Abstract
Background: The oncogenicity of neural precursor cell-expressed developmentally down-regulated 9 (NEDD9) has been demonstrated in multiple cancer types. However, the prognostic value of NEDD9 in some solid cancers remains controversial. Thus, this meta-analysis was conducted to evaluate the relationship between NEDD9 expression survival rates in solid tumors.
Method: Our meta-analysis included studies searched from various search engines with specific inclusion criteria and exclusion criteria. Combined HRs for overall survival (OS) and disease-free survival (DFS) or progression-free survival (PFS) or recurrence-free survival (RFS) or cancer-specific survival (CSS) were assessed using fixed-effects and random-effects models. The source of heterogeneity was identified by subgroup analysis. Additionally, publication bias was assessed using funnel plot and Egger’s regression asymmetry test.
Result: Eighteen studies with a total of 2,476 patients were retrieved for analysis. Pooled HRs and 95% CIs were calculated. Both OS (HR=1.82; 95% CI: 1.43–2.31) and DFS/PFS/RFS/CSS (HR=2.54; 95% CI: 1.93–3.33) indicated that NEDD9 overexpression is associated with poor OS in cancer patients with solid tumors.
Conclusion: NEDD9 overexpression might be a potential marker to predict prognosis in solid cancer patients.
Keywords: NEDD9, solid cancer, prognosis, meta-analysis
Introduction
Epidemiological data show that cancers have become one of the world’s leading causes of mortality.1 From the latest data, solid cancers accounted for over 90% of all types of cancers and the top five of new cases and deaths are lung cancer, breast cancer, prostate cancer, colon cancer and nonmelanoma skin cancer.1 Solid cancers are characterized by malignant tumors that form a discrete tumor mass. By contrast, lymphoproliferative malignancies diffusely infiltrate tissue without forming a mass. Although diagnosis of number of molecular markers and targeted therapies progressed with the efforts of many researchers and clinicians, the outcomes including death rates and overall survival (OS) of the majority of patients remain poor. Thus, more predictive molecular markers of solid cancers must be identified for prevention and individualized cancer treatment.
Neural precursor cell-expressed developmentally down-regulated 9 (NEDD9) , also known as human enhancer of filamentation 1 (HEF1) and Crk-associated substrate lymphocyte type (Cas-L), belongs to the Crk-associated substrate family. NEDD9 coordinates the focal adhesion kinase and SRC signaling cascades that are involved in integrin-dependent adhesion and migration, invasion, cell apoptosis and life cycle, and survival.2–6 NEDD9 overexpression contributes to solid cancer metastasis in lung, liver, breast, ovarian, colon, glioblastoma carcinoma, and cervical cancers.3,7–13 NEDD9 expression levels are a biomarker of cancer aggression and could be a prognostic factor of solid cancers.14 However, prognostic evidence is lacking due to studies with limited sample sizes or cancer types. This is why we evaluated the prognostic value of NEDD9 in solid tumors through meta-analysis to elucidate the clinical implications.
Methods
The meta-analysis was performed according to the PRISMA guideline and the statement for reporting systematic reviews and meta-analyses.15 Previously published studies were summarized and analyzed in this study (ethics board approval was not necessary).
Search strategy and study eligibility
We retrieved studies that measured NEDD9 expression and survival in solid cancer patients between 1995 and January 2019 from PubMed, EMBASE, Web of Science, and Google Scholar. The search terms included “NEDD9” or “neural precursor cell-expressed developmentally down-regulated 9” or “HEF1” or “human enhancer of filamentation 1”or “Cas-L” or “Crk-associated substrate lymphocyte type” and “neoplasms” or “cancer” or “tumor” or “malignancy” or “carcinoma” and “prognosis” or “survival”. Non-English language studies were excluded. Results were restricted to human studies of solid cancer and 402 entries were found.
Inclusion criteria consisted of an evaluation of NEDD9 overexpression associated with OS (date of surgery to date of death as a result of any cause), disease-free survival (DFS) or progression-free survival (PFS) or recurrence-free survival (RFS) or cancer-specific survival (CSS), and immunohistochemistry (IHC) or ELISA analysis of NEDD9 expression. Tumors were classified by NEDD9 expression levels using the cutoffs defined in the studies (Table 1). All references of the included studies were scanned and studies of potential interest were reviewed for further analysis. Reviews, clinical endpoints other than OS or DFS/PFS/RFS/CSS, studies that enrolled less than 15 solid cancer patients, and studies without data that could be used to calculate the HR and 95% CI were excluded from our meta-analysis. Any disagreement was resolved by discussion among all investigators until a final consensus was reached.
Table 1.
