ABSTRACT
Muscle precursors need to be correctly positioned during embryonic development for proper body movement. In zebrafish, a subset of hypaxial muscle precursors from the anterior somites undergo long-range migration, moving away from the trunk in three streams to form muscles in distal locations such as the fin. We mapped long-distance muscle precursor migrations with unprecedented resolution using live imaging. We identified conserved genes necessary for normal precursor motility (six1a, six1b, six4a, six4b and met). These genes are required for movement away from somites and later to partition two muscles within the fin bud. During normal development, the middle muscle precursor stream initially populates the fin bud, then the remainder of this stream contributes to the posterior hypaxial muscle. When we block fin bud development by impairing retinoic acid synthesis or Fgfr function, the entire stream contributes to the posterior hypaxial muscle indicating that muscle precursors are not committed to the fin during migration. Our findings demonstrate a conserved muscle precursor motility pathway, identify dynamic cell movements that generate posterior hypaxial and fin muscles, and demonstrate flexibility in muscle precursor fates.
KEY WORDS: C-met, Skeletal muscle, Sternohyoideus, Lateral line, Limb, Posterior hypaxial muscle, Zebrafish, Six1, Six4
Summary: In vivo examination of long-distance migrations of zebrafish muscle precursors reveals how these cells are specified prior to migration and organized during migration to generate four hypaxial muscles.
INTRODUCTION
Long-range cell migrations are vital to producing the vertebrate body plan. Migratory muscle precursors (MMPs) are specified in the trunk and then migrate to populate hypaxial skeletal muscles including the limb, tongue, neck, chest and diaphragm (Vasyutina and Birchmeier, 2006). We refer to these collectively as ‘MMP-derived muscles’. Mammalian MMPs are specified within the trunk dermomyotome by transcription factors, including Pax3, Six1, Six4, and the Hox genes (Alvares et al., 2003; Bober et al., 1994; Grifone et al., 2005). After specification, MMPs detach from the somite and migrate to their destinations. The limb bud attracts MMPs (Chevallier et al., 1977; Christ and Brand-Saberi, 2004; Christ et al., 1977; Dietrich et al., 1998; Hayashi and Ozawa, 1995) and is required for formation of pectoral girdle and pelvic girdle muscles (Evans et al., 2006; Masyuk et al., 2014; Valasek et al., 2011). Other cues may guide MMPs to non-limb bud locations; for example, occipital MMPs generate a subset of neck and tongue muscles (Dietrich, 1999). The occipital MMPs form far from the limb bud in somites 1-5 (Adachi et al., 2018; Huang et al., 1999; Lours-Calet et al., 2014; Mackenzie et al., 1998; Parada et al., 2012) and do not respond to several molecules that guide limb-level MMPs (Lours-Calet et al., 2014). In zebrafish embryos, three distinct MMP streams originate in somites 1-5, with the second stream populating the fin bud (Haines et al., 2004; Minchin et al., 2013; Neyt et al., 2000). Because all zebrafish MMPs originate near the fin bud, it was unclear whether zebrafish MMPs are guided primarily by fin bud-derived signals, by fin bud-independent mechanisms, or both. Here, we investigate the molecular mechanisms that specify MMPs, facilitate MMP motility, and organize MMP streams in zebrafish.
In mammals, two homeobox genes, Six1 and Six4, are required for formation of the hypaxial dermomyotomal lip, the structure that generates MMPs, and are broadly required for MMP-derived muscle formation (Grifone et al., 2005, 2007; Laclef et al., 2003; Ozaki et al., 2001). Six1 and Six4 are expressed in all muscle precursors and are required for expression of at least three MMP-specification genes: Met, Pax3 and Lbx1 (Grifone et al., 2005). Lbx1 encodes a homeodomain protein present in amniote MMPs (Dietrich, 1999; Mennerich et al., 1998) that is required for MMP migration into limbs and for tongue muscle size (Brohmann et al., 2000; Gross et al., 2000; Masselink et al., 2017; Schäfer and Braun, 1999). Pax3 is a paired homeobox-containing gene required for MMP migration in the mouse (Bober et al., 1994; Daston et al., 1996; Relaix, 2004). Met is a cell surface receptor expressed in MMPs that is essential for mammalian MMP migration (Bladt et al., 1995; Dietrich et al., 1999). The Met ligand, Hgf, is expressed along MMP migratory routes and is essential for MMP migration in mammals (Bladt et al., 1995; Dietrich et al., 1999; Scaal et al., 1999). Thus, Six1 and Six4 are upstream of several genes vital to MMP migration. However, because mammalian Six1 and Six4 function is required to generate the tissue that produces MMPs (Grifone et al., 2005, 2007), it remained unclear whether these genes function in MMPs during migration.
MMP-derived muscle defects are observed in zebrafish injected with a six1b morpholino (MO) (Lin et al., 2009; Nord et al., 2013), met MO (Haines et al., 2004), Hgf antibody (Haines et al., 2004), lbx1a MO, lbx2 MO (Ochi and Westerfield, 2009), or a combination of both pax3a and pax3b MOs (Minchin et al., 2013). These phenotypes resemble those in mouse knockouts and suggest that MMP specification pathways are conserved among bone-forming vertebrates. Early limb development is also conserved between fish and amniotes (Mercader, 2007). Fin bud specification in zebrafish begins during gastrulation, when retinoic acid (RA) induces the fin field (Grandel et al., 2002), a structure that will later develop into the fin bud. RA signaling indirectly activates Fgf signaling in the fin field, leading to fin bud formation (Cunningham et al., 2013; Gibert et al., 2006; Grandel and Brand, 2011). Similarly, in the mouse, RA and Fgf together induce the limb bud (Cunningham et al., 2013; Zhao et al., 2009). Within the zebrafish fin bud, Fgf signaling acts again to generate the fin apical ectodermal ridge, leading to fin bud outgrowth (Masselink et al., 2016; Mercader et al., 2006). RA is also required for sonic hedgehog (shh) expression in the fin field (Alexa et al., 2009). Hedgehog is required for fin bud patterning, but not for fin bud specification (Ahn and Ho, 2008; Neumann et al., 1999), similar to Hedgehog function in mammalian limb development (Robert and Lallemand, 2006). Thus, zebrafish appear to have well-conserved MMP-specification and fin-specification pathways, making them an excellent model to investigate how the fin bud influences MMP migration.
Our study focuses on the formation of four MMP-derived muscles in zebrafish. These muscles are, in anterior-to-posterior order, the sternohyoideus muscle (SHM), the abductor pectoral fin muscle (AbFM), the adductor pectoral fin muscle (AdFM) and the posterior hypaxial muscle (PHM) (Haines et al., 2004; Masselink et al., 2016; Minchin et al., 2013; Neyt et al., 2000; Ochi and Westerfield, 2009). These four muscles are predominantly composed of fast-twitch muscle fibers (Minchin et al., 2013; Patterson et al., 2008). The SHM is homologous to hypobranchial muscles in amniotes, which include the tongue and some neck muscles (Lours-Calet et al., 2014; Minchin et al., 2013; Okamoto et al., 2017). The muscles that lift (AbFM) and lower (AdFM) the pectoral fin are homologous to mammalian forelimb muscles (Mercader, 2007). The PHM is an early-forming ventral body wall muscle, located immediately posterior to the fin and positioned like a chest muscle (Haines et al., 2004; Windner et al., 2011). These four MMP-derived muscles form via three seemingly separate MMP streams (Haines et al., 2004; Masselink et al., 2016; Minchin et al., 2013; Neyt et al., 2000). Previous fate-mapping experiments indicated that somites 1-3 contribute to the SHM, somites 2-4 contribute to fin muscles, and somites 5 and 6 contribute to the PHM (Haines et al., 2004; Hollway and Currie, 2003; Li et al., 2017; Minchin et al., 2013; Neyt et al., 2000). In this work, we use high-resolution imaging to refine and clarify existing zebrafish MMP fate maps and migratory routes.
In this study, we investigate zebrafish MMP migration in vivo. As expected, the anterior and posterior MMP streams contribute to the SHM and PHM, respectively. We find that the middle (second) stream, previously shown to contribute to the AbFM and AdFM fin muscles (Haines et al., 2004; Masselink et al., 2016; Neyt et al., 2000), also contributes to the PHM. When we block fin bud formation, the precursors that would normally form fin muscles instead shift posteriorly and contribute to the PHM. The SHM and PHM streams eventually form even when fin field development is blocked, revealing that they do not require the fin bud. We show that the zebrafish homologs of Six1 and Six4 (six1a, six1b, six4a and six4b) are redundantly required for cell motility in all MMP streams, acting upstream of met. The six1/six4 genes and met also function in the fin bud, where they are required to partition fin bud MMPs into discrete AbFM and AdFM populations. Together, these findings demonstrate a conserved pathway for MMP specification, reveal flexibility in MMP movements, and show how a contiguous precursor population in anterior somites generates four separate muscles.
RESULTS
MMP behaviors suggest they are actively guided along their migratory routes
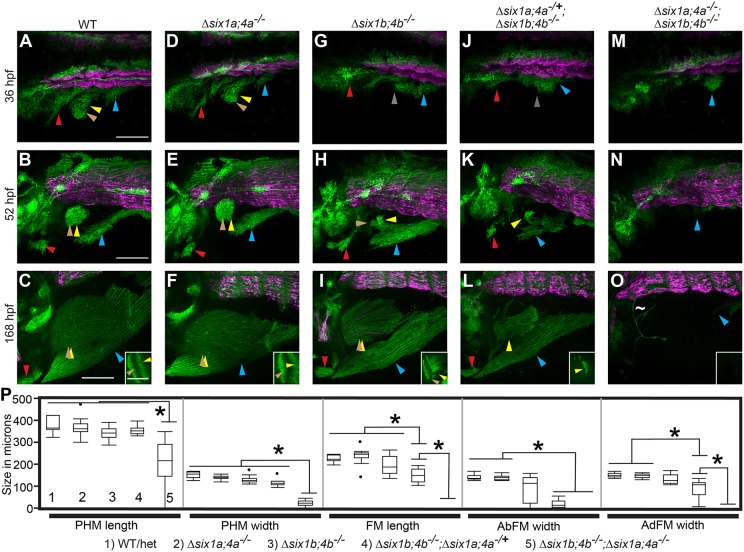
Although previous studies have investigated MMP migration in zebrafish, none examined how the streams form and interact with one another during migration and differentiation. To visualize muscle migration at high resolution, we generated a BAC transgenic line TgBAC(six1b:lyn-GFP)oz5 (hereafter: six1b:lyn-GFP), which expresses membrane-tethered GFP in muscle precursors and mature muscle cells (Fig. 1). This line also labels other six1b-expressing tissues, including the ear and lateral line (Fig. 1, Fig. S1). To distinguish myoblast precursors from maturing fast muscle fibers, we combined the six1b:lyn-GFP transgene with a fast muscle-specific transgene, Tg(mylpfa:mCherry)cz3327 (hereafter: mylpfa:mCherry) (Ignatius et al., 2012). Using these lines, we followed MMP migration and differentiation in fixed samples (Fig. 1A-D′) and in time-lapse of live embryos (Fig. 1E-I′, Movies 1-3). The three MMP streams form between 24 and 36 hours post-fertilization (hpf) (Fig. 1A-B′). As expected, the first stream, emanating from somites 1-3, generates the SHM (Fig. 1A-E, Movie 1). The second stream, which emanates primarily from somite 4, generates the AbFM and AdFM pectoral fin muscles (Fig. 1A-E, Movies 1-3). At 24 hpf, a few cells that will give rise to the AbFM are adjacent to somite 3 (Movie 1); it is presently unclear whether these cells originated in somite 3 or whether they originated in somite 4 and had already migrated adjacent to somite 3. After populating the fin bud, the remaining second stream MMPs alter their course and contribute to the anterior edge of the PHM (Fig. 1E′-I′, Movies 1, 2). Individual MMPs from somite 4 sometimes generate progeny that contribute to both the AdFM and PHM (green lines in Fig. 1E′, green dots in Movies 1, 2). The third stream, which emanates from somite 5, will form the posterior portion of the PHM (Fig. 1E-I′, Movie 1). Later, somite 6 produces ventral extensions that connect the posterior end of the PHM to the trunk (Fig. 1D,D′, Movie 1). The dynamic direction of MMP movement suggests that MMPs may respond to guidance cues during migration. At high resolution, long filopodia extend and retract from every MMP (Movie 3) and resemble filopodia that are specialized to detect guidance cues (Fairchild and Barna, 2014). These filopodia make contact between adjacent streams (Movie 3), suggesting that MMP streams coordinate their movements. Thus, our findings map MMP migration and MMP-derived muscle formation at cellular resolution, suggest that cells are actively guided to their destinations, and reveal that somite 4 MMPs contribute to both fin and PHM.
Fig. 1.
