Summary
Iron-sulfur (Fe-S) clusters, pre-assembled on the ISCU scaffold, are transferred to target proteins or to intermediate scaffolds by a dedicated chaperone-cochaperone system. However, the molecular mechanisms that underlie substrate discrimination and guide delivery of nascent clusters to specific subsets of Fe-S recipients are poorly understood. Here, we identified interacting partners of the cochaperone HSC20, and discovered that LYR motifs are molecular signatures of specific recipient Fe-S proteins or accessory factors that assist Fe-S cluster delivery. In succinate dehydrogenase B, two LYR motifs engage the ISCU-HSC20-HSPA9 complex to aid incorporation of three Fe-S clusters within the final structure of Complex II. Moreover, we show that members of the LYR motif family which assist assembly of Complexes II or III, SDHAF1 and LYRM7 respectively, are HSC20 binding partners. Our studies unveil a network of interactions between HSC20 and LYR motif-containing proteins that are key to the assembly and function of Complexes I-III.
INTRODUCTION
Iron sulfur (Fe-S) clusters are essential cofactors, required for the function of proteins involved in a broad range of cellular processes, including electron transport in respiratory chain complexes, photosynthesis, regulatory sensing and DNA repair. In contrast to the chemical simplicity of Fe-S clusters, their synthesis involves a complex sequence of catalyzed protein-protein interactions and coupled conformational changes between the components of several dedicated multimeric complexes (Rouault, 2012). The proteins involved in Fe-S cluster (ISC) biogenesis are evolutionarily conserved, and many insights into the assembly process have been provided by studies of model organisms, including bacteria, fungi and plants (Couturier et al., 2013; Dos Santos and Dean, 2010; Roche et al., 2013). In mammalian cells, Fe-S clusters are assembled by a complex composed of a cysteine desulfurase, NFS1, its binding partner, ISD11, the ISCU scaffold, and an iron donor or allosteric effector, frataxin (Rouault, 2012). Studies in bacteria and yeast have demonstrated that upon assembly of a nascent [2Fe-2S] cluster, the scaffold protein (IscU in bacteria and Isu in yeast) binds to the J-protein (HscB or Jac1, respectively), through hydrophobic contacts (Ciesielski et al., 2012; Fuzery et al., 2011), and to an HSP70 chaperone (HscA or Ssq1) through a conserved LPPVK motif (Dutkiewicz et al., 2004; Vickery and Cupp-Vickery, 2007). An ATP-driven conformational change of the HSP70 enhances transfer of the ISCU-bound cluster to recipient apoproteins or to secondary scaffolds that then deliver the cluster to specific subsets of final acceptors (Vickery and Cupp-Vickery, 2007).
HSC20 is the sole human DnaJ type III cochaperone dedicated to Fe-S cluster biogenesis (Uhrigshardt et al., 2010). Mutations in HSC20 and in its orthologs cause defects in Fe-S protein activities, mitochondrial iron accumulation, and reduced mitochondrial respiration in human cell lines (Uhrigshardt et al., 2010), and in multiple experimental systems, including yeast (Kim et al., 2001; Voisine et al., 2001) and fly (Uhrigshardt et al., 2013). The importance of Fe-S biogenesis for human health is well established, as mutations that affect proteins involved in the pathway cause several distinctive human diseases (Rouault, 2012). Interestingly, studies of four newly described syndromes caused by mutations in NUBPL, NFU1 or BOLA3, and IBA57 (Ajit Bolar et al., 2013; Rouault, 2012) suggest that transfer of Fe-S clusters from the ISCU-chaperone-cochaperone complex depends on selective downstream pathways, which underscores our lack of knowledge about how discrete subsets of Fe-S recipients are targeted.
We conducted a high-throughput yeast two-hybrid (Y2H) screen to search the human proteome for interacting partners of HSC20, aiming to identify Fe-S proteins and specific molecular recognition motifs that guide targeting of Fe-S clusters to appropriate recipients. J-proteins often determine the specificity of their cognate chaperones (Ciesielski et al., 2012; Kampinga and Craig, 2010; Pukszta et al., 2010), and their C-terminal domains can bind substrates (Perales-Calvo et al., 2010; Szabo et al., 1996), facilitate refolding of denatured proteins, and enhance cell viability (Lee et al., 2002; Li and Sha, 2005). Here we found that direct binding of specific targets to the cochaperone HSC20 is mediated by affinity of its C-terminus for proteins that bear the LYR motif, a tripeptide that constitutes a major molecular signature of distinctive Fe-S recipients.
RESULTS
The Fe-S Protein, SDHB, was among the Binding Partners of HSC20 Identified in the Y2H Screen
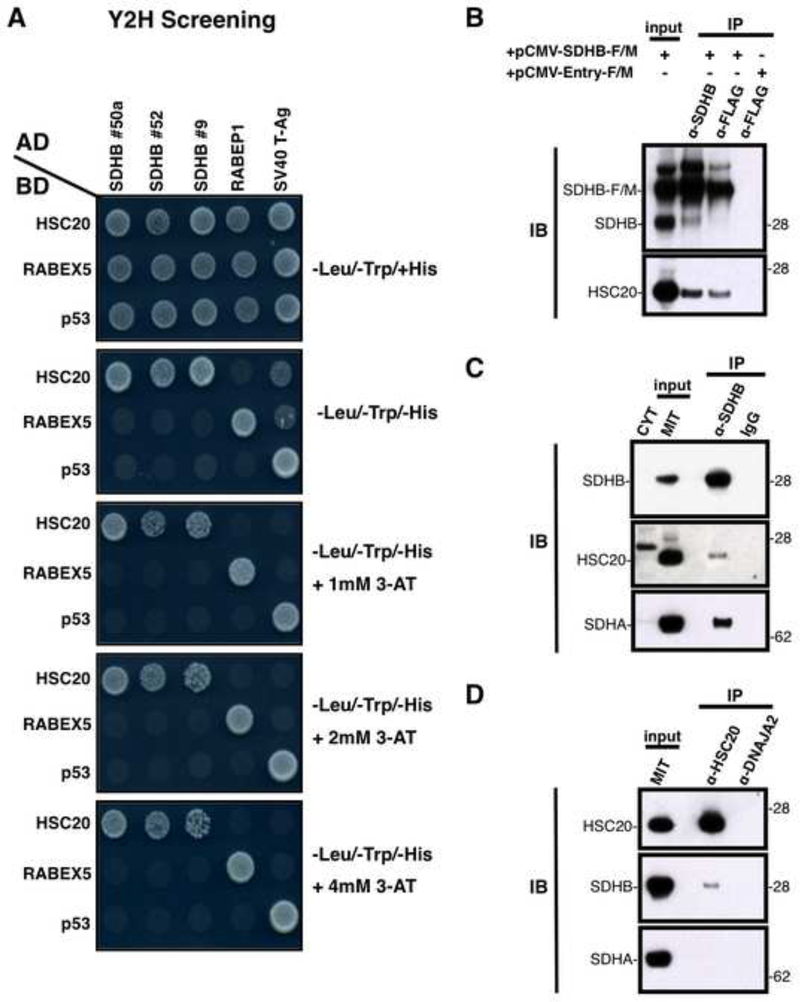
We used a stringent Y2H approach to identify proteins that directly bind to HSC20. In the HSC20-BD-GAL4 clone, residues 29–235 of mature HSC20 were fused to the DNA-binding domain (BD) of the yeast transcriptional factor GAL4 (BD-GAL4), and the fusion protein was used as a bait to screen a human cDNA library (from HeLa cells) from which signal peptides had been excised, and cDNA sequences were fused to the activation domain (AD) of GAL4 (AD-GAL4) to generate “preys”. Specific binary interactions between bait and prey in the nucleus of co-transformed yeast cells reconstituted a functional GAL4 transcription factor, which then switched on the expression of four independent reporter genes, AUR1-C, ADE2, HIS3, and MEL1. We selected for activation of HIS3 using different concentrations of 3-amino-1,2,4-triazole (3-AT) to competitively inhibit histidine synthesis and to select for high HIS3 activation. Approximately 1 × 106 clones were screened under stringent conditions (4 mM 3-AT) and 70 prey plasmids were identified. Some of these represented individual clones that encoded different segments of the same protein, which reduced the total number of candidates to 54 (Table S1). Notably, clones 9, 50a and 52 encoded residues 69–280, 104–280, 115–280, respectively, of succinate dehydrogenase B (SDHB), the Fe-S subunit of the succinate dehydrogenase (SDH) complex (Figure 1A). Growth on plates lacking histidine (-Leu/-Trp/-His) indicated that HSC20 interacted with the protein encoded by the prey construct to activate HIS3 transcription, thus rescuing the histidine-auxotrophy of the yeast strain. As negative controls, the HSC20-BD-GAL4 plasmid was co-transformed with RABEP1 or SV40 T-Ag, and the three SDHB-AD-GAL4 clones with RABEX5 or p53. The expected interactions of RABEX5-RABEP1 and p53-SV40 T-Ag were used as positive controls. Pull down experiments performed with purified HSC20FLAG (HSC20-F) and SDHB-HA further confirmed that HSC20 directly binds to SDHB (Figure S1A).
Figure 1. HSC20 Interacts with SDHB In Vitro and In Vivo.
