Abstract
Background: Kaposi’s sarcoma (KS) is a highly disseminated angiogenic tumour of endothelial cells. Many deregulated miRNAs, including kshv-mir-k12-1-5p, have been identified in KS. kshv-mir-k12-1-5p plays important roles in KS. However, the underlying mechanism is not fully understood. The aim of this study was to investigate the exact functions of kshv-mir-k12-1-5p in KS cells.
Materials and methods: The biological functions of kshv-mir-k12-1-5p were studied using CCK-8, apoptosis, migration and invasion assays. Bioinformatics software was used to identify the target gene (SOCS6) of kshv-mir-k12-1-5p. A dual luciferase assay, Western blot (WB) and quantitative real-time polymerase chain reaction (q-PCR) were performed to further verify the target gene. The underlying molecular mechanisms of kshv-mir-k12-1-5p in KS cells were also explored.
Results: kshv-mir-k12-1-5p can promote the proliferation, migration and invasion of KS cells and inhibit cell apoptosis. Suppressor of cytokine signalling 6 (SOCS6) was identified as a direct target of kshv-mir-k12-1-5p, and kshv-mir-k12-1-5p can downregulate SOCS6 expression. In addition, knockdown of SOCS6 rescued the effects of kshv-mir-k12-1-5p inhibitor. Hence, a direct relationship between kshv-mir-k12-1-5p and SOCS6 was confirmed.
Conclusions: kshv-mir-k12-1-5p promotes the malignant phenotype of KS cells by targeting SOCS6, suggesting that kshv-mir-k12-1-5p could be a potential therapeutic target for KS.
Keywords: kshv-mir-k12-1-5p, SOCS6, Kaposi’s sarcoma
Introduction
KS is a multicentric tumour of mesenchymal origin.1 In China, more than 90% of KS cases, including classic KS (CKS) and AIDS-associated KS (AIDS-KS), have occurred in Xinjiang, especially among Uyghur patients.2 However, little is known about the potential mechanism of KS. Recently, some deregulated miRNAs were found.3,4
miRNAs are a class of small noncoding RNAs with lengths of 22–24 nucleotides.5 The complementary combination of miRNAs and the 3‘ UTR region of its target mRNA can lead to mRNA degradation or protein translation inhibition.5,6 As oncogenes or antioncogenes in different types of cancer,7,8 miRNAs participate in cell proliferation, differentiation, apoptosis, metabolism, migration and invasion9–11 and are closely associated with the occurrence and development of tumours.12–14 They can also serve as cancer diagnostic markers and potential therapeutic targets.15
One study showed that miR-143/145 was an upregulated miRNA biomarker, while miR-221/222, miR-155, and the let-7 family were downregulated in KS.4 In our previous work,16 we studied tumour and peritumoural tissues in KS by using miRNA chips. The results showed that 69 miRNAs were upregulated and 101 miRNAs were downregulated in KS. In particular, kshv-mir-k12-1-5p was significantly upregulated. These results revealed that deregulated miRNAs participate in the tumourigenesis of KS.
Suppressor of cytokine signalling 6 (SOCS6) is an important regulator of survival signalling.17,18 As a negative regulator of cytokines, it is involved in cell proliferation and migration in many human cancers.19 In this paper, we investigated how kshv-mir-k12-1-5p regulates SOCS6 and how it influences the biological behaviour of KS cells . This study provides new insights into the occurrence, development and targeted therapy of KS.
Materials and methods
Cell culture and transfection
SLK cells with KSHV infection (The human KS cells),20–22 at a density of 80% in logarithmic growth phase, were used. The cells were seeded at 2×104 cells per well in 96-well plates and cultured for 24 h. For functional analysis, kshv-mir-k12-1-5p mimics, kshv-mir-k12-1-5p inhibitor, mimics NC (negative control), and inhibitor NC were synthesized by and purchased from GenePharma Co., Ltd. (Shanghai, China). SOCS6 small interfering (si)RNA and control siRNA (siRNA-NC) were synthesized by and purchased from Biofavor Biotech Co., Ltd. (Wuhan, China). Then, The cells were transfected with kshv-mir-k12-1-5p mimics/mimics NC/kshv-mir-k12-1-5p inhibitor/inhibitor NC/SOCS6 siRNA/siRNA-NC/kshv-mir-k12-1-5p inhibitor+SOCS6 siRNA/kshv-mir-k12-1-5p mimics+SOCS6 siRNA by using Lipofectamine 2000 (2 μl) (Invitrogen, USA) in serum-free medium according to the manufacturer’s instructions for 48 h. The mimics/inhibitor/NC/siRNA/siRNA-NC sequences are listed in Table S1.
Table S1.
The sequences of mimics/inhibitor/NC/siRNA/siRNA NC
| Name | Sequences |
|---|---|
| kshv-mir-k12-1-5p mimics | 5ʹ-AUUACAGGAAACUGGGUGUAAGCUUACACCCAGUUUCCUGUAAUUU-3’ |
| kshv-mir-k12-1-5p inhibitor | 5ʹ-GCUUACACCCAGUUUCCUGUAAU-3, |
| mimics NC | 5ʹ-UUCUCCGAACGUGUCACGUTT-3’ |
| inhibitor NC | 5ʹ-CAGUACUUUUGUGUAGUACAA-3’ |
| SOCS6 siRNA | 5ʹ-UUGGUUUAAUCCAUUUACGUA-3’ |
| siRNA NC | 5ʹ-GCUUCAUAAGGCGCAUAGC-3’ |
Abbreviations: NC, negative control; siRNA, small interfering RNA; SOCS6, suppressor of cytokine signalling 6.
Notes: SLK cells are fully permissive for the study of KSHV and KSHV gene expression.23
q-PCR assay
After transfection for 48 h, 1 ml of Trizol (Aidlab Biotechnologies Co., Ltd., Beijing, China) was used to extract total RNA from the cells. mRNA and miRNA expression levels were quantified using the miScript SYBR Green PCR Kit (Qiagen). Approximately 3 μg of total RNA was used for complementary DNA (cDNA) synthesis. Then, 2 μL of cDNA was used for PCR amplification. The PCR conditions were as follows: 10 min at 95 °C for one cycle and 30 s at 95 °C and 30 s at 60 °C for 40 cycles. Primer sequences used are listed in Table S2. The results are presented as the levels of expression following normalization to human U6 small nuclear RNA and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The 2−△△Ct method was used for data analysis. The experiment was repeated three times.