Mapping MMP dynamics during migration. (A-D) MMPs are visualized using the transgenic marker six1b:lyn-GFP (green) and the fast muscle marker mylpfa:mCherry (magenta), fixed at the onset of streaming (24 hpf) (A), during migration (36 hpf) (B), prior to MMP differentiation (48 hpf) (C) and when muscle differentiation is well underway (76 hpf) (D). The inset in A is brightened to compensate for dim transgene expression at 24 hpf. (A′-D′) Schematics of MMP migratory patterns, with a color code depicting MMPs that will contribute to more than one muscle (dark gray), or to SHM (red), AbFM (brown), AdFM (yellow), or the portion of the PHM formed by MMPs from somites 4 and 5 (light blue). Posterior to somite 5 (dark blue), PHM fibers arise via short-range migration and abide by somite boundaries. (E,E′) Stills from a time-lapse (Movie 1) of six1b:lyn-GFP-expressing MMPs, overlaid with cell tracks. Track colors are based on which muscle the cells will eventually contribute to the SHM (red), AbFM (brown), AdFM (yellow), PHM (blue), or to both PHM and AdFM (green). Starting cell positions are marked with squares, current cell positions are marked with dots and intermediate positions are tracked with lines. White lines indicate somite boundaries. (E) Tracks to 36 hpf, showing contributions to SHM and fin muscles. (E′) A later time frame (42 hpf), with cell tracks showing that a single cell (green) can contribute to both the AdFM and the PHM. (F-I) Stills from a time-lapse of six1b:lyn-GFP-expressing MMPs (Movie 2) show that the second MMP stream produces the AbFM and AdFM and contributes to the PHM. (F′-I′) Schematics of time-lapse stills. In all figures and schematics, colored arrowheads and shading indicate different streams and the muscles they form. Dark gray indicates MMPs that will contribute to more than one muscle, red indicates the SHM and its precursors, brown indicates the AbFM and its precursors, yellow indicates the AdFM and its precursors, and light blue indicates the PHM and its precursors. Scale bars: 100 µm (in A for A-D; in E for E,E′; in F for F-I).
MMP-derived muscle formation requires six1a, six4a, six1b and six4b gene function
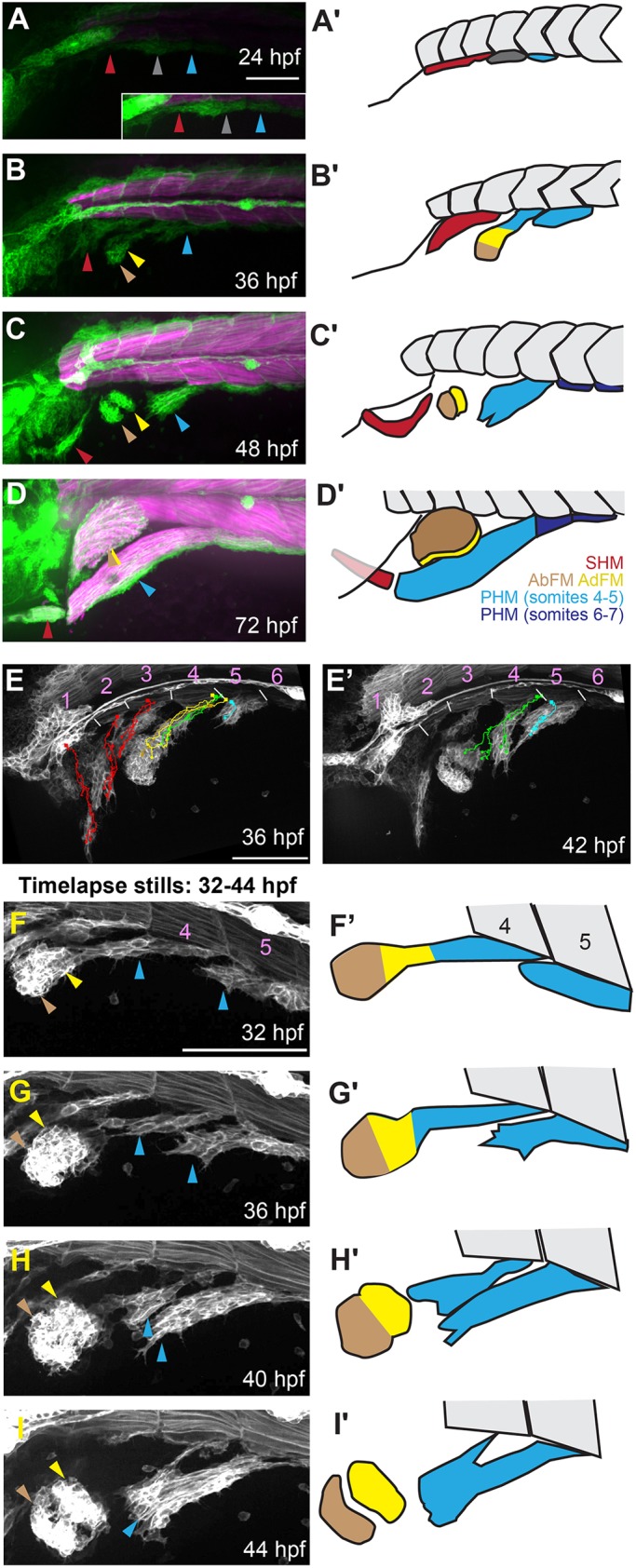
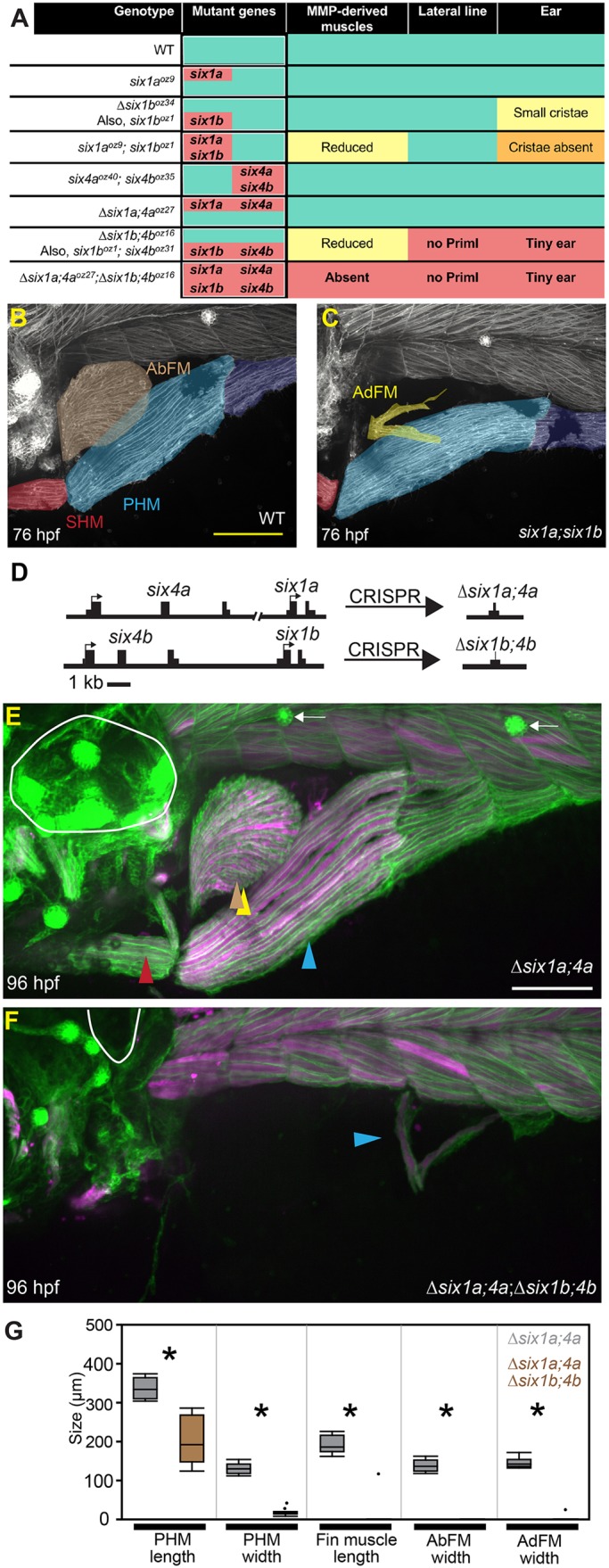
To begin investigating the genetic basis of MMP migration, we re-examined the function of Six family genes. Morpholino-knockdown of six1b causes loss of MMP-derived muscles (Lin et al., 2009; Nord et al., 2013). Surprisingly, when we characterized six1b mutants, we found no muscle defects (Fig. 2A, Fig. S2). We are confident that the zebrafish six1b lesions are null, because the frameshifting allele (six1boz1) and the deletion allele that eliminates the coding sequence (Δsix1boz34) give the same phenotype. The six1b mutants show ear cristae defects that resemble cristae defects in the Six1 hypomorphic mouse (Bosman et al., 2009), suggesting that six1b null alleles only partially eliminate total Six1 function. Embryos double mutant for both zebrafish six1 genes (six1a−/−;six1b−/−) have stronger ear defects and partial loss of MMP-derived muscles (Fig. 2A-C, Fig. S2), consistent with phenotypes in the mouse Six1 knockout (Laclef et al., 2003; Zhang et al., 2017). MMP-derived muscles are lost in the mouse Six1;Six4 double mutant (Grifone et al., 2005), so we knocked out all four zebrafish six1 and six4 genes (six1a, six1b, six4a and six4b) (Fig. 2A). Through combinatorial mutant analysis, we determined that all four genes participate in MMP patterning (Fig. 2A). We mainly used deletion alleles that remove the two bi-gene clusters: six1a-six4a on chromosome 13 and six1b-six4b on chromosome 20 (Fig. 2D). These deficiency alleles are officially named Df(Chr13:six1a,six4a)oz27 and Df(Chr20:six1b,six4b)oz16, and as Δsix1a;4a and Δsix1b;4b for simplicity. Δsix1a;4a;Δsix1b;4b mutant embryos lack nearly all MMP-derived muscle fibers (Fig. 2E,F). The AbFM, AdFM and SHM muscles are consistently absent and the PHM muscle is consistently reduced to a few short muscle fibers (Fig. 2E-G). Despite the severe MMP-derived muscle defects, larval trunk muscle appears normal in Δsix1a;4a;Δsix1b;4b mutants (Fig. 2F, Fig. S3A-L). Δsix1b;4b mutants and Δsix1a;4a;Δsix1b;4b mutants also have severe ear and lateral line defects (Fig. 2A,F, Fig. S3E-T), consistent with sensory defects in the mouse Six1;Six4 mutant (Moody and LaMantia, 2015). These findings indicate that the redundant requirement for Six1 and Six4 family gene function in MMP-derived muscles and sensory tissues is conserved in zebrafish and mouse.
Fig. 2.
six1 and six4 genes are required for proper development of MMP-derived muscles and sensory tissues. (A) Summary of six1 and six4 mutants generated for this study and their phenotypes. Green shading indicates normal phenotype that is indistinguishable from wild type (WT). Salmon shading indicates mutant genotypes and severe phenotypic defects. Milder defects are indicated by yellow or orange shading. ‘No PrimI’ indicates absence of the early-forming lateral line. (B,C) Three-day-old embryos labeled using six1b:lyn-GFP (white), false-colored to highlight different muscles. The SHM is red, the PHM is light blue anteriorly and dark blue posteriorly, and the AbFM is brown when present. The AbFM (brown), which covers the AdFM (yellow) in wild-type embryos (see B), is sometimes lost in six1a;six1b double mutants and in this example only a portion of the AdFM is present. (D) Diagram illustrating the six1-six4 genomic loci and Δsix1a;4a and Δsix1b;4b deletion alleles. (E,F) Four-day-old embryos carrying the six1b:lyn-GFP (green) and fast muscle mylpfa:mCherry (magenta) transgenes, showing morphology of the ear (white circle), trunk neuromasts, if present (white arrows), and MMP-derived muscles (arrowheads). (E) Δsix1a;4a mutants are indistinguishable from wild-type embryos. (F) In contrast, MMP-derived muscles are almost entirely absent when all four Six genes are deleted. Additionally, Δsix1a;4a;Δsix1b;4b mutants lack trunk neuromasts and have severe ear defects (also see Figs S2 and S3). (G) Box plots of 96 hpf Six mutant MMP-derived muscle measurements. For all plots, ∆six1a;4a values are shown in gray and ∆six1a;4a;∆six1b;4b values are shown in brown. Measurements were taken on 11 ∆six1a;4a and 11 ∆six1a;4a;∆six1b;4b individuals. Asterisks indicate significant differences (P<0.01), determined by Tukey–Kramer HSD comparisons after one-way ANOVA. See Materials and Methods for statistical details. Arrowheads are color-coded as described in Fig. 1 legend. Scale bars: 100 μm (in B for B,C; in E for E,F).
