Abstract
Background
Whole-genome sequencing (WGS) of heart transplant recipient- and donor-derived cardiac biopsies may facilitate organ matching, graft failure prediction, and immunotolerance research. The objective of this study was to determine the feasibility of WGS based on formalin-fixed paraffin-embedded endomyocardial biopsies.
Methods and results
The study included serial donor- and recipient samples from patients who had undergone heart transplantation at Skane University Hospital, Lund, Sweden, between 1988 and 2009. DNA extraction and WGS were conducted. Additional WGS sequencing quality metrics and coverage were obtained with the Genome Analysis Toolkit (GATK).
455 endomyocardial samples from 37 heart transplant recipients were acquired from routine rejection monitoring and stored as formalin-fixed paraffin-embedded samples. They were analyzed after 3–26 years of storage. DNA was extracted from 114 samples and WGS was run on 85 samples. DNA extraction yielded 313 ng (IQR 96–601) for all samples. A coverage of 11.3x (IQR 9.0–15.9) was recorded for all WGS samples. Three samples stored for > 25 years yielded a coverage of > 25x. Data were generated for 1.7 billion reads per sample (IQR 1.4–2.7). A Transition/Transversion (TiTv) ratio of 2.09 ± 0.05 was calculated for all WGS samples. No associations were found among storage time, DNA yield, or sequencing quality metrics.
Conclusions
The present study demonstrated the feasibility of whole-genome sequencing based on endomyocardial biopsies. This process could enable large-scale retrospective genomic studies using stored histopathological samples.
Introduction
Post-heart transplantation survival has improved over the last three decades. Median survival now exceeds 12 years [1]. Management of long-term complications such as malignancy underscores the importance of customizing immunosuppression in transplant patients [2]. Graft failure remains the most common cause of death and is often the result of immune-mediated rejection [3].
Whole-genome sequencing (WGS) technologies comprehensively characterizes human leukocyte antigen (HLA) genes located on chromosome 6 and other genomic regions determining immune tolerance. Elucidation of the genetic predisposition to rejection may help tailor immunosuppressive therapy and lower the incidence of malignancies in closely matched patients.
Most next-generation sequencing (NGS) studies have used DNA isolated from whole blood or fresh frozen tissues. However, experiments involving formalin-fixed paraffin-embedded (FFPE) samples have demonstrated that FFPE may be valuable as a DNA source for NGS. These studies have indicated a significant influence of storage time on sequencing quality [4]. Targeted NGS on cardiac biopsies using sequencing panels demonstrated no significant differences between FFPE- and blood samples and adequate sequencing scores for older samples [5].
The endomyocardial biopsies (EMB) obtained from routine rejection monitoring after heart transplantation are FFPE samples. The feasibility of FFPE in WGS of EMB and the effects of storage time on these tissue specimens must be determined. If it is validated, this study design would support large-scale retrospective genomic studies on EMBs derived from FFPE samples of numerous heart transplantations.
The objectives of this study were to determine whether DNA obtained from archived EMB stored as FFPE suffices for WGS and to assess the effects of storage time on sequencing quality.
Results
Study population characteristics
A total of 455 FFPE samples were collected from 37 transplanted patients and used in DNA extraction and WGS. The mean recipient age was 45 ± 12 years with a range of 13–59 years. The mean survival after transplantation was 11.4 ± 6.5 years. The mean donor age was 36 ± 14 years with a range of 13–64 years. Most recipients were male (59%, n = 22) as were most donors (54%, n = 19). Heart failure was caused by non-ischemic cardiomyopathy in 62% of the recipients (n = 23), ischemic cardiomyopathy in 32% of the recipients (n = 12), and cardiac sarcoidosis and tumor cordis in the remaining 5% (n = 2).
