ATP hydrolysis in the KaiC hexamer triggers the exposure of its C-terminal segments into the solvent so as to capture KaiA, providing mechanistic insights into the circadian periodicity regulation.
Abstract
The cyanobacterial clock is controlled via the interplay among KaiA, KaiB, and KaiC, which generate a periodic oscillation of KaiC phosphorylation in the presence of ATP. KaiC forms a homohexamer harboring 12 ATP-binding sites and exerts ATPase activities associated with its autophosphorylation and dephosphorylation. The KaiC nucleotide state is a determining factor of the KaiB–KaiC interaction; however, its relationship with the KaiA–KaiC interaction has not yet been elucidated. With the attempt to address this, our native mass spectrometric analyses indicated that ATP hydrolysis in the KaiC hexamer promotes its interaction with KaiA. Furthermore, our nuclear magnetic resonance spectral data revealed that ATP hydrolysis is coupled with conformational changes in the flexible C-terminal segments of KaiC, which carry KaiA-binding sites. From these data, we conclude that ATP hydrolysis in KaiC is coupled with the exposure of its C-terminal KaiA-binding sites, resulting in its high affinity for KaiA. These findings provide mechanistic insights into the ATP-mediated circadian periodicity.
Introduction
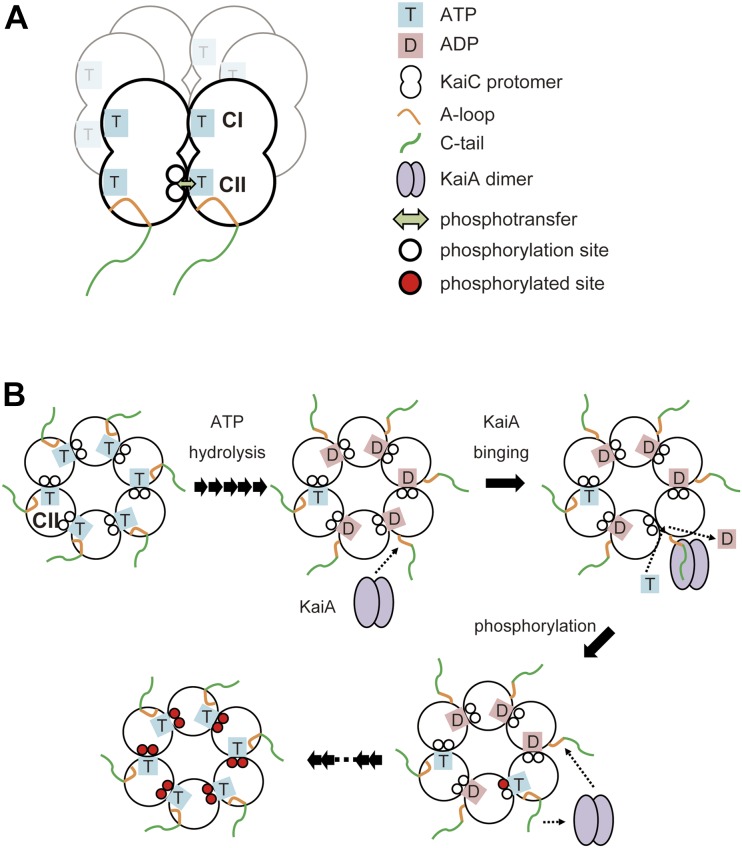
Endogenous, entrainable oscillation with periods of ∼24 h, known as the circadian rhythm, is found in many organisms. In cyanobacteria, the circadian rhythm is regulated by three clock proteins (Nakajima et al, 2005) (i.e., KaiA, KaiB, and KaiC) that autonomously undergo periodic assembly and disassembly irrespective of transcriptional and translational feedback systems (Tomita et al, 2005; Rust et al, 2007). KaiC is an AAA+ ATPase consisting of CI and CII domains and forms a hexameric ring structure, which harbors six ATP-binding sites in the CI ring and another six ATP-binding sites in the CII ring (Pattanayek et al, 2004). The KaiC hexamer experiences autophosphorylation and dephosphorylation cycles in a 24-h period through interactions with KaiA and KaiB in the presence of ATP (Nakajima et al, 2005). Two phosphorylation sites at Ser431 and Thr432 in the KaiC CII domain (simply denoted as S and T, respectively) go through a cycle as follows: S/T → S/pT → pS/pT → pS/T → S/T, where “p” represents the phosphorylated residue (Nishiwaki et al, 2004). Phosphorylation oscillation is controlled via the interplay among KaiA, KaiB, and KaiC. KaiA and KaiB interact with the CII and CI domains of KaiC, respectively, so that its phosphorylation is up-regulated/down-regulated. Dephosphorylated KaiC interacts with KaiA, followed by an increase in the KaiC phosphorylation (Iwasaki et al, 2002; Williams et al, 2002). By contrast, KaiB interacts with phosphorylated KaiC and, thereby, accelerates its dephosphorylation (Kitayama et al, 2003; Xu et al, 2003).
A series of phospho-mimicking KaiC mutants have been widely used for characterizing the Kai protein complex formation in a phosphorylation state–dependent manner (Pattanayek et al, 2009, 2011; Lin et al, 2014; Sugiyama et al, 2016; Tseng et al, 2017; Mori et al, 2018). In these studies, it has been suggested that the interaction of KaiC with KaiB depends not only on the KaiC phosphorylation states but also on the nucleotide states in KaiC. For example, a phosphorylation-mimicking KaiC mutant was reactive to KaiB in the presence of ATP but loses the KaiB-binding affinity when adenylyl imidodiphosphate AMPPNP, a nonhydrolyzable ATP analog, was used instead of ATP (Phong et al, 2013; Mukaiyama et al, 2018). However, little is known about the relationship between the nucleotide state of KaiC and its KaiA-binding activity. Herein, we address this issue through characterizing ATP hydrolysis dependence of the KaiA–KaiC interaction by native mass spectrometry (MS), providing mechanistic insights into the binding of KaiA to the C-terminal segment of KaiC based on nuclear magnetic resonance (NMR) data.
Results and Discussion
KaiC ATP hydrolysis promotes KaiA–KaiC interaction
The phosphorylation states and the bound nucleotide states of KaiC are both governed by its ATP hydrolysis activity (Nishiwaki & Kondo, 2012; Nishiwaki-Ohkawa et al, 2014). To control the phosphorylation states, we used two KaiC mutants, KaiCDD and KaiCAA (in which Ser431 and Thr432 were both substituted with aspartate and alanine residues, respectively), mimicking the phosphorylated and dephosphorylated states of KaiC, respectively. It has been reported that KaiCAA shows enhanced complex formation with KaiA in comparison with KaiCDD (Lin et al, 2014; Tseng et al, 2014). To examine the possible dependence of KaiA–KaiC interaction on KaiC-bound nucleotide states, ATP and its nonhydrolyzable analog, adenylyl imidodiphosphate (AMPPNP), were used for the formation of the mutated KaiC hexamers, which were subjected to native MS analysis.
