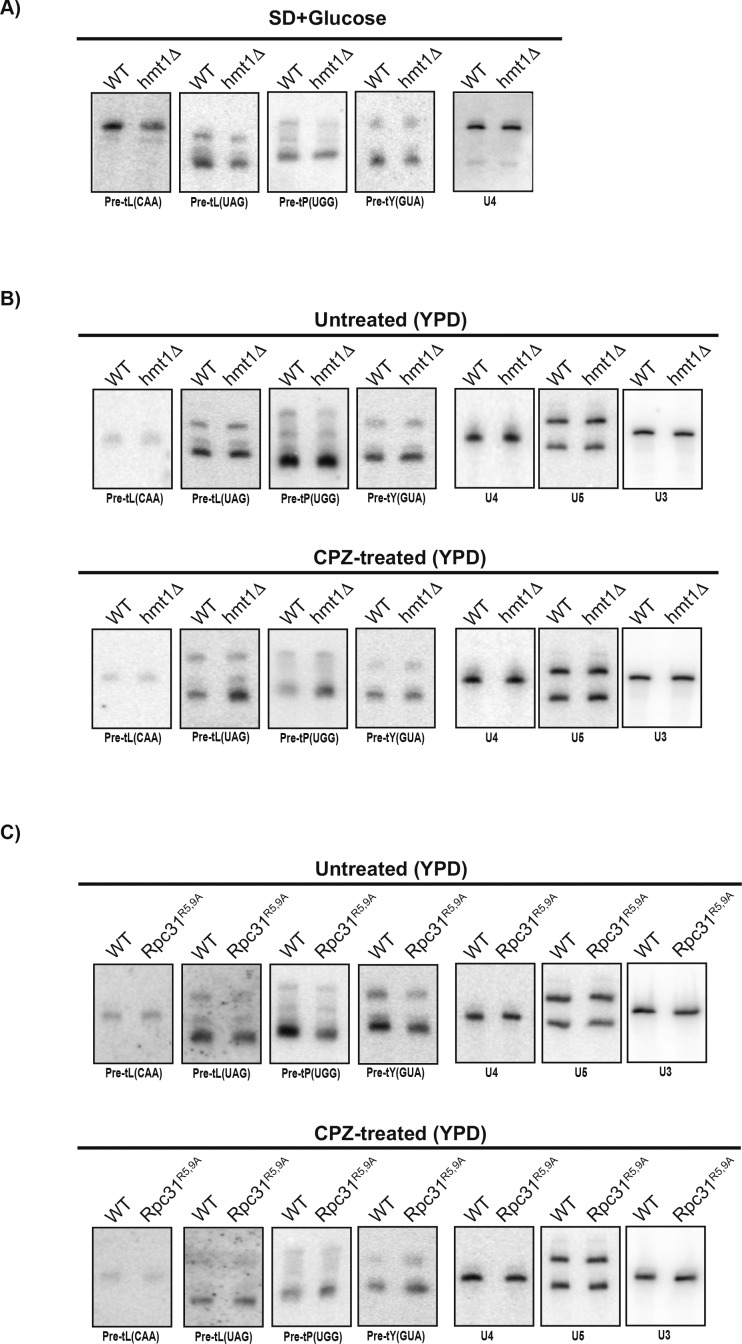
Figure S1. RNA hybridization data from WT, hmt1Δ, and Rpc31R5,9A cells before and after CPZ treatment.
Total RNAs were extracted from WT, hmt1Δ, and Rpc31R5,9A cells before and after CPZ treatment. The media used to grow the cell in each panel are stated. 10 μg of RNA from each biological sample is loaded and resolve by an 8% urea-TBE gel. Resolved RNAs were transferred onto nylon membrane and subjected to RNA hybridization using radiolabeled oligonucleotide probes corresponding to pre-tRNAs of tL(CAA), tL(UAG), tP(UGG), and tY(GUA). (A, B) As a control for loading, the blots were also probed for U3, U4, and U5. Sample hybridization data from WT versus hmt1Δ (A) or versus Rpc31R5,9A (B) before and after CPZ treatment.
To better test the differences among the three strains for significance, we carried out Tukey’s HSD on U4-normalized RNA hybridization data. The results of this analysis is plotted as a violin plot (Fig 3C) where the median point within the cluster of tRNA genes tested in both hmt1Δ and Rpc31R5,9A mutants is lower than the one in the WT strain, indicating that both mutants have a lower fold decrease of pre-tRNA abundance in their treated versus untreated samples. In other words, this plot imply an overall diminished repression in hmt1Δ and Rpc31R5,9A mutants (Fig 3C). Hence, Hmt1-mediated methylation of Rpc31 contributes to the robust repression of pre-tRNA biogenesis in the context of stress, playing a role distinct from that under optimal growth conditions.
Next, we set out to determine whether the attenuated repression of tRNA biogenesis observed in hmt1Δ and Rpc31R5,9A mutants results from a change in Pol III occupancy at the relevant promoters. Using ChIP, we measured the in vivo occupancy of the same four tRNA genes by the Rpc82 and Rpc160 subunits of Pol III before and after treatment with CPZ. In both hmt1Δ and Rpc31R5,9A point mutants under stress, Rpc82 and Rpc160 occupancy at these tRNA genes are higher when compared with their levels seen in WT cells (Fig 4A and B, compare the percentage of change in WT lanes with those in hmt1Δ and Rpc31R5,9A lanes). In addition, we carried out Tukey’s HSD on ChIP data from both Rpc82 and Rpc160 to examine for significance with respect to their occupancy among the three strains. The resulting violin plot (Fig 4C) indicates the median point within the cluster of Rpc82 and Rpc160 occupancy across the tested tRNA genes in both hmt1Δ and Rpc31R5,9A strains is lower than the WT, in the context of fold decrease between treated and untreated samples. What this result implies is that, under CPZ treatment, a higher Rpc82 and Rpc160 occupancy in hmt1Δ and Rpc31R5,9A mutants is observed when compared with the WT (Fig 4C). It is likely that in the context of stress, a higher occupancy of Pol III in hmt1Δ and Rpc31R5,9A mutants is responsible for the higher levels of pre-tRNAs observed in these mutants.