Quantity and quality of DNA extracted
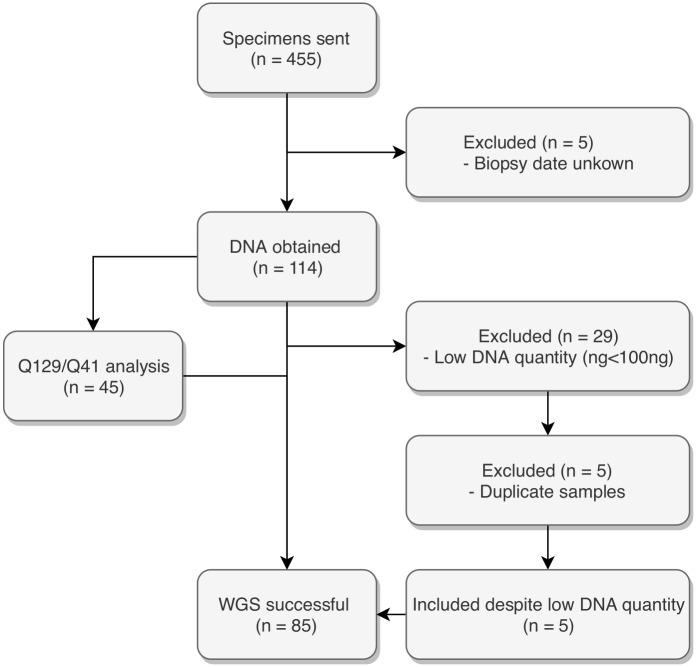
The DNA extraction from the 455 FFPE samples generated 119 unique samples. Of these, 114 had known exact storage times (Fig 1) and their median DNA extraction was 313 ng (IQR 96–601) (Table 1). The median KAPA QC score (Q129/Q41 ratio, n = 45) was 0.003 (IQR 0.0007–0.0178).
Fig 1. Patient flow.
Table 1. DNA quantity (nanogram) following DNA extraction according to sample storage time.
| Variable | All n = 114 |
0–10 y n = 13 |
11–15 y n = 18 |
16–20 y n = 34 |
21–30 y n = 49 |
Min | Max |
|---|---|---|---|---|---|---|---|
| DNA yield | 313 (96–601) | 481 (186–903) | 326 (40–601) | 442 (259–747) | 162 (66–350) | 7 | 3,388 |
Data are expressed as medians and interquartile ranges.
Whole-genome sequencing analysis
The median coverage was 11.3x (IQR 9.0–15.9) for all samples (Table 2). We observed no significant difference in coverage between Illumina HiSeq 2000 (n = 10) and Illumina HiSeq X (n = 75; P = 0.2469). The median percentage of callable bases at Phred score ≥ Q20 was 77.0 (IQR 70.8–82.1). The median percentages of genome covered at 20x, 10x, and 0x for all WGS samples were 15.8 (IQR 12.4–29.4), 40.9 (IQR 25.3–58.4), and 2.9 (IQR 1.9–4.0), respectively. We recorded a mean library size of 1.7 gigabases (IQR 1.4–2.7) for all WGS samples and a Transition/Transversion (TiTv) ratio of 2.09 ± 0.05 (Table 2). We compared the TiTv ratios for our FFPE samples with those from the human genome sample NA12878 which yielded a TiTv ratio of 2.13. Maximum coverage was 55x recorded for a sample with a storage time of 11 years. This sample had 97.2% of the genome covered at 20x and 1.1% of the genome covered at 0x. Three samples with storage times > 25 years yielded coverages > 25x. The sample with the least coverage had a DNA yield of 162 ng and a KAPA QC score of 0.01.
Table 2. Whole-genome sequencing quality metrics according to sample storage time.
