Abstract
Background
Lung cancer is one of the common malignant tumors worldwide with high incidence and mortality. MicroRNA-423-3p (miR-423-3p) acts as an oncogene in several types of cancers. The aim of this study is to reveal the clinical significance and biological function of miR-423-3p in lung cancer.
Methods
The expression of miR-423-3p was detected in lung cancer specimens by reverse transcription-quantitative polymerase chain reaction (qRT-PCR) assay. Kaplan-Meier survival and Cox regression analyses were used to investigate the prognostic significance of miR-423-3p in lung cancer. CCK-8 and Transwell assays were used to determine the functional role of miR-423-3p in lung cancer.
Results
We observed that miR-423-3p was significantly upregulated in lung cancer tissues and cell lines. Overexpression of miR-423-3p was significantly associated with lymph node metastasis, TNM stage, and poor prognosis. Multivariate Cox regression analysis results showed that miR-423-3p was an independent prognostic indicator for lung cancer patients. Results of functional analyses revealed that overexpression of miR-423-3p promoted cell proliferation, migration, and invasion in lung cancer cells.
Conclusions
These results indicated that miR-423-3p acts as an oncogene and promotes cell proliferation migration, and invasion of lung cancer. And miR-423-3p may serve as a potential prognostic biomarker and therapeutic target for the treatment of lung cancer.
Keywords: Lung cancer, microRNA-423-3p, Prognosis, Proliferation, Migration, Invasion
Introduction
Lung cancer is a leading cause of cancer-related deaths worldwide with the highest incidence and mortality [1]. The most effective treatment for patients at an early stage is surgical resection, but the relapse rate after surgery is high. In addition, numbers of patients are diagnosed at an advanced stage and the overall survival rate is poor [2, 3]. Lung cancer is also a severely healthy burden in China, the incidence and mortality rate is still increasing with the accelerated progress of China’s industrial development and changes in lifestyle [4–6]. Because of the poor survival of lung cancer patients in advanced stages, it is important to identify novel effective prognostic biomarkers and therapeutic strategies to improve the treatment of lung cancer.
MicroRNAs (miRNAs or miRs) are a class of small non-coding RNAs, approximately 22 nucleotides in length that negatively regulate gene expression at the post-transcriptional level through binding to the 3′- Untranslated Region(UTR) of target mRNAs [7, 8]. Emerging evidence indicated that miRNAs play critical roles in the development and progression of various kinds of diseases, including cancers [9–11]. Numerous studies have indicated that the aberrant expression of miRNAs is involved in various biological processes, including development, proliferation, migration, invasion, and apoptosis [12–14]. Recent studies demonstrated that miRNAs such as miR-210, miR-577, and miR-141-3p are abnormal expressed in lung cancer pathogenesis [15–17]. A recent study by Zhu et al. showed that ten miRNAs including miR-423-3p are increased in serum of lung cancer patients [18]. However, the expression pattern of miR-423-3p in lung cancer tissues and biological role of miR-423-3p in lung cancer is still unclear.
In the present study, we aimed to investigate the expression and functional role of miR-423-3p in lung cancer and highlight its potential as a biomarker for lung cancer.
Materials and methods
Patients and tissue specimens
A total of 126 patients with lung cancer were included between January 2011 and December 2012 at Weifang Cancer Hospital in this study. The paired lung cancer tissue and matched adjacent normal tissue specimens (all were confirmed by at least two pathologists) were collected during surgery and snap frozen and stored at − 80 °C until RNA extraction. All the patients did not receive any treatment before surgery. 5-year follow-up information was enrolled to collect survival status by telephone. The clinicopathological characters of lung cancer patients were collected and listed in Table 1. All participants signed written informed consent in this study. The research was approved by the Ethics Committee of Weifang Cancer Hospital.
Table 1.
