Abstract
Background: Methicillin-resistant Staphylococcus aureus (MRSA) is a public health threat and a major cause of hospital-acquired and community-acquired infections. This study aimed to investigate the genetic diversity of MRSA isolates from 2015 to 2017 and to characterize the major MRSA clones and anti-biogram trends in Palestine.
Methodology: Isolates were obtained from 112 patients admitted to different hospitals of West Bank and East Jerusalem, originating from different clinical sources. Antibiotic susceptibility patterns, staphylococcal chromosomal cassette mec (SCCmec) typing, and Staphylococcus aureus protein A (spa) typing were determined. Also, a panel of toxin genes and virulence factors was studied, including: Panton-Valentine Leukocidin (PVL), ACME-arcA, Toxic Shock Syndrome Toxin-1 (TSST-1), and Exfoliative Toxin A (ETA).
Results: Of the 112 confirmed MRSA isolates, 100% were resistant to all β-lactam antibiotics. Resistance rates to other non- β-lactam classes were as the following: 18.8% were resistant to trimethoprim-sulfamethoxazole, 23.2% were resistant to gentamicin, 34.8% to clindamycin, 39.3% to ciprofloxacin, and 63.4% to erythromycin. All MRSA isolates were susceptible to vancomycin (100%). Of all isolates, 32 isolates (28.6%) were multidrug- resistant (MDR). The majority of the isolates were identified as SCCmec type IV (86.6%). The molecular typing identified 29 spa types representing 12 MLST-clonal complexes (CC). The most prevalent spa types were: spa type t386 (CC1)/(12.5%), spa type t044 (CC80)/(10.7%), spa type t008 (CC8)/(10.7%), and spa type t223 (CC22)/(9.8%). PVL toxin gene was detected in (29.5%) of all isolates, while ACME-arcA gene was present in 18.8% of all isolates and 23.2% had the TSST-1 gene. The two most common spa types among the TSST-1positive isolates were the spa type t223 (CC22)/(Gaza clone) and the spa type t021 (CC30)/(South West Pacific clone). All isolates with the spa type t991 were ETA positive (5.4%). USA-300 clone (spa type t008, positive for PVL toxin gene and ACME-arcA genes) was found in nine isolates (8.0%).
Conclusions: Our results provide insights into the epidemiology of MRSA strains in Palestine. We report a high diversity of MRSA strains among hospitals in Palestine, with frequent SCCmec type IV carriage. The four prominent clones detected were: t386-IV/ CC1, the European clone (t044/CC80), Gaza clone (t223/CC22), and the USA-300 clone (t008/CC8).
Keywords: MRSA, spa, SCCmec, PVL, TSST-1, arcA, USA-300clone
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is an important bacterial pathogen in both community and healthcare-related settings in different parts of the world. It is one of the major human pathogens that can cause broad variety of human diseases ranging from mild skin and soft tissue infections (SSTIs) to severe life-threatening invasive infections; such as: endocarditis, osteomyelitis, necrotizing pneumonia, bacteremia, septicemia, meningitis, food poisoning, and toxic shock syndrome (1–5). Methicillin resistance is caused by an alteration in the penicillin-binding protein (PBP2a) which has a lower affinity to the β-lactam antibiotics; including: all penicillins, all cephalosporins (except the fifth generation ceftaroline), and carbapenems (6, 7). It is encoded by the mecA gene carried on a staphylococcal chromosomal cassette (SCCmec).Thus; the therapeutic options of MRSA strains are lowered and limited.
Epidemiologic typing and molecular characterization of MRSA are crucial to monitor the occurrence and development of new epidemic clones and to determine intervention policies (8–10). Since hospital-associated (HA-MRSA) clones differ from community-associated (CA-MRSA) clones, clonal characterization is important to determine the source and the transmission routes of MRSA strains. Different typing methods are used to characterize different MRSA strains and clones. Specifically, defining the Staphylococcal Chromosomal Cassette (SCCmec) type is important to suggest the origin of the clone either hospital acquired or community acquired. The SCCmec types: I, II, and III are associated with HA-MRSA strains while types IV and V are considered as community associated MRSA strains (11, 12). Staphylococcus aureus protein A (spa) typing is also frequently used and based on typing of protein A. The spa typing technique depends on DNA sequencing of short sequence repeats (SSRs) of the polymorphic conserved X region of the staphylococcal protein A gene (13). Additionally, defining which virulence factors a strain may harbor is important not only for understanding the virulence of the strain, but also for studying the epidemiology of different common clones. Among these are: Panton-Valentine Leukocidin toxin (PVL); a pore-forming cytotoxin that can highly cause leukocyte destruction and tissue necrosis (14), Toxic Shock Syndrome Toxin-1 (TSST-1); a superantigen that can mediate fever, hypotension, rash, multi-organ dysfunction, and lethal shocks (15), Exfoliative Toxin A (ETA); a toxin that can lead to hydrolysis of the superficial skin layers leading to cutaneous infections and staphylococcal scalded skin syndrome (SSSS) (16) and arcA; a surrogate marker for Arginine Catabolic Mobile Element (ACME) I. The ACME is related with pathogenicity of the MRSA isolates which enhances both virulence and the ability of MRSA to colonize human skin (17). MRSA is a major public health concern in Palestine and little is known and studied about its molecular epidemiology and common epidemic clones (18, 19). This study aimed to investigate the genetic diversity and virulence genes of MRSA isolates identifying the prominent clones causing invasive infections in healthcare settings in the West Bank-Palestine, from 2015 to 2017.
Methodology
Bacterial Strains and Data Collection
In the present cross sectional descriptive study, a total of 112 MRSA isolates were collected from different sources, including: wound, blood, nasal swabs, urine, pus, tissue, abscesses, ear swabs, sputum, and other clinical sources; such as: cerebrospinal fluid (CSF) culture, central venous pressure (CVP) tip culture, skin swabs, synovial fluid culture, and trap. The study period was between 16th of November 2015 and 13th of July 2017 collected from different hospitals distributed in the West Bank-Palestine. Demographic and clinical data including: age, sex, place of residence, date of administration and hospitalization, type of infection, isolate antimicrobial susceptibility testing, specimen origin, and date of isolation were collected from medical records. Age was categorized to four age groups, as the following: infant; from 0 to 1 year, children; from 1 to 10 years, adolescent; from 10 to 19 years and as adult; more than 19 years.
