Abstract
Prostate cancers harboring DNA repair gene alterations are particularly sensitive to PARP inhibitor treatment. We report a case of an advanced prostate cancer patient profiled within the NCT-MASTER (Molecularly Aided Stratification for Tumor Eradication Research) precision oncology program using next-generation sequencing. Comprehensive genomic and transcriptomic analysis identified a pathogenic germline PALB2 variant as well as a mutational signature associated with disturbed homologous recombination together with structural genomic rearrangements. A molecular tumor board identified a potential benefit of targeted therapy and recommended PARP inhibition and platinum-based chemotherapy. Single-agent treatment with the PARP inhibitor olaparib as well as subsequent combination with platinum-based chemotherapy resulted in disease stabilization and substantial improvement of clinical symptoms. Upon progression, we performed whole-exome and RNA sequencing of a liver metastasis, which demonstrated up-regulation of several genes characteristic for the neuroendocrine prostate cancer phenotype as well as a novel translocation resulting in an in-frame, loss-of-function fusion of RB1. We suggest that multidimensional genomic characterization of prostate cancer patients undergoing PARP inhibitor therapy will be necessary to capture and understand predictive biomarkers of PARP inhibitor sensitivity and resistance.
Keywords: prostate cancer
INTRODUCTION
With growing evidence for the clinical activity of PARP inhibitors in several cancer entities, mutations in genes involved in HR are increasingly recognized as possible biomarkers, although the molecular events responsible for the development of resistance are incompletely understood (Lord and Ashworth 2017). Advanced prostate cancer is often characterized by germline and somatic alterations of genes involved in DNA repair via homologous recombination (HR) (Pritchard et al. 2016), which have also been associated with sensitivity to poly(ADP-ribose) polymerase (PARP) inhibition in advanced disease. These data also suggest that a substantial proportion of advanced prostate cancer patients may display “BRCAness” and can benefit from PARP inhibition and/or platinum-based chemotherapy (Lord and Ashworth 2016). Data from a phase 2 study of olaparib in metastatic castration-resistant prostate cancer (CRPC) revealed that 88% of patients that had a response to olaparib harbored one or more deleterious alterations in DNA repair genes (Mateo et al. 2015). Here we report the first case of a pathogenic germline PALB2 (partner and localizer of BRCA2) variant that confers sensitivity to PARP inhibition in a patient with prostate cancer.
Clinical Presentation
The patient was diagnosed with locally advanced prostate adenocarcinoma at the age of 43. His family history was not suggestive of a hereditary cancer predisposition syndrome. The prostate-specific antigen (PSA) level at diagnosis was 76 ng/mL. Because of massive bladder neck infiltration, the patient underwent initial transurethral resection, followed by 6 mo of neoadjuvant combined hormonal therapy with leuprorelin and bicalutamide (PSA nadir 0.6 ng/mL) and radical retropubic prostatectomy. Even though the patient had a PSA response, postoperative staging and grading exposed a large tumor with nodal metastasis and positive surgical margins (pT4, pN1, cM0, R1) and a Gleason score of 5 + 4 = 9. Within a month after radical surgery, and despite ongoing hormonal therapy, the patient became castration-refractory and was treated with six cycles of docetaxel chemotherapy. Unfortunately, prostate-specific membrane antigen positron emission tomography after completion of chemotherapy demonstrated disseminated bone and lymph node metastases. Because of a PSA response during chemotherapy, four additional cycles of docetaxel were administered. Zoledronic acid was added to the treatment regimen. The patient developed fatigue and his general condition worsened because of progressive disease and accumulating side effects of chemotherapy (fatigue and muscular weakness leading to Eastern Cooperative Oncology Group [ECOG] and World Health Organization [WHO] Score 3). Following PSA and radiographic progression with hepatic metastases, therapy with abiraterone acetate and prednisolone was initiated, which could not bring about any treatment benefit.
