Abstract
WNT signaling regulates a variety of ovarian processes, including follicle development, granulosa cell (GC) proliferation and differentiation, steroidogenesis, and ovulation. The secreted frizzled-related proteins (SFRPs) comprise a family of WNT signaling antagonists. Sfrp4 expression was previously reported to be induced in ovarian GCs and cumulus cells in vivo following human chorionic gonadotropin treatment, suggesting that it may play key roles in cumulus expansion, ovulation/luteinization, and corpus luteum (CL) function. In this study, we aimed to define the physiological roles of Sfrp4 in the ovary by gene targeting. Sfrp4-null female mice were found to produce larger litters than did their wild-type littermates. Although previous studies had suggested roles of Sfrp4 in luteal cell survival, no differences in CL formation, morphology, steroidogenesis, involution, or luteal cell apoptosis were found in Sfrp4-null mice. Likewise, cumulus expansion occurred normally in Sfrp4-null mice, with minimal changes in cumulus cell gene expression. Hyperfertility in the Sfrp4-null model was ultimately attributed to decreased antral follicle atresia, leading to an enhanced ovulatory rate. Increased expression of FSH- and LH-responsive genes was found in GCs from Sfrp4-null mice, and GCs isolated from Sfrp4-null mice were found to be hyperresponsive to FSH and LH in vitro. Although Sfrp2 was found to be overexpressed in the GCs of Sfrp4-null mice (suggesting a compensatory mechanism), Sfrp2-null mice had normal fertility and ovulatory rates, and Sfrp2/4 double knockout mice did not differ from Sfrp4-null mice. Taken together, our results suggest that SFRP4 acts to attenuate GC responsiveness to gonadotropins, thereby decreasing follicle survival, ovulatory rate, and fertility.
Ovarian follicle development is controlled by endocrine regulators, as well as by a variety of factors produced within the ovary itself (1). The most important endocrine regulators include the pituitary gonadotropins FSH and LH, which act to regulate the expression and function of different intraovarian factors, including the WNT family of secreted glycoproteins (2, 3). The WNTs then act in turn to modulate and coordinate follicular responses to the gonadotropins, and they therefore represent critical regulators of ovarian function and female fertility (4, 5).
Roles have been proposed for various WNT signaling effectors in processes including the regulation of follicle development, granulosa cell (GC) proliferation and differentiation, steroidogenesis, and ovulation (5). WNT2, for instance, has been shown to positively regulate proliferation in cultured GCs (6). The inactivation of Wnt4 in the GCs of a transgenic mouse model resulted in reduced fertility linked to inadequate antral follicle development. WNT4 has also been shown to regulate the expression of the steroidogenic enzymes Star, Cyp19a1, and Cyp11a1 (7). Likewise, Wnt5a has been shown to be required for normal follicle development and ovulation, as well as to antagonize gonadotropin signaling by downregulating β-catenin (CTNNB1) and cAMP response element–binding protein (CREB) expression (8). In addition to WNT ligands, studies have also shown important roles for the WNT receptor FZD4 in the formation and function of the corpus luteum (CL), as well as for FZD1 in the regulation of genes associated with cumulus expansion (9, 10).
The secreted frizzled-related proteins (SFRPs) are a family of five soluble proteins that all possess a frizzled-like cysteine-rich domain that is capable of binding WNT ligands. The SFRPs therefore inhibit WNT signaling (at least in part) by sequestering WNT ligands in the extracellular space and preventing them from interacting with their receptors (11–14). The expression of Sfrp4 is induced in ovarian GCs and cumulus cells following LH/human chorionic gonadotropin (hCG) stimulation, and high expression is then maintained in the CL (15, 16). Although a study has suggested an association between SFRP4 and apoptosis in the rat CL (17), the physiological roles of SFRP4 remain largely unknown. In the current study, we sought to define the ovarian roles of SFRP4 using a gene targeting approach.
Materials and Methods
Ethics
All animal procedures were approved by the Institutional Animal Care and Use Committee and conformed to the International Guiding Principles for Biomedical Research Involving Animals.
Gene targeting and animal models
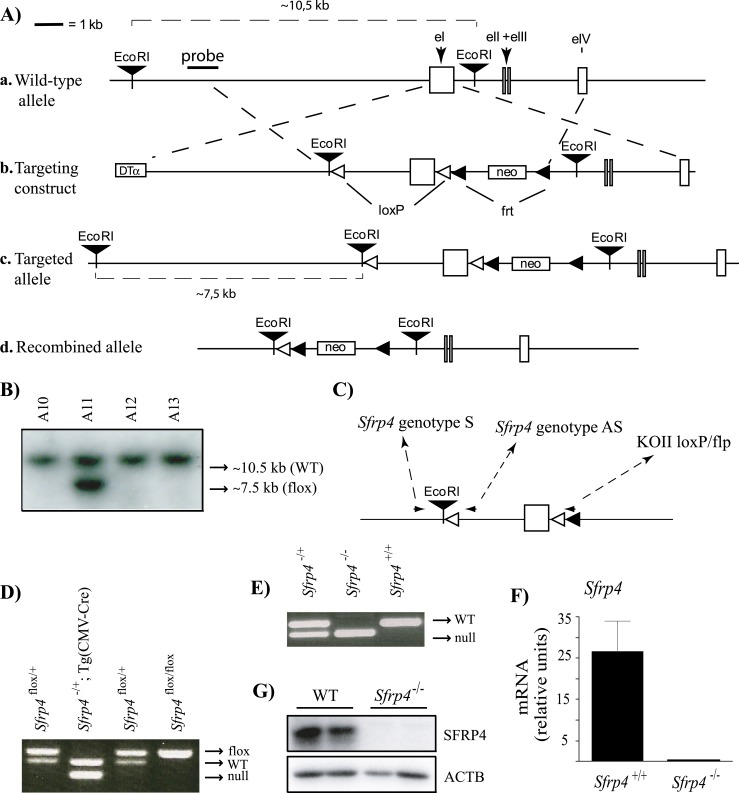
A floxed Sfrp4 allele (Fig. 1Ac) was created using a standard gene targeting approach. Briefly, a targeting construct was built using the pKOII vector (18), into which was inserted a modified genomic fragment containing the first exon of Sfrp4 flanked by two loxP sites. An EcoRI restriction site was also included for Southern blot screening of embryonic stem (ES) cell clones (Fig. 1Ab). All genomic sequences were cloned from R1 ES cell genomic DNA by PCR using the Expand long template PCR system (Roche Molecular Biochemicals, Laval, QC, Canada); LoxP and restriction sites were included in the primers used to amplify the genomic fragments. The targeting vector was linearized prior to electroporation into R1 ES cells. Clonal selection was achieved by growing the cells in a medium containing 400 μg/mL Geneticin (Invitrogen, Burlington, ON, Canada) for 8 to 9 days. Selected colonies were cultured in triplicate, and duplicate sets were analyzed by Southern blotting to screen for proper homologous recombinants as previously described (19). Two of 768 colonies screened showed a second band consistent with the expected size for the targeted allele (Fig. 1B). Both targeted cell lines were subsequently microinjected into blastocysts and transferred to pseudopregnant recipients. Chimeric males derived from both cell lines sired pups heterozygous for the targeted allele.
Figure 1.
