SUMMARY
Correct localization of Hedgehog effectors to the tip of primary cilia is critical for proper signal transduction. The conserved non-motile kinesin Kif7 defines a “cilium-tip compartment” by localizing to the distal ends of axonemal microtubules. How Kif7 recognizes microtubule ends remains unknown. We find that Kif7 preferentially binds GTP-tubulin at microtubule ends over GDP-tubulin in the mature microtubule lattice, and ATP hydrolysis by Kif7 enhances this discrimination. Cryo-EM structures suggest that a rotated microtubule footprint and conformational changes in the ATP-binding pocket underlie Kif7’s atypical microtubule-binding properties. Finally, Kif7 not only recognizes but also stabilizes a GTP-form of tubulin to promote its own microtubule end localization. Thus, unlike the characteristic microtubule-regulated ATPase activity of kinesins, Kif7 modulates the tubulin mechanochemical cycle. We propose that the ubiquitous kinesin fold has been repurposed in Kif7 to facilitate organization of a spatially restricted platform for localization of Hedgehog effectors at the cilium tip.
Keywords: cilia, cilium tip, Hedgehog, Kif7, kinesin, microtubule, tubulin
Introduction
The primary cilium, a solitary antenna-like projection on the surface of most vertebrate cells, has emerged as a major hub for cellular signaling (Singla and Reiter, 2006, Goetz and Anderson, 2010, Briscoe and Therond, 2013). The structural backbone of this organelle is a distinct cylindrical arrangement of parallel microtubules known as the axoneme (Satir and Christensen, 2007). In vertebrates, one important intercellular signaling pathway that depends on the primary cilium is the evolutionarily conserved Sonic Hedgehog (Shh) pathway, which is critical for embryonic development and adult tissue homeostasis, and its mis-regulation is associated with multiple tumor types (Jiang and Hui, 2008, Ingham et al., 2011).
A key step in Hedgehog (Hh) signaling is the translocation of several pathway components, including the transcription factor Gli and its repressor Sufu, to a sub-micron region at the distal cilium tip (Drummond, 2012). The microtubule-associated motor protein Kif7, which is a conserved component of the Shh pathway and member of the kinesin-4 protein family, plays a central role in defining this “cilium-tip compartment” (Liem et al., 2009, He et al., 2014, Liu et al., 2014). Mutations in Kif7 have been linked to developmental birth defects, such as Joubert, fetal hydrolethalus, and acrocallosal syndromes (He et al., 2017). Kif7 localizes to the distal tip of the primary cilium during (i) ciliogenesis, where it is proposed to regulate the length of the axoneme, and (ii) Hh signaling, where it is thought to define the cilium-tip compartment for the recruitment and regulation of the Gli-Sufu complexes (Liem et al., 2009, He et al., 2014, Liu et al., 2014). Therefore, the precise localization of Kif7 to the ends of axonemal microtubules is critical for its cellular functions. In vitro studies have shown that Kif7 neither walks nor diffuses on microtubules but autonomously tracks the growing filament ends, where it regulates tubulin polymerization dynamics (He et al., 2014, Yue et al., 2018); however, the molecular mechanism by which this non-motile kinesin specifically recognizes microtubule plus-ends is unknown.
Microtubule-associated proteins (MAPs) that track growing microtubule tips typically recognize two features of microtubule plus-ends: (i) increased curvature of protofilaments or (ii) presence of GTP-bound tubulin (Jiang and Akhmanova, 2011, Brouhard and Rice, 2014). For example, the tip-tracking kinesins Kif19a and Kip3p recognize curved protofilaments at microtubule ends through family-specific loops (L2 in Kif19a and L11 in Kip3p; (Wang et al., 2016, Arellano-Santoyo et al., 2017). Some kinesins show a small (<3.7-fold) preference for microtubules polymerized with certain GTP analogues, but these pleiotropic effects do not translate to microtubule tip tracking (Nakata et al., 2011, Bechstedt et al., 2014, Morikawa et al., 2015, Guedes-Dias et al., 2019). Selective recognition of GTP-tubulin has been best described for the end-binding protein EB1, which contains a calponin homology domain that binds at the inter-protofilament interface of microtubules and senses structural changes in tubulin associated with GTP hydrolysis (Hayashi and Ikura, 2003, Maurer et al., 2012, Zhang et al., 2015). It is unlikely that Kif7 and EB1 share the same binding site on microtubules as the crystal structure of Kif7 reveals a canonical kinesin fold (Klejnot and Kozielski, 2012). Currently, features at microtubule ends that are recognized by Kif7 and enable tip tracking remain unclear.
A hallmark feature of kinesins is the tight coupling between their chemical and mechanical cycles, allowing these motors to generate motile forces and catalytically regulate microtubule dynamics. Although Kif7 has a 55% sequence similarity with conventional kinesin, adopts the canonical kinesin fold, and retains conserved residues required for ATP hydrolysis (Klejnot and Kozielski, 2012), recent studies have revealed that Kif7 is characterized by atypical chemo-mechanical properties (Yue et al., 2018). First, microtubules do not stimulate the ATP hydrolysis rate of Kif7, as shown by the slow rate of ADP release from Kif7 upon microtubule binding, which is the first step in the kinesin enzymatic cycle. Second, Kif7 binds microtubules in both pre- and post-ATP hydrolysis states, unlike most kinesins for which the ATPase cycle is linked to their microtubule binding-unbinding cycle. Furthermore, the mechanochemical properties of Kif7 are also divergent from other closely related kinesin-4 proteins, including the motile kinesins Kif4A, Kif21, and Kif27 (Bieling et al., 2010, Subramanian et al., 2013, van der Vaart et al., 2013, Muhia et al., 2016, Yue et al., 2018). Currently, the structural basis for the unconventional chemo-mechanical properties of Kif7 has not been described. Further, the contribution of ATP hydrolysis to microtubule tip tracking by Kif7 is unknown.
Here we elucidated the biochemical and structural adaptations that enable Kif7 to specifically recognize microtubule plus-ends for the organization of a cilium-tip compartment. We show that Kif7 exhibits a 15–30-fold greater affinity for GTP-like states of tubulin than for GDP-tubulin, a finding that is unprecedented for a kinesin. ATP hydrolysis by Kif7 fine tunes the ability of Kif7 to discriminate between the GTP and GDP forms of tubulin. Cryo-electron microscopy (cryo-EM) structures of Kif7-microtubule complexes in the pre- and post-hydrolysis states revealed an altered microtubule interaction surface in comparison to other kinesins and provided insight into how mechanochemical coupling is disrupted. Interestingly, while the ATPase activity of Kif7 is not significantly regulated by microtubules, the conformational changes in tubulin associated with GTP hydrolysis are inhibited by Kif7 to promote its own microtubule end binding. Finally, structure-based mutational analyses indicated that the microtubule-binding activity of Kif7 is required for its localization to the distal cilium tip in response to Hh signaling. Together, these findings illuminate a new model in which the interplay between the Kif7 and tubulin mechanochemical cycles underlies the spatially restricted localization of a kinesin to microtubule plus-ends.
RESULTS
Kif7 preferentially binds GTP- over GDP-tubulin within the microtubule lattice
A recombinant dimeric Kif7 construct (aa. 1–560) purified from bacterial cells autonomously tracks the plus-ends of growing microtubules in vitro (He et al., 2014); however, these assays were restricted by low yield, purity, and solubility of the protein. To overcome these limitations, we expressed a shorter dimeric Kif7 construct (aa. 1–543; hereafter referred to as Kif7DM-GFP; Figure S1A) with an N-terminal solubilizing SUMO tag and C-terminal GFP using a baculovirus expression system. Homogenous recombinant Kif7 protein was obtained after removal of purification tags (purity>95%; solubility>10 mg/ml; Figures S1B and S1C). We examined the localization of Kif7DM-GFP on dynamic microtubules in an in vitro total internal reflection fluorescence (TIRF) microscopy-based assay to confirm that it tracked the growing ends of microtubules. Briefly, X-rhodamine and biotin-labeled microtubule seeds, polymerized with the non-hydrolyzable GTP analogue GMPCPP, were immobilized on a glass coverslip with neutravidin-biotin linkages. Kif7DM-GFP, tubulin, GTP, and ATP were added to the chamber, and time-lapse images were acquired to examine Kif7 localization on dynamic microtubules (Figure S1D; hereafter referred to as “dynamic microtubule assay”). Overlay of kymographs from the X-rhodamine and GFP channels showed that Kif7DM-GFP tracks the tips of growing microtubules (Figure S1E).
Kif7 is characterized by an unusually long neck linker (~120 aa.) connecting its motor and coiled-coil dimerization domain. To determine whether the monomeric motor domain of Kif7, without most of the neck linker and the dimerization domain, tracks the ends of growing microtubules, we expressed and purified a GFP-tagged monomeric construct of Kif7 (aa. 1–386; hereafter referred to as Kif7MM-GFP; Figures S1A, S1B, and S1C). The monomeric motor domain of Kif7 was sufficient to track the tips of growing microtubules (Figure S1F); however, a higher concentration of Kif7MM-GFP was required to achieve levels of binding comparable to those of Kif7DM-GFP (1.5 μM vs 150 nM), demonstrating lower microtubule-binding affinity. This finding indicates that microtubule tip tracking is an inherent property of the Kif7 motor domain.
The observation that Kif7 binds to the GMPCPP-bound seeds as well as the plus-ends of microtubules, which contain GTP-tubulin, raises the possibility that Kif7 preferentially binds to the GTP-like form of tubulin in the microtubule lattice (Figure S1E and S1F; (He et al., 2014). To examine this possibility, we visualized Kif7 binding on single microtubules comprising segments of tubulin in different nucleotide states (hereafter referred to as “segmented microtubule assay”). X-rhodamine-labeled GMPCPP-bound microtubule seeds were immobilized on a glass coverslip. Microtubules were grown from the immobilized seed by the addition of HiLyte647-labeled tubulin and GTP. As tubulin polymerizes, GTP is hydrolyzed to GDP, generating a microtubule segment with GDP-tubulin. Polymerization was subsequently halted with the addition of X-rhodamine-labeled tubulin, GTP, and GMPCPP, resulting in the formation of a stable “cap” at the end of the polymer (Figure 1A). Because several tip-tracking kinesins also bind soluble tubulin, this assay has the advantage of allowing quantitative analysis of the Kif7-microtubule interaction at equilibrium in the absence of soluble tubulin. Kif7DM-GFP (150 nM; 1 mM ATP) preferentially localized to regions of the segmented microtubule that contain GMPCPP over those containing GDP (Figure 1B–C). Quantitative analysis of GFP fluorescence showed that the average intensity per pixel was ~15-fold greater on the GMPCPP in comparison to the GDP region (Figure 1D). One difference between the GMPCPP-bound microtubule seed and the GDP-bound lattice is that the seed is immobilized and labeled with biotin. Therefore, we quantified the Kif7DM-GFP fluorescence intensity on the stabilizing cap, which is not attached to the surface or biotinylated. The average intensity per pixel in the cap was an intermediat value, as expected because it contains both GMPCPP and GDP (Figure 1D).
Figure 1. Kif7 preferentially binds GTP- over GDP-tubulin. See also Figure S1.
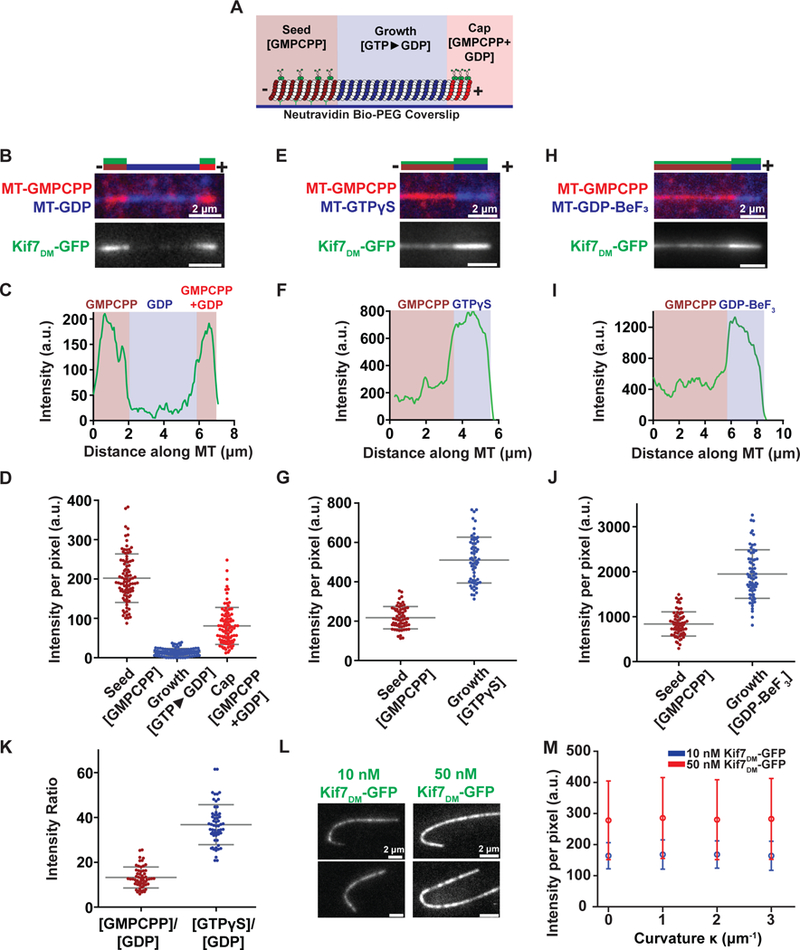
A. Schematic of the segmentation assay used to examine Kif7DM-GFP binding to microtubule segments in different nucleotide states. X-rhodamine-labeled GMPCPP-stabilized microtubule ‘seeds’ (brown) were immobilized on a glass coverslip via a neutravidin-biotin linkage. Microtubule polymerization was initiated by incubating the seeds with HiLyte647 tubulin (blue) and either 1 mM GTP, 1 mM GTPγS or 2 mM GDP-BeF3 (2 mM GTP, 2 mM BeSO4 and 10 mM NaF). The growing microtubules were stably capped by further polymerization in the presence of X-rhodamine-labeled tubulin and a mixture of 2 mM GMPCPP and 1 mM GTP (red). Kif7DM-GFP (green) and 1 mM ATP were subsequently added to examine the association of Kif7.
B. Representative image of microtubule (top) and associated Kif7DM-GFP (bottom) obtained from the experiment described in (A). Assay conditions: 150 nM Kif7DM-GFP and 1 mM ATP and GTP. The schematic above the image indicates the position of the seed (brown), growth (blue), cap (red) and the associated Kif7DM-GFP signal (green).
C. Line scan of GFP intensity from Kif7DM-GFP bound to the microtubule shown in (B).
D. Scatter plot of GFP fluorescence intensity per pixel on the GMPCPP seed (brown), GTP►GDP growth (blue) and GMPCPP+GDP cap (red). Error bars represent standard deviation. Assay conditions: 150 nM Kif7DM-GFP and 1 mM ATP (GMPCPP-seed: I = 202.3 ± 61.7; N = 93, GDP growth: I = 14.1 ± 9.3; N = 93 and GMPCPP+GDP cap: (I = 80.8 ± 47.0; N = 93).
E. Representative image of microtubule (top) and associated Kif7DM-GFP (bottom) obtained from the segmentation assay. Experimental conditions: 150 nM Kif7DM-GFP, 1 mM ATP and 1 mM GTPγS. The schematic above the image indicates the position of the seed (brown), growth (blue) and the associated Kif7DM-GFP signal (green).
F. Line scan of GFP intensity from Kif7DM-GFP bound to the microtubule shown in (E).
G. Scatter plot of GFP fluorescence intensity per pixel on the GMPCPP-seed (brown) and GTPγS growth segment (blue). Error bars represent standard deviation. Assay conditions: 150 nM Kif7DM-GFP and 1 mM ATP. (GMPCPP seed: I = 217.9 ± 56.7; N = 63 and GTPγS growth: I = 510.6 ± 116.6; N = 63).
H. Representative image of microtubule (top) and associated Kif7DM-GFP (bottom) obtained from the segmentation assay. Experimental conditions: 150 nM Kif7DM-GFP and 1 mM ATP and 2 mM GDP-BeF3. The schematic above the image indicates the position of the GMPCPP seed (brown), GDP-BeF3-growth (blue) and the associated Kif7DM-GFP signal (green).
I. Line scan of GFP intensity from Kif7DM-GFP bound to the microtubule shown in (H).
J. Scatter plot of GFP fluorescence intensity per pixel on the GMPCPP seed (brown) and GDP-BeF3 growth segment (blue). Error bars represent standard deviation. Assay conditions: 150 nM Kif7DM-GFP and 1 mM ATP. (GMPCPP-seed: I = 836.4 ± 270.5; N = 65 and GDP-BeF3 growth: I = 1946.8 ± 540.9; N = 65).
K. Scatter plot of GFP fluorescence intensity ratio between GMPCPP/GDP (brown) and GTPγS/GDP (blue). Error bars represent standard deviation. Assay conditions: 150 nM Kif7DM-GFP and 1 mM ATP. (GMPCPP/GDP: 13.2 ± 4.7 and GTPγS/GDP: 36.8 ± 8.9).
L. Representative images of GFP fluorescence signal from Kif7DM-GFP at 10 nM and 50 nM concentrations in the presence of 1 mM ATP associated on curved microtubules.
M. Plot of Kif7DM-GFP fluorescence intensity against microtubule curvature on GMPCPP microtubules. Error bars represent standard deviation (Kif7DM-GFP 10 nM: |κ0| 164 ± 42, |κ1| 168 ± 47, |κ2| 168 ± 44, |κ3| 164 ± 47; 50 nM: |κ0| 278 ± 126, |κ1| 285 ± 130, |κ2| 280 ± 129, |κ3| 283 ± 130). Assay conditions: 10 nM (blue) and 50 nM (red) Kif7DM-GFP and 1 mM ATP.