Bacterial Culture and Identification of MRSA Strains
In the Central Laboratory of Al-Quds Public Health Society, MRSA isolates were identified phenotypically by colony morphology on blood agar and coagulase mannitol salt agar base (CMSA), gram stain, catalase, and coagulase tests. The methicillin resistant strains were defined by the disk agar diffusion method using a cefoxitin disk/30 μg. In addition, the MRSA strains were confirmed by the detection of mecA gene using PCR as described by Geha et al. (20).
Antimicrobial Susceptibility Testing
The antimicrobial susceptibility testing was performed using a disk diffusion method according to CLSI (2017) recommendations (21). Eight antimicrobial agents were tested, as the following: penicillin G (10 units/disc), cefoxitin (30 μg), vancomycin (30 μg), erythromycin (15 μg), clindamycin (10 μg), and gentamicin (10 μg), and trimethoprim-sulfamethoxazole (25 μg), and ciprofloxacin (5 μg) were determined. Isolates were defined as multidrug resistance strains (MDR) when they were resistant to at least three different antibiotic groups in addition to resistance to the β-lactam antibiotics (22). The D-test was performed to test the inducible resistance to clindamycin as needed.
DNA Extraction and Quantification
Genomic DNA was extracted from overnight fresh pure cultures on blood agar using DNA extraction kit (Nucleospin, Macherey-Nagel, Düren, Germany) according to the manufacturer's instructions (23). Isolates were freezed upon collection, and then thawed and sub-cultured on BA overnight to be used, later on, for genomic extraction.
Molecular Characterization of MRSA Strains
PCR Identification of Staphylococcal Cassette Chromosome (SCCmec) Types
SCCmec types (I–V) were identified by multiplex PCR as described by Boye et al. (12). The primers used in this multiplex PCR assay are shown in Table 1.
Table 1.
All target genes and primers used in this study.
| # | Gene | OligoaName | Primer sequence 5′-3′ | Length (bp) | Referenceb |
|---|---|---|---|---|---|
| 1 | mecA | mecA1Fc | GTAGAAATGACTGAACGTCCGATAA | 310 | (20) |
| mecA2Rd | CCAATTCCACATTGTTTCGGTCTAA | ||||
| 2 | SCCmec multiplex ″4 primers″ | β/ccrA2F-B | ATTGCCTTGATAATAGCCYTCT | 937 | (24) |
| α3/ccrA2R-B | TAAAGGCATCAATGCACAAACACT | ||||
| ccrCF/ccrC | CGTCTATTACAAGATGTTAAGGATAAT | 518 | (24) | ||
| ccrCR/ccrC | CCTTTATAGACTGGATTATTCAAAATA | ||||
| 1272F1/IS1272 | GCCACTCATAACATATGGAA | 415 | (12) | ||
| 1272R1/IS1272 | CATCCGAGTGAAACCCAAA | ||||
| 5RmecAF/IS431 | TATACCAAACCCGACAACTAC | 359 | |||
| 5R431R/IS431 | CGGCTACAGTGATAACATCC | ||||
| 3 | Spa | 1095F/spa-F | AGACGATCCTTCGGTGAGC | 200–400 | (25) |
| 1017R/spa-R | GCTTTTGCAATGTCATTTACTG | ||||
| 4 | PVL | luk-PV-1F/PVL | ATCATTAGGTAAAATGTCTGGACATGATCCA | 433 | (26) |
| luk-PV-2R/PVL | GCATCAASTGTATTGGATAGCAAAAGC | ||||
| 5 | TSST-1 | GTSSTR-1F | ACCCCTGTTCCCTTATCATC | 350 | (27) |
| GTSSTR-2R | TTTTCAGTATTTGTAACGCC | ||||
| 6 | ETA | GETAR-1F | GCAGGTGTTGATTTAGCATT | 93 | (27) |
| GETAR-2R | AGATGTCCCTATTTTTGCTG | ||||
| 7 | ACME-arcA | arcA-F | GAGCCAGAAGTACGCGAG | 724 | (28) |
| arcA-R | CACGTAACTTGCTAGAACGAG |
Oligo, Oligonecleotide.
Reference.
F, Forward primer.
R, reverse primer.
Identification of spa Types
All MRSA isolates were characterized by spa sequence-based typing using the PCR primers and cycling as previously described by Shopsin et al. (Tables 1, 2) (25). All spa amplicons were sequenced (Hylabs, Rehovot, Israel) and analyzed using the spa Type Finder/Identifier: (http://spatyper.fortinbras.us/). Then, the Based Upon Repeat Pattern (BURP) algorithm of the Staph-Type™ software (Ridom GmbH, Münster, Germany) was used for clustering of spa types and grouping in spa-clonal complexes (spa-CC).
Table 2.
Thermal cycler programs used for the amplification of the targeted genes and toxins in this study.
| # | Target Gene | PCR program (Tempa°C/Time)-35 cycles | Reference | ||||
|---|---|---|---|---|---|---|---|
| Initial denaturation | Denaturation | Annealing | Extension | Final extension | |||
| 1 | mecA | 95°C/5 minb | 95°C/30 sc | 58°C/30 s | 72°C/80 s | 72°C/10 min | (20) |
| 2 | SCCmec | 95°C /5 min | 94°C/30 s | 55°C/30 s | 72°C/80 s | 72°C/10 min | (12) |
| 3 | spa | 95°C/5 min | 95°C/30 s | 58°C/30 s | 72°C/45 s | 72°C/10 min | (25) |
| 4 | lukS/lukF | 95°C/5 min | 95°C/45 s | 55°C/15 s | 72°C/30 s | 72°C/10 min | (26) |
| 5 | tst | 95°C/5 min | 95°C/2 min | 54°C/2 min | 72°C/2 min | 72°C/10 min | (27) |
| 6 | eta | 95°C/5 min | 95°C/2 min | 54°C/2 min | 72°C/2 min | 72°C/10 min | (27) |
| 7 | arcA | 95°C/5 min | 94°C/20 s | 55°C/30 s | 72°C/30 s | 72°C/10 min | (28) |
Temp, Temperature.
min, Minutes.
s, Seconds.