Targeting of Sfrp4. (A) Illustration of the strategy used to generate Sfrp4−/− mice. (a) Placement of the genomic DNA probe used for Southern blotting. (b) In the targeting construct, LoxP sites were inserted upstream and downstream of the first exon of Sfrp4. (c) Sfrp4flox allele. (d) Sfrp4− (i.e., Cre-recombined) allele. (B) Generation of ES cell lines heterozygous for the targeted Sfrp4 allele. Presence of the targeted allele was detected as a 7.5-kb EcoRI restriction fragment by Southern blotting. One representative positive clone (A11) is shown alongside three negative clones. (C) PCR genotyping analysis strategy. Oligonucleotides (small black arrowheads) were designed to generate PCR products of 249 bp for the Sfrp4+ allele, 143 bp for the Sfrp4flox allele, and 136 bp for the Sfrp4− allele. (D) PCR genotype analysis of pups generated by the mating of Sfrp4flox/+ with Tg(CMV-cre)1Cgn/J mice. (E) PCR genotype analysis of pups generated by backcross of Sfrp4+/− mice. (F) Sfrp4 mRNA levels and (G) SFRP4 protein levels in GCs isolated from immature eCG-treated Sfrp4+/+ and Sfrp4−/− mice 12 h after the administration of an ovulatory dose of hCG, confirming the abrogation of SFRP4 expression in the Sfrp4-null mice. The mRNA levels were measured by RT-qPCR (n = 6 animals per genotype). Data are expressed as means ± SEM. DTα, diphtheria toxin α chain; neo, neomycin resistance cassette.
To then generate a null Sfrp4 allele (Fig. 1Ad), Sfrp4flox/+ mice were mated to the Tg(CMVcre)1Cgn/J “cre deleter” strain (The Jackson Laboratory, Bar Harbor, ME), which expresses Cre in all tissues, including the germline. After PCR verification of the Sfrp4− allele (Fig. 1D), the Cre transgene was eliminated from Sfrp4+/−;Tg(CMV-cre)1Cgn/J mice in the following generation by mating to C57BL/6J [referred to herein as wild-type (WT)] mice. Sfrp4-null mice were then obtained by backcrossing Sfrp4+/− mice (Fig. 1E). Genotyping analyses were done by PCR on DNA obtained from tail biopsies, using the oligonucleotide primers 5′-AACAATGAGCTCTACCCCTCTGT-3′ (Sfrp4 genotype S), 5′-CTCCCCGCACCACACACACTGA-3′ (Sfrp4 genotype AS), and 5′-TAGGAACTTCAATTCCCCGCAAGA-3′ (KOII loxP/flp) using the following PCR conditions; 1 minute at 95°C for 1 cycle, 45 seconds at 95°C, 45 seconds at 63°C and 1 minute at 72°C for 35 cycles, and 7 minutes at 72°C for 1 cycle (Fig. 1C). Primers were designed to generate PCR products of ∼249 bp for the WT Sfrp4 allele, ∼289 bp for the floxed allele, and ∼143 bp for the knockout allele (Fig. 1C–1E). Immunoblotting and quantitative RT-PCR (RT-qPCR) analyses of proteins extracted from GCs confirmed the loss of Sfrp4 gene expression in Sfrp4-null mice (Fig. 1F and 1G).
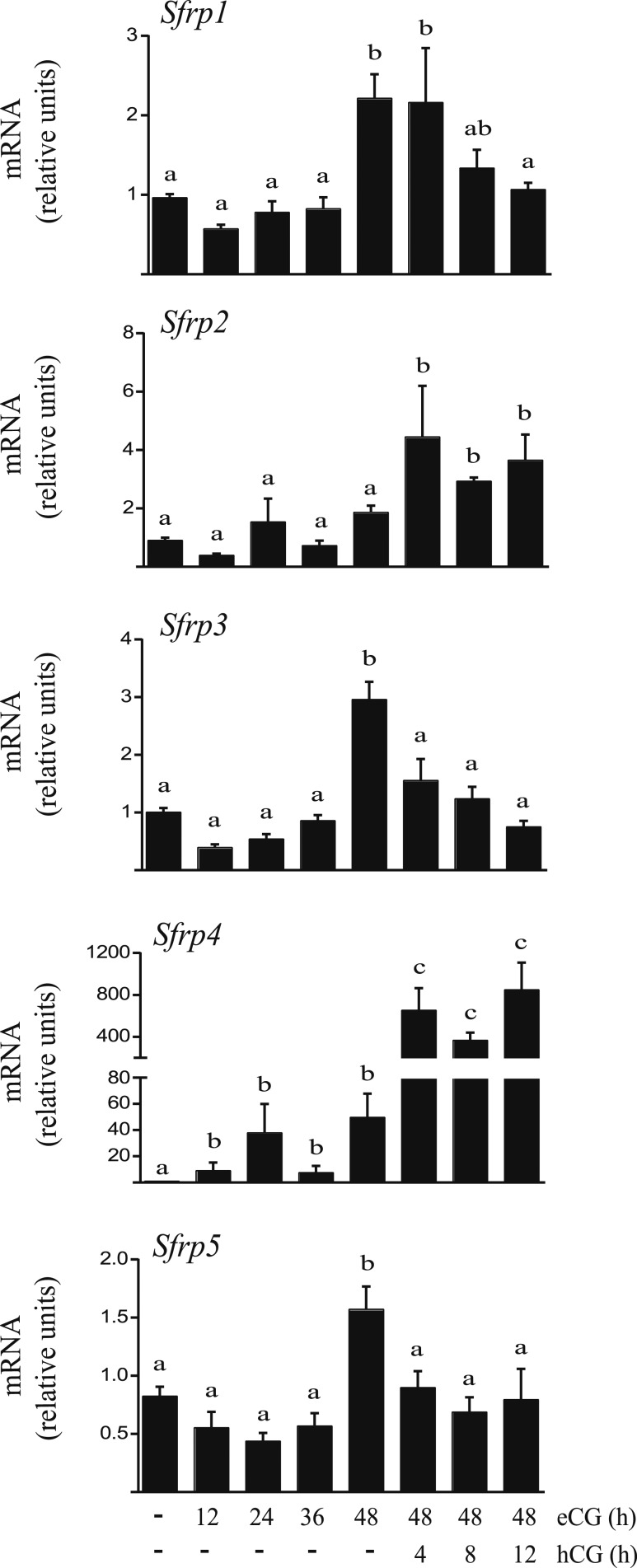
Figure 2.
In vivo regulation of Sfrp mRNA levels in GCs by gonadotropins. Immature (21- to 26-d-old) WT female mice were either injected (or not) with eCG (5 IU, IP) and GCs isolated at the indicated times, or else they were injected with eCG 44 to 48 h prior to the administration of hCG (5 IU, IP) and GCs isolated at the indicated times. mRNA levels for each Sfrp gene were determined by RT-qPCR and normalized to the housekeeping gene Rpl19 (n = 3 to 4 animals per time point). Data are expressed as means ± SEM. Different letters show statistically significant differences between groups (P < 0.05).
WT and B6;129-Sfrp2tm1.Brle/J mice (referred to herein as Sfrp2-null or Sfrp2−/−) were obtained from The Jackson Laboratory. Genotyping analyses for Sfrp2 alleles was done by PCR on DNA obtained from tail biopsies, using the oligonucleotide primers 5′-CGAAACACAAAAGCGAGTGA-3′ (Sfrp2 mutant S), 5′-TGAACTCTCAGGCCAAGTCA-3′ (Sfrp2 WT S), and 5′-CATGGCAGCAAAGAATTGTG-3′ (Sfrp2 common AS) designed to generate PCR products of ∼355 bp for the WT Sfrp2 allele and ∼650 bp for the knockout allele. The following PCR conditions were used: 5 minutes at 95°C for 1 cycle, 30 seconds at 95°C, 30 seconds at 60°C and 45 seconds at 72°C for 35 cycles, and 5 minutes at 72°C for 1 cycle.