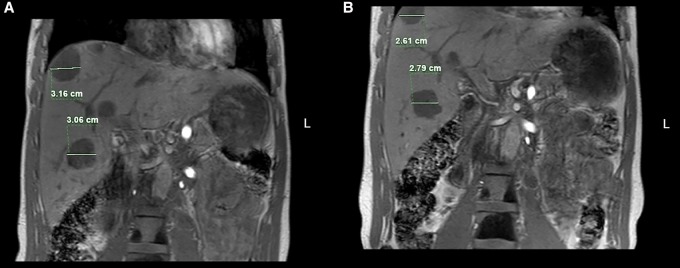
The primary tumor from the radical prostatectomy was analyzed using whole-genome sequencing and made available for clinical interpretation within the NCT-MASTER (Molecularly Aided Stratification for Tumor Eradication Research) program (Horak et al. 2017). We identified a pathogenic, heterozygous germline PALB2 mutation (Table 1) with somatic loss of heterozygosity in the tumor as well as a mutational signature suggestive of defective DNA repair via homologous recombination when analyzing the entirety of somatic SNVs (Alexandrov et al. 2013). This prompted a molecularly informed tumor board recommendation by the NCT MASTER molecular tumor board of PARP inhibitor treatment with or without platinum-based chemotherapy as well as genetic counseling. Single-agent treatment with the PARP inhibitor olaparib resulted in substantial improvement of clinical symptoms, and only moderate side effects (e.g., anemia CTCAE Grade 2) were observed. Remarkably, the patient restarted physical activities and his general condition improved (ECOG/WHO Score 1). However, staging after 6 wk revealed disease progression with 24.1% increase of target lesions. In addition, the PSA value increased during treatment with olaparib monotherapy from 15.2 µg/L to 110 µg/L, whereas LDH decreased from 471 U/L to 297 U/L. Because of the considerable clinical improvement, olaparib treatment was continued and cisplatin (30 mg/m2 i.v. weekly) was added. Magnetic resonance imaging after 2 mo of olaparib/cisplatin therapy showed disease stabilization by Prostate Cancer Clinical Trials Working Group 3 (PCWG3) criteria (Fig. 1). Addition of cisplatin resulted in a mild decrease of PSA to 96.4 µg/L as well as of LDH to 278 U/L. However, worsening anemia (CTCAE Grade 3) required transfusion of red blood cells and we observed decreasing neutrophil counts (CTCAE Grade 2).
Table 1.
Variant table
| Gene | Chromosome | HGVS cDNA | HGVS protein | Variant type | Predicted effect | dbSNP | Genotype | ClinVar |
|---|---|---|---|---|---|---|---|---|
| PALB2 | 16 | NM_024675.3:c.509_510delGA | p.Arg170Ilefs | Frameshift deletion | Loss of function | rs515726123 | Germline heterozygous | 132267 |
Figure 1.
Magnetic resonance imaging of liver metastases (A) before and (B) after 2 mo of olaparib/cisplatin treatment. Two hepatic lesions were selected and their largest diameters measured in comparison according to RECIST 1.1 criteria as defined by PCGW3. Sum of longest diameters of the two target lesions was 6.22 cm before and 5.40 cm after olaparib/cisplatin treatment (13% decrease).
Three months thereafter, the lymphatic and hepatic metastases progressed and clinical symptoms worsened. At time of radiological progression PSA had further decreased to 50.9 µg/L, whereas LDH dramatically increased to 1501 U/L. Given the patient's young age and his desire to continue treatment, several experimental salvage regimens were administered. However, no durable stabilization of disease could be achieved and the patient died 5 mo later.
Germline and Somatic Genomic Alterations
To identify molecular alterations that could inform experimental therapy in this patient, we used whole-genome and transcriptome sequencing data from the prostatectomy specimen that were obtained at diagnosis within the ICGC early-onset prostate cancer project (Weischenfeldt et al. 2013) as well as whole-exome and transcriptome sequencing data from a liver metastasis that was obtained upon progression on PARP Inhibitor therapy two and a half years later. Single nucleotide variants (SNVs), small insertions and deletions (indels), copy-number variations (CNVs), genomic structural variations, and gene fusions were interpreted with regard to their clinical significance and ranked according to therapeutic relevance.
Analysis of the primary tumor revealed 14 somatic nonsynonymous SNVs and two somatic indels including a known TP53 missense variant, NM_000546.5:c.451C>T, p.(Pro151Ser) (rs28934874) in the DNA-binding domain, which has been previously described as a pathogenic mutation in prostate cancer (Supplemental Table S1; Shi et al. 2002).
