ABSTRACT
Impaired osteoblast differentiation may result in bone metabolic diseases such as osteoporosis. It was reported recently that hedgehog (Hh) signaling and autophagy are two important regulators of bone differentiation. In order to further dissect their relationship in bone development, we used a zebrafish larvae model to investigate how disruption of one of these signals affects the function of the other and impacts osteoblast differentiation. Our results showed that activation of Hh signaling negatively regulated autophagy. However, suppression of autophagy by knocking down atg5 expression did not alter Hh signaling, but dramatically upregulated the expression of osteoblast-related genes and increased bone mineralization, especially in the den region. On the contrary, inhibition of the Hh signaling pathway by cyclopamine treatment suppressed the expression of osteoblast-related genes and decreased bone mineralization. In agreement with these findings, blocking Hh signaling through knockdown SHH and Gli2 genes led to defective osteoblast differentiation, while promoting Hh signaling by knockdown Ptch1 was beneficial to osteoblast differentiation. Our results thus support that activation of the Hh signaling pathway negatively regulates autophagy and consequentially promotes osteoblast differentiation. On the contrary, induction of autophagy inhibits osteoblast differentiation. Our work reveals the mechanism underlying Hh signaling pathway regulation of bone development.
KEY WORDS: Zebrafish, Autophagy, Hh signaling, Osteoblast, atg5, Mineralization, Gli2, Ptch1, SHH, BMP2
Summary: Our report of an essential regulation role of hedgehog signaling and autophagy on osteoblast differentiation may contribute to research on bone development biology, hedgehog signaling and the autophagy pathway.
INTRODUCTION
Osteoblasts are responsible for bone formation, and impaired osteoblast development leads to serious bone diseases such as osteoporosis (Edsall and Franz-Odendaal, 2010). Zinc finger-containing transcription factor Osterix (Sp7) is the main regulator of osteoblastgenesis. Loss of Sp7 leads to delay of bone maturation and dysregulates bone formation (Li et al., 2009; Nakashima et al., 2002; Kague et al., 2016). Bone morphogenetic protein 2 (BMP2) is another positive regulator, which stimulates osteoblast differentiation, maturation and osteoblast growth (Zhao et al., 2017; Kuo et al., 2006).
Autophagy is an intracellular degradation system, and is responsible for degrading and recycling aggregated proteins, damaged organelles, misfolded proteins and long-lived proteins (Hocking et al., 2012; Mizushima, 2007). During osteoblast differentiation, rapid synthesis of bone-matrix protein results in accumulation of misfolded proteins, and high autophagy activity is thus required for their removal (Hocking et al., 2012). Autophagy also plays a regulatory role in osteoblast formation and differentiation, although both inhibitory and promoting effects on osteoblast differentiation were observed in different experimental systems. For instance, pneumolysin-induced autophagy inhibited osteoblast differentiation, and treatment with autophagy inhibitors or knockdown of atg5 alleviated the PLY-induced inhibition of differentiation (Kim et al., 2017). Stimulation of autophagy promoted osteoblast differentiation, and suppression of autophagy inhibited osteoblast terminal differentiation in mice (Ha et al., 2014; Liu et al., 2013). Moreover, inhibition of autophagy by depletion of Atg7 in the osteoblast lineage led to low bone mass and fractures associated with reduced numbers of osteoblasts (Piemontese et al., 2016). Promoting autophagy level by the mTOR pathway inhibited osteoblast apoptosis (Yang et al., 2013).
The hedgehog (Hh) signaling pathways, mediated by sonic hedgehog (Shh) and Indian hedgehog (Ihh), are recognized as indispensable regulators for osteoblast differentiation and morphological transition. Specifically, the transmembrane receptor Patched (Ptch1) binding to a secreted ligand initiates Hh signaling, whereas unliganded Ptch1, as a negative regulator of Hh signaling, inhibits the activity of the membrane protein Smoothened (Smo), which regulates downstream Gli transcriptional effectors. Among three Gli proteins, Gli2 is a very important activator for the Hh signaling pathway (Hui and Angers, 2011; Cohen et al., 2015; Sasaki et al., 1997). Activation of both Hh signaling pathways could positively regulate osteoblast differentiation. Shh promotes interactions between epidermal cells and osteoblast progenitors, which affect the shape of regenerated zebrafish bone. Ihh regulates the function of bone morphogenetic protein and further affects chondrocyte and osteoblast differentiation through Gli2 transcription factors. Ihh signaling modulates bone shaping during early morphogenesis of zebrafish craniofacial skeleton (Armstrong et al., 2017; Cai and Liu, 2016; Hojo et al., 2013; Huycke et al., 2012; Marumoto et al., 2017). Although mature osteoblasts were still in long bones of the Ihh deficient mutant mice limbs, growth plate formation was completely lost (Amano et al., 2015). Purmorphamine and other new Hh agonists triggered the Hh signaling pathway in hMSCs, resulting in an increase of osteoblast differentiation (Oliveira et al., 2012; Nakamura et al., 2015). Cyclopamine (cyA), an inhibitor of Hh signaling, decreased bone mass in adult mice (Ohba et al., 2008). However, another study showed that constitutive activation of Hh signaling impaired bone formation in mice (Joeng and Long, 2013).
Interestingly, accumulating evidence suggests that the Hh signaling pathway cross-talks with autophagy pathway. For example, Hh signaling inhibited formation of autophagosome both in basal level and in autophagy-induced conditions (Jimenez-Sanchez et al., 2012). In pancreatic cancer cells and breast cancer cells, activation of Hh signaling inhibits autophagy, while inhibition of Hh signaling promotes autophagy (Xu et al., 2014; Wang et al., 2017). In the Shh intestinal epithelial conditional knockout mouse model, autophagic levels decreased significantly in hippocampal neurons and vascular smooth muscle cells, indicating that the Shh signaling pathway may be involved in autophagy (Gagné-Sansfaçon et al., 2014; Petralia et al., 2013; Li et al., 2012). These findings inspired us to speculate that the Hh signaling and authophagy pathways, two important regulatory pathways of osteoblast development, may coordinate to regulate osteoblast differentiation. We therefore set out to investigate the interaction between Hh signaling and autophagy pathways and their impacts on bone development in zebrafish larvae.
As an animal model of bone development, Danio rerio (zebrafish) has a high similarity with human in bone architecture, bone cell types (osteoblasts and osteoclasts) and matrix proteins (Pasqualetti et al., 2012). Type X collagen (col10a1), a molecular marker of bone tissue, is expressed in both endochondral and intramembranous bones of zebrafish. Kim and colleagues established a col10a1:GFP transgenic zebrafish line that specifically expresses GFP in osteoblasts. This transgenic zebrafish is a useful tool for investigating osteoblast formation and differentiation (Avaron et al., 2006; Kim et al., 2013). In recent years, several reports have shown that Hh signaling promotes osteoblast differentiation in the zebrafish regeneration model (Armstrong et al., 2017; Paul et al., 2016; Blum and Begemann, 2015), however, how the Hh signaling pathway regulates the osteoblasts’ differentiation in zebrafish larvae remains to be investigated. Using the zebrafish model, we demonstrate that the Hh signaling pathway suppresses autophagy, whereas the autophagy pathway didn't affect Hh signaling. Furthermore, we found that whereas autophagy inhibits osteoblast differentiation, activation of the Hh signaling pathway promotes osteoblast differentiation and development. Our results thus favor a hypothesis that suppression of osteoblast differentiation by the Hh signaling pathway inhibits autophagy, which consequentially promotes osteoblast differentiation.