PCR Identification of Staphylococcal Genes for Virulence Factors
Genes for virulence factors, namely: PVL cytotoxin genes (lukS and lukF), TSST-1 (encoded by tst gene), ETA (encoded by eta), and arcA (a surrogate marker for ACME I in the arc gene cluster) were tested using PCR for all isolates. These virulence factors and staphylococcal toxin genes were identified by singular PCRs with amplicons of: 433, 350, 93, and 724 bp, respectively (Table 1) (26–28).
PCR Assay and Visualization
All PCR reactions were optimized and carried by the Basic Gradient Thermocycler (BiometraTProfessional, Jena, Germany)using ready mix (Syntezza, Israel) according to the manufacturer's instructions, (Table 2) (29). Amplicons for all the characterized genes were analyzed electrophoretically in 2% agarose gels and visualized by UV light using a gel documentation system (Bio-Imaging Systems Mini-Lumitransilluminator, Germany).
Statistical Analysis
All data were analyzed using the Statistical Package for Social Sciences (SPSS) version 20, using Chi-square test. A p-value of < 0.05 was considered to be statistically significant.
Results
Bacterial Isolates and Study Population
From November 2015 to July 2017, a total of 112 MRSA isolates were collected from major hospitals in the West Bank-Palestine. The majority of isolates was collected from Al-Makassed Islamic Charitable Hospital (n = 77, 68.8%). Place of residence for all included patients were documented. The most frequent regions of residence were patients from Gaza and Jerusalem (29.5% for each), followed by Hebron (13.4%), Nablus (10.7%), Ramallah (10.7%), Bethlehem (4.5%), Tubas (0.9%), and Tulkarem (0.9%). According to gender, sixty MRSA isolates (53.6%) were obtained from males and 41(36.6%) were obtained from females. Gender of 9.8% of the isolates was not reported. The mean age was 33 years with the oldest case being an 85 years old female and the youngest a 2 months years old female. The most common age group was adults (60.7%), followed by adolescents (14.3%), infants (8.9%), and children (7.1%). Of the adults, most were older than 30 years old, while age was not found for 8.9% of the samples. Samples were collected from different clinical sources. The highest number of isolates were obtained from wound infections (35.7%), followed by: blood culture (12.5%), nasal swabs (8.9%), urine culture (8.9%), pus culture (5.4%), tissue culture (4.5%), abscess (3.6%), ear swabs (2.7%), and sputum culture (2.7%). One MRSA isolate was collected and reported from a CSF culture (0.9%) which is extremely alarming and important in which MRSA strains are reaching the CSF leading to significant bacterial meningitis. Moreover, MRSA was obtained from other sources (4.5%), as the follows: axillary swab (0.9%), CVP culture (0.9%), skin swab culture (0.9%), synovial fluid culture (0.9%), and trap (0.9%). Source of eleven isolates (9.8%) was not identified.
Detection of MRSA by Cefoxitin Disk Diffusion Test and mecA Gene
All isolates (100%) were verified as Methicillin Resistant Staphylococcus aureus using the cefoxitin disc resistance (<22 mm) and confirmed as MRSA using PCR targeting the mecA gene yielding to 310 bp clear band.
Antimicrobial Susceptibility Patterns
Resistance rates to non β-lactam antibiotics were as the following: 18.8% were resistant to trimethoprim-sulfamethoxazole, 23.2% were resistant to gentamicin, 34.8% to clindamycin, 39.3% to ciprofloxacin, and 63.4% to erythromycin. Interestingly, the susceptibility of MRSA isolates to gentamicin and Trimethoprim/sulfamethoxazole (SXT) was the highest (76.8, 81.2%), respectively. All isolates were susceptible to Vancomycin (100%). Of all the isolates, 32 were MDR (Figure 1).
Figure 1.
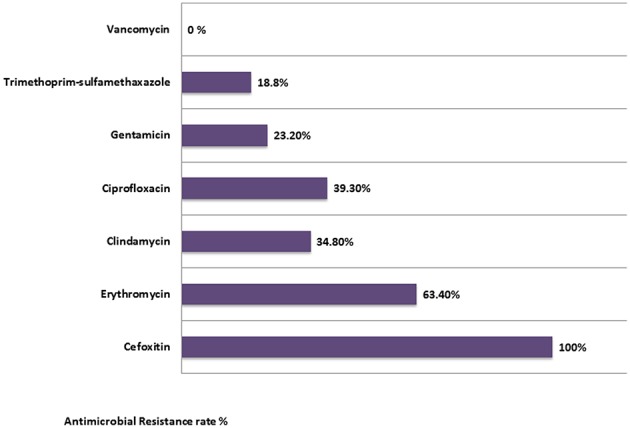
Antimicrobial resistance rates among all MRSA isolates in this study.
The antimicrobial resistance rates among the MRSA spa types were studied. The resistance rates of the MRSA isolates against erythromycin, were significantly high in the spa type t386 (p < 0.005), followed by the spa types: t008 (8.9%), t044 (5.4%), t021 (5.4%), and t223 (4.5%). Similarly, the resistance rates of MRSA isolates against clindamycin were high in the spa types: t386 (7.1%) and t008 (4.5%), followed by t044 (3.6%), and t021 (2.7%). Moreover, the resistance rates of MRSA isolates against ciprofloxacin and gentamicin were found significantly high (p < 0.05) in the spa type t037 (7.1% for each). Also, for the spa types t008 and t044, they have shown a significant high resistance rate to ciprofloxacin (p < 0.05).
Molecular Characterization and Typing of Isolates
SCCmec Typing
Among all MRSA isolates, ninety seven isolates belonged to SCCmec type IV (86.6%) and one belonged to SCCmec type V (0.9%). These isolates identified as CA-MRSA when carrying the SCCmec types IV or V. Ten isolates belonged to SCCmec type I (8.9%) and identified as HA-MRSA. Both SCCmec type II and SCCmec type III were not detected among isolates. Four MRSA isolates (3.6%) could not be SCCmec typed and designated as non-typeable (NT).