Table S2.
Primer sequences used for q-PCR
| Name | Primer sequences |
|---|---|
| kshv-mir-k12-1-5p (stem-loop RT primer) | 5“-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCTTACAC −3” |
| kshv-mir-k12-1-5p (Forward) | 5“-TGCGCATTACAGGAAACTGGGTG −3” |
| Universal reverse primer | 5ʹ-CCAGTGCAGGGTCCGAGGTATT-3’ |
| SOCS6 (Forward) | 5ʹ-TGTGCCTGTCGTTATTGGACT-3’ |
| SOCS6 (Reverse) | 5ʹ-CCAAAGGATACATCCCCTCATCT-3’ |
| U6 (Forward) | 5ʹ-CGCTTCGGCAGCACATATAC-3ʹ |
| U6 (Reverse) | 5ʹ-AAATATGGAACGCTTCACGA-3ʹ |
| GAPDH (Forward) | 5ʹ-TCAAGAAGGTGGTGAAGCAGG-3ʹ |
| GAPDH (Reverse) | 5ʹ-TCAAAGGTGGAGGAGTGGGT-3’ |
Abbreviations: SOCS6, suppressor of cytokine signalling 6; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Cell proliferation assay (CCK-8 assay)
Cell proliferation was detected by using the Cell Counting Kit-8 (CCK-8) (Biosharp, Anhui, China) in accordance with the instruction manual. The density of transfected cells was adjusted to 5×104 cells per well and seeded in 96-well plates. After culture for 48 h, 50 μl of CCK-8 reagent was added, and the cells were incubated for 4 h. Then, a microplate reader was used for detection. Optical density (OD) values were recorded at 570 nm. On this basis, the cell proliferation rate of each group was calculated using the following equation: Cell Proliferation Rate (%) = (As-Ab)/(Ac-Ab)×100%, where Ab, Ac and As represent the ODs of the blank control, normal, and experimental groups, respectively. The experiment was repeated three times.
Cell apoptosis assay
The Annexin V-APC/7-AAD Cell Apoptosis Detection Kit (Key Gen Biotech Co., Ltd., Nanjing, China) was used to detect cell apoptosis according to the instruction manual. After 48 h of transfection, the cells (2×105 cells per well) were obtained and suspended in 100 µl of Annexin V-APC/7-AAD solution. The mixture was then added to a flow cell tube and further transferred to a flow detection tube. After activation, both Annexin V-APC and 7-AAD demonstrated fluorescent signals.The coordinate system was constructed with Annexin-APC as the horizontal axis and 7-AAD as the vertical axis. Cells in the left upper quadrant were defined as dead cells (Annexin V-APC+/7-AAD+). Cells in the right upper quadrant were defined as late apoptotic cells (Annexin V-APC+/7-AAD+). Cells in the left lower quadrant were defined as normal cells (Annexin V-APC -/7-AAD-). Cells in the right lower quadrant were defined as early apoptotic cells (Annexin V-APC +/7-AAD-). The experiment was repeated three times.
Cell migration assay (wound healing assay)
When transfected cells reached 90% confluence, a scratch tester was used to make a wound on the lower centre of each 6-well plate, and the wounds were photographed. Then, the 6-well plates were placed in an incubator (37°C, 5% CO2). After 48 h, the 6-well plates were photographed again. The experiment was repeated three times.
Cell invasion assay (transwell assay)
A transwell chamber with an 8 μm pore size precoated with Matrigel (BD Biosciences, San Jose, CA, USA) was used to determine cell invasion ability. The cell density was adjusted to 2×104/ml. A total of 200 μl of a 2×104/ml single-cell suspension was added to each transwell chamber, and the transwell chambers were incubated for 24 h. The noninvading cells of the upper chamber were removed with a clean cotton swab and then stained with 0.1% crystal violet. The invading cells of the lower chamber were counted under a microscope. The experiment was repeated three times.
Dual luciferase assay
Wild type and mutant type SOCS6 3′ UTRs were amplified and cloned into the pYr-mirTarget vector (Biofavor Biotech Co., Ltd., Wuhan, China). Then, the plasmids (1 μg) kshv-mir-k12-1-5p mimics and mimics NC (50 nM) were cotransfected into 293T cells (Open Biosystems USA; 5×104 cells per well) using Lipofectamine 2000. After transfection for 24 h, luciferase activity was detected using a dual luciferase assay system (Promega Corporation, Fitchburg, WI, USA) according to the instruction manual. The kshv-mir-k12-1-5p binding sites in the 3‘ UTR of SOCS6 are shown in Table S3. The assay was repeated three times.
Table S3.
kshv-mir-k12-1-5p binding sites in the 3‘ UTR of SOCS6
| SOCS6 MUT 3’UTR/SOCS6 WT 3’UTR/kshv-mir-k12-1-5p Seed sequences |
|---|
| 5′UAAUACUUUUUUAAUGCAGAAUA3′ SOCS6 MUT 3’UTR |
| 5′UAAUACUUUUUUAAUCCUGUAAA3′ SOCS6 WT 3’UTR 3′CGAAUGUGGGUCAAAGGACAUUA5′ kshv-mir-k12-1-5p Seed sequences |
Abbreviations: SOCS6, suppressor of cytokine signalling 6; WT, wild type; MUT, mutant type.
Western blot (WB) assay
After transfection for 48 h, 400 μl of RIPA lysis buffer was used to extract total proteins from the cells. According to protein content, a standard curve was plotted. Stacking (5%) and separating (12%) gels were prepared. Then, electrophoresis was performed (1.5 h, 100 V). As bromophenol blue reached the bottom of the gel slab, electrophoresis was stopped. The proteins were transferred onto a membrane, and this process lasted for 70 min. The membrane was blocked with skimmed milk powder (5%) for 2 h and incubated with primary antibodies against SOCS6 (1:1,000, Abcam) and GAPDH (1:1,000, Abcam) overnight at 4 °C. The membrane was washed three times with TBST and then incubated with an HRP-conjugated secondary antibody (Abcam) diluted 1:10,000 for 2 h at 37 °C. Enhanced chemiluminescence imaging was performed. Greyscale values were analysed by using BandScan. The experiment was repeated three times.