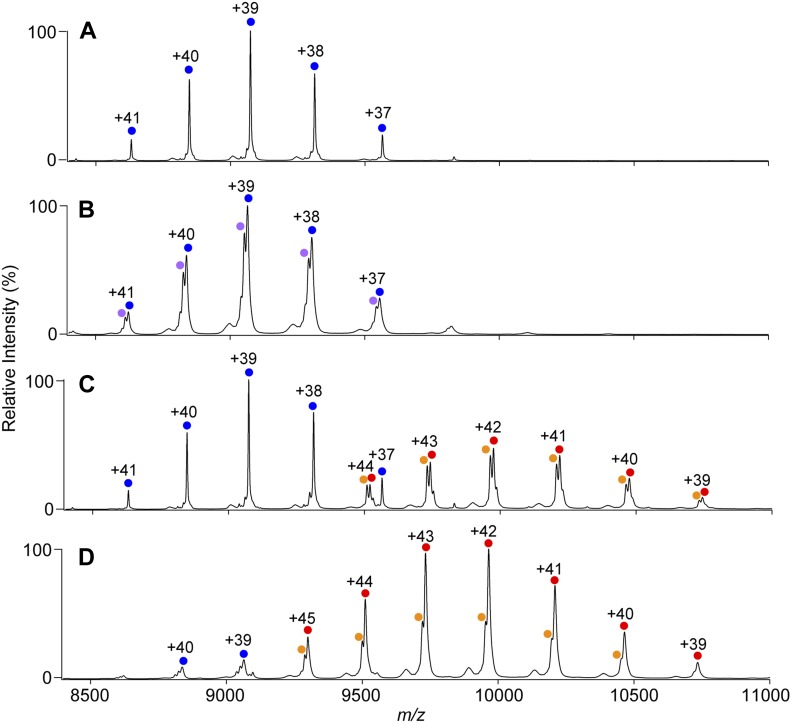
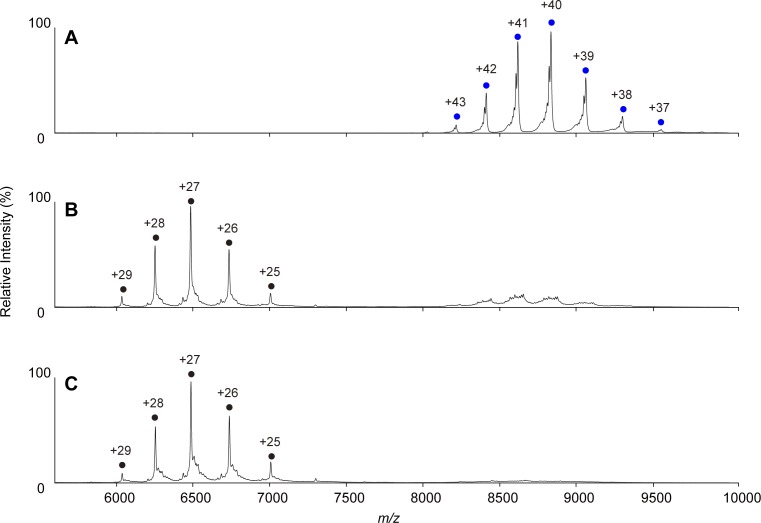
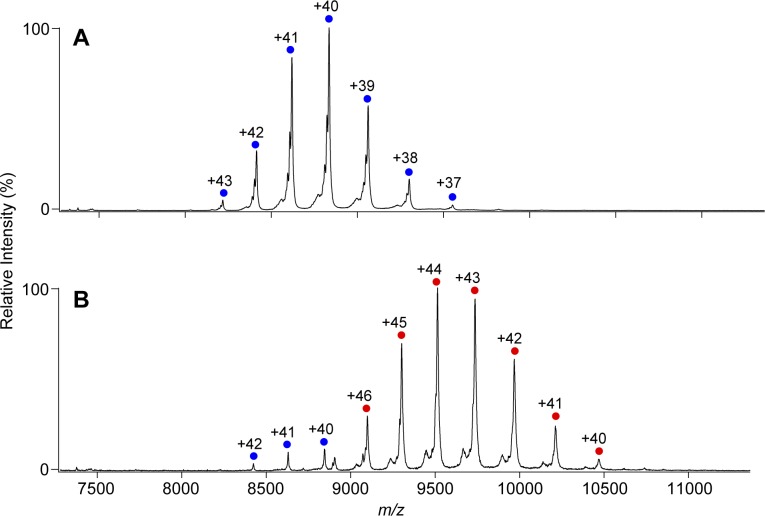
The native MS data confirmed that both KaiCAA and KaiCDD formed hexamers upon the addition of either ATP or AMPPNP (Figs 1A and B, S1A, and B). Whereas the KaiCAA hexamer mediated by AMPPNP had a constant molecular mass of 353,857 ± 10 D holding 12 AMPPNP molecules (Fig 1A and Table 1), the ATP-mediated KaiCAA hexamer preincubated for 5 h in the presence of 1 mM ATP exhibited two series of ion peaks (Fig 1B). The major peaks corresponded to the KaiCAA hexamer containing seven ATP and five ADP molecules, whereas the minor peaks corresponded to the KaiCAA hexamer containing seven ATP and three ADP molecules (Table 2). Native MS data of AMPPNP- or ATP-mediated KaiCAA hexamer incubated for 5 h in the presence of ATP and ADP at varying ratios, indicated that the nucleotide states of KaiCAA did not depend on the external ATP/ADP condition (Fig S2). These data indicate that the KaiCAA hexamer mediated by ATP spontaneously became asymmetric in terms of nucleotide state and that the resultant ADP molecules were releasable. By contrast, the KaiCDD hexamer held at least 11 nucleotides, suggesting its low ATPase activity, which is consistent with a previous report that the ATPase activity of KaiCAA is six times higher than that of KaiCDD (Mutoh et al, 2013). Furthermore, we conducted tryptic fragmentation into CI and CII of the KaiCAA hexamer, followed by native MS analysis. The results revealed that the CI hexamer retained six nucleotides as prehydrolyzed ATP molecules, suggesting that the observed ATP hydrolysis exclusively occurred in the CII ring (Fig S3). This was supported by the observation that a KaiCAA mutant in which a catalytic glutamate (Glu77) in the CI ATPase domain (designated as KaiCAA/E77Q) was substituted by glutamine exhibited virtually identical properties in terms of the nucleotide state and the KaiA binding to those of the KaiCAA hexamer (Fig S4 and Table 2).
Figure 1. KaiA–KaiC interaction depends on ATP hydrolysis.
(A–D) Native mass spectra of (A, B) KaiCAA and (C, D) 6:3 mixtures of KaiCAA and KaiA in the presence of (A, C) 1 mM AMPPNP or (B, D) 1 mM ATP. After 5 h of incubation at 37°C with ATP or AMPPNP, the KaiC solutions with or without KaiA were immediately analyzed by nanoflow electrospray ionization MS. The blue and purple circles show the ion series of the KaiCAA homohexamer, whereas the orange and red circles show the 2:6 KaiA–KaiCAA hetero-octamer complexes. See Tables 1 and 2 for assignment details.