Fertility trials, ovulation rate, and antral follicle counting
To assess fertility, 8-week-old female mice were housed with WT males and monitored daily for litters for 6 (WT vs Sfrp2 vs Sfrp2/4-null) or 12 months (WT vs Sfrp4-null). Litter sizes were recorded at birth, and the experiment was terminated 22 days after removal of the males to allow for a potential final litter. To determine ovulation rates, 8- to 10-week-old Sfrp2-, Sfrp4-, and Sfrp2/4-null females and WT controls were housed with WT males and monitored every morning for the presence of a copulatory plug. Females were euthanized immediately upon identification of the plug and the oviducts were removed and placed in a Petri dish containing sterile saline. Cumulus–oocyte complexes (COCs) were released by tearing open the ampullae of the oviducts with forceps and counted under a dissection microscope. Antral follicle counting (Sfrp4-null vs control mice) was done using formalin-fixed, paraffin-embedded ovaries from 6-week-old mice. In all cases, only the left ovary from each animal was analyzed. Serial sections were prepared at a thickness of 5 μm, and every fifth section was stained with hematoxylin and eosin. Antral follicles in which the oocyte nucleus was visible were counted and scored as healthy or atretic as previously described (7).
Tissue collection and cumulus expansion evaluation
Whole ovaries, GCs, and COCs were obtained from immature (21- to 26-day-old) mice up to 48 hours after administration of equine chorionic gonadotropin (eCG; Folligon, Intervet, Kirkland, QC, Canada; 5 IU, IP) followed or not by hCG (Chorulon, Intervet; 5 IU, IP) at different time points as described for each experiment. Whole ovaries were obtained post-hCG (0 to 48 hours) as well from pregnant mice euthanized 14 days after mating and were frozen at −80°C until the time of RNA extraction. For gene expression analyses in GCs, ovaries obtained from mice euthanized at different time points post-eCG (0 to 48 hours) and 48 hours post-eCG followed by hCG (0 to 12 hours) were placed in Hanks balanced salt solution and punctured with 26-gauge needles to release GCs as previously described (20). GCs were then pooled, centrifuged at 1000g, and frozen at −80°C until RNA extraction. COCs were also collected at different time points post-hCG (0 to 12 hours) by puncture, except for the 20-hour post-hCG time point, for which COCs were flushed from dissected oviducts, collected by pipette, pooled, and frozen at −80°C until RNA extraction.
To evaluate the extent of cumulus expansion in vivo, COCs were collected 20 hours post-hCG by oviduct flushing. COCs were photographed using an Axio Imager M.1 microscope (Zeiss, Toronto, ON, Canada), and diameters were measured using AxioVision 4.6.3 software, with the diameter of each COC being the mean of four separate measurements taken at random points.
GC primary culture
GCs for primary culture were obtained from eCG-treated immature mice (either Sfrp4-null or WT littermates) as described above, pooled, and suspended in MEM containing 1% fetal bovine serum, 0.25 nM sodium pyruvate, and 3 mM l-glutamate. GCs were seeded in 48-well culture plates (divided such that each well received GCs equivalent to 0.5 ovary) and incubated for 3 hours at 37°C with 5% CO2. Culture medium was then replaced with serum-free medium for 2 hours prior to addition (or not) of recombinant human FSH or LH (50 ng/mL; National Hormone and Peptide Program, Torrance, CA) for 2 hours or 1 hour, respectively. GCs were stored at −80°C prior to RNA extraction.
Real-time RT-PCR
Total RNA from whole ovaries, GCs, or COCs was extracted using the RNeasy Mini Kit (Qiagen, Toronto, ON, Canada) according to the manufacturer’s protocol. Reverse transcription was done using 200 ng of RNA and the SuperScript Vilo cDNA synthesis kit (Invitrogen). Real-time PCR was done using SsoAdvanced™ Universal SYBR® Green Supermix (172-5274; Bio-Rad Laboratories, Mississauga, ON, Canada) and a CFX96 Touch™ instrument (Bio-Rad Laboratories). Each PCR reaction consisted of 7.5 μL of SsoAdvanced SYBR Green PCR Master Mix, 2.3 μL of water, 4 μL of cDNA sample, and 0.6 μL (10 pmol) of gene-specific primers. Cycling conditions were 3 minutes at 95°C followed by 40 cycles of 15 seconds at 95°C, 30 seconds at 60°C, and 30 seconds at 72°C. To quantify relative gene expression, the cycle threshold (Ct) of target gene amplification was normalized to the expression level of a housekeeping gene (Rpl19) according to the ratio, R = ECt Rpl19/ECt target, where E is the amplification efficiency for each primer pair.
TUNEL assays
Ovaries from eCG-primed mice were collected 24 hours, 48 hours, 5 days, and 7 days following hCG treatment. Samples were fixed in 4% paraformaldehyde, embedded in paraffin, and serial sections were prepared at a thickness of 7 μm. TUNEL assays were performed using the In Situ Cell Death Detection Kit, TMR red as directed by the manufacturer (Roche Molecular Biochemicals). Slides were mounted using VectaShield with 4′,6′-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA).
Immunoblotting
Immunoblotting was done as previously described (8) using primary antibodies against SFRP4 (catalog no. 15328-1-AP, 1:1000; Proteintech, Rosemont, IL) or ACTB (catalog no. sc-47778, 1:10,000; Santa Cruz Biotechnology, Dallas, TX) diluted in 5% BSA. After washing three times with Tween 20 and Tris-buffered saline, membranes were incubated for 1 hour at room temperature with anti-rabbit horseradish peroxidase–conjugated IgG (GE Healthcare Life Sciences, Baie d’Urfé, QC, Canada) diluted 1:10,000 in 5% nonfat dry milk in Tween 20 and Tris-buffered saline. Protein bands were visualized by chemiluminescence (ECL; Millipore, Billerica, MA) and quantified using a ChemiDoc MP detection system (Bio-Rad Laboratories) and Image Laboratory™ software.
Steroid hormone measurement
Blood samples were collected by cardiac puncture prior to euthanasia. Serum estradiol levels were determined by ELISA, whereas progesterone and FSH levels were determined by RIA. All assays were performed by the Ligand Assay and Analysis Core Laboratory of the University of Virginia (Charlottesville, VA).
Statistical analyses
All statistical analyses were performed using Prism 4.0a (GraphPad Software, La Jolla, CA) software. All of the data sets (fertility measures, ovulatory rates, follicle numbers, mRNA expression, cumulus, diameter and steroid hormone levels) were subjected to the F test to determine equality of variances. Data were transformed to logarithms when they were not normally distributed. Two-tailed t tests were used when two experimental groups were compared, or ANOVA (with a Tukey multiple comparisons posttest) to compare three or more groups. All data are presented as means ± SEM.
