Abstract
Rural households in developing countries rely on communal water supplies and household water frequently becomes contaminated following its collection, transportation and during its storage. Using culture-dependent and -independent techniques, we examined the changes in microbial water quality between communal tap water and household water storage in a rural area of Cameroon, Africa. The culturable fecal indicator bacteria (FIB) were used to assess the potential health risks associated with different household water storage conditions (e.g., type of container and open vs. closed container) and interventions (e.g., water storage days, cleaned on the last day of use, and hygiene practices). Only the amount of days the water was stored significantly differed (p-value < 0.05), which showed that potential health risks increased when water was stored for more than 3 days. The higher abundance of molecular FIB in biofilm than household water suggested that omnipresent biofilm in household water could potential health risk. The high-throughput sequencing revealed that the most abundant phylum was Proteobacteria, followed by Actinobacteria and Bacteroidetes in both the water and the biofilm samples. Bacterial genera seen in biofilm bacteria, such as Pseudomonas, Acinetobacter and Comamonas. Acinetobacter, Chryseobacterium, Stenotrophomonas and Corynebacterium, were relatively more abundant in the biofilm than in the water. Potential bacterial pathogens including Acinetobacter baumannii, Citrobacter freundii, Stenotrophomonas maltophilia and Haemophilus influenza, were detected in household water and biofilm. The microbial quality might be affected by water-storage time and households repeatedly using the same water storage containers without proper sanitization, triggering microbial regrowth and biofilm formation on water containers. Higher bacterial diversity and potentially pathogenic bacteria found in the biofilm samples of a household water supply are unhealthy for the house’s inhabitants. It is important to develop interventions aimed at preventing the formation of these dangerous biofilms in a communal water supply.
Keywords: Drinking water, Household water, Biofilm, Metagenomics, Developing country
Graphical abstract
Highlights
-
•
Using culture-dependent and -independent techniques, microbial water qualities was examined.
-
•
Potential health risks increased when water was stored for more than 3 days.
-
•
Higher bacterial diversity and potentially pathogenic bacteria found in the biofilm samples.
-
•
Poor sanitation and poor hygiene trigger microbial regrowth and biofilm formation.
1. Introduction
There has been significant progress in improving the quality of drinking water worldwide; 91% of the global population uses an improved water source (e.g. piped water, public tap stands, borehole, rainwater, protected spring or dug well), which is an increase from 76% in 1990 (Dos Santos et al., 2017). However, more than 663 million people, mainly living in sub-Saharan Africa and Asia, still lack access to improved sources of drinking water (WHO/UNICEF, 2015). While the water situation has improved in sub-Saharan Africa since 1990, the region as a whole lags behind every other developing region in the world in water and sanitation coverage (Dos Santos et al., 2017). Even in communities with access to potable water, the elderly and children under five years old especially face the risk of drinking contaminated water due to unsafe drinking water, poor sanitation and poor hygiene (Mintz et al., 1995; Wright et al., 2004). In Cameroon, for instance, only 47% of the rural population has access to clean drinking water (WHO/UNICEF, 2015).
In rural areas, microbial contaminations are recognized to be a problem during water collection and transport from the source and then in the subsequent storage of water in a household. To reduce the waterborne disease in household water, environmental health interventions such as household water treatments and safe storage solutions, encouraging inhabitants to wash their hands with soap, and proper sanitation have been highlighted (Davis et al., 2011; Wright et al., 2004). For instance, household water treatment, better sanitation facilities and hygiene education reduced the levels of diarrhea from meta-analysis (Fewtrell et al., 2005). However, the users of the communal water supply often transport their water to private residences to ensure they have enough water to last through non-supply periods. At these residences, the water frequently becomes contaminated during storage due to poor hygienic practices (Mintz et al., 1995). The unsafe storage of potable water and poor hygiene increased the number of coliform inside storage containers possibly due to microbial regrowth and their ability to survive as biofilm (Mellor et al., 2013).
Biofilm formation has been observed on all surfaces of household containers where microorganisms can attach by secreting an extracellular polymer substance (Burkowska-But et al., 2015; Mellor et al., 2013). Biofilm are not sensitive to environmental stressors and more resistant to antimicrobial treatments, creating an environmental reservoir for pathogenic microorganisms (Wingender and Flemming, 2011). Repeated use of the same water storage containers can lead to the regrowth of microorganisms and biofilm formation which poses potential health risks (Bisi-Johnson et al., 2017; Mellor et al., 2013; Stigler-Granados et al., 2014; Thomas et al., 2015; van der Merwe et al., 2013).
To date, most studies of microbial water quality in developing countries have focused on the detection of specific organisms and/or the quantification of coliforms using culturable methods (e.g., the most probable number and membrane filter techniques). Researchers have used the traditional techniques of fecal coliform, heterotrophic bacteria, or E. coli as surrogate bacteria to assess microbial water quality. Recently, molecular techniques, particularly the polymerase chain reaction (PCR)-based method, have been proposed as an alternative method to monitor and track pathogens and fecal indicator bacteria (FIB) because they offer sensitive and quantitative analytical tools (Girones et al., 2010). Also, the advent of the next-generation sequencing (NGS) based on 16s rRNA metagenomics has enabled the analysis of the entire microbial community of a given sample without the need for culturing the bacteria (Tan et al., 2015). A NGS approach can provide a broad-spectrum identification of different microbes along with a high-resolution phylogenetic microbial community profiling, while also indicating the abundance of genetic traits of the pathogens in water samples (Mukherjee et al., 2016). To the best of our knowledge, this is the first study of using a NGS approach to understand the changes in microbial communities of a source water and a household water storage and the effect of hygiene practices on biofilm formation in a water storage container in a developing country. Also, it allow us to gain in-depth knowledge about the comprehensive microbial quality and safety of the water available whereas detection of predetermined microbial indicators are often unable to assess all microbial pathogens. Particularly, NGS approach could reveal the overall microbial composition, ecology, and diversity microorganisms, leading to more comprehensive and representative risk characterization.
Thus, our research questions were designed to investigate how microbial water quality deteriorates when taken from the communal tap water supply to the household water storage container, considering variables such as (1) the type of container used for water storage, (2) the length of time that water is stored in a container, (3) the utensil used to collect water, and (4) the households’ associated hygienic practices. To determine the microbial contamination associated with household water storage, we used traditional culture-based methods, quantitative PCR, and the high-throughput sequencing of 16s rRNA gene amplicon using the Illumina MiSeq platform. The objectives of this study were to (i) determine the FIB concentrations of 20 household water and community tap stands in a gravity-fed water distribution system in Ntisaw, Cameroon, (ii) quantify the molecular FIB of potable water, household water containers and utensils (e.g., cups and buckets) from two households and (iii) assess the microbial diversity and structure associated with the water and containers used in this rural area of Cameroon.
2. Materials and methods
2.1. Field site setting and household selection
The field study was conducted in Ntisaw, a rural community in the Ndu Subdivision of the Donga-Mantung Division in the Northwest Region of Cameroon. The study area is characterized by a monsoon season (from the end of February through November), followed by an intense dry period (December through mid-February). Outside of the town of Ndu, which has a water supply network to pump ground water to the residents, the majority of the rural communities in the subdivision lack improved water sources. Ntisaw has installed a gravity-fed water distribution system, which collects water from a protected spring and distributes the water to 16 public tap stands. The village of Ntisaw is comprised of two ethnic groups: the Mborrorro people, who are primarily herders, and the Wimbum tribe, who are mainly subsistence farmers with an average salary of two groups of approximately 100,000 CFA per year (0.57 USD per day).