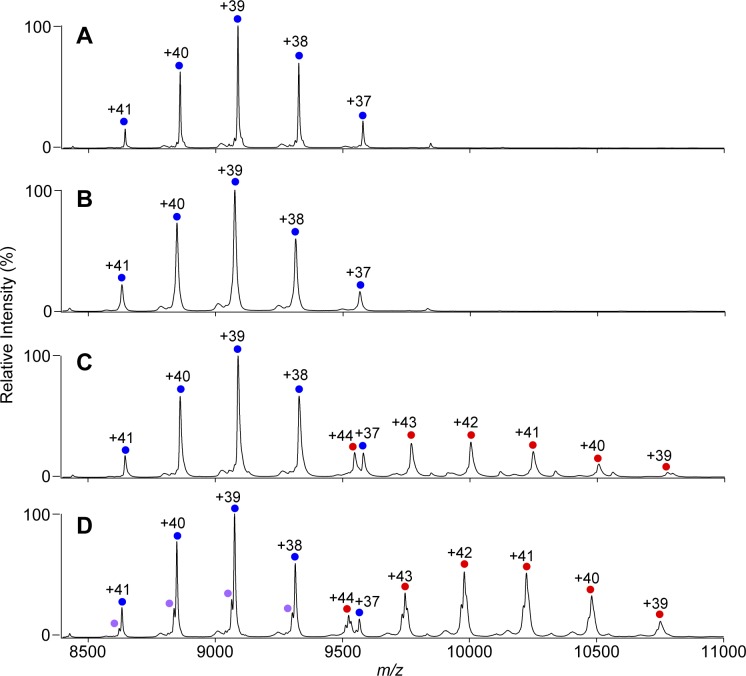
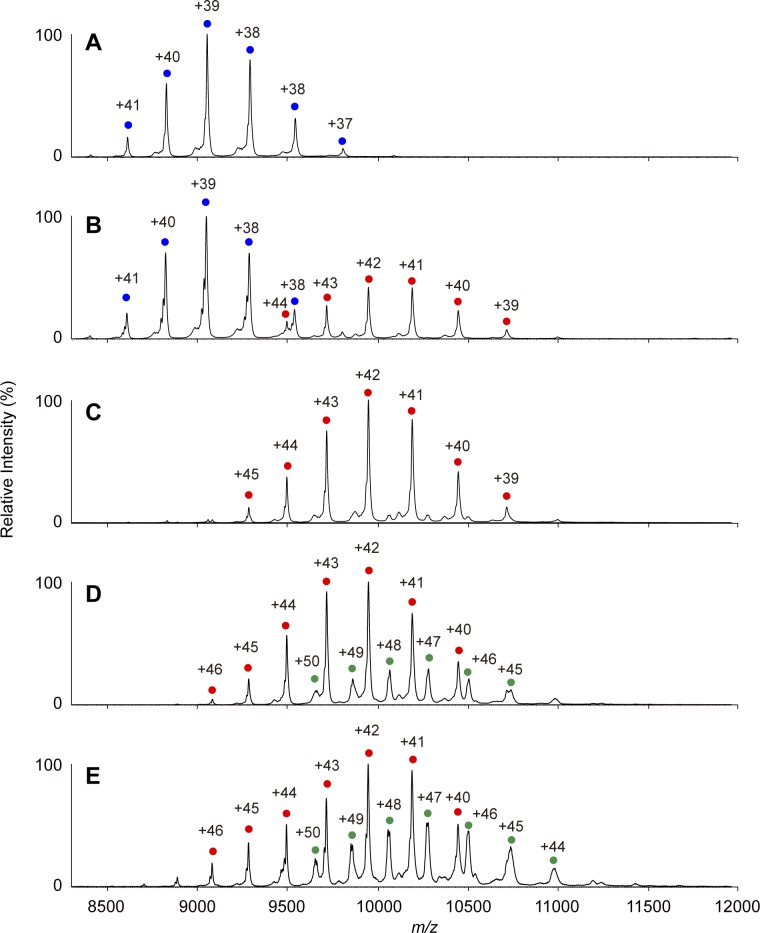
Figure S1. Native MS characterization of KaiA–KaiCDD interaction with ATP or AMPPNP.
(A–D) Native mass spectra of (A, B) KaiCDD and (C, D) 6:3 mixtures of KaiCDD and KaiA in the presence of (A, C) 1 mM AMPPNP or (B, D) 1 mM ATP. After 5 h of incubation at 37°C with the nucleotides, the KaiC solutions with or without KaiA were immediately analyzed by nanoflow electrospray ionization MS. The blue and purple circles show the ion series of the KaiCDD homohexamer, whereas the red circles show the 2:6 KaiA–KaiCAA hetero-octamer complexes. See Tables 1 and 2 for assignment details.
Table 1.
Summary of native MS characterization of KaiC and KaiA–KaiC complex formed in the presence of AMPPNP.
| Figure number | Ion series | Assignment | Theoretical mass (D) | Experimental mass (D) | Δm (D)a | Relative quantity (%) | ||
|---|---|---|---|---|---|---|---|---|
| Protein complex | AMPPNP number | Mg2+ number | ||||||
| Fig 1A | Blue | KaiCAA6 | 12 | 12 | 353,850 | 353,857 ± 10 | −7 | — |
| Fig 1C | Blue | KaiCAA6 | 12 | 12 | 353,850 | 353,855 ± 9 | −5 | — |
| Fig 1C | Red | KaiCAA6/KaiA2 | 11 | 12 | 418,838 | 418,896 ± 27 | −58 | 53b |
| Fig 1C | Orange | KaiCAA6/KaiA2 | 10 | 12 | 418,332 | 418,412 ± 28 | −80 | 47b |
| Fig S2A | Blue | KaiCDD6 | 12 | 12 | 354,396 | 354,375 ± 12 | 21 | — |
| Fig S2C | Blue | KaiCDD6 | 12 | 12 | 354,396 | 354,447 ± 16 | −51 | — |
| Fig S2C | Red | KaiCDD6/KaiA2 | 12 | 12 | 419,890 | 420,076 ± 47 | −186 | — |
Δm is the mass difference between the experimental mass and the theoretical mass.
Relative quantity of two ion series are shown.
Table 2.