Correlation between miR-423-3p expression levels and clinical features in lung cancer patients
| Parameters | Cases No. (n = 126) | miR-423-3p expression | P | |
|---|---|---|---|---|
| Low (n = 61) | High (n = 65) | |||
| Age | ||||
| < 60 | 66 | 29 | 37 | 0.292 |
| ≥ 60 | 60 | 32 | 28 | |
| Gender | ||||
| Male | 75 | 37 | 38 | 0.802 |
| Female | 51 | 24 | 27 | |
| Tumor size | ||||
| < 4 cm | 69 | 37 | 32 | 0.198 |
| ≥ 4 cm | 57 | 24 | 33 | |
| Differentiation | ||||
| Well + Moderate | 77 | 39 | 38 | 0.529 |
| Poor | 49 | 22 | 27 | |
| Lymph node metastasis | ||||
| Negative | 67 | 39 | 28 | 0.019 |
| Positive | 59 | 22 | 37 | |
| TNM stage | ||||
| I - II | 73 | 44 | 29 | 0.002 |
| III - IV | 53 | 17 | 36 | |
Cell lines and transfection
Four lung cancer cell lines (A549, H1299, HCC827, A427) and one human lung epithelial BEAS-2B cell line were purchased from Shanghai Cell Bank of the Chinese Academy of Sciences. All cells were cultured in RPMI 1640 medium (Gibco/Life Technologies, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS) at 37 °C in a humidified atmosphere with 5% CO2. miR-423-3p mimic, mimic negative control (NC), miR-423-3p inhibitor, inhibitor NC (RiboBio, Guangzhou, China) were used for the overexpression and knockdown of miR-423-3p. Lung cancer cells were seeded onto a 6-well plate at 40–50% confluence before indicated transfection. Transfection of cells was conducted using Lipofectamine 2000 Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer’s instructions. Untreated cells were used as control.
RNA extraction and qRT-PCR analysis
Total RNA was extracted from tissues or cells by using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s protocol. RNA concentrations were determined using the NanoDrop. Purified total RNA was reverse transcribed to complementary DNA (cDNA) using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Expression of miR-423-3p was measured by an SYBR Green I Real-Time PCR Kit (GenePharma, Shanghai, China) on an Applied Biosystems 7900 Real-Time PCR system (Applied Biosystems, Foster City, CA). Relative miR-423-3p expression was normalized to the expression of U6 and was quantified with the 2−ΔΔCt methods.
Cell proliferation assay
CCK-8 assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was used to assess the cell proliferation capacity following the manufacturer’s protocol. Transfected cells were seeded into 96-well culture plates (1 × 104 cells/well) at 37 °C in a humidified atmosphere with 5% CO2. Then CCK-8 reagents were added into cells at 0, 24, 48, and 72 h. After incubated for 1 h, the absorbance was measured using a microplate reader (Thermo Fisher Scientific) at 450 nm wavelength. The assay was repeated at least three times at each time point.
Cell migration and invasion assay
Transwell chambers (Multiskan MK3, Thermo, Waltham, MA, USA) with a pore size of 8 μm were used for cell migration and invasion assays. For the migration assay, no Matrigel was used in the top chamber, while for the invasion assay, Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) was pre-coated in the top chamber. After 24 h of transfection, cells (5 × 104 cells/well) were seeded into the upper chamber with serum-free growth medium, and the lower chamber was filled with 500 μl medium with 10% FBS as a chemoattractant. Cells that were adhered to the lower membrane were fixed in 3.7% formaldehyde for 5 min and stained with 0.1% crystal violet for 15 min. Stained cells were counted under a microscope.
Statistical analysis
SPSS 20.0 software (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism 5.0 software (GraphPad Software, Inc., Chicago, USA) were used for statistical analysis. Data are presented as mean ± standard deviation (SD). χ2 test, Student’s t-test or one-way analysis of variance (ANOVA) followed by Tukey’s post-hoc test was applied to evaluate statistical differences. Kaplan-Meier analysis and Cox regression analysis were used to determine the prognostic significance of miR-423-3p. Differences were considered statistically significant when P < 0.05.