RESULTS
Hh pathway suppressed autophagy level in zebrafish larvae
Autophagy had no obvious effect on Hh signaling
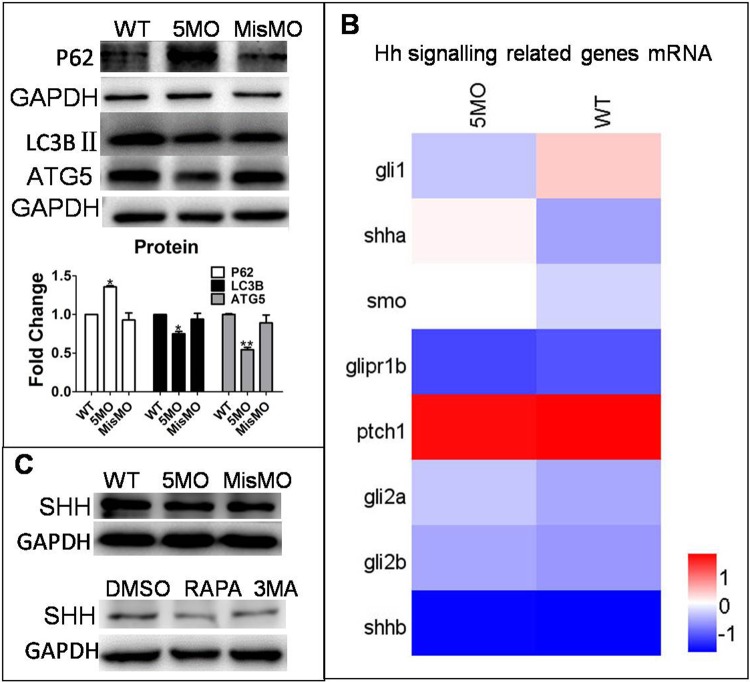
To investigate the relationship between autophagy and the Hh signaling pathway, autophagy-related gene atg5 was knocked down by morpholino oligos, which resulted in down- and upregulation of autophagy marker proteins LC3BII and P62, respectively (Fig. 1A). However, atg5 knockdown did not apparently alter the transcriptional level of Hh signaling pathway-related Shh genes, such as membrane receptors Smo, Ptch1 and downstream transcriptional effectors Gli2, although shha was induced by atg5 knockdown, but shhb had no change, and gli1 was decreased by 5MO, but gli2a and gli2b had no change (Fig. 1B). Furthermore, neither injection of ATG5MO, treatment with autophagy inducer rapamycin (RAPA) nor inhibitor 3MA changed the level of SHH protein expression (Fig. 1C). These results collectively suggest that autophagy has no obvious effect on the Hh signaling pathway.
Fig. 1.
Autophagy did not regulate the Hh signaling pathway. ATG5MO and MisMO were injected into embryos at the 1–4 cell stage. MisMO-injected group as a negative control. The means and s.d. were derived from triple duplicates. (A) Expression of LC3BII and ATG5 proteins was determined by a western blot assay. GAPDH served as a loading control. (B) Heat map of mRNA expression of Hh signaling pathway-related genes Smo, Gli family number Gli1, Gli2a, Gli2b, ptch1 and Shh mRNA from wild-type and ATG5MO-injected embryos was determined by RNAseq assay. (C) Embryos at 1–4 cell stage were either injected with ATG5MO or MisMO or mock-treated and treated with 10 µM of RAPA or 2 mM of 3MA for 3 days from 3 dpf. Expression of SHH protein was determined by a western blot assay. GAPDH served as a loading control.
Hh signaling pathway negatively regulated autophagy activity
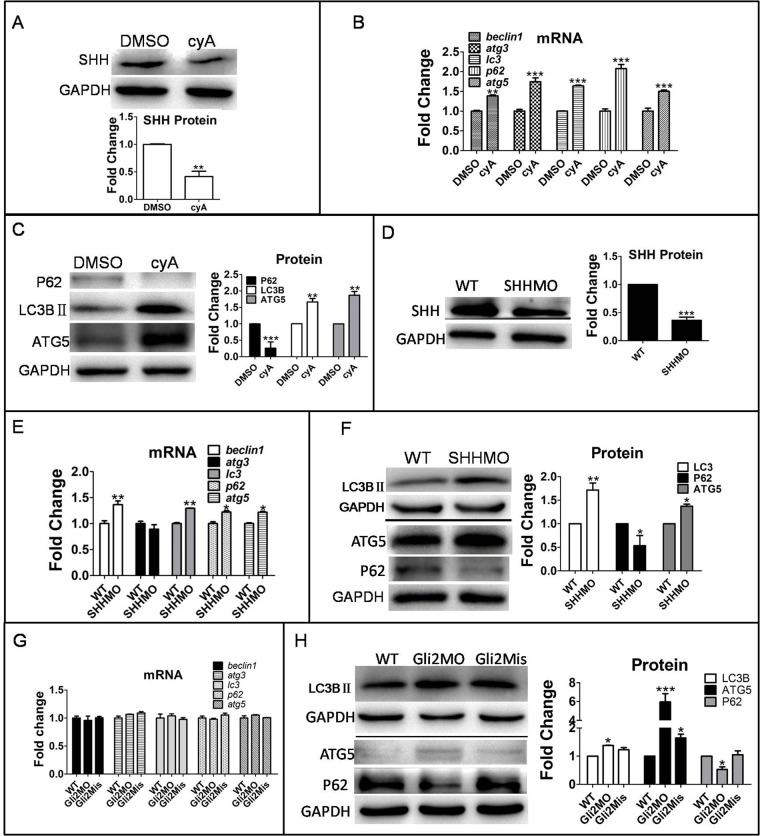
In order to investigate the effects of the Hh signaling pathway on autophagy in zebrafish, we disrupted the Hh signaling pathway via three different approaches. First, treatment of zebrafish larvae with cyclopamine (cyA), an Hh signaling pathway inhibitor, for 3 days at 3 dpf downregulated SHH protein expression (Fig. 2A), but expression of autophagy-related genes, including beclin1, atg3, lc3, p62 and atg5, were upregulated at both mRNA and protein levels (Fig. 2B). Furthermore, LC3BII and ATG5 proteins were upregulated, whereas p62 protein was downregulated by cyA treatment (Fig. 2C). Second, the Hh signaling pathway was downregulated by suppressing the expression of shh and gli2, Hh signaling positively regulated genes by injection of Shh morpholino oligonucleotide (MO) and Gli2MO, respectively. Gli2Mis was used as a negative control. Consistent with results observed under cyA treatment condition, downregulation of SHH and gli2 expression (Fig. 2D) increased the transcripts of autophagy-related genes beclin1, lc3, p62 and atg5 (Fig. 2E) and autophagy-related protein LC3BII and ATG5, but decreased the level of p62 protein (Fig. 2F–H). Those two experiments strongly suggest that inhibition of the Hh signal pathway increases autophagy activity.
Fig. 2.
Autophagy level was promoted by downregulation of the Hh signaling pathway in zebrafish larvae. (A–C) Larvae were exposed to 10 µM cyA and 0.1% DMSO at 3 dpf for 3 days. (D–H) Embryos were injected with SHHMO, Gli2MO and Gli2Mis at the 1–4 cell stage; at 6 dpf, larvae were collected for assays . GAPDH served as a loading control. The means and s.d. were derived from triple duplicates. *P<0.05, **P<0.01, ***P<0.001 versus the WT groups or DMSO control groups (one-way ANOVA). The data were from three independent experiments. (A,D) SHH protein was detected by western blot assay. (B,E,G) Transcriptional level of autophagy-related genes was tested by qPCR assays. (C,F,H) Autophagy-related proteins LC3B II, ATG5 and P62 level were determined by western blot.
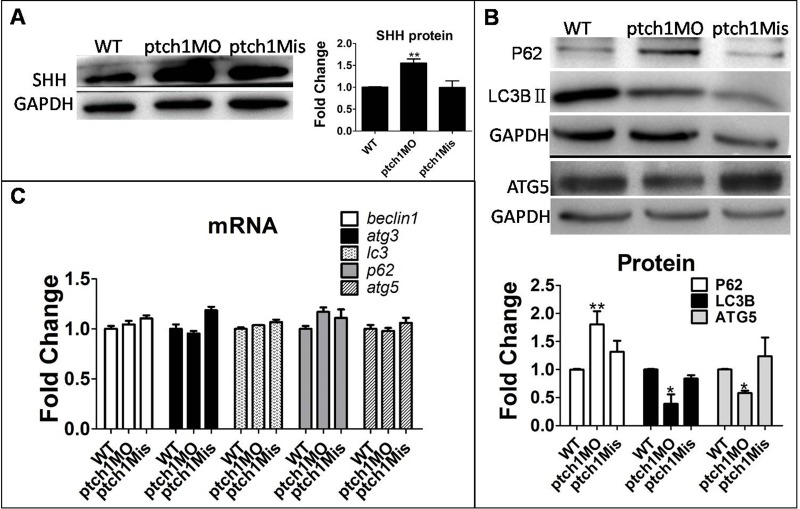
Third, to further confirm the regulatory effect of the Hh signaling pathway on autophagy, we activated the Hh signaling pathway by knocking down Ptch1, the Hh signaling negative regulator, by injection of Ptch1MO or Ptch1Mis into 1–4 cell stage embryos. As anticipated, knockdown of Ptch1 expression increased SHH protein and autophagy protein p62, but decreased the autophagy proteins ATG5 and LC3BII (Fig. 3).