(A) Three clones from the Y2H library encoding SDHB were identified as interactants of HSC20. Growth on plates lacking histidine (-Leu/-Trp/-His) indicated reconstitution of the GAL4 transcription factor by direct interaction of SDHB with HSC20 (first 3 lanes, top row). As negative controls, the HSC20-BD-GAL4 plasmid was co-transformed with RABEP1 or SV40 T-Ag, and the three SDHB-AD-GAL4 clones with RABEX5 or p53. The expected interactions of RABEX5-RABEP1 and p53-SV40 T-Ag promoted growth and were used as positive controls. (B, C, D) HSC20 interacts with SDHB in vivo. (B) Extracts from HeLa cells transfected with pCMV-SDHB-F/M or with empty vector (pCMV-Entry-F/M, as negative control) were subjected to IP with anti-SDHB or anti-FLAG, followed by IB to HSC20. (C) IP of endogenous SDHB, followed by IBs to HSC20 or SDHA on mitochondrial (MIT) or cytosolic (CYT) fractions. (D) IPs of endogenous HSC20 or DNAJA2, followed by IBs to SDHB or SDHA. The cochaperone DNAJA2 did not interact with HSC20 in vivo (IB to HSC20), indicating that its selection in the Y2H study was a false positive. IP of DNAJA2 was used as a control for the specificity of the interaction between HSC20 and SDHB. (A-D, n = 6 biological samples). Inputs were 30% of total used for IPs. See also Figure S1 and Table S1.
We validated multiple putative interacting partners of HSC20 by co-immunoprecipitation (co-IP) (Table S1 and Figure S1). We found that HSC20 co-immunoprecipitated LYRM7 (Figure S1B), the human Rieske Fe-S protein chaperone of Complex III (Sanchez et al., 2013). Co-IP experiments confirmed the interaction of HSC20 with four more candidates from the screening: the succinyl-coA ligase [GDP-forming] subunit beta, SUCLG2, the quinone reductase NQO2, the putative helicase with zinc finger, HELZ (Figures S1C, S1D), and the bifunctional glutamate-proline tRNA ligase (EPRS) (Figure S1E). Interestingly, we found that HSC20 can dimerize, a property shared by several members of the HSP40 family (Kampinga and Craig, 2010; Shi et al., 2005; Terada and Oike, 2010). HSC20 was among the positive clones identified in initial screens (Table S1), and was further validated in vivo, as endogenous HSC20 co-immunoprecipitated recombinant HSC20-FLAG/MYC in transfected cells (Figures S1D, S1E). Future experiments may uncover other processes mediated by HSC20-dependent Fe-S transfer, since proteins involved in DNA/RNA metabolism, energy metabolism, cell cycle control, transcription and translation regulation were identified in our screen (Table S1), and, among these, SPC25 and GTF2E2 were also present in the MMS19 interaction profile which previously identified potential cytosolic Fe-S recipients (Stehling et al., 2012). Interestingly, we detected a prominent pool of HSC20 in the cytosol, where the cochaperone was found to specifically interact with the cytosolic proteins EPRS, NQO2 and HELZ (Figures S1D, S1E).
Binding of HSC20 to SDHB and Formation of the Chaperone-Cochaperone Transfer Complex in the Mitochondrial Matrix is Required for SDH Assembly and Function
SDHB is the sole Fe-S subunit of Complex II (CII), which is situated at the nexus of two essential energy-generating processes of the cell, the citric acid cycle and mitochondrial oxidative phosphorylation. In eukaryotes, mutations in each of the four nuclear-encoded succinate-coQ oxidoreductase (SQR) genes (SDHA, SDHB, SDHC, SDHD) or associated accessory factors (SDHAF1, SDHAF2) account for a variety of clinical phenotypes, including optic atrophy, myopathy, encephalopathy (Birch-Machin et al., 2000; Ghezzi et al., 2009), and tumor formation (Jain-Ghai et al., 2013; Ricketts et al., 2012). We confirmed by reciprocal co-IP experiments that HSC20 interacts with SDHB in vivo (Figures 1B, 1C, 1D). Endogenous HSC20 was efficiently co-immunoprecipitated both with recombinant SDHB-F/M (Figure 1B) and with endogenous SDHB (Figure 1C). Conversely, IP of endogenous HSC20 demonstrated that SDHB was coprecipitated (Figure 1D), confirming that SDHB and HSC20 are binding partners.
HSPA9/GRP75/Mortalin was previously identified as the HSC20 cognate chaperone by a Y2H assay (Uhrigshardt et al., 2010), and in co-IP experiments when the two proteins were overexpressed (Shan and Cortopassi, 2012). We verified that endogenous HSC20 specifically interacted in vivo with HSPA9 (Figure S2A), and with ISCU (Figures S2B, S2C), confirming that the orthologous interactions reported in bacteria and yeast (Vickery and Cupp-Vickery, 2007; Voisine et al., 2001), and recently in an in vitro study employing purified human proteins (Cai et al., 2013), also °Ccur in mammals with HSPA9, but not with its human homologue, HSPA14 (Figures 2A, S2A). Interestingly, the region containing residues 439–590 of HSPA9 shares 97% similarity with the domain of bacterial HscA that binds IscU, and the high degree of sequence homology translates into a remarkable structural conservation between the two domains (Figure S2D).
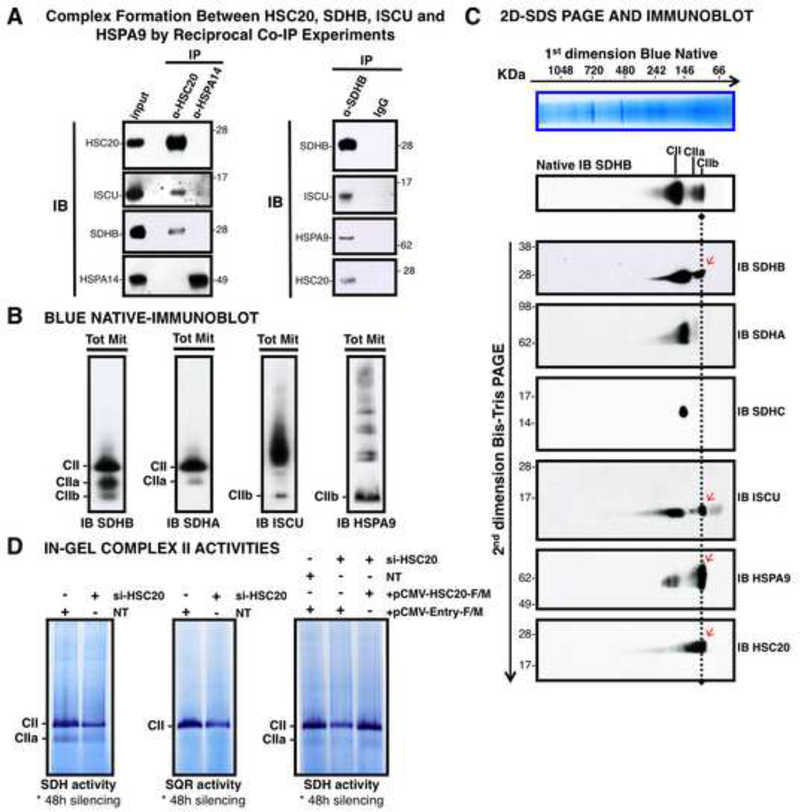
Figure 2. HSC20 Binds SDHB and Forms a Complex with HSPA9 and ISCU.
(A) Complex formation between HSC20, SDHB, ISCU and HSPA9 by reciprocal co-IP experiments. IP of HSC20 or HSPA14, followed by IBs to ISCU, SDHB, HSPA14. HSPA14 did not interact with HSC20 (IB to HSC20), and represented a false positive from the Y2H screen (left panel). IP of SDHB, followed by IBs to ISCU, HSPA9, HSC20 revealed that there was a complex composed of SDHB, ISCU, HSPA9 and HSC20 (right panel), (n= 5 biological samples). (B, C) BN and 2D-BN-SDS IBs of mitochondrial extracts resolved three SDHB-containing complexes of distinctive composition. Red arrows indicate components of the CIIb complex. (D) Response of in-gel SDH and SQR activities of Complex II to silencing of HSC20, and rescue by HSC20-F/M. NT represents a control extract from cells transfected with non-targeting si-RNAs. (B-D, n = 4 biological samples). See also Figure S2.
To address whether HSC20 works with HSPA9 and ISCU to enhance acquisition of Fe-S clusters by SDHB, we analyzed whether SDHB physically interacted with the ISCU-HSC20-HSPA9 complex, and we observed that SDHB coprecipitates ISCU, HSPA9 and HSC20 (Figures 2A, S2E). In contrast, we did not find direct interactions between SDHB and other proposed scaffolds, including ISCA1, ISCA2 (Sheftel et al., 2012), or NFU1 (Ferrer-Cortes et al., 2012) (not shown).
As SDHB likely acquires its Fe-S clusters before entering formation of a multimeric complex with SDHA, SDHC and SDHD, we assessed various stages of assembly of the SDH complex using Blue Native-PAGE (BN-PAGE) analysis and two-dimensional native-SDS-PAGE (2D-SDS PAGE) separations, in combination with immunoblotting (IB). After resolution of mitochondrial extracts by BN-PAGE, three SDHB-containing complexes were identified (Figure 2B), including CII, which conformed to the predicted size of fully assembled Complex II on BN-PAGE (220 KDa) (Hao et al., 2009), CIIa, which appeared to represent an assembly intermediate composed of SDHA and SDHB (Figure 2B), but not SDHC (Figure 2C), and a third pool of SDHB (CIIb) (Figure 2B) of approximately 110 KDa (Figure 2C) which contained SDHB, ISCU, HSPA9 (Figure 2B) and HSC20 (Figure 2C), but not SDHA. CII demonstrated substantial in-gel SDH and succinate-coQ oxidoreductase (SQR) enzymatic activities (Figure 2D). SDH and SQR activities diminished in cells silenced for HSC20 (si-HSC20) for 48 hours, but the phenotype was specifically rescued by re-transfection with pCMV-HSC20-F/M, demonstrating that HSC20 is critical for acquisition of CII enzymatic activity (Figures 2D, S2F). The two bands with SDH activity correspond to the two distinct bands detected in the native immunoblots to SDHB and to SDHA (Figure 2B). As the SDHA/SDHB complex can oxidize succinate into fumarate, CIIa retains SDH activity, though it lacks the SQR function mediated by subunits A-D of fully assembled CII.