Results
Expression of miR-423-3p in lung cancer tissues and cell lines
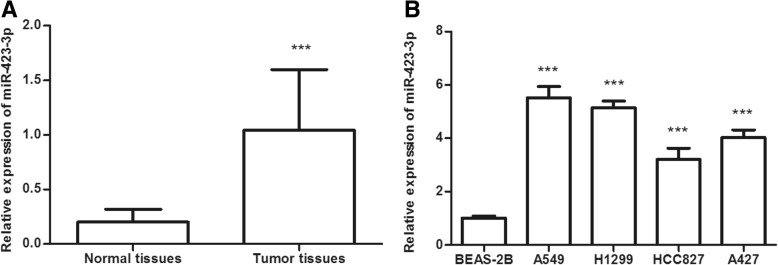
At first, the expression of miR-423-3p was measured in lung cancer tissues and adjacent normal tissues by qRT-PCR analysis. The results revealed that the expression of miR-423-3p was significantly increased in the lung cancer tissues compared with adjacent normal tissues (P < 0.001, Fig. 1a). Then, we assessed the expression of miR-423-3p in four different lung cancer cell lines and one normal cell line. As shown in Fig. 1b, the expression of miR-423-3p was significantly upregulated in all the lung cancer cell lines compared to the normal cell line (P < 0.001). Among the lung cancer cell lines, the highest expression levels of miR-423-3p were observed in A549 cell line, followed by the H1299 cell line. As the two cell lines have a relatively higher expression of miR-423-3p, both of them were used for subsequent experiments.
Fig. 1.
Expression of miR-423-3p in lung cancer tissues and cell lines. a Expression of miR-423-3p was upregulated in lung cancer tissues compared with normal tissues. (***P < 0.001). b Expression of miR-423-3p was increased in lung cancer cell lines (A549, H1299, HCC827, and A427) compared with normal cell line BEAS-2B. (***P < 0.001)
The expression of miR-423-3p was correlated with clinicopathological features of lung cancer patients
In addition, the association between miR-423-3p expression and clinical features of lung cancer patients was analyzed. All the patients were divided into a low expression of the miR-423-3p group and high expression group according to the mean miR-423-3p expression value used as a cutoff. Using the χ2 test, we found upregulation of miR-423-3p was significantly associated with lymph node metastasis (P = 0.019) and tumor, node, and metastasis (TNM) stage (P = 0.002) (Table 1). However, the expression of miR-423-3p has no significant association with other features, such as age, gender, tumor size, and tumor differentiation (all P > 0.05, Table 1).
miR-423-3p was correlated with poor prognosis in lung cancer patients
We then used Kaplan-Meier analysis to evaluate the prognostic significance of miR-423-3p in lung cancer according to the miR-423-3p expression and overall survival information of lung cancer patients. The results showed that the 5-year overall survival rates of lung cancer patients with high expression of miR-423-3p were shorter than those with low expression of miR-423-3p (log rank test P = 0.001, Fig. 2). Next, multivariate Cox’s hazard regression analysis results indicated miR-423-3p expression level (HR = 2.217, 95%CI = 1.344–3.659, P = 0.002) and TNM stage (HR = 0.585, 95%CI = 0.357–0.959, P = 0.034) are independent prognostic factors for 5-year overall survival in lung cancer patients (Table 2).
Fig. 2.
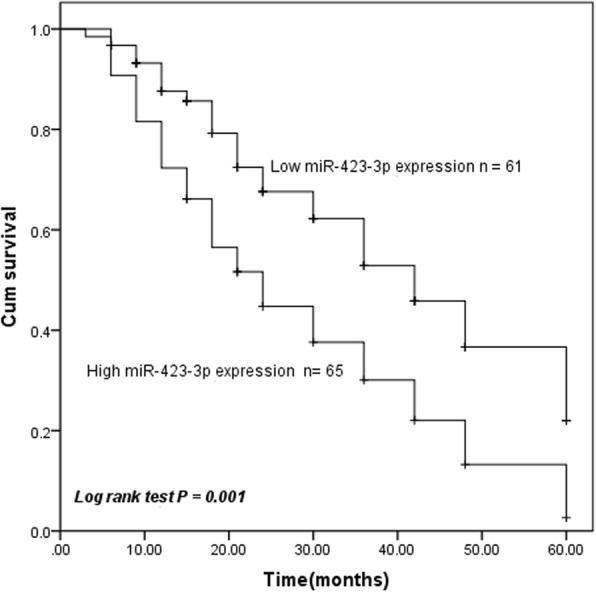
Upregulation of miR-423-3p correlated with poor prognosis of lung cancer patients according to the result of the Kaplan-Meier method
Table 2.