We next determined whether the selectivity of Kif7 for GMPCPP-tubulin reflected recognition of a GTP-like state of tubulin or a structural feature specific to GMPCPP-tubulin. To this end, we assessed whether Kif7 preferentially recognizes microtubules polymerized with other GTP analogues using a microtubule segmentation assay incorporating microtubule segments containing either the slowly hydrolysable GTP analogue GTPγS or the GDP.Pi analogue GDP-BeF3. Microtubules were polymerized from the immobilized GMPCPP-microtubule seed by the addition of HiLyte647-labeled tubulin and GTPγS or a mixture of GTP, BeSO4, and NaF. Microtubule growth was capped by the addition of X-rhodamine-labeled tubulin, GTP, and GMPCPP. Kif7DM-GFP (150 nM; 1 mM ATP) localized to both the GMPCPP and GTPγS segments of the microtubules (Figure 1E–F), and the intensity per pixel on the GTPγS segment was 2-fold greater than that on the GMPCPP-microtubule seed (Figure 1G). Similarly, Kif7DM-GFP (150 nM; 1 mM ATP) localized to both the GMPCPP-microtubule seed and GDP-BeF3-microtubule segments (Figure 1H and I), and the fluorescence intensity per pixel on the GDP-BeF3 segment was ~2-fold greater than that on the GMPCPP-microtubule seed (Figure 1J). Interestingly, the absolute values of the Kif7DM-GFP intensity on the GMPCPP-containing seed in the presence of GDP-BeF3 was 4-fold greater than that for GTP and GTPγS. This difference may result from the higher microtubule-binding affinity of the Kif7-ADP-BeF3 complexes that are likely formed in these experiments. To exclude the possibility that the increased GFP intensity on the different nucleotide segments was due to differences in fluorophore properties (HiLyte647 vs. X-rhodamine), we constructed GMPCPP-GTPγS segmented microtubules with only X-rhodamine-labeled tubulin. Consistent with the experiments containing two fluorophores, higher GFP fluorescence intensity was observed on the GTPγS segment compared to the GMPCPP-containing seed (Figure S1G).
To compare the preference of Kif7DM-GFP for tubulin in the GDP-, GMPCPP-, and GTPγS-states, we normalized the intensity per pixel for GDP and GTPγS against the GMPCPP-seed. Notably, we were unable to reliably perform the normalization with GDP-BeF3 due to the increased binding of Kif7 to the GMPCPP-containing seed in the presence of this analogue (possibly due to the formation of ADP-BeF3). Kif7 exhibited a 37-fold preference for GTPγS-tubulin and a 13-fold preference for GMPCPP-tubulin compared to GDP-tubulin within microtubules (Figure 1K). This degree of discrimination for microtubules composed of GTP- over GDP-tubulin has not been previously reported for a kinesin under similar in vitro conditions.
Several kinesins and non-motor MAPs have been proposed to track microtubule tips by associating with the increased curvature at microtubule plus-ends (Bechstedt et al., 2014, Roostalu et al., 2015, Arellano-Santoyo et al., 2017). These proteins typically show a higher binding affinity to curved over straight microtubules. To determine whether Kif7 binds specifically to curved microtubules similar to proteins such as DCX and Kip3p, we immobilized long X-rhodamine- and biotin-labeled microtubules, which adopt a range of orientations on the coverslip with different curvatures. We examined the localization of Kif7DM-GFP at 10 nM and 50 nM concentrations (1 mM ATP) on curved microtubules. Visual inspection (Figure 1L) and quantitative analysis (Figure 1M) of fluorescence images did not show significantly greater localization of Kif7 to curved regions of microtubules compared to straight regions (N>20). Consistent with this, Kif7DM (1.3μM) with AMPPNP or ADP bound to straight microtubules and not to dolastatin-induced tubulin rings when visualized using negative-stain EM (Figure S1H). Together, these findings demonstrate that Kif7 localizes to the growing microtubule plus-ends by selectively recognizing the GTP and GDP.Pi forms of tubulin.
ATP hydrolysis by Kif7 promotes discrimination between GTP- and GDP-tubulin
The microtubule binding-unbinding cycle of kinesins is closely linked to their ATP hydrolysis cycle. However, Kif7 is characterized by its atypical mechanochemistry (Yue et al., 2018). To quantitatively determine changes in the affinity of Kif7DM-GFP for microtubules during its atypical ATPase cycle, we carried out microtubule co-sedimentation assays in the presence of ATP, ADP, and the non-hydrolyzable ATP-analogue AMPPNP (Figure 2A). Due to the slow rate of ADP release, it is expected that a large fraction of Kif7 will be bound to ADP in experiments conducted with ATP (Yue et al., 2018). Thin Layer Chromatography (TLC) experiments performed in the presence of radio-labeled [α−32P] ATP confirmed that over 95% of the Kif7-associated nucleotide is indeed in the ADP form (Figure S2A). Kif7DM-GFP (1 μM) was incubated with GTP-polymerized Taxol-stabilized microtubules (0.125–5 μM) in the presence of different nucleotides, separated by centrifugation into a supernatant fraction containing unbound Kif7DM-GFP and a pellet fraction containing Kif7DM-GFP-bound microtubules, and then quantified by SDS-PAGE analysis (Figure S2B). We used GDP-Taxol-stabilized microtubules, as high concentrations of Kif7 destabilize GMPCPP microtubules (He et al., 2014). The dissociation constants of the microtubule-Kif7DM-GFP interaction were nearly identical in the presence of ATP and ADP and were 2-fold lower with AMPPNP (Kd for Kif7DM-GFP, ATP: 0.78±0.10 μM, ADP: 0.81±0.16 μM, and AMPPNP: 0.43±0.03 μM). Thus, the changes in the microtubule-Kif7 interaction between different nucleotide states of the ATPase cycle is small in comparison to those observed for conventional kinesin (Crevel et al., 1996, Rosenfeld et al., 1996, Gigant et al., 2013).
Figure 2. Kif7 ATP hydrolysis enhances GTP- and GDP-tubulin discrimination. See also Figure S2.
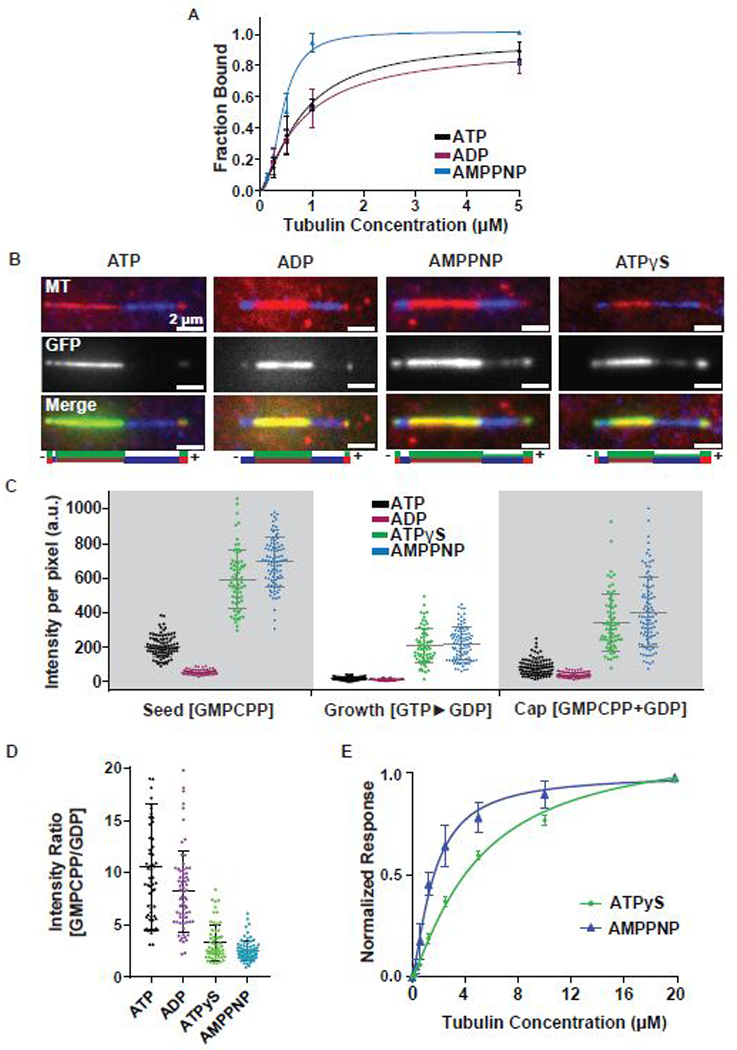
A. Cosedimentation assay to quantitatively examine the binding affinity of Kif7DM-GFP (1 μM) to GDP-taxol stabilized microtubules (0–5 μM) in the presence of 1 mM ATP (black) ADP (purple) and AMPPNP (blue). Error bars represent standard deviation. The plots of fraction of Kif7DM-GFP bound versus microtubule concentration were fit to a Hill equation to determine the equilibrium dissociation constants under different nucleotide conditions. (Kd for Kif7DM-GFP, ATP: 0.78 ± 0.10 μM, ADP: 0.81 ± 0.16 μM and AMPPNP: 0.43 ± 0.03 μM).
B. Representative images showing Kif7DM-GFP (150 nM) associated with segmented microtubules composed of the GMPCPP-seed (red), GDP growth (blue) and GMPCPP+GDP cap (red) in the presence of different adenosine nucleotides (1 mM ATP, ADP, AMPPNP and ATPγS). The schematics under the images indicate the position of the seed (brown), growth (blue), cap (red) and the associated Kif7DM-GFP signal (green).
C. Scatter plot of GFP fluorescence intensity per pixel of microtubule-bound Kif7DM-GFP (150 nM) on the indicated microtubule segments. Error bars represent standard deviation. Assay conditions: 150 nM Kif7DM-GFP and 1 mM ATP (black), ADP (purple), AMPPNP (blue), ATPγS (green) on the GMPCPP-seed, GTP►GDP growth and GMPCPP+GDP cap. (GMPCPP-Seed: ATP: 202.3 ± 61.7; N = 93, ADP: 53.0 ± 14.0; N = 65, AMPPNP: 693.4 ± 141.7; N = 93 and ATPγS: 591.5 ± 172.7; N = 68. GDP growth: ATP: 14.1 ± 9.3; N = 93, ADP: 8.6 ± 5.4; N = 65, AMPPNP: 216.5 ± 96.5; N = 93 and ATPγS: 207.6 ± 102.9; N = 68. GMPCPP+GDP cap: 80.8 ± 47.0; N = 93, ADP: 37.1 ± 15.8; N = 65, AMPPNP: 401.2 ± 203.5; N = 93 and ATPγS: 338.2 ± 165.7; N = 68).
D. Scatter plot of the intensity ratio of microtubule-bound Kif7DM-GFP on the GMPCPP-seed and GTP►GDP growth regions in the presence of different adenosine nucleotides. Error bars represent standard deviation. Assays were performed under Kif7DM-GFP concentrations that resulted in similar average fluorescence intensities on the GMPCPP-seed in the presence of different nucleotides to allow for normalization. Assay conditions: 150 nM Kif7DM-GFP and 1 mM ATP; 250 nM Kif7DM-GFP and 1 mM ADP; 10 nM Kif7DM-GFP and 1 mM AMPPNP; 50 nM Kif7DM-GFP and 1 mM ATPγS. (GMPCPP/GDP: ATP: 10.53 ± 6.04; N = 62, ADP: 8.19 ± 3.84; N = 68, AMPPNP: 2.51 ± 0.96; N = 78 and ATPγS: 3.32 ± 1.71; N = 61).
E. Biolayer interferometry assay to quantitatively examine the binding affinity of Kif7DM to soluble tubulin (0–20 μM) in the presence of 1 mM ATPγS (green) and AMPPNP (blue). Error bars represent standard deviation. The plots of fraction of Kif7DM bound versus soluble tubulin concentration were fit to a Hill equation to determine the equilibrium dissociation constants under different nucleotide conditions. (Kd for Kif7DM, ATPγS: 4.98 ± 0.9 μM and AMPPNP: 1.64 ± 0.2 μM).
The observation that the microtubule affinity of Kif7 is not significantly altered upon ATP hydrolysis spurred us to investigate how the Kif7 ATPase cycle is linked to the discrimination between GTP- and GDP-tubulin. To address this question, we examined the localization of Kif7DM-GFP (150 nM) on segmented microtubules in the presence of ATP, ADP, AMPPNP, or the slowly hydrolysable analogue ATPγS (1 mM). Fluorescence imaging of Kif7DM-GFP binding on segmented microtubules revealed localization of Kif7 preferentially to the GMPCPP-containing segment compared to the GDP-containing segment under all nucleotide conditions (Figure 2B–C). As expected, the stabilizing cap, composed of both GMPCPP and GDP, displayed intermediate values (Figure 2C). In accordance with the results of our cosedimentation assay, the GFP fluorescence intensity on both the GMPCPP and GDP-microtubule segments was lower with ADP than with AMPPNP or ATPγS. Consistent with this finding, in dynamic microtubule assays, we observed that Kif7 tracks microtubule ends in the presence of ATP, ATPγS, and ADP (1 mM; Figure S2C). In the presence of AMPPNP, Kif7DM-GFP localized along the entire length of the polymerizing microtubule as opposed to the GMPCPP-bound seed similar to observations reported by He et al. (Figure S2D (He et al., 2014)). The differences in Kif7 localization in segmentation versus dynamic microtubule assays with AMPPNP are discussed later in this section. Taken together, these findings suggest that ATP hydrolysis does not significantly affect the Kif7-microtubule-binding affinity and Kif7 prefers GTP- over GDP-segments of microtubules in all adenosine nucleotide states.
Next, we used quantitative analysis to elucidate whether the distinction between GTP- and GDP-tubulin is different between the pre- and post-hydrolysis states of Kif7. To reliably measure the ratios of Kif7DM-GFP fluorescence intensities on GMPCPP- and GDP-microtubule segments, we repeated the assay with the different adenosine nucleotides as above and adjusted the Kif7 concentration such that (i) the GFP intensity values in the GMPCPP region were in the same range to facilitate normalization and (ii) the values in the GDP-bound segment were significantly above background (Figure S2E). In the presence of AMPPNP and ATPγS, the ratio of Kif7DM-GFP on the GMPCPP and GDP segments were 2.51 ± 0.96 (N = 78) and 3.32 ± 1.71 (N = 61), respectively (Figure 2D). In contrast, in the presence of ATP and ADP, the ratios of Kif7DM-GFP on the GMPCPP and GDP segments were 10.53 ± 6.04 (N = 62) and 8.19 ± 3.84 (N = 68) These data indicate that ATP hydrolysis enhances the selectivity of Kif7-binding to GTP- over GDP-tubulin.
The contrasting activity of Kif7 in the presence of AMPPNP in the dynamic and segmentation assays suggested that Kif7 may bind soluble tubulin in an adenosine nucleotide-dependent manner. To test this, we used Bio-layer interferometry assay to measure the binding affinity between Kif7 and soluble tubulin in the presence of ATP, ADP, ATPγS and AMPPNP (1 mM). We found that Kif7 bound tubulin with the highest affinity in the presence of AMPPNP (Kd = 1.64 ± 0.2 μM) and with weaker affinity in the presence of ATPγS (Kd = 8.1 ± 0.9 μM) (Figure 2E). In the presence of ATP and ADP, we could not detect binding between Kif7 and tubulin. As previously shown, we expect Kif7 to be largely ADP bound in the presence of ATP (Figure S2A). Additionally, size-exclusion chromatography of Kif7DM-GFP (1 μM) and tubulin (20 μM) in the presence of ATP, ADP, and AMPPNP (1 mM) showed that Kif7DM-GFP forms a stable complex with soluble tubulin in the presence of AMPPNP but not with ATP or ADP (Figure S2F). With these findings in mind, we propose that in a dynamic microtubule assay with soluble tubulin, Kif7-AMPPNP remains bound to the tubulin heterodimer. As a result, Kif7 incorporates into the growing filament as a complex with the tubulin dimer. Hydrolysis releases Kif7 from soluble tubulin, enabling this kinesin to interact specifically with the GTP form of tubulin. Therefore, the ATP hydrolysis cycle of Kif7 is not coupled to microtubule binding-unbinding cycle, but is instead linked to binding and release from soluble tubulin.
We next determined whether microtubules or soluble tubulin could enhance the intrinsic ATPase rate of Kif7. We found that the basal ATPase rate of dimeric Kif7 is stimulated only 3-fold by microtubules (for Kif7 with no microtubules Kcat= 0.01 ± 0.006 s−1site−1; with 4 μM microtubules, Kcat= 0.03 ± 0.005 s−1site−1) (Figure S2G), consistent with the 5-fold enhancement of Kif7’s ATPase reported at a high microtubule concentration of 25 μM (Yue et al., 2018). In contrast, we observed no stimulation of the basal ATPase rate of Kif7 in the presence of soluble tubulin (Figure S2H).
Together, our analyses suggest that ATP hydrolysis thus serves two purposes: (i) it increases the selectivity of the motor for GTP-tubulin over GDP-tubulin, and (ii) it releases Kif7 from soluble tubulin to enable tip tracking.
Structure of Kif7 bound to microtubules
To gain insight into the structural basis of Kif7 function, we determined the structure of microtubule-bound Kif7 using cryo-EM. GMPCPP microtubules were not suitable for these studies, as such structures are prone to breakage at high Kif7 concentrations (He et al., 2014). As an alternative, we examined the binding of Kif7DM-GFP to GTP-polymerized Taxol-stabilized microtubules, which are proposed to be structurally similar to GMPCPP microtubules (Alushin et al., 2014). This analysis revealed that the GFP fluorescence intensity was similar on GDP-Taxol-stabilized and GMPCPP-bound microtubules (Figure S3A). Structural changes upon ATP hydrolysis were captured by obtaining reconstructions of the Kif7-microtubule complex in the presence of AMPPNP and ADP. The reconstructions were at sufficient resolution to reveal secondary structure elements (Table S1).
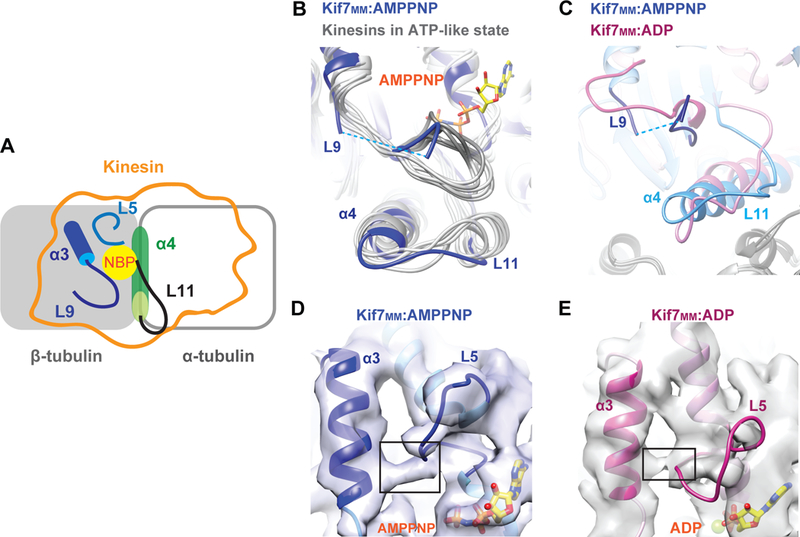
AMPPNP-bound Kif7 adopts a rotated conformation on the microtubule surface compared to kinesin-1
We first obtained three-dimensional (3D) reconstructions of the AMPPNP-Kif7-dimer (pre-hydrolysis state) bound to GDP-Taxol-stabilized microtubules but were unable to improve the resolution beyond 6 Å (data not shown). Reconstructions of Kif7MM bound to GDP-Taxol microtubules extended the resolution to 4.3 Å (Fourier shell correlation [FSC] 0.143 criterion; Figure S3B and Supplemental Table 1; hereafter referred to as Kif7MM:AMPPNP). A reconstruction of the Kif7MM bound to GMPCPP microtubules with a resolution of 7 Å was also generated for comparison (Figure S3C). The microtubule lattice parameters for Kif7-bound microtubules are consistent with those reported earlier (Alushin et al., 2014).
