Abstract
Background: SKP2 is considered an oncogene involved in various malignancies. SKP2 protein is a critical subunit of the SKP1-CUL1-F-box (SCF) E3 ligase complex which affects the cell cycle profoundly by specifically recognizing cell cycle regulators and mediating their ubiquitylation and proteasomal degradation. SKP2 dysfunction is characteristic of many tumor cells. However, its role in uveal melanoma (UM) has not been elucidated.
Materials and methods: We analyzed the expressions of SKP2 in different UM cell lines compared with normal pigment cell by RNA-seq, RT-qPCR and Western blot. We then knocked down SKP2 in OM431 and MUM2B cells and confirmed its roles in cell proliferation via CCK8 assay. The sensitivity of cells to SKP2 inhibitor C1 (SKPin C1) in vitro was evaluated by CCK8 assay and colony formation assay, and the sensitivity of MUM2B cells to SKPin C1 in vivo was estimated using the nude mouse-based xenograft model. Western blot and Immunoprecipitation assay were performed to detect the change of p27 and its ubiquitylation level in UM cells treated with SKPin C1, respectively.
Results: The results showed that SKP2 was significantly highly expressed in UM cells. SKP2 promoted the progression of UM and knockdown of SKP2 inhibited cell proliferation in UM cells. SKP2 inhibitor C1 that targets SKP2 essentially inhibits the growth of UM cells both in vivo and in vitro. SKP2 inhibitor C1 decreased the degradation of p27 by blocking ubiquitylation of p27, resulting in p27 accumulation and cell cycle arrest in UM cells.
Conclusion: Our findings demonstrated that SKP2 targeted inhibition suppresses UM cell proliferation and provides new options and possibilities for targeted therapies in UM.
Keywords: SKP2, p27, uveal melanoma, targeted therapy, small molecular inhibitor
Introduction
Uveal melanoma (UM) is the most common primary intraocular malignancy in adults, accounting for 85–95% of all ocular melanoma cases.1 UM has a high degree of malignancy and a poor outcome following the development of distant disease. Nearly 50% of patients eventually die to metastatic disease, with the most common metastatic site being the liver.2,3 Due to the different genetic backgrounds of UM and cutaneous melanoma,4 the use of cutaneous melanoma-based chemotherapy on patients with UM has yielded disappointing results.5 On the other hand, previous studies have found that UM is characterized by mutations in GNAQ or GNA11 that constitutively activate the MAPK and PI3K/Akt pathways.6–8 Based on this, treatments targeting the downstream effectors of these pathways, such as MEK, Akt and protein kinase C (PKC), are being investigated.9,10 Unfortunately, the results so far have been disappointing, and the response rate has been less than 10%.11 Thus, it is of great significance to reveal molecular alterations in UM and to find novel therapy targets for UM.
Ubiquitylation marks proteins for degradation by the ubiquitin proteasome system (UPS), which is important for maintaining cellular homeostasis during cell cycle progression, proliferation and apoptosis. Dysregulation of the UPS has been implicated in the development of various cancers.12,13 Based on this, different inhibitors targeting UPS have been developed as cancer treatment strategies.14,15 The F-box protein, SKP2, associates with SKP1, RBX1 and CUL1 to form the SCF complex, which mediates specific protein ubiquitylation and degradation. As a kind of E3 ubiquitin ligases, SKP2 specifically recognizes the substrate p21 and p27 in the presence of the accessory protein Cks1.16 It plays major roles in tumorigenesis and cell cycle control. SKP2 has been found to be dysregulated and to exhibit a proto-oncogenic role in the pathogenesis of various human cancers, such as breast cancer,17,18 prostate cancer,19 melanoma,20 pancreatic cancer21 and urothelial carcinoma.22 It is worth noting that the overexpression of SKP2 is associated with poor prognosis in many tumors.23,24 SKP2 inactivation restricts cancer development by targeting cellular senescence in a p27-dependent manner.25 p27 is one of the most critical CDK inhibitors, in that it inhibits the formation of the cyclin E-CDK2 complex. In addition, the excessive degradation of p27 also has been seen in human cancers26,27 and loss of p27 plays a critical role in the aggressiveness of cancers such as gastroenteropancreatic neuroendocrine tumors.28 However, the role that SKP2 plays in UM remains unknown.
In view of this, SKP2, as a molecular star of carcinogenesis, has been selected as a novel and attractive biomarker and a therapeutic target for cancer treatment29-32 and many investigators have produced interesting results, especially in the area of small molecule inhibitors.29,33 High-throughput in silico screens has identified small molecule inhibitors specifically blocking the binding site for p27,34 resulting in reduced SKP2-mediated p27 degradation. Specifically, the accumulation of p27 promotes a cell-type specific block of the G1 or G2/M phase.34 Combined with the current shortcomings of UM treatment, we were inspired to investigate whether SKP2 plays a major role in UM and whether it can be a potential target for UM therapy.
In the current study, we demonstrate that SKP2 expression is upregulated in UM cells. Knockdown of SKP2 in UM cells with high basal levels of this protein promotes cell proliferation. Furthermore, we validated the killing effect of SKP2 inhibitor C1 (SKPin C1), a highly selective inhibitor of SKP2, on UM cells.
Materials and methods
Cell lines and cell culture
Human UM cell lines 92.1, MUM2B, OCM1, OCM1α and OM431 were kindly provided by Professor John F. Marshall (Tumor Biology Laboratory, Cancer Research UK Clinical Center, John Vane Science Centre, London, UK). OMM2.3 was a kind gift from Professor Martine J. Jager (Department of Ophthalmology, Leiden University Medical Center, Leiden, The Netherlands). Human normal melanocyte cell line PIG135 was obtained from the Department of Ophthalmology, Peking University Third Hospital. HEK293t cell line was purchased from ATCC. OCM1, OCM1α and OMM2.3 are derived from distant metastases, and other UM cell lines are derived from primary ocular tumors. The usage and methods of cell lines in cell-based experiments were approved by the ethical and institutional review board of Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine. All cell lines, except 92.1, were cultured in Dulbecco’s modified Eagle medium (DMEM; Gibco, USA) supplemented with 10% certified heat-inactivated fetal bovine serum (FBS; Gibco, USA), penicillin (100 U/ml) and streptomycin (100 μg/ml) at 37°C in a humidified 5% CO2 atmosphere. 92.1 was cultured in RPMI 1640 medium (Gibco, USA).
