Abstract
A 52-yr-old man was found to have a 6.6-cm left frontotemporal mass. Biopsy revealed a low-grade astrocytic neoplasm with significant infiltration and an unusual morphologic appearance. Only rare mitotic figures were seen and the Ki-67 proliferative index was very low. Unexpectedly, the low-grade astrocytoma showed rapid progression within a short time, but subsequent resection showed similar histologic findings to the original biopsy with only slightly more mitoses and a marginally increased Ki-67 proliferative index. Molecular testing performed on the tumor showed no alterations in the IDH1, IDH2, EGFR, or BRAF genes by sequencing, intact 1p/19q by FISH, and a novel BCR-NTRK2 fusion transcript by reverse transcription and anchored multiplex PCR. The patient underwent standard-of-care therapy, both first and second line, for a high-grade glioma because of the aggressive behavior, but the glioma continued to progress despite treatment, and the patient died within 13.5 mo of the original diagnosis. At the time of diagnosis, the BCR-NTRK2 fusion transcript had not been described in solid tumors; however, a recent publication described this fusion transcript in two glioblastomas. Although no approved therapy was available for this patient, FDA approval has now been given for solid tumors with any NTRK gene family fusions. This unexpected molecular finding in a deceptively low-grade-appearing glioma supports the use of expanded molecular testing in gliomas and solid tumors, particularly in instances where targeted therapies are available.
Keywords: astrocytoma, glioblastoma, glioblastoma multiforme, glioma, neoplasm of the nervous system
CASE PRESENTATION
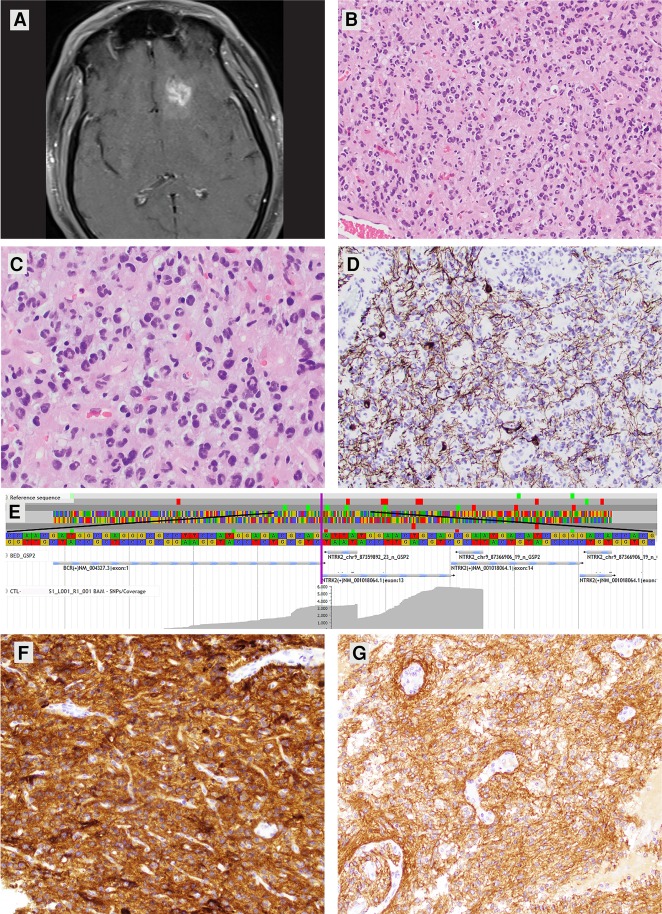
A 52-yr-old male presented to the emergency department after a motor vehicle collision secondary to witnessed, new-onset seizure activity. Imaging (CT and MRI) revealed a 6.6-cm intra-axial mass involving the left frontal and temporal lobes of the brain (Fig. 1A). The mass was partially cystic, showed patchy postcontrast enhancement, and contained calcifications. Initial biopsy showed a low-grade astrocytoma (Fig. 1B) with some features reminiscent of pilocytic astrocytoma (PA) including Rosenthal fibers and eosinophilic granular bodies. However, the cytology and diffusely infiltrative nature of the tumor were not entirely consistent with a PA. The cytology included unusual multinucleated cells with some mild pleomorphism (Fig. 1C). No features of anaplasia were identified; only rare mitotic figures were seen and there was no evidence of necrosis or microvascular proliferation. The Ki-67 proliferative index was <1%. Diffuse infiltration was confirmed with a neurofilament immunostain (Fig. 1D). Immunohistochemical staining for p53 showed nuclear positivity in only rare tumor cells and ATRX nuclear positivity was retained. Overall a final diagnosis of low-grade astrocytoma was rendered. Ancillary testing identified the tumor tissue as negative for hotspot mutations in IDH1, IDH2, EGFR, and BRAF via sequencing and negative for co-deletion of Chromosomes 1p and 19q via fluorescence in situ hybridization (FISH). A subpopulation of cells was noted to show polysomy of 19p and 19q in the range of three to five copies (in ∼39% of nuclei counted). Because of the mild resemblance to a pilocytic astrocytoma, tumor tissue was sent for BRAF fusion testing via reverse transcription and anchored multiplex polymerase chain reaction (AMP) that included multiple other anchored gene targets. Unexpectedly, a BCR-NTRK2 RNA fusion transcript was identified and no alterations were found in the BRAF gene.