All three reconstructions showed several similar features. First, the Kif7 motor domain adopts the canonical kinesin fold and occupies the intradimer region of the tubulin heterodimer (Figure S3C and S3D) as observed in other kinesin-microtubule structures (Klejnot and Kozielski, 2012). Second, we did not observe density on the microtubule lattice at the EB1-binding site (Maurer et al., 2012, Zhang et al., 2015), suggesting that tip tracking by Kif7 and EB1 arise from recognition of different structural features on microtubules (Figures S3D). Third, comparison with the EM reconstruction of monomeric conventional kinesin (kinesin-1 in the ATP state, EMD-6188, and PDB-3J8Y, hereafter referred to as Kin1:ATP; (Shang et al., 2014) surprisingly showed that the density for Kif7MM:AMPPNP was rotated by ~10° on the microtubule surface (Figures 3A C; Kif7MM:AMPPNP in blue, Kin1:ATP in orange). The same rotation was observed in reconstructions from both 14- and 15-protofilament GMPCPP or GDP-Taxol microtubules (Figure S3C). To correlate the rotated conformation of Kif7 with the movement of specific secondary structural elements, we generated a protein model to fit the cryo-EM density data (Figure S3E). Because the Kif7 monomer is sufficient for tip tracking, all structural modeling and analyses were performed using the highest resolution Kif7MM:AMPPNP reconstruction (Table S2). Electron density in the nucleotide-binding pocket (NBP) of Kif7 could be fit with a molecule of AMPPNP.
Figure 3. Kif7 adopts a tilted conformation on the microtubule lattice relative to conventional kinesin. See also Figure S3.
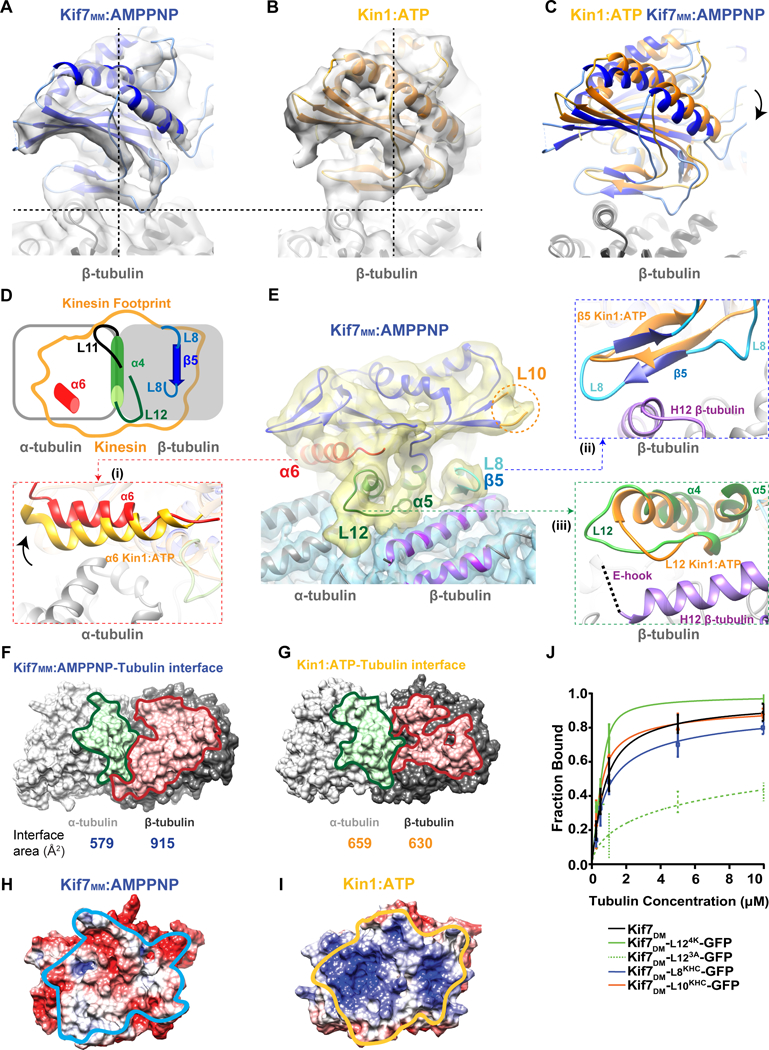
A-C. Comparison of the microtubule-bound conformation of (A) Kif7MM:AMPPNP (blue) and (B) Kin1:ATP (PDB:3J8Y; EMD:6188; orange). Dotted lines are guides for viewing the tilt in Kif7 relative to Kinesin-1. (C) The superposition of pseudo-atomic models of Kif7MM:AMPPNP (blue) and Kin1:ATP (orange) on β-tubulin (gray). Arrow indicates rotation of Kif7MM:AMPPNP with respect to Kin1:ATP.
D. Schematic shows the footprint of a kinesin (orange outline) on α-tubulin (gray unfilled rectangle) and β-tubulin (gray filled rectangle). Secondary structure elements which contact the microtubule lattice are highlighted: α6 (red), L11 (black), α4-L12 (green) and L8-β5 (blue).
E. Cryo-EM reconstruction and structural model of microtubule-bound Kif7MM:AMPPNP. Electron density maps of Kif7MM:AMPPNP and αβ-tubulin are colored yellow and blue respectively. The following secondary structure elements in Kif7 are highlighted as superpositions on Kin1-ATP (orange): (i) α6 (red). Arrow indicates the respective rotation of helix α6 in Kif7MM:AMPPNP compared to Kin1-ATP. (ii) L8-β5 (blue) and (iii) α4-L12 (green). Orange circle indicates the loop L10 region. Helices H11 and H12 of β-tubulin are highlighted in purple. E-hook of β-tubulin is shown as a dotted line. The electron density for Kif7MM:AMPPNP and the tubulin dimer are displayed at different threshold levels.
F&G. Comparison of the binding interface of α- and β-tubulin with (F) Kif7MM:AMPPNP and (G) Kin1:ATP. α- and β-tubulin are shown as surfaces (gray). Residues of α- and β-tubulin that are at <7Å distance from the motor domain are colored green and red respectively. The calculated interaction surface area on tubulin for each kinesin-tubulin interface is below the corresponding image.
H&I. Electrostatic surface potential of the microtubule binding surface of (H) Kif7MM:AMPPNP and (I) Kin1:ATP. Surface potential ranges from −5 (red) to +5 (blue) KT/e. The interface with αβ-tubulin is outlined in light blue for Kif7MM:AMPPNP and in orange for Kin1:ATP.
J. Quantitative analysis of Kif7DM-GFP mutants binding to GTP-polymerized taxol-stabilized microtubules (0 μM to 10 μM) in the presence of 1 mM ATP. The data were fit to a Hill equation to determine the dissociation constants. Error bars represent standard deviation.
To elucidate the influence of the rotated conformation on Kif7-microtubule interaction, we compared the microtubule-binding surfaces of Kif7 and kinesin-1. As shown in Figure 3D, the microtubule footprint of kinesins consists of a central helix α4 (green) anchored on the intradimer cleft of tubulin. This helix is sandwiched between loops L11 (black) and L12 (green) at its N- and C-termini, respectively. This central region is flanked on either side by helix α6 (red) atop the α-tubulin surface and loop L8-strand β5 (blue) atop β-tubulin (Figure 3D). We found that the axis of rotation of Kif7 is coincident with the central helix α4. The rotated conformation of Kif7 moves helix α6 away from α-tubulin (Figure 3E [i]), brings the L8-β5 region closer to the surface of β-tubulin (Figure 3E [ii]), and does not significantly affect the L12 region at the C-terminus of α4 on the tubulin intradimer interface (Figure 3E [iii] and Movie S1).
To quantitate the effects of the rotation on the microtubule footprint of Kif7, we computed the changes in the buried surface area at the α- and β-tubulin interface (assuming side chain orientations). We estimated that the rotation manifests as a ~10% decrease and ~50% increase in the buried surface area on α- and β-tubulin, respectively, for Kif7 compared to kinesin-1 (Figure 3F and 3G). The decrease in surface area at the Kif7-α-tubulin interface occurs mainly due to the loss of molecular interactions with helix α6 of the motor. The gain in surface area at the Kif7-β-tubulin interface arises from two regions: L12-α5 of Kif7 with the C-terminal helix H12 and E-hook of β-tubulin and the L8-β5 region of Kif7 with the helices H4 and H12 of β-tubulin. To further compare the microtubule-binding modes of kinesin-1 and Kif7, we calculated the surface charge potential of the microtubule-binding surfaces of these two proteins. In contrast to most kinesins, the microtubule-binding surface of Kif7 is negatively charged (Figures 3H and 3I). This striking difference arises from non-conserved amino acids in loops L8 and L11 and helices α4 and α6 of Kif7, compared to Kinesin-1. These residues however show a high degree of conservation within the Kif7 family and are conserved in >80% of 135 homologues of human Kif7.
Together, these observations suggest that a global alteration of the microtubule-binding interface through mutations in non-conserved residues allows Kif7 to adopt an altered microtubule footprint while retaining a canonical kinesin fold. In this orientation, the interaction between Kif7 and the C-terminus of β-tubulin, composed of helix H12 and the unstructured E-hook, are enhanced. It has been proposed that upon GTP hydrolysis, helix H12 of β-tubulin undergoes a small conformational change (Alushin et al., 2014). It is therefore possible that this altered footprint on the microtubule surface underlies the ability of Kif7 to discriminate between GTP- and GDP-tubulin.
Contribution of unstructured loops to the Kif7-microtubule interaction
In the tip-tracking kinesins MCAK and Kip3p, loops L2 and L11, respectively, determine their increased affinity for microtubule ends (Hunter et al., 2003, Arellano-Santoyo et al., 2017); however, the sequences of these loops in Kif7 lack the residues implicated in tip tracking (Figure S3F). Therefore, we identified other Kif7 microtubule-proximal loops that are distinct from kinesin-1: (i) a 2-residue Arg-Asp insertion in loop L8, (ii) a 4-residue Pro-Glu-Arg-Arg insertion at the N-terminus of loop L12, and (iii) a 10-residue insertion in loop L10 (loop disordered, no electron density; Figure 3E). Strikingly, all these sequence alterations in Kif7 are on loops that are positioned towards β-tubulin, and close examination of these insertions in the Kif7MM:AMPPNP reconstruction showed that the rotation of the Kif7 motor domain brings the L8 region close to the β-tubulin surface, facilitating the interaction of the two Kif7-specific residues with β-tubulin (Figure S3G, blue). The N-terminus of loop L12, which contains the positively charged “K-loop” in other kinesins and is predicted to bind the negatively charged C-terminus “E-hook” of β-tubulin, is typically not observed in EM structures due to disorder. In Kif7MM:AMPPNP, we observed clear density connecting loop L12 in Kif7, which contains two arginine residues, with the C-terminus of β-tubulin (Figure S3H, green).
The contributions of these loops to microtubule-binding affinity were quantitatively examined using microtubule cosedimentation assays (Figures S3I and S3J). Replacing loop L10 in Kif7 with the corresponding sequence from kinesin-1 (construct Kif7DM-L10KHC-GFP) did not change its microtubule-binding affinity (Kd for Kif7DM-GFP (black): 0.54±0.06 μM and Kif7DM-L10KHC-GFP (red): 0.41±0.06 μM), and substitution of Kif7-L8 with kinesin-1 L8 (construct Kif7DM-L8KHC-GFP) led to a small decrease in microtubule-binding affinity (Kif7DM-L8KHC-GFP (blue): 0.85±0.1 μM; Figure 3J). Increasing the positive charge on loop L12 by mutating all four Kif7-specific residues to lysine resulted in a 4-fold increase in microtubule-binding affinity, while mutation of the three charged residues to alanine (Kif7DM-L123A-GFP) led to an 8-fold decrease compared to Kif7DM-GFP (Kif7DM-L124K-GFP (solid green): 0.13±0.04 μM, Kif7DM-L123A-GFP (dotted green): 4.28±1.36 μM; Figure 3J). Consistently, the fluorescence of Kif7DM-L123A-GFP on microtubules was weak compared to that of Kif7DM-GFP; however, the mutant retained its preference for the GMPCPP-microtubule seed and microtubule tip (500 nM; 1 mM ATP; Figure S3K). These results indicate that among the non-conserved loops of Kif7, L12 significantly contributes to its microtubule-binding affinity but not its ability to track microtubule ends.
Conformational changes in the Kif7-microtubule complex upon ATP hydrolysis
Kif7 tracks the ends of growing microtubules even when bound to ADP (Figure S2C). The best 3D reconstruction of ADP-bound monomeric Kif7 on GDP-Taxol microtubules (referred to as Kif7MM:ADP) at 4.2Å resolution (Figures S4A and S4B, Table S1) was used for model building (Figure 4A and Table S2). The NBP in the Kif7MM:ADP EM reconstruction and the crystal structure of the ADP-Kif7 complex (Klejnot and Kozielski, 2012) are similar, and the density within the NBP indicated the presence of ADP (Figure S4C). A comparison of Kif7MM:ADP and Kif7MM:AMPPNP reconstructions and models showed changes in the secondary structure elements (Figures S4C and S4D) and the electrostatic potential of the NBP (Figures S4E and S4F), indicating that Kif7 responds to the bound adenosine nucleotide.
Figure 4. ADP-bound Kif7 is tilted on the microtubule lattice with respect to AMPPNP-bound Kif7. See also Figure S4.
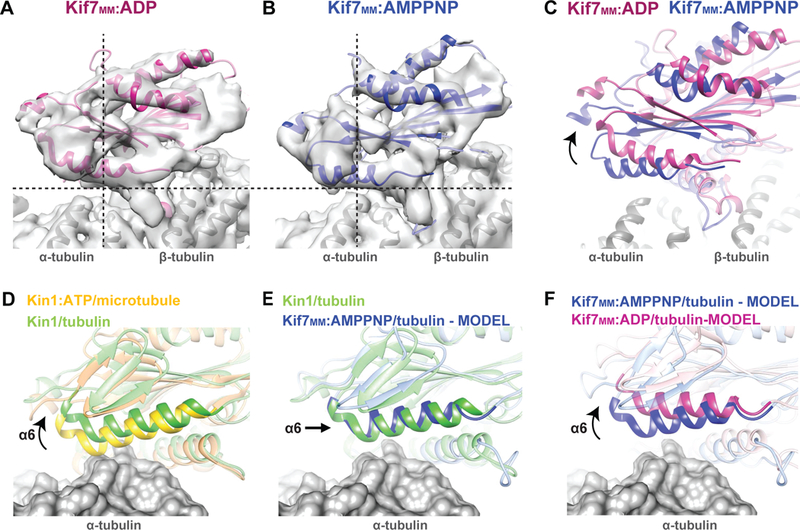
A-C. Comparison of the microtubule-bound conformation of (A) Kif7MM :ADP (pink) and (B) Kif7MM:AMPPNP (blue). Dotted lines are a guide for viewing the tilt in Kif7MM:ADP relative to Kif7MM:AMPPNP. (C) The superposition of pseudo-atomic models of Kif7MM:ADP (pink) and Kif7MM:AMPPNP (blue) on α- and β-tubulin (gray). Arrow indicates the rotated conformation of Kif7MM:ADP with respect to Kif7MM:AMPPNP.
D-F. Comparison of the rotation of helix α6 relative to α-tubulin in different structural models of Kinesin-1 and Kif7. α-tubulin is rendered as a gray surface. Superpositions highlighting helix α6 are shown for (D) Kin1:ATP bound to microtubules (PDB:3J8Y, yellow) and Kin1 bound to soluble tubulin (PDB:4HNA, green). (E) Kin1 bound to soluble tubulin (PDB:4HNA, green) and model of Kif7MM:AMPPNP bound to soluble tubulin (blue). (F) Models of Kif7MM:AMPPNP (blue) and Kif7MM:ADP (pink) bound to soluble tubulin.
The cryo-EM reconstruction of Kif7MM:ADP revealed rotation on the microtubule surface with respect to Kif7MM:AMPPNP (Figures 4A–C, Movie S2). The predominant impact of this change is the 23° rotation of helix α6 of Kif7MM:ADP away from the α-tubulin surface compared to α6 of Kif7MM:AMPPNP (Figure S4G). We found that the density for most of the other microtubule-interacting secondary structural elements of Kif7, such as loop L12, helix α5, and the L8 β5 region, remained largely unchanged (Figure S4H and S4I). Additionally, the central helix α4 remained extended and was only one turn shorter at its N-terminus in the Kif7MM:ADP structure (Figure S4J). Loop L11 displayed well-defined density over its entire length, indicating that this loop is ordered. The estimated interface area of the motor domain of Kif7-ADP on α- and β-tubulin is only 15% less than that for microtubule-bound kinesin-1 in the ATP state. These results suggest that the Kif7 motor domain in both its ADP- and AMPPNP-bound states contacts microtubules through nearly identical secondary structural elements and that the conservation of these interactions underlies its ability to bind microtubules in the ADP-bound state.