Statistical analysis
Statistical calculations were performed using Statistical Program for the Social Sciences (SPSS Inc., Chicago, IL, USA) software 17.0. Data are expressed as the mean ± standard deviation (mean±SD). The results were analysed statistically using one-way analysis of variance or Student's t-test. A P-value <0.05 was considered statistically significant.
Results
Transfection levels of kshv-mir-k12-1-5p
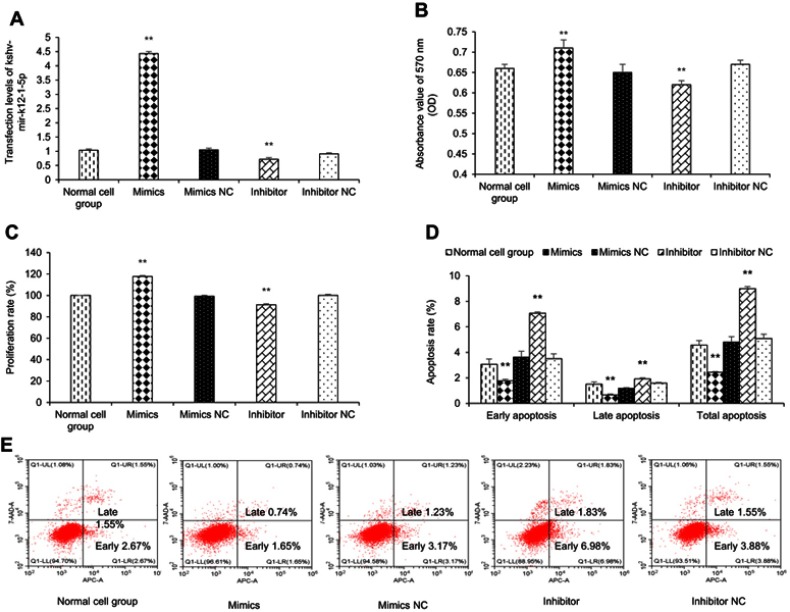
To manipulate kshv-mir-k12-1-5p levels in KS cells, kshv-mir-k12-1-5p mimics/mimics NC/inhibitor/inhibitor NC were transfected into KS cells. After 48 h, q-PCR results showed that the kshv-mir-k12-1-5p expression levels were different in normal cell group (the normal SLK/KS cells with KSHV infection), mimics group, mimics NC group, inhibitor group and inhibitor NC group . kshv-mir-k12-1-5p mimics or inhibitor led to increased or decreased kshv-mir-k12-1-5p expression (Figure 1A).
Figure 1.
kshv-mir-k12-1-5p promotes cell proliferation and inhibits cell apoptosis. (A) The mimics group exhibited increased expression of kshv-mir-k12-1-5p, and the inhibition group exhibited decreased expression of kshv-mir-k12-1-5p. (B and C) Cell proliferation assay was performed by using CCK-8. (D and E) The percentage of apoptosis cells after transfection with kshv-mir-k12-1-5p mimics, mimics NC, inhibitor or inhibitor NC. All data are presented as the mean ± SD. **p<0.01, compared with control groups.
Abbreviation: NC, negative control.
kshv-mir-k12-1-5p promotes cell proliferation
After 48 h of transfection, the CCK-8 assay was used to examine the effect of kshv-mir-k12-1-5p on the proliferation of KS cells. The results showed that the overexpression of kshv-mir-k12-1-5p significantly increased cell proliferation, leading to the highest OD value/proliferation rate. Additionally, the suppression effect was evident after kshv-mir-k12-1-5p inhibitor transfection, which resulted in the lowest OD value/proliferation rate. Therefore, kshv-mir-k12-1-5p can promote cell proliferation . The results are presented in Figure 1B and C.
kshv-mir-k12-1-5p inhibits cell apoptosis
After 48 h of transfection, flow cytometry was performed to detect cell apoptosis. The apoptosis rates of the mimics group were dramatically decreased compared with NC group. The apoptosis rates of the inhibitor group were obviously increased compared with NC group. Therefore, kshv-mir-k12-1-5p can inhibit cell apoptosis. The results are presented in Figure 1D and E.
kshv-mir-k12-1-5p promotes cell migration
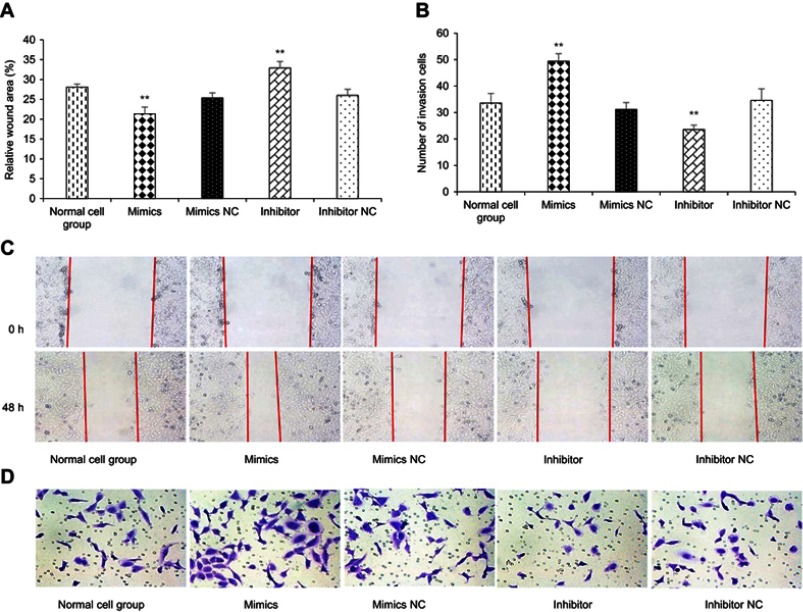
After 48 h of transfection, a wound healing assay was performed. The results revealed that kshv-mir-k12-1-5p mimics can significantly increase the migration ability of KS cells. The wound area accounted for 21.35±1.71% of the total area. The kshv-mir-k12-1-5p inhibitor can significantly decrease the migration ability of KS cells. The wound area accounted for 32.91±1.61% of the total area. Therefore, kshv-mir-k12-1-5p can promote cell migration . The results are shown in Figure 2A and C.