Main characteristics of all studies included in the meta-analysis
| Study | Country | Tumor type | Case number | Age (years) | Gender (M/F) | TNM stage (I/II/III/IV) | Follow-up (months) | Detected method | Survival analysis | Cut-off value | Multivariate analysis | HR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Afsar 201838 | Turkey | Pancrentic cancer | 32 | Media 61 | 20/12 | NR | Max 27 | ELISA | OS/PFS | NR | No | SC |
| El-Babouly 201836 | Egypt | Bladder cancer | 105 | Mean 57 | 69/36 | 32/73(I-II/III-IV) | Mean 25 | IHC | PFS | Hsa >4 | No | SC |
| Erturk 201835 | Turkey | Melanoma | 112 | Mean 52 | NR | NR | Mean 20.8 | ELISA | OS | NR | No | Report |
| Harb 201731 | Egypt | Bladder cancer | 50 | Mean 52 | 35/15 | 24/26 (Ta-T1/T2-T3) (T stage) |
Max 35 | IHC | OS | Hsa >4 | Yes | Report |
| Karabulut 201532 | Turkey | Gastric cancer | 68 | Media 60 | 49/19 | NR | Mean 8 | ELISA | OS/PFS | NR | No | SC |
| Kondo 201223 | Japan | Lung cancer | 60 | Mean 62.5 | 26/34 | 31/5/23/1(I/II/III/IV) | 0–90 | IHC | OS/RFS | >30% of nuclear Stained | Yes | Report |
| Li 201424 | China | Colorectal cancer | 92 | Mean 62 | 52/40 | 15/30/37/10(I/II/III/IV) (UICC stage) | 5–60 | IHC | OS | Hsa >4 | Yse | Report |
| Li 201625 | China | Breast cancer | 226 | Median 56 | 0/226 | 79/83/53/11(I/II/III/IV) | Median 27 | IHC | OS/DFS | Hsb ≥4 | Yes | Report |
| Liu 201439 | China | Gastric cancer | 187 | NR | 104/83 | 77/110(I-II/III-IV) | 0–60 | IHC | OS | >25% of nuclear stained |
No | SC |
| Lu 201526 | China | Hepatocellular cancer | 164 | Median 49 | 133/31 | 32/73(I-II/III-IV) (UICC Stage) | 0–60 | IHC | OS | Hsb ≥4 | Yes | Report |
| Miao 201327 | China | Lung cancer | 105 | Mean 60.4 | 63/42 | 46/59(I-II/III-IV) (UICC stage) | 0–60 | IHC | OS | Postive cell score ≥3 | No | SC |
| Ostojic 201833 | Croatia | Lung cancer | 71 | Median 49.2 | 112/53 | 55/110(I-II/III-IV) (UICC stage) | 0–125 | IHC | OS | Postive cell score ≥3 | No | SC |
| Shi 201428 | China | Gastric cancer | 125 | NR | 61/64 | 73/52(I-II/III-IV) | 0–60 | IHC | OS | Hsa ≥4 | Yes | Report |
| Wang 20148 | China | Ovarian cancer | 129 | NR | 0/129 | 73/52(I-II/III-IV) (FIGO stage) | 0–60 | IHC | OS/PFS | Nuclear staining | Yes | Report |
| Wang 201737 | China | Renal cancer | 68 | NR | 69/39 | 53/15(I-II/III-IV) | 0–60 | IHC | CSS | Hsc ≥5 | No | SC |
| Xue 201329 | China | Pancrentic cancer | 106 | NR | 65/41 | 54/52(I-II/III-IV) | 0–60 | IHC | OS | Hsb ≥4 | No | Report |
| Zhang 201434 | China | Bladder cancer | 175 | Median 61.5 | 117/58 | 61/114(Ta-T1/T2-T3) | 0–60 | IHC | OS | Hsb ≥4 | Yes | Report |
| Zhang 201430 | China | Gastric cancer | 601 | Median 64 | 428/173 | 127/135/231/108(I/II/ III/IV) |
0–60 | IHC | OS | Hsb ≥4 | Yes | Report |
Notes: HSa, histologic score (overall scores = percentage score [0–4] × intensity score [0–3], with the minimum score of 0 and a maximum score of 12); HSb, histologic score (overall scores = percentage score [0–3] × intensity score [0–3], with the minimum score of 0 and a maximum score of 9); HSc, histologic score (overall scores = percentage score [0–3] + intensity score [0–4], with the minimum score of 0 and a maximum score of 7).
Abbreviations: OS, overall survival; RFS, recurrence-free survival; PFS, progression-free survival; DFS, disease-free survival; CSS, cancer-specific survival; HR, hazard ratio; UICC, union for international cancer control; FIGO, international federation of gynecology and obstetrics; NR, not report; SC, survival curve; IHC, immunohistochemistry; ELISA, enzyme-linked immunosorbent assay.
Data collection and quality assessment
The included studies that were assessed independently by two investigators (J.L and Y.G). Disagreements were resolved by consensus. Data retrieved from the studies included the following: author, country, year of publication, patient number, gender, follow-up time, OS, DFS/PFS/RFS/CSS, cutoff value for over/normal NEDD9 expression, histological type, and HR estimation. Some HR data were extracted from tables or Kaplan–Meier curves of NEDD9 overexpression and normal expression groups. We used the Newcastle–Ottawa Non-Randomized scale (NOS) as recommended by the Cochrane Non-Randomized Studies Methods Working Group for a quality assessment of the cohort studies.16,17 The NOS score ranged from 0 to 9 based on three board perspectives: study group selection, study group comparability, and ascertainment of the outcome of interest. A consensus NOS score for each study was obtained by a discussion between all investigators. Studies with a score ≥5 had a high-quality methodology.