Hypaxial muscle precursors are only partially specified in Δsix1a;4a;Δsix1b;4b mutants
The severe Δsix1a;4a;Δsix1b;4b mutant MMP-derived muscle defect suggests that six1/six4 genes are required to generate normal MMPs. However, at the beginning of MMP migration (24 hpf), cells closely resembling MMPs are seen in Δsix1a;4a;Δsix1b;4b mutants (Fig. 3A,B), in the zebrafish equivalent of the hypaxial dermomyotomal lip (Devoto et al., 2006; Feng et al., 2006; Hammond et al., 2007; Stellabotte and Devoto, 2007). High-resolution time-lapse imaging at 30 hpf indicates that these cells have active filopodia but do not migrate (Movie 4). Consistent with a role for six1 and six4 genes in cell migration, we note that migration of the primary lateral line (PrimI) fails in Δsix1b;4b mutants and in Δsix1a;4a;Δsix1b;4b mutants (Movies 4, 5). As Δsix1a;4a;Δsix1b;4b mutants have MMP-like cells that extend filopodia, yet do not migrate, we hypothesize that these cells are only partially specified. The Δsix1a;4a;Δsix1b;4b mutant MMP-like cells also appear to have impaired cell proliferation and survival (Movies 4, 5), which likely contributes to the final phenotype, near-complete absence of MMP-derived muscle. In the mouse, Six1 and Six4 are redundantly required for activation of several MMP-specific genes, including Met, Pax3 and Lbx1 (Grifone et al., 2005). At the beginning of migration, zebrafish MMPs express pax3a, pax3b and lbx2 (Minchin et al., 2013; Ochi and Westerfield, 2009). In contrast, lbx1a and met expression is restricted to an MMP subset, with lbx1a expressed in MMPs at somites 1-4 and met expressed in MMPs at somites 4-5 (Fig. 3C-L, Fig. S4A-J). By 36 hpf, met is also expressed by MMPs in the first migratory stream (Figs S4M, S5), which emanates from somites 1-3. In Δsix1a;4a;Δsix1b;4b mutant embryos, MMP-like cells express lbx2, pax3a and pax3b, but not met or lbx1a (Fig. 3C-L, Fig. S4A-J), even at later times when MMP migration would normally be well underway (36 hpf) (Fig. S4K-N). Thus, we propose that zebrafish six1 and six4 genes stimulate precursor migration by activating a subset of MMP genes.
Fig. 3.
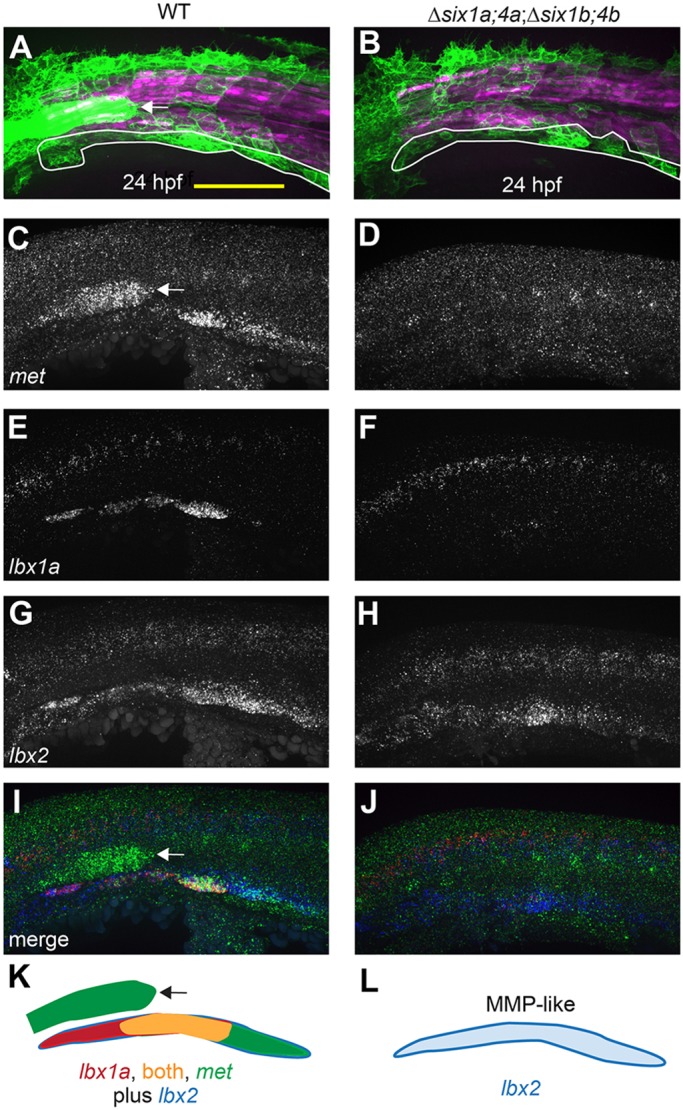
six1/six4 gene function is required to fully specify MMPs. (A,B) Muscle fibers and MMPs, visualized at 24 hpf using six1b:lyn-GFP (green) and mylpfa:mCherry (magenta) transgenes. Prior to MMP migration onset, Δsix1a;4a;Δsix1b;4b mutant trunk muscle appears normal (also see Fig. S3) and MMP-like cells are positioned at the ventral edge of the somite (white outline). (C-J) Fluorescent in situ hybridization showing met (C,D), lbx1a (E,F), lbx2 (G,H), or overlay of met (green), lbx1a (red), and lbx2 (blue) (I,J), at 24 hpf. (K,L) Schematics of gene expression patterns in wild-type (K) and Δsix1a;4a;Δsix1b;4b mutant (L) MMPs at 24 hpf. In 24 hpf wild-type embryos, lbx2 (blue) is broadly expressed whereas lbx1a (red) is prominent in anterior MMPs and met (green) is prominent in posterior MMPs; however, by 36 hpf, met is expressed in all three streams (Fig. S4N, S6E). In contrast, lbx1a and met are not expressed in 24 hpf Δsix1a;4a;Δsix1b;4b mutants, but lbx2-positive MMP-like cells are present. six1b:lyn-GFP and met expression are also seen in the lateral line primordium (arrow in A,C,I,K) of wild type but not quadruple mutants, consistent with the loss of this structure in Δsix1b;4b and Δsix1a;4a;Δsix1b;4b mutants (see also Fig. 2A,F and Fig. S3G,H).Yellow shading represents expression of both lbx1a and met. Scale bar: 100 µm.
The Six1 and Six4 family target gene met is essential for normal MMP migration
We hypothesized that Δsix1a;4a;Δsix1b;4b mutant phenotypes may be explained at least in part by the loss of met expression, consistent with previous mouse Met mutant and zebrafish met knockdown analyses (Bladt et al., 1995; Haines et al., 2004). To block Met function during MMP migration, but after MMP formation, we treated embryos with a potent and specific Met protein inhibitor, SGX523 (Buchanan et al., 2009), at the onset of MMP streaming (24 hpf). SGX523 treatment causes delayed MMP streaming (Fig. 4A-D, Movie 6), and reduced size of all MMP-derived muscles, especially the AbFM (Fig. 4E,F). We also generated a frame-shifting met allele, and show that, like in SGX523-treated embryos, met mutant MMP-derived muscles are shortened (Fig. 4G-K) and the AbFM is especially affected (Fig. 4G,H inset, and K). Similarly, by examining an ‘allelic series’ of Δsix1a;six4a and Δsix1b;six4b heterozygous and mutant combinations, we find that progressive loss of six1 and six4 genes causes delayed MMP streaming, reduced size of all three MMP streams, and reduced size of all MMP-derived muscles (Fig. 5, Figs S2, S5, Movie 5). Furthermore, like met mutants, the AbFM is more sensitive to partial loss of six1/six4 gene function than the AdFM (Fig. 5K,L,P, Fig. S2D,H). These findings support our model that six1 and six4 genes activate MMP migration in part by activating met expression.
Fig. 4.
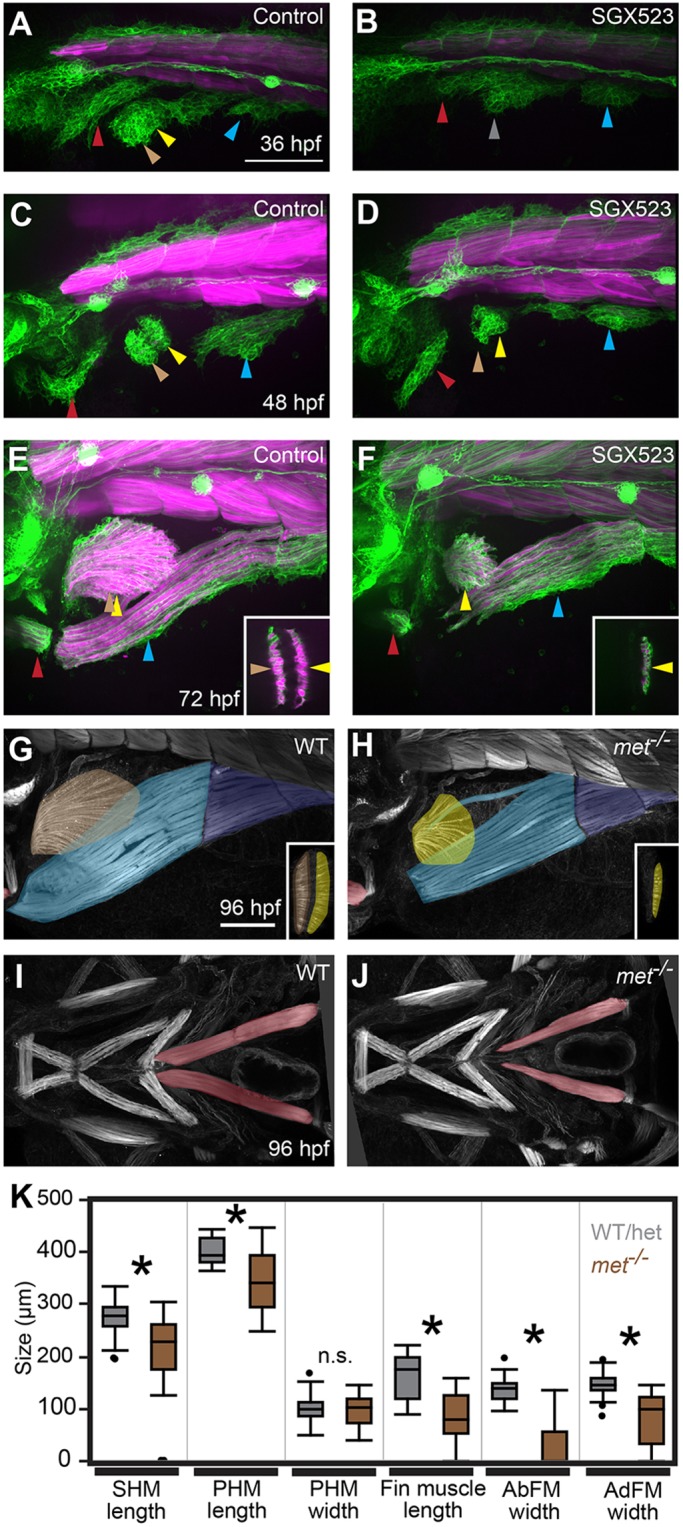
met is required for normal MMP migration. (A-F) Confocal projections of six1b:lyn-GFP (green) and mylpfa:mCherry (magenta) transgene expression in embryos fixed at indicated stages. Compared with DMSO-treated controls, treatment with the Met inhibitor SGX523 causes reduced migratory streams at 36 and 48 hpf and smaller MMP-derived muscles at 72 hpf. (G-J) Confocal projections of phalloidin-labeled wild-type and met mutant embryos at 96 hpf, false-colored to show muscle identity using color code described in Fig. 1. The fin and PHM muscles are shown in lateral view in G and H and the more anteriorly located SHM is shown in ventral view in I and J. Insets in E-H show confocal sections through the fin to distinguish AbFM and AdFM muscles. In met mutants and SGX523-treated embryos, the AbFM is more severely affected than the AdFM and in this example the AbFM is lost entirely. (K) Box plots of 96 hpf wild-type and met mutant MMP-derived muscle measurements. For all plots, WT/Het values are shown in gray and met mutant values are shown in brown. Asterisks indicate significant differences (P<0.01), determined by Tukey–Kramer HSD comparisons after one-way ANOVA; n.s. indicates not significant. See Materials and Methods for statistical details. Measurements were taken on a total of 31 mutants and 29 WT/Het siblings from three separate experiments. Arrowheads are as described in Fig. 1 legend. Scale bars: 100 µm.