Results
The expression of Sfrp2 and Sfrp4 is induced by hCG in GCs
To begin our study of SFRP function in the ovary, we examined the expression patterns of the five Sfrp genes in ovarian GCs from immature mice following treatment with (FSH-mimetic) eCG for up to 48 hours, followed or not by (LH-mimetic) hCG for up to 12 hours. As previously reported (15, 16), Sfrp4 mRNA levels increased dramatically (∼15-fold) in response to hCG (Fig. 2). Unlike previous reports, however, we found Sfrp4 to also be markedly induced by eCG (∼40-fold by 24 hours posttreatment). Sfrp2 was also strongly expressed in GCs, particularly following hCG treatment, but its mRNA levels varied little in response to gonadotropins relative to Sfrp4. Although transcripts encoding Sfrp1, Sfrp3, and Sfrp5 were detectable in these samples, all were present at much lower levels than Sfrp2 or Sfrp4. The highest expression of Sfrp1, Sfrp3, and Sfrp5 was detected 48 hours post-eCG and declined following hCG treatment (Fig. 2). Owing to their expression levels and regulatory patterns during follicle development, we therefore focused our subsequent functional analyses on the Sfrp2 and Sfrp4 genes.
Mice null for Sfrp4 are hyperfertile
To study the functions of Sfrp4 in ovarian physiology, we generated Sfrp4-null mice (Fig. 1). Animals of both sexes were found to be viable, healthy, and otherwise indistinguishable from their WT littermates. Male mice were fertile (data not shown), confirming the results of a recent study (21). To assess the effects of Sfrp4 loss on female fertility, 8-week-old Sfrp4-null females were individually housed with fertile WT males for a period of 12 months, and litter frequency and size were recorded. Whereas Sfrp4−/− females produced a normal number of litters, they were found to be hyperfertile, producing about one additional pup per litter on average relative to their WT sisters (P < 0.05, Table 1).
Table 1.
Fertility Trials
| Control | Sfrp4 −/− | |
|---|---|---|
| n | 8 | 6 |
| No. of litters | 9.38 ± 0.46 | 8.33 ± 1.15 |
| Total pups | 56.75 ± 3.23 | 59.50 ± 9.12 |
| Litter size | 6.05 ± 0.32a | 7.14 ± 0.44b |
Data are means ± SEM.
Different superscript letters indicate significant differences (P < 0.05).
CL function is normal in Sfrp4-null mice
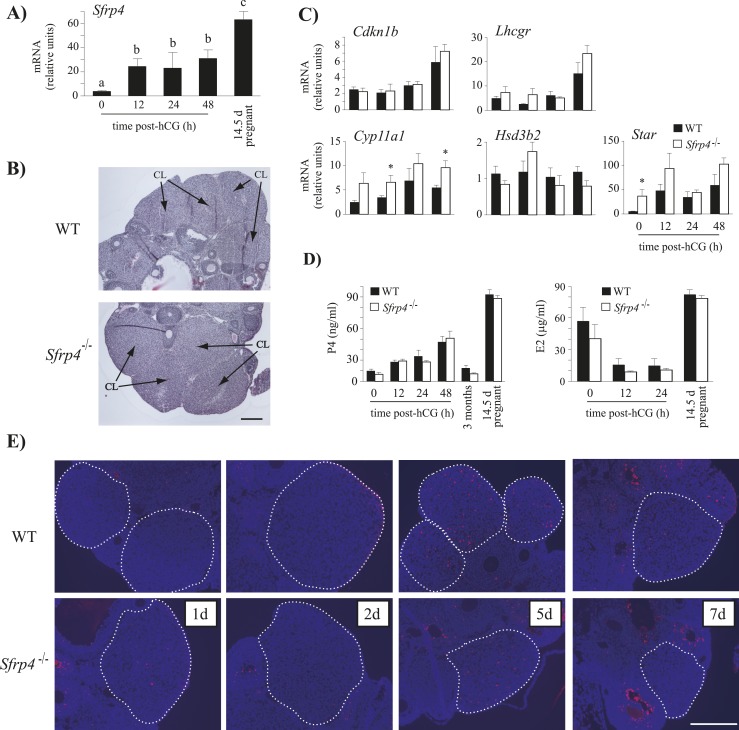
As Sfrp4 expression is maintained at a high level during the formation of the CL and throughout pregnancy (Fig. 3A) (15–17), our initial investigation of the hyperfertility phenotype of Sfrp4-null mice centered on the function of the CL. Histological analyses of ovaries of Sfrp4-null mice collected 48 hours post-hCG showed no obvious differences in CL or luteal cell morphology relative to WT controls (Fig. 3B). The expression of genes with known roles in CL formation and function were then studied in whole ovaries on a time course after hCG treatment. These analyses revealed no differences in mRNA levels of Cdkn1b, Lhcgr, or Hsd3b2 in Sfrp4-null mice relative to controls (P > 0.05, Fig. 3C). However, Star mRNA levels were significantly higher in the ovaries of immature eCG-stimulated Sfrp4-null mice (i.e., prior to hCG treatment), whereas Cyp11a1 mRNA levels were significantly higher 12 hours and 48 hours post-hCG (P < 0.05, Fig. 3C). Although the latter findings suggested that ovarian steroidogenesis could be altered in Sfrp4-null mice, neither serum estradiol nor progesterone levels differed from controls at any time point examined (Fig. 3D). As a previous study suggested that SFRP4 could be involved in luteal cell apoptosis (17), we examined apoptosis in the ovaries of Sfrp4-null and WT mice 1, 2, 5, and 7 days following hCG injection. Neither histopathologic analyses nor TUNEL revealed differences between genotypes at any of the studied time points (Fig. 3E and not shown), suggesting that formation and involution of the CL is normal in Sfrp4-null mice.
Figure 3.
CL function is not impaired in Sfrp4-null mice. (A) Sfrp4 mRNA levels in whole ovaries isolated from immature WT mice treated with eCG (5 IU, IP) followed or not with hCG (5 IU, IP) for 12, 24, or 48 h, and from 14.5-d pregnant mice. Different letters show statistically significant differences between groups (P < 0.05). (B) Representative photomicrographs of ovaries from Sfrp4-null and WT immature eCG-treated mice 48 h post-hCG. Scale bar, 200 μm. (C) Expression pattern of genes known to be involved in the formation or the function of the CL. Ovaries were isolated from immature, eCG-treated mice of the indicated genotypes at different time points post-hCG. (D) Serum progesterone (P4) and estradiol (E2) levels in mice of the indicated genotypes. Blood samples were collected from immature, eCG-treated mice at different time points post-hCG and from 3-mo-old and 14.5-d pregnant mice. n = 8 to 24 animals per genotype per time point or condition. (E) TUNEL staining (red) in ovaries isolated from immature, eCG-treated mice 1, 2, 5, and 7 d after the administration of an ovulatory dose of hCG. The counterstain is 4′,6′-diamidino-2-phenylindole (blue). Scale bar, 200 μm. For clarity, CLs are circumscribed with a dotted white line. For all RT-qPCR analyses, n = 5 to 6 animals per genotype per time point (if applicable), with each sample consisting of one or two ovaries. All data were normalized to the housekeeping gene Rpl19 and are expressed as means ± SEM. *P < 0.05, significant difference vs control.