Statistical analysis
Data were extracted from the primary publications and analyzed with Stata 12 (Stata Corporation, College Station, TX, USA). The HR and corresponding 95% CI estimates were calculated and pooled to determine the association of NEDD9 overexpression with OS and DFS/PFS/RFS/CSS. An HR>1 indicated a poor prognosis in patients with NEDD9 overexpression. The random-effects model and the fixed-effects model were conducted according to DerSimonian and Laird’s method and Mantel-Haenszel’s method, respectively.18,19 Statistical intrastudy heterogeneity was evaluated by the I2 value to quantify the proportion of the total variation. The I2 values of 25%, 50%, and 75% were the cutoff points of low, moderate, and high heterogeneity, respectively.20 We used the fixed-effects model to pool the results if relatively low or moderated heterogeneity existed (I2<50%). The random-effects model was used when the I2 value was ≥50%. If high or moderate heterogeneity existed, a subgroup analysis of the cancer characteristics was conducted to determine possible causes.21 Differences between the subgroups were calculated from the Cochrane Handbook for Systematic Reviews of Interventions. Outcome credibility was validated by sensitivity analysis and the publication bias was assessed by visual inspection of the funnel plots. Funnel plot asymmetry was determined by Egger’s linear regression test.22 A two-sided P<0.05 was considered statistically significant.
Results
Search results and study characteristics
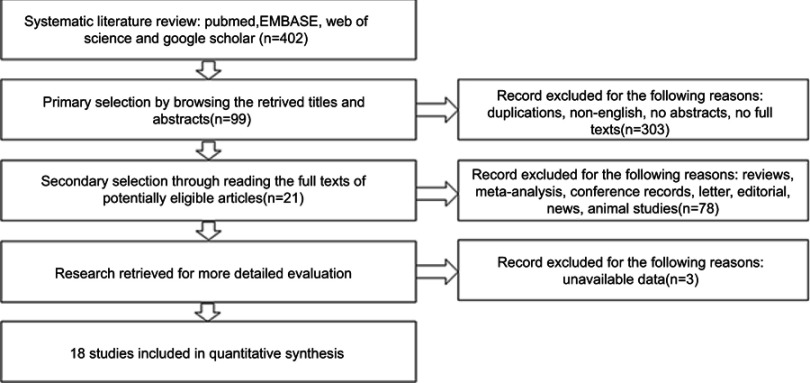
Eighteen studies were eligible for the pooled meta-analysis. Eligible studies were published between 2012 and February 2019 with a total of 2476 patients.8,23–38 A flowchart depicting the selection of literature is shown in Figure 1.
Figure 1.
Flow diagram of the study selection process.
All included studies were retrospectively analyzed. The sample size ranged from 32 to 601 (median 137). NEDD9 overexpression was reported in 1309 (52.9%) of the 2476 included patients. Amongst all study cohorts, 3 evaluated lung cancer,23,27,33 4 assessed gastric cancer,28,30,32,39 3 undertook urinary bladder carcinoma,31,34,36 2 illustrated pancreatic carcinoma,29,38 and single studies focused on breast cancer,25 colorectal cancer,24 hepatocellular carcinoma,26 renal carcinoma,37 melanoma,35 and ovarian cancer.8 Studies were conducted in China (11), Japan (1), Turkey (3), Croatia (1), and Egypt (2). The Newcastle–Ottawa Quality Assessment Scale indicated that the quality scores ranged from 6 to 8 (median 7) with a relatively high study quality. The main characteristics of the 18 eligible publications are reported in Table 1. Seven studies8,23,25,32,36–38 reported that the HR of DFS/PFS/RFS/CSS associated with NEDD9 expression. Sixteen studies8,23–35,38,39 included OS and a follow-up time of 8–125 months. The HR and 95% CI were directly recorded in 11 studies.8,23–26,28–31,34,35 HR was inferred from the remaining studies using the Tierney method to estimate HR from Kaplan–Meier survival curves.
Meta-analysis results
Moderate intrastudy heterogeneity was observed (I2=58.4%) in 16 studies where OS was determined by univariate analysis or multivariate analysis. The random-effects model was used to pool the results. The combined HR estimate of OS was 1.82 (95% CI: 1.43–2.31; P<0.01). NEDD9 overexpression was associated with poor OS (Figure 2).
Figure 2.
Forest plot of NEDD9 overexpression with overall survival.