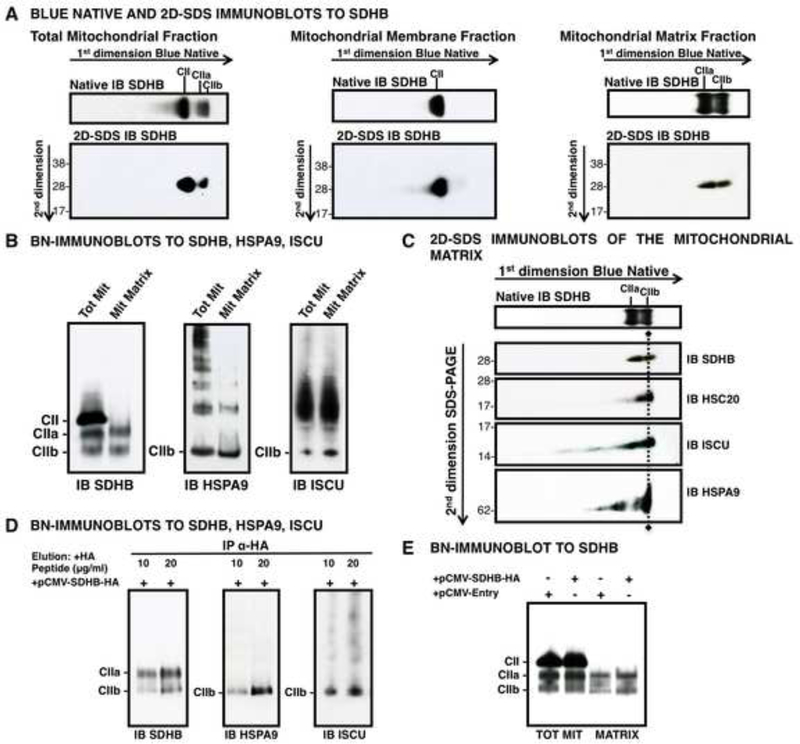
Using mitochondrial sub-fractionations, we further showed that CIIa and CIIb were assembled in the mitochondrial matrix (Figures 3A, 3B, 3C). By immunoprecipitating SDHB-HA from the mitochondrial matrix of HEK293 cells transfected with pCMV-SDHB-HA, we detected SDHB, HSPA9 and ISCU by native IB, further confirming the distinctive composition of the CIIb complex (Figure 3D), which localized to the matrix, unlike CII, which was associated only with mitochondrial membrane lysates (Figures 3A, 3E).
Figure 3. The CIIb Assembly Intermediate of SDH Localizes to the Mitochondrial Matrix.
(A) BN and 2D-SDS IBs to SDHB revealed multiple SDHB-containing complexes in total mitochondrial lysates which localized to either the membrane fraction (CII, middle panel) or to the matrix (CIIa and CIIb, right panel). (B) BN-IBs to SDHB, HSPA9, ISCU on total mitochondrial (Tot Mit) versus mitochondrial matrix fractions (Mit Matrix) verified that CIIa and CIIb localize to the matrix. (C) 2D-SDS IBs of mitochondrial matrix fractions showed that CIIb contains the chaperone-cochaperone transfer complex. (D) BN-IBs to SDHB, HSPA9 and ISCU, after IP of SDHB-HA on mitochondrial matrix fraction from cells transfected with pCMV-SDHB-HA, and elution with 10 or 20 μg/ml HA-peptide, demonstrating that SDHB is found in complexes CIIa and CIIb, which are assembly intermediates of CII. (E) BN-IB to SDHB on total or mitochondrial matrix extracts from cells transfected with pCMV-SDHB-HA, or the negative control, pCMV-Entry, showing that CIIa and CIIb are in the matrix, whereas CII is associated to the mitochondrial membrane extracts. (A-E, n = 3 biological samples).
Complex II Assembly and Enzymatic Activity are Impaired in Cells Silenced for HSC20 or HSPA9
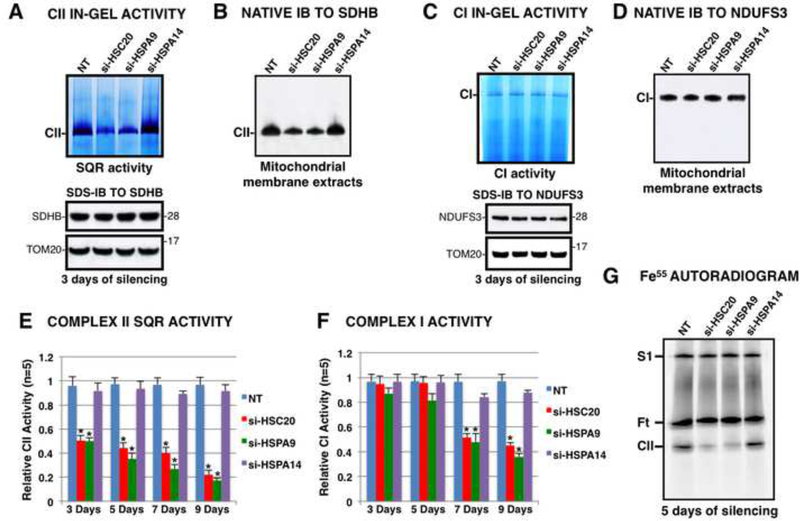
To directly examine the effects of knockdowns of HSC20 or HSPA9 on CII, we measured the SQR activity using an in-gel assay (Figure 4A). The enzymatic activity of CII in cells silenced for HSC20 or HSPA9 for three days decreased significantly, whereas SDHB protein levels in total mitochondrial extracts did not change (Figure 4A, lower panel). However, the fraction of SDHB that was incorporated into CII decreased upon silencing of HSC20 or HSPA9, as shown by the native IB to SDHB on mitochondrial membrane extracts (Figure 4B), indicating defective maturation of SDH. Complex I activity and subunit composition were not affected over the same time period (Figures 4C, 4D). Prolonged knockdowns of HSC20 or HSPA9 for seven or nine days showed early effects on SQR activity (Figure 4E), and late effects on Complex I activity (Figure 4F). SDHB, NDUFS3 (a Fe-S subunit of Complex I) and UQCRFS1 (the Complex III Fe-S subunit also known as the Rieske Fe-S protein) protein levels were significantly reduced after seven or nine days of silencing (Figure S3A), consistent with the known instability of some Fe-S proteins when they fail to incorporate their clusters. In contrast, levels of MTCO1, a Complex IV subunit that lacks a Fe-S cluster, and SDHA did not change.
Figure 4. Knockdown of HSC20 or HSPA9 Inhibits Complex II Assembly and Activity.
(A) In-gel activity assay of CII (upper panel), IB to SDHB (lower panel), and BN-IB (B) after knockdown of HSC20 (si-HSC20), HSPA9 (si-HSPA9), HSPA14 (si-HSPA14), or transfection with non-targeting si-RNAs (NT) for three days showed decreased CII activity and assembly after HSC20 or HSPA9 silencing, but not after knockdown of HSPA14, a negative control. (C) Complex I in-gel activity assay, IB to NDUFS3, and BN-IB to NDUFS3 (D), showed that Complex I was unaffected by silencing of HSC20 or HSPA9 for three days. (E, F) Spectrophotometric measurements of CII and CI activities showed effects of HSC20 or HSPA9 silencing after three days for CII and after seven days for CI. Error bars indicate s. d. from average (A-F, n = 5 biological replicates). (E, F) * t-test P value < 0.001. (G) Fe55 autoradiogram on mitochondrial membrane extracts from silenced cells revealed diminished Fe55 incorporation into CII after silencing of HSC20 or HSPA9. Ft represents cytosolic ferritin, which partially contaminated the mitochondrial preparations, as previously described (Stehling et al., 2009) (n = 3 biological samples). See also Figure S3.
Compromised Fe-S biogenesis due to knockdown of HSC20 or HSPA9 for seven or nine days caused apparent cytosolic iron deficiency as evidenced by changes in IRP1 and IRP2 activities (Figures S3B, S3C) and concomitant increased TFR1 expression (Figure S3D), as has been observed previously when Fe-S biogenesis is impaired (Rouault, 2012).
To examine the effects of knockdown of HSC20 or HSPA9 on SDHB maturation using a complemenatry technique, we analyzed in vivo iron incorporation into CII using the radioisotope Fe55 (Stehling et al., 2009). Upon knockdown of HSC20 or HSPA9 for five days, the Fe55 signal of Complex II was barely detectable (Figure 4G), whereas the levels of Fe55 incorporated into supercomplex S1, which contains Complex I (Stehling et al., 2009), were not diminished.