Fig. 5.
six1/six4 genes function in a largely redundant fashion for normal MMP migration. (A-O) Confocal projections showing six1b:lyn-GFP (green) and slow muscle smyhc1:lyn-tdTomato (magenta) expression in embryos fixed at the indicated time-points. (A-F) Wild-type (A-C) and Δsix1a;4a homozygote (D-F) MMP streams and MMP-derived muscles are similar at all time points. (G-L) MMP streams and MMP-derived muscles are moderately reduced in Δsix1b;4b homozygotes (G-I) and further reduced in Δsix1b;4b homozygotes that are heterozygous for the Δsix1a;4a deficiency (J-L). (M-O) MMP streams and almost all MMP-derived muscle fibers are absent Δsix1a;4aΔsix1b;4b mutant homozygotes. The few GFP-positive cells within the fin of Δsix1a;4a;Δsix1b;4b mutants do not have muscle fiber morphology (tilde in panel O). Insets in C,F,I,L,O show a single z-section through the fin to distinguish the AbFM and AdFM muscles. (P) Box plots showing measurements of muscles at 7 days post-fertilization; asterisks indicate significant differences (P<0.01) as determined by Tukey–Kramer HSD comparisons after one-way ANOVA. For each measurement, genotypes are labeled 1-5 (see key under the plot), with 1 indicating wild type and 5 indicating Δsix1a;4a;Δsix1b;4b mutant. n=12 embryos measured per group. See Materials and Methods for statistical details. Arrowheads are color-coded as described in Fig. 1. Scale bars: 100 µm.
Smoothened function is required for normal AbFM formation
Like AbFM/AdFM separation defects in met mutant zebrafish, limb abductor and adductor streams fail to partition in the Smo mutant mouse (Anderson et al., 2012). To test whether Smoothened function is required for AbFM/AdFM separation in fish, we treated embryos with the Smoothened inhibitor cyclopamine beginning at the onset of MMP migration, 24 hpf. During normal development, MMPs have begun to partition within the fin bud by 36 hpf (Fig. 6A); however, partitioning is impaired in cyclopamine-treated embryos (Fig. 6B, Movie 7). The fin muscle defect becomes more pronounced by 48 hpf in cyclopamine-treated embryos, in which AbFM precursors are reduced compared with AdFM precursors (Fig. 6C,D), similar to met mutants. We thus reasoned that Smoothened might control AbFM/AdFM separation by activating Hgf/Met signaling. Consistent with this idea, we find that the fin buds that form in cyclopamine-treated embryos lack expression of hgfa (Fig. 6E,F) at 36 hpf. Thus, the AbFM/AdFM separation defect in Smoothened-inhibited embryos may be explained by a fin bud-specific loss of Hgf/Met signaling.
Fig. 6.
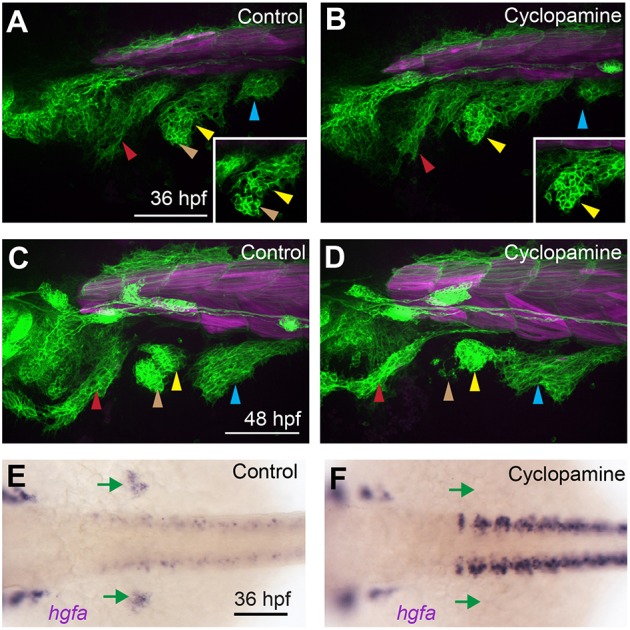
Smo function is required during migration for AbFM/AdFM partitioning. (A-D) Confocal projections of six1b:lyn-GFP (green) and mylpfa:mCherry (magenta) transgene expression in embryos fixed at 36 hpf (A,B) or 48 hpf (C,D). When embryos are treated with cyclopamine beginning at 24 hpf, fin buds form and become populated with MMPs; however, few MMPs contribute to the AbFM. At 36 hpf (A,B), we show an individual confocal plane (inset) that highlights defective AbFM/AdFM partitioning. (E,F) Embryos treated with cyclopamine from 24-36 hpf conspicuously lack hgfa expression in the fin bud domain (green arrows). Arrowheads are as described in Fig. 1 legend. Scale bars: 100 µm.
When the fin bud is absent, the second MMP stream contributes exclusively to the PHM
MMPs enter the fin bud in met mutant embryos and Smoothened-inhibited embryos (Fig. 6), suggesting that other molecules, such as Fgfs, guide and/or recruit MMPs to the fin bud. To investigate how the fin bud influences MMP movements, we blocked fin bud formation by treating embryos with the Fgf/Vegf inhibitor SU5402 at 24 hpf, after MMP formation. Even though fin bud formation is blocked, second stream MMPs initially extend anteriorly towards the area where the fin bud would normally form; later, they change direction and join the third stream (Fig. 7A-E, Movie 8). We narrowed the critical window for Fgf/Vegf function to 24-36 hpf (Fig. 7B,D,E), the period during which the fin bud usually forms and becomes populated with MMPs. To confirm that phenotypes after SU5402 treatment reflect loss of Fgf (not Vegf) signaling, we activated a heat-shock-inducible dominant negative Fgf receptor (dnFgfr) at 24 hpf using the Tg(hsp70l:dnFgfr1-GFP)pd1 transgenic line. As expected, dnFgfr induction at 24 hpf results in pectoral fin loss (Fig. S5A-C), loss of fin muscle, and an expanded PHM stream (Fig. 7F,G). By 72 hpf, the length of the PHM in wild-type and dnFgfr-expressing embryos is equivalent, but PHM girth is larger in wild-type than dnFgfr-expressing embryos (Fig. 7H,I). Similarly, by 72 hpf, the PHM of SU5402-treated embryos is narrower than controls (not shown). Thus, although the PHM stream transiently expands owing to addition of extra MMPs, this expansion does not increase the size of the PHM after myofiber formation. These findings show that Fgfr function is not required for MMP migration and suggest that the fin bud recruits early-migrating second stream MMPs, preventing them from contributing to the PHM.
Fig. 7.
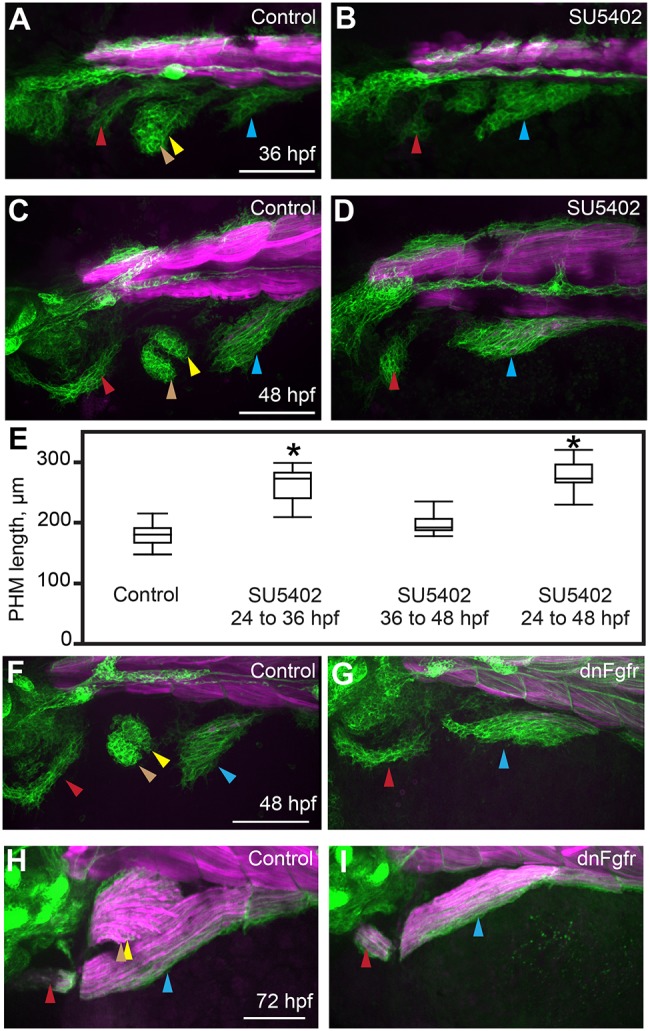
In the absence of the fin bud, the second migratory stream contributes exclusively to the PHM. (A-D) Confocal projections of six1b:lyn-GFP (green) and mylpfa:mCherry (magenta) transgene expression in embryos fixed at 36 or 48 hpf. These embryos were treated either with DMSO (control; A,C) or SU5402 (B,D) from 24 to 36 hpf. (E) Box plots of PHM length in control or SU5402-treated embryos, fixed at 48 hpf, showing that the PHM expands upon Fgfr inhibition. Asterisks indicate significant differences (P<0.01), determined by Tukey–Kramer HSD comparisons after one-way ANOVA. Measurements were obtained from 15 control embryos and 10 inhibitor-treated embryos for each experimental condition. See Materials and Methods for statistical details. (F,G) Similar to Fgfr chemical inhibition, dnFgfr induction (at 24 hpf) results in an initially expanded PHM stream. (H,I) By 72 hpf, the PHM in control wild-type embryos is larger than in dnFgfr-induced embryos. Arrowheads are color-coded as described in Fig. 1 legend. Scale bars: 100 µm.
MMPs are specified and migrate in the absence of a fin field
Surprisingly, MMPs transiently visit the fin-forming region when we block fin bud development. We hypothesized that MMP movements might be influenced by the fin field, an early-forming precursor population that will later become the fin bud. To test this idea, we examined MMP migration in aldh1a2 mutants (Fig. 8), which are deficient in RA synthesis, lack pectoral fin fields, and ultimately fail to form pectoral fins (Fig. S5D, supplementary Materials and Methods; Grandel et al., 2002). At 36 and 48 hpf, six1b-expressing MMPs are present in two small streams in aldh1a2 mutants (Fig. 8A-D) and express the MMP markers lbx1a and met (Fig. S5E-H). The aldh1a2 mutant SHM- and PHM-progenitor streams grow slowly (Fig. 8C-H), and eventually give rise to small SHM and PHM muscles (Fig. 8G,H). The MMPs that emanate from somite 4 in aldh1a2 mutants, which in wild-type embryos would contribute to pectoral fin and PHM muscles, appear to contribute to the PHM. These findings support the hypothesis that the fin field is not required for MMP specification and show a role for aldh1a2 in MMP stream growth.
Fig. 8.
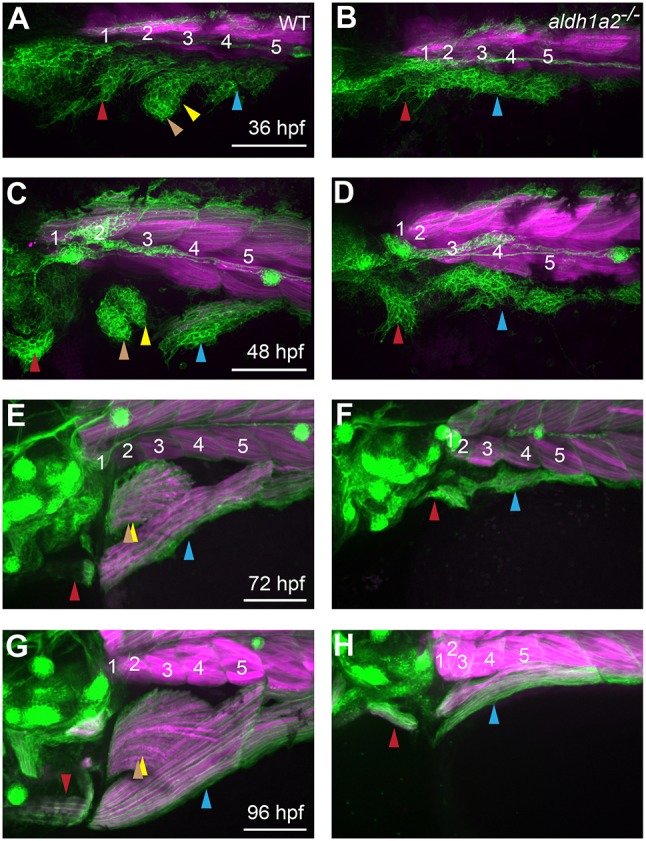
aldh1a2 function is essential for normal MMP migration. (A-H) Expression of six1b:lyn-GFP (green) and mylpfa:mCherry (magenta) in embryos fixed at indicated stages. Compared with wild-type siblings, aldh1a2 mutants have severe MMP migratory defects (A-D), leading to lack of fin muscle formation and reduced SHM and PHM muscles after fiber formation (E-H). Somite numbers are shown in white. Arrowheads are color-coded as described in Fig. 1 legend. Scale bars: 100 µm.