Multivariate Cox analysis of clinical parameters in relation to overall survival
| Characteristics | Multivariate analysis | ||
|---|---|---|---|
| HR | 95% CI | P | |
| miR-423-3p | 2.217 | 1.344–3.659 | 0.002 |
| Age | 0.981 | 0.619–1.553 | 0.933 |
| Gender | 0.945 | 0.597–1.494 | 0.808 |
| Tumor size (cm) | 1.445 | 0.918–2.273 | 0.111 |
| Differentiation | 1.032 | 0.637–1.671 | 0.898 |
| Lymph node metastasis | 1.158 | 0.726–1.847 | 0.539 |
| TNM stage | 0.585 | 0.357–0.959 | 0.034 |
miR-423-3p promoted lung cancer cell proliferation, migration, and invasion
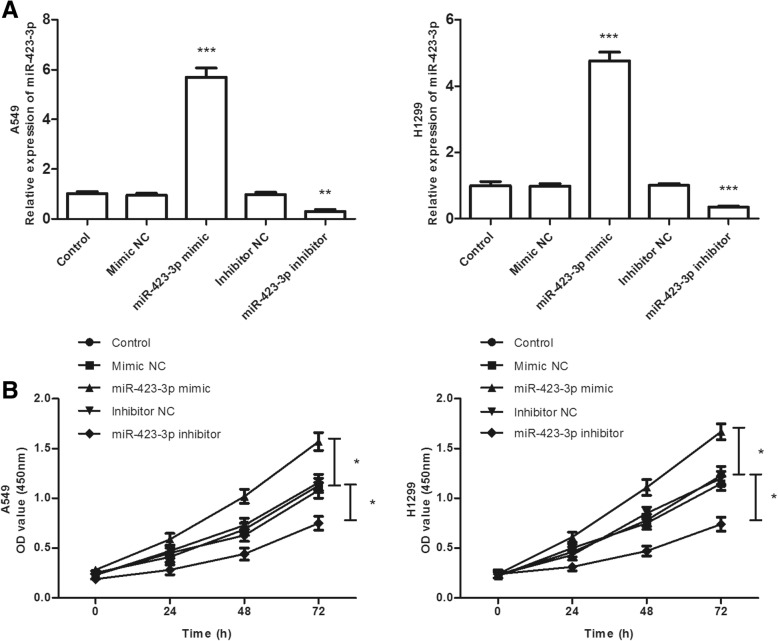
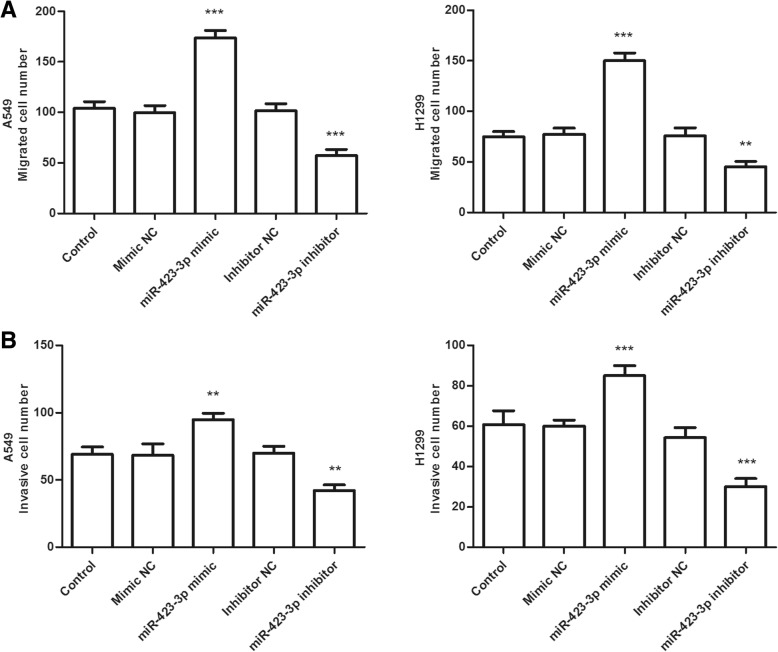
To further explore the functional role of miR-423-3p in lung cancer, miR-423-3p mimic or miR-423-3p inhibitor was transfected into A549 and H1299 cells. As displayed in Fig. 3a, miR-423-3p expression was significantly increased by miR-423-3p mimics and decreased by miR-423-3p inhibitors in A549 and H1299 cells (all P < 0.01). Then we used the CCK-8 assay to determine the cell proliferation capacity. The results showed that the overexpression of miR-423-3p by miR-423-3p mimic promoted cell proliferation, while inhibition of miR-423-3p suppressed cell proliferation (P < 0.05, Fig. 3b). Additionally, Transwell migration and invasion assays results indicated that overexpression of miR-423-3p caused a promotion of the migratory and invasive capacities, whereas downregulation of miR-423-3p caused an inhibition of the migratory and invasive capacities of A549 and H1299 cells (all P < 0.01,Fig. 4a and b).