Figure 1.
(A) Axial MR image postgadolinium administration showing a mass lesion in the left frontal lobe with patchy postcontrast enhancement. (B,C) H&E-stained sections of the initial biopsy specimen showing hypercellularity attributable to irregular and pleomorphic astrocytic-appearing tumor cells without appreciable cytoplasm. Many of the tumor cells had an unusual multinucleated appearance resembling a miniaturized version of “pennies on a plate.” Original magnifications: 200× and 400×. (D) Immunohistochemical staining for neurofilament highlighting background axons and some neurons confirming the infiltrative nature of the astrocytoma. Original magnification: 200×. (E) Screen-captured image taken from JBrowse visualization software. The reference sequence is present at the top. The fusion point is identified by a vertical purple line. The central stacked gray horizontal lines represent individual reads with variable lengths indicative of unique start sites on the left. The flat shelves on the right represent the location where anchored primers for NTRK2 begin. (F) Immunohistochemical staining for TrkB in the patient's low-grade astrocytoma showing diffuse 3+ positivity. Blood vessels serve as internal negative controls. Original magnification: 200×. (G) Immunohistochemical staining for TrkB in a pilocytic astrocytoma with a KIAA1549-BRAF fusion and no alterations in NTRK2 showing positivity as well, but ranging from 2 to 3+. Blood vessels also serve as internal negative controls. Magnification: 200×. (Tissue for both cases were stained on the same slide and images were taken at the same exposure and magnification.)
A follow-up MRI performed 70 d after diagnosis showed a new focus of enhancement in the left frontal lobe suspicious for disease progression. A partial resection was performed showing nearly identical histologic features with the only exception being slightly increased mitotic activity and higher Ki-67 proliferative index (∼2%) compared to the original biopsy. The second specimen maintained low-grade histology overall, not meeting criteria for anaplasia, and the diagnosis remained the same. Because of rapid progression of this otherwise low-grade-appearing glioma, the patient subsequently underwent standard of care therapy for a high-grade glioma. Adjuvant radiation therapy totaling 61.2 Gray, 34 fractions was delivered with concurrent temozolomide over a 1.5-mo period. Two additional cycles of temozolomide at 320 mg daily days 1 through 5 per 28 d were administered after the initial chemo/radiation treatment period. An additional follow-up MRI after therapy showed continued disease progression in the form of increased nodular enhancement in the left frontal lobe adjacent to the resection bed. This prompted a therapy shift to lomustine at 110 mg/m2 and bevacizumab at 10 mg/kg. Two weeks after changing therapy the patient presented with right-sided paralysis. Two weeks later therapy was discontinued and he was discharged to home hospice. He died 13.5 mo after initial diagnosis without definitive benefit from standard-of-care first-line or second-line therapy.