DISCUSSION
Our work has mapped MMP movements over time at single cell resolution in the living embryo and supports a new working model for the control of MMP migration during development. Using time-lapse analysis, we clarified MMP migratory routes. By examining the effect of the fin bud on MMP migratory routes, we identified flexibility between fin muscle and PHM fates. We show that embryos mutant for all six1/six4 genes (six1a, six1b, six4a and six4b) form partially specified MMP-like cells that are unable to migrate, indicating that MMP movements require six1/six4 function. Consistent with previous mammalian findings (Grifone et al., 2005), we show that six1/six4 genes are essential for expression of met and lbx1a. However, in contrast to mouse (Grifone et al., 2005), zebrafish pax3a and pax3b expression does not require six1/six4 function. As expected, the six1/six4 target gene met is essential for normal MMP migration. Although all MMP streams are reduced, some MMPs do, surprisingly, migrate into the fin bud in the zebrafish met mutant. Within the fin bud, met function is required for normal fin muscle formation, especially for the AbFM. Below, we integrate our findings with other studies to offer a new view of MMP migration.
Our time-lapse analyses reveal similarities and differences in some MMP stream origins and fates compared with previous fate-mapping studies that indicated that MMPs from somites 1-3 give rise to the SHM (Minchin et al., 2013), from somites 2-4 to fin muscle (Minchin et al., 2013; Neyt et al., 2000), and from somites 5-6 to the PHM (Neyt et al., 2000) (reviewed by Li et al., 2017). Using time-lapse imaging beginning just prior to MMP migration, we find that MMPs from somites 1-3 give rise to the SHM, somites 3-4 to fin muscle, somites 4-5 to the PHM, and somite 6 and possibly somite 7 contribute to the portion of PHM that connects to myotome either by short-range migration or myotomal extension. Thus, our time-lapse imaging is largely consistent with and extends previous fate-mapping studies, with some differences. For example, we did not observe MMPs from somite 2 cells contributing to fin muscle. Importantly, like Neyt et al. (2000), we confirm that MMPs from somite 4 are the major source of fin muscle. Notably, we find that the second MMP stream, which emanates from somite 4, contributes to three muscles. Second stream MMPs initially populate the AbFM and AdFM fin muscles whereas later-migrating second stream MMPs contribute to the PHM, a previously unrecognized contribution. The slight differences among previous and current work may be due to the different experimental methodologies employed. It is also possible that MMPs can originate in one somite but exit from another. Future work using 3D tracking of all cells in the streams may provide additional insight into MMP origins and cell dynamics during migration.
Using cell tracking, we observed that a single migrating MMP can generate daughters that contribute to different muscles (fin and PHM), showing that MMP fates are not fixed before migration. Thus, although MMP position is a strong indicator of future muscle fate, MMPs retain some plasticity. During normal development, somite 4 MMPs contribute to the fin muscles and to the PHM. However, when fin field or fin bud development is blocked genetically or with signaling inhibitors, all MMPs from the fourth somite default to the PHM fate. For example, in aldh1a2 mutants, which lack the fin field, somite 4 MMPs appear to contribute solely to the PHM. When we block fin bud formation late (e.g. by SU5402 treatment at 24 hpf), somite 4 MMPs move into the fin-forming region, but ultimately contribute to an expanded PHM stream. In wild-type embryos, in which somite 4 MMPs contribute first to the limb muscles and then to the PHM, this shift may be controlled by temporal cues rather than by absolute MMP cell number. We find that somite 4 MMPs shift their migration to the PHM at about the same time even in Δsix1b;4b mutant and Met-inhibited embryos, which populate the fin bud with fewer MMPs than in wild-type embryos. Thus, we propose that MMP guidance cues dynamically change with developmental time, first attracting MMPs to the fin bud and later inactivating or overriding those cues to enable PHM formation.
In addition to fin field- and fin bud-derived guidance cues, our findings also suggest that cell-intrinsic processes could influence MMP stream identity. Prior to migration, we find that lbx1a and met are expressed in a partially overlapping fashion that roughly corresponds to the three MMP streams (Fig. 3). In limb bud-deficient chicken embryos and fin field-deficient medaka embryos, limb-level Lbx1 is still expressed, indicating that the limb is not required for MMP specification (Alvares et al., 2003; Tani-Matsuhana et al., 2018). However, these studies did not test whether Lbx1-positive cells ultimately contribute to muscle in the absence of limb buds. We show that lbx1-positive, met-positive cells are present in zebrafish embryos that lack the fin field. We also show that these cells are true MMPs and that they migrate to produce SHM and PHM muscles. Similarly, in fin field-deficient tbx5a mutants, two MMP-derived muscles, the SHM and PHM, are present (Valasek et al., 2011). This idea is further supported by work in Xenopus laevis tadpoles, which form SHM-like and PHM-like muscles several days before limb bud formation (Martin and Harland, 2001, 2006). Because fin field-deficient zebrafish generate separate SHM and PHM streams, specification of these progenitor populations must not require the fin field. We hypothesize that before migration begins, cell-intrinsic processes distinguish the first stream from the second stream. This model explains why the second and third somites, which are close to the fin bud, generate MMPs that enter the first stream and populate the SHM, whereas the fin bud is populated by the second stream originating from the more distant somite 4. Further work, such as MMP transplantation between anterior somites, could be used to demonstrate the extent that intrinsic and extrinsic processes, and timing, control muscle formation.
It remains unclear which signals directly guide MMPs. Fgf has been described as a potential limb-derived guidance cue in some studies (Itoh et al., 1996; Masselink et al., 2017), but not others (Flanagan-Steet et al., 2000). We find that MMPs transiently visit the fin field when we block Fgf receptor function, indicating that signals other than Fgf draw MMPs toward the fin bud. As in mouse, our work implicates Hedgehog as a potential fin/limb-specific signal. When we inhibit the Hedgehog signal transducer Smoothened, the SHM, PHM and AdFM form, but AbFM formation is disrupted. In mammalian limbs, Hedgehog has also been proposed to guide MMPs and Smoothened is required for abductor and adductor stream separation (Anderson et al., 2012). We also find that the AbFM and AdFM fail to correctly partition in the fin bud of Smoothened-inhibited zebrafish embryos. In mouse, the adductor MMPs are particularly affected in Smo mutants, and distal muscle is lost (Anderson et al., 2012; Hu et al., 2012) whereas in zebrafish the abductor MMPs, which form distal to somites compared with the adductor muscle, are most affected. In both mouse and fish, the limb MMP populations that migrate most distally from the trunk are most sensitive to Smo loss, suggesting conserved function. The effect of Smoothened-inhibition could be explained by fin bud-specific loss of Hgf/Met signaling, or by effects outside the fin bud. Future experiments placing Hgf-soaked beads into the fin bud of Smoothened-inhibited embryos could test the effect of Hgf more directly. Another ligand/receptor pair, Sdf1 (Cxcl12)/Cxcr4, is required to guide MMPs from the limb to the amniote chest (Griffin et al., 2010; Masyuk et al., 2014). Sdf1/Cxcr4 could also potentially control some MMP movements in zebrafish, such as the movement of second stream MMPs into the PHM. Another ligand/receptor pair EphrinA5/EphA4 has been proposed to repulse MMPs from non-myogenic portions of the limb in chick (Swartz et al., 2001). Future work will identify molecules that directly guide zebrafish MMPs to their destinations.
Our findings expand the role of Six1 and Six4 in vertebrate muscle formation. In mouse, Six1/Six4 function is necessary and sufficient for fast-twitch muscle specification (Grifone et al., 2004; Niro et al., 2010; Richard et al., 2011; Wu et al., 2013); and zebrafish six1a and six1b morpholino studies showed a similar result (Bessarab et al., 2008; O′Brien et al., 2014). However, we show here that although deletion of all four six1 and six4 genes (Δsix1a;4a;Δsix1b;4b mutants) severely affects MMPs, and thus fast-twitch MMP-derived muscles, fast muscle in the trunk forms normally. This observation suggests that other genes specify fast-twitch trunk muscles in zebrafish. Similar to mammalian studies (Grifone et al., 2005, 2007), we find that zebrafish six1/six4 genes are required for MMP migration, which happens well before fiber type differentiation. In zebrafish Δsix1a;4a;Δsix1b;4b mutants, MMP-like cells express pax3a, pax3b and lbx2, but not met or lbx1a, and thus are partially specified. Because Δsix1a;4a;Δsix1b;4b mutant MMP-like cells fail to migrate, we propose that zebrafish six1/six4 genes are needed to transition hypaxial muscle precursors from a non-migratory state to a migratory state (MMPs). We hypothesize that six1/six4 genes activate MMP motility in part by activating met, a gene thought to promote MMP motility (Birchmeier et al., 2003; Dietrich et al., 1999; Haines et al., 2004; present study), and in part by activating lbx1a. In the mouse, Lbx1 is required for MMP migration into the limb bud and for normal tongue muscle size (Schäfer and Braun, 1999). six1/six4 genes may also act upstream of other conserved targets (in addition to met and lbx1a) that are important for MMP migration, such as cell guidance receptors. Future work to identify differentially expressed genes in wild-type versus MMP-like cells sorted from Δsix1a;4a;Δsix1b;4b mutants may identify new genes that are vital for MMP migration in both fish and mammals.
Our findings are consistent with previous work implicating Met in cell motility, rather than cell guidance (Birchmeier et al., 2003; Dietrich et al., 1999; Haines et al., 2004; present study). We propose that Met activates MMP cell motility in somites, initiating stream movement and then acts again in the fin bud to promote AbFM/AdFM separation. Because the Met ligand gene hgfa is expressed in the trunk and fin bud (see Fig. 6E), rather than along the entire migratory route, our findings suggest that there are two phases of Met activity. Additionally, we find that all MMP-derived muscles are reduced in met mutants, with the AbFM particularly affected. Time-lapse analysis of Met-inhibited embryos shows delayed streaming of all MMPs, followed by a failure to partition the AbFM from the AdFM within the fin bud. These Met-inhibited phenotypes are surprisingly subtle because Met is considered to be absolutely essential for MMP migration in the mouse limb (Bladt et al., 1995). The subtle defects could be explained if the met+13-3 lesion is not completely null, consistent with a previous study showing that met morphants have a more severe loss of hypaxial muscle (Haines et al., 2004) than what we observe in mutants. Despite the difference in severity, both mutants and morphants have partial loss of MMP-derived muscle (Haines et al., 2004; present study). Disparity between mutant and morphant phenotypes are sometimes explained by morpholino toxicity, maternal contribution, genetic compensation in the mutant, or other factors independent of whether the mutation is null (El-Brolosy et al., 2019; Kok et al., 2015; Rossi et al., 2015; Stainier et al., 2017). However, two lines of evidence suggest that the met+13-3 allele we use in our study is a strong loss-of-function allele. First, the lesion truncates Met prior to the transmembrane and intracellular domains; second, met mutants appear identical to SGX523 Met inhibitor-treated embryos. We suggest that the anterior origins of zebrafish MMPs may explain why met may be only partially required for MMP motility. In mammals, MMPs in the most anterior (occipital) somites do not require several genes, including Met, that are essential for migration into limb buds (Lours-Calet et al., 2014). For instance, in the Met mutant mouse, MMP migration into the limb and diaphragm fails entirely, but Met mutant MMPs do migrate a short distance from the most anterior (occipital) somites to form a subset of neck and tongue muscles (Bladt et al., 1995; Lours-Calet et al., 2014). Thus, we propose that zebrafish met is necessary for some, but not all, MMP motility because the zebrafish MMPs arise from anterior somites and, like amniote occipital MMPs, they are capable of short-range migration independently of met function.
Our findings on zebrafish MMP development have broader implications. Because the fin bud recruits MMPs from somite 4, which also contributes to the PHM, we suggest that these fin/PHM MMPs are partially naïve of their destination when they exit the somite. Such flexibility in development may have provided MMPs with plasticity in form during evolution. Further work is needed to understand how both cell-extrinsic and cell-intrinsic processes produce the vast array of MMP-derived muscles found in vertebrates. Such studies may also shed light on how other cell types migrate, such as the lateral line that we show requires six1b/six4b function for motility. MMP studies may also contribute to understanding of disease; for example, tumor cells undergo epithelial-mesenchymal transitions and migrate in response to SIX1 and MET overexpression (Birchmeier et al., 2003; Christensen et al., 2008; Micalizzi et al., 2009; Xu et al., 2014). Thus, our work on muscle precursor migration may have broad applications to the study of development, evolution and medicine.