2.2. Sampling information
After water leaves the Ntisaw public tap system, there are two primary locations of microbial contamination during domestic storage: the collection and/or storage container, and the utensils used to distribute the water. To assess if water became contaminated, samples were collected from 20 households in Ntisaw (12% of the population) every 1–4 weeks for 4 months (February 2014 to May 2014) between 6:30am-8:30am. A sample was obtained from each household's container using the utensils that the household uses to distribute water for domestic purposes. In our first visits to each households, we surveyed household information (e.g., number of people, water usage, length of water storage, and etc.) and microbial water qualities. Additionally, during 2–3 of the sampling events, a secondary sample was collected directly from the storage containers to determine if there was a difference in the quality of the stored water versus the water that was in contact with a household utensil. All samples were plated within 2 h of collection in duplicate using 3M PetrifilmTM E. coli/coliform (EC) plates.
2.3. Households survey
Households were not told when samples would be collected to prevent any alteration in behavior. However, this meant that some families were not home or did not have water when asked to provide samples, preventing the analysis of their water for that sampling event. One of the twenty households was only sampled from twice, while all others were sampled 5–8 times. All 20 households are included in the analysis unless otherwise stated. When samples were collected, households were asked to provide information on when and where the water was collected and when the container was last cleaned with soap as hygienic practices. We noted how the material of the water storage container looked and if there was a container lid during the sampling events.
2.4. Analysis of E. coli and fecal coliforms
Within the EC plate medium: an indicator dye stains bacterial colonies red, lactose traps gas under the top film to distinguish coliforms from non-coliform bacteria, and 5-bromo-4-chloro-3-indolyl-β-D-glucuronide (BCIG) indicates β-glucuronidase (GUS) activity. When the GUS enzyme cleaves BCIG, it yields a dark blue precipitate at the site of enzymatic activity causing a GUS-producing bacterial colony like Escherichia coli (E. coli) to appear blue. This is referred to as fecal coliform bacteria (FIB) in this paper. However, other coliform colonies are red and closely associated with entrapped gas. The total coliform counts consists of both the red and blue colonies associated with gas.
After 24–36 h of incubation at 36 ± 2 °C, the bacterial growth on the EC plates for each household was enumerated. After classifying coliforms on the EC plate as ‘total coliforms’, ‘non-coliforms’ or ‘fecal coliform’, the sample was then categorized into a microbial contamination category from 0 to 8 based on the criteria shown in Table 1.
Table 1.
The criteria and risk categories of microbial water qualities used in this study.
| Criteria | Risk Category used in this study | WHO Remarks for E. coli or fecal coliform in water supplies |
|---|---|---|
| No bacteria were present | 0 | In conformity with WHO guidelines |
| 1–10 CFU/mL for both total coliform and non-coliforms. No fecal coliform present | 1 | In conformity with WHO guidelines |
| 10–100 CFU/mL for both total coliform and non-coliforms. No fecal coliform present | 2 | Low risk |
| 100–1000 CFU/mL for both total coliform and non-coliforms. No fecal coliform present | 3 | Low risk |
| 10–100 CFU/mL for both total coliform and non-coliforms. 1 CFU/ml fecal coliform present | 4 | Intermediate risk |
| 10–100 CFU/mL for both total coliform and non-coliforms. 2–10 CFU/ml fecal coliform present | 5 | High risk |
| 100–1000 CFU/mL for both total coliform and non-coliforms. 1 CFU/ml fecal coliform present | 6 | Intermediate risk |
| 100–1000 CFU/mL for both total coliform and non-coliforms. 2–10 CFU/ml fecal coliform present | 7 | High risk |
| 100–1000 CFU/mL for both total coliform and non-coliforms. 10–100 CFU/ml fecal coliform present | 8 | Very high risk |
On days in which samples were collected from households, additional samples were collected from some public taps. When tested, bacterial loading rates from the taps were normally in microbial contamination category 0, and never above 1.
2.5. DNA extraction and qPCR
Two of the twenty households participating in water sampling agreed to an additional water sampling event to be used for microbial community analysis. Both houses were located in the center of town and used the same water tap. FLOQSwab, a copan flocked swab (CAT 502CS01), was used to collect samples from: (1) the utensil which the household used to distribute water from the storage container and (2) the bucket used to collect water from the public tap. Researchers brushed the swab around the inside surface of the container. The tip was then broken off into a sterile 15 mL test tube containing 100 μL of PBST (137 mM NaCl, 2.68 mM KCl, 10.14 mM Na2HPO4, 1.76 mM KH2PO4 and 0.1% tween-20).
DNA was extracted from those samples using the PowerWater® DNA Isolation kit (MoBio Laboratories, Carlsbad, CA), following the protocol outlined in the manufacturer’s instructions. Additionally, 0.2–0.5 L of water was collected in sterile 250 mL bottles from (1) the households’ storage containers, (2) the spring box which supplies all water to the village and (3) the tap from which the families collect water. These samples were filtered through a Magna nylon 0.22 μm filter, and the filter was aseptically placed into a Lysing Matrix E tube with the FastPrep® 24 system (MP Biomedicals, Solon, OH, USA). DNA was isolated from these additional 14 samples using the UltraCleanTM Soil DNA Isolation Kit (MO Bio Laboratories, Carlsbad, CA, USA) according to the manufacturer’s instructions. The concentration of the extracted DNA was measured with the NanoDrop spectrophotometer (Thermo Scientific, Foster City). All samples were transported with cold packs and then stored at −20 °C until needed for further analysis.
Amplification was done in 384-well plates in a ViiATM 7 Real-Time PCR system (Applied Biosystems, Foster City, CA). Briefly, each 10 μL PCR reaction contained 5 μL of either Taqman® Universal PCR Matermix (Applied Biosystems, Foster City, CA) or Power SYBR® Green PCR Master Mix (Applied Biosystems, Foster City, CA), 2 μL of DNA template, and the primers/probe for the total coliform (Maheux et al., 2014) and E. coli (Chern et al., 2011). Plates with no DNA template were run as a negative control. The thermal cycle profile consisted of 2 min at 50 °C, followed by 10 min at 95 °C, and 40 cycles of 15 s at 95 °C and 60 s at 60 °C. Triplicate PCR reactions were conducted with Taqman® and SYBR Green qPCR.