TECHNICAL ANALYSIS AND METHODS
All testing described was performed in a CLIA-certified laboratory. The report is exempted from consent protocols by federal and local regulations governing human subject research as it is performed on a deceased subject. The technical protocols were clinically validated except for TrkB immunohistochemistry, which was developed as a research protocol and details are described below. Formalin-fixed, paraffin-embedded (FFPE) tumor tissue from the first diagnostic procedure containing ∼80% tumor tissue was subjected to total nucleic acid extraction using an internally modified protocol performed with the RNeasy kit (QIAGEN 73504). Tumor percentage was estimated microscopically by a molecular pathologist. The modification of the RNeasy kit protocol involves skipping the addition of DNase and the subsequent incubation. This results in intact DNA and RNA in the final elution. Anchored multiplex PCR was performed by an internally validated protocol using the FusionPlex CTL kit (Archer AB0007) RNA-targeted component for fusion, splicing, or exon-skipping variant detection from 19 genes including NTRK1, NTRK2, NTRK3, and BRAF. The procedure involves reverse transcription using random primers and amplification-based library preparation using designed targeted primers for one side and a universal adapter primer for the other. Massively parallel sequencing was performed using the MiSeq platform 300 cycle kit (Illumina, part number: MS-102-2002). Primary data processing to demultiplexed FASTQ files is performed on the instrument. The remaining processing from the FASTQ through annotation was performed using the RNA Fusion and Mutation Archer Analysis Pipeline version 5.0 (Archer). Alignment was performed with reference hg19/GCRh37. The software uses a combination of Bowtie2 and BWA-MEM for alignment. Variant callers including Vision (Archer), FreeBayes, and LoFreq are used to make variant calls. Other in silico interpretive tools used by the pipeline include Ensembl's variant effect predictor (VEP), PolyPhen, SIFT, ClinVar, dbSNP, COSMIC, 1000 Genomes, and ExAc. The BCR-NTRK2 fusion was orthogonally confirmed using RT-PCR performed with the iScript cDNA synthesis kit (BioRad 1708890) that uses oligo(dT) and random hexamers for priming. PCR was performed with AmpliTaq Gold 360 Master Mix (Thermo-Fisher 4398881) and subjected to 2% agarose gel electrophoresis to observe the amplification product. This was followed by dye terminator sequencing using Big Dye Terminator Mix v3.1 (Thermo-Fisher 4337455). A 3130xl genetic analyzer is used for capillary electrophoresis. Interpretation is performed manually comparing electropherograms produced by Sequence Analysis Software version 6.0 (Thermo-Fisher) to the sequence output from the AMP assay. PCR and cycle sequencing were both performed with a BCR forward primer, caacagtccttcgacagcag and an NTRK2 reverse primer cagtgacgtctgtggaagg with an expected PCR product of 236 bp.
Select glioma-related genes were interrogated using the total nucleic acid extraction described above as starting material for massively parallel sequencing for IDH1, IDH2, EGFR, and BRAF hotspots/selected regions. Sequencing was performed using an in-house custom panel designed with AmpliSeq (Thermo-Fisher) amplicon-based library preparation and sequenced on the Ion S5XL (Thermo-Fisher). Data analysis was performed using the Torrent Suite version 5.2 with output of FASTQ, VCF, and BAM files. Further analysis was performed using a custom, in-house variant interpretation pipeline called the VIApipeline version 05.2017.12, which uses masking, filtering, and variant interpretation tools including Ensembl's VEP, PolyPhen, SIFT, ClinVar, dbSNP, COSMIC, and 1000 Genomes. No pathologic variants were detected in IDH1, IDH2, EGFR, or BRAF. Mean read depth for the sample was 1361 and overlying the ho-spot regions for IDH1, IDH2, BRAF, and EGFR ranged from 521 to 5107 reads.
FISH studies were performed on nuclei from FFPE tumor sections from the same block using the DNA probes LSI 1p/1q [1p36/1q25] and LSI 19p/19q [19p13/19q13]. No deletion was detected. There were increased copies of 19p and 19q probes in the range of 3 to 5 per nucleus in 39% of cells counted, nuc ish(LSI1p,LSI1q)x2[100],(LSI19p,LSI19q)x3∼5[39/100].