Fig. 3.
Overexpression of miR-423-3p promotes cell proliferation, while inhibition of miR-423-3p suppresses cell proliferation of A549 and H1299 cells. a Expression of miR-423-3p in mimic NC, miR-423-3p mimic, inhibitor NC, or miR-423-3p inhibitor-transfected cells. (**P < 0.01, ***P < 0.001). b Effects of miR-423-3p on lung cancer cell proliferation in A549 and H1299 cells. (*P < 0.05)
Fig. 4.
Overexpression of miR-423-3p promotes cell migration and invasion, while inhibition of miR-423-3p suppresses cell migration and invasion in A549 and H1299 cells. a Effects of miR-423-3p on lung cancer cell migration. (**P < 0.01, ***P < 0.001). b Effects of miR-423-3p on lung cancer invasion. (**P < 0.01, ***P < 0.001)
Discussion
In this study, miR-423-3p was upregulated in lung cancer tissues compared with that in adjacent normal tissues, and in lung cancer cells compared with normal lung epithelial BEAS-2B cell. Overexpression of miR-423-3p in lung cancer tissues was identified to be significantly associated with lymph node metastasis, TNM stage, and poor prognosis of lung cancer patients. Furthermore, miR-423-3p was revealed to function as an oncogene in lung cancer by promoting cell proliferation, migration, and invasion. Taken together, the present results suggest that miR-423-3p functions as an oncogene in lung cancer and may potentially be utilized as a prognostic indicator.
Although substantive progress has been made in diagnosis and targeted therapy for lung cancer, the 5-year overall survival rate of lung cancer is still fairly low [19, 20]. Searching for effective biomarkers for early diagnosis, prognosis, and therapeutic targets for the treatment of lung cancer is still essential. Increasing studies have identified that molecular markers including miRNAs play an important role in the diagnosis and/or prognosis in various cancers [21–24]. For instance, miR-23a as one of the most extensively studied miRNAs in various cancers plays an essential role in the application of cancer diagnosis, prognosis, and treatment [23]. A study indicated that miR-145 was significantly downregulated in hepatocellular carcinoma tissues and had prognostic value in hepatocellular carcinoma [24]. These studies demonstrated that miRNAs have clinical significance for the treatment of cancers.
Some miRNAs have been identified as an oncogene or suppressor gene and involved in the progression of lung cancer [25]. For instance, miR-1269a acts as an onco-miRNA in non-small cell lung cancer (NSCLC) and promotes cancer cell growth by downregulating the expression of SOX6 [26]. miR-140-3p was indicated as a tumor suppressor and had the prognostic and therapeutic role in squamous cell lung cancer [27]. The expression of miR-218 was found significantly downregulated in lung cancer tissues and it acted as a tumor suppressor in lung cancer by targeting IL-6/STAT3 and negatively correlated with poor prognosis [28]. miR-423-3p, located on chromosome 17q11, was reported as an oncogene in several types of cancers, such as breast cancer, head and neck squamous cell carcinoma, and endometrial cancer [29–31]. In the present study, we examined the expression of miR-423-3p and explored its potential role in lung cancer. The results suggested that miR-423-3p was upregulated in lung cancer tissues and cell lines. Moreover, we found miR-423-3p was associated with lymph node metastasis and TNM stage. These results suggested that miR-423-3p may be an oncogene and is involved in the development and progression of lung cancer, which is consistent with other previous studies.
To explore the clinical prognostic significance of miR-423-3p in lung cancer, we performed Kaplan-Meier curve and Cox regression analyses according to the clinical features and overall survival information of lung cancer patients. The results indicated that lung cancer patients with tumors expressing high profiles of miR-423-3p exhibit poorer survival outcome. And miR-423-3p is an independent prognostic factor for overall survival of lung cancer patients. These results indicated that miR-423-3p is an independent prognostic biomarker for lung cancer.