Spa Typing
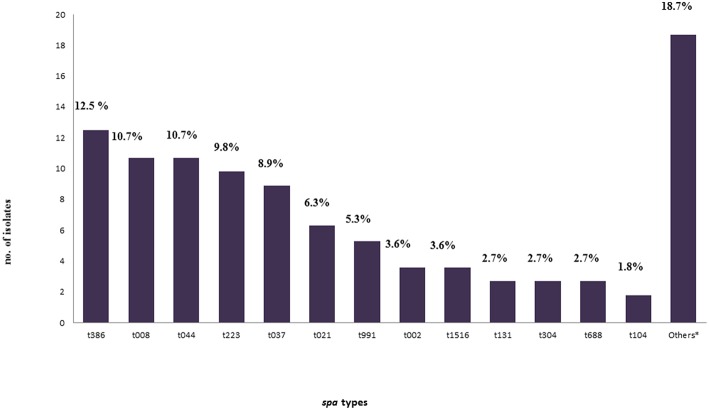
A total of twenty nine spa types were identified. The size of the amplified DNA for spa typing ranged between 200 and 450 bp. Five spa types: t386, t008, t044, t223, and t037 were predominant and represented (12.5%), (10.7%), (10.7%), (9.8%), (8.9%) of isolates, respectively (Figure 2). There was a wide range of clonal varieties, with twelve MLST clonal complexes (CCs) were identified. This was done with regard to the BURP analysis. The 12 identified MLST-CCs were: CC22, CC1, CC8, CC80, CC8/239, CC30, CC5, CC913, CC6, CC121, CC126, and CC15. The CC22 (15.2%), CC1 (13.4%), CC8 13.4%), and CC80 (13.4%) were predominant, followed by CC8/239 (8.9%), CC30 (8.0%), CC5 (8.0%), CC913 (5.4%), CC6 (2.7%), CC121 (0.9%), and CC126 (0.9%).Unfortunately, ten isolates (8.9%) were not found using the BURP analysis. However, the identified MLST-CCs were distributed among the spa types where a single clonal complex could contain more than one spa type as shown in Table 3.
Figure 2.
Frequency of all spa-types found in this study.*Other spa-types: t005, t011, t018, t084, t085, t1094, t121, t1247, t127, t314, t318, t359, t541, t605, t648, and t932 were found in a frequency of one for each isolate (0.9% for each) with four isolates (3.6%) non-typeable (err) isolates.
Table 3.
The MLST-CCs and the related spa types found in this study by the BURP.
| # | MLST-CCas | spa types | No. of isolates (%) |
|---|---|---|---|
| 1 | CC22 | t223 | 11(9.8%) |
| t1516 | 4 (3.6%) | ||
| t005 | 1 (0.9%) | ||
| t541 | 1 (0.9%) | ||
| 2 | CC1 | t386 | 14 (12.5%) |
| t127 | 1 (0.9%) | ||
| 3 | CC8 | t008 | 12 (10.7%) |
| t121 | 1 (0.9%) | ||
| t359 | 1 (0.9%) | ||
| t648 | 1 (0.9%) | ||
| 4 | CC80 | t044 | 12 (10.7%) |
| t131 | 3 (2.7%) | ||
| 5 | CC8/239b | t037 | 10 (8.9%) |
| 6 | CC30 | t021 | 7 (6.3%) |
| t018 | 1 (0.9%) | ||
| t318 | 1 (0.9%) | ||
| 7 | CC5 | t002 | 4 (3.6%) |
| t688 | 3 (2.7%) | ||
| t104 | 2 (1.8%) | ||
| 8 | CC913 | t991 | 6 (5.4%) |
| 9 | CC6 | t304 | 3 (2.7%) |
| 10 | CC121 | t314 | 1 (0.9%) |
| 11 | CC126 | t605 | 1 (0.9%) |
| 12 | CC15 | t084 | 1 (0.9%) |
| 13 | NFc | t011 | 1 (0.9%) |
| t085 | 1 (0.9%) | ||
| t1094 | 1 (0.9%) | ||
| t1247 | 1 (0.9%) | ||
| t932 | 1 (0.9%) | ||
| (*)? | 1 (0.9%) |
(*)?, This spa-type was not identified on the spa database website; (http://spatyper.fortinbras.us/).
Clonal Complexes.
The spa-t037 found to be CC8 or CC239 by the BURP, MLST is recommended for finite identification.
Not Found by BURP.
Toxin Genes Profiling
Detection of PVL Toxin
Of all MRSA isolates, thirty three (29.5%) were positive for PVL toxin gene. All the PVL positive MRSA isolates belonged to SCCmec type IV. PVL was detected among eleven spa types: t044, t008, t223, t021, t002, t131, t104, t084, t121, t318, and t386. Two spa types (t044 and t008) among PVL positive isolates were predominant and represent (11/12 t044, CC80: PVL +; European clone) and (10/12 t008, CC8: PVL +; USA-300 clone) isolates, respectively. Out of 11 isolates of the spa type t223, only one isolate was positive for the PVL toxin gene (1/11 isolates t223, CC22: PVL +) and classified as a “Unique Gaza Strain.”
Detection of ACME-arcA Gene
The gene coding for the ACME-arcA was detected in 18.8 % (21/112) of isolates where 20 isolates harbored the SCCmec type IV, and one isolate harbored the SCCmec type I. It was detected among nine spa types: t008, t991 t037, t104, t1094, t1247, t1516, t223, and t386. Whereas, out of the 12 isolated identified as spa type t008, nine isolates were ACME-arcA positive (9/12 t008, CC8: ACME-arcA). These nine isolate were also PVL positive and classified as: USA-300 clone.