Further evidence for the importance of HSC20 in the biogenesis of SDHB was obtained using a dominant negative mutant of HSC20 in which the HPD tripeptide in the J-domain was replaced by alanines (HSC20HPD-AAA–F/M), to simulate mutations that disrupt function of the yeast ortholog of HSC20, Jac1, by preventing activation of the chaperone (Voisine et al., 2001). By following the import of in vitro synthesized SDHB-HA into mitochondria isolated from cells transfected with pCMV-HSC20-F/M (Figure S3F) or with the mutant (Figure S3G), we found that the ability of HSC20HPD-AAA–F/M to interact with SDHB was not initially impaired (Figure S3G, IB to HSC20), but SDHB did not progress to form a complex with SDHA over the 30 minute time course (Figure S3G, IB to SDHA). Using Fe55 labeling of imported SDHB-HA, we demonstrated that the compromised maturation of SDHB in cells that expressed HSC20HPD-AAA–F/M was due to failure of the HSPA9/ HSC20HPD-AAA–F/M system to assist release and transfer of the nascent cluster from ISCU to SDHB. CIIb did not stably assemble when processing of SDHB was mediated by HSC20HPD-AAA–F/M (Figure S3H) and the weak, transient interaction detected in the SDS-PAGE after IP of SDHB-HA (Figure S3G) did not lead to formation of a productive complex, whereas SDHB-HA progressed from CIIb to CIIa, in mitochondria that expressed wild type HSC20-F/M (Figure S3H).
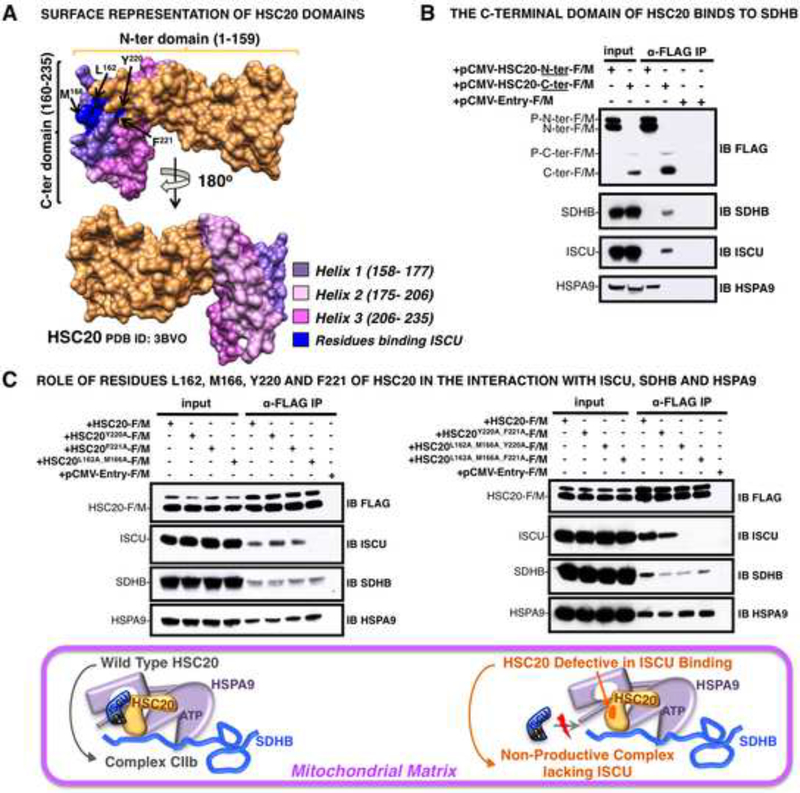
The C-terminal Domain of HSC20 Interacts with SDHB
Using the known crystal structure of human HSC20 (Bitto et al., 2008) for modeling (Figure 5A), we identified the domain of HSC20 that mediates the interaction with SDHB by testing N- and C-terminal constructs of the cochaperone in co-IP experiments. We found that the C-terminus of HSC20 interacted with SDHB, whereas the N-terminal domain that contained the HPD motif bound HSPA9 (Figure 5B), but did not bind SDHB. The C-terminus of HSC20 interacted also with ISCU, and, by testing mutations of HSC20 analogous to those identified as important in IscU/ Isu binding in bacteria or yeast, respectively (Ciesielski et al., 2012; Fuzery et al., 2011), we determined that residues L162 and M166 of HSC20 made the greatest contribution to the binding to ISCU (Figure 5C), whereas Y220 and F221 were less important. Further inspection of the structure (Bitto et al., 2008) revealed that the C-terminus of HSC20 was composed of three alpha-helices (Figure 5A), and helices 1 and 2 were mainly responsible for mediating the interaction with SDHB in Y2H assays (Figure S4 and Table S2).
Figure 5. The C-terminal domain of HSC20 (residues 160– 235) provides the binding surface for SDHB.
(A) 3D-structure of HSC20 (PDB ID: 3BVO). The N-terminal domain (residues 1–159) is in gold; the C-terminus (residues 160–235) is shown in three colors, each corresponding to one of three helices (1–3), as indicated. The residues substituted by alanines in the region involved in the interaction with ISCU, are highlighted in blue and pointed by arrows. (B) Extracts from cells expressing FLAG/MYC-HSC20-N-terminal domain (pCMV-HSC20-N-ter-F/M) or FLAG/MYC-HSC20-C-terminal domain (pCMV-HSC20C-ter-F/M) were subjected to IP with anti-FLAG, followed by IBs of the eluates with antibodies to SDHB, ISCU or HSPA9; results showed that the C-terminus bound to SDHB and ISCU, whereas the N-terminus bound HSPA9. (C) Extracts from cells expressing HSC20-F/M or the mutants, as indicated, were subjected to IP with anti-FLAG, followed by IBs to ISCU, SDHB, or HSPA9; results indicated that residues L162 and M166 were critical for ISCU binding (upper panels). Schematic of the chaperone-cochaperone transfer complex containing HSC20 wild type compared to the mutant defective in binding ISCU (lower panel). (B and C, n = 5 biological replicates). See also Figure S4 and Table S2.
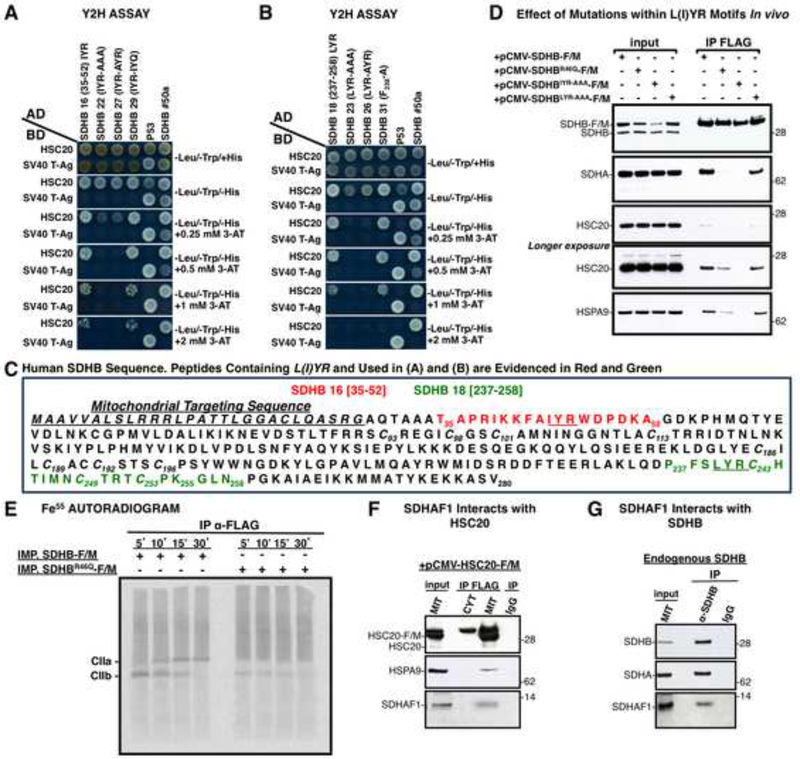
SDHB Contains two L(I)YR Motifs that Bind HSC20
To identify the potential motifs in the SDHB sequence that could bind HSC20, we subdivided SDHB into multiple peptides and used a Y2H approach to screen 50 to 100 clones of varying sizes. Initial deletional analysis of SDHB indicated that two distinct domains (residues 1–147 and 147–280 of SDHB) were able to interact with HSC20 in vivo (Figures S5A, S5B), and in vitro (Figure S5C). Further deletions of SDHB revealed that residues 35–52 (Figure 6A, clone 16) and 237–258 (Figures 6B, clone 18) interacted with HSC20 and each contained a highly conserved L(I)YR motif. To investigate the specific requirement of the L(I)YR motif for binding to HSC20, the motif residues were replaced by alanines. Substitutions of the proximal IYR and the distal LYR motifs into alanines abrogated the interactions in vitro (Figures 6A and 6B, clones 22 and 23), as did mutagenesis of the first residue, I or L of each motif into A, (Figures 6A, 6B, clones 27 and 26). A conserved phenylalanine in SDHB 35–52 (F42) and in SDHB 237–258 (F238) was not important for binding to HSC20 (Figure 6B, clone 31, and Table S3, clone 30). Further experiments were performed in order to systematically analyze the requirement of the L(I)YR consensus at different positions within the primary sequence of SDHB (Figure 6C). Context was important, as insertion of an LYR motif within the region 194–240 of SDHB did not confer ability to interact with HSC20 (Table S3, clone 25). Moreover, en bloc replacement of the tripeptide S198Y199W200 within SDHB 185–240 (Table S3, clone 7) by LYR did not generate interaction with HSC20 (Table S3, clone 28), suggesting that additional elements such as secondary structure could affect the interaction. Notably, the two native L(I)YR consensus sequences in SDHB are located in unstructured loops of the crystal structure. One of the L(I)YR motifs appears in SDHB near the N-terminus, proximal to the first cysteines that ligate the [2Fe-2S] cluster, whereas the second is closer to the C-terminus and the cysteinyl ligands of the [4Fe-4S] and [3Fe-4S] clusters, in positions where binding of the chaperone-cochaperone transfer apparatus can guide release of the cluster from holo-ISCU into the distal Fe-S binding sites of SDHB.