In contrast to microtubules, Kif7 binds soluble tubulin only when in complex with AMPPNP (Figure 2E and S2F). The crystal structure of kinesin-1 bound to soluble α-β-tubulin (PDB:4HNA; (Gigant et al., 2013) revealed that helix α6 of kinesin-1 rotates 7° away from the α-tubulin surface compared to its microtubule-bound conformation (Kin1:ATP) (Shang et al., 2014). This movement of the helix accommodates the increased curvature of soluble α-β-tubulin and prevents steric clashes between α-tubulin and kinesin-1 (Figure 4D). This finding raises the possibility that the helix α6 movements observed in our Kif7 structures could explain the formation of a Kif7-tubulin complex with AMPPNP but not with ADP. As previously noted, when bound to a microtubule, α6 of Kif7MM:AMPPNP is rotated 8° upwards compared to Kin1:ATP (Figure 3E[i]). As a result, α6 in the soluble tubulin-bound kinesin-1 and Kif7MM:AMPPNP superpose on each other (Figure 4E). Upon ATP hydrolysis, α6 of Kif7 rotates even further upwards away from the surface of soluble tubulin (Figure 4F). Thus, while the 8° rotation of α6 of Kif7MM:AMPPNP facilitates its interaction with α-tubulin of soluble tubulin, the 23° rotation of α6 in Kif7MM:ADP releases this domain from contacts with the surface of α-tubulin. These structural changes are likely to underlie the coupling between Kif7 ATP hydrolysis cycle and soluble tubulin binding-unbinding cycle.
Changes in the relay mechanisms that link nucleotide state to microtubule binding underlie the mechanochemical properties of Kif7
Neither microtubules not soluble tubulin regulate the Kif7 ATPase cycle (Figure S2G–H; (Yue et al., 2018)). To gain insights into this atypical feature of Kif7, we inspected secondary structural elements that have been proposed to relay information from the NBP to microtubule binding for other kinesins (Figure 5A). The NBP is flanked by the Switch1, Switch2, and P-loop regions, which are important for ATP binding and hydrolysis in kinesins (Figure S4C and S4D; (Cross, 2016). First, we examined loop L9 that contains the Switch1 region and loop L11 that links the NBP to central helix α4 and encompasses Switch2. The Kif7MM:AMPPNP reconstruction showed no clear density corresponding to the L9 N-terminal half, which precedes Switch1, indicating that the region is disordered (Figure 5B, indicated by the dotted lines). A superposition of seven different EM-derived structures showed that with the exception of Kif7MM:AMPPNP, loop L9 extends out towards L11 in all kinesins in the ATP-bound state (Figure 5B). For example, in kinesin-3 Kif1A, where EM reconstructions of microtubule-bound kinesin are available in both the pre- and post- ATP hydrolysis states, it is observed that in the ADP state, loop L9 forms a tight coil, and L11 is disordered (Figure S5A). Upon ATP binding, L9 extends out towards L11, thus stabilizing L11, which in turn contacts the microtubule-interacting central helix α4 (Figure S5B). These conformational changes thus mediate communication between the NBP and the microtubule surface (Shang et al., 2014). In contrast, comparison of these regions in Kif7 shows that L9 does not extend out to L11 and that loop L11 and α4 are ordered in both ADP- and AMPPNP-bound states (Figure 5C). The absence of the pincer movement between L9 and L11 suggests that the communication between the NBP and microtubule binding is likely to be disrupted in Kif7, permitting tight microtubule interactions with the motor in both pre- and post-ATP hydrolysis states.
Figure 5. Cryo-EM structures in pre- and post-hydrolysis states reveal the structural basis for ATPase-independent microtubule tip-tracking by Kif7. See also Figure S5.
A.Schematic of a kinesin bound to microtubule, showing secondary structure elements flanking the Nucleotide Binding Pocket (NBP): Kinesin footprint (orange outline), helix α3 and loop L9 (dark blue), Loop L5 (light blue), helix α4 (green) and Loop L11 (black).
B. Enlarged view of the L9-L11 region obtained from the superposition of multiple structures of kinesins in the ATP state (gray) with Kif7MM:AMPPNP (blue). AMPPNP is rendered as yellow sticks. Dotted blue line corresponds to the disordered region in Kif7MM:AMPPNP.
C. Enlarged views of the L9-L11 region obtained from the superposition of Kif7MM:ADP (pink) with Kif7MM:AMPPNP (blue). Dotted blue line corresponds to the disordered region in Kif7MM:AMPPNP.
D&E. Enlarged views of α3-L5 region near the nucleotide binding pocket shown for (D) Kif7MM:AMPPNP (blue), and (E) Kif7MM:ADP (pink). Black boxes highlight the density in EM reconstructions, between α3 and L5. AMPPNP and ADP are rendered as yellow sticks.
An alternative pathway that connects microtubule binding and nucleotide hydrolysis in kinesins involves the loop L5 in the vicinity of the NBP and the β-tubulin-contacting strand β5 (Figure 5A). Loop L5 has been proposed to change its conformation in response to the bound nucleotide in the NBP, and this information is then relayed to β5 via helix α3. Furthermore, it has been proposed that upon clamping of the kinesin sub-domain containing β5 and α3 onto the tubulin surface, changes in loop L5 cause NBP opening, facilitating ADP release (Cochran et al., 2005, Cochran et al., 2009). We examined the L5-α3 region of Kin1:ATP and both Kif7 structures. Unlike Kin1:ATP, we noted clear electron density connecting L5 to α3 in both Kif7MM:AMPPNP and Kif7MM:ADP structures (Figures 5D, 5E and S5C). This linkage may limit the conformational flexibility of loop L5 and account for the slow ADP release kinetics of Kif7 (Yue et al., 2018). Together, these observations provide a structural basis for the Kif7-microtubule interaction even in the presence of ADP and underlie the unusually slow ADP release rate in Kif7.
Kif7 stabilizes the GTP form of tubulin to promote its microtubule end binding
We next examined whether Kif7 binding changes the properties of the growing microtubule end. To address this, we analyzed the spatial extent of Kif7DM-GFP fluorescence localization at the ends of dynamic microtubules at 50 nM and 300 nM concentrations (1 mM ATP). Overlay images from the GFP and X-rhodamine channels at a single point in time in combination with a visual inspection of kymographs generated from time-lapse imaging showed that Kif7DM-GFP occupies a larger region of the microtubule tip (hereafter referred to as “spreading”) at the higher protein concentration (Figures 6A and 6B). Line scan analysis of the GFP intensity on the polymerizing microtubule region revealed a decay in the fluorescence intensity extending away from the proximal plus-end of the microtubule (Figure 6C). To quantitatively analyze the extent of Kif7 spreading, GFP intensity profiles along the microtubule were generated at a fixed time point from multiple microtubules (>20) at both concentrations. Profiles were aligned and normalized to the point of maximum GFP intensity (0 μm) for comparison between the two data sets. At 50 nM and 300 nM Kif7DM-GFP, the lengths of the region of motor occupancy extended to an average distance of ~0.75 μm and >3 μm, respectively, from the microtubule plus-end (Figure 6D and S6A).
Figure 6. Kif7 stabilizes the GTP-form of tubulin, thereby extending its binding region at microtubule-ends. See also Figure S6.
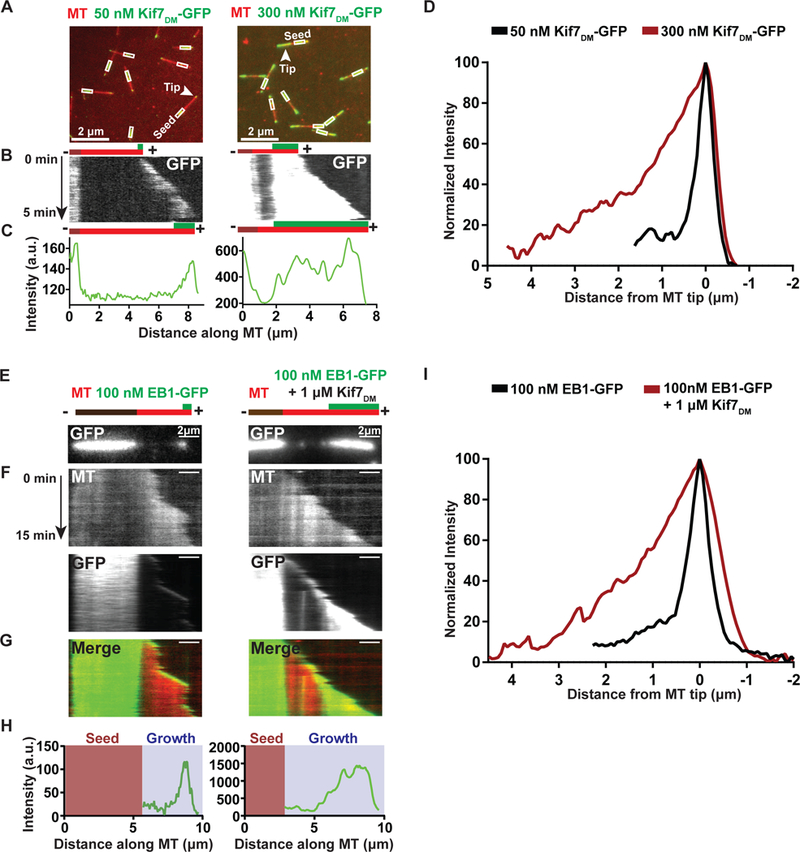
A. Representative fields of view showing the association of Kif7DM-GFP (50 nM and 300 nM; green) on dynamic microtubules (red). Boxes indicate the X-rhodamine-labeled GMPCPP-seed. Arrow indicates the growing tip of the microtubules.
B. Kymographs generated from the microtubules highlighted with an arrowhead in (A) showing the occupancy of Kif7DM-GFP at microtubule-ends. The schematic above (t=0) and below (t=5 mins) the kymographs indicates the position of the seed (brown), growth (red) and the associated Kif7DM-GFP signal (green).
C. Line scan analysis of Kif7DM-GFP fluorescence intensity for the microtubule indicated in (A) and kymograph in (B).
D. Normalized GFP fluorescence intensity profile of Kif7DM-GFP along the growing microtubule-ends at 50 nM (black; N=25) and 300 nM (brown, N=25) concentrations.
E. Snapshots of the GFP fluorescence channel showing the spreading of EB1-GFP on growing microtubules in the absence and in the presence of 1 μM Kif7DM. The schematic above the kymographs indicates the position of the seed (brown), growth (red) and the associated EB1-GFP signal (green).
F. Kymographs generated from the microtubules shown in (E) for the microtubule (MT), GFP and merged channels.
G. Merged kymograph of X-rhodamine microtubule and GFP channels shown in (F).
H. Line scan analysis of EB1-GFP fluorescence intensity for the microtubules indicated in (E).
I. Normalized GFP fluorescence intensity profile of EB1-GFP along the growing microtubule-ends alone (100 nM, black; N=20) and in the presence of 1 μM Kif7DM (brown, N=20).
An increase in Kif7 occupancy at microtubule ends was also observed with higher concentrations of monomeric Kif7MM-GFP that lacks the neck linker and the coiled-coil dimerization domains, indicating that spreading is unlikely to result from cooperative intermolecular interactions between Kif7 molecules at the microtubule end (Figure S6B and S6C). The spreading of Kif7-GFP at microtubule tips increased with decreasing the ionic strength of the buffer, implying that this attribute of Kif7 is dependent on its microtubule binding affinity (Figure S6D and S6E). One possible explanation for Kif7 spreading could be due to low on-rate coupled with slow off-rate of Kif7. To examine this, we visualized single Kif7 molecules in dynamic microtubule assays in the presence of unlabeled Kif7DM (150 nM), spiked with Kif7DM-GFP (1 nM). Our experiments show Kif7 binding and unbinding events along the length of the growing microtubule that are typically just a few frames long (300 ms/frame), suggesting that low protein on-rate coupled with slow off-rate is also unlikely to be responsible for spreading (Figure S6F). In addition, the data does not show evidence for systematically decreasing Kif7 lifetimes away from the microtubule tip. Furthermore, no diffusion of Kif7 molecules were observed from the tip towards lattice (Figure S6F), confirming that biased-diffusion on dynamic microtubules is not responsible for Kif7 tip-tracking.
These results suggest that the spreading of Kif7 signal on the growing microtubule plus-end at increased kinesin concentrations arises from a change in the distribution of the GTP form of tubulin along the microtubule lattice. In other words, Kif7 stabilizes a GTP/GDP.Pi-like form of tubulin at microtubule ends. To test this hypothesis, we performed dynamic microtubule assays with EB1-GFP (100 nM) that tracks microtubule tips by binding the GTP/GDP.Pi-forms of tubulin (Zanic et al., 2009). Overlay images from the GFP and X-rhodamine channels at a single point in time in combination with a visual inspection of kymographs showed that EB1-GFP occupies a larger region of the microtubule tip in the presence of 1 μM Kif7DM (Figures 6E–H and Movie S3). EB1-GFP intensity profiles (>20) were aligned and normalized for comparison between experiments with and without Kif7. The analysis shows that at 1 μM Kif7DM, the average length of microtubule occupied by EB1-GFP increased from ~1 μm to 3 μm (Figure 6I).
In order to interpret this result, we needed to ensure that Kif7 does not directly bind EB1. SxIP sequence motif present in disordered domains of proteins are potential EB1 binding sites. Although, EB1 is not known to directly bind the motor domain of kinesins, bioinformatic analyses have predicted one such SxIP motif in loop L10 of the Kif7 motor domain (Jiang et al., 2012). Currently there is no report of an interaction between Kif7 and EB1 in vitro or in vivo. To rule out an interaction between Kif7 and EB1, we first performed BLI experiments which showed no dose-dependent binding between Kif7DM and EB1 in solution, even at a Kif7 concentration of 10 μM (Figure S6H). Next, we repeated dynamic microtubule assays with EB1-GFP and 1 μM Kif7MM-L10, a monomeric Kif7 construct with loop L10 mutated to that in Kinesin-1, thus eliminating the SxIP motif. Spreading was also observed with this monomeric mutant that lacks the L10 SxIP sequence (Figure S6G). Moreover, we reasoned that if the EB1-Kif7 interaction is important for in vivo tip localization of Kif7, then the SxIP motif in loop L10 must be conserved across organisms with cilia-dependent Hh signaling pathways. We performed sequence alignments of Kif7 homologues from various vertebrates and found that the sequence of loop L10 is highly variable and the SxIP motif is not conserved (Figure S6I). These analyses show that Kif7 is not a EB1 binding protein.
Together, these results indicate that Kif7 not only recognizes the GTP-tubulin lattice at the growing microtubule tip but also further stabilizes the GTP-like state of tubulin to increase the Kif7 occupancy at microtubule ends.
Microtubule-binding activity of Kif7 is required for cilium-tip localization during Hh signaling
In vertebrates, activation of the Hh pathway results in the enrichment of Kif7 at the distal cilium tip (Liem et al., 2009, He et al., 2014, Liu et al., 2014); however, whether cilia tip localization of Kif7 occurs via direct microtubule binding or indirect interaction with other microtubule tip-binding proteins in the cilium is unknown. We transiently transfected C3H10T1/2 cells with full-length GFP-tagged wild-type Kif7 (Kif7FL-GFP) and a construct in which three charged residues (Glu-Arg-Arg) in loop L12 of Kif7 were mutated to alanine (Kif7FL3A -GFP). As shown in Figure 3J, this mutation exhibits an 8-fold lower microtubule-binding affinity compared to wild-type Kif7 but does not impact protein folding. Furthermore, this mutation is not close to the NBP.
We observed that Kif7FL-GFP is enriched at the tip in ~82% of primary cilia (N=97) in the presence of the Hh pathway activator Smoothened Agonist (SAG). In contrast, only ~30% of cells expressing Kif7FL3A -GFP show cilium-tip localization (N=113; Figure 7A and B). In these cells, Kif7 was observed at the base of the cilium, indicating that localization of Kif7 to the cilium base is not affected by reducing its microtubule-binding affinity. To ensure that the observed phenotype with Kif7FL3A-GFP was due to its reduced microtubule-binding affinity, control experiments were performed with Kif7FL4K-GFP, which has an increased positive charge in loop L12 relative to wild-type Kif7 (Figure 3J). We found that this construct has cilium-tip localization similar to wild-type Kif7 (~90%, N=116, Figure 7A and B).
Figure 7. Model of Kif7 interactions at the growing microtubule end.
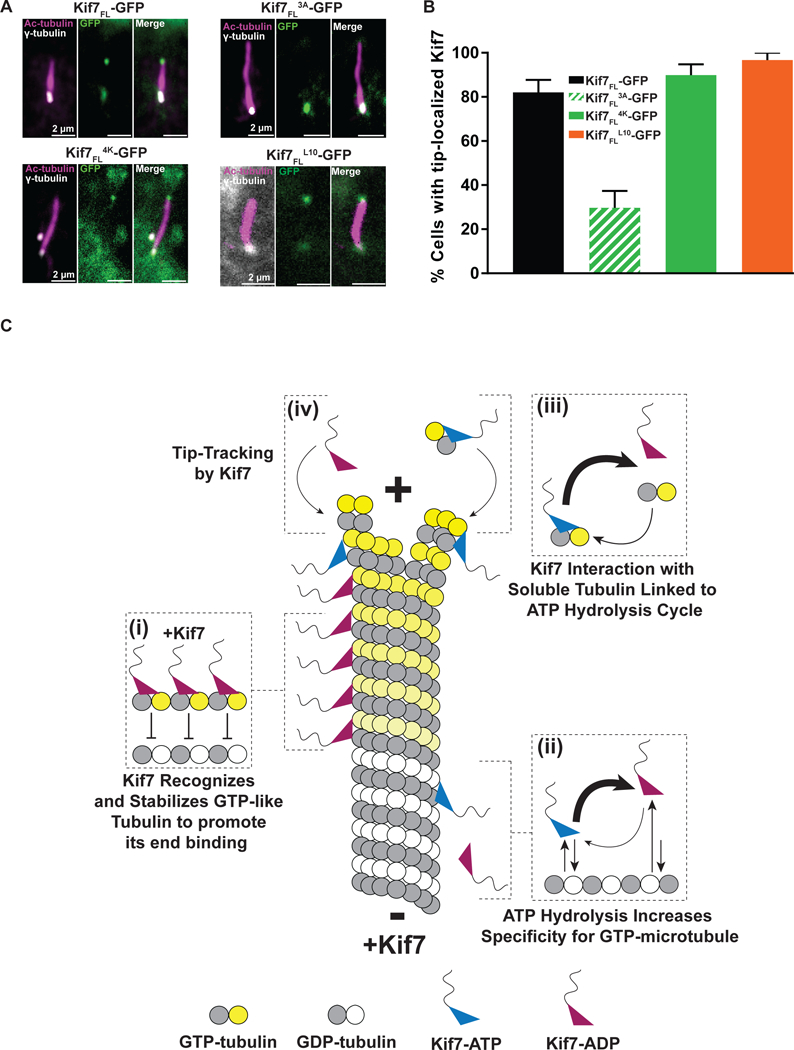
A. Cilia localization of wild-type and mutant Kif7. Immunofluorescence analysis of transiently transfected Kif7-GFP protein in C3H10T1/2 cell upon activation of Hh pathway with SAG. Magnified views of the cilia are shown: acetylated α-tubulin (ciliary axoneme label; pink), γ-tubulin (basal body label; gray), and transfected Kif7-GFP proteins (green).