Detection of TSST-1
Similarly, 26.8% of the MRSA isolates (30/112) tested positive for the presence of the TSST- where 25 isolates belonged to the SCCmec type IV; while the other TSST-1 positive isolate belonged to SCCmec type I. This toxin was detected among eleven spa types: t223, t021, t1516, t002, 1005, t008, t018, t386, t541, t991, and t605. The two most common spa types among TSST-1positive isolates were: the spa type t223 and t021. Out of the 11 identified isolated as spa type t223, nine t223 MRSA isolates were significantly (p < 0.05) TSST-1 positive (9/11 t223, CC22: TSST-1 +; Gaza clone) which was commonly typed previously as “Gaza clone” in Gaza region. Notably, all the seven isolates with spa type t021 were TSST-1 positive (100%) and classified as South West Pacific clone.
Table 4 summarized the common MRSA clones with antibiotic resistance according to their SCCmec types, spa types and toxin genes profile.
Table 4.
Common MRSA clones with antibiotic resistance according to their SCCmec types, spa types and toxin genes profile found in this study.
| MRSA Clone | n | % | spa type/CC | SCCmec | Virulence factors | *% Antibiotic resistance | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVL | TSST-1 | arcA | E | C | CN | CIP | SXT | |||||
| t386-IV | 14 | 12.5 | t386/CC1 | IV | neg | neg | neg | 100 | 57.1 | 21.4 | 21.4 | 0 |
| The European clone | 11 | 9.8 | t044/CC80 | IV | pos | neg | neg | 50 | 33.3 | 16.7 | 41.7 | 16.7 |
| The Gaza clone | 10 | 8.9 | t223/CC1 | IV | neg | pos | neg | 45.5 | 18.2 | 18.2 | 9.1 | 27.3 |
| t037-IV | 10 | 8.9 | t037/ CC8/239 | IV | neg | neg | neg | 40 | 20 | 80 | 80 | 10 |
| USA-300 clone | 9 | 8 | t008/CC8 | IV | pos | neg | pos | 83.3 | 41.7 | 0 | 75 | 8 |
:E, Erythromycin; C, Clindamycin; CN, Gentamicin; CIP, Ciprofloxacin; SXT, Trimethoprim sulfamethoxazole.
Detection of ETA
The gene coding for the exfoliative toxin A (eta) was detected in 5.3% of all the MRSA isolates harboring the spa type t991/CC913. All the other spa types were negative for ETA. Among the 6 isolates with the spa type t991, five isolates were of SCCmec type IV, and one isolate was belonged to SCCmec type I. None of the spa t991; eta positive isolates were harboring the PVL toxin, and only one isolate was positive for ACME-arcA while 4/6 spa t991 isolates harbored the TSST-1.
Discussion
Methicillin Resistant Staphylococcus aureus (MRSA) is a serious life threatening pathogen in hospitals and among healthy populations (29–32). Thus, the characterization of these strains is important for local epidemiology and surveillance studies.
Little is known about MRSA types and clones in Palestine. This study was conducted to characterize the molecular and the antimicrobial profile of MRSA isolates in regions of West Bank, from 2015 to 2017. Most isolates were obtained from Al- Makassed Islamic Charitable Society Hospital (68.8% of all isolates). Al-Makassed is one of the most important leading medical institutions in Palestine that provides medical services to all Palestinians in the West Bank, Gaza Strip and East Jerusalem.
In a recent study conducted at Al-Shifa hospital in Gaza Strip for the nasal colonization of MRSA, a carriage rate of MRSA among the health care workers who are in contact with the vulnerable patient in the hospitals were equal to 25.5% (33).This highlights that there is a high carriage rate of MRSA among the Palestinian populations. Moreover, it has been reported that carriage of MRSA is a major risk factor for transmission and subsequent infections that may develop to systemic or severe infections (34, 35).
Here, 32 MRSA isolates were MDR (28.6%). In comparison with other reports based on the same definition, a higher MDR resistance rate (60.0%) was also reported in Israel (36). However, our highest antibiotic susceptibility patterns were noticed among SXT (81.3%), followed by gentamicin (76.8%), and ciprofloxacin (65.2%). This may aid medical laboratory technologist to test the susceptibility of these antibiotics to be used as major and first line therapeutic options for the treatment of MRSA infections. No Vancomycin Resistance Staphylococcus aureus (VRSA) strains were reported, where the physicians can still prescribe this antibiotic based on empirical therapy when needed, especially for urgent infections. At the molecular level, the predominant MRSA strains in our region are community associated (SCCmec type IV, 86.6%). SCCmec type IV is the smallest structural type among the SCCmec types and believed to be the most mobile version that is associated with CA-MRSA infections (37). About one third of the CA-MRSA strains isolated in our study were harboring the PVL toxin gene (32/97 SCCmec type IV). This indicates that the CA-MRSA strains among Palestinian population are highly virulent with invasive. This can give us an indication that we may have a high carriage of MRSA among healthy population or the health care workers in our region. These CA-MRSA strains could be carried as a normal flora in skin, hands, and groin or in the nasal cavity of the health care workers or the patients themselves, but when these patients are hospitalized or have an opened wound or surgery, their immunity may fall down allowing this normal flora or colonization to cause secondary opportunistic infections. Therefore, hand hygiene and infectious control programs must be applied well in our hospitals. This interface may serve as a causative agent of cross contamination of hospital acquired and community acquired MRSA infections.
This agreed with a study done in Copenhagen where Bartels and his colleagues reported that there is a rapid shift to CA-MRSA, where SCCmec type IV was found in (86%) of isolates (38). Moreover, a significant presence of MRSA was seen. In this study, 62 hospitals were included showing an evidence of endemicity of MRSA in these regions (39). Also, it has been reported that there is an introduction of several MRSA strains with intercontinental exchange of new MRSA clones in the Middle East (40). In a recent study conducted in Gaza by Al Laham and his colleagues, similar results were obtained with a high frequency of the SCCmec type IV, which accounted for (79.3%) of MRSA isolates. Al Laham has found that the most dominant spa types were t223 and t044 where seven isolates were belonged to the spa type t223, in which one of these isolates was called as a “unique Gaza strain” harboring the PVL toxin gene, while all the other isolates with the spa type t223 did not carry the PVL toxin gene (19). This was in a high agreement with our findings with an isolate with a similar pattern as the unique Gaza strain with spa type t223, SCCmec type IV, PVL positive, and TSST-1 negative. Notably, the Gaza clone (t223) with TSST-1 positive, CC22, and SCCmec type IV was detected in the isolates collected from Al- Makassed Islamic Charitable Society Hospital only. Six isolates were from Gaza patient and five isolates were from patients living in Jerusalem and its suburbs. Interestingly, this could be explained due the transmission of the Gaza clone to other Palestinian populations as Gaza patients were referred to Al- Makassed Islamic Charitable Society Hospital in Jerusalem. Moreover, the spa type t223 was also reported in other countries as Kuwait, Egypt and Saudi Arabia. This transmission could be due to the national travels between these countries (41–45).