We detected a germline frameshift deletion in PALB2, NM_024675.3:c.509_510del, p.Arg170Ilefs*14 (rs515726123) (Table 1), which has already been described as a pathogenic Polish founder mutation associated with a predisposition for breast and ovarian cancer (Dansonka-Mieszkowska et al. 2010). This variant demonstrated somatic loss of heterozygosity with a mutational frequency of 70% in the primary tumor tissue. PALB2 is responsible for BRCA2 nuclear localization and DNA damage response (Xia et al. 2006) and serves as a molecular adaptor between BRCA1 and BRCA2 (Sy et al. 2009). Therefore, absence of PALB2 leads to impaired recruitment of BRCA2 and RAD51 to sites of DNA damage, making it a bona fide tumor suppressor. Germline loss-of-function mutations of PALB2 have been primarily associated with hereditary susceptibility to breast, ovarian, and pancreatic cancer (Jones et al. 2009; Antoniou et al. 2014; Southey et al. 2016), whereas homozygous loss of function leads to Fanconi anemia (Reid et al. 2007; Xia et al. 2007).
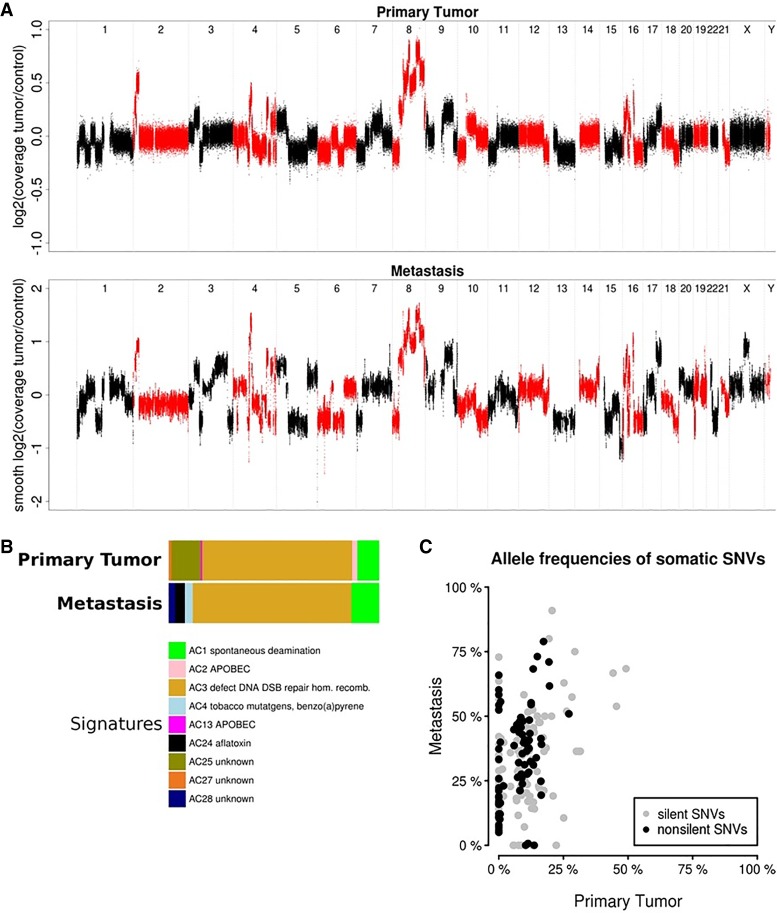
Interestingly, we detected no other genetic hallmarks of prostate cancer, such as a TMPRSS2-ETS fusion or PTEN inactivation. We also observed prominent amplification (total copy number of 6) of the MYC proto-oncogene on Chromosome 8q24 (Fig. 2A), a common alteration in prostate cancer associated with increased aggressiveness (Van Den Berg et al. 1995; Jenkins et al. 1997; Fromont et al. 2013), which was accompanied by increased MYC mRNA expression. Another amplicon on Chromosome 2p24 encompassed the MYC family member MYCN. However, MYCN mRNA expression was not elevated in the primary tumor, in agreement with the observation that MYC and MYCN activation are mutually exclusive events in advanced prostate cancer (Boutros et al. 2015) and MYCN expression is often associated with a neuroendocrine phenotype (Beltran et al. 2016). Of note, MYCN expression was increased 4.8-fold in the therapy-resistant metastasis as compared to the primary tumor. Comparison of the primary and metastatic samples demonstrated no apparent evolution of CNVs contributing to additional oncogenic events, whereas extensive genomic rearrangements were evident at both time points (Fig. 2A). We also performed a supervised analysis of known mutational signatures (Alexandrov et al. 2013) and observed a strong contribution of signature Alexandrov-COSMIC 3, which is associated with defective HR DNA repair, to the mutational catalog in both the primary tumor and the metastasis (Fig. 2B).
