The nucleolus is a subnuclear compartment with key roles in rRNA synthesis and ribosome biogenesis, complex processes that require hundreds of proteins and factors. Alterations in nucleolar morphology and protein content have been linked to the control of cell proliferation and stress responses and, recently, further implicated in cell senescence and ageing.
KEYWORDS: NOL12, nucleolar stress, senescence, p53
ABSTRACT
The nucleolus is a subnuclear compartment with key roles in rRNA synthesis and ribosome biogenesis, complex processes that require hundreds of proteins and factors. Alterations in nucleolar morphology and protein content have been linked to the control of cell proliferation and stress responses and, recently, further implicated in cell senescence and ageing. In this study, we report the functional role of NOL12 in the nucleolar homeostasis of human primary fibroblasts. NOL12 repression induces specific changes in nucleolar morphology, with increased nucleolar area but reduced nucleolar number, along with nucleolar accumulation and increased levels of fibrillarin and nucleolin. Moreover, NOL12 repression leads to stabilization and activation of p53 in an RPL11-dependent manner, which arrests cells at G2 phase and ultimately leads to senescence. Importantly, we found NOL12 repression in association with nucleolar stress-like responses in human fibroblasts from elderly donors, disclosing it as a biomarker in human chronological aging.
INTRODUCTION
The nucleolus is a subnuclear multifunctional compartment with a central role in ribosome biogenesis and RNA-processing events, and it is also involved in the sensing of cellular stress and in cell cycle regulation (1–3). The main function of the nucleolus is the synthesis of rRNA and its complex processing and coassembly with ribosomal proteins into ribosome subunits (4). RNA polymerase I (RNA Pol I) synthesizes a precursor rRNA transcript (47S pre-rRNA in humans) that is processed and modified (including 2′-O-methylation and pseudouridylation) into 28S, 18S, and 5.8S rRNAs. These three rRNAs, together with the 5S rRNA synthesized by RNA Pol III in the nucleoplasm, are assembled with ribosomal proteins into large and small ribosomal subunits to be exported independently to the cytoplasm (4). Ribosome biogenesis is one of the most energy-consuming processes in a cell, and it is highly regulated, so that protein synthesis potential (ribosome levels) matches energy supply to ensure proper cellular proliferation and cell growth (5). Mammalian cells have the ability to sense extreme variations in their internal and external environments and frequently respond accordingly with cell cycle arrest or apoptosis. p53 is a tumor suppressor protein, with crucial functions in protecting genome integrity upon cellular stress (6). Direct and localized micropore UV irradiation of cell nucleoli was shown to induce p53 stabilization. However, cells could tolerate a large amount of DNA damage without inducing a p53 response if this DNA damage was not localized and if the nucleoli were disrupted (7). Furthermore, in addition to DNA damage, several other stresses, including chemotherapeutic drugs and inhibition of RNA Pol I transcription by low doses of actinomycin D (ActD), interfere with nucleolar homeostasis and promote a p53-dependent nucleolar stress response (3). Under normal conditions, MDM2, an E3 ubiquitin ligase, prevents p53 stabilization through proteasome-mediated degradation. If nucleolar stress occurs or ribosome biogenesis is perturbed by ribosomal protein deficiency or oncogenic overstimulation, the intermediate ribosomal assembly complex 5S RNP, comprised of RPL11, RPL5, and 5S rRNA, directly binds and represses MDM2 to stabilize p53 (8–11). Also, both replicative stress (with delayed rRNA processing) and oncogenic stress (with accelerated rRNA transcription) were shown to promote the association of the 5S RNP complex with MDM2, leading to p53 stabilization (12).
Nucleoli have been shown to behave like liquid droplets (13, 14), where proteins and other molecules that participate in ribosome biogenesis dynamically self-segregate into the three microscopically recognized nucleolar regions: dense fibrillar component (DFC), fibrillar center (FC), and granular component (GC). Therefore, alterations in nucleolar structure are expected to closely reflect changes in the nature or dynamic partition of enzymatic nucleolar reactions. For decades, pathologists have recognized the correlation between changes in nucleolar size and/or number and tumor aggressiveness (15). In our previous studies, we identified viriato, the single Drosophila member of the NOL12/Nop25 gene family, as a crucial regulator of nucleolar architecture (16), as also described for rat Nop25 (17). The yeast NOL12 homologue Rrp17 was shown to function as a 5′-to-3′ RNA exonuclease for processing of the internal transcribed spacer 1 (ITS1) region of pre-rRNA during ribosome biogenesis (18, 19). Human NOL12 was shown to be required for pre-rRNA ITS1 processing, in particular for cleavage of site 2 (20, 21), but its putative 5′-to-3′ RNA exonucleolytic activity has not yet been ascertained. Interestingly, NOL12 colocalized with DNA repair proteins, such as Dhx9 and TOPBP1, and was required for HCT116 cells to recover from DNA stress (21). In this colon cancer cell line, p53 stabilization was observed, but it was not required for cell cycle arrest or apoptosis (21). We also previously found that viriato is a novel transcriptional target of Drosophila Myc with a crucial function in ensuring a coordinated nucleolar response to dMyc-induced tissue growth (16). Furthermore, through a retina-targeted double RNA interference (RNAi) screen, we identified a genetic interaction between viriato and several Drosophila transforming growth factor β (TGF-β) signaling gene members (22). This led us to study and implicate TGF-β/activin signaling in the regulation of nucleolar biogenesis and cell growth in Drosophila salivary glands (23). Furthermore, we also disclosed that, during retina development, viriato knockdown induced an increase of p53-independent, caspase-mediated apoptotic cell death (16). Overall, our analysis of Drosophila Viriato suggested a potential novel link between structural/functional changes in the nucleolus and cell proliferation and apoptosis. Nevertheless, the putative role of p53 activation in response to nucleolar stress induced by Viriato/NOL12 knockdown awaited further analysis.
Using primary human fibroblasts to investigate the functional role of human NOL12, we here show that NOL12 is important for nucleolar homeostasis, regulating its structure and the nucleolar levels of the multifunctional fibrillarin and nucleolin proteins. Moreover, we found NOL12 depletion to induce strong p53 activation, which at the mechanistic level requires the function of MDM2 inhibitor 60S ribosomal protein L11 and which causes G2 arrest. Importantly, we show that NOL12 repression, either experimental or age associated, leads to p53-driven senescence, suggesting an important role for NOL12 in replicative and chronological aging and its potential as aging biomarker.
RESULTS
NOL12 regulates nucleolar structure and the protein levels of fibrillarin and nucleolin.
To investigate the functional role of NOL12 at the nucleolus, we started by evaluating the NOL12 localization pattern in human primary dermal fibroblasts (HDFs) from neonatal foreskin by immunostaining (Fig. 1A; see Fig. S1A in the supplemental material). We observed that NOL12 localization is mainly restricted to the nucleolus, partially colocalizing with the fibrillarin RNA methyltransferase at the DFC compartment and with the nucleolin RNA-binding protein that also localizes to the GC (Fig. 1A) (24, 25). To gain insight into the functional role of NOL12 in neonatal HDF, we efficiently depleted NOL12 by about 80% at both transcript and protein levels (Fig. S1B and C). Importantly, the NOL12 nucleolar immunolocalization pattern observed was specific, as it was abolished following NOL12 small interfering RNA (siRNA [siNOL12])-mediated depletion (Fig. S1A).
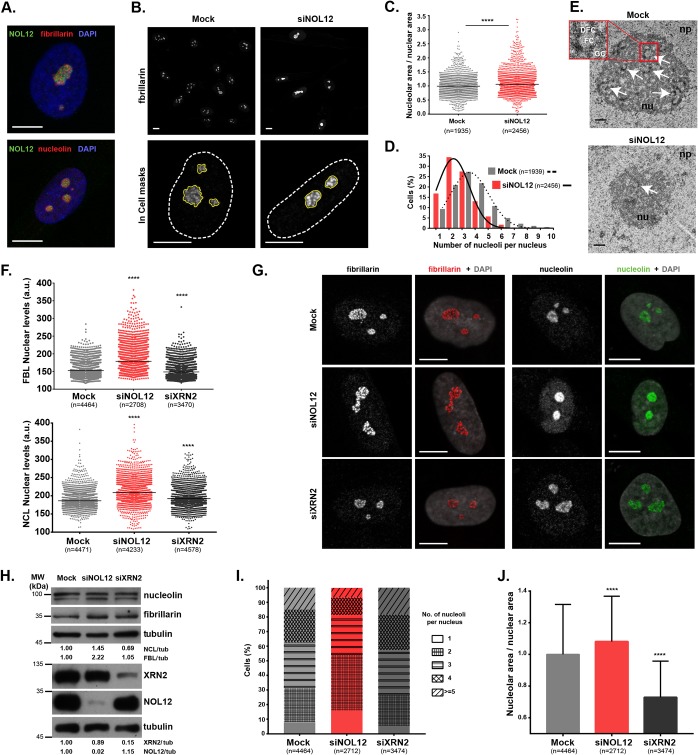
FIG 1.