B. Histogram showing the percentage of cells with the indicated Kif7-GFP construct at the tip of the primary cilium [Kif7FL-GFP: 82% (black; N = 97); Kif7FL3A-GFP: 29.7% (green striped; N = 113); Kif7FL4K-GFP: 89.9% (green solid; N = 116) and Kif7FLL10-GFP: 96.7% (orange; N = 78)].
C. Schematic of Kif7 interactions on a growing microtubule and soluble tubulin. (i) Kif7 recognizes and stabilizes GTP-forms of tubulin in the microtubule lattice. (ii) The ATP hydrolysis cycle of Kif7 increases its selectivity for GTP- over GDP-tubulin. Kif7 in the ATP state dissociates slower from the GDP-tubulin microtubule lattice in comparison to Kif7 in the ADP state. (iii) Kif7 interaction with soluble tubulin is linked to its ATP hydrolysis cycle. Kif7-ATP forms a stable complex with soluble tubulin, while Kif7-ADP does not bind soluble tubulin. However, due to the slow ADP-release rate of Kif7, the major species found in solution is Kif7-ADP and the minor species is Kif7-ATP complexed to soluble tubulin. (iv) Microtubule tip-tracking by Kif7 in the ATP and ADP states. Kif7 is predominantly in the ADP-bound state and recognizes the GTP-like form of tubulin at the microtubule tip. A smaller fraction of Kif7 in the ATP state can potentially form a complex with soluble tubulin and can localize to the microtubule tip via a “ride along” mechanism during microtubule polymerization.
To probe the potential role of EB1 in specifying cilium tip localization of Kif7, we mutated the SxIP motif containing loop L10 of Kif7 to that of kinesin-1 (GRAPSRLPRP to NTQTE, construct Kif7FL-L10-GFP) and examined the cellular localization of this mutant protein. We found that upon Hh activation, this mutant localizes to the cilium tip similar to wild type Kif7 (~97%, N=78; Figures 7A and 7B). The lack of conservation of the EB1-recognition motif in vertebrates, the absence of interaction between Kif7 and EB1 in vitro, the localization of Kif7 to the cilia tip in the absence of an EB1 interaction motif, together with the earlier observation that EB1 is predominantly sequestered at the base and not observed at the tip of the cilium in cells (Schroder et al., 2007), suggests that an EB1-interaction based mechanism is not necessary for the cilium-tip localization of Kif7.
Together, these data show that reduction of the microtubule-binding affinity of Kif7 significantly affects the localization of Kif7 to the primary cilia tip upon activation of Hh signaling.
DISCUSSION
The localization of proteins to the ends of microtubules is required for a diverse range of cellular processes, such as cell division, cell polarization, and cell signaling. Our study uncovered a mechanism by which the ciliary kinesin Kif7, which establishes a precise platform for anchoring Hh pathway proteins at the distal end of the primary cilium, preferentially binds to the plus-ends of microtubules.
How does Kif7 recognize microtubule plus-ends? Our in vitro reconstitution studies suggest that Kif7 localizes to the plus-ends of microtubules by preferentially binding to GTP and GDP-Pi forms of tubulin found at the growing end of the polymer (Figures 1B–J and S1E–F). We found that with increasing Kif7 concentrations, the binding region of Kif7 spreads to longer lengths at microtubule ends (Figure 6A–D). In addition, the EB1 protein, which recognizes GTP and GDP-Pi forms of tubulin, has longer comet length in the presence of Kif7, suggesting that Kif7 stabilizes one or more of the tubulin nucleotide/conformational states recognized by EB1 (Figure 6E–I). Together, these observations indicate that Kif7 both recognizes and stabilizes the GTP-like conformations of tubulin in microtubules (Figure 7C [i]). Our results suggest that a feedback loop between Kif7 and microtubule enables plus-end recognition. GTP/GDP-Pi forms of tubulin at the plus-end of microtubules provides a binding site for Kif7, which in turn regulates the microtubule polymer either through inhibition of the tubulin GTPase activity or suppression of the conformational changes in tubulin that accompany GTP hydrolysis (Geyer et al., 2015). The stabilization of the GTP-form of tubulin at microtubule ends provides a positive feedback to further promote Kif7 binding and enable robust tip-tracking by this non-motile kinesin. These findings suggest that unlike most kinesins, the Kif7-microtubule interaction is mediated by the recognition and regulation of the tubulin GTPase cycle.
The mechanism by which Kif7 preferentially binds the GTP/GDP-Pi-like forms of tubulin is distinct from mechanisms by which some kinesin-1 proteins, such as Kif5C, show some preference (1.5–3.7-fold) for microtubules complexed with certain GTP analogs over GDP (Nakata et al., 2011, Bechstedt et al., 2014, Morikawa et al., 2015). Kif5C is proposed to bind GDP-microtubule and elongate the axial lattice spacing to mimic the GTP-microtubule conformation (Peet et al., 2018, Shima et al., 2018). Such a mechanism would be advantageous for Kif5C’s function in polarized microtubule-based transport over long distances. In contrast, recognition and stabilization of the GTP-like states of tubulin, found largely at growing microtubule ends, is a more suitable mechanism for tip-tracking by Kif7.
Clues to the structural mechanism underlying the selective recognition of GTP- over GDP-tubulin come from our cryo-EM studies, which revealed distinct features of the microtubule-interacting interface of Kif7 in comparison with other kinesins. First, the microtubule-binding surface of Kif7 is more acidic than that of other kinesins, which contain an overall basic surface that has been proposed to promote interaction with the negatively charged surface-exposed residues of tubulin (Figure 3H and 3I; (Woehlke et al., 1997, Atherton et al., 2017)). Second, the motor domain of Kif7 is rotated on the microtubule surface compared to other kinesins (Figure 3C), such as Kif1A, Kif5A, Kif5B, Kif18A, and Eg5 (Kikkawa and Hirokawa, 2006, Goulet et al., 2012, Atherton et al., 2014, Shang et al., 2014, Morikawa et al., 2015, Locke et al., 2017). These properties change the microtubule footprint of Kif7, bringing it in close proximity to helix H12 of β-tubulin (Figure 3D–I). Helix H12 of β-tubulin undergoes a small outward shift when GTP is hydrolyzed to GDP (Alushin et al., 2014). We speculate that this outward shift may sterically hinder Kif7 binding to the GDP form of tubulin. Together, our findings suggest that even though Kif7 shares a protein fold and high sequence similarity with other kinesins, global conformational changes at the Kif7-microtubule interface allow it to specifically bind and stabilize the GTP form of tubulin.
Kif7 has atypical mechanochemical properties for a kinesin. Two features common to many kinesins are: (1) the stimulation of ATP hydrolysis by microtubules (Cross, 2004). In tip-tracking kinesins, such as MCAK and Kip3p, the microtubule ends further regulate ATP hydrolysis rates (Hunter et al., 2003, Hertzer et al., 2006, Friel and Howard, 2011, Arellano-Santoyo et al., 2017). In contrast neither microtubules nor soluble tubulin accelerate the Kif7 ATP hydrolysis cycle (Figures S2G–H). (2) The microtubule-binding affinity of kinesins is typically coupled to their ATP hydrolysis cycle. In contrast, the difference in the dissociation constant of Kif7-microtubule interaction between its AMPPNP and ADP states is only two-fold (Figure 2A and S2B), which is much lower than the usual 20- to 40-fold reported for kinesin-1 (Crevel et al., 1996, Rosenfeld et al., 1996, Gigant et al., 2013). Therefore, Kif7 activity is not coupled to ATP hydrolysis in a manner described for other kinesins, raising the question: what is the purpose of ATP hydrolysis in Kif7? We find that (1) ADP-Kif7 has a greater selectivity for GTP over GDP-microtubule compared to ATP-like states of Kif7 (Figure 2D, 7C(ii)) and (2) ATP hydrolysis cycle is linked to the interaction of Kif7 with dimeric tubulin in solution such that Kif7 binds soluble tubulin tightly with AMPPNP and ATPγS and not in ADP (Figures 2E, S2F and 7C [iii]). During tubulin polymerization in presence of Kif7 and ATP, there are two possibilities. First, ADP-Kif7 in solution selectively binds to GTP-tubulin at microtubule end (Figure 7C [iv]). Second, ATP-Kif7-tubulin in solution co-polymerizes with soluble tubulin on the growing polymer (Figure 7C [iv]). Upon ATP hydrolysis, ADP-Kif7 will dissociate from the GDP-lattice but remain bound to the plus-ends (Figure 7C [iv]). Considering that microtubule-bound Kif7 is predominantly in the ADP-state in experiments with ATP (Figure S2A), we expect that the major fraction of tip-tracking species is post-hydrolysis Kif7-ADP state. Thus, the atypical properties of Kif7, such as microtubule-binding in the ADP state (Figures 2A–C and S2C) and the lack of microtubule-stimulated ADP release (Figure S2G), increase the fraction of Kif7-ADP on microtubules, thereby increasing the selectivity of Kif7 for the microtubule plus-end.
Our cryo-EM reconstructions revealed distinct features of the nucleotide binding region of Kif7 in comparison with other kinesins and provide insight into the structural basis of its atypical chemo-mechanical properties. The observation of a physical linkage between helix α3 and loop L5, which has three additional residues in Kif7 compared to conventional kinesin, hints at the mechanism underlying the slow microtubule-stimulated ADP release. The relative movement of these secondary structural elements, which is likely to be restricted in Kif7, has been proposed to be important for the conformational changes that couple microtubule binding to ADP release in kinesins (Yun et al., 2001, Cochran et al., 2005, Sindelar and Downing, 2007). Interestingly, a similar linkage has been observed in NOD, which also displays slow microtubule-stimulated ADP release (Cochran et al., 2009). The small molecule monastrol has been proposed to restrict the movement between L5 and α3 of the kinesin Eg5 and inhibit ADP release (Cochran et al., 2005). While this property locks NOD and Eg5-monastrol in a weak microtubule-binding state, Kif7 retains microtubule binding when bound to ADP, which in turn provides greater selectivity for GTP over GDP tubulin.
Why is the Kif7-microtubule interaction largely insensitive to the nucleotide state? We gained insight into this question by comparing the AMPPNP- and ADP-bound Kif7 structures. First, in contrast to kinesin-1 and kinesin-3, which have available structures of ADP-bound motors (Atherton et al., 2014), we found that the central helix α4 in Kif7 adopts an extended conformation that contacts microtubules with both AMPPNP and ADP (Figures S4I–J). The extent of the overall microtubule interaction surface area of Kif7 remains high in both the AMPPNP and ADP states, enabling it to retain tight binding to microtubule ends. Second, we found that Kif7 does not undergo conformational changes in loops L9 and L11 in response to nucleotide hydrolysis (Figure 5B–C). Therefore, the mechanism that relays information from the NBP to the microtubule-binding surface of Kif7 is defective. Finally, while the microtubule interaction of Kif7 is insensitive to nucleotide state, its binding to soluble tubulin only occurs in the presence of AMPPNP (Figure 2E). Our results suggest that this preference may arise from the different rotation of helix α6 in the different nucleotide states. The ~8º tilt of helix α6 in AMPPNP-bound Kif7 relative to kinesin-1 is ideal for accommodating the small curvature in soluble tubulin compared to polymerized tubulin (Figure 4E). The binding interactions are lost when helix α6 in Kif7 further tilts in the ADP-bound state of the motor (Figure 4F). Together, these findings reveal the structural design principles that confer Kif7 with unique chemo-mechanical properties.
GTP-tubulin recognition coupled with the lack of motility may reflect an adaptation in Kif7 for increased specificity rather than speed of localizing to microtubule plus-ends. Members of certain tip-tracking kinesins, including kinesin-8 (Kip3p and Kif19a) and kinesin-4 (Kif4A), use directed motility to efficiently arrive at microtubule ends (Gupta et al., 2006, Varga et al., 2006, Bieling et al., 2010, Subramanian et al., 2013, Wang et al., 2016, Arellano-Santoyo et al., 2017). The non-motile kinesin MCAK uses a “diffusion-and-capture” strategy, in which 1-D diffusion of MCAK on the mature GDP-tubulin lattice increases the rate of arrival at microtubule ends (Helenius et al., 2006). These motile and diffusive kinesins interact with both the mature GDP-microtubules and the newly polymerized GTP-microtubules to enable their cellular functions, and therefore, the preferential recognition of GTP- over GDP-tubulin is not a viable strategy. Instead, these tip-tracking kinesins typically recognize other microtubule end structural features, such as tubulin curvature. In contrast, Kif7 neither walks nor diffuses on microtubules (Figure S6F, (He et al., 2014, Yue et al., 2018)). Within the small volume of the cilium, it is expected that the protein-microtubule association rates will be high due to the increased apparent protein concentrations (Nachury, 2014). Therefore, the mechanism of microtubule end-binding by Kif7 may reflect an adaptation for high specificity that is required for its function as a recruitment platform that precisely localizes Hh components to the cilium tip.
Kif7 must enter the primary cilium and accumulate at the distal cilium tip to enable proper localization of Hh components. Therefore, it is functionally distinct from the kinesin-2 family proteins, homodimeric Kif17 and the heterotrimeric KifAB-KAP complex, which play a role in transport within the cilium (Williams et al., 2014, Jiang et al., 2015, Funabashi et al., 2017, Schwarz et al., 2017). Unlike Kif7, Kinesin-2 proteins have hallmarks of transport kinesins with high processivity, fast velocity and microtubule-stimulated ATP hydrolysis (Prevo et al., 2017, Gilbert et al., 2018). Kif17 also accumulates at the cilium tip in cells (Dishinger et al., 2010, Jiang et al., 2015). However, cilium tip localization does not require the motor domain of Kif17. In contrast, our cell-based assays show that motor domain mutations that decrease microtubule-binding affinity disrupt Kif7 localization at the cilium tip (Figure 7A–B). These finding suggests that cilia entry or tip localization of Kif7 is dependent on its own microtubule binding. These distinct properties of Kinesin-2 proteins and Kif7 may reveal functional specialization as transport motors and a cilium-tip scaffold respectively.
Axonemal microtubules in the primary cilium are organized into a specialized 9+0 arrangement of doublet microtubules and heavily post-translationally modified (PTM) (Satir and Christensen, 2007). Currently, it is not experimentally feasible to reconstitute dynamic doublets and in vitro work on ciliary microtubule associated proteins is largely limited to singlets. However, at the tapered tip of the primary cilium, the 9+0 arrangement of axonemal microtubules and their doublet structure is disrupted. Instead a “singlet zone” exists at the distal tip of cilia (Gluenz et al., 2010). Together with the observation that Kif7 binds along a protofilament and microtubule dynamics at axoneme ends is thought to be regulated similar to cytoplasmic singlet microtubules, we expect our findings to be relevant to the physiological mechanisms of microtubule end-binding in the cilium (Avasthi and Marshall, 2012). Similar to the axonal bovine brain tubulin used in our experiments, PTMs including detyrosination, polyglutamylation, acetylation and the Δ2 modification are abundant in ciliary microtubules (Janke and Bulinski, 2011, Song and Brady, 2015, Wloga et al., 2017). However, PTMs that have been characterized for the mature axoneme in the cilia may not be indicative of the PTM at the distal cilia tip, and the in vitro and cell-biological studies on tubulin PTMs and its regulation of the Hh pathway presents an exciting open question for future studies.
On a broader note, the adaptations in the motor domain of Kif7 highlight the generation of a new mechanism of function via repurposing of the ubiquitous kinesin fold. Small changes in microtubule-bound conformations have been reported for two other atypical kinesins, Mklp2 and NOD (Cochran et al., 2009, Atherton et al., 2017), raising a much larger question about the role of small changes in microtubule-binding orientations and their influence on functions of motor and non-motor MAPs.