The spa type t008 was predominant in 12 MRSA isolates. Nine of them were related to the USA-300 clone, harboring both the PVL and the ACME-arcA genes (8% of all isolates). This clone is a common cause of SSTIs and was considered as the most widespread CA-MRSA clone in the United States that emerged in the late 1990s. This clone is a significant and dramatic epidemic clone due to its carriage of virulence and resistance determinants that may enhance severity and pathogenicity of the isolated strain (17). The carriage of PVL toxin gene among the CA-MRSA strains, especially the USA-300 clone, is crucial and make these strains hyper-virulent which may cause occasionally fatal infections (46). To the best of our knowledge, our study is the first study that has characterized the presence of the USA-300 clone in the Palestinian regions. Also, due to the shift of MRSA infections to the community and the high carriage among populations, no individual can be considered not at risk for CA-MRSA infection. This worthy point needs further attention because meaningful results and control programs can be achieved. Regarding the appropriate treatment approaches, culturing is recommended for the MRSA infections because it is the only ideal predictor of the appropriate antibiotics therapy. Educational and public awareness, medical education, professional development, and training are recommended for the public society, parents, pharmacists, medical laboratories and physicians for reliable, appropriate and proper antibiotic therapy decisions. This can help to minimize the development for more complicated resistant MRSA strains in hospitals and may save many immunocompromized hospitalized patients. All together, these points suggest the need for efficient future surveillance studies and infection control strategies. Moreover, these strategies can improve the economic impact because MRSA is considered as a serious economic burden on the healthcare resource and associated with increased hospitalization costs due to the prolonged course on more complex antibiotics and extended hospital days.
The results of this study showed that all the PVL-positive MRSA isolates (29.5% of all isolate) recovered from these hospitals as part of the present investigation were CA which is confirming as an earlier preliminary observation that CA-MRSA is an emerging problem in Palestine. Some reports have suggested that certain strains of CA-MRSA may be more virulent than HA-MRSA (47, 48). The expression of PVL, cytolytic toxin that targets mononuclear and polymorphonuclear cells and causes cell death by necrosisor apoptosis, has been strongly linked with CA-MRSA (49).
Similarly, TSST-1 and ACME-arcA genes were detected in 23.2 and 18.8% of all isolates, respectively. These were relatively high in comparison with a similar study conducted in China where they found the PVL and TSST-1 genes in 11.3 and 18.8%, respectively (50). For the ACME-arcA carriage, this was in agreement with a study conducted in Armenia which found the ACME-arcA in 15% of all MRSA isolates, but contraindicated with the carriage of the PVL toxin gene where they did not detected the PVL toxin gene in any of their MRSA isolates (51). This indicates that there is a high carriage of PVL, ACME-arcA, TSST-1, and ETA genes among MRSA strains in Palestine.
Conclusion
The phenotypic and genotypic diversity with harboring several virulence factors and toxin genes by MRSA isolates in Palestine is crucial. This study indicates that we have unique MRSA strains which may be associated with more complicated infections and associated with more social and economic burdens. Thus, there is an urgent need to develop better measurements among clinical microbiology laboratories for proper detection, identification, and reporting of MRSA isolates with deep knowledge among physicians toward antibiotic stewardship and prescription practices for this multi-drug resistant organism. These together may control and limit the development and spread of new clones and more complicated MRSA infections.
Ethics Statement
The study was approved by the Research Ethical Committee at Al-Quds University. Written informed consents were sent for the participating hospitals and clinics. Written informed consent was obtained from the parents of the participants that were under the age of 16 years old.
Author Contributions
KA designed the study and wrote the final manuscript. EH and KA performed the experiments, analyzed the obtain results. RA and IA participated in the sample collection. ZA contributed to the manuscript revision and overall support of this study.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We greatly acknowledge all hospitals that provided us with the isolates and responded to our contact. Special thanks to Al-Makassed Islamic Charitable Hospital/microbiology laboratory in East Jerusalem; including; Mr. Sabri Baragthithi and Mrs. Suzan Idkaidek. Also, many thanks to Mr. Mamoun Obeideia from the Palestine Medical complex and Mrs. Namir Sabri from the Red Crescent Society in Jerusalem. Many special thanks to Dr. G. Regev-Yochay and Asaf Biber from Sheba Medical Center, assistance. This data were presented as partial requirements for a master degree of Microbiology and Immunology/Al-Quds University under supervision of KA.
Footnotes
Funding. This study was supported financially by grant MERC (M33-014).