NOL12 repression induces a specific nucleolar stress response in human untransformed cells. (A) NOL12 immunolocalization pattern in neonatal dermal fibroblasts (green) and colocalization with fibrillarin and nucleolin nucleolar markers (red). DAPI was used for DNA staining (blue). (B) Fibrillarin immunostaining (grayscale) in control (mock-depleted) and NOL12 siRNA-depleted (siNOL12) cells. In the nuclear magnifications (×63; bottom), the white dashed and the yellow solid lines represent the masks used to define and measure nuclear and nucleolar areas, respectively. (C) Ratios between nucleolar and nuclear areas. Each dot represents the value for a single cell. Horizontal lines represent the mean values normalized to those of mock-treated controls. (D) Histogram and respective distribution curves for the percentages of mock- and siNOL12-treated cells exhibiting total numbers of nucleoli as indicated. (E) Ultrastructures of mock- and siNOL12-treated nucleoli accessed by transmission electron microscopy. Representative micrographs, at 20,000× magnification, are shown. Arrows indicate FC/DFC units. The inset is a 6.6× magnification of a nucleolar unit containing well defined FC, DFC, and GC compartments. Scale bars, 0.5 µm. np, nucleoplasm; nu, nucleolus; FC, fibrillar center; DFC, dense fibrillar component; GC, granular component. (F) Scatter plots of the mean pixel intensities of fibrillarin (FBL) and nucleolin (NCL) nuclear levels in mock-, siNOL12-, and siXRN2-treated cells. Each dot represents the value for a single cell, and horizontal lines represent the mean values. a.u., arbitrary units. (G) Immunostaining of fibrillarin and nucleolin (grayscale/red and grayscale/green, respectively) in mock-, siNOL12-, and siXRN2-treated cells. Nuclei were stained with DAPI (gray). (H) Cell extracts from mock-, siNOL12-, and siXRN2-treated neonatal fibroblasts were immunoblotted for nucleolin, fibrillarin, XRN2, and NOL12. Tubulin levels were used for the loading control. Protein levels were normalized to those of the mock-treated control. MW, molecular weight. (I) Stacked bars showing the distribution of the total numbers of nucleoli per nucleus in mock-, siNOL12-, and siXRN2-treated cells. Different patterns in bars represent different numbers of nucleoli per nucleus. (J) Ratios of nucleolar and nuclear areas in mock-, siNOL12-, and siXRN2-treated cells. Values are means ± standard deviations (SD) normalized to the mean value for the control. Scale bars represent 10 µm in panels A, B, and G. n = total number of cells analyzed. ****, P ≤ 0.0001, by Mann-Whitney and Kruskal-Wallis statistical tests in panels C, F, and J.
Interestingly, we found that NOL12 repression increases both nucleolin and fibrillarin protein levels (Fig. 1B and F to H; Fig. S1D and E). These results are distinct from previous reports showing that fibrillarin, nucleolin, and nucleophosmin repressions do not interfere with the levels of other nucleolar proteins involved in ribosome biogenesis (24, 26, 27). NOL12 knockdown also interfered with nucleolar organization, as evidenced by an increased ratio between nucleolar and nuclear areas (6.2%), along with a decrease in the average number of nucleoli per nucleus (from 3.4 to 2.6) (Fig. 1C and D; Fig. S1F and G). Using transmission electron microscopy (TEM), we observed that, whereas mock-depleted fibroblasts exhibited highly defined nucleoli with the typical amniote tripartite organization (28), nucleoli in NOL12-depleted fibroblasts presented poorly defined FC/DFC units (Fig. 1E).
We then asked if the observed nucleolar phenotype is specific to NOL12 repression or if other regulators of pre-rRNA processing might reveal a similar phenotype. To answer that, we depleted XRN2, a well-characterized nucleolar 5′-to-3′ RNA exonuclease (29–31). However, fibrillarin and nucleolin nuclear levels (Fig. 1F to H), as well as the average number of nucleoli per nucleus (Fig. 1I) were only marginally affected. Unlike the results of NOL12 depletion, XRN2-depleted fibroblasts actually displayed reduced nucleolar-/nuclear-area ratios (Fig. 1J; Fig. S1F and G). No cross-regulation between NOL12 and XRN2 protein levels was observed; therefore, the effects of each repression were specific (Fig. 1H).
Altogether, our results showed that human NOL12 is a nucleolar protein specifically required for the maintenance of nucleolar structure, as well as for the regulation of nucleolar levels of fibrillarin and nucleolin, key players in pre-rRNA processing and ribosome assembly.
NOL12 repression activates the p53-signaling pathway in an RPL11-dependent manner.
Next, we asked whether NOL12 knockdown and the associated disruption of nucleolar structure could induce p53 stabilization, which is elicited by a variety of cellular stresses (32), including RNA Pol I inhibition-driven nucleolar stress (33). Both immunoblot and immunofluorescence quantitative analyses of p53 levels revealed its stabilization upon NOL12 and XRN2 knockdowns, although the level was significantly higher in NOL12 repression (Fig. 2A to C).
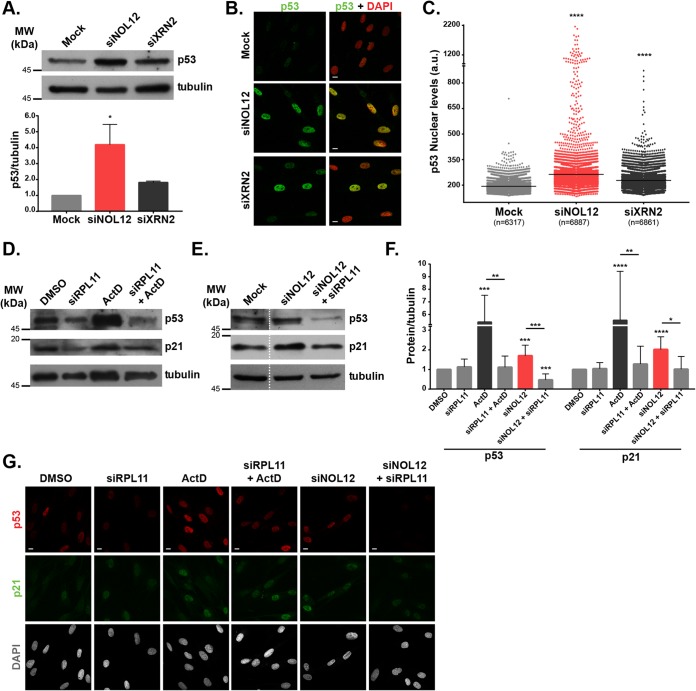
FIG 2.
NOL12 repression activates the p53-signaling pathway in an RPL11-dependent manner. (A) Western blotting of p53 levels in cell extracts from mock-, siNOL12-, and siXRN2-treated neonatal dermal fibroblasts. Tubulin protein levels were used as the loading control. In the graph, bars show mean values ± SD from three independent experiments, normalized to the values for the mock-treated control. *, P ≤ 0.05, by Kruskal-Wallis statistical test. (B) p53 immunostaining (green) of mock-, siNOL12-, and siXRN2-treated human fibroblasts. DNA was stained with DAPI (red). Scale bars, 10 µm. (C) Scatter plot of mean p53 nuclear pixel intensity levels in mock-, siNOL12-, and siXRN2-treated cells. Each dot represents the value for a single cell, and horizontal lines represent the mean values. n = total number of cells analyzed. ****, P ≤ 0.0001, by Kruskal-Wallis statistical test. (D) Western blotting of p53 and p21 protein levels in cell extracts from control or siRPL11-depleted fibroblasts, untreated or treated with 8 nM actinomycin D for 4 h, as indicated. Tubulin was used as the loading control. (E) Western blotting of p53 and p21 protein levels in cell extracts from mock-, siNOL12-, and siNOL12+siRpL11-treated fibroblasts. Tubulin was used as the loading control. (F) p53 and p21 protein levels measured by Western blotting (as shown in panels D and E) from four independent experiments. Bars represent mean values ± SD, normalized to the values for the tubulin loading control and the dimethyl sulfoxide (DMSO)-treated control. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001, by Mann-Whitney statistical test. (G) Immunostaining of p53 (red) and p21 (green) in control, siRpL11-, siNOL12-, and siNOL12+siRpL11-treated fibroblasts, untreated and treated with 8 nM actinomycin D for 4 h. DNA was stained with DAPI (gray). Scale bars, 10 µm.
At low doses, actinomycin D (ActD) specifically inhibits RNA Pol I, preventing the transcription of rDNA into a 47S pre-rRNA primary transcript (34) and inducing alterations in nucleolar structure (2). Mechanistically, it was previously shown in U2OS cells that p53 activation by ActD treatment is dependent on the tripartite complex RPL11-RPL5-5S rRNA binding to MDM2, preventing the MDM2-mediated p53 degradation (11). We confirmed that RPL11 is also required for ActD-induced p53 stabilization in HDF (Fig. 2D, F, and G; Fig. S2). Therefore, we explored the contribution of RPL11 to the p53 activation induced by NOL12 repression. Both immunofluorescence and immunoblot analyses revealed RPL11 to be required for p53 activation upon NOL12 knockdown (Fig. 2E to G). Accordingly, upon NOL12 knockdown, we detected a significant induction of the p53 downstream target p21/CDKN1A (35), which was abolished in the double depletion of NOL12 and RPL11 using siRNAs siNOL12 and siRPL11 (Fig. 2E to G). Altogether, our data suggest that nucleolar stress caused by NOL12 knockdown activates the p53-dependent signaling pathway in an RPL11-dependent manner.
NOL12 repression induces p53-dependent G2 arrest and cellular senescence.