Figure 4. Increased RNA Pol III occupancy at tRNA genes is observed in hmt1Δ and Rpc31R5,9A cells under stress.
(A, B) The in vivo occupancy across the four tRNA genes for Rpc82 (A) and Rpc160 (B) was determined by ChIP. qPCR results for products of ChIP performed on WT, hmt1Δ, orRpc31R5,9A cells before and after treatment of CPZ are displayed as bar graphs. Percentage of input is calculated by ΔCT. The error bars representing SEM of three biological samples (n = 3). P-value as calculated by t test: *<0.05; **<0.01, and ***<0.001. (C) Fold decrease in Rpc82 and Rpc160 occupancy for four candidate tRNA genes in WT, hmt1Δ, or Rpc31R5,9A cells after treatment with CPZ. qPCR was performed for products of ChIP on WT, hmt1Δ, or Rpc31R5,9A cells before and after treatment with CPZ. Percentage of input is calculated as ΔCT. ANOVA on these values yielded significant variation among the three strains (P-value = 0.0015). Post hoc Tukey test revealed significant differences between WT and hmt1Δ cells (adjusted P-value is 5.4 × 10−4), and between WT and Rpc31R5,9A cells (adjusted P-value is 9.8 × 10−4). n = 3 per tRNA gene.
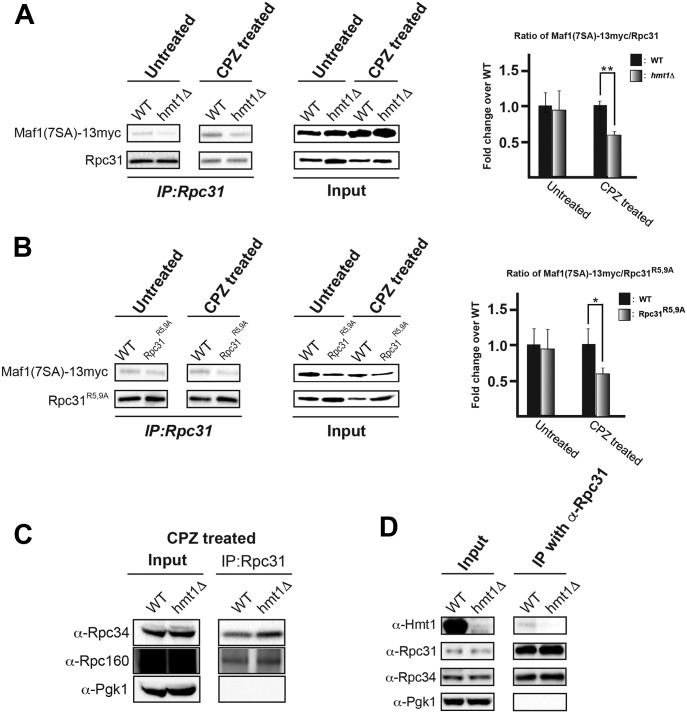
Arginine methylation of Rpc31 is important for its association with Maf1 in the context of stress
Arginine methylation plays a key role in mediating proper protein–protein interactions (Bedford et al, 2000; Côté & Richard, 2005; Cheng et al, 2007), and structural–functional studies have suggested that the recruitment of Pol III to target genes is inhibited by Maf1-mediated rearrangements of the Rpc82/34/31 subcomplex (Vannini et al, 2010). Thus, we hypothesized that Rpc31 methylation plays a key role in the biochemical association of Pol III with Maf1 during stress. We tested this hypothesis by using α-Rpc31 antibodies to immunoprecipitate proteins from yeast lysates prepared from cells expressing a Myc-tagged Maf1-7SA (Fig 5A and B). Maf1-7SA is a functional mutant that cannot be phosphorylated by PKA/Sch9 (Huber et al, 2009). It is constitutively localized in the nucleus (Lee et al, 2009) and, as such, provides an enhanced readout for our co-immunoprecipitation (CoIP) studies. After treatment with CPZ, the association of Maf1-7SA with Rpc31 in hmt1Δ cells was ∼40% lower than that in their WT counterparts (Fig 5A). The same effect was observed with the Rpc31R5,9A mutant (Fig 5B). Our CoIP likely pulled down the entire Pol III complex, as we were able to detect both Rpc34 and Rpc160 in our co-immunoprecipitates (Fig 5C). To determine whether Hmt1 physically associates with the RNA Pol III, we carried out a CoIP experiment using α-Rpc31 antibodies. We were able to capture a weak but clear association between Hmt1 and RNA Pol III complex (Fig 5D). The level of captured Hmt1 association with RNA Pol III may be reflective of Hmt1’s role as an enzyme, where its interaction with a substrate may be too transient to be completely captured. Overall, our data led us to infer that arginine methylation of Rpc31 promotes the interaction between Pol III and Maf1 and that this association is key to achieving robust repression of tRNA gene transcription in the context of stress.
Figure 5. During stress, methylation of Rpc31 is required for the association of Pol III with its negative regulator Maf1.
(A) Rpc31 levels in yeast lysates generated from WT or hmt1Δ cells before and after treatment with CPZ, assessed by CoIP with an α-Rpc31 antibody. The levels of Myc-tagged Maf1-7SA were probed using an α-Myc antibody and the results are displayed in a bar graph. Error bars represent the SEM of three biological replicates (n = 3). P-value as calculated by t test: *<0.05 and **<0.01. (B) Rpc31 and Myc-tagged Maf1-7SA levels in yeast lysates generated from WT or Rpc31R5,9A cells before and after treatment of CPZ, assessed using CoIP as in (A) (including the number of replicates). P-value as calculated by t test: *<0.05 and **<0.01. (C) Rpc160 and Rpc34 levels in complexes immunoprecipitated with α-Rpc31 antibody. The samples were probed with α-Rpc160, α-Rpc34, and α-Pgk1 (as a negative control). (D) Hmt1 physically interacts with Rpc31-containing complex. CoIP of Rpc31 was carried out using cell lysates generated from WT or hmt1Δ cells and resolved on a 4–12% SDS–PAGE followed by immunoblotting using α-Hmt1, α-Rpc31, and α-Rpc34 antibodies to determine the levels of Hmt1, Rpc31, and Rpc34 present in the co-immunoprecipitates. The level of Pgk1 was used as a negative control in the immunoprecipitation experiment.