Figure 6. An L(I)YR Motif that Recurs Twice in SDHB is Recognized by HSC20.
(A, B). Deletional analysis of SDHB in Y2H interactions led to identification of the L(I)YR motif in clones 16 (residues 35–52) and 18 (237–258) of SDHB, and effects of mutagenesis of the motif residues indicated that the SDHB-HSC20 interaction was attributable to the L(I)YR motif. (C) Sequence of human SDHB, in which L(I)YR motifs are underlined in the peptides analyzed by Y2H in A and B. (D) IP of SDHB-F/M, SDHBR46Q–F/M, SDHBIYR-AAA–F/M, SDHBLYR-AAA–F/M with anti-FLAG, followed by IBs to SDHA, HSC20, HSPA9 demonstrated the negative effect of mutagenizing L(I)YR motifs. (A, B and D, n = 5 biological samples). (E) Fe55 labeling of SDHB-F/M or SDHBR46Q–F/M imported into mitochondria isolated from HEK293. The IP performed with anti-FLAG on the matrix fraction showed that the R46Q mutation impaired iron incorporation and Complex II biogenesis. (F) SDHAF1 interacts with HSC20, and with endogenous SDHB (G). (E-G, n = 3 biological samples). Inputs were 30% of total used for IPs. See also Figure S5 and Table S3.
In several reported cases of familial phaeochromocytomas and paragangliomas characterized by selective loss of SDH activity, the third residue in the IYR motif of SDHB, Arg46, was replaced by Gln or Gly (Benn et al., 2003; Ricketts et al., 2012; Takekoshi et al., 2008). Because of its pathogenic significance and the fact that it is part of the L(I)YR motif identified here, we generated an SDHBR46Q for further studies. Although mutation of the IYR tripeptide within SDHB 35–52 into IYQ did not impair the intrinsic competence of this short region of SDHB to bind HSC20 in vitro (Figure 6A, clone 29) in Y2H studies, we found that SDHBR46Q bound poorly to the HSC20 complex or to SDHA in vivo (Figures 6D, S5D), impairing the ability to ultimately form a mature SDH complex. To assess how the mutant was processed, we evaluated Fe55 incorporation into SDHB-F/M or SDHBR46Q–F/M proteins imported into isolated mitochondria. The autoradiogram of the imported proteins revealed that iron incorporation into SDHBR46Q was profoundly diminished in CIIb (Figure 6E). Furthermore, SDHBR46Q did not progress into the CIIa complex at the 15 and 30 minute time-points (Figure 6E, right side of the gel), unlike wild type SDHB-F/M (Figure 6E, left side). These studies suggested that nascent SDHBR46Q did not incorporate the first Fe-S cluster, which likely rendered the protein unstable (Figure S5E).
We next determined how SDHBIYR-AAA and SDHBLYR-AAA mutations affected SDH assembly in vivo. As the L(I)YR motifs are responsible for recruiting the chaperone-cochaperone transfer apparatus, these mutations of the entire motif were expected to impair Fe-S cluster acquisition by SDHB, and to thereby prevent the subsequent association with SDHA. Accordingly, we expressed SDHBIYR-AAA-F/M and SDHBLYR-AAA-F/M in HEK293 cells, immunoprecipitated recombinant proteins with anti-FLAG, and evaluated interactions with the chaperone-cochaperone transfer complex, and with SDHA. Our results showed that substitution of I44Y45R46 into alanines (SDHBIYR-AAA-F/M) completely abrogated the interaction of SDHB with HSC20 and HSPA9 (Figure 6D), and, by compromising Fe-S cluster insertion, was responsible for preventing maturation of SDHB and its subsequent association with SDHA. As SDHBIYR-AAA-F/M retained a wild type L240Y241R242 motif at the C-terminus, but no interaction with the chaperone-cochaperone transfer apparatus was detected, we hypothesized that engagement of the first L(I)YR motif and acquisition of the first Fe-S cluster is a prerequisite for binding of the HSC20-HSPA9-ISCU complex to the second consensus site. Failure to achieve folding of the N-terminal domain of SDHB around its [2Fe-2S] cluster prevented further maturation of the protein, and assembly of the multimeric SDH complex. Mutations of the second LYR motif in SDHBLYR-AAA-F/M showed a less profound defect, since there was a readily detectable interaction with HSC20 and HSPA9 (Figure 6D), suggesting that the first IYR consensus sequence is indispensable for further maturation of SDHB, whereas loss of the second LYR site allowed some SDHB to associate with SDHA. Importantly, mutations of multiple residues in the two L(I)YR motifs of SDHB, including I44, R46, L240 or R242, cause cancer (Amar et al., 2005; Celestino et al., 2012; Neumann et al., 2004; Ricketts et al., 2012).
The LYR motif was present in several proteins selected in the Y2H (i.e. EPRS, HELZ), and also in LYRM7, an annotated member of the LYR family (Conserved Domains Accession: cl05087), which contains at least nine proteins in humans, including SDHAF1, a bona fide CII assembly factor (Ghezzi et al., 2009), and LYRM4, a component of the initial Fe-S assembly complex (Shi et al., 2009).
The presence of the LYR motif in SDHAF1 appeared to be particularly meaningful as SDH activity is adversely affected by inherited mutations in SDHAF1 in human patients (Jain-Ghai et al., 2013). The missense mutation R55P in the LYR motif of SDHAF1 is associated with SDH-defective infantile leukoencephalopathy (Ghezzi et al., 2009). We found that HSC20 interacts with SDHAF1 (Figure 6F). Furthermore, SDHB co-immunoprecipitated SDHAF1 (Figure 6G), and residues 147–280 of SDHB contained an interaction site for SDHAF1 (Figure S6F). En bloc substitution of L240Y241R242 with alanines in SDHB-F/M (SDHBLYR-AAA-F/M) did not affect the interaction with SDHAF1 (Figure S6G), indicating that SDHAF1 interacts with the C-terminal domain of SDHB (residues 147–280) through a non-LYR binding site.
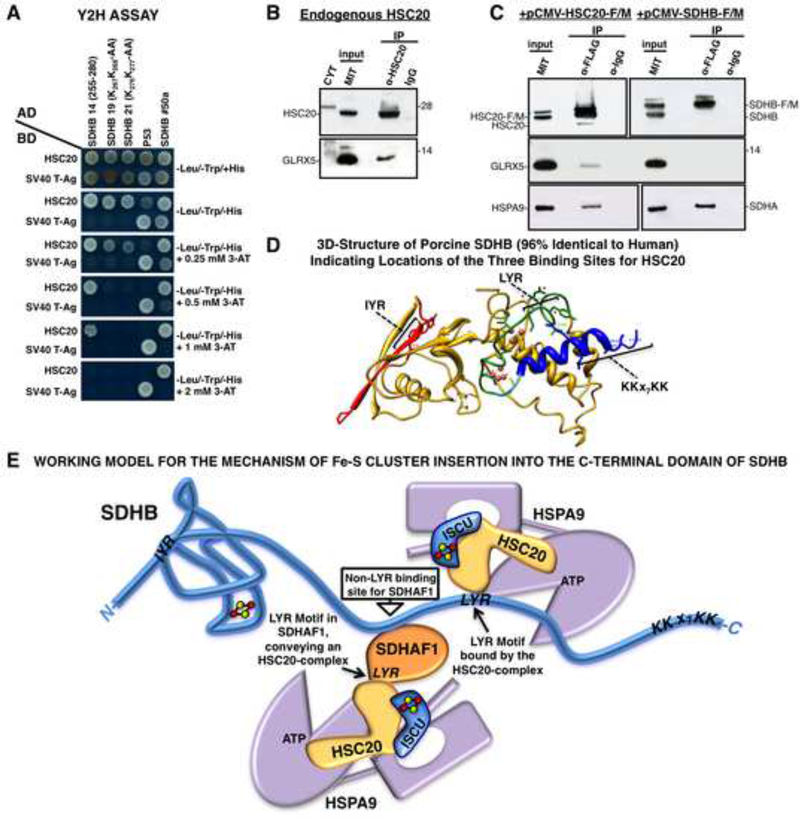
Identification of a KKX(6–10)KK Consensus
Using Y2H assays, we also discovered that a third binding region for HSC20 in SDHB, the sequence K267K268X7K276K277, mediated an interaction with HSC20 (Figure 7A). Interestingly, GLRX5 has a similar pattern of lysines at its C-terminus (K139K140X10K151K151), and we found that HSC20 interacted with GLRX5 in vivo (Figures 7B, 7C, S6). SUCLG2, which was identified in our screen and confirmed by co-IP (Figure S1C), also contains a K423K424X6K431K432 pattern at the C-terminus.
Figure 7. Residues 255–280 in the C-domain of SDHB Contain a Third Binding Site for HSC20.
(A) Y2H on clone 14 of SDHB (255–280), and on peptides in which KK residues of the KKx7KK motif were mutagenized. Mutagenesis of either the first or second KK dipeptide impaired binding to HSC20 (clones 19 and 21). (B, C) Endogenous or overexpressed HSC20 interacts with GLRX5 in vivo (A-C, n = 5 biological samples). (D) 3D-structure of porcine SDHB (96% identical to human SDHB. PBD ID: 3SFD) showing spatial distribution of HSC20 binding motifs. (E) Working model for the possible mechanism of Fe-S cluster insertion into the C-terminal domain of SDHB. See also Figure S6 and Table S3.