References
- 1.Bazzoun DA, Harastani HH, Shehabi AA, Tokajian ST. Molecular typing of Staphylococcus aureus collected from a Major Hospital in Amman, Jordan. J Infect Dev Ctries. (2014) 8:441–7. 10.3855/jidc.3676 [DOI] [PubMed] [Google Scholar]
- 2.Serray B, Oufrid S, Hannaoui I, Bourjilate F, Soraa N, Mliji M, et al. Genes encoding adhesion factors and biofilm formation in methicillin-resistant Staphylococcus aureus in Morocco. J Infect Dev Ctries. (2016) 10:863–9. 10.3855/jidc.8361 [DOI] [PubMed] [Google Scholar]
- 3.Harastani HH, Tokajian ST. Community-associated methicillin-resistant Staphylococcus aureus clonal complex 80 type IV (CC80-MRSA-IV) isolated from the Middle East: a heterogeneous expanding clonal lineage. PLoS ONE. (2014) 9:e103715. 10.1371/journal.pone.0103715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. (2015) 28:603–61. 10.1128/CMR.00134-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobayashi SD, Malachowa N, DeLeo FR. Pathogenesis of Staphylococcus aureus abscesses. Am J Pathol. (2015) 185:1518–27. 10.1016/j.ajpath.2014.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duplessis C, Crum-Cianflone NF. Ceftaroline: a new cephalosporin with activity against methicillin-resistant Staphylococcus aureus (MRSA). Clin Med Rev Ther. (2011) 3. 10.4137/CMRT.S1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.CDC Centers for Disease Control and Prevention. Laboratory Testing for MRSA. (2017). Available online: https://www.cdc.gov/mrsa/lab/index.html
- 8.Sun DD, Ma XX, Hu J, Tian Y, Pang L, Shang H, et al. Epidemiological and molecular characterization of community and hospital acquired Staphylococcus aureus strains prevailing in Shenyang, Northeastern China. Braz J Infect Dis. (2013) 17:682–90. 10.1016/j.bjid.2013.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grubb WB. Genetics of MRSA. Rev Med Microbiol. (1988) 9:153–62. 10.1097/00013542-199807000-00004 [DOI] [Google Scholar]
- 10.Montesinos I, Salido E, Delgado T, Cuervo M, Sierra A. Epidemiologic genotyping of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis at a university hospital and comparison with antibiotyping and protein A and coagulase gene polymorphisms. J Clin Microbiol. (2002) 40:2119–25. 10.1128/JCM.40.6.2119-2125.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naimi TS, LeDell KH, Como-Sabetti K, Borchardt SM, Boxrud DJ, Etienne J, et al. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA. (2003) 290:2976–84. 10.1001/jama.290.22.2976 [DOI] [PubMed] [Google Scholar]
- 12.Boye K, Bartels MD, Andersen IS, Møller JA, Westh H. A new multiplex PCR for easy screening of methicillin-resistant Staphylococcus aureus SCCmec types I-V. Clin Microbiol Infect. (2007) 13:725–7. 10.1111/j.1469-0691.2007.01720.x [DOI] [PubMed] [Google Scholar]
- 13.Oliveira DC, Crisóstomo I, Santos-Sanches I, Major P, Alves CR, Aires-de-Sousa M, et al. Comparison of DNA sequencing of the protein A gene polymorphic region with other molecular typing techniques for typing two epidemiologically diverse collections of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. (2001) 39:574–80. 10.1128/JCM.39.2.574-580.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Talib H, Hasan H, Yean CY, Al-Ashwal SM, Ravichandran M. Fatal necrotizing pneumonia caused by Panton-Valentine leukocidin-producing hospital-acquired Staphylococcus aureus: a case report. Jpn J Infect Dis. (2011) 64:58–60. [PubMed] [Google Scholar]
- 15.Kulhankova K, King J, Salgado-Pabón W. Staphylococcal toxic shock syndrome: superantigen-mediated enhancement of endotoxin shock and adaptive immune suppression. Immunol Res. (2014) 59:182–7. 10.1007/s12026-014-8538-8 [DOI] [PubMed] [Google Scholar]
- 16.Leung AKC, Barankin B, Leong KF. Staphylococcal-scalded skin syndrome: evaluation, diagnosis, and management. World J Pediatr. (2018) 10.1007/s12519-018-0150-x [DOI] [PubMed] [Google Scholar]
- 17.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. (2006) 367:731–9. 10.1016/S0140-6736(06)68231-7 [DOI] [PubMed] [Google Scholar]
- 18.Adwan K, Jarrar N, Abu-Hijleh A, Adwan G, Awwad E, Salameh Y. Molecular analysis and susceptibility patterns of methicillin-resistant Staphylococcus aureus strains causing community- and health care-associated infections in the northern region of Palestine. Am J Infect Control. (2013) 41:195–8. 10.1016/j.ajic.2012.03.040 [DOI] [PubMed] [Google Scholar]
- 19.Al Laham N, Mediavilla JR, Chen L, Abdelateef N, Elamreen FA, Ginocchio CC, et al. MRSA clonal complex 22 strains harboring toxic shock syndrome toxin (TSST-1) are endemic in the primary hospital in Gaza, Palestine. PLoS ONE. (2015) 10:e0120008. 10.1371/journal.pone.0120008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geha DJ, Uhl JR, Gustaferro CA, Persing DH. Multiplex PCR for identification of methicillin-resistant staphylococci in the clinical laboratory. J Clin Microbiol. (1994) 32:1768–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.CLSI Performance Standards for Antimicrobial Susceptibility Testing; 27th Informational Supplement. CLSI document M100-S27 (Wayne, Pa). (2017). [Google Scholar]
- 22.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. (2012) 18:268–81. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 23.NucleoSpin® Genomic DNA from tissue Kit. (2017). Available online at: http://www.mn-net.com/Portals/8/attachments/Redakteure_Bio/Protocols/Genomic%20DNA/UM_gDNATissue_2017.pdf
- 24.Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, et al. (2001). Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 45:1323–36. 10.1128/AAC.45.5.1323-1336.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shopsin B, Gomez M, Montgomery SO, Smith DH, Waddington M, Dodge DE, et al. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol. (1999)37:3556–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. (1999) 29:1128–32. 10.1086/313461 [DOI] [PubMed] [Google Scholar]