Source data are available for this figure.
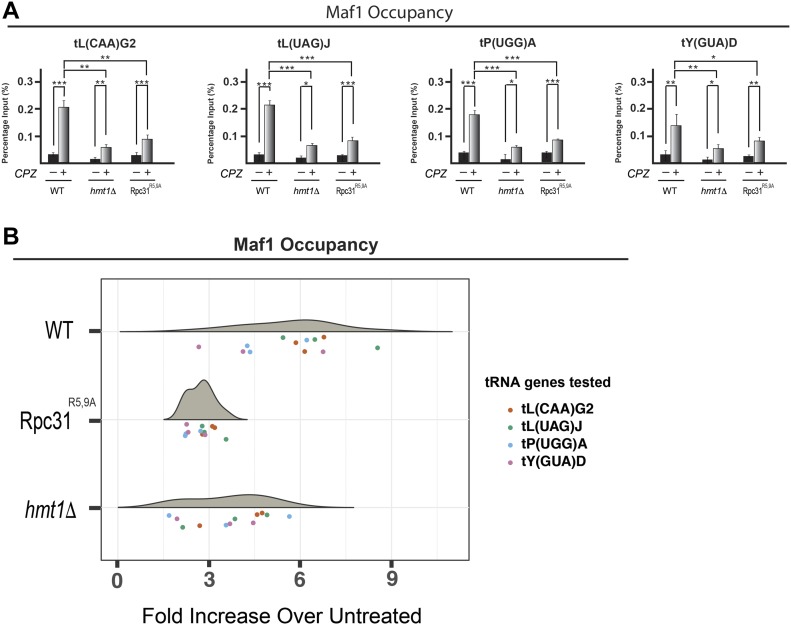
A genome-wide study had previously revealed that Maf1 associates with all Pol III loci in a regulated manner, and that this association is enhanced under conditions of Pol III repression (Roberts et al, 2006). To determine whether this association is changed in hmt1Δ and Rpc31R5,9A mutants, we performed ChIP on the four tRNA genes tested above using a Myc-tagged Maf1 (Fig 6A). In both hmt1Δ and Rpc31R5,9A mutants, we observed a curtailed increase in Maf1 occupancy across the tRNA genes after the CPZ treatment when compared with the WT cells (Fig 6A, compare the percentage input in hmt1Δ and Rpc31R5,9A lanes with WT lanes). To determine the significance of Maf1 occupancy change within these three strains, we carried out Tukey’s HSD on Maf1’s ChIP data in a similar manner as above. The resulting violin plot (Fig 6B) supports the notion that Maf1 occupancy across the tested tRNA genes displays a decreased median point in both hmt1Δ and Rpc31R5,9A mutants, in the context of fold increase between treated versus untreated samples. The results from this analysis implied a lower Maf1 occupancy in hmt1Δ and Rpc31R5,9A mutants in the context of stress when compared with the WT cells (Fig 6B). Taken together with the ChIP data obtained for Rpc82 and Rpc160, our observation of a reduced Maf1 occupancy in the hmt1Δ and Rpc31R5,9A mutants lend support to the notion that Maf1 is not able to fully repress the transcription of tRNA genes in these cells upon exposure to stress. This is likely due to the inability of Maf1 to fully engage in its association with a nonmethylated Rpc31 based on findings from our biochemical data above.
Figure 6. Decreased Maf1 occupancy at tRNA genes is observed in hmt1Δ and Rpc31R5,9A cells under stress.
(A) Maf1 occupancy across the four tRNA genes was determined by ChIP. qPCR results for products of ChIP performed on WT, hmt1Δ, or Rpc31R5,9A cells before and after treatment of CPZ are displayed as bar graphs. Percentage of input is calculated by ΔCT. The error bars representing SEM of three biological samples (n = 3). P-value as calculated by t test: *<0.05; **<0.01, and ***<0.001. (B) Fold increase in Maf1 occupancy of four candidate tRNA genes in WT, hmt1Δ, or Rpc31R5,9A cells after treatment with CPZ. qPCR was performed on products of ChIP in WT, hmt1Δ, or Rpc31R5,9A cells before and after treatment with CPZ. Percentage of input was calculated as ΔCT. ANOVA on these values revealed significant variation among the three strains (P-value = 4.6 × 10−6). Post hoc Tukey test revealed significant difference between WT and hmt1Δ (adjusted P-value is 8.6 × 10−4), and between WT and Rpc31R5,9A (adjusted P-value is 3.7 × 10−6). n = 3 per tRNA gene.