2.6. Microbial community analysis and detection of potential human pathogens
The 16S rRNA genes were amplified from all the DNA extracts using primer sets 515F/909R(Wang and Qian, 2009) for multiplexed 16S amplicon sequencing on the Miseq® system. In brief, PCR preparation and sequencing on the Illumina Miseq® benchtop sequencer using pair-end 250-bp kits was performed. These reads were excluded from the data analysis if the average quality score was lower than 10 during the split library script performed by QIIME (Caporaso et al., 2010). Chimera checking was done in QIIME using USEARCH to remove chimeric sequences. The sequences obtained were processed using QIIME version 1.8 to perform operational taxonomic unit (OTU) clustering. OTUs defined by a 97% sequence similarity were picked using open-reference-based OTU with Uclust, and the representative sequences were submitted to the RDP classifier (Wang et al., 2007) to obtain the taxonomy assignment and relative abundance of each OTU from the Greengenes database (DeSantis et al., 2006). Alpha-diversity (diversity within a given sample) metrics (i.e., Chao1, Shannon index and inverse Simpson) were determined using the R package Phyloseq (McMurdie and Holmes, 2013). Shannon evenness was calculated by dividing observed OTUs by the natural logarithm of the Shannon index. Chao1 is an estimator of species richness (i.e. number of species). The inverse Simpson metric is a species diversity indicator. The differential abundance in a sparse high-throughput microbial marker-gene survey was conducted to determine OTUs that are differentially abundant between two or more groups of multiple samples (Paulson et al., 2013). Beta-diversity (diversity among samples) weighted UniFrac-based principal coordinate analysis (PCoA) plots were calculated using the R package Phyloseq following rarefaction of the data using the QIIME pipeline. To identify potential human pathogens, bioinformatics analysis was performed using QIIME protocol; sequences were clustered against the human pathogen database as described previously (Kumaraswamy et al., 2014; Srinivasan et al., 2015). After the initial preprocessing step, the sequences were clustered against the human pathogenic sequences of clinically relevant pathogenic bacteria, which has 617 sequences of clinical isolates of the 16S rRNA gene sequences and genus/species classification (Srinivasan et al., 2015). The threshold value was set to a 99% similarity. For alignment of the OTUs and taxonomic identification, the closed-reference-based OUT with USEARCH and multiple sequences alignment was done using PyNast. The sequences were filtered with the Human pathogen database. Statistical and metadata analysis of the microbiome data was conducted through Phyloseq (McMurdie and Holmes, 2013) and MicrobiomeAnalyst (Dhariwal et al., 2017).
2.7. Statistical analysis for correlating the data
All statistical analyses were completed using Minitab® for statistical computing. The non-parametric method of Kirskal-Wallis was used to compare the potential risk associated with categorical variables. In addition, the ANOVA statistical test was used to analyze the difference among the abundances of E.coli and total coliform, as measured by two molecular qPCR assays.
3. Results
3.1. Microbial water quality and potential health risks in household water
Data from the survey of 170 households in Ntisaw provides a comprehensive picture of water usage and health in the community (Table S2). In Ntisaw, there are 834 people counted in this census, with a mean household size of 4.9 (family members that do not live in the village for at least two consecutive days every two weeks were not counted).
The concentrations of culturable coliform were monitored in 20 households. Communal water resources, including protected spring and public tap waters, were also monitored. The quality of water at the source water and water tap were of low microbial quality, having risk categories of 0 or 1. The household samples were tested as having risk categories ranging from 0 to 6 (Table 1), which indicates potential health risks. However, the microbial water quality of the household water often deteriorated, with the presence of fecal coliform up to 44 CFU/mL, even though the water quality of the spring and public tap waters met the WHO Guidelines for safe drinking water.
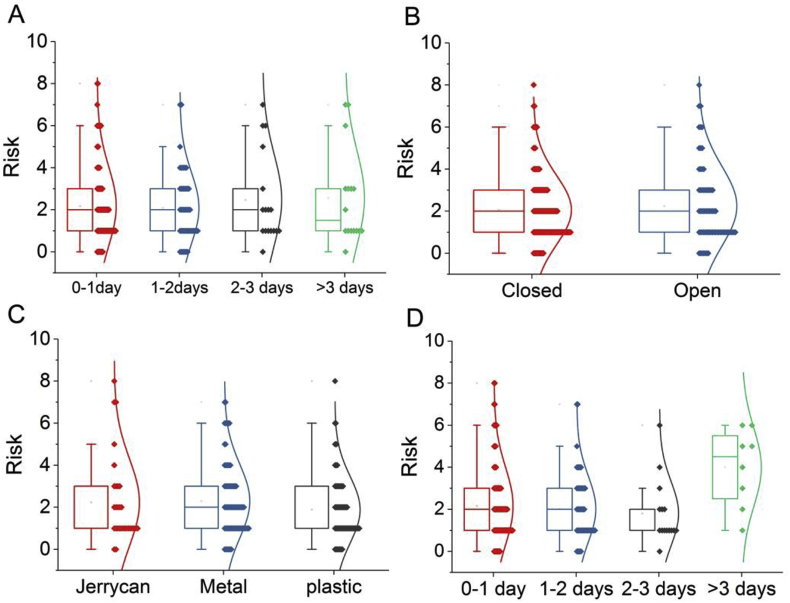
We surveyed the household water storage behaviors linked with health risks (e.g., type of container and open vs. closed container) and potential interventions (e.g., water storage days, cleaned last day, and education). Fig. 1 shows the potential risks associated with household water storage and practices. The containers that the families in Ntisaw use were categorized as either a “jerrycan”, a “metal pot”, or a “plastic bucket”. Statistical tests of Kruskal-wallist indicated that the potential risks posed by the type of containers were insignificant (p-value = 0.263). In addition, the effect of having a closed or open water container on the inhabitants’ health was negligible in this study (p-value = 0.735). The relationship between household storage conditions (e.g., time stored and types of container used) and potential risks were examined because the type of container and the amount of days the water was stored could be associated with microbial regrowth, leading to potential health risks. The potential risks were significantly different depending on the length of time the water was stored (p-value < 0.05), showing that a storage period of more than 3 days increased potential health risks (Fig. 1D). Interestingly, the inhabitants’ hygiene practices were not clearly associated with potential health risks because there was insignificant different among the cleaned last day (p-value = 0.406).
Fig. 1.
The potential health risks associated with household water storage condition and practices such as water cleaned last days (A), opened or closed water containers (B), the type of containers (C), and water storage days (D).
3.2. qPCR analyses of fecal indicator bacteria in a household water system
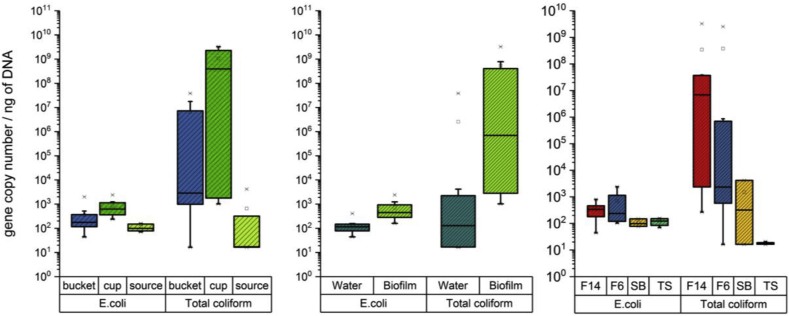
The total coliform and E. coli genes were enumerated using qPCR to determine the level of microbial contamination in the water and biofilm samples from the households’ waters supplies. Two households (F14 and F6) were selected to examine the abundance of FIB and the formation of biofilm in household utensils. Only F14 was instructed to clean cups and buckets before sampling, so the effect of hygienic practices could be compared. As expected, the concentrations of total coliform were significantly higher than those of E. coli in all samples except the source water, where the concentrations of total coliform and E. coli were in the same order of magnitude. The concentrations of total coliform and E. coli from the bucket and cup samples were higher than those of the source water samples, as shown in Fig. 2A. The abundance of total coliform and E. coli, however, more significantly increased in biofilm than in water samples, as shown in Fig. 2B (p-value = 0.02 and 0.05 for E. coli and total coliform, respectively). The abundance of E. coli and total coliform increased in the household water due to bacterial regrowth and biofilm formation. However, the effect of hygiene practices (e.g., cleaning containers) on the reduction of E. coli and total coliform was not significant, implying that biofilm were persistent even after hygiene practices were employed by the household.