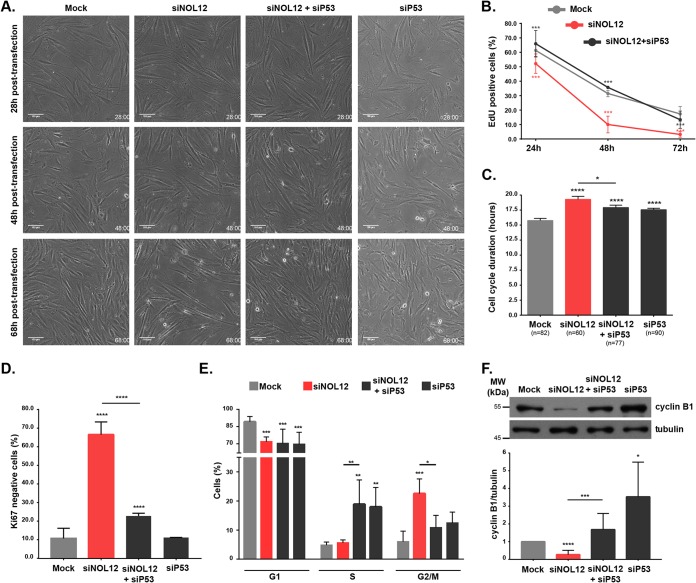
As a consequence of p53 activation, cells can undergo marked phenotypic changes, ranging from increased DNA repair to senescence and apoptosis (32). Independently of the cell fate, p53 activation typically inhibits the cell cycle by inducing the expression of p21/CDKN1A (35). To gain insight into the contribution of stabilized p53 in cell proliferation, we performed time-lapse phase-contrast microscopy of mock-, NOL12-, p53-, and NOL12-and-p53-depleted primary fibroblast cultures (Fig. 3A; Movies S1 to S4). Live-cell imaging revealed a clear reduction in the proliferation rate of NOL12-depleted cell cultures, confirmed by a decreased percentage of living cells that incorporated 5-ethynyl-2′-deoxyuridine (EdU) (Fig. 3B), with cycling cells from NOL12-depleted cell cultures exhibiting a cell cycle delay in comparison to the cycling of the control (Fig. 3C). This cell cycle delay was consistent along three cell generations (from a grandmother cell to the granddaughter cells), although there was a progressive decrease in the number of cycling cells over the recorded period (Fig. S3B). Reduced proliferative capacity was further confirmed by an increased percentage of fixed cells staining negative for the proliferation marker Ki67 (66.5% ± 6.9% [mean ± standard deviation] versus 10.7% ± 5.5% in controls) (Fig. 3D; Fig. S3A). Importantly, the data showed that the proliferation rate of NOL12-depleted cells was rescued by p53 depletion (Fig. 3A to D). To determine which cell cycle phase primarily contributes to the cell cycle delay observed following NOL12 repression, we performed flow cytometry cell cycle profiling. We found a significant increase (3.7-fold) in the G2/M cell subpopulation in NOL12-depleted cell cultures, which was rescued by p53 depletion (Fig. 3E; Fig. S3C). Moreover, cell cultures treated both with siRNA against p53 (siP53) and with siNOL12 and siP53 together (siNOL12+siP53) exhibited an accumulation of S-phase cells, which is likely due to the absence of the breaker role of p53 in cell cycle progression (Fig. 3E). In addition, we measured the levels of cyclin B1, which specifically accumulates at the G2/M transition (36), and surprisingly, we found them to be significantly reduced in NOL12-depleted cell extracts (Fig. 3F). In agreement with the role of p53 as a cell cycle suppressor, p53 repression induced increased levels of cyclin B1 and restored the cyclin B1 levels in NOL12-depleted cell extracts (Fig. 3F).
FIG 3.
NOL12 repression induces p53-dependent G2 arrest. (A) Long-term phase-contrast live-cell imaging of mock-, siNOL12-, siP53-, and siNOL12+siP53-treated fibroblast cell cultures. Representative images captured in movies taken at 28, 48, and 68 h posttransfection are shown. Scale bars,100 µm. (B) Percentages of mock-, siNOL12-, and siNOL12+siP53-treated cells staining positive upon EdU incorporation at 24, 48, and 72 h posttransfection. Data are mean values ± standard errors of the means (SEM) from three independent experiments. ***, P ≤ 0.001, by χ2 statistic test. (C) Cell cycle duration, measured as the interval between mother cell mitosis and daughter cell mitosis, in mock-, siNOL12-, siP53-, and siNOL12+siP53-treated fibroblast cell cultures. Values are means ± SEM; n = total number of cells analyzed. *, P ≤ 0.05; ****, P ≤ 0.0001, by Mann-Whitney statistical test. (D) Percentages of cells staining negative for the proliferation marker Ki67 in mock-, siNOL12-, siP53-, and siNOL12+siP53-treated cell cultures. Values are means ± SD from at least two independent experiments. ****, P ≤ 0.0001, by χ2 statistical test. (E) Percentages of cells in G1, S, and G2/M cell cycle phases in mock-, siNOL12-, siP53-, and siNOL12+siP53-treated fibroblast cultures as determined by flow cytometer cell cycle profiling. Values are means ± SD from three independent experiments. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001, by two-way ANOVA statistical test. (F) Western blot analysis of cyclin B1 levels in cell extracts from mock-, siNOL12-, siP53-, and siNOL12+siP53-treated neonatal fibroblasts. Tubulin levels were used as the loading control, and cyclin B1 levels were normalized to those of mock controls. Values are means ± SD from at least seven independent experiments. *, P ≤ 0.05; ***, P ≤ 0.001; ****, P ≤ 0.0001, by Mann-Whitney statistical test.
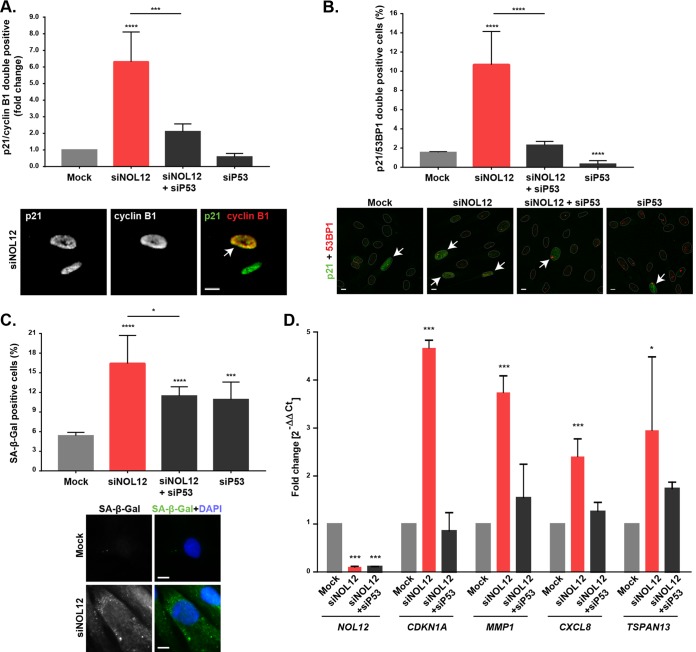
Since the proliferation rate and cyclin B1 levels were significantly decreased following NOL12 repression, we hypothesized that the G2/M subpopulation in the cell cycle profiling most likely reflects a G2 arrest. Interestingly, transient induction of p53 in G2 has been shown to act as the first irreversible step in the onset of senescence, a state of permanent withdrawal from the cell cycle without undergoing cell death (37). Indeed, both live-cell imaging (Fig. 3A; Movies S1 and S2) and immunoblot analysis of cleaved caspase-3 (Fig. S3D) excluded apoptosis as a significant contributor to the decreased proliferation rate in NOL12-depleted cell cultures. Thus, we asked whether NOL12 repression was inducing an accumulation of G2-arrested senescent cells. This permanent arrest is dependent on induction of p53 and p21, resulting in transient nuclear retention of cyclin B1 in the presence of high nuclear p21 levels (37). Additionally, a study in primary fibroblasts showed p21 as being able to retain inactive cyclin B1/Cdk1 complexes in the nucleus (38). Therefore, we stained NOL12-depleted cells for both p21 and cyclin B1 and investigated the presence of double-positive staining. Albeit scarce due to the transient nature of nuclear cyclin B1 retention, we found a 6-fold enrichment of p21/cyclin B1 double-positive nuclei in NOL12 depletion (Fig. 4A). p53 depletion was sufficient to prevent nuclear retention of cyclin B1 in NOL12-depleted cells (Fig. 4A). Furthermore, we quantified the percentages of cells exhibiting senescence-associated markers, namely, β-galactosidase (β-Gal) activity and double positivity for p21 and 53BP1 (a DNA damage response marker) (39). We found significant increases in these markers following NOL12 repression (p21 positive [p21+]/53BP1+, 10.68% ± 3.46% versus 1.55% ± 0.09% in controls; senescence-associated β-Gal positive [SA–β-Gal+], 16.42% ± 4.28% versus 5.38% ± 0.52% in controls), which was rescued upon p53 depletion (Fig. 4B and C). Accordingly, we also found p53-dependent upregulation of the senescence-associated secretome by assessing the mRNA expression of selected genes (Fig. 4D) (39). Taken together, our data strongly support the idea that the cellular senescence induced by NOL12 knockdown is p53 dependent. Notably, we found senescence as the primary cell fate of untransformed cells in response to nucleolar stress, contrary to the induction of p53-dependent and -independent apoptosis previously reported in transformed cells (40).
FIG 4.