Cell transfection
SKP2 siRNA1(CUAAAGGUCUCUGGUGUUUUU), SKP2 siRNA2(GGUAUCGCCUAGCGUCUGAUU) and a control siRNA (GGTTCTCTTATGATGTTACGT) were synthesized by GenePharma Co. (Shanghai, China). A total of 1×105 MUM2B and OM431 cells per well were seeded in 6-well plates and transfected with siRNA at a final concentration of 50 nM using Lipofectamine 2000 (Invitrogen, 11668019, Carlsbad, CA, USA) according to the manufacturer’s protocol in Opti-MEM I Reduced Serum Medium (Gibco, Carlsbad, CA, USA). Six hours later, the supernatant was replaced by a fresh complete medium. Forty-eight hours after the transfection, the cells were harvested for real-time PCR or Western blot analyses and were harvested by trypsinization for tumor assays.
RNA extraction, library construction and illumina sequencing (RNA-seq)
Total RNA was extracted from UM and normal melanocyte cell lines using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. We confirmed RNA integrity using the 2100 Bioanalyzer (Agilent Technologies, USA) and measured RNA concentration in a Qubit 2.0 fluorometer using the Qubit RNA Assay Kit (Life Technologies, Carlsbad, CA, USA). We prepared libraries from 100 ng of total RNA using an Illumina TruSeq RNA Sample Prep Kit (San Diego, CA, USA) following the manufacturer’s protocol. In total, 3 libraries were sequenced using the Illumina Hiseq platform (San Diego, CA, USA). The mRNA levels of the unigenes were identified using TopHat v2.0.9 and Cufflinks and normalized by the Fragments Per Kilobase of exon model per Million mapped reads (FPKM). We used the criteria of false discovery rate (FDR)<0.01 and fold changes <0.5 or >2.0 (<−1 or >1 log2 ratio value, P-value<0.05) to identify differentially expressed genes.
RNA isolation and real-time qPCR
Total RNA from cultured cells was extracted with TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol and complementary DNA (cDNA) was synthesized using PrimeScript RT-PCR Kit (TakaraBio, Otsu, Japan). Real-time qPCR was performed using a SYBR Master Mix (TakaraBio, Otsu, Japan) and an ABI 7500 real-time PCR system (Applied Biosystems, Irvine, CA, USA). PCR products were quantified and normalized using GAPDH as the control. The relative expression fold change of mRNAs was calculated by the 2−ΔΔCt method. The PCR primers for SKP2 were 5ʹ-ATGCCCCAATCTTGTCCATCT-3ʹ and 5ʹ-CACCGACTGAGTGATAGGTGT-3ʹ. The PCR primers for GAPDH were 5ʹ-AGGTCGGTGTGAACGGATTTG-3ʹ and 5ʹ- TGTAGACCATGTAGTTGAGGTCA-3ʹ.
Cell viability assay
SKP2 siRNA-transfected MUM2B and OM431 cells were seeded into 96-well plates at a density of 2000 cells per well with 100 μL of complete medium 6 hrs before the experiments and incubated at 37°C. Cell viability was analyzed by Cell Counting Kit-8 (CCK-8, Dojindo Laboratories, Kumamoto, Japan) reagent at 0, 24, 48 and 72 hrs according to the manufacturer’s instructions. At the indicated time points, 10 μL of CCK-8 reagent was added to each well. After incubation at 37°C for 4 hrs, the absorbance was measured with a plate reader at 450 nm, and the growth curves were examined to determine the growth rates.
Colony formation assay
A colony formation assay was performed in 6-well plates. A total of 2,000 cells were suspended in 2.0 mL of complete medium and seeded into each well. The cultures were replaced every 3 to 4 days for about 2 weeks. For quantification, the colonies grown were stained with 10% crystal violet. Visible colonies were manually counted.
Western blotting
Cells were harvested at the indicated times and rinsed twice with phosphate-buffered saline (PBS). Cell extracts were prepared with RIPA lysis buffer (Biosharp, Hefei, China, BL504A) and centrifuged at 13,000 g for 30 mins at 4°C. Protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 7.5% (w/v) polyacrylamide gels and transferred to polyvinylidene fluoride membranes (Millipore Corporation, Billerica, MA, USA). After blocking with 5% bovine serum albumin (BSA) for 1 hr at room temperature, the membranes were incubated with an appropriate amount of primary antibody at 4°C overnight. The membranes were then incubated with secondary antibodies conjugated to a fluorescent tag (Invitrogen). The band signals were visualized using the Odyssey Infrared Imagining System (LI-COR, USA). Quantification of the band was performed by Image J (National Institutes of Health, US). GAPDH was used as a control. The following antibodies were used: SKP2 p45 Antibody (Santa Cruz, USA, sc-7164, dilution 1:500), GAPDH Antibody (CST, USA, #5174, dilution 1:1000), p27 Kip1 Antibody (CST, USA, #3686, dilution 1:1000), HA-Tag Antibody (CST, USA, #3724, dilution 1:1000).
IC50 assay
For determining the half maximal inhibitory concentration (IC50), cell lines were seeded in 96-well format. The day after plating, if applicable, SKP2 inhibitor C1 (Selleck, S8652) were added. Cells were then drugged with serial dilutions of indicated inhibitors to give final concentrations ranging from 1,000 μmol/L to 1 nmol/L, in half-log increments. Four days later, cellular viability was assessed using CCK-8 (Dojindo Laboratories, Kumamoto, Japan). IC50 calculations were performed in GraphPad Prism.
Cell cycle analysis
Treated UM cells along with control cells were harvested. 5×105 cells of each group were washed twice with cold PBS, fixed in 70% ethanol and stored at 4°C overnight. The next day, the cells were washed twice with cold PBS and incubated with propidium iodide/ribonuclease staining solution (Becton Dickinson, NJ, USA) for 15 mins at room temperature, following the manufacturer’s instructions. Cell cycle distribution was detected and analyzed using the FACScan instrument and CellQuest program (Becton Dickinson, NJ, USA).