Figure 2.
(A) DNA copy-number profiles of the primary tumor (upper panel) and metastasis (lower panel). (B) Relative contributions of mutational signatures (Alexandrov-COSMIC, AC) in the primary tumor (upper panel) and metastasis (lower panel) depicting the prevalence of the AC3 signature. (C) Distribution of measured allele frequencies of all somatic synonymous and nonsynonymous SNVs in both samples.
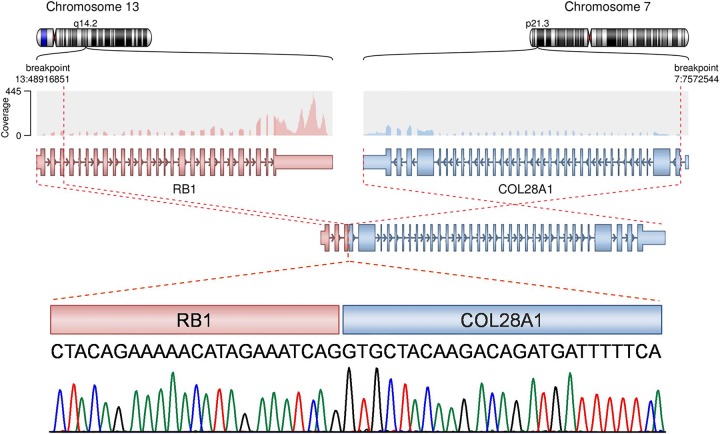
The liver metastasis harbored 69 somatic nonsynonymous SNVs and two somatic indels (Supplemental Table S1). The TP53 mutation as well as five other SNVs and both indels identified in the primary tumor could not be detected in the metastasis, suggesting subclonal selection and evolution of the primary tumor during therapy. However, the distribution of allele frequencies of synonymous and nonsynonymous SNVs in both samples was influenced mainly by the tumor cell content and similar between the two samples (Fig. 2C). Interestingly, the metastasis carried a novel translocation, resulting in an in-frame fusion of RB1 exon 3 and exon 2 of COL28A1, encoding a collagen subtype, and was predicted to cause loss of retinoblastoma protein function (Fig. 3). We also observed discrete changes in the transcriptional profile, in particular up-regulation of several genes characteristic for the neuroendocrine prostate cancer phenotype such as MYCN, AURKA, EZH2, and DNMT1 (Beltran et al. 2016).
Figure 3.
Fusion between exon 3 of RB1 and exon 2 of COL28A1. Upper panels show the sequencing coverage and exon structure of the RB1 and COL28A1 as well as the structure of the putative fusion transcript. Chromatogram of the Sanger sequencing showing the nucleotide sequence of the fusion region is shown below.
DISCUSSION
Our observations suggest that deleterious germline PALB2 variants represent an actionable target in metastatic CRPC and support extended genomic testing in this patient population. CRPC has been reported to be a very heterogeneous entity and several molecular subtypes have recently been defined (Aggarwal et al. 2017). Among these, tumors with DNA repair alterations might be particularly amenable to precision oncology approaches as a plethora of DNA repair genes may be linked to PARP inhibitor sensitivity. Recent studies have established the occurrence of germline PALB2 mutations in prostate cancer as well as the sensitivity of other PALB2 deficient tumor entities to PARP inhibition (Erkko et al. 2007; Mateo et al. 2015). However, not every patient with DNA repair defects may experience clinical benefit, and the identification and detailed characterization of predictive biomarkers is an ongoing effort. A germline missense variant in the FANCA gene has been associated with exceptional response to platinum in neuroendocrine prostate cancer (Beltran et al. 2015). In our case, we observed an initial response to PARP inhibition combined with platinum-based therapy, which was followed by progression of an aggressive tumor resistant to further treatment and displaying newly acquired somatic genetic alterations typically associated with neuroendocrine prostate cancer, such as an inactivating RB1 fusion and elevated MYCN expression (Tan et al. 2014; Beltran et al. 2016). We did not find evidence for restoration of HR proficiency leading to secondary resistance to olaparib. We also hypothesize that resistance to PARP inhibition in prostate adenocarcinoma with “BRCAness” might co-occur with the emergence of neuroendocrine transdifferentiation (Akamatsu et al. 2018).