NOL12 repression induces p53-dependent senescence. (A) Percentages of cells staining positive for p21/cyclin B1 immunofluorescence in mock-, siNOL12-, siP53-, and siNOL12+siP53-treated fibroblast cultures. In the graph, values are means ± SD from >50,000 cells, and values are shown as fold changes in comparison to the values for mock-treated controls. ***, P ≤ 0.001; ****, P ≤ 0.0001, by χ2 statistical test. The panel below shows a representative image of an siNOL12-depleted cell staining double positive for p21/cyclin B1 (arrow). Scale bar, 10 µm. (B) Percentages of cells double positive for p21/53BP1 immunostaining in mock-, siNOL12-, siP53-, and siNOL12+siP53-treated fibroblast cultures. Values are means ± SD from two independent experiments. ****, P ≤ 0.0001, by χ2 statistical test. The panel below shows representative images for each experimental condition. Nuclear masking is shown in white. Arrows indicate p21/53BP1 double-positive cells. Scale bars, 10 µm. (C) Percentages of senescence-associated β-galactosidase (SA-β-Gal)-positive cells in mock-, siNOL12-, siP53-, and siNOL12+siP53-treated fibroblast cultures. Values are means ± SD from two independent experiments. *, P ≤ 0.05; ***, P ≤ 0.001; ****, P ≤ 0.0001, by χ2 statistical test. The panel below shows representative images of mock- and siNOL12-treated cells staining negative and positive, respectively, for SA–β-Gal. DNA was stained with DAPI (blue). Scale bars, 10 µm. (D) qPCR analysis of transcript levels of NOL12, CDKN1A, MMP1, CXCL8, and TSPAN13 in mock-, siNOL12-, and siNOL12+siP53-treated fibroblast cultures. G6PD and TBP were used as housekeeping genes. Values are means ± SD from four independent experiments, normalized to the values for controls. *, P ≤ 0.05; ***, P ≤ 0.001, by ordinary one-way ANOVA statistical test.
NOL12 repression and nucleolar stress during chronological aging.
Our findings showing a link between NOL12 repression and cellular senescence led us to ask whether regulation of NOL12 plays a role in naturally aged cells and in replicative senescence. In agreement with the widely reported accumulation of senescent cells during chronological and replicative aging (41, 42), we found significantly higher levels of senescence-associated biomarkers in the proliferating fibroblast cultures from elderly than from neonatal donors and in high- versus low-passage-number cells, thus validating their suitability as models of aging (Fig. S4A to C).
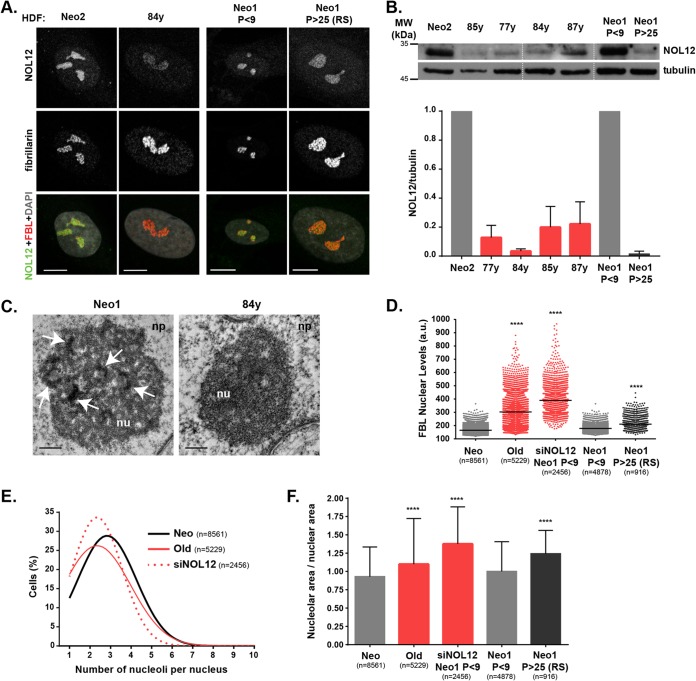
Importantly, we found downregulation of NOL12 protein levels in primary fibroblasts from 77-, 84-, 85-, and 87-year-old donors (<5 early cell culture passages), as well as in replicatively aged cells (Fig. 5A and B). Next, we investigated whether naturally and replicatively aged primary cells with decreased NOL12 levels were under nucleolar stress. Remarkably, nucleoli in elderly donor cells displayed undefined and/or reduced numbers of FC/DFC units (Fig. 5C). Also, both naturally and replicatively senescent cells exhibited alterations in the nucleolar organization similar to those observed in NOL12-depleted cells, namely, upregulation of fibrillarin nuclear levels, decreased numbers of nucleoli per nucleus, and increased ratios between nucleolar/nuclear areas (Fig. 5D to F; Fig. S4D and E). Since NOL12 depletion induces nucleolar stress, ultimately leading to a decreased proliferation rate and accumulation of senescent cells in human primary neonatal fibroblast cultures, we hypothesized that NOL12 overexpression in elderly donor cells should counteract this stress response. Using lentiviral infection, we efficiently overexpressed NOL12 in fibroblasts retrieved from an 84-year-old donor (Fig. S5A). We found NOL12 overexpression to partially rescue the proliferation rate and fibrillarin nuclear levels (Fig. S5B and C). However, other nucleolar-stress-associated phenotypes, such as the ratio between nucleolar/nuclear area and the number of nucleoli per nucleus, were not significantly altered (Fig. S5D and E). Also, the number of senescent cells remained unchanged, as evidenced by quantification of SA–β-Gal-positive cells (Fig. S5F). Thus, by restoring NOL12 protein levels in elderly donor cells, we partially rescued the proliferation rate but not the accumulation of senescent cells. Still, our data disclosed a correlation between NOL12 repression, nucleolar stress, and chronological aging in human primary fibroblasts.
FIG 5.
NOL12 repression and nucleolar stress in human fibroblasts from elderly donors. (A) Immunofluorescence analysis of NOL12 (grayscale/green) and fibrillarin (grayscale/red) levels in human primary dermal fibroblasts (HDFs) from neonatal (Neo2) versus 84-year-old (84y) donors, and low-passage (P)-number (P < 9) versus high-passage-number (P > 25; RS, replicative senescence) neonatal fibroblasts (Neo1). DNA was stained with DAPI (gray). Scale bars, 10 µm. (B) Western blot analysis of NOL12 protein levels in cell extracts from HDFs from neonatal (Neo2) and 85-, 77-, 84-, and 87-year-old donors, as well as from low- and high-passage-number neonatal fibroblasts (Neo1, P < 9, and Neo1, P > 25, respectively). Tubulin was used as the loading control. In the graph, values are means ± SD from two independent experiments, normalized to the values for Neo2 (all red bars) or Neo1 (dark gray bar). (C) Ultrastructural analysis of nucleoli from neonatal and 84-year-old donors by transmission electron microscopy. Representative micrographs, at ×30,000 and ×50,000 magnification, respectively, are shown. Arrows indicate FC/DFC units. Scale bars, 0.5 µm. np, nucleoplasm; nu, nucleolus. (D) Scatter plot shows the mean pixel intensities of fibrillarin nuclear levels in cells from neonatal (Neo1 and Neo2) and elderly (Old; 77-, 84-, 85-, and 87-year-old) donors and NOL12-repressed (siNOL12) and neonatal low- and high-passage-number (Neo1 P < 9 and Neo1 P > 25) cells. Each dot represents the value for a single cell. Horizontal lines represent the mean values. ****, P ≤ 0.0001, by Kruskal-Wallis statistical test. (E) Distribution curves for the total numbers of nucleoli per cell in fibroblast cultures from neonatal (Neo1 and Neo2), elderly (Old; 77-, 84-, 85-, and 87-year-old), and NOL12-repressed (siNOL12) donors. (F) Ratios between nucleolar and nuclear areas in cells from neonatal (Neo1 and Neo2), elderly (Old; 77-, 84-, 85-, and 87-year-old), NOL12-repressed (siNOL12), and neonatal donors at low and high passage numbers (Neo1, P < 9, and Neo1, P > 25, respectively). Values are means ± SD normalized to the value for Neo (red bars) or Neo1 (dark gray bar). ****, P ≤ 0.0001, by Kruskal-Wallis statistical test. RS, replicative senescence.
DISCUSSION
We previously showed that Drosophila Viriato/NOL12 is a nucleolar protein crucial for proper nucleolar structure and dMyc-induced cell growth (16). In this work, we used human primary fibroblasts to further investigate the cellular functions of NOL12.
We examined the nucleolar localization of NOL12 in human primary fibroblasts, finding that it colocalizes with fibrillarin and nucleolin, in accordance with a recent study performed in the HCT116 cell line (21). To investigate the largely elusive role of NOL12 in nucleolar homeostasis, we knocked down NOL12 in neonatal primary fibroblasts, which revealed that NOL12 is required for proper nucleolar organization due to a role in limiting nucleolar size and regulating nucleolar number. Interestingly, repression of NOL11 or hUTP4 in human MCF10A cells was also found to cause a strong reduction in nucleolar number, in correlation with defects in pre-rRNA processing (43). Moreover, a reduction in nucleolar number was recently used in a genome-wide siRNA screen to identify 16 regulators of ribosome biogenesis (44).
In addition, we found NOL12 acting to restrain nucleolar accumulation of nucleolin and fibrillarin. To evaluate whether this pattern of nucleolar stress was specific for NOL12, we knocked down XRN2, another nucleolar 5′-to-3′ RNA exoribonuclease able to localize to the nucleolus (30, 31), which has multiple roles in pre-rRNA cleavage and rRNA maturation (20, 45). We found that in contrast to NOL12 repression, XRN2 depletion actually decreased nucleolar size without modifying the number of nucleoli per cell. Moreover, whereas NOL12 depletion caused significant increases in fibrillarin and nucleolin cellular protein levels, XRN2 did not have that role. Repression of other nucleolar proteins, such as nucleolin (46), fibrillarin (26), and nucleophosmin (27), also did not change the expression levels of different nucleolar proteins.