Summary of native MS characterization of KaiC and KaiA–KaiC complex formed in the presence of ATP.
| Figure number | Ion series | Assignment | Theoretical mass (D) | Experimental mass (D) | Δm (D)a | Relative quantity (%) | |||
|---|---|---|---|---|---|---|---|---|---|
| Protein complex | ATP number | ADP number | Mg2+ number | ||||||
| Fig 1B | Blue | KaiCAA6 | 7 | 5 | 12 | 353,462 | 353,476 ± 18 | −14 | 56b |
| Fig 1B | Purple | KaiCAA6 | 7 | 3 | 12 | 352,608 | 352,593 ± 16 | 15 | 44b |
| Fig 1D | Blue | KaiCAA6 | 7 | 5 | 12 | 353,462 | 353,461 ± 14 | 1 | — |
| Fig 1D | Red | KaiCAA6/KaiA2 | 6 | 5 | 12 | 418,449 | 418,445 ± 20 | 4 | 67b |
| Fig 1D | Orange | KaiCAA6/KaiA2 | 0 | 11 | 12 | 417,969 | 417,963 ± 20 | 6 | 33b |
| KaiCAA6/KaiA2 | 5 | 5 | 12 | 417,942 | 417,963 ± 20 | −21 | |||
| Fig S1B | Blue | KaiCDD6 | 11 | 0 | 12 | 353,901 | 353,902 ± 11 | −1 | — |
| KaiCDD6 | 6 | 6 | 12 | 353,928 | 353,902 ± 11 | 26 | |||
| Fig S1D | Blue | KaiCDD6 | 6 | 6 | 12 | 353,928 | 353,924 ± 9 | 4 | 74b |
| KaiCDD6 | 11 | 0 | 12 | 353,901 | 353,924 ± 9 | −23 | |||
| Fig S1D | Purple | KaiCDD6 | 6 | 5 | 12 | 353,501 | 353,477 ± 9 | 24 | 26b |
| KaiCDD6 | 0 | 12 | 12 | 353,448 | 353,477 ± 9 | −29 | |||
| Fig S1D | Red | KaiCDD6/KaiA2 | 2 | 10 | 12 | 419,102 | 419,102 ± 32 | 0 | — |
| KaiCDD6/KaiA2 | 7 | 4 | 12 | 419,075 | 419,102 ± 32 | −27 | |||
| Fig S4A | Blue | KaiCAA/E77Q6 | 7 | 5 | 12 | 353,396 | 353,383 ± 13 | 13 | — |
| Fig S4B | Blue | KaiCAA/E77Q6 | 12 | 0 | 12 | 353,796 | 353,766 ± 29 | 30 | — |
| Red | KaiCAA/E77Q6/KaiA2 | 7 | 4 | 12 | 418,533 | 418,559 ± 59 | −26 | — | |
Δm is the mass difference between the experimental mass and the theoretical mass.
Relative quantity of two ion series are shown.
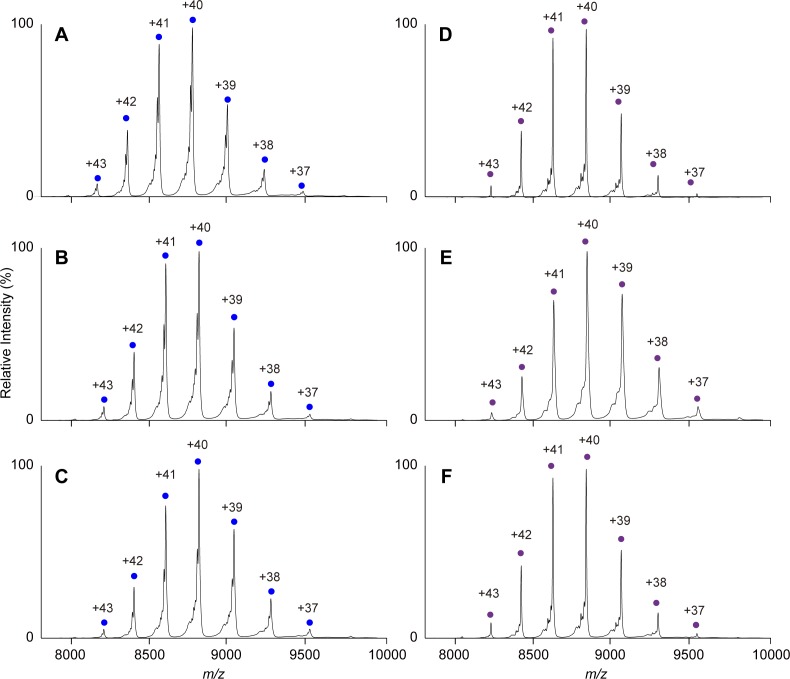
Figure S2. Native MS characterization of KaiCAA nucleotide state depending on external ATP/ADP condition.
(A–F) Native mass spectra of KaiCAA mediated by (A–C) ATP and (D–F) AMPPNP. The KaiCAA hexamers incubated for 5 h at 37°C under (A, D) 100:0, (B, E) 75:25, and (C, F) 50:50 ATP/ADP conditions were immediately analyzed by nanoflow electrospray ionization MS. The blue and purple circles show the ion series of the KaiCAA hexamers containing seven ATP/five ADP molecules and 12 AMPPNP molecules, respectively.
Figure S3. Native MS characterization of nucleotide states of the CI and CII domains on KaiCAA.
(A–C) Native mass spectra of KaiCAA mediated by ATP after trypsin digestion. After 5 h of incubation at 37°C in the presence of 1 mM ATP, KaiCAA was buffer-exchanged into 150 mM aqueous ammonium acetate and digested by 0.02 mg/ml trypsin for (A) 0 min, (B), 30 min, and (C) 60 min. The reaction mixture was directly analyzed by nanoflow electrospray ionization MS. The blue circles show the ion series of the KaiCAA homohexamer containing seven ATP and five ADP molecules, whereas the black circles show the hexameric CI domain (M1–S253) containing six ATP molecules. The CII domain was hardly detected as hexamer under the condition used here.
Figure S4. Native MS characterization of the KaiA–KaiCAA/E77Q interaction.
(A–D) Native mass spectra of (A) KaiCAA/E77Q and (B) a 6:3 mixtures of KaiCAA/E77Q and KaiA. After 5 h of incubation at 37°C in the presence of 1 mM ATP, the KaiCAA/E77Q solutions with or without KaiA were immediately analyzed by nanoflow electrospray ionization MS. The blue circles show the ion series of the KaiCAA/E77Q homohexamer, whereas the red circles show the 2:6 KaiA–KaiCAA/E77Q hetero-octamer complexes. See Table 2 for assignment details.
Upon the addition of KaiA, both KaiCAA and KaiCDD formed ∼420-kD complexes, indicating that two KaiA molecules bind one KaiC hexamer (Fig 1C and D, and Tables 1 and 2, and Fig S1C and D). This stoichiometry is consistent with the previously reported small-angle X-ray scattering and electron microscopy data, indicating that the KaiA dimer is tethered to the KaiC hexamer through its flexible C-terminal region (Pattanayek et al, 2006, 2011). Even in the presence of excess amounts of KaiA, the KaiA–KaiC complex were formed primarily in a 2:6 stoichiometry and in a 4:6 stoichiometry as minor complex (Fig S5). The occurrence of the complex significantly depended on the nucleotide state of KaiC and its phosphorylation mutation. The ATP-mediated KaiCAA hexamer almost exclusively formed the complex with KaiA, whereas the complex formation was compromised in the AMPPNP-mediated KaiCAA hexamer and both the AMPPNP- and ATP-mediated KaiCDD hexamers. These data indicate that the nonphosphorylated KaiC hexamer becomes most reactive with KaiA after ATP hydrolysis. Interestingly, in most cases, the KaiA–KaiC complexes lost one or two nucleotides (Tables 1 and 2). Noteworthy, the AMPPNP-mediated KaiCAA hexamer in complex with the KaiA dimer lacked one and two nucleotides, strongly suggesting that KaiA binding promotes nucleotide release from KaiC.