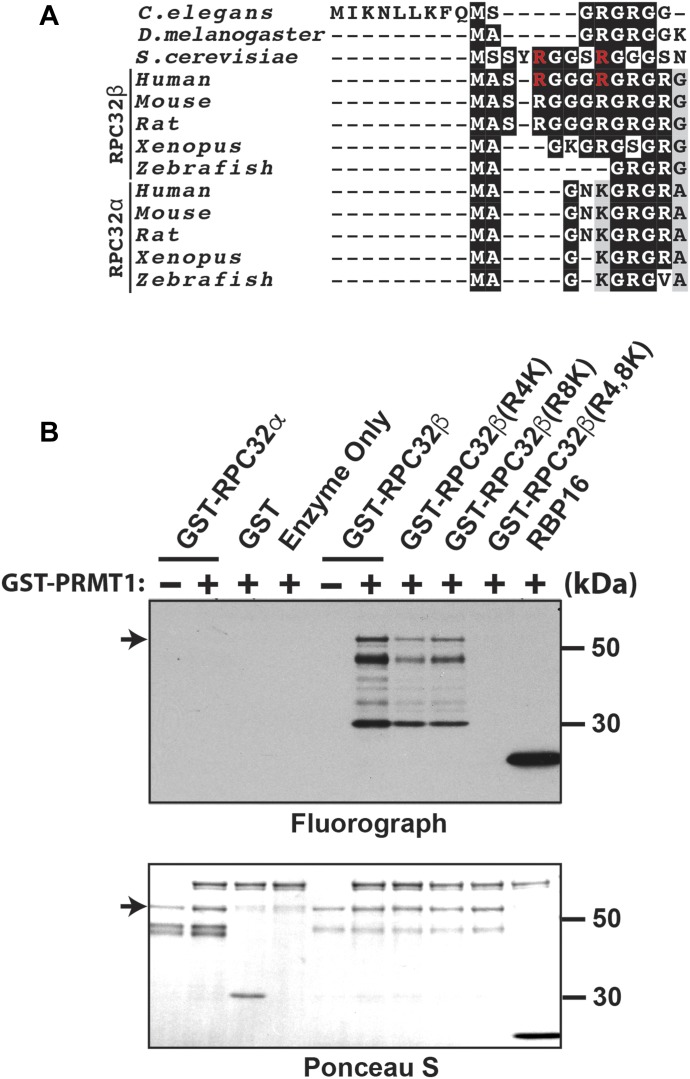
The human ortholog of yeast Rpc31, RPC32β, is methylated by PRMT1 in vitro
The yeast ortholog of Rpc31/34/82 in humans is RPC32/39/62 (Wang & Roeder, 1997; Hu et al, 2002). Of all the subunits of vertebrate RNA Pol III, RPC32 is the only one for which two paralogs exist: RPC32α and RPC32β (Haurie et al, 2010). An alignment of the amino acid sequence of the Rpc31 N terminus in orthologs from various eukaryotic species indicates that its methylarginines are conserved in RPC32β, but not RPC32α (Fig 7A). To determine whether arginine methylation of Rpc31 is conserved, we performed an in vitro methylation assay, using rat PRMT1 to methylate both RPC32α and RPC32β (Fig 7B). A signal corresponding to methylation was observed for RPC32β, but not RPC32α, in the presence of rat PRMT1 (Fig 7B). To determine whether either or both of the conserved arginines on RPC32β are methylated, we generated arginine-to-lysine substitutions in RPC32β to mimic the residues in Rpc31. We observed a significant reduction in the methylation signal from both the RPC32βR4K and RPC32βR8K mutants (Fig 7B). As in the case of the Rpc31R5,9K double mutant, the methylation signal was completely abolished in the RPC32βR4,8K double mutant (Fig 7B). Overall, our data demonstrated that the arginine methylation of Rpc31 is conserved in RPC32β, but not in RPC32α.
Figure 7. Mammalian PRMT1 methylates human Rpc31 homolog RPC32β, but not RPC32α.
(A) Sequence alignment of N-terminal amino acid sequences of yeast Rpc31 with its homologs in Caenorhabditis elegans, Drosophila melanogaster, and various mammalian species. Arginines on yeast Rpc31 that are colored red represent the identified methylated arginine and are conserved in RPC32β. (B) WT RPC32α and RPC32β were purified from E. coli and then subjected to in vitro methylation by rat PRMT1 and [methyl-3H]-SAM. In parallel, methylarginine substitutionmutants of RPC32β (R4K, R8K and R4, 8K) were also tested. The arrow on the fluorograph denotes methylated RPC32β. RBP16 served as a positive control for the in vitro methylation. The protein loading levels for each sample areshownby Ponceau S staining of the same membrane before fluorography, with the arrow denoting the substrate tested.
Discussion
The roles of arginine methylation of Rpc31 in controlling Pol III transcription is dynamic
Determining how arginine methylation is regulated remains the greatest challenge in the field because it requires the existence of enzymes capable of demethylating methylarginines (reviewed in Yang and Bedford (2012), Wesche et al (2017)). There is no evidence that budding yeast has an enzyme that directly removes the methyl moieties from arginines, casting doubt on the reversibility of this modification in yeast. However, one can argue that if methylation of a specific residue on a substrate can allow for distinct outcomes in a biological process (e.g., negative and positive regulation of Pol III transcription) in which the role of methylation is dependent on a specific environmental trigger (e.g., flux in the nutrient or stress), a lack-of-turnover mechanism may actually be better in achieving a higher efficiency and specificity in tuning such biological process. In addition, arginine methylation has a substantial metabolic cost (12 ATPs are required), and thus a rapid reversal of this modification is not energetically favorable (Gary & Clarke, 1998). Given this rationale, it is tantalizing to speculate that an alternative mechanism bypassing the need for a demethylase might be involved, with the same modification able to achieve distinct outcomes in the same process due to different biological signals sensed. Our data show that in cells responding to stress, the methyl marks on Rpc31 lead to robust repression of Pol III, and that they do so by facilitating proper binding between Pol III and Maf1, but that under optimal growth conditions, they ensure high-level transcription by Pol III. Although the underlying molecular mechanism is not yet clear, these marks could potentially influence how Pol III interacts with transcription factors at the pre-initiation complex. Given that Maf1 is present in the cytoplasm in cells undergoing optimal growth, it would not interfere with the interaction between methylated Rpc31 and transcription factors in this context. Thus, it is possible that Rpc31 methylation plays both positive and negative roles in Pol III transcription by participating in distinct biochemical interactions. Given the strong conservation of arginine-methylated substrates in yeast and higher eukaryotes, it is possible that a similar scenario explains the lack of a bona fide demethylase in the latter. Further research is needed to unravel the functional roles of individual methyl marks in these substrates and to better understand the prevalence of such scenarios in yeast and higher eukaryotes.