| Variable | All n = 85 |
0–10 y n = 11 |
11–15 y n = 12 |
16–20 y n = 28 |
21–30 y n = 34 |
Min | Max |
|---|---|---|---|---|---|---|---|
| Coverage (x) | 11.3 (9.0–15.9) | 12.2 (10.7–18.2) | 10.9 (9.6–14.6) | 10.4 (9.2–17.7) | 11.2 (8.4–15.4) | 3.8 | 55.5 |
| % Covered 20x | 15.8 (12.4–29.4) | 16.0 (11.2–29.7) | 13.6 (11.3–22.0) | 14.9 (13.9–31.2) | 17.3 (11.1–29.4) | 0.8 | 97.2 |
| % Covered 10x | 40.9 (25.3–58.4) | 54.7 (43.4–61.5) | 42.0 (28.4–60.9) | 38.2 (26.3–63.2) | 36.0 (20.3–53.2) | 9.3 | 97.9 |
| % Covered 0x | 2.9 (1.9–4.0) | 2.2 (1.9–3.0) | 2.7 (2.0–3.7) | 2.9 (1.9–3.5) | 3.4 (2.2–4.7) | 1.1 | 26.2 |
| % Q20 | 77.0 (70.8–82.1) | 80.8 (75.7–84.0) | 79.4 (73.4–83.1) | 76.8 (73.0–84.0) | 74.2 (67.7–79.8) | 40.5 | 93.9 |
| Library size | 1.7 (1.4–2.7) | 2.8 (1.7–3.2) | 1.7 (1.4–2.0) | 1.7 (1.5–3.0) | 1.6 (1.3–2.4) | 0.87 | 8.61 |
| Ti/Tv Ratio | 2.09 (0.05) | 2.09 (0.03) | 2.09 (0.03) | 2.08 (0.04) | 2.09 (0.07) | 1.94 | 2.28 |
Quantitative data are expressed as medians and interquartile ranges or means and standard deviations. Coverage is expressed as depth of coverage (x). Proportions of observations covered at 20x, 10x and 0x are expressed as percentages (%). Proportions of observations with a Phred quality score of Q20 are expressed as percentages (%). Library sizes are presented in gigabases (billion base pairs). TiTv ratios are expressed as means and standard deviations.
KAPA QC scores and coverage
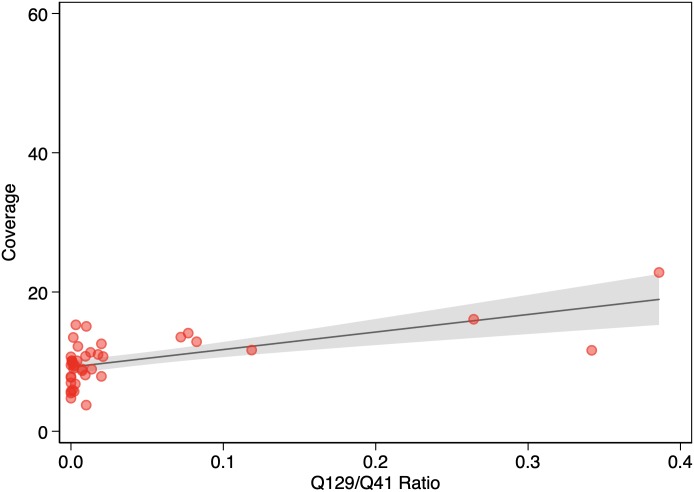
The median KAPA QC score (Q129/Q41 ratio, n = 41) for the WGS samples was 0.004 (IQR 0.0007–0.0178). The KAPA QC score was a good predictor of coverage (P = 0.000012). (Fig 2). Also, KAPA QC score was an independent predictor of coverage (P = 0.000039) in the multivariable regression analysis. In the same analysis, neither DNA yield (P = 0.584) nor storage time (P = 0.254) were predictors of coverage.
Fig 2. Association between Q129/Q41 ratio and coverage (n = 41).
The solid line represents the linear prediction. The shaded area represents the 95% confidence interval for the linear prediction.