Fig. 3.
Promotion of Hh signaling by downregulated Ptch1 gene suppressed autophagy level. Embryos were injected with ptch1MO and ptch1Mis at the 1–4 cell stage and larvae were collected at 6 dpf for qPCR and western blot. GAPDH as a loading control. The means and s.d. were derived from triple duplicates. (A,B) SHH protein and autophagy-related proteins were detected by western blot. (C) Ptch1MO-injected group compared with WT group and ptch1Mis-injected group. Autophagy gene mRNA levels were tested by qPCR. *P<0.05, **P<0.01 versus WT groups (one-way ANOVA).
Taken together, the results presented in this section consistently support the notion that Hh signaling negatively regulates autophagy and may further inhibit autophagy level.
Autophagy negatively regulated osteoblast differentiation in zebrafish
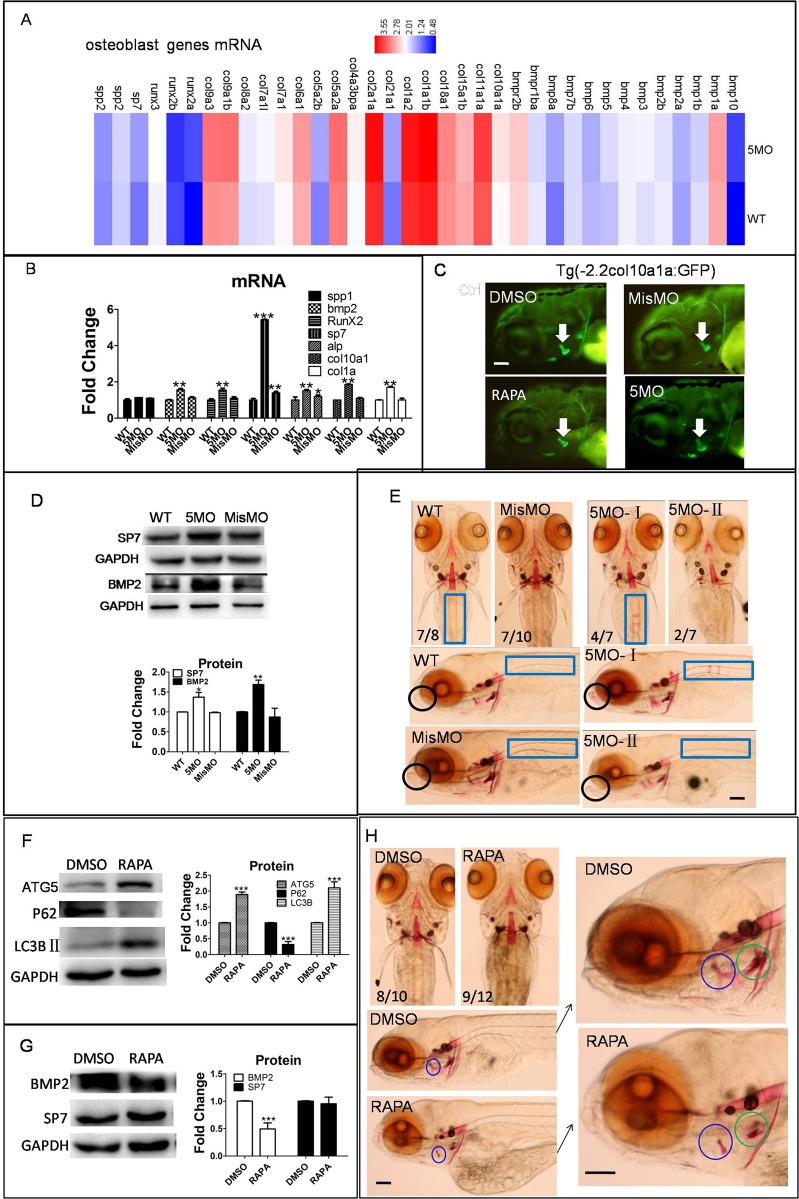
To understand the role of autophagy in osteoblast differentiation, autophagy level was blocked by knockdown autophagy gene atg5. Results showed osteoblast genes runx2a, bmp2, bmp8, bmp10, sp7, col1a, alp and col10a1a were upregulated at transcription level by 5MO. We also showed the regulation of atg5 to many other bmp family genes and col family genes (see details from GEO database, Fig. 4A,B). BMP2 and SP7 protein were also dramatically upregulated by 5MO (Fig. 4D), the protein expression of col10a1a in the op region were increased during the inhibition of autophagy at 4 dpf zebrafish larvae (Fig. 4C). Furthermore, Alizarin Red staining assay demonstrated that osteoblast mineralization of the rib and dentary regions was increased by atg5 knockdown at 6 dpf zebrafish larvae, the den region mineralization was observed even in abnormal phenotype, although the total staining was reduced in 5MO- abnormal group (Fig. 4E).
Fig. 4.
Osteoblast differentiation was regulated by autophagy in zebrafish larvae. (A–E) Embryos or transgenic Tg (-2.2col10a1a:GFP) embryos were injected with 5MO and MisMO at the 1–4 cell stage, and larvae were collected at 6 dpf for transcriptom sequencing, qPCR, western blot and Alizarin Red staining, transgenic larvae were collected at 4 dpf for image capture. (A) Heat map showed osteoblast-related genes mRNA level, data were from transcriptome sequencing. (B) Transcriptional level of osteoblast genes were detected by qPCR. (C) Col10a1a expression pattern in transgenic zebrafish larvae at 4 dpf. (D) Osteoblast proteins SP7 and BMP2 were detected by western blot. (E) Alizarin Red staining showed the mineralization of osteoblast at larvae den and rib. 5MO-II:abnormal phenotype. (F–H) Larvae were exposed to 50 µM RAPA at 3 dpf, larvae were collected at 6 dpf for western blot and staining assay separately. (F,G) Western blot assay showed autophagy proteins and osteoblast proteins were changed by RAPA. (H) Alizarin Red staining showed osteoblast mineralization. Black circle, dentary (den); blue rectangle, rib; blue circle, opercle (op); green circle, ceratobranchial 5 (cb5). The means and s.d. were derived from triple duplicates, *P<0.05, **P<0.01, ***P<0.001 versus WT groups (one-way ANOVA). The data of western blots were from three independent experiments. Scale bars: 50 µm.
Next, RAPA, an inducer of autophagy, was used to treat zebrafish larvae. An increase of autophagy on osteoblasts was observed with upregulation of ATG5 and downregulation of P62 (Fig. 4F). However, col10a1a protein in the op region was reduced by RAPA treatment. Western blot results showed that BMP2 protein was downregulated, but SP7 protein had no obvious change (Fig. 4G). The osteoblast mineralization was reduced by RAPA, especially at the op region (Fig. 4H).
In summary, our results demonstrate that inhibition of autophagy induced osteoblast-related gene upregulation in transcription and protein levels, and promoted osteoblast mineralization. On the contrary, increasing autophagy by RAPA inhibited osteoblast gene expression and reduced osteoblast mineralization. These results imply that autophagy negatively regulates osteoblast differentiation.