MATERIALS AND METHODS
Fish maintenance, husbandry, genotyping and strains
Zebrafish transgenic and mutant strains were maintained on the AB wild-type background, except for the met+13-3 mutants and their control siblings, which were maintained on the TU wild-type background. Standard husbandry and stock maintenance procedures were followed (Westerfield, 2007). All experiments were conducted before zebrafish sex becomes apparent and were replicated on at least two clutches. All animal protocols are approved by the Ohio State University Institutional Animal Care and Use Committee (OSU IACUC) or, for the met mutant experiments, by the Monash University Animal Services Animal Ethics committee. The transgenic lines used are Tg(mylpfa:mCherry)cz3327 (Ignatius et al., 2012), Tg(hsp70l:dnfgfr1-GFP)pd1 (Lee et al., 2005), Tg(smyhc1:lyn-tdTomato)oz29 and Tg(six1b:lyn-GFP)oz5. Transgenic construction used established methods (Suster et al., 2011; Wang et al., 2011); see supplementary Materials and Methods for details. The mutant lines used are aldh1a2i26 (Begemann et al., 2001), six1aoz9 (Talbot and Amacher, 2014), met+13-3, six1boz1, Δsix1boz34, six4aoz40, six4boz31, six4boz35, Df(Chr13:six1a,six4a)oz27 and Df(Chr20:six1b,six4b)oz16. Fish were genotyped using PCR of extracted DNA, followed in some cases by restriction enzyme cleavage. See supplementary Materials and Methods for details on genotyping and mutant construction of the latter eight mutant lines (Talbot and Amacher, 2014; Xiao et al., 2013).
Probe construction and in situ hybridization
Fluorescent RNA in situ hybridization was performed using established protocols (Talbot et al., 2010), available in detail on ZFIN (https://wiki.zfin.org/x/0wHI). Non-fluorescent RNA in situ hybridization was also performed using established protocols (Jowett, 1999). We mounted embryos for imaging as described previously (Morrow et al., 2017). For all in situ experiments, to prevent pigment formation, embryos were treated with 0.003% N-phenylthiourea (PTU) beginning at 24 hpf. In situ hybridization, coloration and imaging was performed side-by-side for compared images. We used previously published probes for lbx1a (Lukowski et al., 2011), lbx2 (Ochi and Westerfield, 2009), hgfa (Haines et al., 2004), met (Haines et al., 2004), and pax3b (Minchin et al., 2013). We generated a new probe covering 1 kb of the pax3a transcript and find expression patterns matching those previously described (Minchin et al., 2013). The six4a probe was kindly provided by Lindsey Barske and Gage Crump (University of Southern California, CA, USA). We generated new probes to six1a, six1b and six4b, complementary to approximately 1 kb of each transcript and validated them by comparison to previously published Six family probes (Bessarab et al., 2004; Kobayashi et al., 2000; Nord et al., 2013); further details on probe construction are available in the supplementary Materials and Methods.
Immunohistochemistry and F-Actin labeling
Immunolabeling used antibodies Rbfox1l (1:500) (Berberoglu et al., 2017), F310 (1:100, Developmental Studies Hybridoma Bank) and A4.1025 (1:1000, Developmental Studies Hybridoma Bank). F-Actin was labeled using Alexa Fluor-conjugated Phalloidin (1:50, Thermo Fisher).
dnFgfr induction
To induce dnFgfr expression, embryos from an outcross of Tg(hsp70l:dnFgfr1-GFP)pd1 (Lee et al., 2005) were transferred to 37°C system water for 30 min. After heat shock, embryos were placed in dishes of 28.5°C system water containing 0.003% PTU to prevent pigment formation. Embryos carrying the Tg(hsp70l:dnFgfr-GFP)pd1 transgene were separated from non-transgenic sibling controls by expression of ubiquitous GFP after heat shock.
Chemical inhibitor treatment
For small molecule treatments, we transferred embryos to new dishes containing either the indicated concentration of chemical inhibitor or the equivalent concentration of solvent as the treatment control. Three protein inhibitors were used in this study: the Fgf/Vegf inhibitor SU5402 (Sun et al., 1999), the Smoothened inhibitor cyclopamine (Chen et al., 2002) and the Met inhibitor SGX523 (Buchanan et al., 2009). Working concentration was 100 µM, except for SU5402, which was 10 µM for Movie 8 and 17 µM for Fig. 7. For SU5402 24-36 hpf treatments (Fig. 7C-E), we transferred embryos to inhibitor-containing water at the beginning of the pulse, and then washed them rapidly and thoroughly in fresh system water at the end of the pulse. For all other inhibitor experiments, embryos were transferred into inhibitor at 24 hpf until fixation.
Imaging
Brightfield images were acquired using an upright Axioplan 2 microscope using Axiovision software. For the met/sibling images (Fig. 4G,H), we used a Zeiss LSM 710 Live Duo confocal microscope with Zen 2011 software. All other confocal images were collected using an inverted Nikon TiE microscope equipped with an Andor Revolution WD spinning disk confocal system. Images were analyzed using Metamorph software, and measurements were taken using ImageJ (Abràmoff et al., 2004). Cell tracking was performed manually using the MTrackJ plugin in ImageJ software (Meijering et al., 2012). All compared images were taken with the same image settings and processed identically. For time-lapse imaging, we mounted embryos in system water containing 0.1% agarose (GeneMate, E-3120-500) that had been boiled and cooled to 42°C before Tricaine was added (0.019% final concentration). Approximately 4 ml of the agarose solution was transferred to 35 mm glass-bottom Petri dishes (World Precision Instruments, FD35-100). Embryos were oriented in the dishes, and the agarose was allowed to set 1 hour before plates were imaged. During imaging, an Okogawa-heated stage and lens collar was used to maintain a temperature of 28.5°C. Petri dishes were covered with lids during imaging to minimize evaporation. Additional time-lapse details are provided in movie legends. For Movie 7, we labeled nuclei by injecting H2B-CFP mRNA into embryos at the one-cell stage (Megason, 2009).
Quantification and statistical analysis
Embryos were selected randomly for confocal imaging, and we included all imaged embryos in statistical analysis. We measured muscles as described in the supplementary Materials and Methods. All statistical analysis was performed using JMP v 11. Significance was defined using Tukey–Kramer Honestly Significantly Different (HSD) comparisons after ANOVA, with a significance threshold of <0.01. n represents the number of fish measured. Graphs of quantification represent data as box and whisker plots, with boxes showing the first through third quartiles, whiskers representing 1.5 times the interquartile range, and outlier data shown as dots.
Supplementary Material
Acknowledgements
We thank the Amacher lab fish facility staff for excellent fish husbandry and the OSU Neuroscience Imaging Core for equipment and advice. We thank the FishCore zebrafish facility (Australian Regenerative Medicine Institute, Monash University) for care and maintenance of the met+13-3 mutant line. We thank Zachary Morrow for imaging many of the embryos shown in Fig. S4 and for genotyping assistance. We thank Ken Poss for sharing the Tg(hsp70l:dnfgfr1-GFP)pd1 line, David Langenau for the Tg(mylpfa:mCherry)cz3327 line, Tom Schilling for the aldh1a2i26 line, Phil Ingham for the plasmid used to generate the Tg(smyhc1:lyn-tdTomato)oz29 line, and Gage Crump, Andrew Waskiewicz and Monte Westerfield for the six4a, lbx1a and lbx2 in situ plasmids, respectively. We thank Zachary Morrow, Anna Newman-Griffis, Lindsey Barske, Kiel Tietz, Joy-El Talbot and Frank Tulenko for comments on this manuscript. The Australian Regenerative Medicine Institute is supported by grants from the State Government of Victoria and the Australian Government.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.C.T., S.L.A.; Methodology: J.C.T.; Investigation: J.C.T., E.M.T., D.R., P.Q.D.; Writing - original draft: J.C.T.; Writing - review & editing: J.C.T., E.M.T., D.R., P.D.C., S.L.A.; Supervision: P.D.C., S.L.A.; Funding acquisition: J.C.T., P.D.C., S.L.A.
Funding
This work was supported by National Institutes of Health (NIH) grants (GM061952, GM088041 and GM117964 to S.L.A.), an NIH T32 training grant (NS077984 to J.C.T.), a National Health and Medical Research Council Senior Principal Research Fellowship (APP1136567 to P.D.C.), an Australian Research Council grant (LP120100281 to P.D.C.), an NIH loan repayment program contract, and a Pelotonia Postdoctoral Fellowship (to J.C.T.). The OSU Neuroscience Imaging Core is funded by NIH grants P30-NS045758, P30-NS104177 and S10-OD010383. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.171421.supplemental
References
- Abràmoff M. D., Magalhães P. J. and Ram S. J. (2004). Image processing with ImageJ. Biophotonics Int. 11, 36-42. [Google Scholar]
- Adachi N., Pascual-Anaya J., Hirai T., Higuchi S. and Kuratani S. (2018). Development of hypobranchial muscles with special reference to the evolution of the vertebrate neck. Zool. Lett. 4, 5 10.1186/s40851-018-0087-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn D. and Ho R. K. (2008). Tri-phasic expression of posterior Hox genes during development of pectoral fins in zebrafish: Implications for the evolution of vertebrate paired appendages. Dev. Biol. 322, 220-233. 10.1016/j.ydbio.2008.06.032 [DOI] [PubMed] [Google Scholar]
- Alexa K., Choe S.-K., Hirsch N., Etheridge L., Laver E. and Sagerström C. G. (2009). Maternal and zygotic aldh1a2 activity is required for pancreas development in zebrafish. PLoS ONE 4, e8261 10.1371/journal.pone.0008261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvares L. E., Schubert F. R., Thorpe C., Mootoosamy R. C., Cheng L., Parkyn G., Lumsden A. and Dietrich S. (2003). Intrinsic, Hox-dependent cues determine the fate of skeletal muscle precursors. Dev. Cell 5, 379-390. 10.1016/S1534-5807(03)00263-6 [DOI] [PubMed] [Google Scholar]
- Anderson C., Williams V. C., Moyon B., Daubas P., Tajbakhsh S., Buckingham M. E., Shiroishi T., Hughes S. M. and Borycki A.-G. (2012). Sonic hedgehog acts cell-autonomously on muscle precursor cells to generate limb muscle diversity. Genes Dev. 26, 2103-2117. 10.1101/gad.187807.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begemann G., Schilling T. F., Rauch G.-J., Geisler R. and Ingham P. W. (2001). The zebrafish neckless mutation reveals a requirement for raldh2 in mesodermal signals that pattern the hindbrain. Development 128, 3081-3094. [DOI] [PubMed] [Google Scholar]
- Berberoglu M. A., Gallagher T. L., Morrow Z. T., Talbot J. C., Hromowyk K. J., Tenente I. M., Langenau D. M. and Amacher S. L. (2017). Satellite-like cells contribute to pax7 -dependent skeletal muscle repair in adult zebrafish. Dev. Biol. 424, 162-180. 10.1016/j.ydbio.2017.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessarab D. A., Chong S.-W. and Korzh V. (2004). Expression of zebrafish six1 during sensory organ development and myogenesis. Dev. Dyn. 230, 781-786. 10.1002/dvdy.20093 [DOI] [PubMed] [Google Scholar]
- Bessarab D. A., Chong S.-W., Srinivas B. P. and Korzh V. (2008). Six1a is required for the onset of fast muscle differentiation in zebrafish. Dev. Biol. 323, 216-228. 10.1016/j.ydbio.2008.08.015 [DOI] [PubMed] [Google Scholar]
- Birchmeier C., Birchmeier W., Gherardi E. and Vande Woude G. F. (2003). Met, metastasis, motility and more. Nat. Rev. Mol. Cell Biol. 4, 915-925. 10.1038/nrm1261 [DOI] [PubMed] [Google Scholar]
- Bladt F., Riethmacher D., Isenmann S., Aguzzi A. and Birchmeier C. (1995). Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature 376, 768-771. 10.1038/376768a0 [DOI] [PubMed] [Google Scholar]