DISCUSSION
Here we have identified a molecular signature, the tripeptide LYR, which defines Fe-S target proteins through their ability to bind the cochaperone HSC20 and acquire their Fe-S clusters from the scaffold protein, ISCU. Upon analyzing SDHB, which binds to HSC20 directly in Y2H assays, and in vivo in coimmunoprecipitations, we identified three distinct HSC20 binding sites that span from the N- to the C-terminus, including two iterations of the motif L(I)YR. Despite its relatively simple subunit composition, the assembly process of SDH has remained relatively enigmatic (Kim and Winge, 2013; Rutter et al., 2010), and two accessory proteins have been identified, SDHAF1 and SDHAF2 (Ghezzi et al., 2009; Hao et al., 2009). In this work, we uncovered molecular details of how SDHB acquires its three Fe-S centers, and how assembly of Complex II is contingent upon successful biogenesis of SDHB. Our co-immunoprecipitation and proteomic approaches demonstrated that HSC20 physically interacts with SDHB, and works together with its cognate chaperone, HSPA9, to enhance transfer of Fe-S clusters from the main scaffold ISCU directly to SDHB. Further evidences for this scenario was provided by mitochondrial subfractionation studies in combination with BN-PAGE and 2D-native/SDS-PAGE analyzes, which enabled us to characterize a novel assembly intermediate of SDH, CIIb. By following the import into isolated mitochondria of in vitro synthesized SDHB, we found that interaction of SDHB with the chaperone-cochaperone complex precedes association with SDHA (Figures 6E, S3F–H, S3L). This result elucidates a key point in the assembly of Complex II. Flavinylation of Sdh1 (the yeast ortholog of the SDHA subunit) was previously recognized as a prerequisite for complex II maturation (Kim and Winge, 2013), and our results have demonstrated that SDHB must acquire its three Fe-S clusters to enter the previously unrecognized CIIa intermediate complex composed of functional SDHA-SDHB subunits. Defective assembly of Complex II °Ccurs early in cells silenced for HSC20 or HSPA9, as shown by reduced levels of SDHB incorporated into CII, reduced SDH and SQR activities upon knockdown for three days, and by decreased incorporation of Fe55 into Complex II.
By pursuing interactions of HSC20 with SDHB, we identified two L(I)YR motifs in the SDHB primary sequence, which were responsible for engaging the chaperone-cochaperone transfer apparatus and facilitating acquisition of Fe-S clusters during primary folding of SDHB. Consistent with our mutagenesis analysis, the motif consists of an aliphatic residue at position 1 (I, L or V), followed by a tyrosine or possibly phenylalanine at position 2, and a basic residue (R or K) at position 3. The two LYR motifs in SDHB are highly conserved throughout the eukaryotic and prokaryotic kingdoms (Table S5), suggesting that these consensus sequences have significant functional importance. Biogenesis of SDHB may require binding of the orthologous cochaperones to the L(I)YR motifs in organisms ranging from eukaryotes, including plants, to prokaryotes. The ability of HSC20 to bind LYR motifs is underscored by interactions of HSC20 with annotated members of the LYR family, namely SDHAF1 (LYRM8), which is involved in SDH assembly, and LYRM7, which is required for the biogenesis of the Rieske Fe-S protein of Complex III (Sanchez et al., 2013).
Our results show that SDHAF1 also associates with SDHB through a non-LYR binding site. Based on the physical association between SDHAF1 and SDHB, we propose that SDHAF1 may promote Fe-S cluster insertion into SDHB through its ability to bind HSC20. In our working model (Figure 7E), SDHAF1 associates with SDHB through a non-LYR binding site, and utilizes its own LYR motif to position an ISCU-HSC20-HSPA9 complex near to chaperone complexes directly associated with the LYR binding site of SDHB. The previously observed propensity of HSC20 to dimerize may allow two holo-ISCU molecules at neighboring binding sites to reorganize their adjacent [2Fe-2S] centers, enabling them to coalesce into the [4Fe-4S] and [3Fe-4S] clusters of mature SDHB (Chandramouli et al., 2007).
Our in vivo and in vitro approaches to inspect the HSC20-mediated insertion of Fe-S clusters into SDHB have shed light on general mechanisms underlying client discrimination, an event of critical importance in the steps that facilitate Fe-S cluster transfer from the main scaffold ISCU to specific Fe-S recipient proteins or potential intermediate scaffolds. We propose that LYR motifs impart specificity by enabling unfolded target proteins to bind the HSC20-dependent Fe-S transfer apparatus and directly acquire their clusters.
Notably, Complex I, which contains eight Fe-S clusters that are deeply buried within the 45 subunit multimeric complex, contains two annotated members of the LYR family, LYRM6 (NDUFA6) and LYRM3 (NDUFB9) and a Fe-S subunit, NDUFS8, that contains a previously unrecognized conserved C-terminal LYR motif and is predicted to ligate two [4Fe-4S] clusters (Table S5). As only 14 of the 45 Complex I subunits have a catalytic function (Fassone and Rahman, 2012), some of the accessory or supernumerary subunits may contribute to the assembly and stability of the complex. Several of the LYR-containing subunits may aid insertion of the Fe-S clusters into the complex.
In our analysis, we also identified a third binding region for HSC20 in residues 255–280 of SDHB. Our mutagenesis analyses demonstrated that the sequence K267K268X7K276K277 was responsible for mediating the interaction with HSC20. SUCLG2, a protein identified in our screen, which may provide succinyl-CoA consumed in the first step of heme biosynthesis, contains a similar consensus at its C-terminus. Moreover, GLRX5, which interacted with HSC20 in vivo, has a similar pattern of lysines in the C-terminal region (K139K140X10K151K151). Notably, Grx5 homologues, including human GLRX5, serve as alternative scaffolds for Fe-S cluster delivery to a subset of target proteins (Shakamuri et al., 2012; Uzarska et al., 2013; Ye et al., 2010).
Our studies unveil a previously unrecognized network of protein-protein interactions that are key to assembly and function of complexes I-III in eukaryotes, and likely also in bacteria, where the same machinery is conserved. Numerous other Fe-S proteins, many of which are not yet recognized, may be revealed by further studies of proteins that contain the LYR motif, or further analyses of other proteins identified in our Y2H screen.
EXPERIMENTAL PROCEDURES
Si-RNA Transfection of Human Cells
On-TARGET Plus SMART Pools against human HSC20 (L-017718–02), human HSPA9 (L-004750–00), human HSPA14 (L-021084–01), an HSPA9 homologue, and the non-targeting pool (D001810–10) were purchased from Thermo Scientific. HeLa or HEK293 cells (purchased from ATCC) were transfected with si-RNAs using Dharmafect 1 (Thermo Scientific) according to the manufacturer’s protocol.
Plasmids and Transfection of Human Cells
Plasmids used for expression in human cells were all pCMV-Entry-based (Origene) to allow the expression of constructs from a CMV-promoter (See Table S4 for a full list). Constructs were C-terminally tagged either with HA or with FLAG-MYC. Point mutations were introduced using the QuikChange II site-directed mutagenesis kit (Stratagene). Plasmid transfections into human cells were routinely performed with lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Plasmids and Transformation in Yeast
The library of proteins (preys) was expressed as fusion to the GAL4 activation domain (GAL4-AD) in pGADT7 and engineered to generate “preys” lacking the mitochondrial targeting sequence to allow the Y2H interactions to take place into the nucleus of yeast cells and to potentially activate the transcription of four independent reporter genes (AUR1-C, ADE2, HIS3, and MEL1). In our experiments we checked for activation of the HIS3 gene which allows for the use of a small inhibitor, 3-amino-1, 2, 4- triazole (3-AT), to modulate the background and to thereby select for high activation of the HIS3 gene. The bait, HSC20 (residues 29– 235), was expressed as a fusion to the GAL4 binding domain (GAL4-BD) in pGBKT7. PGBKT7-P53 and pGADT7-SV40-T-Ag, and PGBKT7-RABEX5 and pGADT7-RABEP1 were used as positive controls. The constructs generated for the Y2H assay were all pGBKT7- or pGADT7- based. The point mutations were introduced by using the QuikChange II site-directed mutagenesis kit (Stratagene). Co-transformations into S. cerevisiae AH109 strain were performed following a polyethylene glycol (PEG)/ LiAc-based method for preparing and transforming competent yeast cells, using the YeastMaker Yeast Transformation System 2 (Clontech).
Cell Extracts and Fractionation
Whole cell lysates were prepared with lysis buffer I: 25 mM Tris, 0.15 M NaCl, 1 mM EDTA, 1% NP-40, 5% glycerol pH 7.4, protease and phosphatase inhibitors. Cellular fractionation into cytosol and intact mitochondria was done essentially as described (Frezza et al., 2007). The mitochondrial matrix fraction was prepared after swelling of the mitochondrial outer membrane by hypotonic shock, followed by sonication (4 repeats of 5 sec- bursts at high settings with 30 sec pauses), supplementation with 150 mM NaCl and 5% glycerol and centrifugation at 20000 × g 30 min. The supernatant was saved as soluble mitochondrial matrix fraction. The pellet was washed twice with 10 mM Tris/MOPS, 1 mM EGTA/Tris, 0.2 M sucrose, finally resuspended in 20 mM TrisHCl, pH 7.8, 0.4 M NaCl, 15% glycerol, 1 mM DTT, 1 mM PMSF, 0.5% digitonin, incubated on ice for 15 min and centrifuged at 20000 × g 30 min. The supernatant was saved as the mitochondrial membrane proteins.