STAR Methods
Key Resource Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| α Tubulin Antibody (B-7) Alexa Fluor®647 | Santa Cruz Biotechnology | sc-5286 AF647 |
| gamma Tubulin Polyclonal Antibody, Cyanine3 | Thermo Fisher | PA1–28043 |
| Bacterial and Virus Strains | ||
| E. coli Rosetta (DE3) | Millipore | 70954 |
| Bac-to-Bac Baculovirus Expression System | Thermo Fischer Scientific | 10359016 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| TEV Protease | This Paper | N/A |
| HyCloneTM CCM3 SFM | GE Life Sciences | SH30065.02 |
| Thrombin Protease | GE Life Sciences | 27084601 |
| PreScission Protease | This Paper | N/A |
| isopropylthiogalactoside | Sigma Aldrich | I675 |
| TWEEN® 20 | Sigma Aldrich | P1379 |
| IGEPAL® CA-630 | Sigma Aldrich | I8896 |
| ATP | Jena Bioscience | NU-1010 |
| ADP | Sigma Aldrich | A2754 |
| ATPyS | Jena Bioscience | NU-406 |
| AMPPNP | Jena Bioscience | NU-407 |
| GMPCPP | Jena Bioscience | NU-405 |
| GTP | Sigma Aldrich | G8877 |
| Beryllium Sulfate | Sigma Aldrich | 202789 |
| Sodium Fluoride | Sigma Aldrich | 201154 |
| Pierce™ Bond-Breaker™ TCEP Solution | Thermo Fischer Scientific | PI-77720 |
| Tubulin | PurSolutions | 32005 |
| Tubulin | Cytoskeleton | T240 |
| Tubulin X-Rhodamine Labeled | Cytoskeleton | TL620M |
| Tubulin HiLyte647 Labeled | Cytoskeleton | TL670M |
| Biotin-PEG-SVA | Laysan Bio | #Biotin-PEG-SVA-5000 |
| mPEG-Succinimidyl Valerate | Laysan Bio | #NH2-PEG-VA-5K |
| 1,4-piperazinediethanesulfonic acid | Sigma Aldrich | P6757 |
| Ethylene glycol tetraacetic acid | Sigma Aldrich | 3777 |
| Methyl cellulose | Sigma Aldrich | M0512 |
| Paclitaxel | Sigma Aldrich | T7402 |
| 2-Mercaptoethanol | Sigma Aldrich | M-6250 |
| Neutravidin | Thermo Fischer Scientific | A2666 |
| GelCodeTM Blue Stain | Thermo Fischer Scientific | PI24592 |
| SAG | Fischer Scientific | 56–666 |
| ProLong™ Diamond Antifade Mountant | Thermo Fischer Scientific | P36961 |
| [a-32P] ATP | PerkinElmer | BLU003H250UC |
| Cellulose PEI TLC | Millipore | 105579 |
| 1.2/1.3 400-mesh grids | Electron Microscopy Services | CF413 |
| Catalase | Sigma Aldrich | C40 |
| Glucose Oxidase | Sigma Aldrich | G2133 |
| 6His-TEV-Kif7DM1–543-GFP | This paper | N/A |
| 6His-TEV-Kif7DM1–543 | This paper | N/A |
| 6His-TEV-Kif7DM-L124K-GFP | This paper | N/A |
| 6His-TEV-Kif7DM-L123A-GFP | This paper | N/A |
| 6His-TEV-Kif7DM-L8KHC-GFP | This paper | N/A |
| 6His-TEV-Kif7DM-L10KHC-GFP | This paper | N/A |
| 6His-TEV-Kif7MM1–386 | This paper | N/A |
| 6His-TEV-Kif7MM1–386-GFP | This paper | N/A |
| 6His-TEV-Kif7MM-L10 | This paper | N/A |
| Strep-PreScission-Clasp1(654–1455)-GFP-TEV-6His | This paper | N/A |
| 6His-Thrombin-EB1-GFP | Laboratory of Mike Blower | N/A |
| Critical Commercial Assays | ||
| Q5® Site-Directed Mutagenesis Kit | NEB | E0554S |
| EnzChek™ Phosphate Assay Kit | Invitrogen | E6646 |
| Anti-Penta-HIS biosensor Chip | ForteBio | His1K |
| Amine Reactive Second-Generation Biosensors | ForteBio | AR2G |
| Deposited Data | ||
| ADP-bound Kif7 atomic model | Protein Data Bank Archive | 4A14 |
| Taxol stabilized Sus scrofa tubulin atomic model | Protein Data Bank Archive | 3J6G |
| Taxol stabilized Sus scrofa tubulin electron density | Electron Microscopy Data Bank | EMD-5897 |
| GmpCpp stabilized Sus scrofa tubulin electron density | Electron Microscopy Data Bank | EMD-5895 |
| ATP analog-bound kinesin-1 H. Sapiens on tubulin atomic model | Protein Data Bank Archive | 3J8Y |
| ATP analog-bound kinesin-1 H. Sapiens on tubulin electron density | Electron Microscopy Data Bank | EMD-6188 |
| ADP-Mg-AlFx-bound kinesin motor domain in complex with tubulin atomic model | Protein Data Bank Archive | 4HNA |
| HsMCAK motor domain complex atomic model | Protein Data Bank Archive | 4UBF |
| Hs Kinesin-1 motor domain atomic model | Protein Data Bank Archive | 1BG2 |
| ADP-bound kinesin-3 on microtubules atomic model | Protein Data Bank Archive | 4UXS |
| ADP-bound kinesin-3 on microtubules electron density | Electron Microscopy Data Bank | EMD-2768 |
| AMPPNP-bound kinesin-3 on microtubules atomic model | Protein Data Bank Archive | 4UXP |
| AMPPNP-bound kinesin-3 on microtubules electron density | Electron Microscopy Data Bank | EMD-2766 |
| AMPPNP-bound Hs Kif7 motor domain on GDP-taxol microtubules atomic model | This work | 6MLR |
| AMPPNP-bound Hs Kif7 motor domain on GDP-taxol microtubules electron density | This work | EMD-9141 |
| ADP-bound Hs Kif7 motor domain on GDP-taxol microtubules atomic model | This work | 6MLQ |
| ADP-bound Hs Kif7 motor domain on GDP-taxol microtubules electron density | This work | EMD-9140 |
| Experimental Models: Cell Lines | ||
| C3H10T1/2 | Lab of Stephane Angers | |
| Recombinant DNA | ||
| pI-SUMOstar-6His-TEV-Kif7DM1–543-GFP | This paper | RS177 (Subramanian Lab Collection) |
| pI-SUMOstar-6His-TEV-Kif7DM1–543 | This paper | RS168 (Subramanian Lab Collection) |
| pI-SUMOstar-6His-TEV-Kif7DM-L124K-GFP | This paper | SJ034 (Subramanian Lab Collection) |
| pI-SUMOstar-6His-TEV-Kif7DM-L123A-GFP | This paper | SJ035 (Subramanian Lab Collection) |
| pI-SUMOstar-6His-TEV-Kif7DM-L8KHC-GFP | This paper | SJ045 (Subramanian Lab Collection) |
| pI-SUMOstar-6His-TEV-Kif7DM-L10KHC-GFP | This paper | SJ042 (Subramanian Lab Collection) |
| pET28a-Kif7MM1–386 | This paper | SJ029 (Subramanian Lab Collection) |
| pET30a-Kif7MM1–386-GFP | This paper | SJ051 (Subramanian Lab Collection) |
| pET28a-Kif7MM-L10 | This paper | SJ055 (Subramanian Lab Collection) |
| pFastBac-Strep-PreScission-Clasp1(654–1455)-GFP-TEV-6His | This paper | NM503 (Subramanian Lab Collection) |
| pET30a-EB1-GFP | Laboratory of Mike Blower | pMB283 (Blower Lab Collection) |
| CMV-Kif7-GFP | This paper | pk13 (Subramanian Lab Collection) |
| CMV-Kif7–3A-GFP | This paper | pk28 (Subramanian Lab Collection) |
| CMV-Kif7–4K-GFP | This paper | pk13 (Subramanian Lab Collection) |
| CMV-Kif7-L10-GFP | This paper | pk13 (Subramanian Lab Collection) |
| Software and Algorithms | ||
| Matlab | Mathworks | https://www.mathworks.com |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| NIS Elements | Nikon | https://www.nikoninstruments.com/Products/Software |
| Octet Analysis | ForteBio | http://www.fortebio.com/octet-platform.html |
| UCSF Chimera | RBVI | https://www.cgl.ucsf.edu/chimera/ |
| Appion | Lander et al., 2009 |
http://emg.nysbc.org/redmine/ projects/appion/wiki/Appion_Home |
| Leginon | Suloway et al., 2005 |
http://emg.nysbc.org/redmine/ projects/leginon/wiki/Leginon_Homepage |
| MotionCor | Li et al., 2013, Hirschi et al., 2017 | http://msg.ucsf.edu/em/software/motioncor2.html |
| CTFFIND4 | Rohou and Grigorieff, 2015 | http://grigoriefflab.janelia.org/ctffind4 |
| EMAN2 | Tang et al., 2007 | https://blake.bcm.edu/emanwiki/EMAN2 |
| FREALIGN | Grigorieff, 2007 | |
| Image Studio Lite | LI-COR |
https://www.licor.com/bio/products/ software/image_studio_lite/ |
| Octet Data Analysis | ForteBio | https://www.fortebio.com/octet-software.html |
| phenix.real_space_refine | Afonine et al., 2013 |
https://www.phenix-online.org/documentation/reference/ real_space_refine.html |
| Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Coot | Emsley and Cowtan, 2004 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Contact | Winn et al., 2011 | http://www.ccp4.ac.uk/html/contact.html |
| PDBSUM | de Beer et al., 2014 | www.ebi.ac.uk/pdbsum |
| APBS | Jurrus et al., 2018 | http://nbcr-222.ucsd.edu/pdb2pqr_2.1.1/ |
| Clustal Omega | Sievers et al., 2011 | https://www.ebi.ac.uk/Tools/msa/clustalo/ |
| Curvature Analysis Algorithm | Bechstedt et al., 2014 | Lab of Gary Brouhard |
Contact for Reagent and Resource Sharing
Further information and reagent requests may be made to, and will be fulfilled by the lead contact, Radhika Subramanian (radhika@molbio.mgh.harvard.edu).
Experimental Model and Subject Details
Recombinant proteins were overexpressed in SF9 cells in accordance with the Bac-to-Bac Baculovirus virus expression system as provided in the key resource table. Recombinant proteins expressed in E. coli Rosetta (DE3) cells were induced with the addition of 1 mM IPTG after an OD of 0.6. Cells were grown for 18 hours at 4 °C. Expression of recombinant proteins in C3H10T1/2 cells have been detailed in the section headed “Cell Culture and Immunofluorescence”.
Method Details
Protein Expression and purification
The N-terminal fragment of all dimeric forms of Kif7 (Uniprot ID: Q2M1P5) (Kif7DM-GFP, Kif7DM, Kif7DM-L124K-GFP, Kif7DM-L123A-GFP, Kif7DM-L8KHC-GFP and Kif7DM-L10KHC-GFP; amino acids 1–543) containing the N-terminal motor and the first coiled-coil domains were cloned into a modified pFastBac expression vector (Thermo Fischer Scientific) that included a tobacco etch virus (TEV) protease cleavable N-terminal 6x His-tag and SUMOstar solubilization tag and C-terminal GFP-tag. Mutations were made using the Q5® Site-Directed Mutagenesis Kit (NEB). Kif7 dimers were expressed in the Sf9 insect cell line using the Bac-to-Bac® Baculovirus Expression System (Thermo Fischer Scientific) with cells grow in HyClone™ CCM3 SFM (GE Life Sciences). The N-terminal fragment of the monomeric Kif7 containing only the N-terminal motor domain (Kif7MM, amino acids 1–386) was cloned into a modified pET28a expression vector (Novagen) that contained a TEV protease cleavable N-terminal 6x His-tag. The N-terminal fragment of the monomeric Kif7 containing the N-terminal motor domain and C-terminal GFP-tag (Kif7MM-GFP, amino acids 1–386), and EB1-GFP were each cloned into a modified pET30a expression vector (Novagen) that contained a thrombin protease cleavable N-terminal 6x His-tag and C-terminal GFP-tag. Kif7 monomers and EB1-GFP were expressed in BL21(DE3) Rosetta (Millipore) E. coli at 18 °C with 0.25 mM isopropylthiogalactoside for 18 – 20 h. All pellets were lysed by a short sonication in buffer A (50 mM Phosphate buffer pH 8.0, 300 mM NaCl, 5 % glycerol, 1 mM MgCl2 and 30 mM imidazole) supplemented with 0.15 % tween, 0.5 % Igepal 630, 100 μM ATP, 2 mM TCEP, 1 mM PMSF, 75 U Benzonase and protease inhibitor cocktail (Roche)). The lysate was clarified by ultracentrifugation and supernatant was incubated with Ni-NTA for 1 h. The resin was washed with buffer A supplemented with 20 μM ATP and 0.5 mM TCEP and eluted with 50 mM Phosphate buffer pH 8.0, 300 mM NaCl, 5 % glycerol, 400 mM imidazole and 100 μM ATP. Peak protein fractions were pooled and incubated with protease (1:30 w/w) overnight at 4 °C. The proteins were further purified by size exclusion chromatography (Kif7 dimer constructs: Superose 6 increase 10/300 GL; Kif7 monomeric constructs: Superdex 75 10/300 GL, EB1-GFP: Superdex 200 16/600, Amersham Pharmacia Biotech) in 50 mM HEPES pH 7.4, 300 nM NaCl, 5 % glycerol, 5 mM β-mercaptoethanol, 1 mM MgCl2 and 100 μM ATP and frozen in liquid nitrogen. EB1-GFP was purified in buffer without ATP. Elution volumes for all Kif7 constructs, which are likely to be elongated in shape, are greater than expected in comparison with globular molecular weight markers. Tubulin was purified from bovine brain tissue (Hyman et al., 1991) or purchased from Cytoskeleton, Inc and PurSolutions, LLC. Microscopy assays were performed interchangeably with lab-purified and cytoskeleton tubulin. Tubulin was labeled in 1:10 proportions with biotin or X-rhodamine according to published protocols or mixed with prelabeled tubulin purchased from Cytoskeleton, Inc. Cosedimentation experiments were performed with PurSolutions tubulin.
Total Internal Reflection Fluorescence Microscopy Assays
In vitro TIRF-based microscopy experiments were carried out as described in Subramanian, et al., 2013 (Subramanian et al., 2013). Microscope chambers were constructed using a 24 × 60 mm PEG-Biotin coated glass slide and 18 × 18 mm PEG coated glass slide separated by double-sided tape to create two channels for exchange of solutions. Standard assay buffer was 1 x BRB80 (80 mM K-PIPES at pH 6.8, 2 mM MgCl2 and 1 mM EGTA at pH 6.8), 1 mM ATP, 1 mM GTP, 0.1 % methylcellulose and 3 % sucrose. Images were acquired using NIS-Elements (Nikon) and analyzed using ImageJ.
Dynamic Microtubule Assay:
Experiments with dynamic microtubules were carried out as described in He et al, 2014 (He et al., 2014). X-rhodamine (1:10 labeled to unlabeled) and biotin (1:10 labeled to unlabeled) labeled microtubules were polymerized in the presence of GMPCPP, a non-hydrolysable GTP-analogue, and immobilized on a neutravidin coated glass coverslip. Coverslips were briefly incubated with casein to block non-specific surface binding before addition of 16 μM tubulin (1 X-rhodamine-labelled tubulin:10 unlabeled tubulin) in assay buffer and antifade reagent along with Kif7 protein and nucleotide. Imaging of EB1 localization on growing microtubules was carried out by addition of 100 nM EB1-GFP with and without non-fluorescent Kif7. Images were recorded every 10 seconds for 20 minutes.
Single molecule imaging:
For single molecule imaging, dynamic microtubule assays were performed with altered conditions: Kif7DM (150 nM) was spiked with Kif7DM-GFP (1 nM). Images were collected at 300 ms intervals on an ANDOR iXon Ultra EMCCD camera.
Segmented Microtubule Assay:
To assemble segmented microtubules and test the localization of Kif7, X-rhodamine (1:10 labeled to unlabeled) and biotin (1:10 labeled to unlabeled) labeled microtubules were polymerized in the presence of GMPCPP, a non-hydrolysable GTP-analogue, and immobilized on a neutravidin coated glass coverslip. To grow out the microtubule, coverslips were briefly incubated with casein to block non-specific surface binding before addition of assay buffer containing 16 μM tubulin (1 HiLyte647-labelled tubulin:10 unlabeled tubulin to initiate microtubule growth for 5 minutes. Growth was capped by the addition of assay buffer containing 16 μM tubulin (1 X-rhodamine labelled tubulin:10 unlabeled tubulin) and 1 mM GMPCPP for 2 minutes. Kif7 was subsequently added to the chamber along with an antifade reagent. For microtubules containing GTPγS segments, 1 mM GTPγS was used as a substitute for GTP during the growth step. For microtubules containing GDP-BeF3 segments, 2 mM BeS04 and 10 mM NaF were added along with Kif7 in the final assay mix. For microtubule containing GDP-taxol microtubule segments, 20 μM taxol was added along with Kif7 in the final assay mix. A segmented microtubule containing GMPCPP and GTPγS segments constructed from only X-Rhodamine-labeled microtubules was used as a fluorophore control. Samples were allowed to equilibrate for 1 minute before image acquisition. ATP was substituted with 1 mM ADP, ATPγS and AMPPNP in experiments comparing different adenosine nucleotide analogs. For analysis of ratios between intensities on GMPCPP/GDP microtubules, GFP-fluorescence intensity on the GMPCPP-seed and GDP-regions that were <20 A.U. from background were eliminated as analysis showed these intensities could not be accurately measured.
Analysis of Curved Microtubules:
X-rhodamine (1:10 labeled to unlabeled) and biotin (1:10 labeled to unlabeled) labeled microtubules were polymerized in the presence of GMPCPP, left overnight, and immobilized on a neutravidin coated glass coverslip. Kif7DM-GFP was subsequently added to the chamber at 10 nM and 50 nM in the presence of ATP and antifade reagent. Images of Kif7DM-GFP on curved microtubules were collected and analysis of fluorescence intensity as a function of microtubule curvature was carried out as described in Bechstedt et al, 2014.
Cosedimentation of Kif7 on GDP-taxol stabilized microtubules
GDP-taxol stabilized microtubules were prepared from purified pre-cleared bovine tubulin as described in (Subramanian et al., 2010). Kif7 and Kif7 mutants at 1 μM concentrations were incubated with microtubules (0 – 10 μM) for 15 minutes at room temperature in 1 × BRB80 supplemented with 0.1 μg/μL BSA and then subjected to sedimentation by ultracentrifugation at 90,000 rpm for 15 minutes at 27 °C for 15 minutes in a TLA 120.1 rotor (Beckman Coulter). Pellets were subsequently resuspended and the amount of protein in each pellet and supernatant was analyzed by SDS-PAGE. Proteins were stained using GelCode™ Blue Stain (Thermo Fischer Scientific). For cosedimentation of Kif7 with ATP, ADP and AMPPNP, 1 mM was used for each. Dissociation constants were calculated by fitting of experimental data to the Hill equation using GraphPad Prism. Cooperative binding between Kif7 and microtubules was observed in the presence of AMPPNP and best fit with a sigmoidal curved (Hill coefficient of 2.4).
Thin Layer Chromatography (TLC)
Microtubules were polymerized with GTP or GMPCPP and stabilized with taxol. 2 uM Kif7 was mixed with 18–28 uM microtubules and 1 mM ATP containing a trace amount of [a-32P] ATP, for 5 mins at room temperature. The mixture was layered over a glycerol cushion and centrifuged for 10 mins at 90,000 RPM in a TLA120.1 rotor. The pellet was quickly washed twice with 1× BRB80 containing 10 uM taxol. The pellet was denatured with 20 μL formic acid to release bound nucleotide and spotted onto a Cellulose PEI TLC plate. TLC was performed in buffer containing 0.6 M KH2PO4 pH 3.4. The plate was air dried and exposed to an X-ray film. No hydrolysis of the radioactive ATP was observed under these conditions in the absence of Kif7. ATP and ADP were used as markers and imaged using a UV lamp.