Stabilization and/or activation of p53 is central in the nucleolar stress response to defects in rRNA synthesis/processing and ribosomal subunit assembly (47), even though p53-independent mechanisms have also been suggested (40, 48, 49). Thus, we assessed p53 protein levels upon NOL12 or XRN2 repression in human fibroblasts. Interestingly, we found significantly increased p53 protein levels in NOL12-depleted cells compared with the levels in mock- or XRN2-depleted cells. The transcriptional activity of p53 also increased, as p21 target protein levels were upregulated. p53 has been described to repress fibrillarin expression at the transcriptional level (50), so we were intrigued by the concomitant increase of both p53 and fibrillarin upon NOL12 knockdown. Nevertheless, we cannot exclude the possibility that in the context of NOL12 repression, p53 activation actually helps to limit an otherwise stronger upregulation of nucleolar fibrillarin. Given that high fibrillarin expression levels are associated with poor prognosis in breast cancer (50), more studies are needed to clarify the mechanisms behind fibrillarin upregulation.
Mechanistically, our results show that activation of the p53 signaling pathway by NOL12 repression is dependent on RPL11 expression. In response to nucleolar stress, RPL11 interacts with MDM2 (or HDM2 in human), inhibiting its ability to regulate p53 degradation. This has been reported for several stress conditions, including the inhibition of the activity of RNA Pol I by low-level actinomycin D treatment (51, 52) or the perturbation of ribosome biogenesis by RPL29 and RPL30 knockdown (53).
We next explored the cellular consequences of p53 activation in NOL12-depleted cells. Phase-contrast live-cell-imaging analysis first indicated enlarged-cell morphology and reduced proliferative capacity in NOL12-depleted cell cultures. These effects were rescued by NOL12 and p53 codepletion, suggesting defective cell proliferation, as previously described for Drosophila vitoRNAi (16). This is not due to apoptosis, as we did not detect cleaved caspase-3 or an increased sub-G1 population by flow cytometry. This is in contrast to nucleolar stress caused by depletion of TIF-IA, a basal transcription initiation factor for RNA Pol I, found to induce p53-dependent apoptosis in embryonic fibroblasts (54). One possibility is that the increase in fibrillarin nucleolar levels in NOL12-depleted cells contributes to biased translation for internal ribosomal entry site (IRES)-containing mRNAs encoding important antiapoptotic proteins, as previously described (50, 55). With apoptosis excluded from contributing to the low proliferation rate of NOL12-depleted cell cultures, we evaluated senescence. Cell cycle profiling and immunodetection of cyclin B1 levels allowed us to conclude that NOL12-depleted cells are arrested in G2 phase in a p53-dependent manner. Interestingly, the increased nucleolar area observed in NOL12-depleted cells is consistent with cells at the G2 phase (56–58). Previous studies also found the repression of nucleolin and nucleostemin to induce G2 arrest (24, 59), even though p53 activation was not investigated. Remarkably, an irreversible G2 arrest, characterized by a transient nuclear accumulation of p21 and cyclin B1, was shown to be the first step in the onset of senescence (37). Accordingly, we found that NOL12-depleted cell cultures are enriched in p21/cyclin B1 double-positive cells. Moreover, Nol12-depleted cell cultures displayed increased numbers of cells positive for senescence-associated biomarkers, SA–β-galactosidase activity, and p21/53BP1 immunostaining and exhibited the senescence-associated secretory phenotype. Therefore, our results show that senescence, rather than apoptosis, is the outcome for untransformed cells in response to NOL12 depletion-driven nucleolar stress. Interestingly, therapeutic inhibition of Pol I transcription by treatment with CX-5461, a specific small-molecule inhibitor of RNA Pol I, was shown to induce cell death in malignant B cell lymphomas but not in normal B cells (60). This bias was also reported in mouse epidermis upon p53 activation induced by MDM2 deletion (61). In contrast, in transformed cells, it was reported that apoptosis mainly configures the response to nucleolar stress (40). During preparation of our manuscript, increased apoptosis upon NOL12 depletion in colon cancer HCT116 cells was reported (21). In contrast to our results, apoptosis was associated with p53-independent G1 arrest and ATR-Chk1 activation (21). It has been shown that, in response to nucleostemin depletion, embryonic fibroblasts and HCT116 cells have different p53-regulated responses (59). At the mechanistic level, whereas transient induction of p53 in untransformed G2 cells is sufficient to induce the onset of senescence (37), HCT116 cancer cells can overcome the G2 arrest 18 h after a DNA-damaging insult, upon checkpoint recovery mediated by the function of the Wip1 phosphatase (62). Therefore, we hypothesize that a similar differential response to p53 activation could explain the distinct phenotypes induced by NOL12 knockdown in primary fibroblasts and HCT116 cells.
After establishing a connection between NOL12 repression and increased senescence, we next asked whether regulation of NOL12 expression plays a role in aging-associated senescence. Remarkably, we found that both low-passage-number fibroblast cultures retrieved from elderly donors or high-passage-number human neonatal fibroblasts (replicative senescence) exhibit significant downregulation of NOL12 expression. Similar to the nucleolar phenotypes observed following NOL12 repression, both replicatively and naturally aged cells exhibited increased nucleolar area, lower numbers of nucleoli per nucleus, higher nuclear fibrillarin levels, and altered nucleolar ultrastructure. In parallel, recent studies characterized fibrillarin upregulation and increased nucleolar size as hallmarks of aging across species (63, 64). In Caenorhabditis elegans, fibrillarin knockdown reduced nucleolar size and ribosome biogenesis, and these alterations correlated with extended life span (64). Furthermore, prematurely aged cells from HGPS (Hutchinson-Gilford progeria syndrome) donors exhibited bigger but fewer nucleoli (63). By restoring NOL12 protein levels in fibroblasts from elderly donors, we expected to rescue their senescent phenotype. Although we found a partial rescue in the number of proliferative cells and a mild reduction of fibrillarin levels, the number of senescent cells and the nucleolar organization remained unchanged, likely due to the complexity of the senescence process. Eventually, cells would need more time in culture in order to allow the dilution of the senescent cells and their paracrine effects by the more fit proliferating cells expressing NOL12. While the recovery of NOL12 protein levels was not sufficient to revert senescence in cell cultures from elderly donors, FoxM1, a transcription factor involved in G2/M transition (65, 66), was recently reported as able to do it (39). Interestingly, RNA sequencing profiling of neonatal FoxM1-depleted cells and of octogenarian FoxM1-overexpressing cells showed that NOL12 expression is responsive to FoxM1 modulation (39), suggesting that cell proliferation is closely linked to nucleolar homeostasis. Further work will be needed to address the functions of NOL12 during aging, in particular the mechanism responsible for its aging-associated repression and its contribution to ribosome biogenesis and p53-dependent senescence.
MATERIALS AND METHODS
Cell culture.
Human dermal fibroblasts retrieved from skin biopsy specimens of Caucasian males reported as healthy were acquired from cell biobanks as summarized in Table S1 in the supplemental material. Cells were seeded at 1 to 1.5 × 104 cells per cm2 of growth area in minimal essential medium Eagle-Earle (MEM) supplemented with 15% fetal bovine serum (FBS), 2.5 mM l-glutamine, and antibiotic-antimycotic (1:100) (all Gibco; Life Technologies/Thermo Scientific, Carlsbad, CA). Only early-passage dividing fibroblasts (up to passage 3 to 5) with a cumulative population doubling level (PDL) of <24, well below replicative senescence, were used in all experiments. Cells were grown at 37°C in a humidified atmosphere with 5% CO2. For immunostaining experiments, cells were cultured on coverslips coated with 50 µg/ml fibronectin (Sigma-Aldrich, St. Louis, MO).
Drug treatments.
For RNA polymerase I inhibition, fibroblasts were incubated for 4 h in medium containing 8 nM actinomycin D (Sigma-Aldrich, MO). For siRNA-depleted cells, actinomycin D was added 4 h before cell harvesting or fixation. For apoptosis induction, cells were incubated for 4 h in medium with 5 µM staurosporine (LC Laboratories).
Senescence-associated–β-galactosidase (SA–β-Gal) assay.
Cells were incubated in medium with 100 nM bafilomycin A1 (Sigma-Aldrich, MO) for 90 min to induce lysosomal alkalinization. A concentration of 33 μM of the fluorogenic substrate for β-galactosidase (β-Gal), fluorescein di-β-d-galactopyranoside (Sigma-Aldrich, MO), was then added to the cell culture medium, and incubation carried out for 90 min. Cells were fixed in 4% paraformaldehyde for 15 min, rinsed with phosphate-buffered saline (PBS), and permeabilized for 15 min with 0.1% Triton X-100 in PBS. Finally, cells were counterstained with 1 µg/ml DAPI (4′,6-diamidino-2-phenylindole; Sigma-Aldrich, MO).
siRNA transfection.
For NOL12 depletion, we tested 3 different siRNAs from Sigma (SASI_Hs01_00047858, SASI_Hs01_00047859, and SASI_Hs01_00047860). SASI_Hs01_00047859 depleted NOL12 the most efficiently. The sequences of all siRNAs used in this study (from Sigma-Aldrich) are summarized in Table S2. For efficient depletion of XRN2, we used a mixture of two previously reported siRNAs (20, 29). One hour after their plating, cells were transfected with a final concentration of 45 nM siRNA using Lipofectamine RNAiMax transfection reagent (Thermo Scientific, CA) according to the manufacturer’s instructions. Cells treated under the same conditions with transfection reagent without siRNA were used as controls (mock-depleted cells). Six hours after transfection, the medium was replaced by fresh medium supplemented with 5% FBS, and the next day, by complete MEM. Phenotypes were analyzed and quantified 72 h posttransfection.