Implication of Hmt1 recruitment on the conformational changes of RNA Pol III at tRNA genes
We have previously identified a physical association between Hmt1 and Bdp1 (Milliman et al, 2012), a subunit of TFIIIB. Our present work further demonstrates a physical association between Hmt1 and RNA Pol III, and that Hmt1 methylates Rpc31, a subunit of RNA Pol III. Together, these observations suggest that Bdp1 is instrumental in recruiting Hmt1 to the tRNA genes to allow for methylation of Rpc31 by Hmt1. Together with Rpc82 and Rpc34, this heterotrimeric subcomplex is key for promoter opening (Brun et al, 1997). Structural studies of RNA Pol III transcription initiation shows that upon TFIIIB binding, structural rearrangements leads to a shift of the Rpc82/34/31 subcomplex towards the cleft and this subcomplex undergoes further structural rearrangement that result in the stabilization of the C-terminal segment of Rpc34 and Rpc31 (Abascal-Palacios et al, 2018). Rpc31 is predicted to be disordered in the elongating Pol III structures (Hoffmann et al, 2015). Upon the formation of RNA Pol III open complex, the “stalk bridge” of Rpc31 is able to fold into a α-helix that directs this bridge to contact the RNA Pol III stalk (Abascal-Palacios et al, 2018). This contact promotes the “stalk bridge” of Rpc31 to be locked at a defined angle within the Rpc82/34/31 subcomplex (Abascal-Palacios et al, 2018). It is possible that methylation of Rpc31 plays a key role in this process by modulating the degree of disorder within Rpc31. Recent work has shown that arginine methylation has emerged as a key post-translational modification can modulate low complexity domains within the disordered regions in a protein, thereby fine-tune the degree of interactions with other molecular partners (reviewed in Chong et al (2018), Hofweber and Dormann (2019)). Thus, methylation of Rpc31 may provide a similar role in this instance to change the degree by which Rpc31’s “stalk bridge” contacts RNA Pol III stalk and this, in turn, may impact the ability of RNA Pol III to carry out transcription at its maximum capacity.
Biological significance stemming from a change in tRNA biogenesis
tRNAs are among the most abundant molecules in a cell. They are heavily modified by a large network of proteins that collectively process the pre-tRNA into a stable, mature tRNA (reviewed in Phizicky and Hopper (2010)). In addition, some tRNA modifications can be altered by changing Pol III activity, and this can lead to non-uniform changes in tRNA function that might have an impact on the translation profiles of specific mRNAs (Arimbasseri et al, 2016). Whereas a lack of Rpc31 methylation does not seem to be detrimental under conditions optimal for growth, it is possible that it alters the tRNA modifications and the translational profile that is required for an appropriate response to stress. On the other hand, an increase in abundance of a tRNA due to a defect in arginine methylation of Rpc31 could potentially lead to the futile cycling of tRNAs, thereby leading to wasted expenditure of energy as was previously observed in mice lacking the repressor MAF1 (Bonhoure et al, 2015).
A working model for how RPC32β methylation controls cellular homeostasis
Under normal growth conditions, RPC32β is ubiquitously expressed but not RPC32α (Haurie et al, 2010). In undifferentiated and transformed cells, however, RPC32α expression is the highest (Haurie et al, 2010). The data from our yeast work show that Rpc31 methylation promotes its repression by Maf1 during stress. Therefore, the existence of two mammalian Rpc31 isoforms in which one can be methylated (RPC32β) but not the other (RPC32α) suggests a potential interplay between these isoforms in orchestrating a desired functional outcome in cellular homeostasis. One possible scenario for this interplay is that the level of methylated versus unmethylated RPC32 shifts the levels of Pol III isoforms available for MAF1-mediated repression. In other words, cells that express a high level of RPC32α, which lacks the methylation motif required for robust repression by MAF1, will dilute the overall level of RPC32β-incorporated Pol III isoforms available. As a consequence, there will be an increase in the level of RPC32α-incorporated Pol III isoforms in these cells. Isoforms of Pol III that incorporate either RPC32α or RPC32β target the same genes (Renaud et al, 2014), but having RPC32α-incorporated Pol III isoforms will make this isoform less susceptible to regulation by MAF1. In tumor cells, having more RPC32α-incorporated Pol III isoform will render these isoforms refractory to MAF1-mediated repression, thereby meeting the high demands of RNA Pol III transcription in tumor cells. Indeed, MAF1 expression and activity inversely correlates with the oncogenic activity (Shor et al, 2010; Palian et al, 2014). Given the key roles of RPC32α on cell differentiation and transformation, future experiments dissecting at how methylation contributes to RPC32 function in these processes may offer novel avenues of therapeutics targeting tumorigenesis.
Materials and Methods
Yeast strains used in this study
All yeast strains used are listed in Table S1. All plasmids used are listed in Table S2. All primers used are listed in Table S3. Cells were grown at 30°C on YPD medium except for the RNA hybridization assay of hmt1Δ cells depicted in Fig 2F, in which these cells were grown at 30°C on SC + glucose. Genomic deletions were generated, and epitope tag cassettes were integrated, as previously described (Yu et al, 2004).
Table S1 Yeast strains used in this study. (29.5KB, xlsx)
Table S2 Plasmids used in this study. (29.5KB, xlsx)
Table S3 Primers and their sequences used in this study. (29.5KB, xlsx)
Tandem-affinity purification of Rpc82
Tandem-affinity purification of TAP-tagged Rpc82 from hmt1Δ cells was carried out exactly as described previously (Jackson et al, 2012). The purified protein was dialyzed in 1× PBS/15% glycerol overnight, and the dialyzed fraction was then concentrated in a 10-kD MWCO Amicon Ultra 0.5 ml protein concentrator (Millipore). The concentrated samples were stored at −80°C before use.