For TrkB immunohistochemistry, tissue sections were cut from FFPE blocks from the original biopsy specimen and, for comparison, from two additional pilocytic astrocytomas with known KIAA1549-BRAF fusions and no alterations in NTRK2. Four-micron-thick sections on positively charged slides were stained with a rabbit monoclonal antibody to TrkB (80G2; 1:200) from Cell Signaling Technology. Slides were stained on an Autostainer Link 48 (Agilent Dako) after PT Link (Agilent Dako) heat-induced epitope retrieval in Target Retrieval Solution (pH 9; Agilent Dako). The primary antibody and subsequent polymer-based EnVision FLEX + detection system (Agilent Dako) used were each incubated for 15 min. Neocortical brain parenchyma was used as a positive control.
VARIANT INTERPRETATION
An RNA fusion transcript (Chr 9:g.87359888::Chr 22:g.23524426) was detected that included exon 1 of BCR (NM_004327.3) and exon 13 of NTRK2 (NM_001018064.1) and was predicted to arise from a t(9;22)(q21;q11) rearrangement (Table 1; Fig. 1E). This fusion product was abundantly expressed with 1790 reads (41% of total reads at this locus) and 277 unique starts sites. The fusion product included the sequence that codes for the amino-terminal oligomerization portion of the BCR protein (codons 1–72 in exon 1). This is the region theorized to be necessary for the productive product of the BCR-ABL oncogenic fusion. The fusion product also contains the region that codes for the carboxy-terminal tyrosine kinase domain of the NTRK2 protein (predominantly in exons 21–23) that has been reported as necessary for increased activity in previously reported productive NTRK2 fusions (Zhao et al. 2002; Yeo et al. 2004). At the time of the clinical diagnosis, the fusion was novel; however, it has subsequently been identified in two cases of glioblastoma (Gatalica et al. 2019). This newly identified fusion transcript has not been previously reported in solid organ tumors. Other fusions containing the kinase domain of NTRK2 (e.g., NACC2-NTRK2 and QKI-NTRK2) have been reported in solid tumors and pilocytic astrocytomas (Jones et al. 2013). The neurotrophic tropomyosin receptor kinase 2 (NTRK2) gene encodes for the TrkB receptor tyrosine kinase expressed in neuronal tissue. Ligand binding results in activation of the Ras-ERK, PI3K, and PLCγ pathways leading to cellular differentiation and survival. NTRK2 gene rearrangements in cancer have been identified as pathogenic, tipping the homeostatic scales toward proliferation and tumorigenesis. Tumors with NTRK gene rearrangements have been shown to overexpress Trk by immunohistochemistry (Hechtman et al. 2017; Murphy et al. 2017; Hung et al. 2018). At the time that this case was clinically active, no approved NTRK targeted therapies were available; however, clinical trials were active including those for larotrectinib and entrectinib, which was indicated in the clinical molecular diagnostics report (ClinicalTrials.gov identification numbers NCT02637687, NCT02568267, and NCT02097810).
Table 1.
Genomic findings
| Gene/genomic location | Chr | HGVS DNA ref | HGVS protein ref | Variant type | Predicted effect | Allele frequency | Target coverage |
|---|---|---|---|---|---|---|---|
| BCR- NTRK2 | 9, 22 | t(9;22)(q21;q11) (hg 19 Chr 9:g.87359888::Chr 22:g.23524426) | n/a | BCR-NTRK2 fusion | Pathogenic, activating | 41% of reads at this locus of NTRK2 | 4567 reads |
The BCR-NTRK2 fusion product was predicted to be activating based on the prediction of an in-frame product, the protein domains that were predicted to be retained, and by similarity with other fusions involving the BCR and NTRK2 genes (Zhao et al. 2002; Yeo et al. 2004). This led us to investigate the protein expression of TrkB in this patient's glioma. We also stained control tissue in the form of normal brain parenchyma and two pilocytic astrocytomas that both harbored a KIAA1549-BRAF fusion and showed no alterations in NTRK2 based on the same FusionPlex CTL kit. Immunohistochemical staining for TrkB in this patient's glioma showed diffuse 3+ positivity (Fig. 1F). However, normal brain and both pilocytic astrocytomas all showed diffuse positivity, ranging from 2+ to 3+ (Fig. 1G).