Immunoprecipitation assay
HEK293t cells were plated in 60 mm dishes and grew until 70% confluence. Two groups of cells were transfected with SKP2, Flag-p27 and HA-Ub plasmids; the first group of cells was treated with SKPin C1 (5 μM) 4 hrs later, and the second group of cells was treated with an equivalent volume of DMSO (negative control). In the other two groups of 2,93t cells, SKP2, Flag-p27 and empty vector (pcDNA3.1) plasmids were transfected; likewise, one group of cells was treated with SKPin C1 (5 μM), and the other group was treated with an equivalent volume of DMSO. After 24 hrs, cells were harvested and lysed in RIPA buffer (50 mM Tris-Cl, pH7.4; 150 mM NaCl; 5 mM EDTA; 1% (v/v) Triton X-100; 0.5% sodium pyrophosphate; 0.1% SDS; protease inhibitor cocktail (Roche)). The lysates were cleared by centrifugation (15,000× g, 20 mins, 4°C) and incubated overnight with anti-FLAG Magnetic Beads (Sigma, M8823). The beads were gently washed 3 times with Tris-buffered saline (TBS), pH 7.5 and the complexes were eluted by boiling the samples before running the SDS-PAGE. These antibodies were used in the following immunoblotting: SKP2 p45 Antibody (Santa Cruz, USA, sc-7164, dilution 1:500), p27 Kip1 Antibody (CST, USA, #3686, dilution 1:1,000), HA-Tag Antibody (CST, USA, #3724, dilution 1:1,000).
Xenograft model
Animal experiments were approved by Shanghai Jiao Tong University animal care and use committee and conducted following The Shanghai Jiao Tong University animal policy in accordance with the Ministry of Science and Technology of the People’s Republic of China guidelines. Cells were harvested by trypsinization and washed twice with PBS. Cells (1×106) were injected subcutaneously into the left flank of 5-week-old male Balb/c (nu/nu) mice (n=5 per group) in a PBS volume of 100 µL. The experimental group was treated with SKPin C1 20 mg/kg by intraperitoneal injection one week after the transplantation, and the control group was injected with an equivalent volume of solvent. After 4 weeks, the tumors were harvested and weighed and prepared for HE staining and histological analysis.
Statistical analysis
All of the experiments were performed in triplicate, and the data were expressed as the mean ± standard deviation (SD). Statistical analyses were performed in SPSS 23.0 statistical software (SPSS Inc., Chicago, IL, USA). The comparative threshold cycle method was applied in the quantitative real-time PCR assay according to the ΔΔ threshold cycle method. The differences between two groups were analyzed with the unpaired two-sided Student’s t-test. A P-value of less than 0.05 was considered statistically significant and is indicated with asterisks, as described in the figure legends.
Results
Aberrant SKP2 expression in uveal melanoma cells
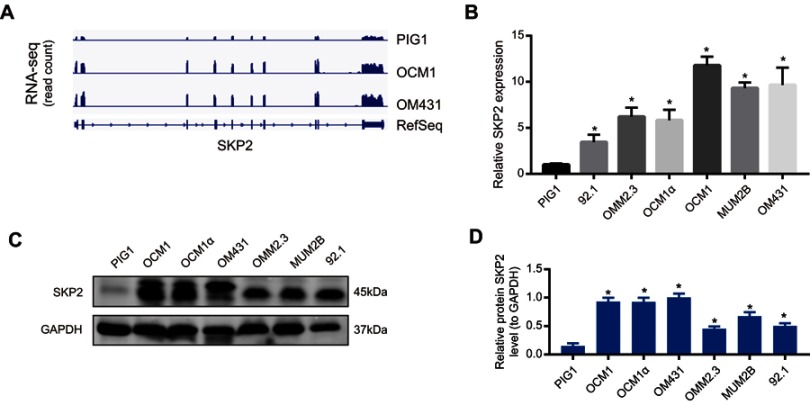
To identify important genes that may be dysregulated in UM cells, genome-wide RNA-seq was carried out to detect changes in the transcriptome of UM cells. We found that SKP2 was highly expressed in UM cell lines OCM1 and OM431 compared to normal melanocyte cell line PIG1 (Figure 1A). Thereafter, we verified the mRNA level of SKP2 in UM cells by real-time PCR. We found that the endogenous expression of SKP2 in UM cells increased significantly compared to PIG1, especially MUM2B, OCM1 and OM431 cells, which was consistent with the high-throughput sequencing results (Figure 1B). In addition, we investigated the SKP2 protein expression level in UM cells by Western blotting. As expected, its protein expression was increased significantly in UM cells compared with PIG1 (Figure 1C and D). In summary, SKP2 was aberrantly overexpressed in UM cells, suggesting that it plays roles in the progression of UM.
Figure 1.
Aberrant SKP2 expression in uveal melanoma. (A) RNA-sequence analysis was performed to evaluate the transcriptome in two uveal melanoma cell samples and normal pigment cell samples and the genomic sequence of SKP2 is shown. (B) SKP2 expression in six UM cell lines (OCM1, OCM1α, OM431, MUM2B, OMM2.3 and 92.1) and normal pigment cell (PIG1) was measured by RT-qPCR analysis. The SKP2 expression was normalized to that in PIG1. The relative expression fold change of mRNAs was calculated by the 2−ΔΔCt method. *P<0.05. (C) Western blot showed that the SKP2 protein is overexpressed in UM cells (OCM1, OCM1α, OM431, MUM2B, OMM2.3 and 92.1) compared to normal cells. (D) Densitometric analysis was performed to quantify and statistically compare the protein levels of SKP2. *P < 0.05.
Abbreviations: SKP2, S-phase kinase-associated protein 2; UM, uveal melanoma.