Effects of storage time
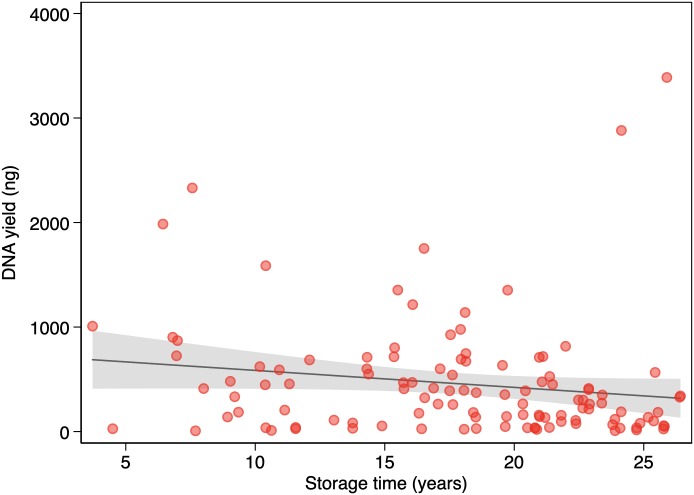
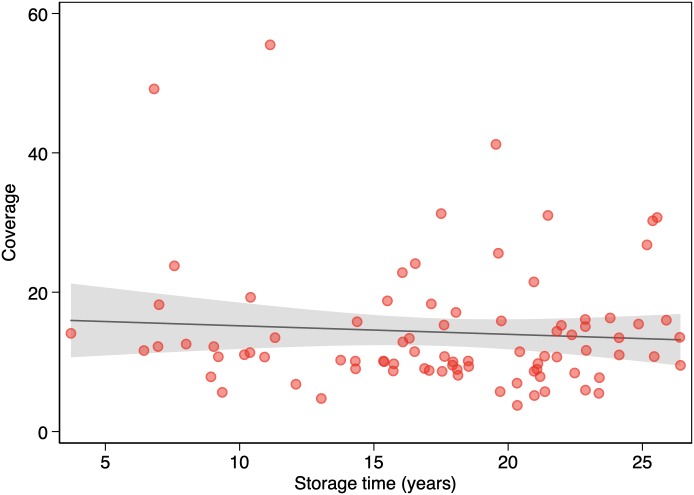
Table 1 shows DNA yields for all samples and for samples grouped by 5-year or 10-year storage intervals. We found no association between DNA yield and storage time (P = 0.08) (Fig 3). Table 2 shows WGS metrics for all samples and for samples grouped by 5-year or 10-year storage intervals. We found no association between coverage and storage time (P = 0.495) (Fig 4). There were no apparent statistically significant differences in storage time according to linear regression (including log-transformed variables) or among storage groups according to the two-sided t-test or the Mann-Whitney U test. We found no association between library size and storage time (P = 0.065) (Fig 5) or between coverage and DNA yield (P = 0.671). There were no associations among coverage, storage time (P = 0.539), and DNA yield (P = 0.757) according to multivariable regression analysis.
Fig 3. Association between storage time and DNA yield (n = 114).
The solid line represents the linear prediction. The shaded area represents the 95% confidence interval for the linear prediction.
Fig 4. Association between storage time and coverage (n = 85).
The solid line represents the linear prediction. The shaded area represents the 95% confidence interval for the linear prediction.
Fig 5. Association between storage time and library size (n = 85).
The solid line represents the linear prediction. The shaded area represents the 95% confidence interval for the linear prediction.
Discussion
This first of its kind study investigates the practicality of WGS analysis and the effects of storage time on formalin-fixed paraffin-embedded endomyocardial biopsies obtained from heart transplant recipients. DNA yield, coverage, and library size did not significantly decrease with FFPE storage time even up to 26 years. Previous studies have investigated the practicality of applying NGS to FFPE samples [4–9]. To the best of our knowledge, the present study is the first to test the feasibility of WGS in biopsies derived from heart transplant recipients.
Appropriate coverage is a prerequisite for accurate variant calling in human genome studies. However, the results of studies estimating the coverage required for accurate variant calling are highly variable. Ajay et al. showed that 95% of the genome was callable at 35x coverage. They also demonstrated that the increase in the proportion of callable genome rapidly declines at coverages > 25x. Therefore, increasing coverage above this level does not necessarily improve variant calling. Nevertheless, a coverage of 50x was recommended to call all single nucleotide polymorphisms (SNPs) and indels [10]. A more recent study on WGS coverage requirements used a different variant calling approach and determined that a mean coverage of 14x was necessary to call 95% of the SNPs accurately [11]. In the present study, many samples did not meet the 30x target coverage but nonetheless yielded sufficient coverage for variant analysis. Numerous samples can be sequenced from one biopsy. Moreover, the cost of WGS has markedly decreased. Therefore, sequencing can be repeated when the coverage is inadequate. In practice, coverage requirements depend on the objective of the analysis. Retrospective studies require less accuracy than diagnostic investigations.
Several samples yielded disappointing Q129/Q41 ratios from the KAPA QC assay (Q129/Q41 ratio). Nevertheless, Q129/Q41 may still predict coverage and many samples with poor Q129/Q41 ratios still yielded sufficient coverage. According to the present study, Q129/Q41 is not recommended as a WGS gatekeeper.