These immunohistochemical results are not surprising given publicly available gene expression data in normal tissues and immunohistochemistry data in brain tissue and tumors. Both the Human Protein Atlas and GTEx data set show the highest NTRK2 mRNA expression in brain tissue. Muragaki and colleagues developed pan-Trk immunohistochemistry and demonstrated widespread protein expression in the developing and adult central and peripheral nervous systems (Muragaki et al. 1995). Trk-isoform-specific antibodies have also been evaluated in gliomas. Wang and colleagues demonstrated TrkA, B, and C reactivity in 100% of 4 low-grade astrocytomas and 12 glioblastomas and in 0% of 15 oligodendrogliomas (Wang et al. 1998). In this same study, weak staining was seen in normal astrocytes, strong staining was seen in reactive astrocytes, and no staining was seen in oligodendroglia. Wadhwa and colleagues detected TrkA and TrkB immunoreactivity in 100% of astrocytomas and glioblastomas (n = 5 for each), with stronger staining in lower-grade tumors (Wadhwa et al. 2003). Unfortunately, given widespread Trk protein expression in the brain, Trk immunohistochemistry appears to have a limited role in the detection of NTRK-fusion positive gliomas.
SUMMARY
We described a case of a clinically aggressive glioma with deceptively low-grade morphologic appearance. The malignancy showed progression at interval follow-up and did not show a response to conventional therapy including radiation and chemotherapy including alkylating agents and a VEGF inhibitor. The case ended in death only 13.5 mo after diagnosis. A novel RNA BCR-NTRK2 fusion transcript was identified in the tumor tissue by reverse transcription and anchored multiplex PCR that is favored to be activating and a driver mutation. It is currently unknown whether targeted therapy in the form of NTRK inhibition will be effective in treating gliomas and other brain tumors, particularly given the high expression of Trk proteins in normal brain. However, a promising report was recently published in which a single patient harboring an EML4-NTRK3 fusion-positive glioblastoma was treated with larotrectinib (a targeted small molecular inhibitor directed at NTRK1, NTRK2, and NTRK3) and showed a partial response (Schram et al. 2017). Since the conclusion of this case, larotrectinib was approved by the FDA for use in advanced solid tumors with fusions involving any of the three NTRK genes. Gliomas are not specifically referenced in the limited available data related to the trial that led to FDA approval for larotrectinib. But an expanded access trial and clinical trials including general primary brain neoplasms and gliomas specifically are open at this time (ClinicalTrials.gov identification numbers NCT03025360, NCT02576431, NCT02465060). The report of this case, variant, and deceptive morphology suggests utility in expanded testing for potentially clinically actionable variants to include interrogation for NTRK fusions. Because NTRK fusions are present in gliomas at a rate of ∼1.4%, testing at the time of diagnosis or primary treatment failure could be considered (Gatalica et al. 2019). The presented patient was unable to benefit from targeted therapy; however, the approved NTRK inhibitor may provide an option for future similar patients.
ADDITIONAL INFORMATION
Data Deposition and Access
The variant described in this manuscript was deposited in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and assigned the accession number SCV000882596.
Ethics Statement
This report is presented on a deceased patient. Per the electronic code of federal regulations title 45, subtitle A, subchapter A, part 46 (46.102e1) and the guide for investigators representing the policies of the University of Iowa Human Subjects Office/IRB approval processes are necessary for studies pertaining to a “living individual” or “live subject.” Material reported on and information gathered from deceased subjects is exempt from consent protocols.
Acknowledgments
We would like to thank the members of the University of Iowa Hospitals and Clinics Immunopathology Laboratory, and specifically Ellen Abusada for performing the immunohistochemical staining for TrkB.
Author Contributions
A.N.S. acquired clinical data, molecular images, performed literature review, and participated in composition and editing of the manuscript. K.A.J. acquired clinical data, created figures, composed portions of the manuscript, edited the manuscript, and performed literature review. A.M.B. assisted with performing IHC, performed literature review, composed portions of the manuscript, and edited the manuscript. A.D.B. composed portions of the manuscript and edited the manuscript.