Knockdown of SKP2 suppresses uveal melanoma growth in vitro
Considering that SKP2 was highly expressed in UM cells, we further verified the function of SKP2 in UM cells by investigating whether silencing SKP2 would affect the growth and proliferation of UM cells. Based on the expression level of SKP2 and the characteristics of UM cells, we chose OM431 and MUM2B cells for this experiment, and SKP2 was successfully knocked down via the classical siRNA method (Figure 2A and B), of which the siRNA sequence was referenced to the literature published by Hu R. et al.36 Thereafter, we performed CCK8 assays, which showed that cell growth was significantly inhibited after SKP2 knockdown compared to control cells (Figure 2C and D). In addition, plate colony formation assays showed a similar trend; we observed roughly 60% fewer colonies after SKP2 knockdown (Figure 2E–G). Taken together, these data indicated that knockdown of SKP2 inhibited UM proliferation in vitro, suggesting that SKP2 might play a tumorigenic regulatory role in UM.
Figure 2.
Knocking down SKP2 suppresses uveal melanoma cells growth. (A and B) Real-time qPCR assay showed that SKP2-specifc siRNA 1 and 2 reduced endogenous SKP2 mRNA levels in 48 hrs post-transfection OM431 and MUM2B cells and Western blot showed the siRNA efficiency at the protein level. *P<0.05. (C and D) CCK8 assay was performed to measure 3-day cell growth rate of OM431 and MUM2B cells after SKP2 knock down by siRNA. *P<0.05. (E) Representative results of colony formation assays of OM431 and MUM2B cells transfected with ctrl siRNA, SKP2 siRNA-1/2. (F and G) Quantification of visible colonies. The colony number in the NC group was set as 100%. All the experiments were performed in triplicate, and the relative colony formation rates are shown as the mean ± SD. *P<0.05.
Abbreviations: SKP2, S-phase kinase-associated protein 2; NC, negative control.
SKPin C1 inhibits uveal melanoma growth in vitro and in vivo
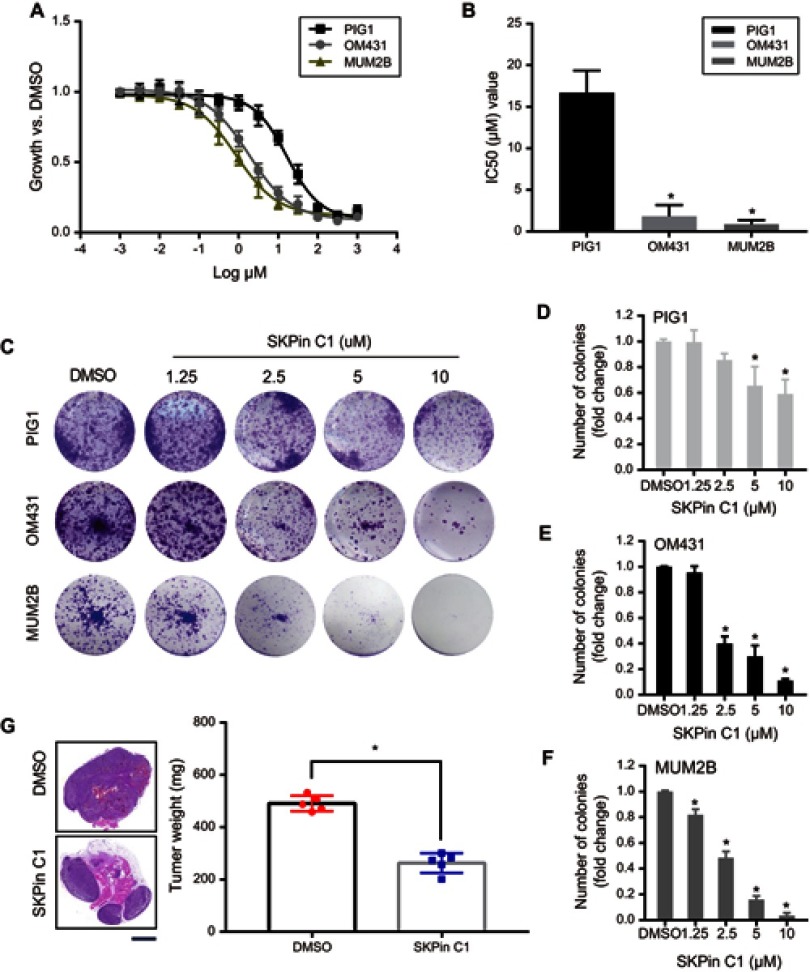
To further verify whether targeted inhibition of SKP2 can affect UM growth, we selected the most potent compound, SKP2 inhibitor C1 (SKPin C1), among the reported SKP2 inhibitors for the following experiments. Similarly, we used CCK8 and plate colony assays to investigate UM cell growth in the presence of SKPin C1 with multiple concentration gradients. At the same time, we included normal control cells, PIG1, to exclude the potentially harmful effects of the drug on normal cell growth. The results showed that UM cell growth was inhibited with the increase in the drug concentration (Figure 3A and C), which was consistent with the results of the SKP2 knockdown experiment. In addition, we drew the survival curves of UM cells and normal cells treated with serial dilutions of indicated inhibitors to give final concentrations ranging from 1000 μmol/L to 1 nmol/L, in half-log increments. Then, the half maximal inhibitory concentration (IC50) of SKPin C1 was calculated. We found that the IC50 value of normal cell (16.71 μM) was significantly higher than that of MUM2B (0.86 μM) and OM431 (1.83 μM) (Figure 3B). These findings confirmed that SKPin C1 could inhibit the growth of UM cells and revealed the positive effect of SKP2 on UM cell growth. The same trend was observed in the plate colony formation assays (Figure 3C). As the drug concentration increased, the colony counts of both cell types became lower (Figure 3D–F). It is worth noting that normal cells were significantly less inhibited than UM cells under the same concentration of the drug (Figure 3C and D), thereby supporting the potential application of this small molecule inhibitor as a drug for use in clinical settings.
Figure 3.