Immunostaining.
Fibroblasts were grown on sterilized glass coverslips coated with 50 µg/ml fibronectin (Sigma-Aldrich, MO). Cells were fixed in freshly prepared 2% paraformaldehyde in PBS for 20 min or in 4% paraformaldehyde in PBS for 15 min in the case of 53BP1 and p21 markers. Following fixation, cells were rinsed in PBS, permeabilized with 0.3% Triton X-100 in PBS for 7 min, and blocked in 0.05% Tween 20 in PBS (PBS-T) supplemented with 10% FBS for 1 h at room temperature. Cells were then incubated overnight at 4°C with primary antibodies (Abs) diluted in PBS-T supplemented with 5% FBS as follows: mouse anti-NOL12 Ab (sc-374257; Santa Cruz Biotechnology, CA), 1:800; rabbit antifibrillarin Ab (ab5821; Abcam, Shanghai, China), 1:1,000; rabbit anti-p53 Ab (sc-6243; Santa Cruz Biotechnology), 1:1,000; mouse antifibrillarin Ab (ab4566; Abcam, Shanghai, China), 1:3,000; rabbit antinucleolin Ab (sc-13057; Santa Cruz Biotechnology), 1:500; rabbit anti-53BP1 Ab (#4937; Cell Signaling Technology, Danvers, MA), 1:100; mouse anti-p21 Ab (sc-6246; Santa Cruz Biotechnology, CA), 1:1,000; rat anti-α-tubulin Ab (Sigma-Aldrich), 1:100; rabbit anti-Ki67 Ab (ab15580; Abcam, Shanghai, China), 1:1,500; and rabbit anti-cyclin B1 Ab (#4138; Cell Signaling Technology, USA), 1:100. Secondary antibodies conjugated with Alexa Fluor 488, 568, and 647 were diluted 1:1,500 in PBS-T supplemented with 5% FBS, and coverslips were incubated for 45 min at room temperature. DAPI (Sigma-Aldrich) was used at 1 µg/ml for nuclear staining, and coverslips were then mounted in slides with mounting solution (90% glycerol, 0.5% N-propyl gallate, and 20 nM Tris, pH 8).
EdU incorporation assay.
To assess the proliferative activity of human primary fibroblasts, 5-ethynyl-2′-deoxyuridine (EdU) (20 μM/well; Invitrogen, Darmstadt, Germany) was added to cells for 3 h before each time point monitored (24, 48, and 72 h). Coverslips were fixed in 4% paraformaldehyde (PFA) for 15 min, washed with 10% FBS, and permeabilized with 0.3% Triton X-100 in PBS for 7 min. Instructions from the Click-iT EdU Alexa Fluor 594 imaging kit (Invitrogen) were then followed.
Microscopy and image analysis.
Slides were analyzed under an HC PL APO CS 63×/1.30-numeric-aperture (NA) glycerin (21°C) objective in a Leica TCS SP5 II laser scanning confocal microscope (Leica Microsystems, Germany). Images were acquired at 1,024- by 1,024-pixel resolution and edited using Adobe Photoshop CS4 extended version 11.0. All image fields used for quantitative analyses were acquired in an IN Cell Analyzer 2000 (GE Healthcare, United Kingdom) equipped with a Photometrics CoolSnap K4 camera. IN Cell Investigator software was used for measuring nucleolus and nucleus parameters (numbers and areas); nuclear area was measured in the DAPI channel, whereas the total nucleolar area and the number of nucleoli per nucleus were measured in the fibrillarin channel. The mean pixel intensities of fibrillarin, nucleolin, and p53 (referred to as “nuclear protein levels”) were measured based on masking defined by the DAPI channel. In the SA–β-Gal activity assay, cells displaying >5 fluorescent granules were considered positive.
Western blotting.
Cell pellets were resuspended in lysis buffer (150 nM NaCl, 10 nM Tris-HCl, pH 7.4, 1 nM EDTA, 1 nM EGTA, 0.5% IGEPAL) and instantaneously frozen in liquid nitrogen. Clarified lysates were quantified for protein content by the Lowry method (DC protein assay; Bio-Rad, CA). Amounts of 20 μg of total extract (except for the detection of cleaved caspase-3, for which we used 40 μg of total extract) were loaded in SDS-PAGE gels and transferred into nitrocellulose membranes for Western blot analysis. Membranes were blocked with TBS-T containing 5% low-fat milk for 1 h at room temperature. Primary antibodies were diluted in TBS-T containing 2% low-fat milk as follows: mouse anti-NOL12 Ab (sc-374257), 1:1,000; rabbit anti-p53 Ab (sc-6243), 1:1,500; mouse anti-p21 Ab (sc-6246), 1:1,000; mouse anti-cyclin B1 Ab (#4135; Cell Signaling Technology, MA), 1:1,500; rabbit anti-RPL11 Ab (ab79352; Abcam), 1:10,00; rabbit antinucleolin Ab (sc-13057), 1:3,000; rabbit anti-cleaved caspase-3 Ab (#9661; Cell Signaling Technology, MA), 1:1,000; mouse antifibrillarin Ab (ab4566), 1:1,000; rabbit anti-XRN2 Ab (A301-103A-T; Bethyl Laboratories, Montgomery, TX), 1:1,000; mouse anti-α-tubulin Ab (Sigma-Aldrich, CA), 1:100,000; and mouse anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) Ab (Protein Tech Group, Inc., IL), 1:30,000. Horseradish peroxidase (HRP)-conjugated goat anti-mouse and anti-rabbit antibodies (sc-2005 and sc-2004, respectively; Santa Cruz Biotechnology, CA) were used at a dilution of 1:3,000 in TBS-T containing 2% low-fat milk. Signals were detected using Clarity Western ECL substrate reagent (Bio-Rad) according to the manufacturer’s instructions. A GS-800 calibrated densitometer (Bio-Rad Laboratories, CA) was used for quantitative analysis of protein levels.
Phase-contrast live-cell imaging.
Fibroblasts were cultured in glass-bottom 35-mm µ-dishes (Ibidi GmbH, Germany) coated with 50 µg/ml fibronectin (Sigma-Aldrich, MO) and transfected with siRNAs. At 24 h posttransfection, cell cultures were imaged on a Zeiss Axiovert 200 M inverted microscope (Carl Zeiss, Germany) equipped with a CoolSnap camera (Photometrics, Tucson, AZ), xy motorized stage, and NanoPiezo z stage under controlled temperature, atmosphere, and humidity. Neighbor fields (20–25) were imaged every 2.5 min for 42 h, using a 20×/0.3-NA A-Plan objective. Stitching of neighboring fields was done using the plug-in Stitch Grid (Stephan Preibisch) from ImageJ/Fiji software. The time between two consecutive mitoses (referred to as “cell cycle duration”) was manually quantified using ImageJ/Fiji software.
Flow cytometry.
Cells were trypsinized, washed twice in cold PBS, and fixed in 70% ethanol at −20°C overnight. Cells were washed twice in cold PBS and then incubated at 37°C for 6 h in propidium iodide (20 µg/ml)–RNase (40 µg/ml) solution. Finally, cells were filtrated into a fluorescence-activated cell sorting (FACS) tube. Data were recorded using the FACSCanto II and analyzed with FlowJo software using the Watson pragmatic algorithm to generate the cell cycle profiles.
Transmission electron microscopy.
Cells were fixed with 2.5% glutaraldehyde (Electron Microscopy Sciences, Hatfield, PA) and 2% paraformaldehyde (Merck, Darmstadt, Germany) in 0.1 M cacodylate buffer (pH 7.4), dehydrated, and embedded in Epon resin (TAAB, Aldermaston, Berkshire, England). Ultrathin sections (40 to 60 nm) were prepared on an RMC ultramicrotome (PowerTome, USA) using diamond knives (DDK, Wilmington, DE). The sections were mounted on 200-mesh copper or nickel grids, stained with uranyl acetate and lead citrate for 15 min each, and examined under a JEOL JEM 1400 TEM (Tokyo, Japan). Images were digitally recorded using an Orious 1100W charge-coupled device (CCD) digital camera (Tokyo, Japan).
qRT-PCR.
Total RNA was isolated from cultured cells using the RNeasy minikit (Qiagen) according to the manufacturer’s instructions. An amount of 1 µg of RNA was reverse transcribed (RT) using the SuperScript III first-strand synthesis supermix for reverse transcription-quantitative PCR (qRT-PCR) (Invitrogen). Quantitative real-time PCR analysis was performed in duplicate in 20-µl reaction mixtures containing iQ SYBR green supermix (Bio-Rad), each gene-specific primer at 125 nM (100 nM for the G6PD primer), and 1 µl of cDNA template previously diluted at 1:5. Cycling conditions in the iCycler iQ5 (Bio-Rad Laboratories) were 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 20 s, and a final extension step at 55°C for 10 s. The cycle threshold (2−ΔΔCT) method was used to quantify the transcript levels of NOL12, CDKN1A, MMP1, CXCL8, and TSPAN13 against the transcript levels of the housekeeping genes TBP and G6PD. Primers were designed to span at least one exon-intron junction (Table S3). Data were analyzed using Bio-Rad iQ5 standard edition (Bio-Rad Laboratories).
Lentiviral plasmids.
NOL12 was amplified from pENTR223.1-NOL12 (clone OCABo5050F031D; ImaGenes) as a BamHI-NOL12-NotI fragment by using the following primers: 5′-CGGGATCCATGGGCCGCAACAAGAAG-3′ and 5′-ATAAGAATGCGGCCGCTCACTCCCCGCTGTGC-3′, forward and reverse, respectively. This BamHI-NOL12-NotI fragment was cloned into pLVX–Tight-Puro (Clontech, CA) digested with BamHI and NotI to generate pLVX–Tight-Puro-NOL12.