Fig. 2.
Enumeration of E.coli and total coliform genes from the source water (SB and TS), the two household water from two families (F 4 and F6) and biofilm collected from buckets and cups in F14 and F6. The two households of F14 (Family #14) and F6 (Family #6) were selected among twenty households. Only F14 was instructed to clean containers before sampling. SB and TS indicates spring box and tap stand, respectively.
3.3. Analysis of microbial communities in the household water
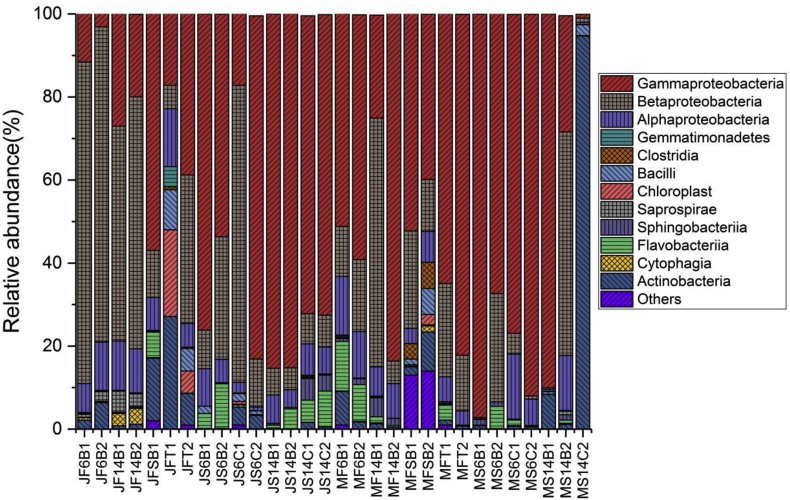
Taxonomic composition of water and biofilm samples revealed that the most abundant phylum was Proteobacteria in the household water, except for one biofilm sample from a cup (MS14C2). Fig. 3 shows the relative abundance of microbial diversity, indicating that Alpha-, Beta- and Gamma-proteobacteria are consistently the most abundant among clades on the class level. Others included Actinobacteria, Cytophagia, Bacilli, Flavobacteriia, and Clostridia. Under the Proteobacteria phylum, Burkholderiales, Pseudomonadales, Enterobacteriales and Xanthomonadales were the most predominant bacterial families. At the genus level, the most abundant genera included unassigned groups from Pseudomonas, Acinetobacter, Enterobacteriaceae, Comamonadaceae, and Xanthomonadaceae (Figs. S1 and S2).
Fig. 3.
Relative abundance of microbial community obtained from the household water systems and source water. Two sampling events (January(J) and May(M)) were conducted to collect water (F) and biofilm (S) samples in this project. Among twenty families, two households (family #14 (F14) and #16 (F6)) were chosen to collect water and biofilm samples from different containers such as buckets(B) and cup(C). SB and T indicate spring box (SB) and tap stand (T), respectively.
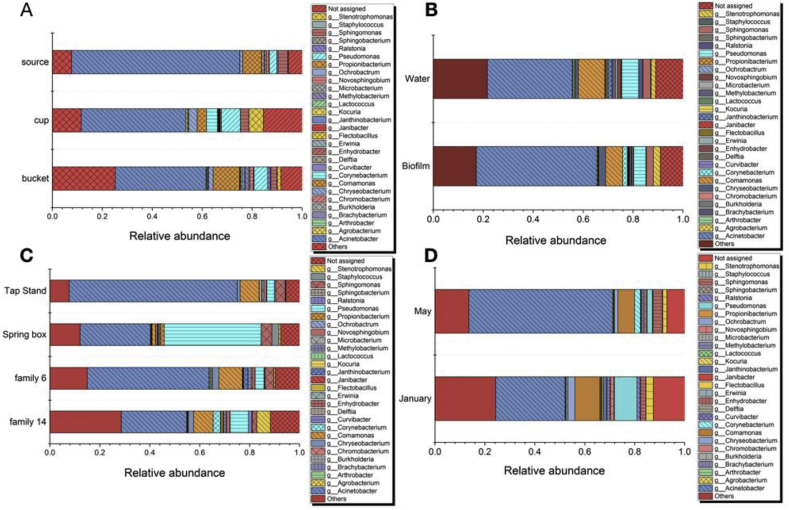
To understand the changes in microbial communities caused by the households’ water storage and hygiene practices, the relative abundances of microbial communities were clustered by the type of water storage containers and sample type (biofilm vs. water), sampling location, and seasonal variation. When the types of containers were examined, Acinetobacter was most abundant in the bucket (36%), cup (42%) and source water (67%), but Methylobacterium, Erwinia, Sphingobacterium, Brachybacterium were present in the buckets and cups, only as shown in Fig. 4. The second most abundant genera were Comamonas (bucket and source water) and Pseudomonas (cup). The microbial distribution observed in the water and biofilm samples were similar, even though Acinetobacter, Chryseobacterium, Stenotrophomonas and Corynebacterium were relatively more abundant in biofilm than in water. Also, the most abundant genus in the source water (spring box) was Pseudomonas, while Acinetobacter was the most abundant in the other samples (tap stand, Family 6, and Family 14), indicating that microbial compositions changed during transport and storage.
Fig. 4.
Relative abundance of microbial communities at the genus level, clustering by the type of containers (A), type of samples (B), locations (C) and season (D).
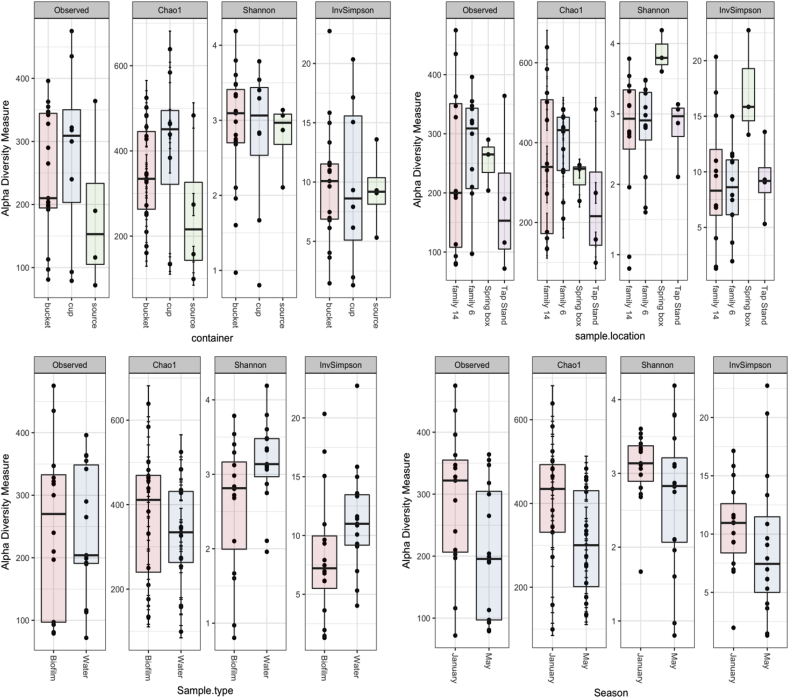
Alpha-diversities were calculated within a given community to understand how the type of containers, the water/biofilm samples, locations or season altered the species’ richness and evenness. In Fig. 5, alpha-diversity metrics of the type of containers demonstrated that the microbial communities observed in the household container were more diverse than those observed in the source water, implying that microbial communities originated not only from the source water but also from other sources, such as the soil, humans, and the air. The Observed and Chao 1 indices in biofilm and water showed that species richness did not significantly differ (p-value >0.05), while the Shannon and inverse Simpson indices from biofilm were significantly different than those of the water (p-value <0.05). Since Shannon and inverse Simpson indices measure the distribution of an individual among groups, high indices show a low homogeneity of species, suggesting specific OTUs were more abundant than others in the biofilm. Taxonomically less diverse biofilm on the surface of buckets and cups implied that biofilm colonizers would persist in the household water storage containers. For instance, the differential abundance analysis using MicrobiomeAnalyst showed that genera of Brachybacterium, Erwinia, Enhydrobacter, Citrobacter, Deinococcus were differentially abundant in the biofilm (p-value <0.05).