Figure 2.
kshv-mir-k12-1-5p promotes cell migration and invasion. (A and B) Cell migration and invasion assays were performed after transfection with kshv-mir-k12-1-5p mimics, mimics NC, inhibitor or inhibitor NC. (C) Micrographs (100×) of 0 and 48 h wound areas were analysed by ImageJ software. (D) Micrographs (200×) of crystal violet-stained cells (invasion cells) were shown . All data are presented as the mean ± SD. **p<0.01, compared with control groups.
Abbreviation: NC, negative control.
kshv-mir-k12-1-5p promotes cell invasion
After 48 h of transfection, the cell invasion assay showed that the number of invasion cells was obviously increased in the mimics group compared with NC group, with the largest cell invasion number(49.40±2.88). The number of invasion cells was obviously decreased in the inhibitor group compared with NC group, with the smallest cell invasion number(23.60±1.67). Therefore, kshv-mir-k12-1-5p can promote cell invasion, as shown in Figure 2B and D.
kshv-mir-k12-1-5p directly targets the SOCS6 gene
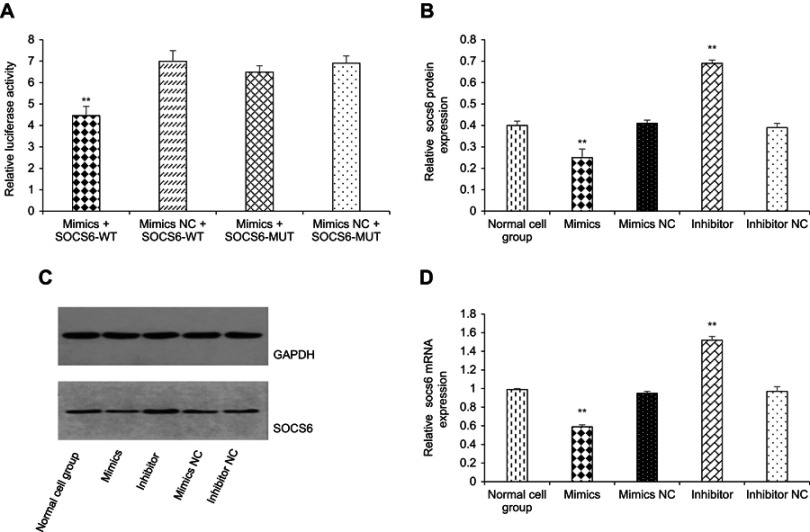
Bioinformatics analysis (http://www.mirbase.org) revea-led the potential target genes of kshv-mir-k12-1-5p. We selected to study SOCS6 among hundreds of genes because it is closely involved in cell migration, proliferation, and survival.19 We synthesized the sequences of wild type and mutant type SOCS6. Subsequently, 293T cells were transfected with kshv-mir-k12-1-5p mimics + SOCS6-WT, mimics NC + SOCS6-WT, kshv-mir-k12-1-5p mimics + SOCS6-MUT and mimics NC + SOCS6-MUT. After 24 h, the activity of luciferase was determined. After co-transfection with kshv-mir-k12-1-5p mimics and SOCS6- WT, relative luciferase activity decreased (4.46±0.43) . After co-transfection with kshv-mir-k12-1-5p mimics and SOCS6-MUT, however, relative luciferase activity changed only slightly (6.49±0.30). Co-transfection with mimics NC and SOCS6-WT/SOCS6-MUT led to no obvious change in relative luciferase activity (6.99±0.50/6.91±0.34). Therefore, SOCS6 is a direct target gene of kshv-mir-k12-1-5p (Figure 3A). Western blot and q-PCR results showed that the overexpression of kshv-mir-k12-1-5p significantly decreased SOCS6 expression at both the protein and mRNA levels . In contrast, following transfection with the kshv-mir-k12-1-5p inhibitor, SOCS6 expression increased significantly at both the protein and mRNA levels . The results are presented in Figure 3B, C and D.
Figure 3.
kshv-mir-k12-1-5p directly targets the SOCS6 gene. (A) Dual luciferase assay was used to demonstrate the target binding relationship between kshv-mir-k12-1-5p and SOCS6 . (B and C) kshv-mir-k12-1-5p decreased SOCS6 expression at the protein level. (D) kshv-mir-k12-1-5p decreased SOCS6 expression at the mRNA level . All data are presented as the mean ± SD. **p<0.01, compared with control groups.