In summary, this comprehensive genomic and transcriptomic analysis indicates that pathogenic germline mutations in PALB2 are associated with hallmarks of defective HR, including extensive structural genomic rearrangements and enrichment of specific mutational signatures, and might predict response to PARP inhibition as well as platinum-based chemotherapy. We propose that multidimensional genomic characterization of prostate cancer patients undergoing PARP inhibitor therapy will be necessary to define the entire spectrum of predictive biomarkers.
METHODS
Next-Generation Sequencing, Bioinformatic Analysis, and Target Validation
Whole-genome, whole-exome, and RNA sequencing were performed as described previously (Weischenfeldt et al. 2013; Kordes et al. 2016). The fusion transcript was validated by Sanger sequencing. SNVs and indels were identified using previously reported bioinformatics pipelines (Weischenfeldt et al. 2013). Structural variants were called with CREST (Wang et al. 2011). CNVs were extracted from whole-exome sequencing data with CNVkit (version 0.8.3.dev0). For the extraction of CNVs from whole-genome sequencing data we used our in-house CNV calling pipeline ACEseq (version 1.0.189) (Kleinheinz et al. 2017). A supervised analysis of mutational signatures to determine the contributions of known mutational signatures to the mutational catalog of individual samples was performed as described previously (Dieter et al. 2017). RNA-sequencing reads were mapped to the GRCh37.p13 assembly of the human reference genome using STAR 2.4.1c with the following alignment parameters:–outFilterMultimapNmax 10 –outFilterScoreMinOverLread 0.66 –outFilterMatchNmin 0 –outFilterMatchNminOverLread 0.66 –outFilterMismatchNmax 10 –outFilterMismatchNoverLmax 0.3 –clip5pNbases 0 –clip3pNbases 0 –clip3pAfterAdapterNbases 0 –clip3pAdapterSeq AGATCGGAAGAG –clip3pAdapterMMp 0.1 –seedSearchLmax 0 –seedSearchStartLmax 50 –alignIntronMin 21 –alignIntronMax 0 –alignMatesGapMax 0 –alignEndsType Local –alignSoftClipAtReferenceEnds No –chimSegmentMin 15 –chimScoreMin 1 –chimJunctionOverhangMin 15. The expression of genes annotated in the GencodeV19 gene model was quantified with featureCounts (version 1.4.6-p2) (Liao et al. 2014). High-confidence gene fusion predictions were extracted from chimeric alignments produced by STAR using our in-house pipeline Arriba (version 0.8), which removes recurrent alignment artifacts, transcript variants also observed in normal tissue, and reads with low sequence complexity as well as events with short anchors or breakpoints in close proximity or a low number of supporting reads relative to the overall number of predicted events in a gene. Sequencing coverage metrics are provided in Supplemental Table S2.
ADDITIONAL INFORMATION
Data Deposition and Access
The raw analytical data have been deposited in the European Genome-phenome Archive (https://www.ebi.ac.uk/ega/datasets) under accession numbers EGAS00001000383 (primary tumor, patient ID EOPC-125) and EGAS00001003133 (metastasis). Oncological outcome data were collected via the Progether PROM's (patient reported outcome measurement) interface (www.progether.com).
Ethics Statement
Prostate tumor samples and a matched normal blood sample were obtained following written informed consent under an institutional review board–approved protocol covering all aspects relevant to clinical cancer genome sequencing. This study was approved and conducted in accordance with the Declaration of Helsinki.
Author Contributions
P.H., S.U., L.F., B.H., H.G., S.F., and T.S. were involved in the conception and design of the study. P.H., J.W., G.v.A., Bu.B., A.S., S.U., L.F., B.K., L.G., D.H., S.K., C.H., B.H., and R.P. collected and assembled the data. All authors performed data analysis and interpretation. P.H., J.W., S.U., B.K., L.G., S.F., and T.S. wrote the manuscript, and all authors approved the final version and are accountable for all aspects of the work.