Figure S5. Native MS analysis of KaiCAA-KaiA complex formation.
(A–E) Native mass spectra of (A) KaiCAA and (B) 6:1, (C) 6:3, (D) 6:6, and, (E) 6:9 mixtures of KaiCAA and KaiA in the presence of 1 mM ATP. After 5 h of incubation at 37°C in the presence of 1 mM ATP, the KaiC solutions with or without KaiA were immediately analyzed by nanoflow electrospray ionization MS. The blue circles show the ion series of the KaiCAA homohexamer, whereas the red and green circles show the 2:6 and 4:6 KaiA–KaiCAA complexes, respectively.
On the basis of our data and previous reports that KaiA binding stimulates the ATPase activity of KaiC (Terauchi et al, 2007; Murakami et al, 2008), we conclude that ATP hydrolysis of KaiC promotes its interaction with KaiA, which is coupled with ADP release and, in turn, enhances the KaiC ATPase activity.
KaiC ATP hydrolysis triggers exposure of its KaiA-binding segments
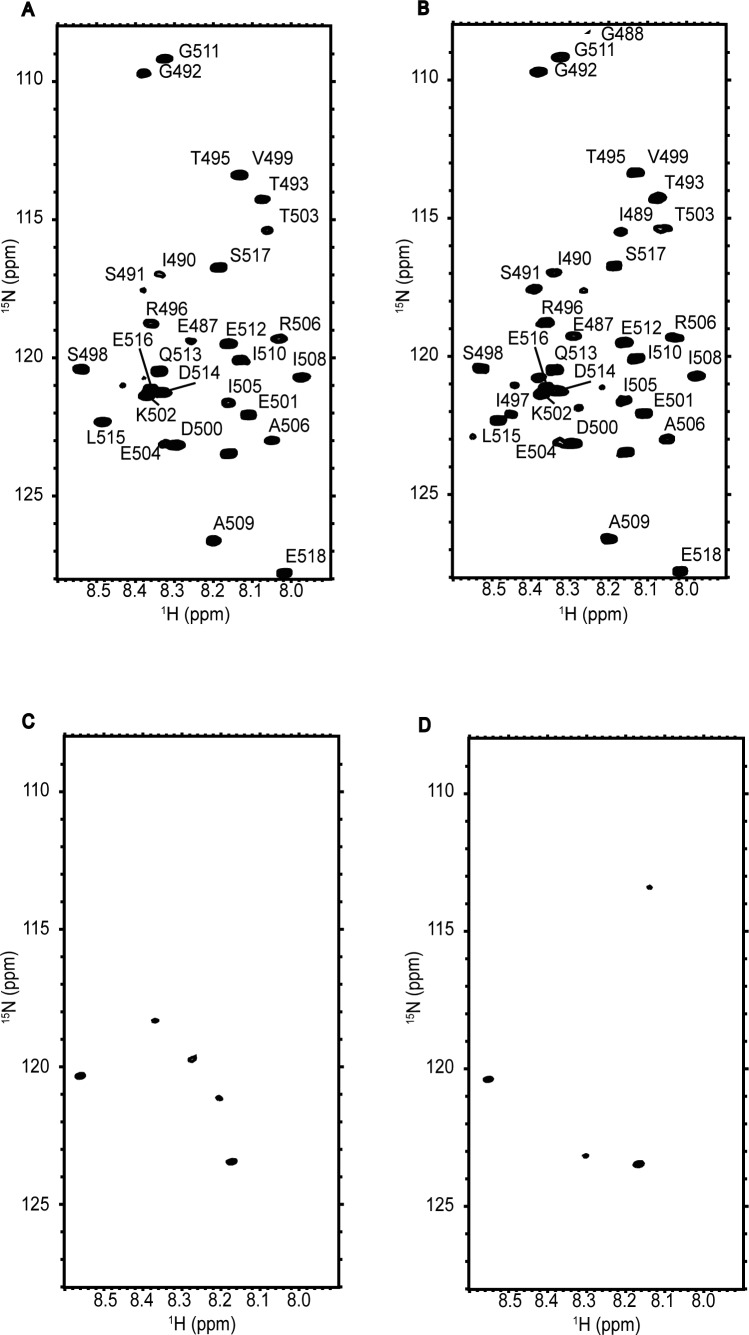
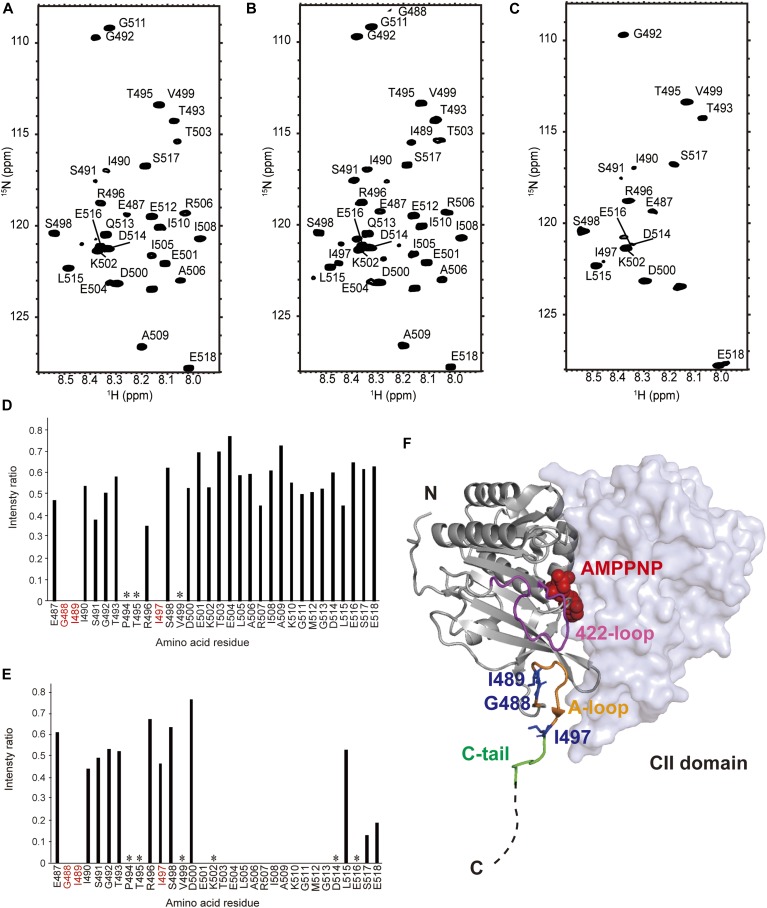
Using NMR-based structural characterization, we attempted to address how ATP hydrolysis in KaiC affects its binding to KaiA. Because of the fact that KaiA was reported to interact with the A-loop (Glu487-Ile497) and C-tail (Ser498-Ser518) of KaiC (Vakonakis & LiWang, 2004; Pattanayek et al, 2006, 2011; Pattanayek & Egli, 2015), we focused on these C-terminal segments. We compared the 1H-15N HSQC spectra of KaiCAA and its mutant with the deletion of the C-terminal region 487–518 in the presence of AMPPNP or ATP (Fig S6). Despite its high molecular mass (347 kD), the KaiCAA hexamer yielded ∼30 observable peaks originating from the backbone amide groups with high flexibility, most of which disappeared in the spectrum of the truncated mutant and, therefore, were assigned to the C-terminal region. The signals derived from the C-terminal region were assigned to individual amino acid residues located in the A-loop and C-tail by a series of 1H/13C/15N multinuclear NMR experiments (Fig 2A and B). In the KaiCAA hexamer mediated by AMPPNP, these peaks demonstrated enhanced peak broadening in comparison with those from the ATP-mediated hexamer and, importantly, Gly488, Ile489, and Ile497 yielded no observable peak, indicating that its A-loop became mobile upon ATP hydrolysis (Fig 2D and F). Furthermore, the peaks originating from the C-terminal segments, including Gly488 and Ile489 in the A-loop, were attenuated or even disappeared upon the addition of KaiA in the presence of ATP, indicating that both the A-loop and the C-tail are involved in the interaction with KaiA (Fig 2C and E). This result is consistent with the X-ray crystal structure, which shows that the full-length Synechococcus elongatus KaiA homodimer is in complex with two KaiC C-tail peptides (Asp500-Ser519) (Pattanayek & Egli, 2015) and also that the NMR solution structure of the C-terminal domain of Thermosynechococcus elongatus KaiA (residues 180–283) interacting with a C-terminal peptide includes the A-loop and C-tail (residues 488–518) of KaiC. In the latter case, the KaiC-derived peptide binds a KaiA surface through its eight hydrophobic side chains in the A-loop (Ile490, Thr493, and Thr495) and in the C-tail (Val499, Leu505, Ala506, Ile508, and Met512) (Vakonakis & LiWang, 2004).
Figure S6. 1H-15N HSQC spectra of KaiCAA and its C-terminally truncated mutant.
(A–D) 1H-15N HSQC spectra of (A, B) KaiCAA and (C, D) the mutated KaiCAA lacking the C-terminal segment 487–518 in the presence of (A, C) 1 mM AMPPNP and (B, D) 1 mM ATP.
Figure 2. ATP hydrolysis–dependent conformational change of the C-terminal KaiA-binding region of KaiC.