Abbreviations: SOCS6, suppressor of cytokine signalling 6; NC, negative control; WT, wild type; MUT, mutant type; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
kshv-mir-k12-1-5p promotes KS cell growth and metastasis by targeting SOCS6
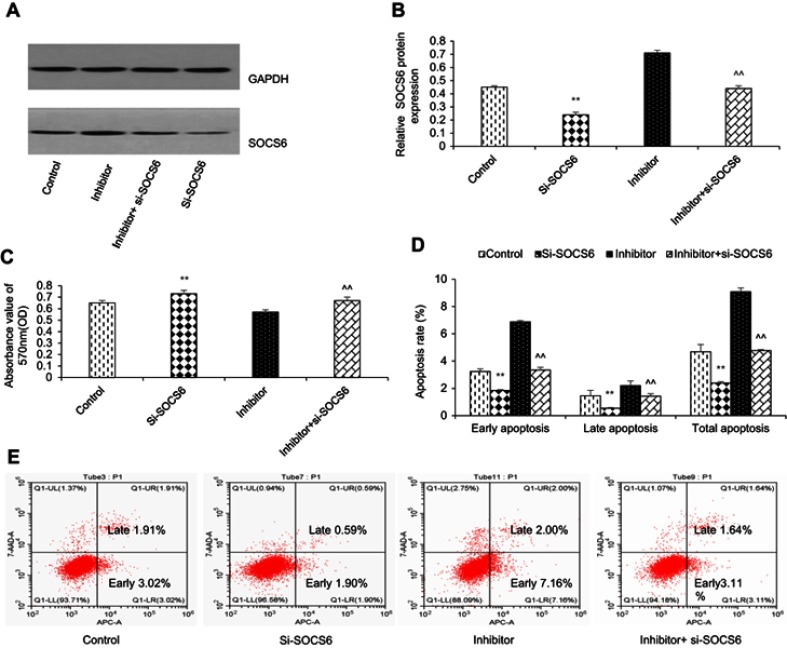
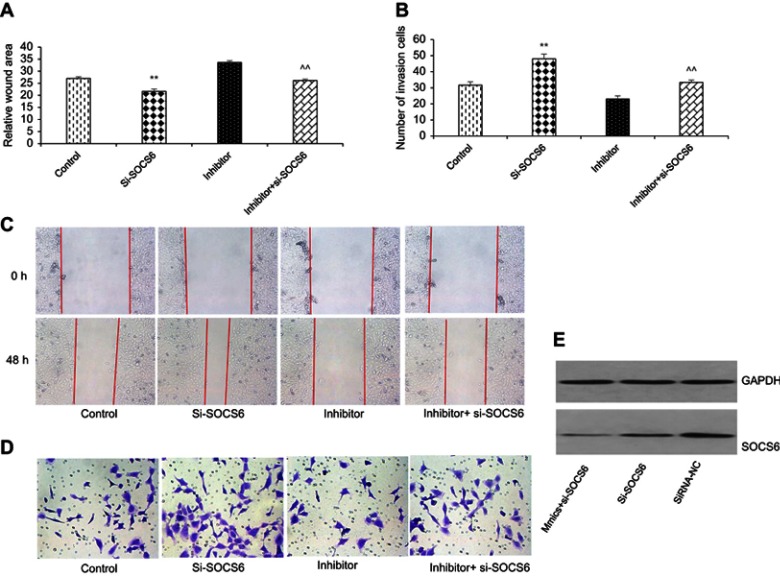
To further study the functional relationship between kshv-mir-k12-1-5p and SOCS6, we explored whether kshv-mir-k12-1-5p enhanced KS cell growth and metastasis by targeting SOCS6. For this purpose, KS cells were transfected with SOCS6 siRNA, kshv-mir-k12-1-5p inhibitor, co-transfection with kshv-mir-k12-1-5p inhibitor and SOCS6 siRNA . As demonstrated in Western blot (Figure 4A and B), SOCS6 expression was downregulated by SOCS6 siRNA. However, the protein level of SOCS6 was significantly upregulated after transfection with kshv-mir-k12-1-5p inhibitor, which was reversed by co-transfection with SOCS6 siRNA. Cell function assays showed that SOCS6 siRNA promoted cell proliferation, migration and invasion abilities and inhibited cell apoptosis, similar to the effects of the overexpression of kshv-mir-k12-1-5p. Knockdown of SOCS6 expression by SOCS6 siRNA in kshv-mir-k12-1-5p inhibitor-transfected cells reversed the effects of kshv-mir-k12-1-5p inhibitor (Figures 4–5). In addition, treatment with SOCS6 siRNA in kshv-mir-k12-1-5p mimics-transfected cells significantly decreased SOCS6 expression compared with the SOCS6 siRNA group (Figure 5E). Therefore, co-transfection with SOCS6 siRNA and kshv-mir-k12-1-5p mimics can increase the growth and metastasis capabilities of KS cells compared with the SOCS6 siRNA . Collectively, these data indicate that kshv-mir-k12-1-5p promotes KS cell growth and metastasis by targeting SOCS6.
Figure 4.
kshv-mir-k12-1-5p promotes cell growth by targeting SOCS6. (A and B) Western blot was used to detect the protein level of SOCS6 in KS cells transfected with SOCS6 siRNA, kshv-mir-k12-1-5p inhibitor, co-transfected with kshv-mir-k12-1-5p inhibitor and SOCS6 siRNA, respectively. Control: The cells without any transfection. (C) CCK-8 and (D and E) flow cytometry assays were performed to evaluate the proliferation, apoptosis of KS cells. All data are presented as the mean ± SD. **P<0.01 for SOCS6 siRNA vs Control, ^^P<0.01 for kshv-mir-k12-1-5p inhibitor vs co-transfection with kshv-mir-k12-1-5p inhibitor and SOCS6 siRNA.
Abbreviations: siRNA, small interfering RNA; CCK-8, cell counting kit-8.
Figure 5.
kshv-mir-k12-1-5p promotes cell metastasis by targeting SOCS6. (A and C) Wound healing and (B and D)Transwell assays were performed to evaluate the migration and invasion of KS cells. All data are presented as the mean ± SD. **P<0.01 for SOCS6 siRNA vs Control, ^^P<0.01 for kshv-mir-k12-1-5p inhibitor vs co-transfection with kshv-mir-k12-1-5p inhibitor and SOCS6 siRNA. (E) Western blot was used to detect the protein level of SOCS6 in KS cells transfected with SOCS6 siRNA, siRNA-NC or co-transfected with kshv-mir-k12-1-5p mimics and SOCS6 siRNA respectively.
Abbreviations: siRNA-NC, small interfering RNA- negative control.
Discussion
As important genes in the post-transcriptional regulation system, miRNAs have attracted much attention in the medical field around the world.24–26 miRNAs are involved in a wide range of biological processes.27 Previous studies have demonstrated that miRNAs participate in a number of human diseases, particularly different types of cancer.28–30 KS is a highly disseminated angiogenic tumour of endothelial cells most commonly found in untreated HIV-AIDS or immunocompromised individuals.31,32 Kaposi’s sarcoma-associated herpesvirus (KSHV) is the aetiological agent of KS.33,34 KSHV is closely linked to KS and two other B-cell lymphoproliferative diseases, primary effusion lymphoma (PEL) and a plasmablastic variant of multicentric Castleman’s disease (MCD).32,35 Similar to other DNA viruses, KSHV also encodes many miRNAs.36 Wu et al16 analysed the expression profiles of miRNAs in KS. They found that 13 miRNAs encoded by KSHV were all upregulated, among them, kshv-mir-k12-1-5p was the most upregulated. Catrina et al37 analysed 17 KS specimens and 3 cases of normal skin. They found that 76 miRNAs were upregulated and 109 miRNAs were downregulated. The miR-99a and miR-200 families were the most significantly downregulated. In contrast, kshv-mir-k12-4, kshv-mir-k12-1, kshv-mir-k12-2 and kshv-mir-k12-8 were the most significantly upregulated. Therefore, the high expression of KSHV miRNAs, especially kshv-mir-k12-1-5p, might play an important role in KS, but the detailed mechanism remains elusive.