Funding
This work was supported by grant H021 from the DKFZ-Heidelberg Center for Personalized Oncology.
Competing Interest Statement
The authors have declared no competing interest.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Aggarwal RR, Feng FY, Small EJ. 2017. Emerging categories of disease in advanced prostate cancer and their therapeutic implications. Oncology 31: 467–474. [PubMed] [Google Scholar]
- Akamatsu S, Inoue T, Ogawa O, Gleave ME. 2018. Clinical and molecular features of treatment-related neuroendocrine prostate cancer. Int J Urol 25: 345–351. 10.1111/iju.13526 [DOI] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale AL, et al. 2013. Signatures of mutational processes in human cancer. Nature 500: 415–421. 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou AC, Casadei S, Heikkinen T, Barrowdale D, Pylkäs K, Roberts J, Lee A, Subramanian D, De Leeneer K, Fostira F, et al. 2014. Breast-cancer risk in families with mutations in PALB2. N Engl J Med 371: 497–506. 10.1056/NEJMoa1400382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran H, Eng K, Mosquera JM, Sigaras A, Romanel A, Rennert H, Kossai M, Pauli C, Faltas B, Fontugne J, et al. 2015. Whole-exome sequencing of metastatic cancer and biomarkers of treatment response. JAMA Oncol 1: 466–474. 10.1001/jamaoncol.2015.1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BV, Varambally S, et al. 2016. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 22: 298–305. 10.1038/nm.4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros PC, Fraser M, Harding NJ, de Borja R, Trudel D, Lalonde E, Meng A, Hennings-Yeomans PH, McPherson A, Sabelnykova VY, et al. 2015. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat Genet 47: 736–745. 10.1038/ng.3315 [DOI] [PubMed] [Google Scholar]
- Dansonka-Mieszkowska A, Kluska A, Moes J, Dabrowska M, Nowakowska D, Niwinska A, Derlatka P, Cendrowski K, Kupryjanczyk J. 2010. A novel germline PALB2 deletion in Polish breast and ovarian cancer patients. BMC Med Genet 11: 20 10.1186/1471-2350-11-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieter SM, Heining C, Agaimy A, Huebschmann D, Bonekamp D, Hutter B, Ehrenberg KR, Fröhlich M, Schlesner M, Scholl C, et al. 2017. Mutant KIT as imatinib-sensitive target in metastatic sinonasal carcinoma. Ann Oncol 28: 142–148. 10.1093/annonc/mdw446 [DOI] [PubMed] [Google Scholar]
- Erkko H, Xia B, Nikkilä J, Schleutker J, Syrjäkoski K, Mannermaa A, Kallioniemi A, Pylkäs K, Karppinen SM, Rapakko K, et al. 2007. A recurrent mutation in PALB2 in Finnish cancer families. Nature 446: 316–319. 10.1038/nature05609 [DOI] [PubMed] [Google Scholar]
- Fromont G, Godet J, Peyret A, Irani J, Celhay O, Rozet F, Cathelineau X, Cussenot O. 2013. 8q24 amplification is associated with Myc expression and prostate cancer progression and is an independent predictor of recurrence after radical prostatectomy. Hum Pathol 44: 1617–1623. 10.1016/j.humpath.2013.01.012 [DOI] [PubMed] [Google Scholar]
- Horak P, Klink B, Heining C, Gröschel S, Hutter B, Fröhlich M, Uhrig S, Hübschmann D, Schlesner M, Eils R, et al. 2017. Precision oncology based on omics data: the NCT Heidelberg experience. Int J Cancer 141: 877–886. 10.1002/ijc.30828 [DOI] [PubMed] [Google Scholar]
- Jenkins RB, Qian J, Lieber MM, Bostwick DG. 1997. Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. Cancer Res 57: 524–531. [PubMed] [Google Scholar]
- Jones S, Hruban RH, Kamiyama M, Borges M, Zhang X, Parsons DW, Lin JC, Palmisano E, Brune K, Jaffee EM, et al. 2009. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science 324: 217 10.1126/science.1171202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinheinz K, Bludau I, Huebschmann D, Heinold M, Kensche P, Gu Z, Lopez C, Hummel M, Klapper W, Moeller P, et al. 2017. ACEseq—allele specific copy number estimation from whole genome sequencing. bioRxiv 10.1101/210807 [DOI] [Google Scholar]