(A–C) 1H-15N HSQC spectrum of KaiCAA in the presence of (A) AMPPNP, (B) ATP, and (C) KaiA and ATP. NMR experiments were set up to take a total time of 3 h using the KaiC hexamer incubated with AMPPNP or ATP for 9 h. Assignments of the peaks from the C-terminal region are given in each spectrum. (D) Plot of relative peak intensity for KaiCAA resonances in the presence of AMPPNP versus ATP. (E) Plot of relative peak intensity for KaiCAA resonances in the presence versus absence of KaiA under the ATP condition. In (D) and (E), the residues that yielded no observable peaks under the AMPPNP condition are highlighted in red, whereas the asterisks indicate the proline residues and residues whose chemical shift perturbation data could not be obtained because of severe peak overlapping. (F) Crystal structure of two KaiC protomers in cartoon and surface representation, respectively, in the KaiC homohexameric ring mediated by AMPPNP (PDB ID code: 4O0M). In the crystal structure,the C-terminal region comprises a U-shaped A-loop (Glu487-Ile497) (orange) and a solvent-exposed C-tail (S498-S518), in which only the Ser498-Glu504 part (green) was modeled. The three residues (i.e., Gly488, Ile489, and Ile497) located in the A-loop, whose HSQC peaks were unobserved under the AMPPNP condition, are colored blue. The A-loop and AMPPNP molecule (red) are mediated by a loop comprising residues 415–430 (termed 422-loop, magenta).
To date, the crystal structures of the wild-type KaiC hexamer and its phosphorylated mutants have been solved as AMPPNP-bound states (Pattanayek et al, 2004, 2009, 2014). In these crystal structures, the A-loop forms a U-shaped hairpin, which is accommodated in a cleft proximal to the nucleotide-binding site and is followed by the C-tail, which is exposed to the solvent and gives no interpretable electron density for the segment beyond E504 (Fig 2F) (Pattanayek & Egli, 2015). Under this circumstance, the interaction of KaiA with the KaiC hexamer is compromised because of the inaccessibility of the A-loop. Our MS and NMR data indicate that, upon ATP hydrolysis, the A-loop is released and becomes reactive with KaiA, leading to the enhanced KaiA–KaiC interaction.
The crystal structure revealed that AMPPNP molecules are located at the intersubunit interfaces of the KaiC hexamer (Pattanayek et al, 2014). In the CII ring, a loop comprising residues 415–433 (termed “422-loop”) makes contact with both AMPPNP and the A-loop (Fig 2F) (Vakonakis & LiWang, 2004; Kim et al, 2008; Egli et al, 2013). Based on our findings in conjunction with the crystallographic data, we propose a mechanistic model in which ATP hydrolysis in the CII ring triggers a conformational rearrangement of the 422-loop, resulting in “leaping out” of the A-loop so as to interact with KaiA. Reciprocally, it is possible that KaiA pulls out the A-loop and, thereby, causes a microenvironmental rearrangement surrounding the nucleotide-binding site of the KaiC CII ring, facilitating ADP release from it. This is supported by a recently reported molecular dynamics simulation of the KaiC hexamer (Hong et al, 2018).
The present study experimentally reveals that ATP hydrolysis in the KaiC hexamer triggers the exposure of its C-terminal segments into the solvent so as to capture KaiA, which in turn facilitates ADP release. These findings imply that the ATPase-dependent KaiA interaction promotes ADP/ATP turnover on KaiC, leading to the up-regulation of its autophosphorylation (Fig 3). Our findings provide mechanistic insights into the circadian periodicity mediated by the unique AAA+ ATPase.
Figure 3. The “fishing a line” mechanism coupling ATP hydrolysis and KaiA-mediated up-regulation of autophosphorylation in the KaiC hexamer.
(A) While both CI and CII domains harbor nucleotide-binding sites and ATPase-active sites at the subunit interfaces, the autokinase activity is exerted only in the CII domain. This is because the autophosphorylation sites (i.e., Ser431 and Thr432) are spatially proximal to the ATP molecule accommodated in the CII domain of the neighboring protomer. (B) In the CII AAA+ ring hexamer, ATP hydrolysis releases the A-loop, which thereby becomes reactive with KaiA. KaiA binding to the C-terminal segments of KaiC facilitates ADP release and ATP incorporation. The rapid ATP/ADP turnover leads to the up-regulation of autophosphorylation of KaiC.