Previous studies also demonstrated that miR-423-3p is closely associated with tumor progression of several types of cancers [32–35]. For instance, in gastric cancer, miR-423-3p is upregulated in gastric cancer tissues and promotes cell proliferation, migration, and invasion in cell lines and animal models [32]. To explore the biological function of miR-423-3p in the progression of lung cancer, we investigated the effects of miR-423-3p on cell proliferation, migration, and invasion of lung cancer cell lines. We found that overexpression of miR-423-3p significantly promoted lung cancer cell proliferation, migration, and invasion, while inhibition of miR-423-3p suppressed cell proliferation, migration, and invasion, compared with controls. In addition, a study by Li et al. showed miR-423-3p is significantly upregulated in colorectal cancer tissues and enhances cell growth through inhibition of p21Cip1/Waf1 in colorectal cancer [33]. Another study by Guan et al. demonstrated miR-423-3p plays an important oncogenic role in laryngeal carcinoma progression via modulation of AdipoR2 [34]. A recent study showed that overexpression of AdipoR1 and AdipoR2 inhibited migration and invasion in NSCLC [36]. In our current study, we observed that miR-423-3p is involved in the development and progression of lung cancer. Therefore, We speculate miR-423-3p may also function as an oncogene role in lung cancer progression through regulating expression of AdipoR2. The detailed molecular mechanisms of miR-423-3p in lung cancer will be investigated in further investigation.
Taken together, this study indicated miR-423-3p is upregulated in lung cancer and promotes cell proliferation, migration, and invasion. Our data suggested that miR-423-3p functions as an oncogene and may act as a prognostic biomarker and therapeutic target for lung cancer.
Acknowledgments
Not applicable.
Abbreviations
- ANOVA
Analysis of Variance
- FBS
Fetal bovine serum
- NC
Negative control
- NSCLC
Non-small cell lung cancer
- qRT-PCR
Reverse transcription-quantitative polymerase chain reaction
- SD
Standard deviation
- TNM
Tumor, node, and metastasis
- UTR
Untranslated Region
Authors’ contributions
RW and ZS contributed significantly to the design of the experiments manuscript preparation, the analysis of data, and was a major contributor in writing and revising the manuscript. GL, GZ, SS performed the experiments and acquired data. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Ethics approval and consent to participate
All participants signed written informed consent in this study. The research was approved by the Ethics Committee of Weifang Cancer Hospital.
Consent for publication
Not applicable.
Competing interests
The authors declare that they had no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Niksic M, et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391(10125):1023–1075. doi: 10.1016/S0140-6736(17)33326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toh CK, Ong WS, Tan DS, Ng QS, Kanesvaran R, Fong KW, et al. Improved Survival of Advanced Lung Cancer in Singapore Over the Past Decade. Ann Acad Med Singap. 2017;46(9):333–338. [PubMed] [Google Scholar]
- 3.Walters S, Maringe C, Coleman MP, Peake MD, Butler J, Young N, et al. Lung cancer survival and stage at diagnosis in Australia, Canada, Denmark, Norway, Sweden and the UK: a population-based study, 2004-2007. Thorax. 2013;68(6):551–564. doi: 10.1136/thoraxjnl-2012-202297. [DOI] [PubMed] [Google Scholar]