miRNAs participate in cell growth and apoptosis, inhibit the differentiation of tumour cells, and accelerate the migration and invasion of tumours.38,39 Likewise, KSHV miRNAs contribute to KS development via complex mechanisms.40 Unlimited proliferation of cells is the crucial characteristic of malignant tumours.41 The imbalance between cell proliferation and apoptosis has been implicated in key roles in cancer progression.24 In the present study, the overexpression of kshv-mir-k12-1-5p significantly increased cell proliferation and decreased cell apoptosis. The suppression of kshv-mir-k12-1-5p expression decreased cell proliferation and increased cell apoptosis. Invasion and metastasis are considered the most significant biological behaviours of malignant tumours.42 They are dynamic, multistage and complex processes that are the major factors of tumour-related deaths.43 Our results showed that the invasion and migration abilities of KS cells were obviously improved after transfection with kshv-mir-k12-1-5p mimics. In contrast, cell migration and invasion were inhibited by transfection with kshv-mir-k12-1-5p inhibitor.
In previous studies, several viral miRNAs were shown to stabilize viral latency or promote cell growth, migration and/or invasion.8,44–47 Suffert et al48 reported that miR-K12-1, 3 and 4-3p directly participated in the inhibition of apoptosis by the virus and were thus likely to play a role in KSHV-induced oncogenesis. In this study, all experimental results indicated that kshv-mir-k12-1-5p can contribute to the malignant biological behaviour of KS cells by accelerating cell proliferation and increasing cell migration and invasion abilities, as well as inhibiting cell apoptosis.
Because miRNAs exert their biological functions by inhibiting the translation of their target gene, the core research on miRNA functions is identifying target genes.49 For instance, the potential target genes of miRNAs may play a key role in the pathogenesis of KSHV infection-related lymphoma.50 SOCS6 belongs to the family of suppressor of cytokine signalling (SOCS) proteins.51 Reduced expression of SOCS6 has been observed in many human cancers.52,53 including lung, colorectal, ovarian, stomach, thyroid, hepatocellular, and pancreatic cancers. Recent studies have shown that its expression is modulated by several miRNAs in different cancers. miR-424-5p can downregulate SOCS6 expression in pancreatic cancer.54 High miR-17-5p expression in gastric cancer inhibited SOCS6 expression.55 As a tumour suppressor, its downregulation is correlated with tumour aggression and poor prognosis, suggesting the importance of SOCS6 in tumour growth and progression.55 Bioinformatics analysis revealed that kshv-mir-k12-1-5p can specifically bind to the 3ʹ UTR of SOCS6. Our dual luciferase assay showed that the SOCS6 gene was a target gene of kshv-mir-k12-1-5p. The results of WB and q-PCR showed that the protein and mRNA levels of SOCS6 were negatively regulated by kshv-mir-k12-1-5p. Finally, rescue experiments confirmed that the downregulation of SOCS6 was essential for kshv-mir-k12-1-5p to be effective in the malignant phenotypes of KS cells. Knockdown of SOCS6 significantly reversed the effects of kshv-mir-k12-1-5p inhibitor.
In conclusion, our study demonstrates that kshv-mir-k12-1-5p can exert oncogenic functions to promote growth and metastasis in KS cells by targeting SOCS6. These findings contribute to our understanding of the molecular mechanism of KS .
Conclusions
We provide evidence that kshv-mir-k12-1-5p functions as an oncogene in KS cells by targeting SOCS6. kshv-mir-k12-1-5p accelerated cell proliferation, increased cell migration and invasion abilities, and inhibited cell apoptosis. The suppression of kshv-mir-k12-1-5p expression might be a potential target therapy for KS.
Acknowledgments
This work was supported by the hospital level project of affiliated Traditional Chinese Medicine Hospital, Xinjiang Medical University, China (Grant No.ZYY201702).
Ethical approval
This study does not contain any research on human participants or animals. This study was approved by the Ethics Committee of the Affiliated Traditional Chinese Medicine Hospital, Xinjiang Medical University.