Lentiviral production and infection.
Lentiviruses were produced according to the protocol described for the Lenti-X Tet-On advanced inducible expression system (Clontech). Lentiviruses carrying pLVX–Tight-Puro–NOL12 and lentiviruses carrying pLVX–Tet-On advanced (which expresses reverse tetracycline-controlled transactivator [rtTA]) were generated in HEK293T helper cells transfected with packaging plasmids (pMd2.G and psPAX2) using Lipofectamine 2000 (Life Technologies, Thermo Scientific, CA). Human fibroblasts were coinfected for 12 to 16 h with responsive and trans-activator lentiviruses at a 2:1 ratio in the presence of 8 μg/ml Polybrene (AL-118; Sigma-Aldrich, MO). A concentration of 375 ng/ml doxycycline (stock keeping unit [SKU] number D9891; Sigma-Aldrich, MO) was added for 1 day to induce cotransduction and washed out for 2 days. This cyclic induction scheme was repeated 7 times before phenotype quantitative analysis. Western blot analysis was used to monitor the efficiency of transduction.
Statistical analysis.
P values were obtained using Prism version 7.00 (GraphPad, San Diego, CA). Data were tested for parametric versus nonparametric distribution using the D’Agostino and Pearson omnibus normality test. The Mann-Whitney, paired t, Kruskal-Wallis, or two-tailed χ2 test or one-way analysis of variance (ANOVA) was then applied according to the experiment.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge technical support from the BioSciences Screening and Histology and Electron Microscopy i3S Scientific Platforms, granted by the Portuguese Platform of Bioimaging (PPBI-POCI-01-0145-FEDER-022122).
E.L. holds an FCT Investigator grant, and M.P. holds an FCT Fellowship (SFRH/BD/110668), both from FCT/MCTES (Fundação para a Ciência e a Tecnologia/Ministério da Ciência, Tecnologia e Ensino Superior). The following project grants supported this work: Norte-01-0145-FEDER-000029 supported by North Portugal Regional Operational Programme (NORTE 2020) under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (FEDER); National Funds through FCT under project PTDC/BEX-BCM/2090/2014; European Regional Development Fund (FEDER) through the COMPETE 2020—Operational Program for Competitiveness and Internationalization (POCI), Portugal 2020, and National funds through FCT in the framework of project POCI-01-0145-FEDER-031120 (PTDC/BIA-CEL/31120/2017); and POCI-01-0145-FEDER-007274 i3S framework project cofunded by COMPETE 2020/PORTUGAL 2020 and by FCT.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00099-19.
REFERENCES
- 1.Boisvert F-M, van Koningsbruggen S, Navascues J, Lamond AI. 2007. The multifunctional nucleolus. Nat Rev Mol Cell Biol 8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 2.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. 2010. The nucleolus under stress. Mol Cell 40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsai RY, Pederson T. 2014. Connecting the nucleolus to the cell cycle and human disease. FASEB J 28:3290–3296. doi: 10.1096/fj.14-254680. [DOI] [PubMed] [Google Scholar]
- 4.Henras AK, Plisson-Chastang C, O’Donohue MF, Chakraborty A, Gleizes PE. 2015. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA 6:225–242. doi: 10.1002/wrna.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lempiainen H, Shore D. 2009. Growth control and ribosome biogenesis. Curr Opin Cell Biol 21:855–863. doi: 10.1016/j.ceb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 6.Woods SJ, Hannan KM, Pearson RB, Hannan RD. 2015. The nucleolus as a fundamental regulator of the p53 response and a new target for cancer therapy. Biochim Biophys Acta 1849:821–829. doi: 10.1016/j.bbagrm.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Rubbi CP, Milner J. 2003. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J 22:6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horn HF, Vousden KH. 2008. Cooperation between the ribosomal proteins L5 and L11 in the p53 pathway. Oncogene 27:5774–5784. doi: 10.1038/onc.2008.189. [DOI] [PubMed] [Google Scholar]
- 9.Bursac S, Brdovcak MC, Pfannkuchen M, Orsolic I, Golomb L, Zhu Y, Katz C, Daftuar L, Grabusic K, Vukelic I, Filic V, Oren M, Prives C, Volarevic S. 2012. Mutual protection of ribosomal proteins L5 and L11 from degradation is essential for p53 activation upon ribosomal biogenesis stress. Proc Natl Acad Sci U S A 109:20467–20472. doi: 10.1073/pnas.1218535109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donati G, Peddigari S, Mercer CA, Thomas G. 2013. 5S ribosomal RNA is an essential component of a nascent ribosomal precursor complex that regulates the Hdm2-p53 checkpoint. Cell Rep 4:87–98. doi: 10.1016/j.celrep.2013.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sloan KE, Bohnsack MT, Watkins NJ. 2013. The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep 5:237–247. doi: 10.1016/j.celrep.2013.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishimura K, Kumazawa T, Kuroda T, Katagiri N, Tsuchiya M, Goto N, Furumai R, Murayama A, Yanagisawa J, Kimura K. 2015. Perturbation of ribosome biogenesis drives cells into senescence through 5S RNP-mediated p53 activation. Cell Rep 10:1310–1323. doi: 10.1016/j.celrep.2015.01.055. [DOI] [PubMed] [Google Scholar]
- 13.Brangwynne CP, Mitchison TJ, Hyman AA. 2011. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci U S A 108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP. 2016. Coexisting liquid phases underlie nucleolar subcompartments. Cell 165:1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derenzini M, Montanaro L, Trere D. 2009. What the nucleolus says to a tumour pathologist. Histopathology 54:753–762. doi: 10.1111/j.1365-2559.2008.03168.x. [DOI] [PubMed] [Google Scholar]
- 16.Marinho J, Casares F, Pereira PS. 2011. The Drosophila Nol12 homologue viriato is a dMyc target that regulates nucleolar architecture and is required for dMyc-stimulated cell growth. Development 138:349–357. doi: 10.1242/dev.054411. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki S, Fujiwara T, Kanno M. 2007. Nucleolar protein Nop25 is involved in nucleolar architecture. Biochem Biophys Res Commun 358:1114–1119. doi: 10.1016/j.bbrc.2007.05.069. [DOI] [PubMed] [Google Scholar]
- 18.Oeffinger M, Zenklusen D, Ferguson A, Wei KE, El Hage A, Tollervey D, Chait BT, Singer RH, Rout MP. 2009. Rrp17p is a eukaryotic exonuclease required for 5′ end processing of pre-60S ribosomal RNA. Mol Cell 36:768–781. doi: 10.1016/j.molcel.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sahasranaman A, Dembowski J, Strahler J, Andrews P, Maddock J, Woolford JL Jr. 2011. Assembly of Saccharomyces cerevisiae 60S ribosomal subunits: role of factors required for 27S pre-rRNA processing. EMBO J 30:4020–4032. doi: 10.1038/emboj.2011.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sloan KE, Mattijssen S, Lebaron S, Tollervey D, Pruijn GJ, Watkins NJ. 2013. Both endonucleolytic and exonucleolytic cleavage mediate ITS1 removal during human ribosomal RNA processing. J Cell Biol 200:577–588. doi: 10.1083/jcb.201207131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scott DD, Trahan C, Zindy PJ, Aguilar LC, Delubac MY, Van Nostrand EL, Adivarahan S, Wei KE, Yeo GW, Zenklusen D, Oeffinger M. 2017. Nol12 is a multifunctional RNA binding protein at the nexus of RNA and DNA metabolism. Nucleic Acids Res 45:12509–12528. doi: 10.1093/nar/gkx963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marinho J, Martins T, Neto M, Casares F, Pereira PS. 2013. The nucleolar protein Viriato/Nol12 is required for the growth and differentiation progression activities of the Dpp pathway during Drosophila eye development. Dev Biol 377:154–165. doi: 10.1016/j.ydbio.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Martins T, Eusebio N, Correia A, Marinho J, Casares F, Pereira PS. 2017. TGFβ/Activin signalling is required for ribosome biogenesis and cell growth in Drosophila salivary glands. Open Biology 7:160258. doi: 10.1098/rsob.160258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ugrinova I, Monier K, Ivaldi C, Thiry M, Storck S, Mongelard F, Bouvet P. 2007. Inactivation of nucleolin leads to nucleolar disruption, cell cycle arrest and defects in centrosome duplication. BMC Mol Biol 8:66. doi: 10.1186/1471-2199-8-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ochs RL, Lischwe MA, Spohn WH, Busch H. 1985. Fibrillarin: a new protein of the nucleolus identified by autoimmune sera. Biol Cell 54:123–133. doi: 10.1111/j.1768-322X.1985.tb00387.x. [DOI] [PubMed] [Google Scholar]
- 26.Amin MA, Matsunaga S, Ma N, Takata H, Yokoyama M, Uchiyama S, Fukui K. 2007. Fibrillarin, a nucleolar protein, is required for normal nuclear morphology and cellular growth in HeLa cells. Biochem Biophys Res Commun 360:320–326. doi: 10.1016/j.bbrc.2007.06.092. [DOI] [PubMed] [Google Scholar]