Immunoprecipitation (IP)
The lysis buffer used for the IP experiments was prepared as lysis buffer I supplemented with 2 mM ADP. FLAG- immunoprecipitations were performed using M2-FLAG beads (Sigma). IPs of endogenous proteins or HA-immunoprecipitations were done by covalently coupling the proper antibodies onto an amine-reactive resin (Pierce Co-IP kit, Cat. No. 26149), following the manufacturer’s instructions. Whole cell lysates or subcellular fractions, as indicated, were incubated with beads for 4 h at 4 °C, and then washed extensively with lysis buffer I. Bound proteins were either eluted by incubation of beads with lysis buffer I containing 100 μg/ml 3 × FLAG peptide for 1 h at 4 °C, or with lysis buffer I containing 10 or 20 μg/ml HA peptide. Alternatively, acidic elution with Tris-Glycine pH 2.8 for 10 min at 4 °C was performed for samples to be analyzed by SDS-PAGE and immunoblot (IB). Aliquots, corresponding to 30– 40% of total or subcellular lysates were run alongside the IP fractions onto the gels as inputs.
Native PAGE (BN-PAGE) and Native Immunoblot
The NativePAGE Novex Bis-Tris gel system (Invitrogen) was used for the analysis of native membrane protein complexes and native mitochondrial matrix complexes, with the following modifications: only the Light Blue Cathode Buffer was used; 20 μg of membrane protein extracts were loaded/ well; the electrophoresis was performed at 150 V for 1 h and 250 V for 3.5 h. For the native IB, PVDF was used as the blotting membrane. The transfer was performed at 25 V for 4 h at 4 °C. After transfer, the membrane was washed with 8% acetic acid for 20 min to fix the proteins, and then rinsed with water before air-drying. The dried membrane was washed 5– 6 times with methanol (to remove residual Coomassie Blue G-250), rinsed with water and then blocked for 2h at room temperature in 5% milk, before incubating with the desired antibodies overnight at 4 °C. In order to avoid strip and reprobing of the same membrane, which might allow detection of signals from the previous IBs, samples were loaded and run in replicates on adjacent wells of the same gel, and probed independently with different antibodies.
Two-Dimensional Native/ SDS-PAGE
Two-dimensional Native/ SDS-PAGE was performed by resolving the mitochondrial protein complexes in the first dimension, by BN-PAGE. Each lane of the gel was excised, equilibrated in SDS buffer supplemented with reducing agent, and then immersed in the alkylating solution for 15 min. For the second dimension, the gel strip was fixed horizontally onto the NuPAGE 4– 12% Bis-Tris Zoom Gel (Invitrogen), and the SDS-PAGE was performed.
Complex I and Complex II activities
In- gel Complex I and Complex II activities were performed as described (Diaz et al., 2009; Wittig et al., 2007). See Supplemental Experimental Procedures.
Iron incorporation assay
The Fe55 incorporation assay into the mitochondrial respiratory chain complexes was done essentially as described (Stehling et al., 2009) with several modifications (see Supplemental Experimental Procedures).
In vitro Transcription/ Translation and Import into Isolated Mitochondria
The cell-free E. coli expression system (Life Technologies), which couples transcription/ translation reactions to produce high yields of soluble, functionally active proteins, was used to synthesize HA- tagged SDHB to be imported into isolated mitochondria, following the manufacturer’s instructions. The mitochondrial binding and insertion assay was performed as described (Yang et al., 2012). Briefly, mitochondria were isolated from HEK293 transfected with wild type HSC20-F/M or with HSC20HPD-AAA-F/M. Equal amounts of in vitro synthesized SDHB-HA and isolated mitochondria were incubated, for the designated time points, with ATP, creatine phosphate, creatine phosphokinase, methionine, MgCl2, KCl, DTT, sucrose and HEPES buffered to a pH of 7.5 with KOH. The reaction mixture was kept at 37 °C for the indicated time points, and then placed on ice. Mitochondria were washed and treated with proteinase K before lysis (in order to remove the amount of SDHB-HA that was not imported into mitochondria). Extracts were then prepared, followed by IP with an anti-HA to immunoprecipitate SDHB-HA. Elution was performed with HA peptide. IBs to HSC20, ISCU, HSPA9, or SDHA were then performed. Alternatively, FLAG/MYC- tagged SDHB wild type or R46Q mutant were synthesized in vitro, as previously described, and used for the import into mitochondria isolated from HEK293. Extracts were prepared after allowing the import to proceed for the indicated time points and the IPs were performed with anti-FLAG.
Supplementary Material
HIGHLIGHTS.
The cochaperone HSC20 binds to LYR motifs in iron sulfur cluster recipient proteins
Two LYR motifs and a KKX7KK sequence in SDHB independently bind the HSC20 complex
SDHB acquires Fe-S clusters from the ISCU/HSC20/HSPA9 complex before binding SDHA
The LYR proteins SDHAF1 and LYRM7, assembly factors of complexes II-III, bind HSC20
ACKNOWLEDGEMENTS
We are grateful to members of our lab for constructive discussions, to Drs. J.S. Bonifacino, R. Mattera and Y. Prabhu for providing advice on the Y2H assay, to Dr. A. Banerjee for insights into the structural aspects of our work, to A. Sharma for the gift of antibodies to the epitope tags. This work was supported by the Eunice Kennedy Shriver NICHD Intramural Research Program.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, six figures, five tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ajit Bolar N, Vanlander AV, Wilbrecht C, Van der Aa N, Smet J, De Paepe B, Vandeweyer G, Kooy F, Eyskens F, De Latter E, Delanghe G, Govaert P, Leroy JG, Loeys B, Lill R, Van Laer L, and Van Coster R (2013). Mutation of the ironsulfur cluster assembly gene IBA57 causes severe myopathy and encephalopathy. Human molecular genetics 22, 2590–2602. [DOI] [PubMed] [Google Scholar]
- Amar L, Bertherat J, Baudin E, Ajzenberg C, Bressac-de Paillerets B, Chabre O, Chamontin B, Delemer B, Giraud S, Murat A, Niccoli-Sire P, Richard S, Rohmer V, Sadoul JL, Strompf L, Schlumberger M, Bertagna X, Plouin PF, Jeunemaitre X, and Gimenez-Roqueplo AP (2005). Genetic testing in pheochromocytoma or functional paraganglioma. J Clin Oncol 23, 8812–8818. [DOI] [PubMed] [Google Scholar]
- Benn DE, Croxson MS, Tucker K, Bambach CP, Richardson AL, Delbridge L, Pullan PT, Hammond J, Marsh DJ, and Robinson BG (2003). Novel succinate dehydrogenase subunit B (SDHB) mutations in familial phaeochromocytomas and paragangliomas, but an absence of somatic SDHB mutations in sporadic phaeochromocytomas. Oncogene 22, 1358–1364. [DOI] [PubMed] [Google Scholar]
- Birch-Machin MA, Taylor RW, Cochran B, Ackrell BA, and Turnbull DM (2000). Late-onset optic atrophy, ataxia, and myopathy associated with a mutation of a complex II gene. Ann Neurol 48, 330–335. [PubMed] [Google Scholar]
- Bitto E, Bingman CA, Bittova L, Kondrashov DA, Bannen RM, Fox BG, Markley JL, and Phillips GN Jr. (2008). Structure of human J-type co-chaperone HscB reveals a tetracysteine metal-binding domain. The Journal of biological chemistry 283, 30184–30192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai K, Frederick RO, Kim JH, Reinen NM, Tonelli M, and Markley JL (2013). Human mitochondrial chaperone (mtHSP70) and cysteine desulfurase (NFS1) bind preferentially to the disordered conformation, whereas co-chaperone (HSC20) binds to the structured conformation of the iron-sulfur cluster scaffold protein (ISCU). The Journal of biological chemistry 288, 28755–28770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celestino R, Lima J, Faustino A, Maximo V, Gouveia A, Vinagre J, Soares P, and Lopes JM (2012). A novel germline SDHB mutation in a gastrointestinal stromal tumor patient without bona fide features of the Carney-Stratakis dyad. Fam Cancer 11, 189–194. [DOI] [PubMed] [Google Scholar]
- Chandramouli K, Unciuleac MC, Naik S, Dean DR, Huynh BH, and Johnson MK (2007). Formation and properties of [4Fe-4S] clusters on the IscU scaffold protein. Biochemistry 46, 6804–6811. [DOI] [PubMed] [Google Scholar]
- Ciesielski SJ, Schilke BA, Osipiuk J, Bigelow L, Mulligan R, Majewska J, Joachimiak A, Marszalek J, Craig EA, and Dutkiewicz R (2012). Interaction of Jprotein co-chaperone Jac1 with Fe-S scaffold Isu is indispensable in vivo and conserved in evolution. J Mol Biol 417, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier J, Touraine B, Briat JF, Gaymard F, and Rouhier N (2013). The ironsulfur cluster assembly machineries in plants: current knowledge and open questions. Frontiers in plant science 4, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz F, Barrientos A, and Fontanesi F (2009). Evaluation of the mitochondrial respiratory chain and oxidative phosphorylation system using blue native gel electrophoresis. Curr Protoc Hum Genet Chapter 19, Unit19 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos PC, and Dean DR (2010). Bioinorganic chemistry: electrons in Fe-S protein assembly. Nat Chem Biol 6, 700–701. [DOI] [PubMed] [Google Scholar]
- Dutkiewicz R, Schilke B, Cheng S, Knieszner H, Craig EA, and Marszalek J (2004). Sequence-specific interaction between mitochondrial Fe-S scaffold protein Isu and Hsp70 Ssq1 is essential for their in vivo function. The Journal of biological chemistry 279, 29167–29174. [DOI] [PubMed] [Google Scholar]
- Fassone E, and Rahman S (2012). Complex I deficiency: clinical features, biochemistry and molecular genetics. Journal of medical genetics 49, 578–590. [DOI] [PubMed] [Google Scholar]