Supplementary materials
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Hengge UR, Ruzicka T, Tyring SK, et al. Update on Kaposi‘s sarcoma and other HHV8 associated diseases. Part 1: epidemiology, environmental predispositions, clinical manifestations, and therapy. Lancet Infect Dis. 2002;2:281–292. [DOI] [PubMed] [Google Scholar]
- 2.Wu XJ, Pu XM, Kang XJ, et al. One hundred and five Kaposi sarcoma patents: a clinical study in Xinjiang, Northwest of China. Acad Dermatol Venereol. 2014;28(11):1545–1552. doi: 10.1111/jdv.12349 [DOI] [PubMed] [Google Scholar]
- 3.Wang X, He B, Zhang Z, et al. Human herpesvirus-8 in northwestern China: epidemiology and characterization among blood donors. Virol J. 2010;7:62. doi: 10.1186/1743-422X-7-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O‘Hara AJ, Wang L, Dezube BJ, et al. Tumor suppressor microRNAs are underrepresented in primary effusion lymphoma and Kaposi sarcoma. Blood. 2009;113:5938–5941. doi: 10.1182/blood-2008-09-179168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruksha TG, Komina AV, Palkina NV. MicroRNA in skin diseases. Eur J Dermatol. 2017;27(4):343–352. doi: 10.1684/ejd.2017.3024 [DOI] [PubMed] [Google Scholar]
- 6.Engels BM, Hutvagner G. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene. 2006;25(46):6163–6169. doi: 10.1038/sj.onc.1209909 [DOI] [PubMed] [Google Scholar]
- 7.Gottwein E, Mukherjee N, Sachse C, et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450(7172):1096–1099. doi: 10.1038/nature05992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Sun R, Lin X, Liang D, Deng Q Lan K. Kaposi‘s sarcoma-associated herpesvirus-encoded microRNA miR-K12-11 attenuates transforming growth factor beta signaling through suppression of SMAD5. J Virol. 2012;86(3):1372–1381. doi: 10.1128/JVI.06245-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tiifekci KU, Meuwissen PAL, Genc S. The role of microRNAs in biological processes. Methods Mol Biol. 2014;1107:15–31. doi: 10.1007/978-1-62703-748-8_2 [DOI] [PubMed] [Google Scholar]
- 10.Zhang W, Han S, Sun K. Combined analysis of gene expression, miRNA expression and DNA methylation profiles of osteosarcoma. Oncol Rep. 2017;37(2):1175–1181. doi: 10.3892/or.2016.5324 [DOI] [PubMed] [Google Scholar]
- 11.Kim J, Yao F, Xiao Z, Sun Y, Ma L. MicroRNAs and metastasis: small RNAs play big roles. Cancer Metastasis Rev. 2018;37(1):5–15. doi: 10.1007/s10555-017-9712-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felekkis K, Touvana E, Stefanou C, Deltas C. microRNAs: a newly described class of encoded molecules that play a role in health and disease. Hippokratia. 2010;14(4):236–240. [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- 14.Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs down regulate large numbers of target mRNAs. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315 [DOI] [PubMed] [Google Scholar]
- 15.Mouillet JF, Ouyang Y, Coyne CB, Sadovsky Y. MicroRNAs in placental health and disease. Am J Obstet Gynecol. 2015;213(4 Suppl):S163–S172. doi: 10.1016/j.ajog.2015.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu XJ, Pu XM, Zhao ZF, et al. The expression profiles of microRNAs in Kaposi’s sarcoma. Tumour Biol. 2015;36(1):437–446. doi: 10.1007/s13277-014-2626-1 [DOI] [PubMed] [Google Scholar]
- 17.Kabir NN, Sun J, Ronnstrand L, Kazi JU. SOCS6 is a selective suppressor of receptor tyrosine kinase signaling. Tumour Biol. 2014;35(11):10581–10589. doi: 10.1007/s13277-014-2542-4 [DOI] [PubMed] [Google Scholar]
- 18.Xue X, Liu Y, Wang Y, et al. MiR-21 and MiR-155 promote non-small cell lung cancer progression by downregulating SOCS1, SOCS6, and PTEN. Oncotarget. 2016;7(51):84508–84519. doi: 10.18632/oncotarget.13022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bellezza I, Neuwirt H, Nemes C, et al. Suppressor of cytokine signaling-3 antagonizes cAMP effects on proliferation and apoptosis and is expressed in human prostate cancer. Am J Pathol. 2006;169(6):2199–2208. doi: 10.2353/ajpath.2006.060171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arias C, Weisburd B, Stern-Ginossar N, et al. KSHV 2.0: a comprehensive annotation of the Kaposi’s sarcoma-associated herpesvirus genome using next- generation sequencing reveals novel genomic and functional features. PLoS Pathog. 2014;10(1):e1003847. doi: 10.1371/journal.ppat.1003847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viollet C, Davis DA, Reczko M, et al. Next-generation sequencing analysis reveals differential expression profiles of MiRNA-mRNA target pairs in KSHV-infected cells. PLoS One. 2015;10(5):e0126439. doi: 10.1371/journal.pone.0126439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosario SA, Santiago GE, Mesri EA, Verdun RE. Kaposi’s sarcoma-associated herpesvirus-encoded viral IL-6 (vIL-6) enhances immunoglobulin class-switch recombination. Front Microbiol. 2018;9:3119. doi: 10.3389/fmicb.2018.03119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stürzl M, Gaus D, Dirks WG, Ganem D, Jochmann R. Kaposi‘s sarcoma-derived cell line SLKK is not of endothelial origin, but is a contaminant from a known renal carcinoma cell line. Int J Cancer. 2013;132(8):1954–1958. doi: 10.1002/ijc.27849 [DOI] [PubMed] [Google Scholar]
- 24.Wang W, Yang J, Yu F, et al. MicroRNA-122-3p inhibits tumor cell proliferation and induces apoptosis by targeting Forkhead box O in A549 cells. Oncol Lett. 2018;15(2):2695–2699. doi: 10.3892/ol.2017.7577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Xu J, Shao T, Zhang Y, Chen H, Li X. RNA function prediction. Methods Mol Biol. 2017;1654:17–28. doi: 10.1007/978-1-4939-7231-9_2 [DOI] [PubMed] [Google Scholar]
- 26.Zhao Y, Song Y, Yao L, Song G, Teng C. Circulating microRNAs: promising biomarkers involved in several cancers and other diseases. DNA Cell Biol. 2017;36(2):77–94. doi: 10.1089/dna.2016.3426 [DOI] [PubMed] [Google Scholar]