- 27.Mehrotra M, Wang G, Johnson WM. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J Clin Microbiol. (2000) 38:1032–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vento TJ, Calvano TP, Cole DW, Mende K, Rini EA, Tully CC, et al. Staphylococcus aureus colonization of healthy military service members in the United States and Afghanistan. BMC Infect Dis. (2013) 13: 325. 10.1186/1471-2334-13-325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Syntezza PCR-Ready Products™. (2004). Available online at: http://www.syntezza.com/pcr-ready/
- 30.Cardona AF, Wilson SE. Skin and soft-tissue infections: a critical review and the role of telavancin in their treatment. Clin Infect Dis. (2015) 61(Suppl_2):S69–78. 10.1093/cid/civ528 [DOI] [PubMed] [Google Scholar]
- 31.Reddy PN, Srirama K, Dirisala VR. An update on clinical burden, diagnostic tools, and therapeutic options of Staphylococcus aureus. Infect Dis. (2017) 10:1179916117703999. 10.1177/1179916117703999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boswihi SS, Udo EE. Methicillin-resistant Staphylococcus aureus: an update on the epidemiology, treatment options and infection control. Curr Med Res Pract. (2018) 8:18–24. 10.1016/j.cmrp.2018.01.001 [DOI] [Google Scholar]
- 33.El Aila NA, Al Laham NA, Ayesh BM. Nasal carriage of methicillin resistant Staphylococcus aureus among health care workers at Al Shifa hospital in Gaza Strip. BMC Infect Dis. (2017) 17:28. 10.1186/s12879-016-2139-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuehnert MJ, Kruszon-Moran D, Hill HA, McQuillan G, McAllister SK, Fosheim G, et al. Prevalence of Staphylococcus aureus nasal colonization in the United States, 2001-2002. J Infect Dis. (2006) 193:172–9. 10.1086/499632 [DOI] [PubMed] [Google Scholar]
- 35.Piechowicz L, Garbacz K, Wiśniewska K, Da̧browska-Szponar M. Screening of Staphylococcus aureus nasal strains isolated from medical students for toxin genes. Folia Microbiol. (2011) 56:225–9. 10.1007/s12223-011-0041-1 [DOI] [PubMed] [Google Scholar]
- 36.Biber A, Parizade M, Taran D, Jaber H, Berla E, Rubin C, et al. Molecular epidemiology of community-onset methicillin-resistant Staphylococcus aureus infections in Israel. Eur J Clin Microbiol Infect Dis. (2015) 34:1603–13. 10.1007/s10096-015-2395-9 [DOI] [PubMed] [Google Scholar]
- 37.Milheiriço C, Oliveira DC, de Lencastre H. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: 'SCCmec IV multiplex'. J Antimicrob Chemother. (2007) 60:42–8. 10.1093/jac/dkm112 [DOI] [PubMed] [Google Scholar]
- 38.Bartels MD, Boye K, Rhod Larsen A, Skov R, Westh H. Rapid increase of genetically diverse methicillin-resistant Staphylococcus aureus, Copenhagen, Denmark. Emerg Infect Dis. (2007) 13:1533–40. 10.3201/eid1310.070503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borg MA, de Kraker M, Scicluna E, van de Sande-Bruinsma N, Tiemersma E, Monen J, et al. Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in invasive isolates from southern and eastern Mediterranean countries. J Antimicrob Chemother. (2007) 60:1310–5. 10.1093/jac/dkm365 [DOI] [PubMed] [Google Scholar]
- 40.Tokajian S. New epidemiology of Staphylococcus aureus infections in the Middle East. Clin Microbiol Infect. (2014) 20:624–8. 10.1111/1469-0691.12691 [DOI] [PubMed] [Google Scholar]
- 41.Aqel AA, Alzoubi HM, Vickers A, Pichon B, Kearns AM. Molecular epidemiology of nasal isolates of methicillin-resistant Staphylococcus aureus from Jordan. J Infect Public Health. (2015) 8:90–7. 10.1016/j.jiph.2014.05.007 [DOI] [PubMed] [Google Scholar]
- 42.Udo EE, Boswihi SS, Al-Sweih N. High prevalence of toxic shock syndrome toxin-producing epidemic methicillin-resistant Staphylococcus aureus 15 (EMRSA-15) strains in Kuwait hospitals. N Microb N Infect. (2016) 12:24–30. 10.1016/j.nmni.2016.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khairalla AS, Wasfi R, Ashour HM. Carriage frequency, phenotypic, and genotypic characteristics of methicillin-resistant Staphylococcus aureus isolated from dental health-care personnel, patients, and environment. Sci Rep. (2017) 7:7390. 10.1038/s41598-017-07713-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Udo EE, Al-Sweih N. Dominance of community-associated methicillin-resistant Staphylococcus aureus clones in a maternity hospital. PLoS ONE. (2017) 12:e0179563. 10.1371/journal.pone.0179563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abou Shady HM, Bakr AE, Hashad ME, Alzohairy MA. Staphylococcus aureus nasal carriage among outpatients attending primary health care centers: a comparative study of two cities in Saudi Arabia and Egypt. Braz J Infect Dis. (2015) 19:68–76. 10.1016/j.bjid.2014.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Otto M. Basis of virulence in community-associated methicillin-resistant Staphylococcus aureus. Annu Rev Microbiol. (2010) 64:143–62. 10.1146/annurev.micro.112408.134309 [DOI] [PubMed] [Google Scholar]
- 47.Vindel A, Cuevas O, Cercenado E, Marcos C, Bautista V, Castellares C, et al. Methicillin-resistant Staphylococcus aureus in Spain: molecular epidemiology and utility of different typing methods. J Clin Microbiol. (2009) 47:1620–7. 10.1128/JCM.01579-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. Lancet. (2010) 375:1557–68. 10.1016/S0140-6736(09)61999-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boyle-Vavra S, Daum RS. Community-acquired methicillin-resistant Staphylococcus aureus: the role of Panton-Valentine leukocidin. Lab Invest. (2007) 87:3–9. 10.1038/labinvest.3700501 [DOI] [PubMed] [Google Scholar]
- 50.Wang M, Zheng Y, Mediavilla JR, Chen L, Kreiswirth BN, Song Y, et al. Hospital dissemination of tst-1-positive clonal complex 5 (CC5) methicillin-resistant Staphylococcus aureus. Front Cell Infect Microbiol. (2017) 7:101. 10.3389/fcimb.2017.00101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mkrtchyan HV, Xu Z, Yacoub M, Ter-Stepanyan MM, Karapetyan HD, Kearns AM, et al. Detection of diverse genotypes of Methicillin-resistant Staphylococcus aureus from hospital personnel and the environment in Armenia. Antimicrob Resist Infect Control. (2017) 6:19. 10.1186/s13756-017-0169-0 [DOI] [PMC free article] [PubMed] [Google Scholar]