- Bober E., Franz T., Arnold H.-H., Gruss P. and Tremblay P. (1994). Pax-3 is required for the development of limb muscles: a possible role for the migration of dermomyotomal muscle progenitor cells. Development 120, 603-612. [DOI] [PubMed] [Google Scholar]
- Bosman E. A., Quint E., Fuchs H., Hrabé de Angelis M. and Steel K. P. (2009). Catweasel mice: a novel role for Six1 in sensory patch development and a model for branchio-oto-renal syndrome. Dev. Biol. 328, 285-296. 10.1016/j.ydbio.2009.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohmann H., Jagla K. and Birchmeier C. (2000). The role of Lbx1 in migration of muscle precursor cells. Development 127, 437-445. [DOI] [PubMed] [Google Scholar]
- Buchanan S. G., Hendle J., Lee P. S., Smith C. R., Bounaud P.-Y., Jessen K. A., Tang C. M., Huser N. H., Felce J. D., Froning K. J. et al. (2009). SGX523 is an exquisitely selective, ATP-competitive inhibitor of the MET receptor tyrosine kinase with antitumor activity in vivo. Mol. Cancer Ther. 8, 3181-3190. 10.1158/1535-7163.MCT-09-0477 [DOI] [PubMed] [Google Scholar]
- Chen J. K., Taipale J., Cooper M. K. and Beachy P. A. (2002). Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 16, 2743-2748. 10.1101/gad.1025302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevallier A., Kieny M. and Mauger A. (1977). Limb-somite relationship: origin of the limb musculature. J. Embryol. Exp. Morphol. 41, 245-258. [PubMed] [Google Scholar]
- Christ B. and Brand-Saberi B. (2004). Limb muscle development. Int. J. Dev. Biol. 46, 905-914. [PubMed] [Google Scholar]
- Christ B., Jacob H. J. and Jacob M. (1977). Experimental analysis of the origin of the wing musculature in avian embryos. Anat. Embryol. (Berl.) 150, 171-186. 10.1007/BF00316649 [DOI] [PubMed] [Google Scholar]
- Christensen K. L., Patrick A. N., McCoy E. L. and Ford H. L. (2008). The six family of homeobox genes in development and cancer. Adv. Cancer Res. 101, 93-126. 10.1016/S0065-230X(08)00405-3 [DOI] [PubMed] [Google Scholar]
- Cunningham T. J., Zhao X., Sandell L. L., Evans S. M., Trainor P. A. and Duester G. (2013). Antagonism between retinoic acid and fibroblast growth factor signaling during limb development. Cell Rep. 3, 1503-1511. 10.1016/j.celrep.2013.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daston G., Lamar E., Olivier M. and Goulding M. (1996). Pax-3 is necessary for migration but not differentiation of limb muscle precursors in the mouse. Development 122, 1017-1027. [DOI] [PubMed] [Google Scholar]
- Devoto S. H., Stoiber W., Hammond C. L., Steinbacher P., Haslett J. R., Barresi M. J. F., Patterson S. E., Adiarte E. G. and Hughes S. M. (2006). Generality of vertebrate developmental patterns: evidence for a dermomyotome in fish. Evol. Htmlent Glyphamp Asciiamp Dev. 8, 101-110. 10.1111/j.1525-142X.2006.05079.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich S. (1999). Regulation of hypaxial muscle development. Cell Tissue Res. 296, 175-182. 10.1007/s004410051278 [DOI] [PubMed] [Google Scholar]
- Dietrich S., Schubert F. R., Healy C., Sharpe P. T. and Lumsden A. (1998). Specification of the hypaxial musculature. Development 125, 2235-2249. [DOI] [PubMed] [Google Scholar]
- Dietrich S., Abou-Rebyeh F., Brohmann H., Bladt F., Sonnenberg-Riethmacher E., Yamaai T., Lumsden A., Brand-Saberi B. and Birchmeier C. (1999). The role of SF/HGF and c-Met in the development of skeletal muscle. Development 126, 1621-1629. [DOI] [PubMed] [Google Scholar]
- El-Brolosy M., Rossi A., Kontarakis Z., Kuenne C., Guenther S., Fukuda N., Takacs C., Lai S.-L., Fukuda R., Gerri C. et al. (2019). Genetic compensation is triggered by mutant mRNA degradation. Nature 568, 193-197. 10.1038/s41586-019-1064-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D. J. R., Valasek P., Schmidt C. and Patel K. (2006). Skeletal muscle translocation in vertebrates. Brain Struct. Funct. 211, 43-50. 10.1007/s00429-006-0121-1 [DOI] [PubMed] [Google Scholar]
- Fairchild C. L. and Barna M. (2014). Specialized filopodia: at the ‘tip’ of morphogen transport and vertebrate tissue patterning. Curr. Opin. Genet. Dev. 27, 67-73. 10.1016/j.gde.2014.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X., Adiarte E. G. and Devoto S. H. (2006). Hedgehog acts directly on the zebrafish dermomyotome to promote myogenic differentiation. Dev. Biol. 300, 736-746. 10.1016/j.ydbio.2006.08.056 [DOI] [PubMed] [Google Scholar]
- Flanagan-Steet H., Hannon K., McAvoy M. J., Hullinger R. and Olwin B. B. (2000). Loss of FGF receptor 1 signaling reduces skeletal muscle mass and disrupts myofiber organization in the developing limb. Dev. Biol. 218, 21-37. 10.1006/dbio.1999.9535 [DOI] [PubMed] [Google Scholar]
- Gibert Y., Gajewski A., Meyer A. and Begemann G. (2006). Induction and prepatterning of the zebrafish pectoral fin bud requires axial retinoic acid signaling. Development 133, 2649-2659. 10.1242/dev.02438 [DOI] [PubMed] [Google Scholar]
- Grandel H. and Brand M. (2011). Zebrafish limb development is triggered by a retinoic acid signal during gastrulation. Dev. Dyn. 240, 1116-1126. 10.1002/dvdy.22461 [DOI] [PubMed] [Google Scholar]
- Grandel H., Lun K., Rauch G. J., Rhinn M., Piotrowski T., Houart C., Sordino P., Küchler A. M., Schulte-Merker S., Geisler R. et al. (2002). Retinoic acid signalling in the zebrafish embryo is necessary during presegmentation stages to pattern the anterior-posterior axis of the CNS and to induce a pectoral fin bud. Development 129, 2851-2865. [DOI] [PubMed] [Google Scholar]
- Griffin C. A., Apponi L. H., Long K. K. and Pavlath G. K. (2010). Chemokine expression and control of muscle cell migration during myogenesis. J. Cell Sci. 123, 3052-3060. 10.1242/jcs.066241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifone R., Laclef C., Spitz F., Lopez S., Demignon J., Guidotti J.-E., Kawakami K., Xu P.-X., Kelly R., Petrof B. J. et al. (2004). Six1 and Eya1 expression can reprogram adult muscle from the slow-twitch phenotype into the fast-twitch phenotype. Mol. Cell. Biol. 24, 6253-6267. 10.1128/MCB.24.14.6253-6267.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifone R., Demignon J., Houbron C., Souil E., Niro C., Seller M. J., Hamard G. and Maire P. (2005). Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. Development 132, 2235-2249. 10.1242/dev.01773 [DOI] [PubMed] [Google Scholar]
- Grifone R., Demignon J., Giordani J., Niro C., Souil E., Bertin F., Laclef C., Xu P.-X. and Maire P. (2007). Eya1 and Eya2 proteins are required for hypaxial somitic myogenesis in the mouse embryo. Dev. Biol. 302, 602-616. 10.1016/j.ydbio.2006.08.059 [DOI] [PubMed] [Google Scholar]
- Gross M. K., Moran-Rivard L., Velasquez T., Nakatsu M. N., Jagla K. and Goulding M. (2000). Lbx1 is required for muscle precursor migration along a lateral pathway into the limb. Development 127, 413-424. [DOI] [PubMed] [Google Scholar]
- Haines L., Neyt C., Gautier P., Keenan D. G., Bryson-Richardson R. J., Hollway G. E., Cole N. J. and Currie P. D. (2004). Met and Hgf signaling controls hypaxial muscle and lateral line development in the zebrafish. Development 131, 4857-4869. 10.1242/dev.01374 [DOI] [PubMed] [Google Scholar]
- Hammond C. L., Hinits Y., Osborn D. P. S., Minchin J. E. N., Tettamanti G. and Hughes S. M. (2007). Signals and myogenic regulatory factors restrict pax3 and pax7 expression to dermomyotome-like tissue in zebrafish. Dev. Biol. 302, 504-521. 10.1016/j.ydbio.2006.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K. and Ozawa E. (1995). Myogenic cell migration from somites is induced by tissue contact with medial region of the presumptive limb mesoderm in chick embryos. Development 121, 661-669. [DOI] [PubMed] [Google Scholar]
- Hollway G. E. and Currie P. D. (2003). Myotome meanderings. Cellular morphogenesis and the making of muscle. EMBO Rep. 4, 855-860. 10.1038/sj.embor.embor920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. K.-H., McGlinn E., Harfe B. D., Kardon G. and Tabin C. J. (2012). Autonomous and nonautonomous roles of Hedgehog signaling in regulating limb muscle formation. Genes Dev. 26, 2088-2102. 10.1101/gad.187385.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R., Zhi Q., Izpisua-Belmonte J.-C., Christ B. and Patel K. (1999). Origin and development of the avian tongue muscles. Anat. Embryol. (Berl.) 200, 137-152. 10.1007/s004290050268 [DOI] [PubMed] [Google Scholar]
- Ignatius M. S., Chen E., Elpek N. M., Fuller A. Z., Tenente I. M., Clagg R., Liu S., Blackburn J. S., Linardic C. M., Rosenberg A. E. et al. (2012). In vivo imaging of tumor-propagating cells, regional tumor heterogeneity, and dynamic cell movements in embryonal rhabdomyosarcoma. Cancer Cell 21, 680-693. 10.1016/j.ccr.2012.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N., Mima T. and Mikawa T. (1996). Loss of fibroblast growth factor receptors is necessary for terminal differentiation of embryonic limb muscle. Development 122, 291-300. [DOI] [PubMed] [Google Scholar]
- Jowett T. (1999). Analysis of protein and gene expression. Methods Cell Biol. 59, 63-85. 10.1016/S0091-679X(08)61821-X [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Osanai H., Kawakami K. and Yamamoto M. (2000). Expression of three zebrafish Six4 genes in the cranial sensory placodes and the developing somites. Mech. Dev. 98, 151-155. 10.1016/S0925-4773(00)00451-2 [DOI] [PubMed] [Google Scholar]
- Kok F. O., Shin M., Ni C.-W., Gupta A., Grosse A. S., van Impel A., Kirchmaier B. C., Peterson-Maduro J., Kourkoulis G., Male I. et al. (2015). Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev. Cell 32, 97-108. 10.1016/j.devcel.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laclef C., Hamard G., Demignon J., Souil E., Houbron C. and Maire P. (2003). Altered myogenesis in Six1-deficient mice. Development 130, 2239-2252. 10.1242/dev.00440 [DOI] [PubMed] [Google Scholar]
- Lee Y., Grill S., Sanchez A., Murphy-Ryan M. and Poss K. D. (2005). Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development 132, 5173-5183. 10.1242/dev.02101 [DOI] [PubMed] [Google Scholar]
- Li M., Hromowyk K. J., Amacher S. L. and Currie P. D. (2017). Chapter 14-Muscular dystrophy modeling in zebrafish. In Methods in Cell Biology (ed. Detrich H. W., Westerfield M. and Zon L. I.), pp. 347-380. Academic Press. [DOI] [PubMed] [Google Scholar]
- Lin C.-Y., Chen W.-T., Lee H.-C., Yang P.-H., Yang H.-J. and Tsai H.-J. (2009). The transcription factor Six1a plays an essential role in the craniofacial myogenesis of zebrafish. Dev. Biol. 331, 152-166. 10.1016/j.ydbio.2009.04.029 [DOI] [PubMed] [Google Scholar]
- Lours-Calet C., Alvares L. E., El-Hanfy A. S., Gandesha S., Walters E. H., Sobreira D. R., Wotton K. R., Jorge E. C., Lawson J. A., Kelsey Lewis A. et al. (2014). Evolutionarily conserved morphogenetic movements at the vertebrate head–trunk interface coordinate the transport and assembly of hypopharyngeal structures. Dev. Biol. 390, 231-246. 10.1016/j.ydbio.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowski C. M., Drummond D. L., Waskiewicz A. J. and Danzmann R. (2011). Pbx-dependent regulation of lbx gene expression in developing zebrafish embryos. Genome 54, 973-985. 10.1139/g11-061 [DOI] [PubMed] [Google Scholar]
- Mackenzie S., Walsh F. S. and Graham A. (1998). Migration of hypoglossal myoblast precursors. Dev. Dyn. 213, 349-358. [DOI] [PubMed] [Google Scholar]
- Martin B. L. and Harland R. M. (2001). Hypaxial muscle migration during primary myogenesis in Xenopus laevis. Dev. Biol. 239, 270-280. 10.1006/dbio.2001.0434 [DOI] [PubMed] [Google Scholar]
- Martin B. L. and Harland R. M. (2006). A novel role for lbx1 in Xenopus hypaxial myogenesis. Development 133, 195-208. 10.1242/dev.02183 [DOI] [PubMed] [Google Scholar]