- Kordes M, Röring M, Heining C, Braun S, Hutter B, Richter D, Geörg C, Scholl C, Gröschel S, Roth W, et al. 2016. Cooperation of BRAFF595L and mutant HRAS in histiocytic sarcoma provides new insights into oncogenic BRAF signaling. Leukemia 30: 937–946. 10.1038/leu.2015.319 [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lord CJ, Ashworth A. 2016. BRCAness revisited. Nat Rev Cancer 16: 110–120. 10.1038/nrc.2015.21 [DOI] [PubMed] [Google Scholar]
- Lord CJ, Ashworth A. 2017. PARP inhibitors: synthetic lethality in the clinic. Science 355: 1152–1158. 10.1126/science.aam7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, Nava Rodrigues D, Robinson D, Omlin A, Tunariu N, et al. 2015. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med 373: 1697–1708. 10.1056/NEJMoa1506859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard CC, Mateo J, Walsh MF, De Sarkar N, Abida W, Beltran H, Garofalo A, Gulati R, Carreira S, Eeles R, et al. 2016. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med 375: 443–453. 10.1056/NEJMoa1603144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid S, Schindler D, Hanenberg H, Barker K, Hanks S, Kalb R, Neveling K, Kelly P, Seal S, Freund M, et al. 2007. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet 39: 162–164. 10.1038/ng1947 [DOI] [PubMed] [Google Scholar]
- Shi XB, Nesslinger NJ, Deitch AD, Gumerlock PH, deVere White RW. 2002. Complex functions of mutant p53 alleles from human prostate cancer. Prostate 51: 59–72. 10.1002/pros.10072 [DOI] [PubMed] [Google Scholar]
- Southey MC, Goldgar DE, Winqvist R, Pylkäs K, Couch F, Tischkowitz M, Foulkes WD, Dennis J, Michailidou K, van Rensburg EJ, et al. 2016. PALB2, CHEK2 and ATM rare variants and cancer risk: data from COGS. J Med Genet 53: 800–811. 10.1136/jmedgenet-2016-103839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy SM, Huen MS, Chen J. 2009. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci 106: 7155–7160. 10.1073/pnas.0811159106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan HL, Sood A, Rahimi HA, Wang W, Gupta N, Hicks J, Mosier S, Gocke CD, Epstein JI, Netto GJ, et al. 2014. Rb loss is characteristic of prostatic small cell neuroendocrine carcinoma. Clin Cancer Res 20: 890–903. 10.1158/1078-0432.CCR-13-1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Berg C, Guan XY, Von Hoff D, Jenkins R, Bittner, Griffin C, Kallioniemi O, Visakorpi, McGill, Herath J, et al. 1995. DNA sequence amplification in human prostate cancer identified by chromosome microdissection: potential prognostic implications. Clin Cancer Res 1: 11–18. [PubMed] [Google Scholar]
- Wang J, Mullighan CG, Easton J, Roberts S, Heatley SL, Ma J, Rusch MC, Chen K, Harris CC, Ding L, et al. 2011. CREST maps somatic structural variation in cancer genomes with base-pair resolution. Nat Methods 8: 652–654. 10.1038/nmeth.1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Simon R, Feuerbach L, Schlangen K, Weichenhan D, Minner S, Wuttig D, Warnatz HJ, Stehr H, Rausch T, et al. 2013. Integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancer. Cancer Cell 23: 159–170. 10.1016/j.ccr.2013.01.002 [DOI] [PubMed] [Google Scholar]
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM. 2006. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell 22: 719–729. 10.1016/j.molcel.2006.05.022 [DOI] [PubMed] [Google Scholar]
- Xia B, Dorsman JC, Ameziane N, de Vries Y, Rooimans MA, Sheng Q, Pals G, Errami A, Gluckman E, Llera J, et al. 2007. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet 39: 159–161. 10.1038/ng1942 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw analytical data have been deposited in the European Genome-phenome Archive (https://www.ebi.ac.uk/ega/datasets) under accession numbers EGAS00001000383 (primary tumor, patient ID EOPC-125) and EGAS00001003133 (metastasis). Oncological outcome data were collected via the Progether PROM's (patient reported outcome measurement) interface (www.progether.com).