Activation of the Hh signaling pathway promoted osteoblast differentiation in zebrafish larvae
Hh signaling pathway inhibitor cyA suppressed osteoblast differentiation in zebrafish larvae
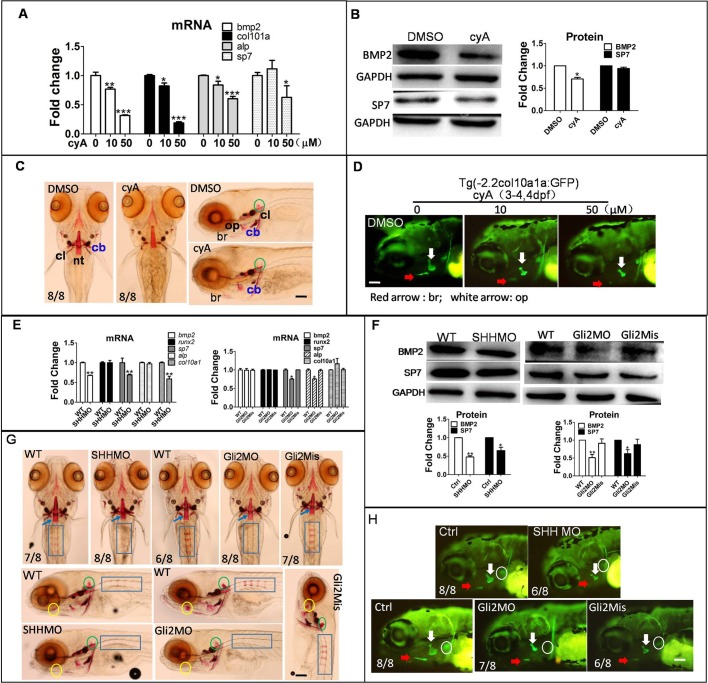
To investigate the effect of Hh signaling inhibitor cyA on the osteoblast differentiation, zebrafish larvae were exposed to cyA at 3 dpf. qPCR results showed osteoblast-related genes bmp2, col10a, alp and sp7 mRNA were downregulated in a dose-dependent manner (Fig. 5A). col10a1a: GFP transgenic zebrafish line was used to detect col10a1a protein location and expression and, after cyA treatment, col10a1a protein was inhibited in the br and op regions in a dose-independent manner (Fig. 5D) and BMP2 and SP7 protein were downregulated by cyA (Fig. 5B). Alizarin Red staining showed mineralization of osteoblasts was obviously suppressed by cyA (Fig. 5C), particularly in the cb region as well as in the cl, nt and op regions, indicating that osteoblast differentiation was decreased, which is consistent with the results in Fig. 5D.
Fig. 5.
Reduction of Hh signaling inhibited osteoblast differentiation. Larvae were exposed to 10 µM and 50 µM cyA at 3 dpf for 3 days, and were collected to measure osteoblast-related gene mRNA, protein level and mineralization of osteoblasts with DMSO treated as a control. (A) qPCR detected osteoblast genes in transcription level. (B) BMP2 and SP7 proteins were detected by western blot. (C) Alizarin Red staining showed mineralization of osteoblasts. Note that the DMSO panel on the right is reproduced from middle WT panel in Fig. 4E. (D) Transgene larvae Tg (-2.2col10a1a:GFP) were treated with 10 µM and 50 µM cyA at 3 dpf for 1 day, images were obtained by fluorescence microscope at 4 dpf. Embryos were injected with SHHMO, Gli2MO and Gli2Mis at the 1–4 cell stage. Lateral view. (E–H) At 6 dpf, larvae were collected for qPCR, western blot and Alizarin Red staining. (E) Osteoblast genes bmp2, sp7, col10a1, runx2 and alp mRNA were detected by qPCR. (F) Osteoblast-related proteins BMP2 and SP7 were tested by western blot. (G) Alizarin Red staining showed mineralization of osteoblasts including nt (notochord tip, blue arrow), rib (blue rectangle), hm (hyomandibular, yellow circle) and regions delayed in SHHMO- and Gli2MO-injected groups. Green circle, bop (basioccipital articulatory process). The difference in osteoblast name and abbreviation were derived from previous sources (Laue et al., 2008; Aceto et al., 2015). (H) In Tg (-2.2col10a1a:GFP) transgene zebrafish, col10a1a signal was shown. White circle, cb (ceratobranchial 5); red arrow, br; white arrow, op. *P<0.05, **P<0.01, ***P<0.001 versus untreated groups (one-way ANOVA). The data were from three independent assays. Scale bars: 50 µm.
Osteoblastic differentiation was reduced after knocking down SHH and Gli2 genes in zebrafish larvae
To examine the function of the Hh signaling pathway in osteoblasts, we chose to decrease Hh signaling by injecting SHHMO and Gli2MO into zebrafish embryos at the 1–4 cell stage. In SHHMO-injected groups, results showed that BMP2, SP7 and col10a1a were downregulated in mRNA and protein levels (Fig. 5E,F,H), Furthermore, knockdown of SHH reduced col10a1a protein in the op and br regions (Fig. 5H). Positive positions in rib and bop were reduced by SHHMO (Fig. 5G). Transcriptional factor Gli2 in Hh signaling downstream did not affect osteoblast-related gene mRNA, but downregulated BMP2 and SP7 protein abundance (Fig. 5E,F). Col10a1a in the op, cl and cb regions were reduced by Gli2MO (Fig. 5H), and in Gli2MO-injected groups, Alizarin Red staining at rib and hm were decreased, indicating that Gli2MO inhibited osteoblast mineralization.
The above results suggest that osteoblast mineralization is blocked or decreased after suppressing Hh signaling by compound cyA or by knockdown Shh and Gli2 genes.
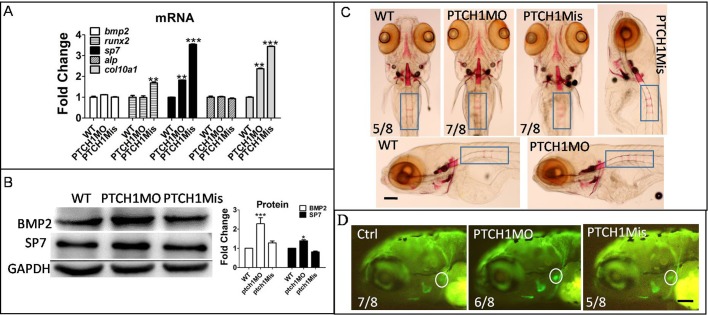
Upregulation of Hh signaling by ptch1 MO-induced osteoblast differentiation
To further explore the role of Hh signaling in osteoblast differentiation, we activated Hh signaling by knockdown ptch1. Results showed sp7 and col10a1 mRNAs were upregulated in ptch1MO-injected groups, but bmp2, runx2 and alp mRNA did not change (Fig. 6A). BMP2 and SP7 proteins increased in the ptch1MO-injected group (Fig. 6B). The col10a1 protein located in the cb region was increasing in the ptch1MO-injected group (Fig. 6D). The mineralization of larvae rib was promoted in ptch1 group compared to wild-type (WT) and ptch1Mis groups (Fig. 6C).
Fig. 6.
Increasing Hh signaling by Ptch1 knockdown might be beneficial for osteoblast development. Embryos were injected with Ptch1MO and Ptch1Mis at the 1–4 cell stage, at 6 dpf, larvae were collected for qPCR, western blot and Alizarin Red staining respectively. (A) QPCR showed osteoblast genes mRNA level. (B) Western blot results showed BMP2 and SP7 protein level. (C) Alizarin Red staining showed bone mineralization; blue rectangle, rib. (D) Col10a1a protein expression in cb5 (ceratobranchial 5, white circle) region was observed in Ptch1MO-injected group. *P<0.05, **P<0.01, ***P<0.001 versus the untreated groups (one-way ANOVA). The data were from three independent assays.