Studies were divided into five groups by cancer location for subgroup analysis: (1) digestive system cancer (n=8);24,26,28–30,32,38,39 (2) respiratory system cancer (n=3);23,27,33 (3) breast and ovarian cancer (n=2);8,25 (4) urinary system cancer (n=2);31,34 and (5) melanoma (n=1).35 The combined HR estimate for OS in the digestive system cancer group was 1.61 (95% CI: 1.32–1.95; I2=8.4%). The HR estimate for OS in the respiratory system cancer group was 1.96 (95% CI: 0.68–5.70; I2=85%), 3.96 (95% CI: 2.26–6.94; I2=0%) for breast and ovarian cancer groups, and 2.44 (95% CI: 1.54–3.86; I2=0%) for urinary system groups.
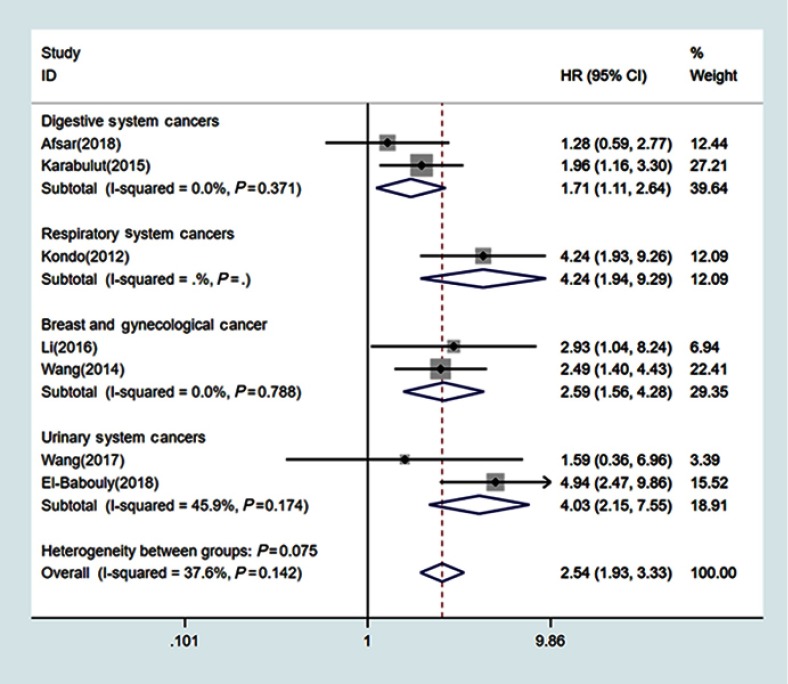
There was no apparent intrastudy heterogeneity (I2=0%) among 7 studies where DFS/PFS/RFS/CSS was determined by univariate analysis. The fixed-effects model was used to pool the results. The combined HR estimate was 2.54 (95% CI: 1.93–3.33; P=0.142), which indicated that NEDD9 overexpression was associated with poor DFS/PFS/RFS/CSS (Figure 3). The studies were subdivided by cancer location: digestive system (n=2),32,38 respiratory system (n=1),23 breast and ovarian cancer (n=2),8,25 and urinary system cancer (n=2).36,37The combined HR estimate for DFS/PFS/RFS/CSS were 1.71 (95% CI: 1.11–2.64; I2=0%) in the group digestive system cancer and 2.59 (95% CI: 1.56–4.28; I2=0%) in the group breast and ovarian cancers. The HR estimate for DFS/PFS/RFS/CSS in urinary cancer was 4.03 (95% CI: 2.15–7.55; I2=45.9%).
Figure 3.
Forest plot of NEDD9 overexpression with DFS/PFS/RFS/CSS.
Abbreviations: DFS, disease-free survival; PFS, progression-free survival; RFS, recurrence-free survival; CSS, cancer-specific survival.