- 4.Ni D, Xiao Z, Zhong B, Feng X. Multiple Human-Behaviour Indicators for Predicting Lung Cancer Mortality with Support Vector Machine. Sci Rep. 2018;8(1):16596. doi: 10.1038/s41598-018-34945-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao JK, Wu M, Kim CH, Jin ZY, Zhou JY, Han RQ, et al. Jiangsu Four Cancers Study: a large case-control study of lung, liver, stomach, and esophageal cancers in Jiangsu Province, China. Eur J Cancer Prev. 2017;26(4):357–364. doi: 10.1097/CEJ.0000000000000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Wanqing, Zheng Rongshou, Baade Peter D., Zhang Siwei, Zeng Hongmei, Bray Freddie, Jemal Ahmedin, Yu Xue Qin, He Jie. Cancer statistics in China, 2015. CA: A Cancer Journal for Clinicians. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Malka Y, Steiman-Shimony A, Rosenthal E, Argaman L, Cohen-Daniel L, Arbib E, et al. Post-transcriptional 3 -UTR cleavage of mRNA transcripts generates thousands of stable uncapped autonomous RNA fragments. Nat Commun. 2017;8(1):2029. doi: 10.1038/s41467-017-02099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu X, Meng X, Tan F, Jiao Z, Zhang X, Tong H, et al. The regulatory mechanism of miR-543-3p on GLT-1 in a mouse model of Parkinson's disease. ACS chemical neuroscience. 2019. [DOI] [PubMed] [Google Scholar]
- 10.Zhang M, Li H, Zhang Y. Oncogenic miR-744 promotes prostate cancer growth through direct targeting of LKB1. Oncol Lett. 2019;17(2):2257–2265. doi: 10.3892/ol.2018.9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruan L, Chen J, Yang T, Wang P. MicroRNA-186 suppresses lung cancer progression by targeting SIRT6. Cancer biomarkers : section A of Disease markers. 2018;21(2):415–423. doi: 10.3233/CBM-170650. [DOI] [PubMed] [Google Scholar]
- 12.Yu H, Sun J, Jiang S, Xu Y MicroRNA- 490-3p regulates cell proliferation and apoptosis in gastric cancer via direct targeting of AKT1. Experimental and therapeutic medicine. 2019;17(2):1330–6. Epub 2019/01/27. [DOI] [PMC free article] [PubMed] [Retracted]
- 13.Yao R, Zheng H, Wu L, Cai P. miRNA-641 inhibits the proliferation, migration, and invasion and induces apoptosis of cervical cancer cells by directly targeting ZEB1. OncoTargets and therapy. 2018;11:8965–8976. doi: 10.2147/OTT.S190303. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Irmak-Yazicioglu MB. Mechanisms of MicroRNA Deregulation and MicroRNA Targets in Gastric Cancer. Oncology research and treatment. 2016;39(3):136–139. doi: 10.1159/000443224. [DOI] [PubMed] [Google Scholar]
- 15.Xie S, Liu G, Huang J, Hu HB, Jiang W. miR-210 promotes lung adenocarcinoma proliferation, migration, and invasion by targeting lysyl oxidase-like 4. J Cell Physiol. 2019; Epub 2019/01/12. [DOI] [PubMed]
- 16.Men L, Nie D, Nie H. microRNA577 inhibits cell proliferation and invasion in nonsmall cell lung cancer by directly targeting homeobox A1. Mol Med Rep. 2019; Epub 2019/01/11. [DOI] [PubMed]
- 17.Li W, Cui Y, Wang D, Wang Y, Wang L. MiR-141-3p functions as a tumor suppressor through directly targeting ZFR in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;509(3):647–656. doi: 10.1016/j.bbrc.2018.12.089. [DOI] [PubMed] [Google Scholar]
- 18.Zhu Y, Li T, Chen G, Yan G, Zhang X, Wan Y, et al. Identification of a serum microRNA expression signature for detection of lung cancer, involving miR-23b, miR-221, miR-148b and miR-423-3p. Lung Cancer. 2017;114:6–11. doi: 10.1016/j.lungcan.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 19.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 20.Torre LA, Siegel RL, Jemal A. Lung Cancer Statistics. Adv Exp Med Biol. 2016;893:1–19. doi: 10.1007/978-3-319-24223-1_1. [DOI] [PubMed] [Google Scholar]
- 21.Townsend MH, Ence ZE, Felsted AM, Parker AC, Piccolo SR, Robison RA, et al. Potential new biomarkers for endometrial cancer. Cancer Cell Int. 2019;19:19. doi: 10.1186/s12935-019-0731-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu W, Ouyang S, Zhou Z, Wang M, Wang T, Qi Y, et al. Identification of genes associated with cancer progression and prognosis in lung adenocarcinoma: Analyses based on microarray from Oncomine and The Cancer Genome Atlas databases. Molecular genetics & genomic medicine. 2018; Epub 2018/12/18. [DOI] [PMC free article] [PubMed]