Rpc31-MORF overexpression
For MORF plasmid-containing yeast strains, the cells were first cultured SC minus uracil, and then the expression of MORF-tagged Rpc31 was induced for 6 h by the addition of 2× YEP with 2% sucrose and 2% galactose. To determine the relative levels of MORF-Rpc31 to endogenous Rpc31, diluted lysates made from induced yeast cultures were resolved by the SDS–PAGE, transferred to a nitrocellulose membrane, and then blotted with antibodies against Rpc31.
In vitro methylation assay
Yeast Rpc31 was expressed using the yeast MORF collection (Open BioSystems) and purified as previously described (Gelperin et al, 2005). Methylarginine mutants of Rpc31 were expressed as GST-tagged fusion proteins and purified as previously described (Muddukrishna et al, 2017). The GST tag was cleaved off with thrombin and in vitro methylation assays were performed as previously described (Jackson & Yu, 2014).
RNA hybridization assay
Yeast strains were grown to log phase (OD600 ≈ 1.8) and one-half of the culture was treated with CPZ at a final concentration of 500 μM for 1 h. Both treated and untreated samples were harvested, and total RNA was extracted and RNA hybridization was carried out as previously described (Milliman et al, 2012).
ChIP
Yeast strains were grown to log phase (OD600 ≈ 1.8), and one-half of the culture was treated with CPZ at a final concentration of 500 μM for 1 h. ChIP was performed on both treated and untreated cells as previously described (Muddukrishna et al, 2017), with the exception of the sonication conditions (Branson Digital Sonifier 450, 3 mm tapered microtip, 20% amplitude, 20 s pulse/55 s pause/15× cycles). For each IP, anti-Myc (MS127P; Thermo Fisher Scientific) or anti-IgG (209-005-082; Jackson ImmunoResearch) antibodies were precoupled to protein A-Sepharose beads. qPCR was performed as previously described (Muddukrishna et al, 2017).
RT–qPCR
RT–qPCR is carried out as previously described (Muddukrishna et al, 2017). Statistical testing was performed using ANOVA and post hoc Tukey’s test, using the R statistical analysis software. All values reported are the mean of three biological replicates (n = 3).
Yeast CoIP
Yeast strains were grown to log phase (OD600 ≈ 1.8) and one-half of the culture was treated with CPZ for 1 h. The α-Rpc31 antibody was cross-linked to magnetic beads (Dynabeads Cat. No. 14203) using the manufacturer’s protocol. Lysates were generated as previously described (Muddukrishna et al, 2017). The lysates were brought to a total protein concentration of 10 mg/ml and incubated with α-Rpc31–cross-linked beads for 2 h at 4°C, followed by three washes with PBS, 0.5% Triton X-100, and 2.5 mM MgCl2. Bound protein was eluted in SDS gel loading buffer and resolved on a 4–12% Bis-Tris gradient gel (Life Technologies). The protein was transferred to nitrocellulose membrane, which was probed with relevant antibodies, developed using the Clarity Western ECL kit (Bio-Rad), imaged using the Bio-Rad ChemiDoc, and quantified using the Bio-Rad ImageQuant.
Supplementary Material
Acknowledgements
We thank Ian Willis for the Maf1-7SA and Maf1-Myc plasmids, and other reagents; Hung-Ta Chen for Rpc34 and Rpc160 antibodies; and Seok Kooi Khoi for helpful discussions. This work was supported by a National Science Foundation award (MCB-1051350) to MC Yu.
Author Contributions
RB Davis: conceptualization, data curation, formal analysis, validation, investigation, methodology, and writing—original draft, review, and editing.
N Likhite: data curation, formal analysis, investigation, and methodology.
CA Jackson: data curation, formal analysis, validation, investigation, methodology, and writing—review and editing.
T Liu: formal analysis and methodology.
MC Yu: conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, methodology, project administration, and writing—original draft, review, and editing.
Conflict of Interest Statement
The authors declare that they have no conflict of interest.
References
- Abascal-Palacios G, Ramsay EP, Beuron F, Morris E, Vannini A (2018) Structural basis of RNA polymerase III transcription initiation. Nature 553: 301 10.1038/nature25441 [DOI] [PubMed] [Google Scholar]
- Arimbasseri AG, Blewett NH, Iben JR, Lamichhane TN, Cherkasova V, Hafner M, Maraia RJ (2016) RNA polymerase III output is functionally linked to tRNA dimethyl-G26 modification. PLoS Genet 11: e1005671 10.1371/journal.pgen.1005671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Frankel A, Yaffe MB, Clarke S, Leder P, Richard S (2000) Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J Biol Chem 275: 16030–16036. 10.1074/jbc.m909368199 [DOI] [PubMed] [Google Scholar]
- Bedford MT, Richard S (2005) Arginine methylation an emerging regulatorof protein function. Mol Cell 18: 263–272. 10.1016/j.molcel.2005.04.003 [DOI] [PubMed] [Google Scholar]
- Blanc RS, Richard S (2017) Arginine methylation: The coming of age. Mol Cell 65: 8–24. 10.1016/j.molcel.2016.11.003 [DOI] [PubMed] [Google Scholar]
- Boguta M. (2013) Maf1, a general negative regulator of RNA polymerase III in yeast. Biochim Biophys Acta 1829: 376–384. 10.1016/j.bbagrm.2012.11.004 [DOI] [PubMed] [Google Scholar]