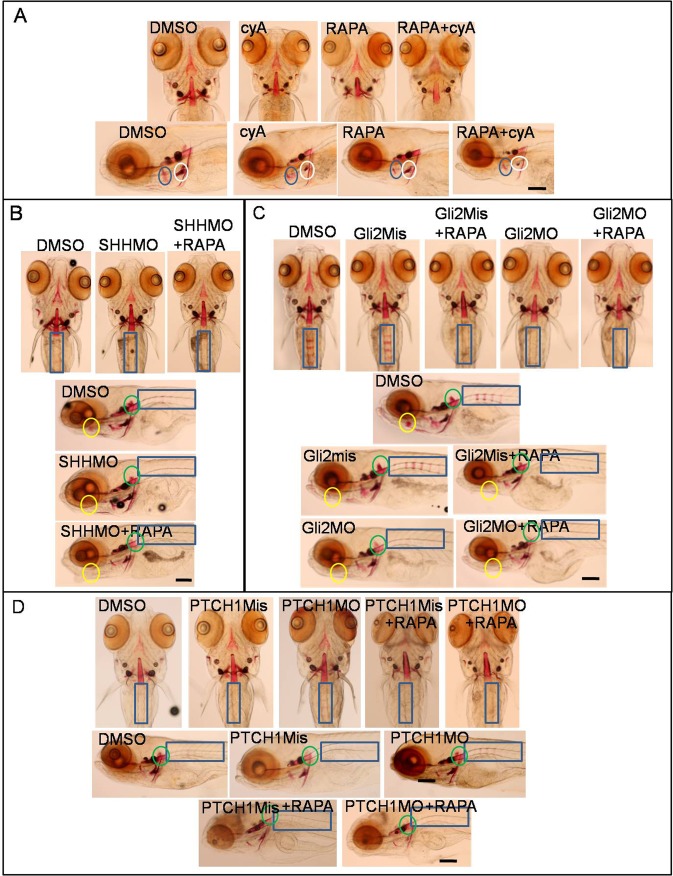
Activation of autophagy and inhibition of Hh signaling synergistically impair bone development in zebrafish larvae
To further reveal the role of autophagy level and Hh signaling in osteoblast mineralization, larvae at 3 dpf were treated by cyA, RAPA and cyA plus RAPA for 3 days, staining by Alizarin Red showed cyA mainly inhibited cb5 region osteoblast mineralization, RAPA suppressed osteoblast mineralization in the op area. However, combination treatment of RAPA and cyA inhibited osteoblast mineralization in broad areas, including the op, cb5, nt, hm regions and so on (Fig. 7A). Moreover, with suppression of Hh signaling by knockdown of SHH and Gli2 genes, at 3 dpf, larvae were exposed to RAPA for 3 days, and then were collected for Alizarin Red staining. Results showed osteoblast mineralization of the rib and hm regions is less in the SHHMO-injected group than in DMSO control, however RAPA-treated SHHMO-injected group intensified osteoblast mineralization loss in the rib and hm regions compared to SHHMO-injected group. Osteoblast staining reduced in rib, hm and bop regions in RAPA plus Gli2MO group compared with WT, Gli2MO, Gli2Mis and Gli2mis plus RAPA groups (Fig. 7B,C). While promotion of Hh signal by knockdown PTCH1 enhanced osteoblast mineralization, it was blocked by autophagy inducer RAPA (Fig. 7D). In this section, data suggest that the inhibition of the Hh signaling pathway and activation of autophagy synergistically suppressed bone development and osteoblast mineralization. However, the bone mineralization enhancement, dependent on Hh signal activation, was blocked by activation of autophagy.
Fig. 7.
Autophagy inducer RAPA and block of Hh signaling co-inhibited osteoblast mineralization. (A) Larvae at 3 dpf were exposed to 10 µM cyA, 10 µM RAPA or cyA plus RAPA, with larvae collected for Alizarin Red staining at 6 dpf. Blue circle, op; white circle, cb. (B–D) Embryos were injected with 50 µM SHHMO, Gli2MO, Gli2Mis, PTCH1MO and PTCH1Mis at the 1–4 cell stage, DMSO control group and injected group larvae were treated with 10 µM RAPA, with larvae collected at 6 dpf for Alizarin Red staining. Yellow circle, bop; green circle, hm; blue rectangle, rib. Scale bars: 50 µm.
DISCUSSION
Reports about the relationship between Hh signaling and autophagy exist paradoxically in cell models. For instance, in hippocampal neurons and intestinal secretory cells, reports indicate the Hh signaling pathway promotes autophagy (Petralia et al., 2013; Gagné-Sansfaçon et al., 2014), but studies in human hepatocellular carcinoma cells, pancreatic ductal adenocarcinoma cells and cancer cells suggest that inhibition of Hh signaling induces autophagy and that autophagy is a key factor for the Hh signaling pathway to regulate a variety of biological functions (Wang et al., 2013; Xu et al., 2014; Li et al., 2012; Tang et al., 2015; Jimenez-Sanchez et al., 2012). It is probable that Hh signaling has completely different roles on autophagy depending on cell types and patho-physiological conditions. In this study, we dissected the interaction between Hh signaling and autophagy in vivo zebrafish model. Our results clearly indicate that autophagy has no influence on Hh signaling, whereas Hh signaling negatively regulates autophagy (Fig. 1). Specifically, inhibition of Hh signaling by pharmacological and knockdown approaches consistently promoted autophagy activity, while activation of Hh signaling by knocking down the expression of its negative regulator Ptch1 protein inhibited autophagy. Interestingly, inhibition or activation of Hh signaling was associated with up- or downregulation of autophagy-related protein ATG5, respectively (Figs 2 and 8). Our results suggest that the Hh signaling pathway is an upstream regulator of autophagy, at least in zebrafish osteoblasts. This notion is in agreement with an early report that the autophagy upstream inducer beclin1 was regulated by the Hh signaling pathway through Gli2 expression (Won et al., 2015). In this study, Gli2 knockdown could indeed increase autophagy proteins ATG5 and LC3B. Whether BECLIN1 mediates the Hh signaling pathway regulation of autophagy in zebrafish remains to be determined.
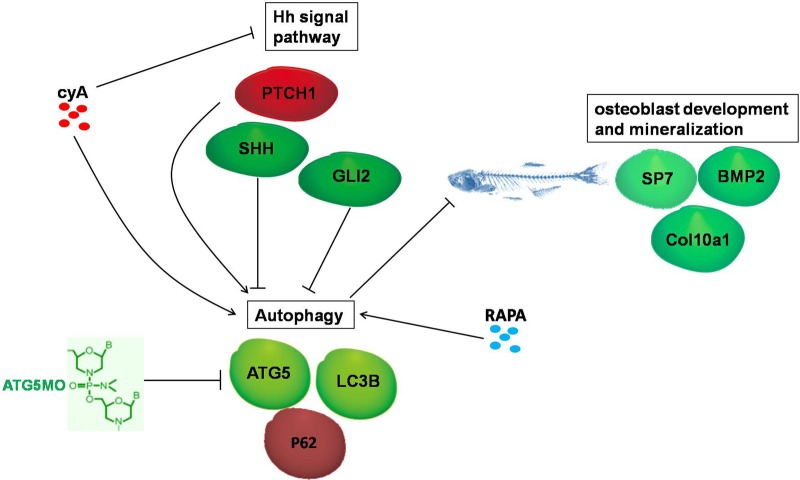
Fig. 8.
Schematic illustrates the interaction between Hh signaling and autophagy, and their regulated roles on osteoblast differentiation and mineralization.
Autophagy was reported to play an essential role in bone development. For instance, deleting ATG7, an essential factor in autophagy level, in mice led to low bone mass and reduction of osteoblast number (Piemontese et al., 2016; Bartolomé et al., 2013). However, in agreement with our findings reported herein, other studies showed that autophagy played a negatively regulatory role in osteoblast differentiation. Specifically, suppression of autophagy by inhibitors or autophagy-related gene knockdown could increase osteoblast differentiation and activity (Kim et al., 2017; Whitehouse et al., 2010). On the other hand, autophagy inducer RAPA dramatically inhibited osteoblast differentiation and mineralization (Singha et al., 2008; Isomoto et al., 2007). In this study, we obtained several lines of evidence suggesting that autophagy does regulate osteoblast differentiation in zebrafish larvae. Knockdown autophagy gene atg5 promoted osteoblast genes and proteins expression, and increased osteoblast mineralization. Also, autophagy inducer RAPA treatment led to BMP2 reduction, while osteoblast mineralization in the op and cb regions were decreased, especially in the op region of zebrafish larvae (Fig. 4).
RAPA is an mTORC1 inhibitor, mTORC1 had been demonstrated to be an important kinase for bone formation (Sugamori et al., 2016). As an autophagy inducer, RAPA also promotes ATG5 and LC3B expression (Hu et al., 2011) and our data indicate as such. Also we report that RAPA inhibits P62 protein level in zebrafish larvae (Fig. 4). It is thus possible that RAPA suppresses osteobalstic differentiation and mineralization through both the mTORC1 and autophagy pathways.