Materials and Methods
Protein expression and purification for native MS
KaiA and KaiC mutants originating from thermophilic cyanobacteria, T. elongatus BP-1, were expressed in Escherichia coli and purified as previously described (Ishii et al, 2014). The expression plasmids of the KaiC mutants (KaiCAA, KaiCAA/E77Q, and KaiCDD) were also constructed according to a previous study (Murakami et al, 2012; Mutoh et al, 2013).
Protein expression and purification for NMR spectroscopy
The expression plasmids of KaiCAA and its mutant with the deletion of the C-terminal segment 487–518 were constructed according to a previous study (Ishii et al, 2014). For NMR analyses, the protein was expressed in M9 minimal medium containing [15N]NH4Cl and/or [13C]glucose. The protein was purified according to a previous report (Ishii et al, 2014). The NMR samples were prepared by dissolving the mutated KaiC (53 μM final concentration) in 10% D2O containing 20 mM Tris–HCl (pH 7.0), 150 mM NaCl, 5 mM MgCl2, 0.5 mM EDTA, and 1 mM DTT in the presence of 1 mM ATP or AMPPNP.
Native MS analyses
For KaiC hexamerization, the purified monomeric KaiC mutants (KaiCAA, KaiCAA/E77Q, and KaiCDD) (20 μM) were incubated in the presence of 1 mM nucleotide (ATP or AMPPNP) for 30 min on ice. The KaiC solutions were incubated at 37°C for 5 h in the presence of ATP or AMPPNP. In the examination of possible effects of the external ATP/ADP variation, the AMPPNP- or ATP-mediated KaiCAA hexamer was incubated in the presence of ATP and ADP at varying ratios. Subsequently, these KaiC solutions, in the absence or presence of KaiA (10 μM), were buffer-exchanged into 150 mM ammonium acetate (pH 6.8) by passing the proteins through a Bio-Spin 6 column (Bio-Rad) and were then immediately subjected to nanoflow electrospray ionization MS using gold-coated glass capillaries made in-house (∼2–5 μl sample loaded per analysis). For the tryptic fragmentation into CI and CII of the KaiCAA hexamer, KaiCAA incubated at 37°C in the presence of 1 mM ATP for 5 h was buffer-exchanged into 150 mM aqueous ammonium acetate and then digested with 0.02 mg/ml trypsin for up to 60 min, followed directly by nanoflow electrospray ionization MS measurements. Spectra were recorded on a SYNAPT G2-Si HDMS mass spectrometer (Waters) in positive ionization mode at 1.33 kV with a 150 V sampling cone voltage and source offset voltage, 0 V trap and transfer collision energy, and 5 ml/min trap gas flow. Spectra were calibrated using 1 mg/ml cesium iodide and analyzed using MassLynx software (Waters) (Sugiyama et al, 2016).
NMR analyses
All NMR spectra were acquired at 37°C using Bruker DMX-500 and Bruker AVANCE 800 US spectrometers. The chemical shifts of KaiC peaks were assigned on the basis of 2D 1H-15N HSQC, 3D 1H-13C-15N HSQC, 3D HNCO, and 3D HNCA spectral data in the presence of AMPPNP or ATP. The purified monomeric KaiC with isotope labeling (53 μM) was buffer-exchanged into 20 mM Tris buffer (pH 7.0) containing 150 mM NaCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM nucleotide (ATP or AMPPNP), and 10% (vol/vol) D2O and was then incubated at 4°C for 30 min for hexamerization. The KaiC hexamers formed by AMPPNP were used for the NMR assignment experiment. After 9 h of incubation at 37°C, the KaiC solutions were subjected to 1H-15N HSQC measurements in the absence and presence of a KaiA dimer (53 μM). All NMR data were processed using NMRPipe software and analyzed with Sparky software (Delaglio et al, 1995; Lee et al, 2009).
Supplementary Material
Acknowledgements
We thank Dr. T Suzuki (ExCELLS) for his assistance in MS analysis. This work was supported by grants (JP18J21063 to Y Yunoki, JP15K18492 to R Murakami, JP18K14671 to K Ishii, and JP25102001 and JP25102008 to K Kato) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan, by the Okazaki ORION project, and by the Joint Research by ExCELLS. We acknowledge the assistance of the Research Equipment Sharing Center at the Nagoya City University.
Author Contributions
Y Yunoki: conceptualization, data curation, formal analysis, funding acquisition, investigation, visualization, methodology, and writing—original draft.
K Ishii: data curation, formal analysis, validation, investigation, and methodology.
M Yagi-Utsumi: data curation, formal analysis, investigation, methodology, and project administration
R Murakami: investigation.
S Uchiyama: supervision and project administration.
H Yagi: conceptualization, formal analysis, supervision, visualization, project administration, and writing—original draft, review, and editing.
K Kato: conceptualization, resources, supervision, funding acquisition, project administration, and writing—review and editing.
Conflict of Interest Statement
The authors declare no conflict of interest.