Fig. 5.
Alpha-diversity analyses from types of container (cup, bucket and source water), sample location (Tap stand, Spring box, Family #6 and Family #14), sample type (water and biofilm) and season (January and May).
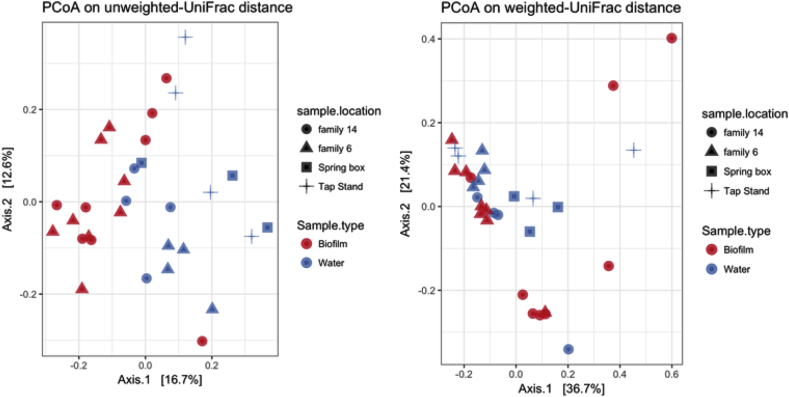
A principal coordinate analysis (PCoA) was conducted to visualize similarities or dissimilarities in the microbial community in Fig. 6. Although no obvious clustering was apparent between the biofilm and water samples, biofilm samples clustered separately from the spring box and tap stand samples, suggesting that the biofilm community was relatively distinct and far from the water microbiome. No distinct clustering between family 14 and family 6 indicated that hygiene practices might not have a strong effect on the microbial communities in the houses’ water systems.
Fig. 6.
A principal coordinates analysis (PCoA) showing similar and dissimilar relation among 31 bacterial community samples using the unweighted and weighted UniFrac distance matrix.
3.4. Estimation of potential human pathogens in the household water system using high-throughput 16s rRNA gene analysis
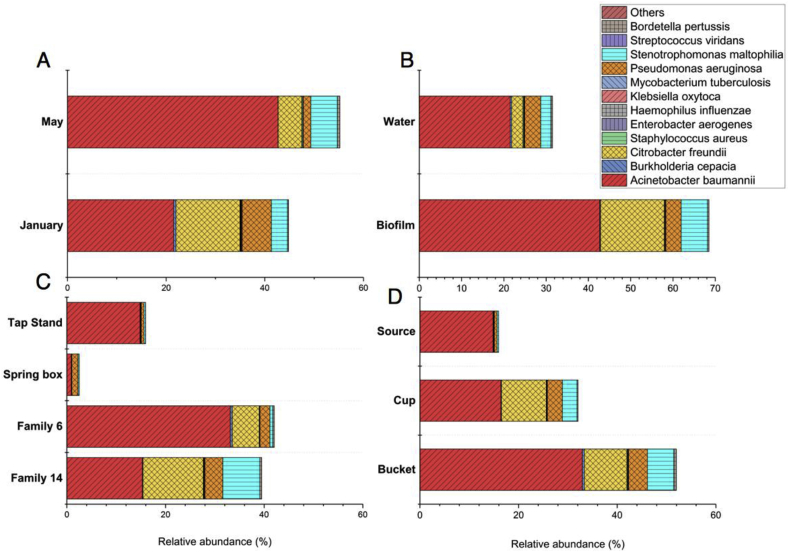
The 16s rRNA gene sequences were compared against the human pathogenic sequences using closed-reference OUT calling. The 16s rRNA gene sequences matched 10 different pathogenic sequences belonging to various genera and species within the database of clinically relevant bacterial pathogens. The most sequences of all the gram-negative pathogens identified were Acinetobacter baumannii, followed by strains of Pseudomonas aeruginosa, Citrobacter freundii, Stenotrophomonas maltophilia and Haemophilus influenzae. Among the Gram-positive bacteria matched with human pathogenic sequences, Staphylococcus epidermidis and Streptococcus viridans were present in the most samples (Fig. S3). Fig. 7 displays the relative abundance of the different clustering based on seasons (A), type of samples (B), sample location (C), and type of containers (D). The human pathogenic bacteria were more abundant in the samples taken in May rather than those taken in January. Also, several other potential human pathogens such as Acinetobacter baumannii, Citrobacter freundii and Stenotrophomonas maltophilia constituted a significantly higher proportion of microbial communities in biofilm samples as compared to the source water. The relative abundance of bacterial pathogens increased up to 40% when compared to the source water (Spring box), suggesting that the microbial contamination may have originated from non-source water. Citrobacter freundii, Stenotrophomonas maltophilia, and Enterobacter aerogenes were observed in non-source water only. Similarly, bacterial pathogen from the containers exhibited a different abundance than those of the source water (Fig. 7). Alpha-diversity analyses proved that higher diversity indices were present in the household water systems, implying that abundant and diverse pathogenic bacteria could increase the potential health risks in the household systems (Fig. 4S).
Fig. 7.
Relative abundance of potential human pathogenic bacteria using 16s rRNA sequencing analysis against human pathogenic bacterial database. The relative abundances were clustered by the type of containers (A), type of samples (B), locations (C) and season (D).
4. Discussion
The study of microbial pollutants in potable water has generally been limited in developing countries, and microbial communities, including their potential pathogenic bacteria, in household water have not yet been fully investigated. In this study, we investigated microbial water quality in household water in rural regions of sub-Saharan Africa using culture-dependent and -independent techniques such as qPCR and high-throughput 16S rRNA gene amplicon sequencing. This study could be the first comprehensive attempt to assess the changes in microbial communities associated with hygiene practices (e.g., cleaning a container) and sampling campaigns (e.g., type of container, location, and season) that could interfere with the microbial structures of the household water storage system in a developing country.
In this study, the potential health risks observed were higher in the household containers and utensils, than in the source water; this is consistent with the findings of previous studies (Ahmed et al., 2013; Momba and Kaleni, 2002; Oloruntoba et al., 2016). The microbial water quality declined after being collected from the community tap and transported to the home. Hygiene practice (cleaning water containers) and the water vessel type (opened/closed container and the type of material used for the containers) did not reduce the health risk significantly. Interestingly, employing hygiene practices did not result in a substantial reduction of the concentration of fecal indicator bacteria in the water samples. One possible explanation for this might be that the short duration of the study limits our ability to draw a conclusion about more complicated hygiene practices, such as larger behavioral changes and informational interventions. It has been shown that hygiene practices and time-intensive behaviors like cleaning the vessels more frequently are difficult for subjects to maintain (Hamoudi et al., 2012). A socio-psychological approach of behavioral change might be required to test the effectiveness of household water treatment practices (Davis et al., 2011; Lilje and Mosler, 2017). However, the amount of time the water was stored in a household increased potential health risks plausibly due to biofilm growth and formation on the surface of the containers. Another possible explanation might be that the repeated use of the same containers for water storage resulted in the formation of biofilm containing bacterial cells, because of the presence of biofilms on walls and sediments within the household water storage (Burkowska-But et al., 2015; Mellor et al., 2013; Wingender and Flemming, 2011).