Publication bias
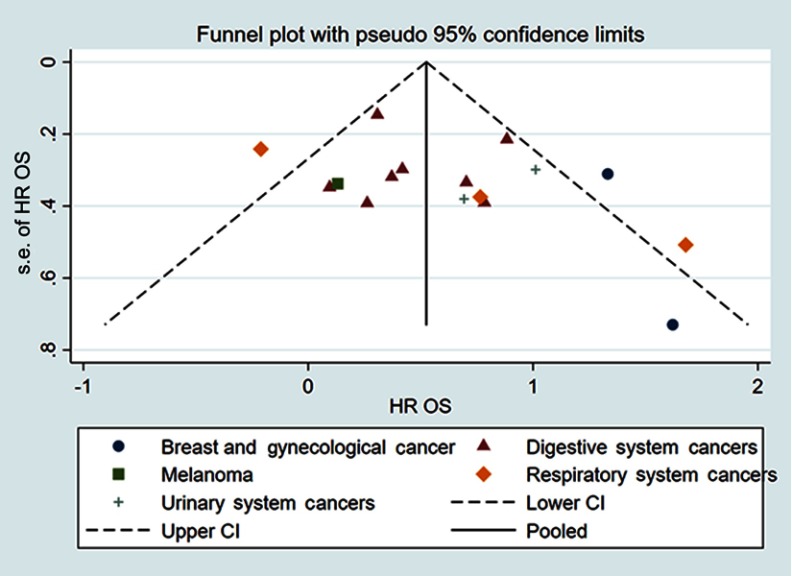
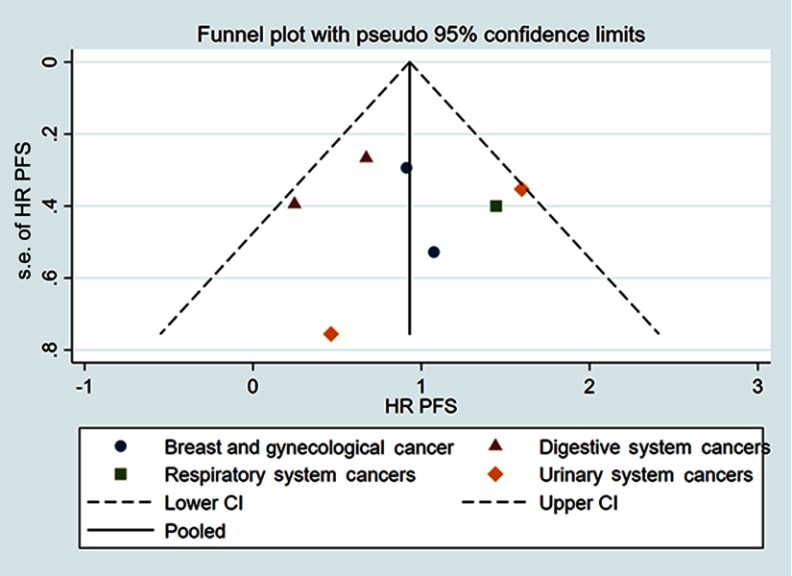
Funnel plot and Egger’s test were used to evaluate the publication bias. The shape of the funnel plots did not show obvious asymmetry for OS determined by univariate analysis or multivariate analysis (Figure 4). Egger’s test also failed to provide evidence of significant publication bias. Publication bias was also investigated for DFS/PFS/RFS/CSS obtained by univariate analysis and the funnel plots showed basic symmetry (Figure 5). Our results suggested that publication bias was not evident.
Figure 4.
Funnel plots for OS.
Abbreviation: OS, overall survival.
Figure 5.
Funnel plots for DFS/PFS/RFS/CSS.
Abbreviations: DFS, disease-free survival; PFS, progression-free survival; RFS, recurrence-free survival; CSS, cancer-specific survival.
Discussion
Regarding prognostic value of level of NEDD9 and survival rates of solid tumors patients, we use the tool of meta-analysis to demonstrate that the high expression of NEDD9 acts as a prediction of the poor outcome for patients with solid tumors.
NEDD9 contains multiple functional modules for protein interaction, yet lacks any known enzymatic function. Adult lung and kidney tissues, which are rich in immature lymphoid cells, as well as the fetal brain, contain the highest levels of NEDD9.40–42 NEDD9 is crucial in integrin-dependent signaling cascades to activate cell migration and is involved in lateral communication to Ras signaling cascades.7 NEDD9 can drive mesenchymal-type movement by activating RAC2 GTPase and WAVE in complex with GEF DOCK3 to inhibit GTPase Rho and amoeboid movement.43 Invasion is accompanied by ECM proteolysis through activation of MMP14, MMP2, and MMP9 metalloproteinases.44 Epithelial–mesenchymal transition (EMT) is a transdifferentiation strategy where carcinoma cells acquire the ability to execute the steps of invasion-metastasis cascade.45 Epithelial cells express mesenchymal markers and cytoskeletal changes that increase motility and invasiveness during EMT. NEDD9 also interacts with key proteins to coordinate the signaling cascades controlling cell cycle transitions, apoptosis, migration, and invasion.4,5,46,47 NEDD9 expression has been linked to tumorigenesis of various malignancies, including cancers of the breast,25 colon,25,48,49 pancreas,29 head and neck,50 ovaries,8 gastric,24,30,32 lung,23,27,51 liver,52 kidney,53 glioblastoma,54 and neuroblastoma.55 However, some studies have the opposite point of view. One example is a study from Ostojić et al (2018) that is also included in our meta-analysis.33 It is interesting that they concluded the relationship between NEDD9 expression and survival did not reach statistically significant difference and there was a trend toward the correlation of higher NEDD9 to longer survival. The explanation is that expression of NEDD9 over certain level inevitably leads to apoptosis.56 Furthermore, it is also possible that elevated expression of NEDD9 may indicate better responses to therapy.23 Although there are some different opinions, our findings through meta-analysis suggested that elevated NEDD9 expression was correlated with poor survival in most solid cancers. Further studies are needed to elucidate the mechanisms of NEDD9 in cancer pathogenesis.