BioLayer Interferometry assays
BLI experiments were performed in an Octet Red 96 instrument (ForteBio). To quantify Kif7-soluble tubulin binding, experiments were performed in assay buffer containing 1× BRB80 supplemented with 1mM MgCl2, 1mM GDP, 1 mM adenosine nucleotide, 0.2% Tween and 1 mM β-ME. Dimeric, non GFP-tagged Kif7 at a concentration of 150 nM was immobilized on an anti-penta-HIS biosensor chip (HIS1K, fortebio). Unbound Kif7 was washed away with assay buffer. The Kif7-bound biosensors were then dipped into tubulin diluted in assay buffer, at concentrations ranging from 0–20 μM. After the binding reached steady-state, tubulin-off rates were measured by switching the biosensors to assay buffer. Data was analyzed with the ForteBio software. The response of the sensor from three independent measurements at each tubulin concentration were normalized from 0 (at 0 tubulin) to 1 (at 20 μM tubulin), plotted against tubulin concentration and fitted with a Hill curve to obtain the Kd.
EB1-Kif7 binding experiments were performed in assay buffer containing 1× BRB80 supplemented with 1mM MgCl2, 1 mM ATP, 0.05% Tween and 1% BSA. EB1 at a concentration of 70 nM in acetate buffer at pH 5 was covalently linked to an amine reactive biosensor (AR2G, Fortebio). Empty sites on the biosensor were quenched with ethanolamine. The EB1-bound biosensors were then dipped into dimeric Kif7 diluted in assay buffer, at concentrations ranging from 0–10 μM, for 800 seconds. The biosensors were switched to assay buffer for a further 800 sec to measure off-rates. Double referencing was performed to subtract the contribution to signal from non-specific binding of Kif7 to the sensor. To ensure that EB1 could bind SxIP-motif containing proteins under these experimental conditions, CLASP1, a known EB1 binder was used as a positive control. The response of the sensor from three independent measurements at each Kif7 and CLASP1 concentration was plotted and binding Kd was determined.
Measurement of ATPase rates
ATPase rates were measured using the EnzChek phosphate assay kit (Molecular probes, ThermoFisher Scientific). All ATPase assays were performed in assay buffer containing 1XBRB80 with 1 mM ATP and 1 μM Kif7 dimer. The phosphate released upon ATP hydrolysis was monitored at OD360 in a SpectraMax microplate reader, every minute for 2 hours. All reactions were initiated by the addition of ATP. The initial reaction velocity was used to determine the rate of the reaction. For measurements of microtubule stimulated ATPase, taxol stabilized microtubules containing 40 μM of taxol were used. Reaction mixture containing microtubule and ATP and no Kif7 was used as a blank. For measurements of tubulin-stimulated ATPase, assay buffer was supplemented with 1 mM GDP and 2 mM MgCl2. Reaction mixture containing tubulin at the highest concentration, ATP and no Kif7 was used as a blank. The mean and standard deviation of the ATPase rate from 3 independent experiments were plotted for each concentration of microtubule and tubulin.
Size-exclusion chromatography to probe Kif7-tubulin interaction
Kif7DM-GFP (1 μM) was incubated with unlabeled-tubulin (16 μM) in the presence of ATP, ADP and AMPPNP (1mM) for 15 minutes. Samples were separated by size exclusion chromatography (Superdex 200 increase 10/300 GL; Amersham Pharmacia Biotech) and analyzed by SDS-PAGE.
Cryo-EM sample Preparation
For grid preparation, all Kif7 preparations were diluted using BRB80 (80 mM 1,4-piperazinediethanesulfonic acid [PIPES], pH 6.8, 1 mM MgCl2, 1 mM ethylene glycol tetraacetic acid [EGTA]). Porcine brain microtubules were prepared by polymerizing 5 mg/ml tubulin (Cytoskeleton, Denver, CO) in polymerization buffer (80 mM PIPES, pH 6.8, 1 mM EGTA, 4 mM MgCl2, 2 mM GTP or GMP-CPP, 9% dimethyl sulfoxide) for 30 min at 36º C. Paclitaxel was added at 250 μM before further incubation of 30 min at 36ºC. The polymerized microtubules were then incubated at room temperature for several hours or overnight before use.
All microtubule samples were prepared on 1.2/1.3 400-mesh grids (Electron Microscopy Services) Grids were glow-discharged before sample application. The cryosamples were prepared using a manual plunger, which was placed in a homemade humidity chamber that varied between 80 and 90% relative humidity. A 4-μl amount of the microtubules at ∼0.5 μM in 80 mM PIPES, pH 6.8, 4 mM MgCl2, and 1 mM EGTA supplemented with 20 μM Taxol was allowed to absorb for 2 min, and then 4 μl of the Kif7 preparations (0.5mg/mL in BRB80 plus AMPPNP (2mM) or ADP (2mM)) was added to the grid. After a short incubation of 2 min, the sample was blotted (from the back side of the grid) and plunged into liquid ethane. For samples used for reconstructions of ADP-bound Kif7, high concentrations of ADP were maintained during protein purification, and in the storage and EM buffers. The Kif7 protein was not exposed to ATP during purification, storage or EM sample preparation.
Kif7 binding to curved protofilaments
Kif7DM-GFP at a concentration of 1 μM in BRB80, with 2 mM AMPPNP or 2 mM ADP was incubated in the presence of dolostatin-induced microtubule rings and straight microtubule at a Kif7: microtubule ratio of 1:10.8 μM. 4 μL aliquots of the samples were then incubated for 1–2 min on glow-discharged EM carbon coated grids, blotted and stained with 1% Uranyl Acetate. Images were collected on a Morgagni TEM (FEI, Hillsboro, OR).
EM image acquisition and data processing
Images of frozen-hydrated Kif7-microtubule complexes (see Table S1) were collected on a Titan Krios (FEI, Hillsboro, OR) operating at 300 keV equipped with a K2 Summit direct electron detector (Gatan, Pleasanton, CA). The data were acquired using the Leginon automated data acquisition (Suloway et al., 2005). Image processing was performed within the Appion processing environment (Lander et al., 2009). Movies were collected at a nominal magnification of 22500× with a physical pixel size of 1.31 Å/pixel. Movies were acquired using a dose rate of ~4.7 electrons/pixel/second over 12 seconds yielding a cumulative dose of ~63 electrons/Å2. The MotionCor frame alignment program (Li et al., 2013, Hirschi et al., 2017) was used to motion-correct. Aligned images were used for CTF determination using CTFFIND4 (Rohou and Grigorieff, 2015) and only micrographs yielding CC estimates better than 0.5 at 4 Å resolution were kept. Microtubule segments were manually selected, and overlapping segments were extracted with a spacing of 80 Å along the filament. The boxed segments (1.34Å/pixel, 384 pixel box size) were then subjected to reference-free 2D classification using multivariate statistical analysis (MSA) and multi-reference alignment (MRA) (Ogura et al., 2003, Hirschi et al., 2017). Particles in classes that did not clearly show Kif7 density were excluded from further processing.
Cryo-EM 3D reconstruction
Undecorated 13,14- and 15-protofilament microtubule densities (Sui and Downing, 2010) were used as initial models for all preliminary reconstructions. We used the IHRSR procedure (Egelman, 2007) for multimodel projection matching of microtubule specimens with various numbers of protofilaments (Alushin et al., 2014), using libraries from the EMAN2 image processing package (Tang et al., 2007). After each round of projection matching, an asymmetric backprojection is generated of aligned segments, and the helical parameters (rise and twist) describing the monomeric tubulin lattice are calculated. These helical parameters are used to generate and average 13, 14 and 15 symmetry-related copies of the asymmetric reconstruction, and the resulting models were used for projection matching during the next round of refinement. The number of particles falling into the different helical families varied. Helical families that had enough segments were further refined. Final refinement of microtubule segment alignment parameters was performed in FREALIGN (Grigorieff, 2007) without further refinement of helical parameters. FSC curves were used to estimate the resolution of each reconstruction, using a cutoff of 0.143. To better display the high-resolution features, we applied a B-factor of 200 Å, using the program bfactor (http://grigoriefflab.janelia.org).
Model building
The crystal structure of the Kif7 motor domain (residues 9–344, PDB:4A14) and the EM-derived structure of GDP-taxol stabilized microtubules (PDB:3J6G) were fit as rigid bodies into the EM-density of either Kif7MM:ADP or Kif7MM:AMPPNP, using the ‘Fit in Map’ utility in UCSF Chimera (Pettersen et al., 2004). Flexible fitting of the resulting structure was performed with phenix.real_space_refine (Afonine et al., 2013). The map and refined structure were viewed in Coot and missing loops in Kif7 were manually modelled into the density (Emsley and Cowtan, 2004). We then performed iterative rounds of model building and refinement. The final Kif7MM:AMPPNP and Kif7MM:ADP models have model-map correlations of 0.81 and 0.83, bond length RMSDs of 0.011 and 0.006 Å, and bond angle RMSDs of 1.37 and 1.30°, respectively. Both models have no Ramachandran outliers. A superposition of the starting model (PDB: 3J6G) with the refined tubulin dimers from Kif7MM:AMPPNP and Kif7MM:ADP yielded low RMSDs of 1Å and 0.97Å respectively. Therefore, at the resolution of our maps, the final models of α- and β-tubulin in the Kif7-bound structures are not significantly different from the starting model. For both GMPCPP and GDP-taxol microtubules (EMDB IDs: 5895 and 5897), the twist of the microtubule lattices reported in the EMDB is −25.75°. For Kif7-bound microtubules, we calculated the twist to be −25.75° and −25.76° for GMPCPP and GDP-taxol microtubules respectively. The rise reported for the GMPCPP and GDP-taxol microtubules lattices are 8.9Å and 8.96Å respectively. For Kif7 bound to GMPCPP and GDP-taxol microtubules lattices, the calculated rise is 8.82Å and 8.83Å respectively.
Analysis of protein structures
To superpose the different EM reconstructions and their associated models, the EM maps were first superposed using the UCSF Chimera “Fit in map” tool. The protein chains were subsequently fit into their reconstructions using the same tool (Pettersen et al., 2004). In order to analyze the Kinesin:tubulin interface, residues on α- and β-tubulin which were <7Å away from the kinesin were identified using the program ‘Contact’ in the CCP4 suite (Winn et al., 2011). The interface areas were calculated by the PDBSUM webserver (de Beer et al., 2014). Electrostatic potential of the surfaces were generated from the APBS server (Jurrus et al., 2018). All sequence alignments were generated by the Clustal Omega program (Sievers et al., 2011). Residue conservation among Kif7 homologues was calculated by the MultAlign program in UCSF Chimera.
Cell Culture and immunofluorescence
C3H10T1/2 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), sodium pyruvate (1 mM) and L-glutamine (2 mM). Cells were transfected with 2.5 μg of various constructs of Kif7-GFP using Lipofectamine LPX (Invitrogen) at 18 hours after seeding (1.2×105 cells/well in 35 mm plates with 18×18mm coverslip). 36 hours post transfection, the medium was replaced with low serum of DMEM (0.2% FBS) mixed with 6 μl of 500 μM SAG (Smoothened Agonist; EMD Millipore). After another 36 hours of serum starvation and Hh-signaling activation, the sample was fixed using a mixture of methanol and acetone (1:1 in volume) for 10 minutes in −20°C, washed with washing buffer (PBS+0.05% Tween 20) for 3 times, blocked with blocking buffer (TBS+1%BSA; OmniPur BSA; EMD Millipore) for one hour at room temperature. Samples were probed overnight in 4°C with the following primary antibodies (diluted in blocking buffer): Alexa Fluor 647nm labeled acetylated α-tubulin antibody (1:100) (Santa Cruz Biotechnology) and Cyanine3 labeled polyclonal γ-tubulin antibody (1:200) (Thermo Fisher). Samples were mounted on a 25mmX75mm glass with ProLong Diamond Antifade Mountant (Thermo Fisher). Z-stacks were acquired on an inverted Nikon microscope in the epifluorescence imaging mode using a SpectraX illumination source (488nm, 561nm and 647nm channels corresponding to Kif7-GFP, γ-tubulin-Cy3and acetylated α-tubulin-Alexa647). Maximum projection images were generated form planes that included the entire cilium. Images were collected over several days due to the low transfection efficiency of this cell line.
Quantification and Statistical Analysis
ImageJ was used to assess GFP fluorescence intensities on microtubules. For all average intensity per pixel values recorded, a rectangular area along the microtubule was selected with a width of 5 pixels. Background intensities were also subtracted locally from regions of interest using the same principle around the selected microtubule. Intensities were not analyzed for microtubules found at the edges of the camera’s field of view. Plotting of intensities was carried out in GraphPad Prism. “N” numbers in all experiments refer to the unique number of microtubules used for the dataset and standard deviations correspond to deviations from the mean. Statistical details can be found in the results section and corresponding figure legends.
Intensity per pixel analysis on curved microtubules were analyzed on ImageJ and plotted on MatLab as per Bechstedt et al., 2014 (Bechstedt et al., 2014).
Li-Cor Image Studio Lite was used to analyze intensity of protein bands for cosedimentation assays in figures 2, 3, S2 and S3. A rectangular box was drawn around the protein band of interest for intensity readout. Local background was subtracted using a 3 pixel radius around the region of interest. Analysis of intensities and a binding curve was fit to a Hill curve using GraphPad Prism.
Octet Data Analysis software was used to extract the binding response for BLI data shown in figures 2E and S6H. The responses were plotted as a function of protein concentration and fit to Hill curves using GraphPad Prism. In figure 2E, the binding responses were normalized between 0 and 1 prior to curve fitting.
For structure fitting, atomic models were fit into the cryo-EM density using UCSF Chimera and model to map correlation coefficients were calculated using the ‘Fit in Map’ utility.
For normalized intensity measurements from the tip of the microtubule in figure 6 and S6, the intensity along a growing microtubule was taken at the 10 minute timepoint. The tip of the microtubule was determined by the highest intensity point at the tip of the microtubule. Intentisity profiles of different lengths were aligned by their highest intensity value at the tip of the microtubule and averaged. Normalized intensities were calculated using GraphPad Prism.
For cell biology, images of cilia were analyzed on ImageJ and numbers of cilia with and without GFP fluorescence were plotted on GraphPad prism.
Supplementary Material
Comparison of Kif7:AMPPNP and Kin1:ATP (Related to Figure 3) Comparison of microtubule-bound conformations of Kif7MM:AMPPNP (blue) and Kin1:ATP (orange), superposed by their tubulin dimers (gray), highlighting helices α4, α6, strand β5 and loop L12.
Comparison of Kif7 ADP and AMPPNP (Related to Figure 4) Comparison of microtubule-bound conformations of Kif7MM:ADP (pink) and Kif7MM:AMPPNP (blue), superposed by their tubulin dimers (gray), highlighting helices α4, α6, strand β5 and loop L12.
EB1-GFP Expansion in the presence of Kif7 (Related to Figure 6) Localization of EB1-GFP (100 nM) on growing microtubules in the presence of 1 μM Kif7DM. Microtubules are in red, EB1-GFP in green. The movie was taken over 20 min (120 frames) and displayed at a rate of 25 frames/sec.
Acknowledgements
This work was supported by a grant from the NIH (GM 52468) to RAM and EMW-K and funds from the Pew Charitable Trusts and the Smith Family Foundation to RS. We thank Dr. Mike Blower (MGH) for EB1 plasmid and protein reagents, Dr. Mike Blower and Dr. Li Li (MGH) for help with TLC assays, Dr. Gabe Lander and members of his laboratory (The Scripps Institute, San Diego, CA) for discussions and help with imaging and image analysis. We also thank Sithara Wijeratne (MGH and HMS, Boston, MA) and Shreyas Gokhale (MIT, Cambridge, MA) for help with image processing of curved microtubules.