Loss of Sfrp4 does not affect cumulus expansion
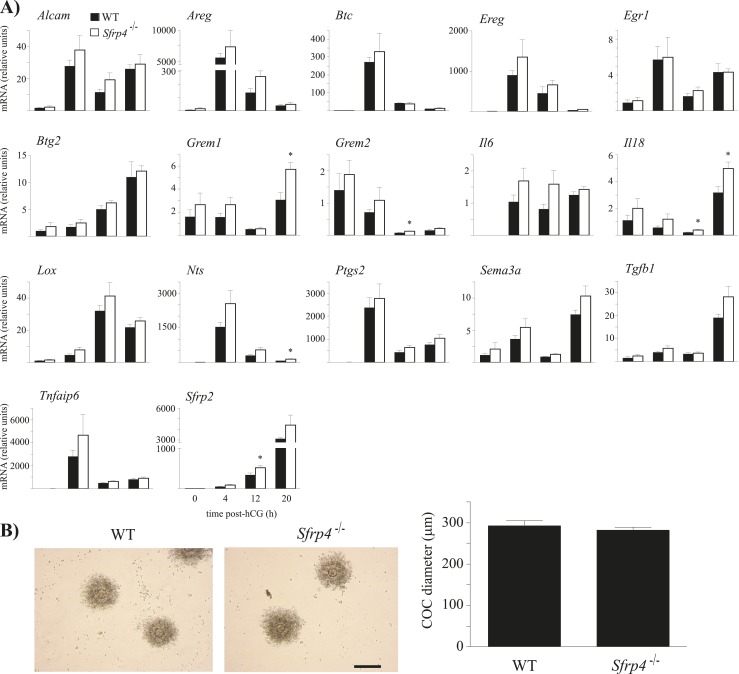
As Sfrp4 was previously shown to be induced by hCG in mouse COCs (15), we assessed the potential effects of Sfrp4 loss on the cumulus expansion process. The expression of genes associated with cumulus expansion was determined in COCs at different stages of the expansion process, from 0 hours post-hCG (unexpanded COCs present in preovulatory follicles) to 20 hours post-hCG (fully expanded COCs recovered from the oviduct). Grem1, Grem2, Il18, and Nts mRNA levels were significantly higher in COCs from Sfrp4−/− mice, but only late in the expansion process (12 to 20 hours post-hCG) (Fig. 4A). To study the functional consequences of the alteration of COC gene expression, we evaluated cumulus expansion in COCs recovered from oviducts 20 hours post-hCG. COCs from Sfrp4−/− mice expanded to the same extent as did controls (P > 0.05), and without any obvious morphological differences (Fig. 4B). These results indicate that the loss of Sfrp4 does not have a major impact on the cumulus expansion process.
Figure 4.
Loss of Sfrp4 does not affect cumulus expansion. (A) Expression pattern of genes known to be regulated during COC expansion. Immature mice of the indicated genotypes were treated with eCG for 48 h, followed (or not) by hCG for up to 20 h. mRNA levels were determined in isolated COCs by RT-qPCR and normalized to the housekeeping gene Rpl19. For all analyses, n = 5 samples per genotype per time point, with each sample consisting of a pool of COCs collected from two to five animals. All data are expressed as means ± SEM. *P < 0.05, significant difference vs control at the same time point. (B) Representative photomicrographs and diameter measurements of COCs collected by flushing the oviducts of mice of the indicated genotypes 20 h following an ovulatory dose of hCG. n = 10 to 11 COCs per genotype. Scale bar, 100 μm. Data are expressed as means ± SEM.
Enhanced ovulatory rate, follicle survival, and gonadotropin responsiveness in Sfrp4-null mice
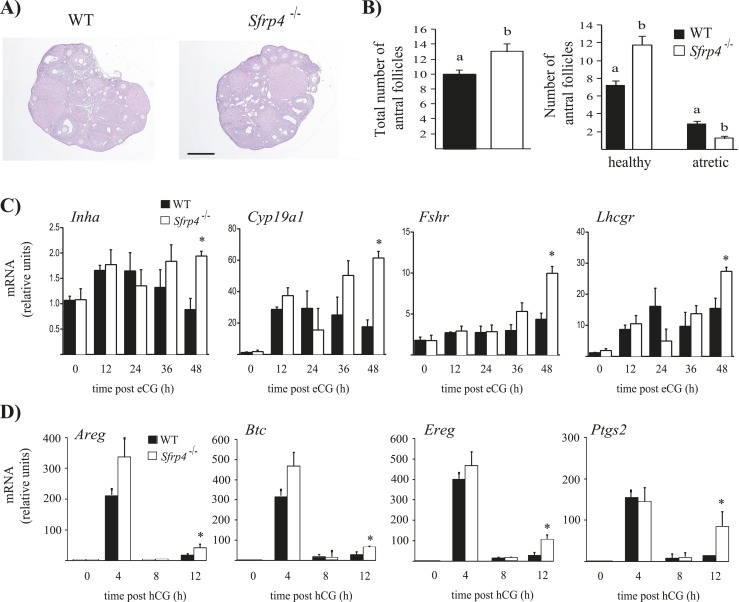
As neither CL function nor cumulus expansion was altered by Sfrp4 ablation, we next evaluated the ovulation rate of Sfrp4-null mice. Interestingly, Sfrp4−/− females ovulated approximately threefold more COCs following natural mating than did WT littermate controls (P < 0.05, Table 2), suggesting that hyperfertility in these mice could be due to an enhanced follicular response to LH, enhanced follicle development, or a combination of both. To assess follicle development in Sfrp4-null mice, we counted antral follicles in serial histologic sections and scored follicles as healthy or atretic. Although no obvious morphological alterations (Fig. 5A) or ovary weight differences (data not shown) were detected in the ovaries of Sfrp4-null mice, the latter had increased total numbers of antral follicles (P < 0.05, Fig. 5B), apparently due (at least in part) to a decrease in follicular atresia (P < 0.05, Fig. 5B). Serum FSH levels in Sfrp4-null mice were comparable to controls (Table 3), suggesting that the enhanced follicle survival/growth phenotype was ovary-autonomous.
Table 2.
Ovulation Rates
| WT | Sfrp4 −/− | Sfrp2 −/− | Sfrp2/4-null | |
|---|---|---|---|---|
| n | 6 | 6 | 4 | 6 |
| No. of COCs | 6.66 ± 0.61a | 10.17 ± 0.31b | 7.51 ± 0.64a | 10.33 ± 0.42b |
Data are means ± SEM.
Different superscript letters indicate significant differences (P < 0.05).
Figure 5.
Mice null for Sfrp4 have enhanced follicle survival and increased expression of LH and FSH target genes in GCs. (A) Representative images of Sfrp4-null and control WT mice ovaries from 42-d-old animals. Paraformaldehyde-fixed, paraffin-embedded ovaries were sectioned at a thickness of 4 μm. Slides were stained with hematoxylin and eosin prior to mounting. Scale bar, 100 μm. (B) Ovaries from 42-d-old mice (n = 4 to 5 per genotype) were serially sectioned, and all antral follicles from every fifth section were counted and scored as either healthy or atretic. Data (means ± SEM) represent raw follicle count numbers and were not adjusted to estimate the total ovarian follicle population. Different letters above histograms indicate statistically significant differences between strains (P < 0.05). (C and D) GCs were isolated from immature mice of the indicated genotypes following the indicated hormonal treatments. mRNA levels of the indicated genes were determined by RT-qPCR and normalized to the housekeeping gene Rpl19. For all analyses, n = 3 to 4 animals per genotype per time point. All data are expressed as means ± SEM *P < 0.05, significant difference vs control at the same time point.
Table 3.