In the present study, we enumerated the total coliform and E. coli from the household water and containers using the qPCR technique. Although a PCR-based method could not differentiate between live and dead cells in given samples, molecular microbiological testing could provide insight into the fecal contamination of the household water. The concentration of genomic E.coli and total coliform in the containers was significantly higher than the concentrations in the source water, indicating that bacteria were able to persist and even grow in such oligotrophic environments by forming biofilm. Also, the higher concentration of genomic E.coli and total coliform might be explained by the presence of the viable but nonculturable cell (VBNC) inside biofilm (Girones et al., 2010; Lee and Bae, 2017). It has been shown that biofilms from water samples do not reflect the total cell count. At least a part of the biofilm populations of bacteria persists in a viable but non-culturable (VBNC) state and remains undetectable by the conventional methods. (Liu et al., 2008; Wingender and Flemming, 2011). More importantly, many bacterial species enter a transient state of “dormancy” as a strategy to survive unfavorable conditions (Lennon and Jones, 2011). When in this state, bacteria cannot be detected using culture-based methods, but they remain viable and retain virulence under environmental stressors (Lennon and Jones, 2011). Thus, molecular microbiological testing could elucidate the relationship between hygiene practices and the concentration of fecal indicator bacteria in a household water storage system.
Past studies have reported that the microbial community structure of a drinking water system is governed by the source water, the drinking water treatment process and the type of pipe used in the urban water system (Berry et al., 2006; Proctor and Hammes, 2015). To date, however, little is known about the changes that happen in water’s microbial communities when taken from the communal source to a household water storage container. In a developing country, a household’s potable water supply must be collected from a single public well, spring or stand pipe, transported to the home, and then stored for future household use (Oloruntoba et al., 2016). By understanding the changes in microbial communities in a household water supply, researchers can gain insight into the inhabitants’ hygiene practices and the quality of water storage. The water is frequently contaminated by unhygienic conditions and/or biofilm formation in the home. In our study, the ubiquitous and persistent genera were observed; these were mainly members of phylum Proteobacteria including Pseudomonas, Acinetobacter, Comamonas and unassigned genus from Enterobacteriaceae. The Pseudomonas genus organisms are mostly free-living bacteria widely distributed in soil, water, marine environments and on the skin of animals, including humans (Mann and Wozniak, 2012). Also, Acinetobacter was ubiquitously detected in the samples. Acinetobacter can be isolated from freshwater, estuaries, sewages, sea water, drinking water and biofilms in drinking water distribution systems (Doughari et al., 2011). Both bacteria are able to form highly structured biofilm with distinct properties that colonize new surfaces or join existing biofilm. One of most abundant bacterial families in this study was Enterobacteriaceae which is a large family of Gram-negative bacteria that includes many of the more familiar pathogens, such as Salmonella, Escherichia coli, Yersinia pestis, Klebsiella, Shigella, Proteus, Enterobacter, Serratia, and Citrobacter.
We further interrogated the presence of potential human pathogens using metagenomic sequencing data. Initially, after quality filtering, the raw sequences were compared with the Greengenes database using the QIIME OTUs classification. Later, the sequences were compared with the databases of a broad range of clinically relevant bacteria pathogens (Srinivasan et al., 2015). Using this approach, we identified 10 different potential pathogenic bacteria in the source water and household samples. The relative abundance of those potential pathogens indicated that they were pervasive in the household water and biofilm samples. These pathogens included Acinetobacter baumannii, Citrobacter freundii, and Stenotrophomonas maltophilia. The sequence matched bacteria are mostly opportunistic pathogen bacteria, but they are often isolated from environments. For instance, Citrobacter freundii is an opportunistic pathogen, but it is also a resident of the human gastrointestinal tract and is often found in drinking water samples (Payne et al., 2010). Stenotrophomona. maltophilia is an environmental bacterium found in aqueous habitats, including plant rhizospheres, animals, foods, and water sources (Brooke, 2012). Interestingly, Acinetobacter baumannii is an opportunistic bacterial pathogen primarily associated with hospital-acquired infections (Peleg et al., 2008). It was reported, however, that the environmental Acinetobacter baumannii strain was similar to a clinical isolate and originated from paleosol (Hrenovic et al., 2014). These potential pathogenic bacteria found in biofilm could be recovered from soil and water, implying that household water could be contaminated during collection, transport and storage. Also, Haemophilus influenzae and Citrobacter freundii could come from human microbiota because they are commensal bacteria in gut and the upper respiratory (Arce et al., 2009; Payne et al., 2010). Overall, the potential pathogen bacteria tended to be persistent in the containers and utensils as a biofilm on the water’s surface. The observed bacteria are known to be efficient at forming biofilm (Faille et al., 2003; Longo et al., 2014; Pompilio et al., 2008).
Based on the findings presented here, this study reveals several practical implications for control microbial water quality in a household water system. First, consistent and multiple hygiene interventions at household level required for inhibiting and removing biofilm from the containers and utensils in a household. Biofilm formation on containers and utensils were persistent after hygiene practices (e.g., closed vessel and cleaning water storage container with soap). The use of chlorine or bleach could be an appropriate method to remove biofilm from the water container. Second, fecal indicator bacteria such as fecal coliform and E. coli might not be a good proxy for assessing microbial water qualities in a household water system. Since potential pathogenic microorganisms were not enteric pathogen, fecal indicator bacteria may not assess microbial water qualities where reintroduced or regrown pathogens exit in a household water system. Finally, NGS techniques reveals microbial composition, ecology, diverse microorganisms, leading to more representative microbial characterization and risk assessment of water quality. The potential human pathogenic bacteria such as Acinetobacter baumannii, Pseudomonas aeruginosa, Citrobacter freundii and Stenotrophomonas maltophilia were related to antimicrobial resistant bacteria (Blasco et al., 2008; Brooke, 2012; Karumathil et al., 2014; McKeon et al., 1995). Since self-medication is a common practice in developing countries where patients often get antimicrobials without prescription, potential health risks associated with antimicrobial resistant bacteria and prevalence of those bacteria in water resources in a developing country should be examined. Thus, household hygiene practices (e.g., point-of-use chemical disinfection) and more comprehensive and representative risk characterization should be required to reduce health risks in water storage condition in a developing country.
The 16s rRNA gene could prove to be a useful diagnostic tool for identifying the presence of pathogens in a water sample (Kumaraswamy et al., 2014; Srinivasan et al., 2015). However, some genera need further validation in addition to the 16S rRNA sequences for accurate pathogen identification, because even though the sequences have a more than 99% similarity to the reference database, they do not conclusively prove the presence of pathogenic bacteria. Also, the high similarity of 16s rRNA sequences of species within the genera makes the NGS approach less discriminatory for pathogen identification. While the presence of pathogens could be confirmed with qPCR with suitable maker genes or culturable methods, it would not be feasible to screen the lists of potential human pathogens that are important in a developing country. Using the clinically relevant human pathogen database, we could assess the overall risk of the microbial contaminants found in a household’s water storage system and the inhabitants’ hygiene practices. The findings of our approach provide a potential way to identify potential human pathogens in household water, especially in a developing country where alternate suitable microbial monitoring methods are limited.