SKPin C1 inhibits UM growth in vitro and in vivo. (A) Survival curves of PIG1, OM431 and MUM2B cells according to the sensitivity to SKPin C1. Data are represented as mean ± SD, n =3. (B) IC50 value of SKPin C1 in PIG1, OM431 and MUM2B cells. Data are represented as mean ± SD, n =3. *P<0.05. (C) Colony formation assays were performed to determine the colony formation ability of UM cells treated with different concentration SKPin C1. (D–F) Quantification of visible colonies. The colony number in the DMSO group was set as 100%. All the experiments were performed in triplicate, and the relative colony formation rates are shown as the mean ± SD. *P<0.05. (G) Tumorigenesis ability in a xenograft in vivo assay model. UM cells were injected to the left flank of five-week-old male nude mice in this assay (n=5 per group). Mice were killed at four weeks after injection of cells and SKPin C1. Tumor weight was measured and was presented as the mean ± SD. *P<0.05. The tumors were prepared for HE staining and histological analysis. Representative images were shown. scale bar: 2 mm.
Abbreviations: SKPin C1, SKP2 inhibitor C1; DMSO, Dimethyl Sulfoxide; HE, hematoxylin-eosin.
To investigate the ability of SKP2 to alter the characteristics of the tumor in vivo, we established a xenograft model in nude mice. We performed subcutaneous injections of UM cells into two groups of nude mice. The experimental group was treated with SKPin C1 20 mg/kg by intraperitoneal injection one week after the transplantation, and the control group was injected with an equivalent volume of solvent. After 4 weeks, the tumors were harvested and prepared for histological analysis.
As expected, we found that both the tumor size and weight of the experimental group were significantly lower than that of the control group (Figure 3G), indicating that the drug inhibited the growth of tumors in mice, further supporting the treatment of UM with SKP2 inhibitors.
SKPin C1 induces G1 phase arrest in uveal melanoma cells
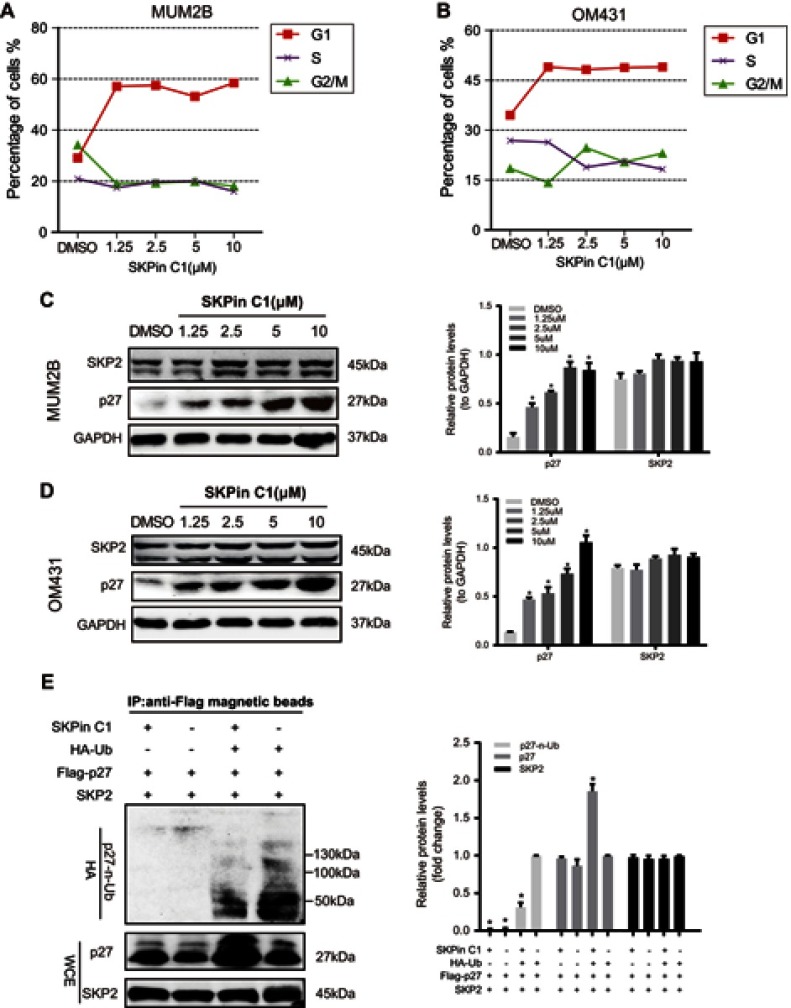
In order to further explore and confirm the mechanism of SKPin C1 inhibition in UM cells, flow cytometry was used to determine the percentage of cells at the different phases of the cell cycle. The results showed that the proportion of UM cells at the G1 phase increased significantly after SKP2 inhibitor treatment, whereas the proportion of cells at the G2/M phase decreased (Figure 4A and B), indicating that the cell cycle was arrested at the G1 phase. Previous research has shown that SKP2 recognizes p27 within the UPS, and SKP2 inhibition results in p27-dependent arrest at the G1 phase in colorectal cancer,25 breast cancer34 and prostate cancer cells.19 Considering this, we hypothesized that there may be a similar mechanism in UM cells. We prepared lysates from the cells treated with different concentrations of inhibitors for Western blot analysis. Interestingly, the results of Western blotting showed that there was no significant change in the expression level of SKP2 in UM cells treated with increasing inhibitor concentrations, whereas the expression level of p27 showed a dose-dependent increase, indicating that p27 accumulated in cells after SKP2 inhibition (Figure 4C and D). To validate the change of p27 ubiquitylation in UM cells treated with SKPin C1, we performed immunoprecipitation experiments. The results showed that the level of ubiquitinated p27 in the SKPin C1-treated group (Figure 4E, line 3) was significantly lower than that in the vehicle-treated group (Figure 4E, line 4). Moreover, p27 was not significantly ubiquitinated in the results of the two groups without HA-Ub transfection (Figure 4E, lines 1, 2), which might be related to the background ubiquitin level of the tool cell. Our results suggest that SKP2 targeted inhibition interferes with ubiquitylation and degradation of p27, resulting in accumulation of p27, and induces G1 phase arrest in UM cells.
Figure 4.