Serum FSH Levels
| WT | Sfrp4 −/− | Sfrp2 −/− | Sfrp2/4-null | |
|---|---|---|---|---|
| n | 6 | 5 | 6 | 6 |
| FSH, ng/mL | 20.17 ± 7.71 | 18.03 ± 5.74 | 17.66 ± 2.12 | 14.34 ± 3.81 |
FSH levels were determined in 8- to 10-wk-old mice. Data are means ± SEM. No significant differences were found.
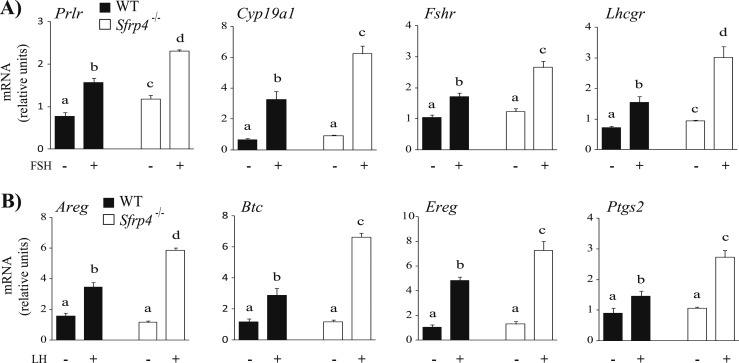
To study gonadotropin-regulated gene expression in the GCs of Sfrp4-null mice, mRNA levels of key FSH-responsive genes were determined in GCs isolated from treated immature mice 0, 12, 24, 36, and 48 hours following eCG treatment. Interestingly, the expression of the FSH-responsive genes Inha, Cyp19a1, Fshr, and Lhcgr was increased in Sfrp4 knockout mice in comparison with controls, particularly at 48 hours post-eCG (P < 0.05, Fig. 5C). mRNA levels of critical LH-responsive genes were also determined in GCs isolated from eCG-primed immature mice 0, 4, 8, and 12 hours after administration of hCG. Areg, Ereg, Btc, and Ptgs2 were found to be more highly expressed in GCs from Sfrp4 knockout mice relative to WT controls around the time of ovulation (12 hours post-hCG) (P < 0.05, Fig. 5D). These results therefore suggested that loss of Sfrp4 results in enhanced gonadotropin responsiveness. To test this more directly, GCs were isolated from Sfrp4-null and WT littermate mice and treated (or not) with FSH or LH. As shown in Fig. 6A, FSH induced a significantly greater increase in Prlr, Cyp19a1, Fshr, and Lhcgr mRNA levels in GCs from Sfrp4-null animals relative to WT mice. In the case of Prlr and Lhcgr, basal (i.e., unstimulated) mRNA levels were also significantly higher in the Sfrp4-null group. As for FSH, LH also produced a significantly greater increase in Areg, Btc, Ereg, and Ptgs2 mRNA levels in GCs from Sfrp4-null mice (Fig. 6B).
Figure 6.
GCs of Sfrp4-null mice are hyperresponsive to gonadotropins. GCs were isolated from immature (21- to 26-d-old) eCG-primed (5 IU, IP for 44 to 48 h) mice of the indicated genotypes and placed in culture prior to addition (or not) of 50 ng/mL recombinant human (A) FSH or (B) LH for 2 h and 1 h, respectively. mRNA levels of the indicated genes were determined by RT-qPCR and normalized to the housekeeping gene Rpl19. For all analyses, n = 3 to 4 replicates per time point. All data are expressed as means ± SEM. Different letters show statistically significant differences between groups (P < 0.05).
Taken together, these results suggest that SFRP4 acts to attenuate the follicular response to FSH, negatively regulating FSH target gene expression, follicle development, and survival. SFRP4 also antagonizes LH action, negatively regulating the expression of its target genes in the luteinizing GCs of periovulatory follicles, in cumulus cells during expansion, and in the CL (Figs. 3C, 4A, and 5C).
Sfrp2 is not functionally redundant with Sfrp4 and is dispensable for ovarian function
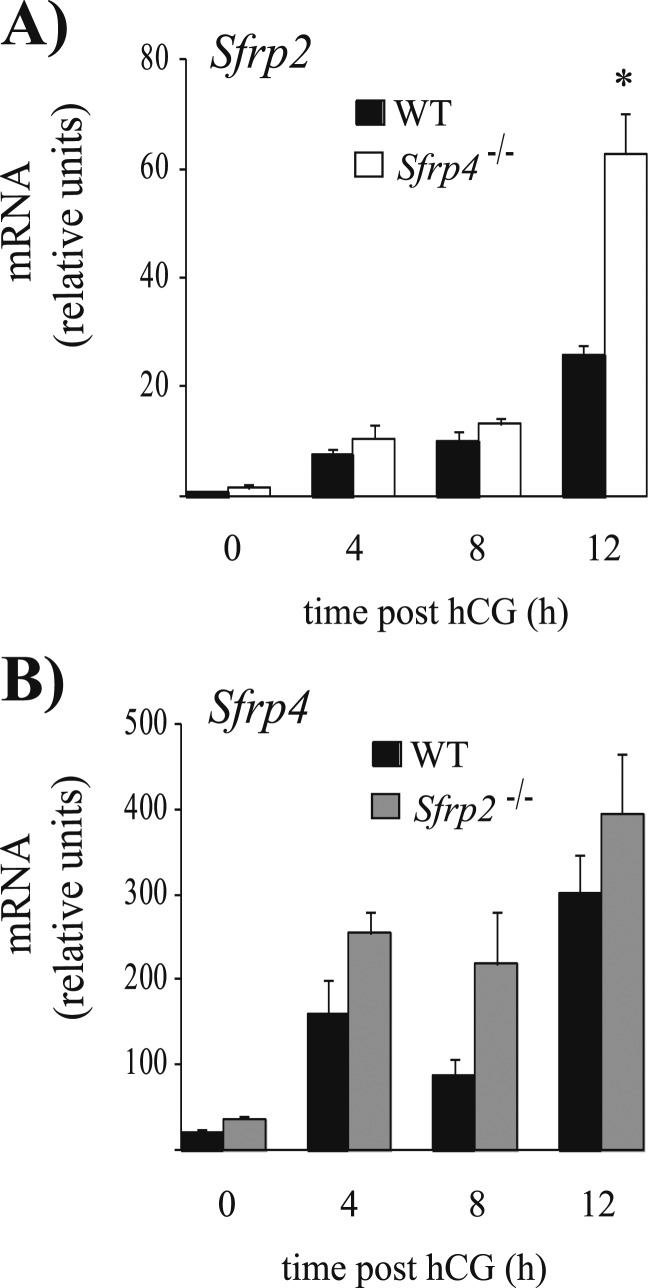
Sfrp2 and Sfrp4 are both strongly expressed in ovarian GCs and encode proteins predicted to have similar functions. Additionally, we found increased Sfrp2 mRNA levels in cumulus cells and GCs of Sfrp4-null mice at 12 hours post-hCG (Figs. 4A and 7A), suggesting a potential compensatory mechanism to offset the loss of Sfrp4. Hypothesizing that Sfrp2 and Sfrp4 could be functionally redundant in the ovary, we characterized Sfrp2-null and Sfrp2/4 double-knockout mice. Mice of both genotypes were viable, developed normally, and were fertile and otherwise indistinguishable from their WT littermates. A 6-month fertility trial showed no difference in the fertility of Sfrp2-null females relative to controls, whereas Sfrp2/4-null mice had an increase in number of pups per litter comparable to that observed in Sfrp4-null mice (P < 0.05, Table 4). Likewise, ovulation rates following natural mating were normal in Sfrp2-null females, and they increased in Sfrp2/4-null mice to the same extent as in Sfrp4-null mice (P < 0.05, Table 2). Furthermore, we were unable to detect an increase in Sfrp4 mRNA levels in the GCs of Sfrp2-null mice (Fig. 7B). Taken together, these results indicate that Sfrp2 and Sfrp4 do not exert redundant roles in ovarian function, and that Sfrp2 is dispensable in this context.