- 23.Wang Ning, Tan Hor-Yue, Feng Yi-Gang, Zhang Cheng, Chen Feiyu, Feng Yibin. microRNA-23a in Human Cancer: Its Roles, Mechanisms and Therapeutic Relevance. Cancers. 2018;11(1):7. doi: 10.3390/cancers11010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li P, Fan H, He Q. Investigation of the clinical significance and prognostic value of microRNA-145 in human hepatocellular carcinoma. Medicine. 2018;97(51):e13715. doi: 10.1097/MD.0000000000013715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Zi-Qing, Wu Chang-An, Cheng Yi-Xing. Prognostic Value of microRNA-133a Expression and Its Clinicopathologic Significance in Non-Small Cell Lung Cancer: A Comprehensive Study Based on Meta-Analysis and the TCGA Database. Oncology Research and Treatment. 2018;41(12):762–768. doi: 10.1159/000492343. [DOI] [PubMed] [Google Scholar]
- 26.Jin RH, Yu DJ, Zhong M. MiR-1269a acts as an onco-miRNA in non-small cell lung cancer via down-regulating SOX6. Eur Rev Med Pharmacol Sci. 2018;22(15):4888–4897. doi: 10.26355/eurrev_201808_15625. [DOI] [PubMed] [Google Scholar]
- 27.Huang Haitao, Wang Yuxuan, Li Qin, Fei Xiaoyan, Ma Haitao, Hu Rongkuan. miR-140-3p functions as a tumor suppressor in squamous cell lung cancer by regulating BRD9. Cancer Letters. 2019;446:81–89. doi: 10.1016/j.canlet.2019.01.007. [DOI] [PubMed] [Google Scholar]
- 28.Yang Y, Ding L, Hu Q, Xia J, Sun J, Wang X, et al. MicroRNA-218 functions as a tumor suppressor in lung cancer by targeting IL-6/STAT3 and negatively correlates with poor prognosis. Mol Cancer. 2017;16(1):141. doi: 10.1186/s12943-017-0710-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao H, Gao A, Zhang Z, Tian R, Luo A, Li M, et al. Genetic analysis and preliminary function study of miR-423 in breast cancer. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36(6):4763–4771. doi: 10.1007/s13277-015-3126-7. [DOI] [PubMed] [Google Scholar]
- 30.Hui A. B.Y., Lenarduzzi M., Krushel T., Waldron L., Pintilie M., Shi W., Perez-Ordonez B., Jurisica I., O'Sullivan B., Waldron J., Gullane P., Cummings B., Liu F.-F. Comprehensive MicroRNA Profiling for Head and Neck Squamous Cell Carcinomas. Clinical Cancer Research. 2010;16(4):1129–1139. doi: 10.1158/1078-0432.CCR-09-2166. [DOI] [PubMed] [Google Scholar]
- 31.Boren T, Xiong Y, Hakam A, Wenham R, Apte S, Wei Z, et al. MicroRNAs and their target messenger RNAs associated with endometrial carcinogenesis. Gynecol Oncol. 2008;110(2):206–215. doi: 10.1016/j.ygyno.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 32.Kong P, Zhu X, Geng Q, Xia L, Sun X, Chen Y, et al. The microRNA-423-3p-Bim Axis Promotes Cancer Progression and Activates Oncogenic Autophagy in Gastric Cancer. Molecular therapy : the journal of the American Society of Gene Therapy. 2017;25(4):1027–1037. doi: 10.1016/j.ymthe.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li HT, Zhang H, Chen Y, Liu XF, Qian J. MiR-423-3p enhances cell growth through inhibition of p21Cip1/Waf1 in colorectal cancer. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology. 2015;37(3):1044–1054. doi: 10.1159/000430230. [DOI] [PubMed] [Google Scholar]
- 34.Guan G, Zhang D, Zheng Y, Wen L, Yu D, Lu Y, et al. microRNA-423-3p promotes tumor progression via modulation of AdipoR2 in laryngeal carcinoma. Int J Clin Exp Pathol 2014;7(9):5683–5691. Epub 2014/10/23. [PMC free article] [PubMed]
- 35.Lin J, Huang S, Wu S, Ding J, Zhao Y, Liang L, et al. MicroRNA-423 promotes cell growth and regulates G(1)/S transition by targeting p21Cip1/Waf1 in hepatocellular carcinoma. Carcinogenesis. 2011;32(11):1641–1647. doi: 10.1093/carcin/bgr199. [DOI] [PubMed] [Google Scholar]
- 36.Cui E, Guo H, Shen M, Yu H, Gu D, Mao W, et al. Adiponectin inhibits migration and invasion by reversing epithelialmesenchymal transition in nonsmall cell lung carcinoma. Oncol Rep. 2018;40(3):1330–1338. doi: 10.3892/or.2018.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.