Hh signaling plays a vital role in osteoblast differentiation and mineralization (Bae et al., 2016; Nakamura et al., 2015). Shh has the ability to induce ectopic cartilage and bone formation in vivo (Iwamoto et al., 1999) and stimulates osteoblastic differentiation mainly through upregulation of SP7 gene expression in osteoblastic cells (Tian et al., 2012), Hh signaling-related zinc finger transcription factor Gli2 mediates BMP2 expression in osteoblasts (Hojo et al., 2013; Zhao et al., 2006). Our data supports that Hh signaling promotes osteoblastic differentiation and mineralization in the zebrafish larvae model. Firstly, SHH dysfunction downregulated BMP2, SP7 and col10a1 expression, in addition osteoblastic mineralization located in rib, hm, op and cb5 regions of zebrafish larvae were damaged. Secondly, Gli2 dysfunction resulted in downregulation of BMP2 and SP7 protein, and reduction of bone mineralization area. The conclusion was identified in a zebrafish fin regeneration study by Armstrong et al. (2017). Thirdly, Hh signaling inhibitor cyA downregulated osteoblast gene BMP2, SP7 and col10a1 in transcription and translation levels – following osteoblastic mineralization – decreased, especially the mineralization located in the cb5 region, which was strongly reduced in zebrafish larvae. Fourthly, promoting Hh signaling by knockdown ptch1 gene, resulted in upregulation of osteoblast-related proteins.
In summary, using the zebrafish larvae model, we illustrated that the autophagy pathway has no obvious role in regulating the Hh signaling pathway, but the Hh signaling pathway negatively regulates autophagy, including inhibited autophagy protein ATG5, which implies that autophagy might be a downstream pathway of Hh signaling. Interestingly, both the Hh signaling pathway and autophagy are important regulators of osteoblastic differentiation and mineralization. Hh signaling was a positive regulator, whereas autophagy was a negative regulator signal on osteoblast differentiation. Most importantly, the Hh signaling pathway inducing osteoblast differentiation seems to occur by suppressing the autophagy pathway (Fig. 8). Our work reveals the mechanism underlying Hh signaling pathway regulation of bone development, and thus establishes a molecular basis for development of therapeutics for metabolic bone disorders.
MATERIALS AND METHODS
Fish handling and embryo preparation
Zebrafish (Danio rerio) wild-type AB strain was originally obtained from the College of Life Sciences and Technology, Tsinghua University. Fish feeding, breeding and maintenance were according to the literature (Kimmel et al., 1995). Embryos were obtained by natural mating; synchronous embryos at the appropriate stage were collected. Transgenic zebrafish lines Tg (-2.2col10a1a:GFP) were bought from CZRC (China Zebrafish Resource Center), fluorescence images were taken using a fluorescence microscope (Olympus, IX 51). This research was reviewed and approved by the Laboratory Animal Management and Animal Welfare Committee at the Institute of Medicinal Biotechnology of the Chinese Academy of Medical Sciences and all efforts were made to minimize the animals' suffering.
Synthesis of MOs and microinjection
All morpholino oligos were synthesized and bought from Gene-tools LLC (http://www.gene-tools.com), 5MO and MisMO sequences were synthesized according to our previous published data (Hu et al., 2011; Hu et al., 2017). SHHMO sequence ′5-GCAGCACTCTCGTCAAAAGCCGCAT-3′ was according to Nasevicius and Ekker, (2000). Gli2MO/Ptch1MO was used to target the initiation site of Gli2/Ptch1 mRNA and inhibit translation, Gli2Mis and Ptch1Mis was used as control, and designed by Gene Tools, LLC. The sequences were as follows. Gli2MO-5′ CTCCATGATGAGACTTCTTGGATGA3′; Gli2Mis:5′CTCgATcATcAGACTTgTTcGATGA3′; Ptch1MO-5′ACATTAACAGCCGAGGCCATGTTGC3′; Ptch1Mis-5′ACATaAAgAcCCGAGcCCATcTTGC3′. The 50 µM MOs were injected into 1–4 cell stage embryos.
Drug treatment
10 mM stocking RAPA (provided by National Institutes for Food and Drug Control) solution in DMSO was added to the embryo media at final concentrations of 10 µM from 3 dpf to 6 dpf, and cyA (Sigma-Aldrich, C4116), were dissolved into DMSO, and then was diluted to 10 µM and 50 µM. 0.1% DMSO treated larvae was used as negative control. 3MA (Sigma-Aldrich, M9281) was dissolved into heated ddH2O.
qPCR and RNA-seq
Larvae at 6 dpf were collected into Trizol Reagent (Sigma-Aldrich). Total RNA was extracted following the Trizol Reagent RNA extraction kit manual. First-strand cDNAs were synthesized by reverse transcription using the M-MLV RTase cDNA Synthesis Kit (Promega). qPCR primer pairs are listed in Table 1, parameters were 95°C 5 min, (95°C 15 s, 68°C 40 s), for 40 cycles. β-actin was amplified as a template loading control. Then the ct values were analyzed using a statistical calculation according to the instrument manual. WT group and 5MO-injected group larvae (30 larvae each group) at 6 dpf were sent to Compass biotechnology company for transcriptome sequencing, the mRNA-related Hh signaling and osteoblast were analyzed. RNA-seq data had been deposited with GEO under accession number GSE120170.
Table 1.
qPCR primers and their sequences

Western blot
The procedure has been previously described in our previous literature (Hu et al., 2011). Briefly, zebrafish larvae total protein was extracted with the RIPA Lysis Kit (Applygen Technologies Inc, C1053), different protein samples were separated in 12% SDS-PAGE gel, then transferred to NC (nitrocellulose) membrane, and blocked with 5% milk in TBS for 40 min–1 h at room temperature. Then, the membranes were incubated with primary antibody at 4°C overnight, TBS washed three times and incubated with horseradish peroxidase-conjugated secondary antibodies 1 h at room temperature. The membranes were exposed to the Tanon 5200 Imaging System and we acquired the images. The primary antibody information is as follows: mouse anti-human β-GAPDH antibody (Zhongshan Goldbridge, TA08); anti-human ATG5 (Novus, NB110-53818), P62 rabbit polyclonal antibody (MBL, PM045), LC3B mouse monoclonal antibody (MBL, M186-3), SP7 (osterix) goat polyclonal antibody (Santa Cruz, sc-22538) and BMP2 Mouse Monoclonal antibody (Novus, MAB 1128).
Alizarin Red staining
Larvae at 6 dpf were collected and fixed for 2 h by 4% paraformaldehyde at room temperature, fixed larvae were stained with 0.1% Alizarin Red (Sigma-Aldrich, A5533) in 0.5% KOH overnight. Stained larvae were gradient rehydrated with 75% ethanol (100 mM Tris pH 7.5, 10 mM MgCl2), 50% ethanol and 25% ethanol, bleached with 3% H2O2 containing 1% KOH for 20 min (Luo et al., 2016), and rinsed six times with ddH2O. Samples were then placed in 90% glycerol for imaging (Olympus, IX 51).
Statistical analysis
Data in figures represent mean±s.d., derived from at least three experiments. Statistical analyses were performed using one-way ANOVA tests.
Acknowledgements
We thank professor Anming Meng (Tsinghua University), who provided zebrafish AB stain seedling; and Jie Meng for fish husbandry. We also thank Prof. Jutao Guo and Dr Liudi Tang for revising this manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: Z.H.; Methodology: Z.H., B.C.; Software: B.C.; Formal analysis: Z.H., Q.Z.; Investigation: Z.H.; Data curation: Z.H.; Writing - original draft: Z.H.; Writing - review & editing: B.C., Q.Z.; Project administration: Z.H.; Funding acquisition: Z.H.
Funding
This study was supported by The National Natural Science Foundation of China (No. 81402986).
Data availability
RNA-seq data has been deposited in the GEO database under accession number GSE120170.