Footnotes
Data and Software Availability
Data deposition
The cryo-EM maps and models have been deposited in the EMDB and PDB with the following accession numbers:
AMPPNP-bound Kif7 monomer on GDP-Taxol stabilized microtubules: EMDB 9141, PDB 6MLR
ADP-bound Kif7 monomer on GDP-Taxol stabilized microtubules: EMDB 9140, PDB 6MLQ
References
- AFONINE PV, HEADD JJ, TERWILLIGER TC & ADAMS PD 2013. New tool: phenix.real_space_refine. Computational Crystallography Newsletter, 4, 43–44. [Google Scholar]
- ALUSHIN GM, LANDER GC, KELLOGG EH, ZHANG R, BAKER D & NOGALES E 2014. High-resolution microtubule structures reveal the structural transitions in alphabeta-tubulin upon GTP hydrolysis. Cell, 157, 1117–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ARELLANO-SANTOYO H, GEYER EA, STOKASIMOV E, CHEN GY, SU X, HANCOCK W, RICE LM & PELLMAN D 2017. A Tubulin Binding Switch Underlies Kip3/Kinesin-8 Depolymerase Activity. Dev Cell, 42, 37–51 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ATHERTON J, FARABELLA I, YU IM, ROSENFELD SS, HOUDUSSE A, TOPF M & MOORES CA 2014. Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins. Elife, 3, e03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ATHERTON J, YU IM, COOK A, MURETTA JM, JOSEPH A, MAJOR J, SOURIGUES Y, CLAUSE J, TOPF M, ROSENFELD SS, HOUDUSSE A & MOORES CA 2017. The divergent mitotic kinesin MKLP2 exhibits atypical structure and mechanochemistry. Elife, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AVASTHI P & MARSHALL WF 2012. Stages of ciliogenesis and regulation of ciliary length. Differentiation, 83, S30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BECHSTEDT S, LU K & BROUHARD GJ 2014. Doublecortin recognizes the longitudinal curvature of the microtubule end and lattice. Curr Biol, 24, 2366–75. [DOI] [PubMed] [Google Scholar]
- BIELING P, TELLEY IA & SURREY T 2010. A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell, 142, 420–32. [DOI] [PubMed] [Google Scholar]
- BRISCOE J & THEROND PP 2013. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol, 14, 416–29. [DOI] [PubMed] [Google Scholar]
- BROUHARD GJ & RICE LM 2014. The contribution of alphabeta-tubulin curvature to microtubule dynamics. J Cell Biol, 207, 323–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COCHRAN JC, GATIAL JE 3RD, KAPOOR TM & GILBERT SP 2005. Monastrol inhibition of the mitotic kinesin Eg5. J Biol Chem, 280, 12658–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COCHRAN JC, SINDELAR CV, MULKO NK, COLLINS KA, KONG SE, HAWLEY RS & KULL FJ 2009. ATPase cycle of the nonmotile kinesin NOD allows microtubule end tracking and drives chromosome movement. Cell, 136, 110–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CREVEL IM, LOCKHART A & CROSS RA 1996. Weak and strong states of kinesin and ncd. J Mol Biol, 257, 66–76. [DOI] [PubMed] [Google Scholar]
- CROSS RA 2004. The kinetic mechanism of kinesin. Trends Biochem Sci, 29, 301–9. [DOI] [PubMed] [Google Scholar]
- CROSS RA 2016. Review: Mechanochemistry of the kinesin-1 ATPase. Biopolymers, 105, 476–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE BEER TA, BERKA K, THORNTON JM & LASKOWSKI RA 2014. PDBsum additions. Nucleic Acids Res, 42, D292–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DISHINGER JF, KEE HL, JENKINS PM, FAN S, HURD TW, HAMMOND JW, TRUONG YN, MARGOLIS B, MARTENS JR & VERHEY KJ 2010. Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-beta2 and RanGTP. Nat Cell Biol, 12, 703–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DRUMMOND IA 2012. Cilia functions in development. Curr Opin Cell Biol, 24, 24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EGELMAN EH 2007. The iterative helical real space reconstruction method: surmounting the problems posed by real polymers. J Struct Biol, 157, 83–94. [DOI] [PubMed] [Google Scholar]
- EMSLEY P & COWTAN K 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr, 60, 2126–32. [DOI] [PubMed] [Google Scholar]
- FRIEL CT & HOWARD J 2011. The kinesin-13 MCAK has an unconventional ATPase cycle adapted for microtubule depolymerization. EMBO J, 30, 3928–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FUNABASHI T, KATOH Y, MICHISAKA S, TERADA M, SUGAWA M & NAKAYAMA K 2017. Ciliary entry of KIF17 is dependent on its binding to the IFT-B complex via IFT46-IFT56 as well as on its nuclear localization signal. Mol Biol Cell, 28, 624–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GEYER EA, BURNS A, LALONDE BA, YE X, PIEDRA FA, HUFFAKER TC & RICE LM 2015. A mutation uncouples the tubulin conformational and GTPase cycles, revealing allosteric control of microtubule dynamics. Elife, 4, e10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GIGANT B, WANG W, DREIER B, JIANG Q, PECQUEUR L, PLUCKTHUN A, WANG C & KNOSSOW M 2013. Structure of a kinesin-tubulin complex and implications for kinesin motility. Nat Struct Mol Biol, 20, 1001–7. [DOI] [PubMed] [Google Scholar]
- GILBERT SP, GUZIK-LENDRUM S & RAYMENT I 2018. Kinesin-2 motors: Kinetics and biophysics. J Biol Chem, 293, 4510–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GLUENZ E, HOOG JL, SMITH AE, DAWE HR, SHAW MK & GULL K 2010. Beyond 9+0: noncanonical axoneme structures characterize sensory cilia from protists to humans. FASEB J, 24, 3117–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOETZ SC & ANDERSON KV 2010. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet, 11, 331–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOULET A, BEHNKE-PARKS WM, SINDELAR CV, MAJOR J, ROSENFELD SS & MOORES CA 2012. The structural basis of force generation by the mitotic motor kinesin-5. J Biol Chem, 287, 44654–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRIGORIEFF N 2007. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol, 157, 117–25. [DOI] [PubMed] [Google Scholar]
- GUEDES-DIAS P, NIRSCHL JJ, ABREU N, TOKITO MK, JANKE C, MAGIERA MM & HOLZBAUR ELF 2019. Kinesin-3 Responds to Local Microtubule Dynamics to Target Synaptic Cargo Delivery to the Presynapse. Curr Biol, 29, 268–282 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUPTA ML JR., CARVALHO P, ROOF DM & PELLMAN D 2006. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol, 8, 913–23. [DOI] [PubMed] [Google Scholar]
- HAYASHI I & IKURA M 2003. Crystal structure of the amino-terminal microtubule-binding domain of end-binding protein 1 (EB1). J Biol Chem, 278, 36430–4. [DOI] [PubMed] [Google Scholar]
- HE M, AGBU S & ANDERSON KV 2017. Microtubule Motors Drive Hedgehog Signaling in Primary Cilia. Trends Cell Biol, 27, 110–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HE M, SUBRAMANIAN R, BANGS F, OMELCHENKO T, LIEM KF JR., KAPOOR TM & ANDERSON KV 2014. The kinesin-4 protein Kif7 regulates mammalian Hedgehog signalling by organizing the cilium tip compartment. Nat Cell Biol, 16, 663–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HELENIUS J, BROUHARD G, KALAIDZIDIS Y, DIEZ S & HOWARD J 2006. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature, 441, 115–9. [DOI] [PubMed] [Google Scholar]
- HERTZER KM, EMS-MCCLUNG SC, KLINE-SMITH SL, LIPKIN TG, GILBERT SP & WALCZAK CE 2006. Full-length dimeric MCAK is a more efficient microtubule depolymerase than minimal domain monomeric MCAK. Mol Biol Cell, 17, 700–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HIRSCHI M, HERZIK MA JR., WIE J, SUO Y, BORSCHEL WF, REN D, LANDER GC & LEE SY 2017. Cryo-electron microscopy structure of the lysosomal calcium-permeable channel TRPML3. Nature, 550, 411–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUNTER AW, CAPLOW M, COY DL, HANCOCK WO, DIEZ S, WORDEMAN L & HOWARD J 2003. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol Cell, 11, 445–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HYMAN A, DRECHSEL D, KELLOGG D, SALSER S, SAWIN K, STEFFEN P, WORDEMAN L & MITCHISON T 1991. Preparation of modified tubulins. Methods Enzymol, 196, 478–85. [DOI] [PubMed] [Google Scholar]
- INGHAM PW, NAKANO Y & SEGER C 2011. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet, 12, 393–406. [DOI] [PubMed] [Google Scholar]
- JANKE C & BULINSKI JC 2011. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat Rev Mol Cell Biol, 12, 773–86. [DOI] [PubMed] [Google Scholar]
- JIANG J & HUI CC 2008. Hedgehog signaling in development and cancer. Dev Cell, 15, 801–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JIANG K & AKHMANOVA A 2011. Microtubule tip-interacting proteins: a view from both ends. Curr Opin Cell Biol, 23, 94–101. [DOI] [PubMed] [Google Scholar]
- JIANG K, TOEDT G, MONTENEGRO GOUVEIA S, DAVEY NE, HUA S, VAN DER VAART B, GRIGORIEV I, LARSEN J, PEDERSEN LB, BEZSTAROSTI K, LINCE-FARIA M, DEMMERS J, STEINMETZ MO, GIBSON TJ & AKHMANOVA A 2012. A Proteome-wide screen for mammalian SxIP motif-containing microtubule plus-end tracking proteins. Curr Biol, 22, 1800–7. [DOI] [PubMed] [Google Scholar]
- JIANG L, TAM BM, YING G, WU S, HAUSWIRTH WW, FREDERICK JM, MORITZ OL & BAEHR W 2015. Kinesin family 17 (osmotic avoidance abnormal-3) is dispensable for photoreceptor morphology and function. FASEB J, 29, 4866–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JURRUS E, ENGEL D, STAR K, MONSON K, BRANDI J, FELBERG LE, BROOKES DH, WILSON L, CHEN J, LILES K, CHUN M, LI P, GOHARA DW, DOLINSKY T, KONECNY R, KOES DR, NIELSEN JE, HEAD-GORDON T, GENG W, KRASNY R, WEI GW, HOLST MJ, MCCAMMON JA & BAKER NA 2018. Improvements to the APBS biomolecular solvation software suite. Protein Sci, 27, 112–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIKKAWA M & HIROKAWA N 2006. High-resolution cryo-EM maps show the nucleotide binding pocket of KIF1A in open and closed conformations. EMBO J, 25, 4187–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KLEJNOT M & KOZIELSKI F 2012. Structural insights into human Kif7, a kinesin involved in Hedgehog signalling. Acta Crystallogr D Biol Crystallogr, 68, 154–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LANDER GC, STAGG SM, VOSS NR, CHENG A, FELLMANN D, PULOKAS J, YOSHIOKA C, IRVING C, MULDER A, LAU PW, LYUMKIS D, POTTER CS & CARRAGHER B 2009. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol, 166, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI X, MOONEY P, ZHENG S, BOOTH CR, BRAUNFELD MB, GUBBENS S, AGARD DA & CHENG Y 2013. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods, 10, 584–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIEM KF JR., HE M, OCBINA PJ & ANDERSON KV 2009. Mouse Kif7/Costal2 is a cilia-associated protein that regulates Sonic hedgehog signaling. Proc Natl Acad Sci U S A, 106, 13377–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU YC, COUZENS AL, DESHWAR AR, LD BM-C, ZHANG X, PUVIINDRAN V, SCOTT IC, GINGRAS AC, HUI CC & ANGERS S 2014. The PPFIA1-PP2A protein complex promotes trafficking of Kif7 to the ciliary tip and Hedgehog signaling. Sci Signal, 7, ra117. [DOI] [PubMed] [Google Scholar]
- LOCKE J, JOSEPH AP, PENA A, MOCKEL MM, MAYER TU, TOPF M & MOORES CA 2017. Structural basis of human kinesin-8 function and inhibition. Proc Natl Acad Sci U S A, 114, E9539–E9548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAURER SP, FOURNIOL FJ, BOHNER G, MOORES CA & SURREY T 2012. EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell, 149, 371–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORIKAWA M, YAJIMA H, NITTA R, INOUE S, OGURA T, SATO C & HIROKAWA N 2015. X-ray and Cryo-EM structures reveal mutual conformational changes of Kinesin and GTP-state microtubules upon binding. EMBO J, 34, 1270–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MUHIA M, THIES E, LABONTE D, GHIRETTI AE, GROMOVA KV, XOMPERO F, LAPPE-SIEFKE C, HERMANS-BORGMEYER I, KUHL D, SCHWEIZER M, OHANA O, SCHWARZ JR, HOLZBAUR ELF & KNEUSSEL M 2016. The Kinesin KIF21B Regulates Microtubule Dynamics and Is Essential for Neuronal Morphology, Synapse Function, and Learning and Memory. Cell Rep, 15, 968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NACHURY MV 2014. How do cilia organize signalling cascades? Philos Trans R Soc Lond B Biol Sci, 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAKATA T, NIWA S, OKADA Y, PEREZ F & HIROKAWA N 2011. Preferential binding of a kinesin-1 motor to GTP-tubulin-rich microtubules underlies polarized vesicle transport. J Cell Biol, 194, 245–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OGURA T, IWASAKI K & SATO C 2003. Topology representing network enables highly accurate classification of protein images taken by cryo electron-microscope without masking. J Struct Biol, 143, 185–200. [DOI] [PubMed] [Google Scholar]
- PEET DR, BURROUGHS NJ & CROSS RA 2018. Kinesin expands and stabilizes the GDP-microtubule lattice. Nat Nanotechnol, 13, 386–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PETTERSEN EF, GODDARD TD, HUANG CC, COUCH GS, GREENBLATT DM, MENG EC & FERRIN TE 2004. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem, 25, 1605–12. [DOI] [PubMed] [Google Scholar]
- PREVO B, SCHOLEY JM & PETERMAN EJG 2017. Intraflagellar transport: mechanisms of motor action, cooperation, and cargo delivery. FEBS J, 284, 2905–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROHOU A & GRIGORIEFF N 2015. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol, 192, 216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROOSTALU J, CADE NI & SURREY T 2015. Complementary activities of TPX2 and chTOG constitute an efficient importin-regulated microtubule nucleation module. Nat Cell Biol, 17, 1422–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROSENFELD SS, RENER B, CORREIA JJ, MAYO MS & CHEUNG HC 1996. Equilibrium studies of kinesin-nucleotide intermediates. J Biol Chem, 271, 9473–82. [DOI] [PubMed] [Google Scholar]
- SATIR P & CHRISTENSEN ST 2007. Overview of structure and function of mammalian cilia. Annu Rev Physiol, 69, 377–400. [DOI] [PubMed] [Google Scholar]
- SCHRODER JM, SCHNEIDER L, CHRISTENSEN ST & PEDERSEN LB 2007. EB1 is required for primary cilia assembly in fibroblasts. Curr Biol, 17, 1134–9. [DOI] [PubMed] [Google Scholar]
- SCHWARZ N, LANE A, JOVANOVIC K, PARFITT DA, AGUILA M, THOMPSON CL, DA CRUZ L, COFFEY PJ, CHAPPLE JP, HARDCASTLE AJ & CHEETHAM ME 2017. Arl3 and RP2 regulate the trafficking of ciliary tip kinesins. Hum Mol Genet, 26, 2480–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHANG Z, ZHOU K, XU C, CSENCSITS R, COCHRAN JC & SINDELAR CV 2014. High-resolution structures of kinesin on microtubules provide a basis for nucleotide-gated force-generation. Elife, 3, e04686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHIMA T, MORIKAWA M, KANESHIRO J, KAMBARA T, KAMIMURA S, YAGI T, IWAMOTO H, UEMURA S, SHIGEMATSU H, SHIROUZU M, ICHIMURA T, WATANABE TM, NITTA R, OKADA Y & HIROKAWA N 2018. Kinesin-binding-triggered conformation switching of microtubules contributes to polarized transport. J Cell Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIEVERS F, WILM A, DINEEN D, GIBSON TJ, KARPLUS K, LI W, LOPEZ R, MCWILLIAM H, REMMERT M, SODING J, THOMPSON JD & HIGGINS DG 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol, 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SINDELAR CV & DOWNING KH 2007. The beginning of kinesin’s force-generating cycle visualized at 9-A resolution. J Cell Biol, 177, 377–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SINGLA V & REITER JF 2006. The primary cilium as the cell’s antenna: signaling at a sensory organelle. Science, 313, 629–33. [DOI] [PubMed] [Google Scholar]
- SONG Y & BRADY ST 2015. Post-translational modifications of tubulin: pathways to functional diversity of microtubules. Trends Cell Biol, 25, 125–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUBRAMANIAN R, TI SC, TAN L, DARST SA & KAPOOR TM 2013. Marking and measuring single microtubules by PRC1 and kinesin-4. Cell, 154, 377–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUBRAMANIAN R, WILSON-KUBALEK EM, ARTHUR CP, BICK MJ, CAMPBELL EA, DARST SA, MILLIGAN RA & KAPOOR TM 2010. Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein. Cell, 142, 433–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUI H & DOWNING KH 2010. Structural basis of interprotofilament interaction and lateral deformation of microtubules. Structure, 18, 1022–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SULOWAY C, PULOKAS J, FELLMANN D, CHENG A, GUERRA F, QUISPE J, STAGG S, POTTER CS & CARRAGHER B 2005. Automated molecular microscopy: the new Leginon system. J Struct Biol, 151, 41–60. [DOI] [PubMed] [Google Scholar]
- TANG G, PENG L, BALDWIN PR, MANN DS, JIANG W, REES I & LUDTKE SJ 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol, 157, 38–46. [DOI] [PubMed] [Google Scholar]
- VAN DER VAART B, VAN RIEL WE, DOODHI H, KEVENAAR JT, KATRUKHA EA, GUMY L, BOUCHET BP, GRIGORIEV I, SPANGLER SA, YU KL, WULF PS, WU J, LANSBERGEN G, VAN BATTUM EY, PASTERKAMP RJ, MIMORI-KIYOSUE Y, DEMMERS J, OLIERIC N, MALY IV, HOOGENRAAD CC & AKHMANOVA A 2013. CFEOM1-associated kinesin KIF21A is a cortical microtubule growth inhibitor. Dev Cell, 27, 145–160. [DOI] [PubMed] [Google Scholar]
- VARGA V, HELENIUS J, TANAKA K, HYMAN AA, TANAKA TU & HOWARD J 2006. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol, 8, 957–62. [DOI] [PubMed] [Google Scholar]
- WANG D, NITTA R, MORIKAWA M, YAJIMA H, INOUE S, SHIGEMATSU H, KIKKAWA M & HIROKAWA N 2016. Motility and microtubule depolymerization mechanisms of the Kinesin-8 motor, KIF19A. Elife, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILLIAMS CL, MCINTYRE JC, NORRIS SR, JENKINS PM, ZHANG L, PEI Q, VERHEY K & MARTENS JR 2014. Direct evidence for BBSome-associated intraflagellar transport reveals distinct properties of native mammalian cilia. Nat Commun, 5, 5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WINN MD, BALLARD CC, COWTAN KD, DODSON EJ, EMSLEY P, EVANS PR, KEEGAN RM, KRISSINEL EB, LESLIE AG, MCCOY A, MCNICHOLAS SJ, MURSHUDOV GN, PANNU NS, POTTERTON EA, POWELL HR, READ RJ, VAGIN A & WILSON KS 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr, 67, 235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WLOGA D, JOACHIMIAK E, LOUKA P & GAERTIG J 2017. Posttranslational Modifications of Tubulin and Cilia. Cold Spring Harb Perspect Biol, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WOEHLKE G, RUBY AK, HART CL, LY B, HOM-BOOHER N & VALE RD 1997. Microtubule interaction site of the kinesin motor. Cell, 90, 207–16. [DOI] [PubMed] [Google Scholar]
- YUE Y, BLASIUS TL, ZHANG S, JARIWALA S, WALKER B, GRANT BJ, COCHRAN JC & VERHEY KJ 2018. Altered chemomechanical coupling causes impaired motility of the kinesin-4 motors KIF27 and KIF7. J Cell Biol, 217, 1319–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YUN M, ZHANG X, PARK CG, PARK HW & ENDOW SA 2001. A structural pathway for activation of the kinesin motor ATPase. EMBO J, 20, 2611–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZANIC M, STEAR JH, HYMAN AA & HOWARD J 2009. EB1 recognizes the nucleotide state of tubulin in the microtubule lattice. PLoS One, 4, e7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG R, ALUSHIN GM, BROWN A & NOGALES E 2015. Mechanistic Origin of Microtubule Dynamic Instability and Its Modulation by EB Proteins. Cell, 162, 849–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of Kif7:AMPPNP and Kin1:ATP (Related to Figure 3) Comparison of microtubule-bound conformations of Kif7MM:AMPPNP (blue) and Kin1:ATP (orange), superposed by their tubulin dimers (gray), highlighting helices α4, α6, strand β5 and loop L12.
Comparison of Kif7 ADP and AMPPNP (Related to Figure 4) Comparison of microtubule-bound conformations of Kif7MM:ADP (pink) and Kif7MM:AMPPNP (blue), superposed by their tubulin dimers (gray), highlighting helices α4, α6, strand β5 and loop L12.
EB1-GFP Expansion in the presence of Kif7 (Related to Figure 6) Localization of EB1-GFP (100 nM) on growing microtubules in the presence of 1 μM Kif7DM. Microtubules are in red, EB1-GFP in green. The movie was taken over 20 min (120 frames) and displayed at a rate of 25 frames/sec.