- Ferrer-Cortes X, Font A, Bujan N, Navarro-Sastre A, Matalonga L, Arranz JA, Riudor E, Del Toro M, Garcia-Cazorla A, Campistol J, Briones P, Ribes A, and Tort F (2012). Protein expression profiles in patients carrying NFU1 mutations. Contribution to the pathophysiology of the disease. J Inherit Metab Dis. [DOI] [PubMed] [Google Scholar]
- Frezza C, Cipolat S, and Scorrano L (2007). Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc 2, 287–295. [DOI] [PubMed] [Google Scholar]
- Fuzery AK, Oh JJ, Ta DT, Vickery LE, and Markley JL (2011). Three hydrophobic amino acids in Escherichia coli HscB make the greatest contribution to the stability of the HscB-IscU complex. BMC Biochem 12, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzi D, Goffrini P, Uziel G, Horvath R, Klopstock T, Lochmuller H, D’Adamo P, Gasparini P, Strom TM, Prokisch H, Invernizzi F, Ferrero I, and Zeviani M (2009). SDHAF1, encoding a LYR complex-II specific assembly factor, is mutated in SDH-defective infantile leukoencephalopathy. Nature genetics 41, 654–656. [DOI] [PubMed] [Google Scholar]
- Hao HX, Khalimonchuk O, Schraders M, Dephoure N, Bayley JP, Kunst H, Devilee P, Cremers CW, Schiffman JD, Bentz BG, Gygi SP, Winge DR, Kremer H, and Rutter J (2009). SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science 325, 1139–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain-Ghai S, Cameron JM, Al Maawali A, Blaser S, MacKay N, Robinson B, and Raiman J (2013). Complex II deficiency--a case report and review of the literature. Am J Med Genet A 161A, 285–294. [DOI] [PubMed] [Google Scholar]
- Kampinga HH, and Craig EA (2010). The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol 11, 579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, and Winge DR (2013). Emerging concepts in the flavinylation of succinate dehydrogenase. Biochimica et biophysica acta 1827, 627–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R, Saxena S, Gordon DM, Pain D, and Dancis A (2001). J-domain protein, Jac1p, of yeast mitochondria required for iron homeostasis and activity of Fe-S cluster proteins. The Journal of biological chemistry 276, 17524–17532. [DOI] [PubMed] [Google Scholar]
- Lee S, Fan CY, Younger JM, Ren H, and Cyr DM (2002). Identification of essential residues in the type II Hsp40 Sis1 that function in polypeptide binding. The Journal of biological chemistry 277, 21675–21682. [DOI] [PubMed] [Google Scholar]
- Li J, and Sha B (2005). Structure-based mutagenesis studies of the peptide substrate binding fragment of type I heat-shock protein 40. Biochem J 386, 453–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann HP, Pawlu C, Peczkowska M, Bausch B, McWhinney SR, Muresan M, Buchta M, Franke G, Klisch J, Bley TA, Hoegerle S, Boedeker CC, Opocher G, Schipper J, Januszewicz A, and Eng C (2004). Distinct clinical features of paraganglioma syndromes associated with SDHB and SDHD gene mutations. JAMA 292, 943–951. [DOI] [PubMed] [Google Scholar]
- Perales-Calvo J, Muga A, and Moro F (2010). Role of DnaJ G/F-rich domain in conformational recognition and binding of protein substrates. The Journal of biological chemistry 285, 34231–34239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukszta S, Schilke B, Dutkiewicz R, Kominek J, Moczulska K, Stepien B, Reitenga KG, Bujnicki JM, Williams B, Craig EA, and Marszalek J (2010). Coevolution-driven switch of J-protein specificity towards an Hsp70 partner. EMBO Rep 11, 360–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricketts CJ, Shuch B, Vocke CD, Metwalli AR, Bratslavsky G, Middelton L, Yang Y, Wei MH, Pautler SE, Peterson J, Stolle CA, Zbar B, Merino MJ, Schmidt LS, Pinto PA, Srinivasan R, Pacak K, and Linehan WM (2012). Succinate dehydrogenase kidney cancer: an aggressive example of the Warburg effect in cancer. J Urol 188, 2063–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche B, Aussel L, Ezraty B, Mandin P, Py B, and Barras F (2013). Iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity. Biochimica et biophysica acta 1827, 455–469. [DOI] [PubMed] [Google Scholar]
- Rouault TA (2012). Biogenesis of iron-sulfur clusters in mammalian cells: new insights and relevance to human disease. Disease models & mechanisms 5, 155–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter J, Winge DR, and Schiffman JD (2010). Succinate dehydrogenase - Assembly, regulation and role in human disease. Mitochondrion 10, 393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez E, Lobo T, Fox JL, Zeviani M, Winge DR, and Fernandez-Vizarra E (2013). LYRM7/MZM1L is a UQCRFS1 chaperone involved in the last steps of mitochondrial Complex III assembly in human cells. Biochimica et biophysica acta 1827, 285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakamuri P, Zhang B, and Johnson MK (2012). Monothiol glutaredoxins function in storing and transporting [Fe2S2] clusters assembled on IscU scaffold proteins. J Am Chem Soc 134, 15213–15216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Y, and Cortopassi G (2012). HSC20 interacts with frataxin and is involved in iron-sulfur cluster biogenesis and iron homeostasis. Human molecular genetics 21, 1457–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheftel AD, Wilbrecht C, Stehling O, Niggemeyer B, Elsasser HP, Muhlenhoff U, and Lill R (2012). The human mitochondrial ISCA1, ISCA2, and IBA57 proteins are required for [4Fe-4S] protein maturation. Mol Biol Cell 23, 1157–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Ghosh MC, Tong WH, and Rouault TA (2009). Human ISD11 is essential for both iron-sulfur cluster assembly and maintenance of normal cellular iron homeostasis. Human molecular genetics 18, 3014–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YY, Hong XG, and Wang CC (2005). The C-terminal (331–376) sequence of Escherichia coli DnaJ is essential for dimerization and chaperone activity: a small angle X-ray scattering study in solution. The Journal of biological chemistry 280, 22761–22768. [DOI] [PubMed] [Google Scholar]
- Stehling O, Sheftel AD, and Lill R (2009). Chapter 12 Controlled expression of ironsulfur cluster assembly components for respiratory chain complexes in mammalian cells. Methods Enzymol 456, 209–231. [DOI] [PubMed] [Google Scholar]
- Stehling O, Vashisht AA, Mascarenhas J, Jonsson ZO, Sharma T, Netz DJ, Pierik AJ, Wohlschlegel JA, and Lill R (2012). MMS19 assembles iron-sulfur proteins required for DNA metabolism and genomic integrity. Science 337, 195–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo A, Korszun R, Hartl FU, and Flanagan J (1996). A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. EMBO J 15, 408–417. [PMC free article] [PubMed] [Google Scholar]
- Takekoshi K, Isobe K, Suzuki H, Nissato S, Kawakami Y, Kawai K, and Yamada N (2008). R46Q mutation in the succinate dehydrogenase B gene (SDHB) in a Japanese family with both abdominal and thoracic paraganglioma following metastasis. Endocr J 55, 299–303. [DOI] [PubMed] [Google Scholar]
- Terada K, and Oike Y (2010). Multiple molecules of Hsc70 and a dimer of DjA1 independently bind to an unfolded protein. The Journal of biological chemistry 285, 16789–16797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrigshardt H, Rouault TA, and Missirlis F (2013). Insertion mutants in Drosophila melanogaster Hsc20 halt larval growth and lead to reduced iron-sulfur cluster enzyme activities and impaired iron homeostasis. J Biol Inorg Chem 18, 441–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrigshardt H, Singh A, Kovtunovych G, Ghosh M, and Rouault TA (2010). Characterization of the human HSC20, an unusual DnaJ type III protein, involved in ironsulfur cluster biogenesis. Human molecular genetics 19, 3816–3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzarska MA, Dutkiewicz R, Freibert SA, Lill R, and Muhlenhoff U (2013). The mitochondrial Hsp70 chaperone Ssq1 facilitates Fe/S cluster transfer from Isu1 to Grx5 by complex formation. Mol Biol Cell 24, 1830–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickery LE, and Cupp-Vickery JR (2007). Molecular chaperones HscA/Ssq1 and HscB/Jac1 and their roles in iron-sulfur protein maturation. Crit Rev Biochem Mol Biol 42, 95–111. [DOI] [PubMed] [Google Scholar]
- Voisine C, Cheng YC, Ohlson M, Schilke B, Hoff K, Beinert H, Marszalek J, and Craig EA (2001). Jac1, a mitochondrial J-type chaperone, is involved in the biogenesis of Fe/S clusters in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America 98, 1483–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig I, Carrozzo R, Santorelli FM, and Schagger H (2007). Functional assays in high-resolution clear native gels to quantify mitochondrial complexes in human biopsies and cell lines. Electrophoresis 28, 3811–3820. [DOI] [PubMed] [Google Scholar]
- Yang C, Matro JC, Huntoon KM, Ye DY, Huynh TT, Fliedner SM, Breza J, Zhuang Z, and Pacak K (2012). Missense mutations in the human SDHB gene increase protein degradation without altering intrinsic enzymatic function. FASEB J 26, 45064516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Jeong SY, Ghosh MC, Kovtunovych G, Silvestri L, Ortillo D, Uchida N, Tisdale J, Camaschella C, and Rouault TA (2010). Glutaredoxin 5 deficiency causes sideroblastic anemia by specifically impairing heme biosynthesis and depleting cytosolic iron in human erythroblasts. J Clin Invest 120, 1749–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.