5. Conclusion
-
•
A microbial survey of the household water in a rural community in Cameroon indicated that water was frequently contaminated during the storage period before consumption.
-
•
Culture-dependent and culture-independent techniques of qPCR and high-throughput sequences analyses revealed more complex microbial communities exhibited in the household water containers and utensils than source water.
-
•
The high-throughput sequencing of 16s rRNA gene amplicon analysis revealed a rich bacterial diversity and higher abundance of potential pathogenic bacteria in biofilm, which may pose a health risk to the houses’ inhabitants.
-
•
The qPCR and high-throughput sequences analyses confirmed microbial regrowth and biofilm formation even after cleaning the container and utensils, implying that health risk could be minimized and avoided by consistent and multiple hygiene interventions.
-
•
Next-generation sequencing for assessment of microbial water quality can serve as a tool of estimating potential health risk in a household water system.
Conflicts interest
None.
Acknowledgements
A special thank you to our host and translator, Yusinyu Julius Ngayinkyu, and our mentor, Dr. Richard Cook, ithout whom this project would not have been possible. Thank you to the entire village of Ntisaw for welcoming us into their community during the length of this project. Thank you also to Aimee Gall and Kenneth Long for fruitful discussions and help in preparing materials. Also thank you to Juan Pedro Maestro for helping to prepare the sampling equipment, Ralph Albano, Laurie Dehaan and Katie Badskey for helping to prepare the education materials, EWB-USA UIUC for collaborating with us on this project, and to Dean Bruce Litchfield for making this study in Ntisaw possible. A very special thanks to 3M’s University program for donating 3M PetrifilmTM E. coli/coliform plates. Funding provided by personal scholarships from British Petroleum (NO), Illinois and Central Texas Water Environment Association (CL), American Water Works Association – Austin branch (CL), Susan E. Stutz Foundation (CL) and NSF-GRFP (CL). This research was partly supported by start-up funds from the National University of Singapore (R-302-000-137-133).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.wroa.2019.100026.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Ahmed D., Islam M.S., Begum Y.A., Janzon A., Qadri F., Sjoling A. Presence of enterotoxigenic Escherichia coli in biofilms formed in water containers in poor households coincides with epidemic seasons in Dhaka. J. Appl. Microbiol. 2013;114(4):1223–1229. doi: 10.1111/jam.12109. [DOI] [PubMed] [Google Scholar]
- Arce F.T., Carlson R., Monds J., Veeh R., Hu F.Z., Stewart P.S., Lal R., Ehrlich G.D., Avci R. Nanoscale structural and mechanical properties of nontypeable Haemophilus influenzae biofilms. J. Bacteriol. 2009;191(8):2512–2520. doi: 10.1128/JB.01596-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry D., Xi C.W., Raskin L. Microbial ecology of drinking water distribution systems. Curr. Opin. Biotechnol. 2006;17(3):297–302. doi: 10.1016/j.copbio.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Bisi-Johnson M.A., Adediran K.O., Akinola S.A., Popoola E.O., Okoh A.I. Comparative physicochemical and microbiological qualities of source and stored household waters in some selected communities in southwestern Nigeria. Sustainability. 2017;9(3) [Google Scholar]
- Blasco M.D., Esteve C., Alcaide E. Multiresistant waterborne pathogens isolated from water reservoirs and cooling systems. J. Appl. Microbiol. 2008;105(2):469–475. doi: 10.1111/j.1365-2672.2008.03765.x. [DOI] [PubMed] [Google Scholar]
- Brooke J.S. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin. Microbiol. Rev. 2012;25(1):2–41. doi: 10.1128/CMR.00019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkowska-But A., Kalwasinska A., Brzezinska M.S. Bacterial growth and biofilm formation in household-stored groundwater collected from public wells. J. Water Health. 2015;13(2):353–361. doi: 10.2166/wh.2014.097. [DOI] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., McDonald D., Muegge B.D., Pirrung M., Reeder J., Sevinsky J.R., Turnbaugh P.J., Walters W.A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern E.C., Siefring S., Paar J., Doolittle M., Haugland R.A. Comparison of quantitative PCR assays for Escherichia coli targeting ribosomal RNA and single copy genes. Lett. Appl. Microbiol. 2011;52(3):298–306. doi: 10.1111/j.1472-765X.2010.03001.x. [DOI] [PubMed] [Google Scholar]
- Davis J., Pickering A.J., Rogers K., Mamuya S., Boehm A.B. The effects of informational interventions on household water management, hygiene behaviors, stored drinking water quality, and hand contamination in peri-urban Tanzania. Am. J. Trop. Med. Hyg. 2011;84(2):184–191. doi: 10.4269/ajtmh.2011.10-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhariwal A., Chong J., Habib S., King I.L., Agellon L.B., Xia J.G. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017;45(W1):W180–W188. doi: 10.1093/nar/gkx295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos S., Adams E.A., Neville G., Wada Y., de Sherbinin A., Mullin Bernhardt E., Adamo S.B. Urban growth and water access in sub-Saharan Africa: progress, challenges, and emerging research directions. Sci. Total Environ. 2017;607–608:497–508. doi: 10.1016/j.scitotenv.2017.06.157. [DOI] [PubMed] [Google Scholar]
- Doughari H.J., Ndakidemi P.A., Human I.S., Benade S. The ecology, biology and pathogenesis of acinetobacter spp.: an overview. Microb. Environ. 2011;26(2):101–112. doi: 10.1264/jsme2.me10179. [DOI] [PubMed] [Google Scholar]
- Faille C., Fontaine F., Lelievre C., Benezech T. Adhesion of Escherichia coli, Citrobacter freundii and Klebsiella pneumoniae isolated from milk: consequence on the efficiency of sanitation procedures. Water Sci. Technol. 2003;47(5):225–231. [PubMed] [Google Scholar]
- Fewtrell L., Kaufmann R.B., Kay D., Enanoria W., Haller L., Colford J.M., Jr. Water, sanitation, and hygiene interventions to reduce diarrhoea in less developed countries: a systematic review and meta-analysis. Lancet Infect. Dis. 2005;5(1):42–52. doi: 10.1016/S1473-3099(04)01253-8. [DOI] [PubMed] [Google Scholar]
- Girones R., Ferrus M.A., Alonso J.L., Rodriguez-Manzano J., Calgua B., Correa Ade A., Hundesa A., Carratala A., Bofill-Mas S. Molecular detection of pathogens in water--the pros and cons of molecular techniques. Water Res. 2010;44(15):4325–4339. doi: 10.1016/j.watres.2010.06.030. [DOI] [PubMed] [Google Scholar]
- Hamoudi A., Jeuland M., Lombardo S., Patil S., Pattanayak S.K., Rai S. The effect of water quality testing on household behavior: evidence from an experiment in rural India. Am. J. Trop. Med. Hyg. 2012;87(1):18–22. doi: 10.4269/ajtmh.2012.12-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrenovic J., Durn G., Goic-Barisic I., Kovacic A. Occurrence of an environmental acinetobacter baumannii strain similar to a clinical isolate in paleosol from Croatia. Appl. Environ. Microbiol. 2014;80(9):2860–2866. doi: 10.1128/AEM.00312-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karumathil D.P., Yin H.B., Kollanoor-Johny A., Venkitanarayanan K. Effect of chlorine exposure on the survival and antibiotic gene expression of multidrug resistant acinetobacter baumannii in water. Int. J. Environ. Res. Publ. Health. 2014;11(2):1844–1854. doi: 10.3390/ijerph110201844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaraswamy R., Amha Y.M., Anwar M.Z., Henschel A., Rodriguez J., Ahmad F. Molecular analysis for screening human bacterial pathogens in municipal wastewater treatment and reuse. Environ. Sci. Technol. 2014;48(19):11610–11619. doi: 10.1021/es502546t. [DOI] [PubMed] [Google Scholar]