- Masselink W., Cole N. J., Fenyes F., Berger S., Sonntag C., Wood A., Nguyen P. D., Cohen N., Knopf F., Weidinger G. et al. (2016). A somitic contribution to the apical ectodermal ridge is essential for fin formation. Nature 535, 542-546. 10.1038/nature18953 [DOI] [PubMed] [Google Scholar]
- Masselink W., Masaki M., Sieiro D., Marcelle C. and Currie P. D. (2017). Phosphorylation of Lbx1 controls lateral myoblast migration into the limb. Dev. Biol. 430, 302-309. 10.1016/j.ydbio.2017.08.025 [DOI] [PubMed] [Google Scholar]
- Masyuk M., Abduelmula A., Morosan-Puopolo G., Ödemis V., Rehimi R., Khalida N., Yusuf F., Engele J., Tamamura H., Theiss C. et al. (2014). Retrograde migration of pectoral girdle muscle precursors depends on CXCR4/SDF-1 signaling. Histochem. Cell Biol. 142, 473-488. 10.1007/s00418-014-1237-7 [DOI] [PubMed] [Google Scholar]
- Megason S. G. (2009). In toto imaging of embryogenesis with confocal time-lapse microscopy. In Zebrafish (ed. Lieschke G. J., Oates A. C. and Kawakami K.), pp. 317-332. Totowa, NJ: Humana Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering E., Dzyubachyk O. and Smal I. (2012). Methods for cell and particle tracking. In Methods in Enzymology (ed. P.M. Conn), pp. 183-200. Elsevier. [DOI] [PubMed] [Google Scholar]
- Mennerich D., Schäfer K. and Braun T. (1998). Pax-3 is necessary but not sufficient for lbx1 expression in myogenic precursor cells of the limb. Mech. Dev. 73, 147-158. 10.1016/S0925-4773(98)00046-X [DOI] [PubMed] [Google Scholar]
- Mercader N. (2007). Early steps of paired fin development in zebrafish compared with tetrapod limb development: Paired fin development in the zebrafish. Dev. Growth Differ. 49, 421-437. 10.1111/j.1440-169X.2007.00942.x [DOI] [PubMed] [Google Scholar]
- Mercader N., Fischer S. and Neumann C. J. (2006). Prdm1 acts downstream of a sequential RA, Wnt and Fgf signaling cascade during zebrafish forelimb induction. Development 133, 3949-3949. 10.1242/dev.02455 [DOI] [PubMed] [Google Scholar]
- Micalizzi D. S., Christensen K. L., Jedlicka P., Coletta R. D., Barón A. E., Harrell J. C., Horwitz K. B., Billheimer D., Heichman K. A., Welm A. L. et al. (2009). The Six1 homeoprotein induces human mammary carcinoma cells to undergo epithelial-mesenchymal transition and metastasis in mice through increasing TGF-β signaling. J. Clin. Invest. 119, 2678-2690. 10.1172/JCI37815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minchin J. E. N., Williams V. C., Hinits Y., Low S., Tandon P., Fan C.-M., Rawls J. F. and Hughes S. M. (2013). Oesophageal and sternohyal muscle fibres are novel Pax3-dependent migratory somite derivatives essential for ingestion. Development 140, 4296-4296. 10.1242/dev.103184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody S. A. and LaMantia A.-S. (2015). Transcriptional regulation of cranial sensory placode development. Curr. Top. Dev. Biol. 111, 301-350. 10.1016/bs.ctdb.2014.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow Z. T., Maxwell A. M., Hoshijima K., Talbot J. C., Grunwald D. J. and Amacher S. L. (2017). tbx6l and tbx16 are redundantly required for posterior paraxial mesoderm formation during zebrafish embryogenesis: Zebrafish T-Box Genes and Somitogenesis. Dev. Dyn. 246, 759-769. 10.1002/dvdy.24547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann C. J., Grandel H., Gaffield W., Schulte-Merker S. and Nusslein-Volhard C. (1999). Transient establishment of anteroposterior polarity in the zebrafish pectoral fin bud in the absence of sonic hedgehog activity. Development 126, 4817-4826. [DOI] [PubMed] [Google Scholar]
- Neyt C., Jagla K., Thisse C., Thisse B., Haines L. and Currie P. D. (2000). Evolutionary origins of vertebrate appendicular muscle. Nature 408, 82-86. 10.1038/35040549 [DOI] [PubMed] [Google Scholar]
- Niro C., Demignon J., Vincent S., Liu Y., Giordani J., Sgarioto N., Favier M., Guillet-Deniau I., Blais A. and Maire P. (2010). Six1 and Six4 gene expression is necessary to activate the fast-type muscle gene program in the mouse primary myotome. Dev. Biol. 338, 168-182. 10.1016/j.ydbio.2009.11.031 [DOI] [PubMed] [Google Scholar]
- Nord H., Nygard Skalman L. and von Hofsten J. (2013). Six1 regulates proliferation of Pax7-positive muscle progenitors in zebrafish. J. Cell Sci. 126, 1868-1880. 10.1242/jcs.119917 [DOI] [PubMed] [Google Scholar]
- O'Brien J. H., Hernandez-Lagunas L., Artinger K. B. and Ford H. L. (2014). MicroRNA-30a regulates zebrafish myogenesis through targeting the transcription factor Six1. J. Cell Sci. 127, 2291-2301. 10.1242/jcs.143677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochi H. and Westerfield M. (2009). Lbx2 regulates formation of myofibrils. BMC Dev. Biol. 9, 13 10.1186/1471-213X-9-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto E., Kusakabe R., Kuraku S., Hyodo S., Robert-Moreno A., Onimaru K., Sharpe J., Kuratani S. and Tanaka M. (2017). Migratory appendicular muscles precursor cells in the common ancestor to all vertebrates. Nat. Ecol. Evol. 1, 1731-1736. 10.1038/s41559-017-0330-4 [DOI] [PubMed] [Google Scholar]
- Ozaki H., Watanabe Y., Takahashi K., Kitamura K., Tanaka A., Urase K., Momoi T., Sudo K., Sakagami J., Asano M. et al. (2001). Six4, a putative myogenin gene regulator, is not essential for mouse embryonal development. Mol. Cell. Biol. 21, 3343-3350. 10.1128/MCB.21.10.3343-3350.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada C., Han D. and Chai Y. (2012). Molecular and cellular regulatory mechanisms of tongue myogenesis. J. Dent. Res. 91, 528-535. 10.1177/0022034511434055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson S. E., Mook L. B. and Devoto S. H. (2008). Growth in the larval zebrafish pectoral fin and trunk musculature. Dev. Dyn. 237, 307-315. 10.1002/dvdy.21400 [DOI] [PubMed] [Google Scholar]
- Relaix F. (2004). Divergent functions of murine Pax3 and Pax7 in limb muscle development. Genes Dev. 18, 1088-1105. 10.1101/gad.301004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard A.-F., Demignon J., Sakakibara I., Pujol J., Favier M., Strochlic L., Le Grand F., Sgarioto N., Guernec A., Schmitt A. et al. (2011). Genesis of muscle fiber-type diversity during mouse embryogenesis relies on Six1 and Six4 gene expression. Dev. Biol. 359, 303-320. 10.1016/j.ydbio.2011.08.010 [DOI] [PubMed] [Google Scholar]
- Robert B. and Lallemand Y. (2006). Anteroposterior patterning in the limb and digit specification: contribution of mouse genetics. Dev. Dyn. 235, 2337-2352. 10.1002/dvdy.20890 [DOI] [PubMed] [Google Scholar]
- Rossi A., Kontarakis Z., Gerri C., Nolte H., Hölper S., Krüger M. and Stainier D. Y. R. (2015). Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524, 230-233. 10.1038/nature14580 [DOI] [PubMed] [Google Scholar]
- Scaal M., Bonafede A., Dathe V., Sachs M., Cann G., Christ B. and Brand-Saberi B. (1999). SF/HGF is a mediator between limb patterning and muscle development. Development 126, 4885-4893. [DOI] [PubMed] [Google Scholar]
- Schäfer K. and Braun T. (1999). Early specification of limb muscle precursor cells by the homeobox gene Lbx1h. Nat. Genet. 23, 213-216. 10.1038/13843 [DOI] [PubMed] [Google Scholar]
- Stainier D. Y. R., Raz E., Lawson N. D., Ekker S. C., Burdine R. D., Eisen J. S., Ingham P. W., Schulte-Merker S., Yelon D., Weinstein B. M. et al. (2017). Guidelines for morpholino use in zebrafish. PLoS Genet. 13, e1007000 10.1371/journal.pgen.1007000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellabotte F. and Devoto S. H. (2007). The teleost dermomyotome. Dev. Dyn. 236, 2432-2443. 10.1002/dvdy.21253 [DOI] [PubMed] [Google Scholar]
- Sun L., Tran N., Liang C., Tang F., Rice A., Schreck R., Waltz K., Shawver L. K., McMahon G. and Tang C. (1999). Design, synthesis, and evaluations of substituted 3-[(3- or 4-Carboxyethylpyrrol-2-yl)methylidenyl]indolin-2-ones as inhibitors of VEGF, FGF, and PDGF receptor tyrosine kinases. J. Med. Chem. 42, 5120-5130. [DOI] [PubMed] [Google Scholar]
- Suster M. L., Abe G., Schouw A. and Kawakami K. (2011). Transposon-mediated BAC transgenesis in zebrafish. Nat. Protoc. 6, 1998-2021. 10.1038/nprot.2011.416 [DOI] [PubMed] [Google Scholar]
- Swartz M. E., Eberhart J., Pasquale E. B. and Krull C. E. (2001). EphA4/ephrin-A5 interactions in muscle precursor cell migration in the avian forelimb. Development 128, 4669-4680. [DOI] [PubMed] [Google Scholar]
- Talbot J. C. and Amacher S. L. (2014). A streamlined CRISPR pipeline to reliably generate zebrafish frameshifting alleles. Zebrafish 11, 583-585. 10.1089/zeb.2014.1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot J. C., Johnson S. L. and Kimmel C. B. (2010). hand2 and Dlx genes specify dorsal, intermediate and ventral domains within zebrafish pharyngeal arches. Development 137, 2507-2517. 10.1242/dev.049700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani-Matsuhana S., Kusakabe R. and Inoue K. (2018). Developmental mechanisms of migratory muscle precursors in medaka pectoral fin formation. Dev. Genes Evol. 228, 189-196. 10.1007/s00427-018-0616-9 [DOI] [PubMed] [Google Scholar]
- Valasek P., Theis S., DeLaurier A., Hinits Y., Luke G. N., Otto A. M., Minchin J., He L., Christ B., Brooks G. et al. (2011). Cellular and molecular investigations into the development of the pectoral girdle. Dev. Biol. 357, 108-116. 10.1016/j.ydbio.2011.06.031 [DOI] [PubMed] [Google Scholar]
- Vasyutina E. and Birchmeier C. (2006). The development of migrating muscle precursor cells. Brain Struct. Funct. 211, 37-41. 10.1007/s00429-006-0118-9 [DOI] [PubMed] [Google Scholar]
- Wang X., Ono Y., Tan S. C., Chai R. J., Parkin C. and Ingham P. W. (2011). Prdm1a and miR-499 act sequentially to restrict Sox6 activity to the fast-twitch muscle lineage in the zebrafish embryo. Development 138, 4399-4404. 10.1242/dev.070516 [DOI] [PubMed] [Google Scholar]
- Westerfield M. (2007). The Zebrafish Book: A guide for the laboratory use of zebrafish (Danio rerio). Eugene: University of Oregon Press. [Google Scholar]
- Windner S. E., Steinbacher P., Obermayer A., Kasiba B., Zweimueller-Mayer J. and Stoiber W. (2011). Distinct modes of vertebrate hypaxial muscle formation contribute to the teleost body wall musculature. Dev. Genes Evol. 221, 167-178. 10.1007/s00427-011-0369-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Ren Z., Zhang L., Liu Y., Li H. and Xiong Y. (2013). Overexpression of Six1 gene suppresses proliferation and enhances expression of fast-type muscle genes in C2C12 myoblasts. Mol. Cell. Biochem. 380, 23-32. 10.1007/s11010-013-1653-3 [DOI] [PubMed] [Google Scholar]
- Xiao A., Wang Z., Hu Y., Wu Y., Luo Z., Yang Z., Zu Y., Li W., Huang P., Tong X. et al. (2013). Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res. 41, e141-e141. 10.1093/nar/gkt464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Zhang Y., Altomare D., Peña M. M., Wan F., Pirisi L. and Creek K. E. (2014). Six1 promotes epithelial-mesenchymal transition and malignant conversion in human papillomavirus type 16-immortalized human keratinocytes. Carcinogenesis 35, 1379-1388. 10.1093/carcin/bgu050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Xu J., Maire P. and Xu P.-X. (2017). Six1 is essential for differentiation and patterning of the mammalian auditory sensory epithelium. PLoS Genet. 13, e1006967 10.1371/journal.pgen.1006967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Sirbu I. O., Mic F. A., Molotkova N., Molotkov A., Kumar S. and Duester G. (2009). Retinoic acid promotes limb induction through effects on body axis extension but is unnecessary for limb patterning. Curr. Biol. 19, 1050-1057. 10.1016/j.cub.2009.04.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.