Figure 7.
Compensatory Sfrp expression in the GCs of (A) Sfrp4-null and (B) Sfrp2-null mice. Immature mice of the indicated genotypes were treated with eCG for 48 h, followed (or not) by hCG for the indicated times. mRNA levels were determined in isolated GCs by RT-qPCR and normalized to the housekeeping gene Rpl19. For all analyses, n = 3 to 4 animals per genotype per time point. All data are expressed as means ± SEM. *P < 0.05, significant difference vs control at the same time point.
Table 4.
Fertility Trials
| Control | Sfrp2 −/− | Sfrp2/4-null | |
|---|---|---|---|
| n | 6 | 6 | 6 |
| No. of litters | 6.50 ± 0.34 | 6.66 ± 0.55 | 6.00 ± 0.36 |
| Total pups | 48.16 ± 3.24 | 48.50 ± 6.04 | 49.83 ± 4.15 |
| Litter size | 7.38 ± 0.21a | 7.19 ± 0.56a | 8.29 ± 0.43b |
Data are means ± SEM.
Different superscript letters indicate significant differences (P < 0.05).
Discussion
A growing body of evidence now indicates that the canonical WNT signaling pathway is a key mediator of gonadotropin action, and therefore an important positive regulator of follicle development, GC proliferation, steroidogenesis, ovulation, and luteinization (4–6, 8, 22). The SFRPs comprise a family of WNT signaling antagonists that are expressed in the ovary, and thus they represent potential locally acting factors that could serve to attenuate gonadotropin signaling in certain contexts. In the present study, we tested the latter hypothesis by generating and characterizing an Sfrp4 knockout mouse model. We found these mice to be hyperfertile, at least in part due to enhanced follicle survival, and to have an increased ovulation rate, increased expression of LH and FSH target genes in follicular granulosa and cumulus cells and in the CL, and enhanced responsiveness of GCs to both FSH and LH. These findings represent evidence that Sfrp4 acts to antagonize gonadotropin signaling and apparently serves as a negative feedback regulator of gonadotropin action.
Sfpr4 expression in ovarian follicles was initially reported to be very low in immature and eCG-treated mice, and to increase significantly only after hCG stimulation (16). We therefore expected phenotypic abnormalities in Sfrp4-null mice to occur mainly (if not exclusively) in post-LH surge phases of follicle development. However, we found the LH-driven processes of luteinization, progesterone secretion, and cumulus expansion to occur normally in Sfrp4-null mice, with only mild increases in LH target gene expression occurring in cumulus cells and in the CL, suggesting that neither is a main target of SFRP4 action. Conversely, we found much more dramatic changes in the FSH-driven processes of follicle development and survival, as well as in the responsiveness of GCs to FSH both in vivo and in vitro. Importantly, and contrary to previous reports, we found that Sfrp4 mRNA levels increase markedly in GCs in response to eCG, which is also consistent with it playing an important role during follicle growth. The exact reasons why an increased ovulation rate occurs in the Sfrp4-null model remain to be determined, but the increase in healthy antral follicle numbers in these mice would appear to be the main cause, as more LH-responsive follicles would be available to ovulate with every cycle. We cannot, however, exclude that the increased ovulation rate was also due, at least in part, to a more robust response to LH. Increased Lhcgr expression occurred in the preovulatory follicles of Sfrp4-null mice, as well as an increase in LH response genes 12 hours following administration of hCG, clearly indicating an LH hyperresponsiveness around the time of ovulation that may have contributed to the increased ovulation rate and resultant hyperfertility.
Although ovulation rates following natural mating were substantially higher (approximately three oocytes per cycle) in Sfrp4-null mice relative to their WT siblings, this only resulted in an increase of about one pup per litter. This apparent discrepancy could be explained in several ways, but we presume that the carrying capacity of the uterus was the limiting factor (i.e., the uterus in the mutant mice could not establish and/or maintain pregnancies of 10+ pups). Additional experiments would be needed to determine whether the excess embryos failed to implant or were lost at some point during pregnancy. Our study therefore illustrates that breeding trials are not ideal tools for the characterization of female hyperfertility phenotypes in mice. Indeed, our ovulation rate and follicle count data would appear to be more robust indicators of hyperfertility in our model.
Given the expression patterns of Sfrp2 and Sfrp4 in the ovary, we anticipated that they would be functionally redundant, and therefore that the Sfrp2/4-null model would have a more dramatic phenotype than when each gene was inactivated individually. However, not only did the Sfrp2/4-null mouse simply phenocopy the Sfrp4-null mouse, the inactivation of Sfrp2 alone had no effect on ovulation rates or fertility. This unexpected result indicates that Sfrp2 and Sfrp4 are functionally unrelated, at least in the ovary. What the ovarian functions of Sfrp2 may be, as well as why and how they differ from those of Sfrp4, remains to be resolved. Note, however, that Sfrp2 mRNA levels did not change in response to eCG in GCs, unlike Sfrp4, which was markedly induced. As most of our data suggest a more critical role for Sfrp4 in antagonizing FSH activity rather than LH, Sfrp2 may simply not be sufficiently highly expressed in GCs during FSH-driven follicle development to antagonize FSH signaling and act in a redundant manner with Sfrp4.
One obvious question raised by our study is the mechanism whereby SFRP4 may regulate gonadotropin signaling. By sequestering canonical WNTs, SFRP4 may act to reduce levels of the signaling effector CTNNB1 (β-catenin), which is a known transcriptional coregulator of FSH target genes, including Cyp19a1 and Lhcgr (23, 24). Another possibility was raised in a recent study of Wnt5a function in the ovarian follicle (8). WNT5a is a noncanonical WNT that, in the ovary and in other tissues, can act to suppress canonical WNT signaling, possibly by competitively displacing canonical WNTs from their receptors (25). Treatment of GCs with WNT5a reduced gonadotropin responsiveness not only by reducing CTNNB1 expression, but also by causing CREB levels to decrease via a mechanism involving glycogen synthase kinase 3β (8). As CREB is a key effector of the protein kinase A signaling pathway, which transduces both the FSH and LH signals, it is likely the main effector through which WNT5a antagonizes gonadotropin signaling. If SFRP4 antagonizes WNT signaling by sequestering canonical WNTs, it may therefore act via a mechanism similar to that of WNT5a. Indeed, we have recently presented data that are consistent with the latter hypothesis (26).
In summary, we report the ovarian functions of Sfrp4. We provide evidence herein that SFRP4 acts as a negative regulator of the follicular response to the gonadotropins, thereby limiting follicle development, ovulation, and female fertility. These findings further define the role of WNT signaling in ovarian follicle development, and they highlight its role as a major regulator of gonadotropin action.
Acknowledgments
The authors thank Meggie Girard for assistance with mouse colony management and Marie-Noëlle Laguë for technical support. The R1 ES cell line was provided by Dr. Andras Nagy, Reka Nagy, Dr. Janet Rossant, and Dr. Wanda Abramow-Newerly (University of Toronto).