- 27.Goto Y, Kurozumi A, Arai T, et al. Impact of novel miR-145-3p regulatory networks on survival in patients with castration-resistant prostate cancer. Br J Cancer. 2017;117(3):409–420. doi: 10.1038/bjc.2017.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han C, Seebacher NA, Hornicek FJ, Kan Q, Duan Z. Regulation of microRNAs function by circular RNAs in human cancer. Oncotarget. 2017;8:64622–64637. doi: 10.18632/oncotarget.19930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Celano M, Rosignolo F, Maggisano V, et al. MicroRNAs as biomarkers in thyroid carcinoma. Int J Enomics. 2017;2017:6496570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shomali N, Mansoori B, Mohammadi A, Shirafkan N, Ghasabi M, Baradaran B. MiR-146a functions as a small silent player in gastric cancer. Biomed Pharmacother. 2017;96:238–245. doi: 10.1016/j.biopha.2017.09.138 [DOI] [PubMed] [Google Scholar]
- 31.Hu M, Wang C, Li W, et al. A KSHV microRNA directly targets G protein-coupled receptor kinase 2 to promote the migration and invasion of endothelial cells by inducing CXCR2 and activating AKT signaling. PLoS Pathog. 2015;11:e1005171. doi: 10.1371/journal.ppat.1005171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purushothaman P, Uppal T, Sarkar R, et al. KSHV-mediated angiogenesis in tumor progression. Viruses. 2016;8(7):198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gay LA, Sethuraman S, Thomas M, Turner PC, Renne R. Modified cross-linking, ligation, and sequencing of hybrids (qCLASH) identifies Kaposi‘s sarcoma-associated herpesvirus MicroRNA Targets In Endothelial Cells. J Virol. 2018;92(8):e02138–17. doi: 10.1128/JVI.02138-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haecker I, Gay LA, Yang Y, et al. Ago HITS-CLIP expands understanding of Kaposi‘s sarcoma-associated herpesvirus miRNA function in primary effusion lymphomas. PLoS Pathog. 2012;8(8):e1002884. doi: 10.1371/journal.ppat.1002884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaplan LD. Human herpesvirus-8: Kaposi sarcoma, multicentric Castleman disease, and primary effusion lymphoma. Hematology Am Soc Hematol Educ Program. 2013;2013(1):103–108.. doi: 10.1182/asheducation-2013.1.103 [DOI] [PubMed] [Google Scholar]
- 36.Grundhoff A, Sullivan CS. Virus-encoded microRNAs. Virology. 2011;411(2):325–343. doi: 10.1016/j.virol.2011.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Catrina Ene AM, Borze I, Guled M, et al. MicroRNA expression profiles in Kaposi‘s sarcoma. Pathol Oncol Res. 2014;20(1):153–159. doi: 10.1007/s12253-013-9678-1 [DOI] [PubMed] [Google Scholar]
- 38.Yu F, Yao H, Zhu P, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;13l(6):1109–1123. doi: 10.1016/j.cell.2007.10.054 [DOI] [PubMed] [Google Scholar]
- 39.Yu F, Jiao Y, Zhu Y, et al. MicroRNA 34c gene down-regulation via DNA methylation promotes self-renewal and epithelial-mesenchymal transition in breast tumor-initiating cells. J Biol Chem. 2012;287(1):465–473. doi: 10.1074/jbc.M111.280768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin J, Li W, Gao SJ, Lu C. KSHV microRNAs: tricks of the devil. Trends Microbiol. 2017;25(8):648–661. doi: 10.1016/j.tim.2017.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandal T. Molecular aspects of the mammalian cell cycle and cancer. Oncologist. 2002;7:73–81. [DOI] [PubMed] [Google Scholar]
- 42.Wu J, Ji A, Wang X, et al. MicroRNA-195-5p, a new of Fra-1, suppresses the migration and invasion regulator of prostate cancer cells. J Transl Med. 2015;13:289. doi: 10.1186/s12967-015-0541-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nola S, Sin S, Bonin F, Lidereau R, Driouch K. A methodological approach to unravel organ-specific breast cancer metastasis. J Mammary Gland Biol Neoplasia. 2012;17:135–145. doi: 10.1007/s10911-012-9256-2 [DOI] [PubMed] [Google Scholar]
- 44.Dahlke C, Maul K, Christalla T, et al. A microRNA encoded by Kaposi sarcoma-associated herpesvirus promotes B-cell expansion in vivo. PLoS One. 2012;7:e49435. doi: 10.1371/journal.pone.0049435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Forte E, Raja AN, Shamulailatpam P, et al. MicroRNA-mediated transformation by the Kaposi‘s sarcoma-associated herpesvirus Kaposin locus. J Virol. 2015;89(4):2333–2341. doi: 10.1128/JVI.03317-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lei X, Zhu Y, Jones T, Bai Z, Huang Y, Gao S-J. A Kaposi‘s sarcoma-associated herpesvirus microRNA and its variants target the transforming growth factor β pathway to promote cell survival. J Virol. 2012;86:11698–11711. doi: 10.1128/JVI.06855-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin Z, Dai L, Defee M, et al. Kaposi‘s sarcoma-associated herpesvirus suppression of DUSP1 facilitates cellular pathogenesis following de novo infection. J Virol. 2013;87:621–635. doi: 10.1128/JVI.01441-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suffert G, Malterer G, Hausser J, et al. Kaposi‘s sarcoma herpesvirus microRNAs target caspase 3 and regulate apoptosis. PLoS Pathog. 2011;7(12):e1002405. doi: 10.1371/journal.ppat.1002405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim HS, Shen Q, Nam SW. Histone deacetylases and their regulatory MicroRNAs in hepatocarcinogenesis. J Korean Med Sci. 2015;30(10):1375–1380. doi: 10.3346/jkms.2015.30.10.1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quan L, Qiu T, Liang J, Li M, Zhang Y, Tao K. Identification of target genes regulated by KSHV miRNAs in KSHV-infected lymphoma cells. Pathol Oncol Res. 2015;21(4):875–880. doi: 10.1007/s12253-015-9902-2 [DOI] [PubMed] [Google Scholar]
- 51.Trengove MC, Ward AC. SOCS proteins in development and disease. Am J Clin Exp Immunol. 2013;2(1):1–29. [PMC free article] [PubMed] [Google Scholar]
- 52.Qi X, Li J, Zhou C, Lv C, Tian M. MiR-142-3p suppresses SOCS6 expression and promotes cell proliferation in nasopharyngeal carcinoma. Cell Physiol Biochem. 2015;36(5):1743–1752. doi: 10.1159/000430147 [DOI] [PubMed] [Google Scholar]
- 53.Tanaka T, Arai M, Jiang X, et al. Downregulation of microRNA-431 by human interferon-beta inhibits viability of medulloblastoma and glioblas- toma cells via upregulation of SOCS6. Int J Oncol. 2014;44(5):1685–1690. doi: 10.3892/ijo.2014.2317 [DOI] [PubMed] [Google Scholar]
- 54.Wu K, Hu G, He X, et al. MicroRNA-424-5p suppresses the expression of SOCS6 in pancreatic cancer. Pathol Oncol Res. 2013;19(4):739–748. doi: 10.1007/s12253-013-9637-x [DOI] [PubMed] [Google Scholar]
- 55.Wu Q, Luo G, Yang Z, et al. miR-17-5p promotes proliferation by targeting SOCS6 in gastric cancer cells. FEBS Lett. 2014;588(12):2055–2062. doi: 10.1016/j.febslet.2014.04.036 [DOI] [PubMed] [Google Scholar]