The prognostic value of NEDD9 in solid cancer is controversial despite previous investigations of this marker in multiple cancers. This is the first meta-analysis to analyze NEDD9 overexpression in relation to OS or DFS/PFS/RFS/CSS of solid cancer patients. We systematically evaluated the survival data of 2393 solid cancer patients from 18 eligible studies. We demonstrated that NEDD9 overexpression was associated with poor survival in solid cancer patients. The detrimental effect of NEDD9 overexpression on survival was statistically significant in OS or DFS/PFS/RFS/CSS. NEDD9 could be a prognostic biomarker and novel therapeutic target of solid cancer.
Several limitations should be considered when interpreting the findings of our meta-analysis. Differences among dilution solubility, antibodies, and cutoff values influenced the assessment of NEDD9 overexpression. A large multicenter clinical study using consistent antibodies and cutoff values is needed to validate our results. Language bias may exist in our meta-analysis because the search strategy was limited to English. Most eligible studies included Asian populations, especially Chinese, leading to population homogeneity. Some studies did not report HR and 95% CI directly. Data extracted by Tierney’s methods may introduce bias to the original data.
Conclusion
Here, we provide evidence that NEDD9 overexpression is associated with poor OS or DFS/PFS/RFS/CSS in solid cancer patients. Further investigation is required to elucidate the role of NEDD9 overexpression and provide new insights into tumor metastasis and progression for the development of target-specific therapies.
Author contributions
Conception/Design: Fei Zheng. Provision of study materials: Yang Gu, Jingjing Lu, Chen Chen. Collection and extract data: Yang Gu, Jingjing Lu. Data analysis and statistical guidance: Yang Gu. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424 [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. [DOI] [PubMed] [Google Scholar]
- 3.Natarajan M, Stewart J, Golemis E, et al. HEF1 is a necessary and specific downstream effector of FAK that promotes the migration of glioblastoma cells. Oncogene. 2006;25(12):1721. doi: 10.1038/sj.onc.1209199 [DOI] [PubMed] [Google Scholar]
- 4.O’neill GM, Golemis EA. Proteolysis of the docking protein HEF1 and implications for focal adhesion dynamics. Mol Cell Biol. 2001;21(15):5094–5108. doi: 10.1128/MCB.21.15.5094-5108.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seventer G, Salmen HJ, Law SF, et al. Focal adhesion kinase regulates β1 integrin‐dependent T cell migration through an HEF1 effector pathway. Eur Jo Immunol. 2001;31(5):1417–1427. doi: [DOI] [PubMed] [Google Scholar]
- 6.Tikhmyanova N, Golemis EA. NEDD9 and BCAR1 negatively regulate E-cadherin membrane localization, and promote E-cadherin degradation. PloS One. 2011;6(7):e22102. doi: 10.1371/journal.pone.0022102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Izumchenko E, Singh MK, Plotnikova OV, et al. NEDD9 promotes oncogenic signaling in mammary tumor development. Cancer Res. 2009;69(18):7198–7206. doi: 10.1158/0008-5472.CAN-08-3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, Mu X, Zhou S, et al. NEDD9 overexpression is associated with the progression of and an unfavorable prognosis in epithelial ovarian cancer. Hum Pathol. 2014;45(2):401–408. doi: 10.1016/j.humpath.2013.10.005 [DOI] [PubMed] [Google Scholar]
- 9.Chang J-X, Gao F, Zhao G-Q, Zhang G-J. Expression and clinical significance of NEDD9 in lung tissues. Med Oncol. 2012;29(4):2654–2660. doi: 10.1007/s12032-012-0213-0 [DOI] [PubMed] [Google Scholar]
- 10.Budhu A, Forgues M, Ye Q-H, et al. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell. 2006;10(2):99–111. doi: 10.1016/j.ccr.2006.06.016 [DOI] [PubMed] [Google Scholar]
- 11.Kong C, Wang C, Wang L, et al. NEDD9 is a positive regulator of epithelial-mesenchymal transition and promotes invasion in aggressive breast cancer. PloS One. 2011;6(7):e22666. doi: 10.1371/journal.pone.0022666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sima N, Cheng X, Ye F, Ma D, Xie X, Lü W. The overexpression of scaffolding protein NEDD9 promotes migration and invasion in cervical cancer via tyrosine phosphorylated FAK and SRC. Plos One. 2013;8(9):e74594. doi: 10.1371/journal.pone.0074594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei H, Li J, Xie M, Lei R, Hu B. Comprehensive analysis of metastasis-related genes reveals a gene signature predicting the survival of colon cancer patients. Peer J. 2018;6:e5433. doi: 10.7717/peerj.5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shagisultanova E, Gaponova AV, Gabbasov R, Nicolas E, Golemis EA. Preclinical and clinical studies of the NEDD9 scaffold protein in cancer and other diseases. Gene. 2015;567(1):1–11. doi: 10.1016/j.gene.2015.04.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med. 2009;6(7):e1000100. doi: 10.1371/journal.pmed.1000100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z [DOI] [PubMed] [Google Scholar]