References
- Aceto J., Nourizadeh-Lillabadi R., Marée R., Dardenne N., Jeanray N., Wehenkel L., Aleström P., van Loon J. J. W. A. and Muller M. (2015). Zebrafish bone and general physiology are differently affected by hormones or changes in gravity. PLoS ONE 10, e0126928 10.1371/journal.pone.0126928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano K., Densmore M. J. and Lanske B. (2015). Conditional deletion of indian hedgehog in limb mesenchyme results in complete loss of growth plate formation but allows mature osteoblast differentiation. J. Bone Miner. Res. 30, 2262-2272. 10.1002/jbmr.2582 [DOI] [PubMed] [Google Scholar]
- Armstrong B. E., Henner A., Stewart S. and Stankunas K. (2017). Shh promotes direct interactions between epidermal cells and osteoblast progenitors to shape regenerated zebrafish bone. Development 144, 1165-1176. 10.1242/dev.143792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avaron F., Hoffman L., Guay D. and Akimenko M. A. (2006). Characterization of two new zebrafish members of the hedgehog family: atypical expression of a zebrafish indian hedgehog gene in skeletal elements of both endochondral and dermal origins. Dev. Dyn. 235, 478-489. 10.1002/dvdy.20619 [DOI] [PubMed] [Google Scholar]
- Bae W.-J., Auh Q.-S., Lim H.-C., Kim G.-T., Kim H.-S. and Kim E.-C. (2016). Sonic hedgehog promotes cementoblastic differentiation via activating the BMP pathways. Calcif. Tissue Int. 99, 396-407. 10.1007/s00223-016-0155-1 [DOI] [PubMed] [Google Scholar]
- Bartolomé A., López-Herradón A., Portal-Núñez S., García-Aguilar A., Esbrit P., Benito M. and Guillén C. (2013). Autophagy impairment aggravates the inhibitory effects of high glucose on osteoblast viability and function. Biochem. J. 455, 329-337. 10.1042/BJ20130562 [DOI] [PubMed] [Google Scholar]
- Blum N. and Begemann G. (2015). Retinoic acid signaling spatially restricts osteoblasts and controls ray-interray organization during zebrafish fin regeneration. Development 142, 2888-2893. 10.1242/dev.120212 [DOI] [PubMed] [Google Scholar]
- Cai H. and Liu A. (2016). Spop promotes skeletal development and homeostasis by positively regulating Ihh signaling. Proc. Natl. Acad. Sci. USA 113, 14751-14756. 10.1073/pnas.1612520114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M., Kicheva A., Ribeiro A., Blassberg R., Page K. M., Barnes C. P. and Briscoe J. (2015). Ptch1 and Gli regulate Shh signalling dynamics via multiple mechanisms. Nat. Commun. 6, 6709 10.1038/ncomms7709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edsall S. C. and Franz-Odendaal T. A. (2010). A quick whole-mount staining protocol for bone deposition and resorption. Zebrafish 7, 275-280. 10.1089/zeb.2009.0641 [DOI] [PubMed] [Google Scholar]
- Gagné-Sansfaçon J., Allaire J. M., Jones C., Boudreau F. and Perreault N. (2014). Loss of Sonic hedgehog leads to alterations in intestinal secretory cell maturation and autophagy. PLoS ONE 9, e98751 10.1371/journal.pone.0098751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S.-W., Weitzmann M. N. and Beck G. R. Jr. (2014). Bioactive silica nanoparticles promote osteoblast differentiation through stimulation of autophagy and direct association with LC3 and p62. ACS Nano 8, 5898-5910. 10.1021/nn5009879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocking L. J., Whitehouse C. and Helfrich M. H. (2012). Autophagy: a new player in skeletal maintenance? J. Bone Miner. Res. 27, 1439-1447. 10.1002/jbmr.1668 [DOI] [PubMed] [Google Scholar]
- Hojo H., Ohba S., Taniguchi K., Shirai M., Yano F., Saito T., Ikeda T., Nakajima K., Komiyama Y., Nakagata N. et al. (2013). Hedgehog-Gli activators direct osteo-chondrogenic function of bone morphogenetic protein toward osteogenesis in the perichondrium. J. Biol. Chem. 288, 9924-9932. 10.1074/jbc.M112.409342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Zhang J. and Zhang Q. (2011). Expression pattern and functions of autophagy-related gene atg5 in zebrafish organogenesis. Autophagy 7, 1514-1527. 10.4161/auto.7.12.18040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z.-Y., Chen B., Zhang J.-P. and Ma Y.-Y. (2017). Up-regulation of autophagy-related gene 5 (ATG5) protects dopaminergic neurons in a zebrafish model of Parkinson's disease. J. Biol. Chem. 292, 18062-18074. 10.1074/jbc.M116.764795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui C.-C. and Angers S. (2011). Gli proteins in development and disease. Annu. Rev. Cell Dev. Biol. 27, 513-537. 10.1146/annurev-cellbio-092910-154048 [DOI] [PubMed] [Google Scholar]
- Huycke T. R., Eames B. F. and Kimmel C. B. (2012). Hedgehog-dependent proliferation drives modular growth during morphogenesis of a dermal bone. Development 139, 2371-2380. 10.1242/dev.079806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isomoto S., Hattori K., Ohgushi H., Nakajima H., Tanaka Y. and Takakura Y. (2007). Rapamycin as an inhibitor of osteogenic differentiation in bone marrow-derived mesenchymal stem cells. J. Orthop. Sci. 12, 83-88. 10.1007/s00776-006-1079-9 [DOI] [PubMed] [Google Scholar]
- Iwamoto M., Enomoto-Iwamoto M. and Kurisu K. (1999). Actions of hedgehog proteins on skeletal cells. Crit. Rev. Oral Biol. Med. 10, 477-486. 10.1177/10454411990100040401 [DOI] [PubMed] [Google Scholar]
- Jimenez-Sanchez M., Menzies F. M., Chang Y.-Y., Simecek N., Neufeld T. P. and Rubinsztein D. C. (2012). The Hedgehog signalling pathway regulates autophagy. Nat. Commun. 3, 1200 10.1038/ncomms2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joeng K. S. and Long F. (2013). Constitutive activation of Gli2 impairs bone formation in postnatal growing mice. PLoS ONE 8, e55134 10.1371/journal.pone.0055134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kague E., Roy P., Asselin G., Hu G., Simonet J., Stanley A., Albertson C. and Fisher S. (2016). Osterix/Sp7 limits cranial bone initiation sites and is required for formation of sutures. Dev. Biol. 413, 160-172. 10.1016/j.ydbio.2016.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-I., Lee S., Jung S.-H., Kim H.-T., Choi J.-H., Lee M.-S., You K.-H., Yeo S.-Y., Yoo K.-W., Kwak S. et al. (2013). Establishment of a bone-specific col10a1:GFP transgenic zebrafish. Mol. Cells 36, 145-150. 10.1007/s10059-013-0117-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Lee H.-W., Rhee D. K., Paton J. C. and Pyo S. (2017). Pneumolysin-induced autophagy contributes to inhibition of osteoblast differentiation through downregulation of Sp1 in human osteosarcoma cells. Biochim. Biophys. Acta 1861, 2663-2673. 10.1016/j.bbagen.2017.07.008 [DOI] [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B. and Schilling T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253-310. 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Kuo P.-L., Huang Y.-T., Chang C.-H. and Chang J.-K. (2006). Bone morphogenetic protein-2 and −4 (BMP-2 and −4) mediates fraxetin-induced maturation and differentiation in human osteoblast-like cell lines. Biol. Pharm. Bull. 29, 119-124. 10.1248/bpb.29.119 [DOI] [PubMed] [Google Scholar]
- Laue K., Janicke M., Plaster N., Sonntag C. and Hammerschmidt M. (2008). Restriction of retinoic acid activity by Cyp26b1 is required for proper timing and patterning of osteogenesis during zebrafish development. Development 135, 3775-3787. 10.1242/dev.021238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Felber K., Elks P., Croucher P. and Roehl H. H. (2009). Tracking gene expression during zebrafish osteoblast differentiation. Dev. Dyn. 238, 459-466. 10.1002/dvdy.21838 [DOI] [PubMed] [Google Scholar]