- 27.Amin MA, Matsunaga S, Uchiyama S, Fukui K. 2008. Depletion of nucleophosmin leads to distortion of nucleolar and nuclear structures in HeLa cells. Biochem J 415:345–351. doi: 10.1042/BJ20081411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thiry M, Lafontaine DL. 2005. Birth of a nucleolus: the evolution of nucleolar compartments. Trends Cell Biol 15:194–199. doi: 10.1016/j.tcb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 29.West S, Gromak N, Proudfoot NJ. 2004. Human 5′ → 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature 432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- 30.Coccia M, Rossi A, Riccio A, Trotta E, Santoro MG. 2017. Human NF-kappaB repressing factor acts as a stress-regulated switch for ribosomal RNA processing and nucleolar homeostasis surveillance. Proc Natl Acad Sci U S A 114:1045–1050. doi: 10.1073/pnas.1616112114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Memet I, Doebele C, Sloan KE, Bohnsack MT. 2017. The G-patch protein NF-kappaB-repressing factor mediates the recruitment of the exonuclease XRN2 and activation of the RNA helicase DHX15 in human ribosome biogenesis. Nucleic Acids Res 45:5359–5374. doi: 10.1093/nar/gkx013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joerger AC, Fersht AR. 2016. The p53 pathway: origins, inactivation in cancer, and emerging therapeutic approaches. Annu Rev Biochem 85:375–404. doi: 10.1146/annurev-biochem-060815-014710. [DOI] [PubMed] [Google Scholar]
- 33.Choong ML, Yang H, Lee MA, Lane DP. 2009. Specific activation of the p53 pathway by low dose actinomycin D: a new route to p53 based cyclotherapy. Cell Cycle 8:2810–2818. doi: 10.4161/cc.8.17.9503. [DOI] [PubMed] [Google Scholar]
- 34.Hadjiolova KV, Hadjiolov AA, Bachellerie JP. 1995. Actinomycin D stimulates the transcription of rRNA minigenes transfected into mouse cells. Implications for the in vivo hypersensitivity of rRNA gene transcription. Eur J Biochem 228:605–615. doi: 10.1111/j.1432-1033.1995.0605m.x. [DOI] [PubMed] [Google Scholar]
- 35.Wang S, El-Deiry WS. 2005. P53, cell cycle arrest and apoptosis, p 141–163. In Hainaut P, Wiman KG (ed), 25 Years of p53 research doi: 10.1007/978-1-4020-2922-6_6 Springer, Dordrecht, Netherlands. [DOI] [Google Scholar]
- 36.Pines J, Hunter T. 1989. Isolation of a human cyclin cDNA: evidence for cyclin mRNA and protein regulation in the cell cycle and for interaction with p34cdc2. Cell 58:833–846. doi: 10.1016/0092-8674(89)90936-7. [DOI] [PubMed] [Google Scholar]
- 37.Krenning L, Feringa FM, Shaltiel IA, van den Berg J, Medema RH. 2014. Transient activation of p53 in G2 phase is sufficient to induce senescence. Mol Cell 55:59–72. doi: 10.1016/j.molcel.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 38.Charrier-Savournin FB, Chateau MT, Gire V, Sedivy J, Piette J, Dulic V. 2004. p21-mediated nuclear retention of cyclin B1-Cdk1 in response to genotoxic stress. Mol Biol Cell 15:3965–3976. doi: 10.1091/mbc.e03-12-0871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macedo JC, Vaz S, Bakker B, Ribeiro R, Bakker PL, Escandell JM, Ferreira MG, Medema R, Foijer F, Logarinho E. 2018. FoxM1 repression during human aging leads to mitotic decline and aneuploidy-driven full senescence. Nat Commun 9:2834. doi: 10.1038/s41467-018-05258-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.James A, Wang Y, Raje H, Rosby R, DiMario P. 2014. Nucleolar stress with and without p53. Nucleus 5:402–426. doi: 10.4161/nucl.32235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A 92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, Kirkland JL, van Deursen JM. 2011. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Freed EF, Prieto JL, McCann KL, McStay B, Baserga SJ. 2012. NOL11, implicated in the pathogenesis of North American Indian childhood cirrhosis, is required for pre-rRNA transcription and processing. PLoS Genet 8:e1002892. doi: 10.1371/journal.pgen.1002892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farley-Barnes KI, McCann KL, Ogawa LM, Merkel J, Surovtseva YV, Baserga SJ. 2018. Diverse regulators of human ribosome biogenesis discovered by changes in nucleolar number. Cell Rep 22:1923–1934. doi: 10.1016/j.celrep.2018.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sloan KE, Bohnsack MT, Schneider C, Watkins NJ. 2014. The roles of SSU processome components and surveillance factors in the initial processing of human ribosomal RNA. RNA 20:540–550. doi: 10.1261/rna.043471.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma N, Matsunaga S, Takata H, Ono-Maniwa R, Uchiyama S, Fukui K. 2007. Nucleolin functions in nucleolus formation and chromosome congression. J Cell Sci 120:2091–2105. doi: 10.1242/jcs.008771. [DOI] [PubMed] [Google Scholar]
- 47.Deisenroth C, Franklin DA, Zhang Y. 2016. The evolution of the ribosomal protein-MDM2-p53 pathway. Cold Spring Harb Perspect Med 6:a026138. doi: 10.1101/cshperspect.a026138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holmberg Olausson K, Nistér M, Lindström MS. 2012. p53-dependent and -independent nucleolar stress responses. Cells 1:774–798. doi: 10.3390/cells1040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jayaraman S, Chittiboyina S, Bai Y, Abad PC, Vidi PA, Stauffacher CV, Lelievre SA. 2017. The nuclear mitotic apparatus protein NuMA controls rDNA transcription and mediates the nucleolar stress response in a p53-independent manner. Nucleic Acids Res 45:11725–11742. doi: 10.1093/nar/gkx782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marcel V, Ghayad SE, Belin S, Therizols G, Morel AP, Solano-Gonzalez E, Vendrell JA, Hacot S, Mertani HC, Albaret MA, Bourdon JC, Jordan L, Thompson A, Tafer Y, Cong R, Bouvet P, Saurin JC, Catez F, Prats AC, Puisieux A, Diaz JJ. 2013. p53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell 24:318–330. doi: 10.1016/j.ccr.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. 2003. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3:577–587. doi: 10.1016/S1535-6108(03)00134-X. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y. 2003. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol 23:8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun XX, Wang YG, Xirodimas DP, Dai MS. 2010. Perturbation of 60 S ribosomal biogenesis results in ribosomal protein L5- and L11-dependent p53 activation. J Biol Chem 285:25812–25821. doi: 10.1074/jbc.M109.098442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan X, Zhou Y, Casanova E, Chai M, Kiss E, Grone HJ, Schutz G, Grummt I. 2005. Genetic inactivation of the transcription factor TIF-IA leads to nucleolar disruption, cell cycle arrest, and p53-mediated apoptosis. Mol Cell 19:77–87. doi: 10.1016/j.molcel.2005.05.023. [DOI] [PubMed] [Google Scholar]
- 55.Marcel V, Catez F, Diaz JJ. 2015. Ribosome heterogeneity in tumorigenesis: the rRNA point of view. Mol Cell Oncol 2:e983755. doi: 10.4161/23723556.2014.983755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Junera HR, Masson C, Geraud G, Hernandez-Verdun D. 1995. The three-dimensional organization of ribosomal genes and the architecture of the nucleoli vary with G1, S and G2 phases. J Cell Sci 108:3427–3441. [DOI] [PubMed] [Google Scholar]
- 57.Maszewski J, Kwiatkowska M. 1984. Number, size, and transcriptional activity of nucleoli during different periods of interphase in antheridial filaments of Chara vulgaris L. Folia Histochem Cytobiol 22:9–19. [PubMed] [Google Scholar]
- 58.Hernandez-Verdun D. 2011. Assembly and disassembly of the nucleolus during the cell cycle. Nucleus 2:189–194. doi: 10.4161/nucl.2.3.16246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang G, Meng L, Tsai RY. 2015. p53 configures the G2/M arrest response of nucleostemin-deficient cells. Cell Death Discov 1:15060. doi: 10.1038/cddiscovery.2015.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bywater MJ, Poortinga G, Sanij E, Hein N, Peck A, Cullinane C, Wall M, Cluse L, Drygin D, Anderes K, Huser N, Proffitt C, Bliesath J, Haddach M, Schwaebe MK, Ryckman DM, Rice WG, Schmitt C, Lowe SW, Johnstone RW, Pearson RB, McArthur GA, Hannan RD. 2012. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell 22:51–65. doi: 10.1016/j.ccr.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gannon HS, Donehower LA, Lyle S, Jones SN. 2011. Mdm2-p53 signaling regulates epidermal stem cell senescence and premature aging phenotypes in mouse skin. Dev Biol 353:1–9. doi: 10.1016/j.ydbio.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lindqvist A, de Bruijn M, Macurek L, Bras A, Mensinga A, Bruinsma W, Voets O, Kranenburg O, Medema RH. 2009. Wip1 confers G2 checkpoint recovery competence by counteracting p53-dependent transcriptional repression. EMBO J 28:3196–3206. doi: 10.1038/emboj.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buchwalter A, Hetzer MW. 2017. Nucleolar expansion and elevated protein translation in premature aging. Nat Commun 8:328. doi: 10.1038/s41467-017-00322-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tiku V, Jain C, Raz Y, Nakamura S, Heestand B, Liu W, Spath M, Suchiman HED, Muller RU, Slagboom PE, Partridge L, Antebi A. 2017. Small nucleoli are a cellular hallmark of longevity. Nat Commun 8:16083. doi: 10.1038/ncomms16083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sadasivam S, Duan S, DeCaprio JA. 2012. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev 26:474–489. doi: 10.1101/gad.181933.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fischer M, Grossmann P, Padi M, DeCaprio JA. 2016. Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res 44:6070–6086. doi: 10.1093/nar/gkw523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.