SKP2 inhibition induces G1 phase arrest in UM cell cycle. (A and B) Cell cycle arrest of MUM2B and OM431 after treated with SKPin C1 in different concentration. (C and D) Western blot showed the accumulation of p27 in MUM2B and OM431 treated with SKPin C1 in different concentration. Densitometric analysis was performed to quantify and statistically compare the protein levels of p27 and SKP2. *P < 0.05. (E) IP experiments confirmed that SKP2 inhibition led to a decrease in the level of ubiquitylation of p27: IB analysis of anti-Flag magnetic beads pull-down and WCEs derived from HEK293t cells transfected with indicated plasmids. The experimental groups were treated with SKPin C1 with a concentration of 5 μM. Densitometric analysis was performed to quantify and statistically compare the relative protein levels of p27-n-Ub, SKP2 and p27. *P < 0.05.
Abbreviations: SKP2, S-phase kinase-associated protein 2; UM, uveal melanoma; IP, Immunoprecipitation; HA-Ub, HA-Ubiquitin; WCE, whole-cell extract.
Discussion
In the present study, we investigated an important component protein of the UPS, SKP2, whose level was significantly up-regulated in UM cell lines after analyzing RNA-Seq data and performing RT-qPCR and Western blot analysis. Subsequent functional and mechanistic experiments showed that knockdown or inhibition of SKP2 caused UM cell cycle arrest, primarily through the SKP2-p27 axis, thereby significantly inhibiting the proliferation of UM cells. Our results revealed the carcinogenic effects of SKP2 in UM cells, which promoted an understanding of the progression of UM.
The dysfunction or dysregulation of the UPS is closely related to the development and progression of various cancers.37 Recently, SKP2, one of the component proteins of the UPS, was confirmed to be a proto-oncogene in many cancers by influencing the development of cancer through different ways.20,38 In fact, increasing evidence suggests that SKP2 is overexpressed in various cancers, such as lymphoma, prostate cancer and breast cancer.19,39,40 Additionally, the overexpression of SKP2 is correlated with the poor prognosis of osteosarcoma, hepatocellular carcinoma and breast cancer.24,41,42 In some of these studies, the relationship between SKP2 and melanoma has been implicated. For example, SKP2 protein expression is implicated in the progression of cutaneous melanoma, and it can also be used as a biomarker to detect premalignant and malignant lesions.43 One study showed an increase in copy number at the SKP2 locus in 6/14 (43%) metastatic cell lines and 9/22 (41%) human metastatic tissues, which correlates with overexpression of SKP2 protein.44 Moreover, overexpression of SKP2 protein in human tissues is associated with poor survival in a multivariate model controlling metastatic sites.45 Interestingly, the increase in SKP2 and the loss of p27 protein expression are associated with melanoma progression, where the level of p27 can be regulated by targeted proteolysis of SKP2.43 However, the function of SKP2 in UM remains largely obscure in previous study.
Here, we found that the SKP2–p27 axis played a positive role in promoting tumorigenesis in UM cells. Furthermore, p27 is a cell cycle regulator and considered to be a tumor suppressor.27 SKP2 is up-regulated in various cancers and can promote p27 proteolysis.25,46 As one of the components of the SCF complex, SKP2 can specifically recognize and bind to p27. Wu et al identified small molecule inhibitors specific to SCF–SKP2 activity using in silico screens that targeted the binding site for p27.34 In some cancer cells, such as those from breast cancer and prostate cancer, the compounds induced p27 accumulation in a SKP2-dependent manner and promoted cell cycle arrest. Based on previous studies, we hypothesized that SKP2 inhibitors might provide better options for the treatment of UM. Interestingly, we found that SKPin C1, a small molecule drug, could specifically inhibit the growth of UM cells, whereas the effect on normal cells was insignificant. Moreover, our results showed that SKPin C1 induced accumulation of p27 in UM cells, whereas there was no significant change in the expression level of SKP2, which were consistent with those of previous studies that reported SKPin C1 to interfere with the binding site between SKP2 and p27, rather than affecting the expression of SKP2.34 These results provide the insights needed to develop SKP2 inhibitors as a new therapy for UM. In fact, quite a few SKP2 inhibition methods have shown great potential for clinical application as anti-tumor strategies. Compound #25 identified by Chan et al interferes with the combination of SKP2 and SKP1, thereby inhibiting the survival of cancer cells, and it exhibited potent anti-tumor activity in animal experiments.33 A research by HY Hsieh et al has shown that the bacterial bioactive substance prodigiosin mediates transcriptional inhibition of SKP2 and lead to subsequent accumulation of p27, which is associated with prodigiosin-induced anti-proliferation active to many cancers.47 Furthermore, the inhibition of SKP2 not only directly inhibits tumors, but also enhances the sensitivity of tumors to some existing treatments. Ce Li et al40 reported that SKP2 promoted radiation tolerance by PDCD4 degradation via ubiquitination and radiotherapy combined with SKP2 targeted inhibition may improve breast cancer patient survival in clinical medicine. This study suggests that SKP2 may have different substrates and associated cancer pathways that contribute differently to tumor behaviors in various cancers. Combined with our findings, we believe that SKP2 targeted inhibition to treat UM is promising. In addition, the relationship between SKP2 and UM metastasis or drug resistance can be further studied.
However, it should be pointed out that our investigation on SKPin C1 was rather preliminary, and further studies are needed to improve the detection of its effects and toxicity. SKP2 targeted therapies may one day be used to treat patients with UM. We call for the research and development of more specific and effective SKP2 small molecule inhibitors to provide new treatment options for UM.
Conclusion
We identified the SKP2-p27 axis as a novel cancer-associated pathway that plays the main role in UM cell proliferation and regulates the UM cell cycle, and we demonstrate the ability of a SKP2 small molecule inhibitor to kill UM cells both in vitro and in vivo. These data provide evidence supporting SKP2-targeted treatment of UM.
Acknowledgments
This study was supported by National Key R&D Program of China (2018YFC1106100), Scientific Research Program of National Health and Family Planning Commission of China (201402014) and the National Natural Science Foundation of China (No. 81702781).