- Li H., Li J., Li Y., Singh P., Cao L., Xu L.-J., Li D., Wang Y., Xie Z., Gui Y. et al. (2012). Sonic hedgehog promotes autophagy of vascular smooth muscle cells. Am. J. Physiol. Heart Circ. Physiol. 303, H1319-H1331. 10.1152/ajpheart.00160.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Fang F., Yuan H., Yang D., Chen Y., Williams L., Goldstein S. A., Krebsbach P. H. and Guan J.-L. (2013). Suppression of autophagy by FIP200 deletion leads to osteopenia in mice through the inhibition of osteoblast terminal differentiation. J. Bone Miner. Res. 28, 2414-2430. 10.1002/jbmr.1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S.-Y., Chen J.-F., Zhong Z.-G., Lv X.-H., Yang Y.-J., Zhang J.-J. and Cui L. (2016). Salvianolic acid B stimulates osteogenesis in dexamethasone-treated zebrafish larvae. Acta Pharmacol. Sin. 37, 1370-1380. 10.1038/aps.2016.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marumoto A., Milani R., DA Silva R. A., da Costa Fernandes C. J., Granjeiro J. M., Ferreira C. V., Peppelenbosch M. P. and Zambuzzi W. F. (2017). Phosphoproteome analysis reveals a critical role for hedgehog signalling in osteoblast morphological transitions. Bone 103, 55-63. 10.1016/j.bone.2017.06.012 [DOI] [PubMed] [Google Scholar]
- Mizushima N. (2007). Autophagy: process and function. Genes Dev. 21, 2861-2873. 10.1101/gad.1599207 [DOI] [PubMed] [Google Scholar]
- Nakamura T., Naruse M., Chiba Y., Komori T., Sasaki K., Iwamoto M. and Fukumoto S. (2015). Novel hedgehog agonists promote osteoblast differentiation in mesenchymal stem cells. J. Cell. Physiol. 230, 922-929. 10.1002/jcp.24823 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Zhou X., Kunkel G., Zhang Z., Deng J. M., Behringer R. R. and DE Crombrugghe B. (2002). The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108, 17-29. 10.1016/S0092-8674(01)00622-5 [DOI] [PubMed] [Google Scholar]
- Nasevicius A. and Ekker S. C. (2000). Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet. 26, 216-220. 10.1038/79951 [DOI] [PubMed] [Google Scholar]
- Ohba S., Kawaguchi H., Kugimiya F., Ogasawara T., Kawamura N., Saito T., Ikeda T., Fujii K., Miyajima T., Kuramochi A. et al. (2008). Patched1 haploinsufficiency increases adult bone mass and modulates Gli3 repressor activity. Dev. Cell 14, 689-699. 10.1016/j.devcel.2008.03.007 [DOI] [PubMed] [Google Scholar]
- Oliveira F. S., Bellesini L. S., Defino H. L. A., da Silva Herrero C. F., Beloti M. M. and Rosa A. L. (2012). Hedgehog signaling and osteoblast gene expression are regulated by purmorphamine in human mesenchymal stem cells. J. Cell. Biochem. 113, 204-208. 10.1002/jcb.23345 [DOI] [PubMed] [Google Scholar]
- Pasqualetti S., Banfi G. and Mariotti M. (2012). The zebrafish scale as model to study the bone mineralization process. J. Mol. Histol. 43, 589-595. 10.1007/s10735-012-9425-z [DOI] [PubMed] [Google Scholar]
- Paul S., Schindler S., Giovannone D., de Millo Terrazzani A., Mariani F. V. and Crump J. G. (2016). Ihha induces hybrid cartilage-bone cells during zebrafish jawbone regeneration. Development 143, 2066-2076. 10.1242/dev.131292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia R. S., Schwartz C. M., Wang Y.-X., Kawamoto E. M., Mattson M. P. and Yao P. J. (2013). Sonic hedgehog promotes autophagy in hippocampal neurons. Biol. Open 2, 499-504. 10.1242/bio.20134275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piemontese M., Onal M., Xiong J., Han L., Thostenson J. D., Almeida M. and O'Brien C. A. (2016). Low bone mass and changes in the osteocyte network in mice lacking autophagy in the osteoblast lineage. Sci. Rep. 6, 24262 10.1038/srep24262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H., Hui C., Nakafuku M. and Kondoh H. (1997). A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development 124, 1313-1322. [DOI] [PubMed] [Google Scholar]
- Singha U. K., Jiang Y., Yu S., Luo M., Lu Y., Zhang J. and Xiao G. (2008). Rapamycin inhibits osteoblast proliferation and differentiation in MC3T3-E1 cells and primary mouse bone marrow stromal cells. J. Cell. Biochem. 103, 434-446. 10.1002/jcb.21411 [DOI] [PubMed] [Google Scholar]
- Sugamori Y., Mise-Omata S., Maeda C., Aoki S., Tabata Y., Murali R., Yasuda H., Udagawa N., Suzuki H., Honma M. et al. (2016). Peptide drugs accelerate BMP-2-induced calvarial bone regeneration and stimulate osteoblast differentiation through mTORC1 signaling. BioEssays 38, 717-725. 10.1002/bies.201600104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Deng L., Chen Q., Wang Y., Xu R., Shi C., Shao J., Hu G., Gao M., Rao H. et al. (2015). Inhibition of Hedgehog signaling pathway impedes cancer cell proliferation by promotion of autophagy. Eur. J. Cell Biol. 94, 223-233. 10.1016/j.ejcb.2015.03.003 [DOI] [PubMed] [Google Scholar]
- Tian Y., Xu Y., Fu Q. and Dong Y. (2012). Osterix is required for Sonic hedgehog-induced osteoblastic MC3T3-E1 cell differentiation. Cell Biochem. Biophys. 64, 169-176. 10.1007/s12013-012-9369-7 [DOI] [PubMed] [Google Scholar]
- Wang Y., Han C., Lu L., Magliato S. and Wu T. (2013). Hedgehog signaling pathway regulates autophagy in human hepatocellular carcinoma cells. Hepatology 58, 995-1010. 10.1002/hep.26394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wei S., Zhao Y., Shi C., Liu P., Zhang C., Lei Y., Zhang B., Bai B., Huang Y. et al. (2017). Anti-proliferation of breast cancer cells with itraconazole: Hedgehog pathway inhibition induces apoptosis and autophagic cell death. Cancer Lett. 385, 128-136. 10.1016/j.canlet.2016.10.034 [DOI] [PubMed] [Google Scholar]
- Whitehouse C. A., Waters S., Marchbank K., Horner A., McGowan N. W. A., Jovanovic J. V., Xavier G. M., Kashima T. G., Cobourne M. T., Richards G. O. et al. (2010). Neighbor of Brca1 gene (Nbr1) functions as a negative regulator of postnatal osteoblastic bone formation and p38 MAPK activity. Proc. Natl. Acad. Sci. USA 107, 12913-12918. 10.1073/pnas.0913058107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won K. Y., Kim G. Y., Lim S.-J., Sung J.-Y., Kim Y. W., Park Y.-K., Lee J. and Choi H. S. (2015). Autophagy is related to the hedgehog signaling pathway in human gastric adenocarcinoma: prognostic significance of Beclin-1 and Gli2 expression in human gastric adenocarcinoma. Pathol. Res. Pract. 211, 308-315. 10.1016/j.prp.2014.11.005 [DOI] [PubMed] [Google Scholar]
- Xu Y., An Y., Wang X., Zha W. and Li X. (2014). Inhibition of the Hedgehog pathway induces autophagy in pancreatic ductal adenocarcinoma cells. Oncol. Rep. 31, 707-712. 10.3892/or.2013.2881 [DOI] [PubMed] [Google Scholar]
- Yang Y.-H., Chen K., Li B., Chen J.-W., Zheng X.-F., Wang Y.-R., Jiang S.-D. and Jiang L.-S. (2013). Estradiol inhibits osteoblast apoptosis via promotion of autophagy through the ER-ERK-mTOR pathway. Apoptosis 18, 1363-1375. 10.1007/s10495-013-0867-x [DOI] [PubMed] [Google Scholar]
- Zhao M., Qiao M., Harris S. E., Chen D., Oyajobi B. O. and Mundy G. R. (2006). The zinc finger transcription factor Gli2 mediates bone morphogenetic protein 2 expression in osteoblasts in response to hedgehog signaling. Mol. Cell. Biol. 26, 6197-6208. 10.1128/MCB.02214-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Han Y., Li J., Cai B., Gao H., Feng W., Li S., Liu J. and Li D. (2017). BMP-2 immobilized PLGA/hydroxyapatite fibrous scaffold via polydopamine stimulates osteoblast growth. Mater. Sci. Eng. C Mater. Biol. Appl. 78, 658-666. 10.1016/j.msec.2017.03.186 [DOI] [PubMed] [Google Scholar]