- Bonhoure N, Byrnes A, Moir RD, Hodroj W, Preitner F, Praz V, Marcelin G, Chua SC Jr, Martinez-Lopez N, Singh R, et al. (2015) Loss of the RNA polymerase III repressor MAF1 confers obesity resistance. Genes Dev 29: 934–947. 10.1101/gad.258350.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briand JF, Navarro F, Gadal O, Thuriaux P (2001) Cross talk between tRNA and rRNA synthesis in Saccharomyces cerevisiae. Mol Cell Biol 21: 189–195. 10.1128/mcb.21.1.189-195.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun I, Sentenac A, Werner M (1997) Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J 16: 5730–5741. 10.1093/emboj/16.18.5730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D, Côté J, Shaaban S, Bedford MT (2007) The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol Cell 25: 71–83. 10.1016/j.molcel.2006.11.019 [DOI] [PubMed] [Google Scholar]
- Chong PA, Vernon RM, Forman-Kay JD (2018) RGG/RG motif regions in RNA binding and phase separation. J Mol Biol 430: 4650–4665. 10.1016/j.jmb.2018.06.014 [DOI] [PubMed] [Google Scholar]
- Chymkowitch P, Enserink JM (2018) Regulation of tRNA synthesis by posttranslational modifications of RNA polymerase III subunits. Biochim Biophys Acta 1861: 310–319. 10.1016/j.bbagrm.2017.11.001 [DOI] [PubMed] [Google Scholar]
- Clarke EM, Peterson CL, Brainard AV, Riggs DL (1996) Regulation of the RNA polymerase I and III transcription systems in response to growth conditions. J Biol Chem 271: 22189–22195. 10.1074/jbc.271.36.22189 [DOI] [PubMed] [Google Scholar]
- Côté J, Richard S (2005) Tudor domains bind symmetrical dimethylated arginines. J Biol Chem 280: 28476–28483. 10.1074/jbc.m414328200 [DOI] [PubMed] [Google Scholar]
- Frankel A, Clarke S (1999) RNase treatment of yeast and mammalian cell extracts affects in vitro substrate methylation by type I protein arginine N-methyltransferases. Biochem biophysical Res Commun 259: 391–400. 10.1006/bbrc.1999.0779 [DOI] [PubMed] [Google Scholar]
- Gary JD, Clarke S (1998) RNA and protein interactions modulated by protein arginine methylation. Prog Nucleic Acid Res Mol Biol 61: 65–131. 10.1016/s0079-6603(08)60825-9 [DOI] [PubMed] [Google Scholar]
- Gary JD, Lin WJ, Yang MC, Herschman HR, Clarke S (1996) The predominant protein-arginine methyltransferase from Saccharomyces cerevisiae. J Biol Chem 271: 12585–12594. 10.1074/jbc.271.21.12585 [DOI] [PubMed] [Google Scholar]
- Geiduschek EP, Tocchini-Valentini GP (1988) Transcription by RNA polymerase III. Annu Rev Biochem 57: 873–914. 10.1146/annurev.biochem.57.1.873 [DOI] [PubMed] [Google Scholar]
- Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ, Lopez-Hoyo N, Jiang L, Piccirillo S, Yu H, et al. (2005) Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev 19: 2816–2826. 10.1101/gad.1362105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan V, Guo A, Trudgian D, Thomas B, Acuto O (2015) Comprehensive identification of arginine methylation in primary T cells reveals regulatory roles in cell signalling. Nat Commun 6: 6758 10.1038/ncomms7758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graczyk D, Ciesla M, Boguta M (2018) Regulation of tRNA synthesis by the general transcription factors of RNA polymerase III - TFIIIB and TFIIIC, and by the MAF1 protein. Biochim Biophys Acta Gene Regul Mech 1861: 320–329. 10.1016/j.bbagrm.2018.01.011 [DOI] [PubMed] [Google Scholar]
- Haurie V, Durrieu-Gaillard S, Dumay-Odelot H, Da Silva D, Rey C, Prochazkova M, Roeder RG, Besser D, Teichmann M (2010) Two isoforms of human RNA polymerase III with specific functions in cell growth and transformation. Proc Natl Acad Sci USA 107: 4176–4181. 10.1073/pnas.0914980107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry MF, Silver PA (1996) A novel methyltransferase (Hmt1p) modifies poly(A)+-RNA-binding proteins. Mol Cell Biol 16: 3668–3678. 10.1128/mcb.16.7.3668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann NA, Jakobi AJ, Moreno-Morcillo M, Glatt S, Kosinski J, Hagen WJ, Sachse C, Muller CW (2015) Molecular structures of unbound and transcribing RNA polymerase III. Nature 528: 231–236. 10.1038/nature16143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofweber M, Dormann D (2019) Friend or foe-Post-translational modifications as regulators of phase separation and RNP granule dynamics. J Biol Chem 294: 7137–7150. 10.1074/jbc.tm118.001189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Wu S, Sun Y, Yuan CC, Kobayashi R, Myers MP, Hernandez N (2002) Characterization of human RNA polymerase III identifies orthologues for Saccharomyces cerevisiae RNA polymerase III subunits. Mol Cell Biol 22: 8044–8055. 10.1128/mcb.22.22.8044-8055.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A, Bodenmiller B, Uotila A, Stahl M, Wanka S, Gerrits B, Aebersold R, Loewith R (2009) Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev 23: 1929–1943. 10.1101/gad.532109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CA, Yadav N, Min S, Li J, Milliman EJ, Qu J, Chen YC, Yu MC (2012) Proteomic analysis of interactors for yeast protein arginine methyltransferase Hmt1 reveals novel substrate and insights into additional biological roles. Proteomics 12: 3304–3314. 10.1002/pmic.201200132 [DOI] [PubMed] [Google Scholar]
- Jackson CA, Yu MC (2014) Detection of protein arginine methylation in Saccharomyces cerevisiae. Methods Mol Biol 1163: 229–247. 10.1007/978-1-4939-0799-1_18 [DOI] [PubMed] [Google Scholar]