Funding
Department of Pathology funding was used to perform immunohistochemistry.
Competing Interest Statement
The authors have declared no competing interest.
Referees
Toni Rose Jue
S. Zheng
Stephen Yip
REFERENCES
- Gatalica Z, Xiu J, Swensen J, Vranic S. 2019. Molecular characterization of cancers with NTRK gene fusions. Mod Pathol 32: 147–153. 10.1038/s41379-018-0118-3 [DOI] [PubMed] [Google Scholar]
- Hechtman JF, Benayed R, Hyman DM, Drilon A, Zehir A, Frosina D, Arcila ME, Dogan S, Klimstra DS, Ladanyi M, et al. 2017. Pan-Trk immunohistochemistry is an efficient and reliable screen for the detection of NTRK fusions. Am J Surg Pathol 41: 1547–1551. 10.1097/PAS.0000000000000911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung YP, Fletcher CDM, Hornick JL. 2018. Evaluation of pan-TRK immunohistochemistry in infantile fibrosarcoma, lipofibromatosis-like neural tumour and histological mimics. Histopathology 73: 634–644. 10.1111/his.13666 [DOI] [PubMed] [Google Scholar]
- Jones DT, Hutter B, Jäger N, Korshunov A, Kool M, Warnatz HJ, Zichner T, Lambert SR, Ryzhova M, Quang DA, et al. 2013. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet 45: 927–932. 10.1038/ng.2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muragaki Y, Timothy N, Leight S, Hempstead BL, Chao MV, Trojanowski JQ, Lee VM. 1995. Expression of trk receptors in the developing and adult human central and peripheral nervous system. J Comp Neurol 356: 387–397. 10.1002/cne.903560306 [DOI] [PubMed] [Google Scholar]
- Murphy DA, Ely HA, Shoemaker R, Boomer A, Culver BP, Hoskins I, Haimes JD, Walters RD, Fernandez D, Stahl JA, et al. 2017. Detecting gene rearrangements in patient populations through a 2-step diagnostic test comprised of rapid IHC enrichment followed by sensitive next-generation sequencing. Appl Immunohistochem Mol Morphol 25: 513–523. 10.1097/PAI.0000000000000360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schram AM, Taylor BS, Hechtman JF, Benayed R, Wang L, Hanusch B, Young R, Grommes C, Ku N, Hyman DM, et al. 2017. Abstract LB-302: potential role of larotrectinib (LOXO-101), a selective pan-TRK inhibitor, in NTRK fusion-positive recurrent glioblastoma. Cancer Res 77: LB-302 10.1158/1538-7445.AM2017-LB-302 [DOI] [Google Scholar]
- Wadhwa S, Nag TC, Jindal A, Kushwaha R, Mahapatra AK, Sarkar C. 2003. Expression of the neurotrophin receptors Trk A and Trk B in adult human astrocytoma and glioblastoma. J Biosci 28: 181–188. 10.1007/BF02706217 [DOI] [PubMed] [Google Scholar]
- Wang Y, Hagel C, Hamel W, Müller S, Kluwe L, Westphal M. 1998. Trk A, B, and C are commonly expressed in human astrocytes and astrocytic gliomas but not by human oligodendrocytes and oligodendroglioma. Acta Neuropathol 96: 357–364. 10.1007/s004010050906 [DOI] [PubMed] [Google Scholar]
- Yeo GS, Connie Hung CC, Rochford J, Keogh J, Gray J, Sivaramakrishnan S, O'Rahilly S, Farooqi IS. 2004. A de novo mutation affecting human TrkB associated with severe obesity and developmental delay. Nat Neurosci 7: 1187–1189. 10.1038/nn1336 [DOI] [PubMed] [Google Scholar]
- Zhao X, Ghaffari S, Lodish H, Malashkevich VN, Kim PS. 2002. Structure of the Bcr-Abl oncoprotein oligomerization domain. Nat Struct Biol 9: 117–120. 10.1038/nsb747 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The variant described in this manuscript was deposited in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and assigned the accession number SCV000882596.