- 17.Maxwell L, Santesso N, Tugwell PS, Wells GA, Judd M, Buchbinder R. Method guidelines for Cochrane Musculoskeletal Group systematic reviews. J Rheumatol. 2006;33(11):2304–2311. [PubMed] [Google Scholar]
- 18.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 19.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2 [DOI] [PubMed] [Google Scholar]
- 20.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Br Med J. 2003;327(7414):557. doi: 10.1136/bmj.327.7414.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson SG, Higgins J. How should meta‐regression analyses be undertaken and interpreted? Stat Med. 2002;21(11):1559–1573. doi: 10.1002/(ISSN)1097-0258 [DOI] [PubMed] [Google Scholar]
- 22.Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. Bmj. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kondo S, Iwata S, Yamada T, et al. Impact of the integrin signaling adaptor protein NEDD9 on prognosis and metastatic behavior of human lung cancer. Clin Cancer Res. 2012. doi: 10.1158/1078-0432.CCR-11-2162 [DOI] [PubMed] [Google Scholar]
- 24.Li P, Zhou H, Zhu X, et al. High expression of NEDD9 predicts adverse outcomes of colorectal cancer patients. Int J Clin Exp Pathol. 2014;7(5):2565. [PMC free article] [PubMed] [Google Scholar]
- 25.Li P, Sun T, Yuan Q, Pan G, Zhang J, Sun D. The expressions of NEDD9 and E-cadherin correlate with metastasis and poor prognosis in triple-negative breast cancer patients. Onco Targets Ther. 2016;9:5751. doi: 10.2147/OTT.S113768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu P, Wang Z-P, Dang Z, et al. Expression of NEDD9 in hepatocellular carcinoma and its clinical significance. Oncol Rep. 2015;33(5):2375–2383. doi: 10.3892/or.2015.3863 [DOI] [PubMed] [Google Scholar]
- 27.Miao Y, Li A-L, Wang L, et al. Overexpression of NEDD9 is associated with altered expression of E-cadherin, β-catenin and N-cadherin and predictive of poor prognosis in non-small cell lung cancer. Pathol Oncol Res. 2013;19(2):281–286. doi: 10.1007/s12253-012-9580-2 [DOI] [PubMed] [Google Scholar]
- 28.Shi R, Wang L, Wang T, Xu J, Wang F, Xu M. NEDD9 overexpression correlates with the progression and prognosis in gastric carcinoma. Med Oncol. 2014;31(3):852. doi: 10.1007/s12032-014-0852-4 [DOI] [PubMed] [Google Scholar]
- 29.Xue Y-Z, Sheng -Y-Y, Liu Z-L, et al. Expression of NEDD9 in pancreatic ductal adenocarcinoma and its clinical significance. Tumor Biol. 2013;34(2):895–899. doi: 10.1007/s13277-012-0624-8 [DOI] [PubMed] [Google Scholar]
- 30.Zhang Q, Wang H, Ma Y, et al. Overexpression of Nedd9 is a prognostic marker of human gastric cancer. Med Oncol. 2014;31(7):33. doi: 10.1007/s12032-014-0033-5 [DOI] [PubMed] [Google Scholar]
- 31.Harb OA, Haggag R, Ali MM, et al. The prognostic role of NEDD9 and P38 protein expression levels in urinary bladder transitional cell carcinoma. J Oncol. 2017;2017. doi: 10.1155/2017/6095205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karabulut M, Alis H, Afsar CU, et al. Serum neural precursor cell-expressed, developmentally down regulated 9 (NEDD9) level may have a prognostic role in patients with gastric cancer. Biomed Pharmacother. 2015;73:140–146. doi: 10.1016/j.biopha.2015.06.010 [DOI] [PubMed] [Google Scholar]
- 33.Ostojić J, Brčić L, Hrabač P, Seiwerth S. Expression of NEDD9 in transbronchial biopsies of lung adenocarcinoma. Acta Clinica Croatica. 2018;57(2):251–256. doi: 10.20471/acc.2018.57.02.04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Q, Wang H-J, Zhang D-H, Ru G-Q, He X-J, Ma -Y-Y. High expression of HEF1 is associated with poor prognosis in urinary bladder carcinoma. Onco Targets Ther. 2014;7:1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erturk K, Tas F, Serilmez M, Bilgin E, Duranyildiz D. Significance of serum neural precursor cell‑expressed developmentally downregulated protein 9 in melanoma. Mol Clin Oncol. 2018;8(1):204–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El-Babouly IM, Desoky EA, El Sayed D, et al. The role of neural precursor cell-expressed developmentally down-regulated protein 9 in predicting bacillus Calmette-Guerin response in nonmuscle invasive bladder cancer. Urol Oncol 2018;36(5): 242. e9-242. e14. [DOI] [PubMed]