Author contributions
Hongbo Zhao, Hui Pan and Huixue Wang are co-first authors of this report. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Yang J, Manson DK, Marr BP, Carvajal RD. Treatment of uveal melanoma: where are we now? Ther Adv Med Oncol. 2018;10:1758834018757175. doi: 10.1177/1758834018757175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rietschel P, Panageas KS, Hanlon C, Patel A, Abramson DH, Chapman PB. Variates of survival in metastatic uveal melanoma. J Clin Oncol. 2005;23(31):8076–8080. doi: 10.1200/JCO.2005.02.6534 [DOI] [PubMed] [Google Scholar]
- 3.Ossowski L, Aguirre-Ghiso JA. Dormancy of metastatic melanoma. Pigment Cell Melanoma Res. 2010;23(1):41–56. doi: 10.1111/j.1755-148X.2009.00647.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pandiani C, Beranger GE, Leclerc J, Ballotti R, Bertolotto C. Focus on cutaneous and uveal melanoma specificities. Genes Dev. 2017;31(8):724–743. doi: 10.1101/gad.296962.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaliki S, Shields CL. Uveal melanoma: relatively rare but deadly cancer. Eye (Lond). 2017;31(2):241–257. doi: 10.1038/eye.2016.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Raamsdonk CD, Griewank KG, Crosby MB, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363(23):2191–2199. doi: 10.1056/NEJMoa1000584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X, Wu Q, Depeille P, et al. RasGRP3 mediates MAPK pathway activation in GNAQ mutant uveal melanoma. Cancer Cell. 2017;31(5):685–696.e686. doi: 10.1016/j.ccell.2017.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X, Wu Q, Tan L, et al. Combined PKC and MEK inhibition in uveal melanoma with GNAQ and GNA11 mutations. Oncogene. 2014;33(39):4724–4734. doi: 10.1038/onc.2013.418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alvarez-Rodriguez B, Latorre A, Posch C, Somoza A. Recent advances in uveal melanoma treatment. Med Res Rev. 2017;37(6):1350–1372. doi: 10.1002/med.21460 [DOI] [PubMed] [Google Scholar]
- 10.Khalili JS, Yu X, Wang J, et al. Combination small molecule MEK and PI3K inhibition enhances uveal melanoma cell death in a mutant GNAQ- and GNA11-dependent manner. Clin Cancer Res. 2012;18(16):4345–4355. doi: 10.1158/1078-0432.CCR-11-3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hutchenreuther J, Leask A. Why target the tumor stroma in melanoma? J Cell Commun Signal. 2018;12(1):113–118. doi: 10.1007/s12079-017-0419-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt M, Finley D. Regulation of proteasome activity in health and disease. Biochim Biophys Acta. 2014;1843(1):13–25. doi: 10.1016/j.bbamcr.2013.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rousseau A, Bertolotti A. Regulation of proteasome assembly and activity in health and disease. Nat Rev Mol Cell Biol. 2018;19(11):697–712. doi: 10.1038/s41580-018-0040-z [DOI] [PubMed] [Google Scholar]
- 14.Yuan T, Yan F, Ying M, et al. Inhibition of ubiquitin-specific proteases as a novel anticancer therapeutic strategy. Front Pharmacol. 2018;9:1080. doi: 10.3389/fphar.2018.01080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skaar JR, Pagan JK, Pagano M. SCF ubiquitin ligase-targeted therapies. Nat Rev Drug Discovery. 2014;13(12):889–903. doi: 10.1038/nrd4432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hao B, Zheng N, Schulman BA, et al. Structural basis of the Cks1-dependent recognition of p27(Kip1) by the SCF(SKP2) ubiquitin ligase. Mol Cell. 2005;20(1):9–19. doi: 10.1016/j.molcel.2005.09.003 [DOI] [PubMed] [Google Scholar]
- 17.Sonoda H, Inoue H, Ogawa K, Utsunomiya T, Masuda TA, Mori M. Significance of SKP2 expression in primary breast cancer. Clin Cancer Res. 2006;12(4):1215–1220. doi: 10.1158/1078-0432.CCR-05-1709 [DOI] [PubMed] [Google Scholar]
- 18.Jang W, Kim T, Koo JS, Kim SK, Lim DS. Mechanical cue-induced YAP instructs SKP2-dependent cell cycle exit and oncogenic signaling. Embo J. 2017;36(17):2510–2528. doi: 10.15252/embj.201696089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Gao D, Fukushima H, et al. Skp2: a novel potential therapeutic target for prostate cancer. Biochim Biophys Acta. 2012;1825(1):11–17. doi: 10.1016/j.bbcan.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang JY, Liu GZ, Wilmott JS, et al. SKP2-mediated stabilization of MTH1 promotes survival of melanoma cells upon oxidative stress. Cancer Res. 2017;77(22):6226–6239. doi: 10.1158/0008-5472.CAN-17-1965 [DOI] [PubMed] [Google Scholar]
- 21.Schuler S, Diersch S, Hamacher R, Schmid RM, Saur D, Schneider G. SKP2 confers resistance of pancreatic cancer cells towards TRAIL-induced apoptosis. Int J Oncol. 2011;38(1):219–225. [PubMed] [Google Scholar]
- 22.Kawakami K, Enokida H, Tachiwada T, Nishiyama K, Seki N, Nakagawa M. Increased SKP2 and CKS1 gene expression contributes to the progression of human urothelial carcinoma. J Urol. 2007;178(1):301–307. doi: 10.1016/j.juro.2007.03.002 [DOI] [PubMed] [Google Scholar]
- 23.Liang Y, Hou X, Cui Q, et al. Skp2 expression unfavorably impacts survival in resectable esophageal squamous cell carcinoma. J Transl Med. 2012;10:73. doi: 10.1186/1479-5876-10-233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calvisi DF, Ladu S, Pinna F, et al. SKP2 and CKS1 promote degradation of cell cycle regulators and are associated with hepatocellular carcinoma prognosis. Gastroenterology. 2009;137(5):1816–1826.e1811-1810. doi: 10.1053/j.gastro.2009.08.005 [DOI] [PubMed] [Google Scholar]