- Lee S., Bae S. Molecular viability testing of viable but non-culturable bacteria induced by antibiotic exposure. Microb. Biotechnol. 2017 doi: 10.1111/1751-7915.13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon J.T., Jones S.E. Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 2011;9(2):119–130. doi: 10.1038/nrmicro2504. [DOI] [PubMed] [Google Scholar]
- Lilje J., Mosler H.J. Socio-psychological determinants for safe drinking water consumption behaviors: a multi-country review. J. Water, Sanit. Hyg. Dev. 2017;7(1):13–24. [Google Scholar]
- Liu Y.M., Gilchrist A., Zhang J., Li X.F. Detection of viable but nonculturable Escherichia coli O157 : H7 bacteria in drinking water and river water. Appl. Environ. Microbiol. 2008;74(5):1502–1507. doi: 10.1128/AEM.02125-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo F., Vuotto C., Donelli G. Biofilm formation in Acinetobacter baumannii. New Microbiol. 2014;37(2):119–127. [PubMed] [Google Scholar]
- Maheux A.F., Boudreau D.K., Bisson M.A., Dion-Dupont V., Bouchard S., Nkuranga M., Bergeron M.G., Rodriguez M.J. Molecular method for detection of total coliforms in drinking water samples. Appl. Environ. Microbiol. 2014;80(14):4074–4084. doi: 10.1128/AEM.00546-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann E.E., Wozniak D.J. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol. Rev. 2012;36(4):893–916. doi: 10.1111/j.1574-6976.2011.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon D.M., Calabrese J.P., Bissonnette G.K. Antibiotic-resistant gram-negative bacteria in rural groundwater supplies. Water Res. 1995;29(8):1902–1908. [Google Scholar]
- McMurdie P.J., Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J.E., Smith J.A., Samie A., Dillingham R.A. Coliform sources and mechanisms for regrowth in household drinking water in Limpopo, South Africa. J. Environ. Eng. 2013;139(9):1152–1161. doi: 10.1061/(ASCE)EE.1943-7870.0000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz E.D., Reiff F.M., Tauxe R.V. Safe water treatment and storage in the home. A practical new strategy to prevent waterborne disease. J. Am. Med. Assoc. 1995;273(12):948–953. [PubMed] [Google Scholar]
- Momba M.N.B., Kaleni P. Regrowth and survival of indicator microorganisms on the surfaces of household containers used for the storage of drinking water in rural communities of South Africa. Water Res. 2002;36(12):3023–3028. doi: 10.1016/s0043-1354(02)00011-8. [DOI] [PubMed] [Google Scholar]
- Mukherjee N., Bartelli D., Patra C., Chauhan B.V., Dowd S.E., Banerjee P. Microbial diversity of source and point-of-use water in rural Haiti - a pyrosequencing-based metagenomic survey. PLoS One. 2016;11(12):e0167353. doi: 10.1371/journal.pone.0167353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oloruntoba E.O., Babalola T.F., Morakinyo O.M., Mumuni A. Effects of improved storage containers on the bacteriological quality of household drinking water in low-income urban communities in Ibadan, Nigeria. Water Sci. Technol. Water Supply. 2016;16(2):378–387. [Google Scholar]
- Paulson J.N., Stine O.C., Bravo H.C., Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods. 2013;10(12) doi: 10.1038/nmeth.2658. 1200-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne S.J., Besner M.C., Lavoie J., Krentz C., Hansen L.T., Friedman M., LeChevallier M.W., Prevost M., Gagnon G.A. Molecular techniques and data integration: investigating distribution system coliform events. J. Water Supply Res. Technol. - Aqua. 2010;59(5):298–311. [Google Scholar]
- Peleg A.Y., Seifert H., Paterson D.L. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 2008;21(3):538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompilio A., Piccolomini R., Picciani C., D'Antonio D., Savini V., Di Bonaventura G. Factors associated with adherence to and biofilm formation on polystyrene by Stenotrophomonas maltophilia: the role of cell surface hydrophobicity and motility. FEMS Microbiol. Lett. 2008;287(1):41–47. doi: 10.1111/j.1574-6968.2008.01292.x. [DOI] [PubMed] [Google Scholar]
- Proctor C.R., Hammes F. Drinking water microbiology - from measurement to management. Curr. Opin. Biotechnol. 2015;33:87–94. doi: 10.1016/j.copbio.2014.12.014. [DOI] [PubMed] [Google Scholar]
- Srinivasan R., Karaoz U., Volegova M., MacKichan J., Kato-Maeda M., Miller S., Nadarajan R., Brodie E.L., Lynch S.V. Use of 16S rRNA gene for identification of a broad range of clinically relevant bacterial pathogens. PLoS One. 2015;10(2):22. doi: 10.1371/journal.pone.0117617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stigler-Granados P., Quintana P.J.E., Gersberg R., Zuniga M.L., Novotny T. Comparing health outcomes and point-of-use water quality in two rural indigenous communities of Baja California, Mexico before and after receiving new potable water infrastructure. J. Water, Sanit. Hyg. Dev. 2014;4(4):672–680. [Google Scholar]
- Tan B., Ng C., Nshimyimana J.P., Loh L.L., Gin K.Y., Thompson J.R. Next-generation sequencing (NGS) for assessment of microbial water quality: current progress, challenges, and future opportunities. Front. Microbiol. 2015;6:1027. doi: 10.3389/fmicb.2015.01027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas K., McBean E., Shantz A., Murphy H.M. Comparing the microbial risks associated with household drinking water supplies used in peri-urban communities of Phnom Penh, Cambodia. J. Water Health. 2015;13(1):243–258. doi: 10.2166/wh.2014.214. [DOI] [PubMed] [Google Scholar]
- van der Merwe V., Duvenage S., Korsten L. Comparison of biofilm formation and water quality when water from different sources was stored in large commercial water storage tanks. J. Water Health. 2013;11(1):30–40. doi: 10.2166/wh.2012.014. [DOI] [PubMed] [Google Scholar]
- Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Qian P.Y. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS One. 2009;4(10):e7401. doi: 10.1371/journal.pone.0007401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO/UNICEF . (JMP) for Water Supply and Sanitation; Geneva, Switzerland: 2015. WHO/UNICEF Joint Monitoring Programme. [Google Scholar]
- Wingender J., Flemming H.C. Biofilms in drinking water and their role as reservoir for pathogens. Int. J. Hyg Environ. Health. 2011;214(6):417–423. doi: 10.1016/j.ijheh.2011.05.009. [DOI] [PubMed] [Google Scholar]
- Wright J., Gundry S., Conroy R. Household drinking water in developing countries: a systematic review of microbiological contamination between source and point-of-use. Trop. Med. Int. Health. 2004;9(1):106–117. doi: 10.1046/j.1365-3156.2003.01160.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.