- 37.Wang J, Wang S, Luan Y, et al. Overexpression of NEDD9 in renal cell carcinoma is associated with tumor migration and invasion. Oncol Lett. 2017;14(6):8021–8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Afsar CU, Karabulut M, Karabulut S, et al. Clinical significance of serum NEDD9 levels in patients with pancreatic cancer. Biomolecules. 2018;8:4. doi: 10.3390/biom8020035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y, Wang D, Zhao K-L, et al. NEDD9 overexpression correlates with poor prognosis in gastric cancer. Tumor Biol. 2014;35(7):6351–6356. doi: 10.1007/s13277-014-1839-7 [DOI] [PubMed] [Google Scholar]
- 40.Minegishi M, Tachibana K, Sato T, Iwata S, Nojima Y, Morimoto C. Structure and function of Cas-L, a 105-kD Crk-associated substrate-related protein that is involved in beta 1 integrin-mediated signaling in lymphocytes. J Expl Med. 1996;184(4):1365–1375. doi: 10.1084/jem.184.4.1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar S, Tomooka Y, Noda M. Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem Biophys Res Commun. 1992;185(3):1155–1161. doi: 10.1016/0006-291X(92)91747-E [DOI] [PubMed] [Google Scholar]
- 42.Law S, Estojak J, Wang B. Human Enhancer of Filamenta-tion 1 (HEF1/NEDD9/CAS-L), a novel pl3OCas-Like docking protein, associates with FAK, and induces pseudohyphal growth in yeast. Mol Cell Biol. 1996;16(7):3327–3337. doi: 10.1128/MCB.16.7.3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanz-Moreno V, Gadea G, Ahn J, et al. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135(3):510–523. doi: 10.1016/j.cell.2008.09.043 [DOI] [PubMed] [Google Scholar]
- 44.McLaughlin SL, Ice RJ, Rajulapati A, et al. NEDD9 depletion leads to MMP14 inactivation by TIMP2 and prevents invasion and metastasis. Mol Cancer Res. 2014;12(1):69–81. doi: 10.1158/1541-7786.MCR-13-0300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2(6):442. doi: 10.1038/nrc822 [DOI] [PubMed] [Google Scholar]
- 46.Fashena SJ, Einarson MB, O’Neill GM, Patriotis C, Golemis EA. Dissection of HEF1-dependent functions in motility and transcriptional regulation. J Cell Sci. 2002;115(1):99–111. [DOI] [PubMed] [Google Scholar]
- 47.Dadke D, Jarnik M, Pugacheva EN, Singh MK, Golemis EA. Deregulation of HEF1 impairs M-phase progression by disrupting the RhoA activation cycle. Mol Biol Cell. 2006;17(3):1204–1217. doi: 10.1091/mbc.e06-04-0311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y, Bavarva JH, Wang Z, et al. HEF1, a novel target of Wnt signaling, promotes colonic cell migration and cancer progression. Oncogene. 2011;30(23):2633. doi: 10.1038/onc.2010.632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim S-H, Xia D, Kim S-W, Holla V, Menter DG, DuBois RN. Human enhancer of filamentation 1 is a mediator of hypoxia-inducible factor-1α–mediated migration in colorectal carcinoma cells. Cancer Res. 2010;70(10):4054–4063. doi: 10.1158/0008-5472.CAN-09-2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lucas JT Jr, Salimath BP, Slomiany MG, Rosenzweig SA. Regulation of invasive behavior by vascular endothelial growth factor is HEF1-dependent. Oncogene. 2010;29(31):4449. doi: 10.1038/onc.2010.378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jin Y, Li F, Zheng C, et al. NEDD9 promotes lung cancer metastasis through epithelial–mesenchymal transition. Int Jo Cancer. 2014;134(10):2294–2304. doi: 10.1002/ijc.28568 [DOI] [PubMed] [Google Scholar]
- 52.Xia L, Huang W, Tian D, et al. Overexpression of forkhead box C1 promotes tumor metastasis and indicates poor prognosis in hepatocellular carcinoma. Hepatology. 2013;57(2):610–624. doi: 10.1002/hep.26029 [DOI] [PubMed] [Google Scholar]
- 53.Lu R, Ji Z, Li X, et al. miR-145 functions as tumor suppressor and targets two oncogenes, ANGPT2 and NEDD9, in renal cell carcinoma. J Cancer Res Clin Oncol. 2014;140(3):387–397. doi: 10.1007/s00432-013-1577-z [DOI] [PubMed] [Google Scholar]
- 54.Speranza MC, Frattini V, Pisati F, et al. NEDD9, a novel target of miR-145, increases the invasiveness of glioblastoma. Oncotarget. 2012;3(7):723. doi: 10.18632/oncotarget.547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Merrill RA, See AWM, Wertheim ML, Clagett‐Dame M. Crk‐associated substrate (Cas) family member, NEDD9, is regulated in human neuroblastoma cells and in the embryonic hindbrain by all‐trans retinoic acid. Dev Dyn. 2004;231(3):564–575. doi: 10.1002/dvdy.20159 [DOI] [PubMed] [Google Scholar]
- 56.Pugacheva EN, Golemis EA. HEF1-aurora A interactions: points of dialog between the cell cycle and cell attachment signaling networks. Cell Cycle. 2006;5(4):384–391. doi: 10.4161/cc.5.4.2439 [DOI] [PMC free article] [PubMed] [Google Scholar]