- 25.Bochis OV, Irimie A, Pichler M, Berindan-Neagoe I. The role of SKP2 and its substrate CDKN1B (p27) in colorectal cancer. J Gastrointestin Liver Dis. 2015;24(2):225–234. doi: 10.15403/jgld.2014.1121.242.skp2 [DOI] [PubMed] [Google Scholar]
- 26.Wander SA, Zhao D, Slingerland JM. p27: a barometer of signaling deregulation and potential predictor of response to targeted therapies. Clin Cancer Res. 2011;17(1):12–18. doi: 10.1158/1078-0432.CCR-10-0752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu IM, Hengst L, Slingerland JM. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat Rev Cancer. 2008;8(4):253–267. doi: 10.1038/nrc2347 [DOI] [PubMed] [Google Scholar]
- 28.Grabowski P, Schrader J, Wagner J, et al. Loss of nuclear p27 expression and its prognostic role in relation to cyclin E and p53 mutation in gastroenteropancreatic neuroendocrine tumors. Clin Cancer Res. 2008;14(22):7378–7384. doi: 10.1158/1078-0432.CCR-08-0698 [DOI] [PubMed] [Google Scholar]
- 29.Lough L, Sherman D, Ni E, Young LM, Hao B, Cardozo T. Chemical probes of SKP2-mediated p27 ubiquitylation and degradation. Med Chem Comm. 2018;9(7):1093–1104. doi: 10.1039/c8md00140e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pavlides SC, Huang KT, Reid DA, et al. Inhibitors of SCF-SKP2/Cks1 E3 ligase block estrogen-induced growth stimulation and degradation of nuclear p27kip1: therapeutic potential for endometrial cancer. Endocrinology. 2013;154(11):4030–4045. doi: 10.1210/en.2013-1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uddin S, Hussain A, Ahmed M, et al. S-phase kinase protein 2 is an attractive therapeutic target in a subset of diffuse large B-cell lymphoma. J Pathol. 2008;216(4):483–494. doi: 10.1002/path.2433 [DOI] [PubMed] [Google Scholar]
- 32.Khan AQ, Siveen KS, Prabhu KS, et al. Curcumin-mediated degradation of S-Phase kinase protein 2 induces cytotoxic effects in human papillomavirus-positive and negative squamous carcinoma cells. Front Oncol. 2018;8:399. doi: 10.3389/fonc.2018.00399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan CH, Morrow JK, Li CF, et al. Pharmacological inactivation of SKP2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell. 2013;154(3):556–568. doi: 10.1016/j.cell.2013.06.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu L, Grigoryan AV, Li Y, Hao B, Pagano M, Cardozo TJ. Specific small molecule inhibitors of SKP2-mediated p27 degradation. Chem Biol. 2012;19(12):1515–1524. doi: 10.1016/j.chembiol.2012.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Poole IC, van den Berg FM, van den Wijngaard RM, et al. Generation of a human melanocyte cell line by introduction of HPV16 E6 and E7 genes. In Vitro Cell Dev Biol Anim. 1997;33(1):42–49. doi: 10.1007/s11626-997-0021-6 [DOI] [PubMed] [Google Scholar]
- 36.Hu R, Aplin AE. Skp2 regulates G2/M progression in a p53-dependent manner. Mol Biol Cell. 2008;19(11):4602–4610. doi: 10.1091/mbc.e07-11-1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458(7237):438–444. doi: 10.1038/nature07960 [DOI] [PubMed] [Google Scholar]
- 38.Wang G, Chan CH, Gao Y, Lin HK. Novel roles of SKP2 E3 ligase in cellular senescence, cancer progression, and metastasis. Chin J Cancer. 2012;31(4):169–177. doi: 10.5732/cjc.011.10319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kulinski M, Achkar IW, Haris M, Dermime S, Mohammad RM, Uddin S. Dysregulated expression of SKP2 and its role in hematological malignancies. Leuk Lymphoma. 2018;59(5):1051–1063. doi: 10.1080/10428194.2017.1359740 [DOI] [PubMed] [Google Scholar]
- 40.Li C, Du L, Ren Y, et al. SKP2 promotes breast cancer tumorigenesis and radiation tolerance through PDCD4 ubiquitination. J Exp Clin Cancer Res. 2019;38(1):76. doi: 10.1186/s13046-019-1069-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Zvi YS, Batko B, et al. Down-regulation of SKP2 expression inhibits invasion and lung metastasis in osteosarcoma. Sci Rep. 2018;8(1):14294. doi: 10.1038/s41598-018-32428-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang C, Nan H, Ma J, et al. High SKP2/Low p57(Kip2) expression is associated with poor prognosis in human breast carcinoma. Breast Cancer. 2015;9(Suppl 1):13–21. doi: 10.4137/BCBCR.S30101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woenckhaus C, Maile S, Uffmann S, et al. Expression of SKP2 and p27KIP1 in naevi and malignant melanoma of the skin and its relation to clinical outcome. Histol Histopathol. 2005;20(2):501–508. doi: 10.14670/HH-20.501 [DOI] [PubMed] [Google Scholar]
- 44.Rose AE, Wang G, Hanniford D, et al. Clinical relevance of SKP2 alterations in metastatic melanoma. Pigment Cell Melanoma Res. 2011;24(1):197–206. doi: 10.1111/j.1755-148X.2010.00784.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Q, Murphy M, Ross J, Sheehan C, Carlson JA. Skp2 and p27kip1 expression in melanocytic nevi and melanoma: an inverse relationship. J Cutan Pathol. 2004;31(10):633–642. doi: 10.1111/j.0303-6987.2004.00243.x [DOI] [PubMed] [Google Scholar]
- 46.Masuda TA, Inoue H, Sonoda H, et al. Clinical and biological significance of S-phase kinase-associated protein 2 (SKP2) gene expression in gastric carcinoma: modulation of malignant phenotype by SKP2 overexpression, possibly via p27 proteolysis. Cancer Res. 2002;62(13):3819–3825. [PubMed] [Google Scholar]
- 47.Hsieh HY, Shieh JJ, Chen CJ, et al. Prodigiosin down-regulates SKP2 to induce p27(KIP1) stabilization and antiproliferation in human lung adenocarcinoma cells. Br J Pharmacol. 2012;166(7):2095–2108. doi: 10.1111/j.1476-5381.2012.01921.x [DOI] [PMC free article] [PubMed] [Google Scholar]