References
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6: 277–293. 10.1007/BF00197809 [DOI] [PubMed] [Google Scholar]
- Egli M, Pattanayek R, Sheehan JH, Xu Y, Mori T, Smith JA, Johnson CH (2013) Loop-loop interactions regulate KaiA-stimulated KaiC phosphorylation in the cyanobacterial KaiABC circadian clock. Biochemistry 52: 1208–1220. 10.1021/bi301691a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L, Vani BP, Thiede EH, Rust MJ, Dinner AR (2018) Molecular dynamics simulations of nucleotide release from the circadian clock protein KaiC reveal atomic-resolution functional insights. Proc Natl Acad Sci USA 115: E11475–E11484. 10.1073/pnas.1812555115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Terauchi S, Murakami R, Valencia Swain J, Mutoh R, Mino H, Maki K, Arata T, Ishiura M (2014) Site-directed spin labeling-electron spin resonance mapping of the residues of cyanobacterial clock protein KaiA that are affected by KaiA-KaiC interaction. Genes Cells 19: 297–324. 10.1111/gtc.12130 [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T (2002) KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc Natl Acad Sci USA 99: 15788–15793. 10.1073/pnas.222467299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YI, Dong G, Carruthers CW Jr, Golden SS, LiWang A (2008) The day/night switch in KaiC, a central oscillator component of the circadian clock of cyanobacteria. Proc Natl Acad Sci USA 105: 12825–12830. 10.1073/pnas.0800526105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T (2003) KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J 22: 2127–2134. 10.1093/emboj/cdg212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Westler WM, Bahrami A, Eghbalnia HR, Markley JL (2009) PINE-SPARKY: Graphical interface for evaluating automated probabilistic peak assignments in protein NMR spectroscopy. Bioinformatics 25: 2085–2087. 10.1093/bioinformatics/btp345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Chew J, Chockanathan U, Rust MJ (2014) Mixtures of opposing phosphorylations within hexamers precisely time feedback in the cyanobacterial circadian clock. Proc Natl Acad Sci USA 111: E3937–E3945. 10.1073/pnas.1408692111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Sugiyama S, Byrne M, Johnson CH, Uchihashi T, Ando T (2018) Revealing circadian mechanisms of integration and resilience by visualizing clock proteins working in real time. Nat Commun 9: 3245 10.1038/s41467-018-05438-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukaiyama A, Furuike Y, Abe J, Yamashita E, Kondo T, Akiyama S (2018) Conformational rearrangements of the C1 ring in KaiC measure the timing of assembly with KaiB. Sci Rep 8: 8803 10.1038/s41598-018-27131-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami R, Miyake A, Iwase R, Hayashi F, Uzumaki T, Ishiura M (2008) ATPase activity and its temperature compensation of the cyanobacterial clock protein KaiC. Genes Cells 13: 387–395. 10.1111/j.1365-2443.2008.01174.x [DOI] [PubMed] [Google Scholar]
- Murakami R, Mutoh R, Iwase R, Furukawa Y, Imada K, Onai K, Morishita M, Yasui S, Ishii K, Valencia Swain JO, et al. (2012) The roles of the dimeric and tetrameric structures of the clock protein KaiB in the generation of circadian oscillations in cyanobacteria. J Biol Chem 287: 29506–29515. 10.1074/jbc.M112.349092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh R, Nishimura A, Yasui S, Onai K, Ishiura M (2013) The ATP-mediated regulation of KaiB-KaiC interaction in the cyanobacterial circadian clock. PLoS One 8: e80200 10.1371/journal.pone.0080200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T (2005) Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308: 414–415. 10.1126/science.1108451 [DOI] [PubMed] [Google Scholar]
- Nishiwaki T, Kondo T (2012) Circadian autodephosphorylation of cyanobacterial clock protein KaiC occurs via formation of ATP as intermediate. J Biol Chem 287: 18030–18035. 10.1074/jbc.M112.350660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaki T, Satomi Y, Nakajima M, Lee C, Kiyohara R, Kageyama H, Kitayama Y, Temamoto M, Yamaguchi A, Hijikata A, et al. (2004) Role of KaiC phosphorylation in the circadian clock system of Synechococcus elongatus PCC 7942. Proc Natl Acad Sci USA 101: 13927–13932. 10.1073/pnas.0403906101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaki-Ohkawa T, Kitayama Y, Ochiai E, Kondo T (2014) Exchange of ADP with ATP in the CII ATPase domain promotes autophosphorylation of cyanobacterial clock protein KaiC. Proc Natl Acad Sci USA 111: 4455–4460. 10.1073/pnas.1319353111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayek R, Egli M (2015) Protein-protein interactions in the cyanobacterial circadian clock: Structure of KaiA dimer in complex with C-terminal KaiC peptides at 2.8 A resolution. Biochemistry 54: 4575–4578. 10.1021/acs.biochem.5b00694 [DOI] [PubMed] [Google Scholar]
- Pattanayek R, Mori T, Xu Y, Pattanayek S, Johnson CH, Egli M (2009) Structures of KaiC circadian clock mutant proteins: A new phosphorylation site at T426 and mechanisms of kinase, ATPase and phosphatase. PLoS One 4: e7529 10.1371/journal.pone.0007529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayek R, Wang J, Mori T, Xu Y, Johnson CH, Egli M (2004) Visualizing a circadian clock protein: Crystal structure of KaiC and functional insights. Mol Cell 15: 375–388. 10.1016/j.molcel.2004.07.013 [DOI] [PubMed] [Google Scholar]
- Pattanayek R, Williams DR, Pattanayek S, Xu Y, Mori T, Johnson CH, Stewart PL, Egli M (2006) Analysis of KaiA-KaiC protein interactions in the cyano-bacterial circadian clock using hybrid structural methods. EMBO J 25: 2017–2028. 10.1038/sj.emboj.7601086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayek R, Williams DR, Rossi G, Weigand S, Mori T, Johnson CH, Stewart PL, Egli M (2011) Combined SAXS/EM based models of the S. elongatus post-translational circadian oscillator and its interactions with the output His-kinase SasA. PLoS One 6: e23697 10.1371/journal.pone.0023697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayek R, Xu Y, Lamichhane A, Johnson CH, Egli M (2014) An arginine tetrad as mediator of input-dependent and input-independent ATPases in the clock protein KaiC. Acta Crystallogr D Biol Crystallogr 70: 1375–1390. 10.1107/s1399004714003228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phong C, Markson JS, Wilhoite CM, Rust MJ (2013) Robust and tunable circadian rhythms from differentially sensitive catalytic domains. Proc Natl Acad Sci USA 110: 1124–1129. 10.1073/pnas.1212113110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Markson JS, Lane WS, Fisher DS, O’Shea EK (2007) Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science 318: 809–812. 10.1126/science.1148596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama M, Yagi H, Ishii K, Porcar L, Martel A, Oyama K, Noda M, Yunoki Y, Murakami R, Inoue R, et al. (2016) Structural characterization of the circadian clock protein complex composed of KaiB and KaiC by inverse contrast-matching small-angle neutron scattering. Sci Rep 6: 35567 10.1038/srep35567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terauchi K, Kitayama Y, Nishiwaki T, Miwa K, Murayama Y, Oyama T, Kondo T (2007) ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc Natl Acad Sci USA 104: 16377–16381. 10.1073/pnas.0706292104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita J, Nakajima M, Kondo T, Iwasaki H (2005) No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science 307: 251–254. 10.1126/science.1102540 [DOI] [PubMed] [Google Scholar]
- Tseng R, Chang YG, Bravo I, Latham R, Chaudhary A, Kuo NW, Liwang A (2014) Cooperative KaiA-KaiB-KaiC interactions affect KaiB/SasA competition in the circadian clock of cyanobacteria. J Mol Biol 426: 389–402. 10.1016/j.jmb.2013.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng R, Goularte NF, Chavan A, Luu J, Cohen SE, Chang YG, Heisler J, Li S, Michael AK, Tripathi S, et al. (2017) Structural basis of the day-night transition in a bacterial circadian clock. Science 355: 1174–1180. 10.1126/science.aag2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakonakis I, LiWang AC (2004) Structure of the C-terminal domain of the clock protein KaiA in complex with a KaiC-derived peptide: Implications for KaiC regulation. Proc Natl Acad Sci USA 101: 10925–10930. 10.1073/pnas.0403037101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SB, Vakonakis I, Golden SS, LiWang AC (2002) Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: A potential clock input mechanism. Proc Natl Acad Sci USA 99: 15357–15362. 10.1073/pnas.232517099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Mori T, Johnson CH (2003) Cyanobacterial circadian clockwork: Roles of KaiA, KaiB and the kaiBC promoter in regulating KaiC. EMBO J 22: 2117–2126. 10.1093/emboj/cdg168 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.