Financial Support: This work was supported by Canadian Institutes of Health Research Grant MOP-102508 (to D.B.) and by National Institute of Child Health and Human Development Grant U54-HD28934.
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- CL
corpus luteum
- COC
cumulus–oocyte complex
- CREB
cAMP response element–binding protein
- Ct
cycle threshold
- eCG
equine chorionic gonadotropin
- ES
embryonic stem
- GC
granulosa cell
- hCG
human chorionic gonadotropin
- RT-qPCR
quantitative RT-PCR
- SFRP
secreted frizzled-related protein
- WT
wild-type
References
- 1. Edson MA, Nagaraja AK, Matzuk MM. The mammalian ovary from genesis to revelation. Endocr Rev. 2009;30(6):624–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Conti M, Hsieh M, Park JY, Su YQ. Role of the epidermal growth factor network in ovarian follicles. Mol Endocrinol. 2006;20(4):715–723. [DOI] [PubMed] [Google Scholar]
- 3. Richards JS, Pangas SA. The ovary: basic biology and clinical implications. J Clin Invest. 2010;120(4):963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boyer A, Goff AK, Boerboom D. WNT signaling in ovarian follicle biology and tumorigenesis. Trends Endocrinol Metab. 2010;21(1):25–32. [DOI] [PubMed] [Google Scholar]
- 5. Lapointe E, Boerboom D. WNT signaling and the regulation of ovarian steroidogenesis. Front Biosci (Schol Ed). 2011;3:276–285. [DOI] [PubMed] [Google Scholar]
- 6. Wang HX, Li TY, Kidder GM. WNT2 regulates DNA synthesis in mouse granulosa cells through beta-catenin. Biol Reprod. 2010;82(5):865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boyer A, Lapointe E, Zheng X, Cowan RG, Li H, Quirk SM, DeMayo FJ, Richards JS, Boerboom D. WNT4 is required for normal ovarian follicle development and female fertility. FASEB J. 2010;24(8):3010–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Abedini A, Zamberlam G, Lapointe E, Tourigny C, Boyer A, Paquet M, Hayashi K, Honda H, Kikuchi A, Price C, Boerboom D. WNT5a is required for normal ovarian follicle development and antagonizes gonadotropin responsiveness in granulosa cells by suppressing canonical WNT signaling. FASEB J. 2016;30(4):1534–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hsieh M, Boerboom D, Shimada M, Lo Y, Parlow AF, Luhmann UF, Berger W, Richards JS. Mice null for Frizzled4 (Fzd4−/−) are infertile and exhibit impaired corpora lutea formation and function. Biol Reprod. 2005;73(6):1135–1146. [DOI] [PubMed] [Google Scholar]
- 10. Lapointe E, Boyer A, Rico C, Paquet M, Franco HL, Gossen J, DeMayo FJ, Richards JS, Boerboom D. FZD1 regulates cumulus expansion genes and is required for normal female fertility in mice. Biol Reprod. 2012;87(5):104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bovolenta P, Esteve P, Ruiz JM, Cisneros E, Lopez-Rios J. Beyond Wnt inhibition: new functions of secreted Frizzled-related proteins in development and disease. J Cell Sci. 2008;121(Pt 6):737–746. [DOI] [PubMed] [Google Scholar]
- 12. Espada J, Calvo MB, Díaz-Prado S, Medina V. Wnt signalling and cancer stem cells. Clin Transl Oncol. 2009;11(7):411–427. [DOI] [PubMed] [Google Scholar]
- 13. Finch PW, He X, Kelley MJ, Uren A, Schaudies RP, Popescu NC, Rudikoff S, Aaronson SA, Varmus HE, Rubin JS. Purification and molecular cloning of a secreted, Frizzled-related antagonist of Wnt action. Proc Natl Acad Sci USA. 1997;94(13):6770–6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rattner A, Hsieh JC, Smallwood PM, Gilbert DJ, Copeland NG, Jenkins NA, Nathans J. A family of secreted proteins contains homology to the cysteine-rich ligand-binding domain of frizzled receptors. Proc Natl Acad Sci USA. 1997;94(7):2859–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hernandez-Gonzalez I, Gonzalez-Robayna I, Shimada M, Wayne CM, Ochsner SA, White L, Richards JS. Gene expression profiles of cumulus cell oocyte complexes during ovulation reveal cumulus cells express neuronal and immune-related genes: does this expand their role in the ovulation process? Mol Endocrinol. 2006;20(6):1300–1321. [DOI] [PubMed] [Google Scholar]
- 16. Hsieh M, Mulders SM, Friis RR, Dharmarajan A, Richards JS. Expression and localization of secreted frizzled-related protein-4 in the rodent ovary: evidence for selective up-regulation in luteinized granulosa cells. Endocrinology. 2003;144(10):4597–4606. [DOI] [PubMed] [Google Scholar]
- 17. Drake JM, Friis RR, Dharmarajan AM. The role of sFRP4, a secreted frizzled-related protein, in ovulation. Apoptosis. 2003;8(4):389–397. [DOI] [PubMed] [Google Scholar]
- 18. Bardeesy N, Sinha M, Hezel AF, Signoretti S, Hathaway NA, Sharpless NE, Loda M, Carrasco DR, DePinho RA. Loss of the Lkb1 tumour suppressor provokes intestinal polyposis but resistance to transformation. Nature. 2002;419(6903):162–167. [DOI] [PubMed] [Google Scholar]
- 19. Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20. Zeleznik AJ, Midgley AR Jr, Reichert LE Jr. Granulosa cell maturation in the rat: increased binding of human chorionic gonadotropin following treatment with follicle-stimulating hormone in vivo. Endocrinology. 1974;95(3):818–825. [DOI] [PubMed] [Google Scholar]
- 21. Christov M, Koren S, Yuan Q, Baron R, Lanske B. Genetic ablation of Sfrp4 in mice does not affect serum phosphate homeostasis. Endocrinology. 2011;152(5):2031–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hsieh M, Johnson MA, Greenberg NM, Richards JS. Regulated expression of Wnts and Frizzleds at specific stages of follicular development in the rodent ovary. Endocrinology. 2002;143(3):898–908. [DOI] [PubMed] [Google Scholar]
- 23. Hernandez Gifford JA, Hunzicker-Dunn ME, Nilson JH. Conditional deletion of beta-catenin mediated by Amhr2cre in mice causes female infertility. Biol Reprod. 2009;80(6):1282–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Parakh TN, Hernandez JA, Grammer JC, Weck J, Hunzicker-Dunn M, Zeleznik AJ, Nilson JH. Follicle-stimulating hormone/cAMP regulation of aromatase gene expression requires β-catenin. Proc Natl Acad Sci USA. 2006;103(33):12435–12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bryja V, Andersson ER, Schambony A, Esner M, Bryjová L, Biris KK, Hall AC, Kraft B, Cajanek L, Yamaguchi TP, Buckingham M, Arenas E. The extracellular domain of Lrp5/6 inhibits noncanonical Wnt signaling in vivo. Mol Biol Cell. 2009;20(3):924–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zamberlam G, Lapointe E, Abedini A, Rico C, Paquet M, Boerboom D SFRP4 inhibits gonadotropin action in mouse granulosa cells by blocking GSK3B-CREB signaling. In: Proceedings of the 50th Annual Meeting of the Society for the Study of Reproduction; 13–16 July 2017; Washington, DC. Abstract 355. [Google Scholar]