- Lee J, Moir RD, Willis IM (2009) Regulation of RNA polymerase III transcription involves SCH9-dependent and SCH9-independent branches of the target of rapamycin (TOR) pathway. J Biol Chem 284: 12604–12608. 10.1074/jbc.c900020200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesniewska E, Boguta M (2017) Novel layers of RNA polymerase III control affecting tRNA gene transcription in eukaryotes. Open Biol 7: 170001 10.1098/rsob.170001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Moir RD, Sethy-Coraci IK, Warner JR, Willis IM (2000) Repression of ribosome and tRNA synthesis in secretion-defective cells is signaled by a novel branch of the cell integrity pathway. Mol Cell Biol 20: 3843–3851. 10.1128/mcb.20.11.3843-3851.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milliman EJ, Hu Z, Yu MC (2012) Genomic insights of protein arginine methyltransferase Hmt1 binding reveals novel regulatory functions. BMC Genomics 13: 728 10.1186/1471-2164-13-728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir RD, Lee J, Haeusler RA, Desai N, Engelke DR, Willis IM (2006) Protein kinase A regulates RNA polymerase III transcription through the nuclear localization of Maf1. Proc Natl Acad Sci USA 103: 15044–15049. 10.1073/pnas.0607129103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales Y, Cáceres T, May K, Hevel JM (2016) Biochemistry and regulation of the protein arginine methyltransferases (PRMTs). Arch Biochem Biophys 590: 138–152. 10.1016/j.abb.2015.11.030 [DOI] [PubMed] [Google Scholar]
- Muddukrishna B, Jackson CA, Yu MC (2017) Protein arginine methylation of Npl3 promotes splicing of the SUS1 intron harboring non-consensus 5’ splice site and branch site. Biochim Biophys Acta 1860: 730–739. 10.1016/j.bbagrm.2017.04.001 [DOI] [PubMed] [Google Scholar]
- Palian BM, Rohira AD, Johnson SAS, He L, Zheng N, Dubeau L, Stiles BL, Johnson DL (2014) Maf1 is a novel target of PTEN and PI3K signaling that negatively regulates oncogenesis and lipid metabolism. PLoS Genet 10: e1004789 10.1371/journal.pgen.1004789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky EM, Hopper AK (2010) tRNA biology charges to the front. Genes Dev 24: 1832–1860. 10.1101/gad.1956510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud M, Praz V, Vieu E, Florens L, Washburn MP, l’Hôte P, Hernandez N (2014) Gene duplication and neofunctionalization: POLR3G and POLR3GL. Genome Res 24: 37–51. 10.1101/gr.161570.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DN, Stewart AJ, Huff JT, Cairns BR (2003) The RNA polymerase III transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc Natl Acad Sci USA 100: 14695–14700. 10.1073/pnas.2435566100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DN, Wilson B, Huff JT, Stewart AJ, Cairns BR (2006) Dephosphorylation and genome-wide association of Maf1 with Pol III-transcribed genes during repression. Mol Cell 22: 633–644. 10.1016/j.molcel.2006.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethy-Coraci I, Moir RD, Lopez-de-Leon A, Willis IM (1998) A differential response of wild type and mutant promoters to TFIIIB70 overexpression in vivo and in vitro. Nucleic Acids Res 26: 2344–2352. 10.1093/nar/26.10.2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shor B, Wu J, Shakey Q, Toral-Barza L, Shi C, Follettie M, Yu K (2010) Requirement of the mTOR kinase for the regulation of Maf1 phosphorylation and control of RNA polymerase III-dependent transcription in cancer cells. J Biol Chem 285: 15380–15392. 10.1074/jbc.m109.071639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuillier V, Stettler S, Sentenac A, Thuriaux P, Werner M (1995) A mutation in the C31 subunit of Saccharomyces cerevisiae RNA polymerase III affects transcription initiation. EMBO J 14: 351–359. 10.1002/j.1460-2075.1995.tb07009.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turowski TW, Tollervey D (2016) Transcription by RNA polymerase III: Insights into mechanism and regulation. Biochem Soc Trans 44: 1367–1375. 10.1042/bst20160062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhya R, Lee J, Willis IM (2002) Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol Cell 10: 1489–1494. 10.1016/s1097-2765(02)00787-6 [DOI] [PubMed] [Google Scholar]
- Vannini A, Ringel R, Kusser AG, Berninghausen O, Kassavetis GA, Cramer P (2010) Molecular basis of RNA polymerase III transcription repression by Maf1. Cell 143: 59–70. 10.1016/j.cell.2010.09.002 [DOI] [PubMed] [Google Scholar]
- Wang Z, Roeder RG (1997) Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev 11: 1315–1326. 10.1101/gad.11.10.1315 [DOI] [PubMed] [Google Scholar]
- Warner JR. (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24: 437–440. 10.1016/s0968-0004(99)01460-7 [DOI] [PubMed] [Google Scholar]
- Werner M, Hermann-Le Denmat S, Treich I, Sentenac A, Thuriaux P (1992) Effect of mutations in a zinc-binding domain of yeast RNA polymerase C (III) on enzyme function and subunit association. Mol Cell Biol 12: 1087–1095. 10.1128/mcb.12.3.1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesche J, Kuhn S, Kessler BM, Salton M, Wolf A (2017) Protein arginine methylation: A prominent modification and its demethylation. Cell Mol Life Sci 74: 3305–3315. 10.1007/s00018-017-2515-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis IM, Moir RD (2018) Signaling to and from the RNA polymerase III transcription and processing machinery. Annu Rev Biochem 87: 75–100. 10.1146/annurev-biochem-062917-012624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Bedford MT (2012) Protein arginine methyltransferases and cancer. Nat Rev Cancer 13: 37–50. 10.1038/nrc3409 [DOI] [PubMed] [Google Scholar]
- Yu MC, Bachand F, McBride AE, Komili S, Casolari JM, Silver PA (2004) Arginine methyltransferase affects interactions and recruitment of mRNA processing and export factors. Genes Dev 18: 2024–2035. 10.1101/gad.1223204 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Source Data for Figure 5LSA-2018-00261_Sdata5A,B.tif (83.2MB, tif) LSA-2018-00261_Sdata5C,D.tif (40.4MB, tif)
Table S1 Yeast strains used in this study. (29.5KB, xlsx)
Table S2 Plasmids used in this study. (29.5KB, xlsx)
Table S3 Primers and their sequences used in this study. (29.5KB, xlsx)