Detailed characterization of the long-term impact of variations in HLA and other genomic regions on outcomes requires sequencing and variant calling of FFPE samples stored for > 10 years. Potential applications of WGS on serial donor- and recipient samples include donor-recipient match/mismatch analysis, HLA interaction analysis and GWAS (genomewide association studies) including functional effect prediction analysis. Studies on the impact of FFPE storage time on NGS quality metrics reported decreases in library size and coverage with storage time (4). In the present study, WGS quality metrics decreased with increasing storage time but these changes were not statistically significant either by comparison between storage groups or by linear regression. However, statistical significance might have been detected if a larger number of biopsies been sampled. In any case, the WGS quality metrics for many older samples were adequate.
The TiTv ratios reported in the present study suggested that there were no artificial variants introducing bias. The TiTv ratios were within the expected range and were comparable to the reference sample NA12878. Therefore, the endomyocardial samples stored as FFPE were not genetically altered by FFPE processing [12].
Immune cell differentiation and function may depend on epigenetic mechanisms. Nevertheless, the influences of DNA methylation profiles on transplant acceptance, transplant rejection, and other post-transplant complications are unknown. The candidate genes contributing to this phenomenon can be identified by NGS and gene expression profiling. It is known that FFPE is suitable for use in DNA methylation analysis [13, 14]. DNA methylation analysis also has clinical potential in the early diagnosis of post-transplant complications [15]. Patients may be segregated into low-, intermediate-, and high rejection risk groups. New diagnostic and therapeutic methods could be developed by identifying genomic variants and aberrant DNA methylation. Moreover, immunosuppressive therapy can be adapted by using non-invasive blood sampling to isolate and quantify DNA methylation. Like other NGS studies, most DNA methylation studies are performed on fresh frozen tissue. Studies have shown comparable results when comparing NGS on fresh frozen samples to NGS on FFPE samples [16, 17]. Likewise, a study comparing DNA methylation using targeted sequencing on matched fresh frozen tissue and FFPE tissue found no effect on methylation calling and concordant results for several loci [18]. In this study we demonstrate the practicality of WGS on stored EMB samples. We therefore hypothesize that DNA extracted from EMB samples can be utilized in methylation analysis using NGS approaches and suggest that this be investigated further.
A limitation of the present study was the lack of sequencing techniques for comparison. The TiTv ratios calculated in this study were based on data from chromosome 6 alone because variant calling is very time-consuming. However, this limitation was partially offset by comparing the results for chromosome 6 to a reference genome. A strength of the present study was the ability to examine cardiac biopsies after heart transplantation, especially since all patients were transplanted at Skane University Hospital and all biopsies were processed by the same pathology unit.
Conclusion
This study demonstrated the feasibility of whole-genome sequencing on formalin-fixed paraffin-embedded endomyocardial biopsies and indicated that this process can facilitate large-scale studies based on stored histopathological samples.
Materials and methods
Ethics statement
The study protocol was approved by the Regional Ethical Committee in Lund, Sweden (Nos. 2012/824, 2013/263, and 2016/112). The data were anonymized and de-identified prior to analysis.
Study population
The study involved recipients who had undergone heart transplantation at Skane University Hospital, Lund, Sweden between 1988 and 2009. EMB samples were obtained from routine rejection monitoring and stored as FFPE samples. FFPE tissue samples were acquired before transplant (recipient heart), shortly after transplant (donor heart), and around the time of death (donor heart). If the DNA yield was inadequate, multiple fragments were taken from different FFPE tissue samples at the same time points and analyzed as a single sample. All patients transplanted at the center were followed up, according to routine protocols for post-transplant care with at least yearly controls until the time of death.
FFPE fixation and storage process
Fixation, dehydration, and embedding were performed at the Department of Pathology, Skane University Hospital, Lund, Sweden. Tissues were fixed in 4% formaldehyde, dehydrated with Tissue-Tek Xpress x120 (Sakura Finetek Co. Ltd., Tokyo, Japan), and manually embedded in Histowax (Histolab Products AB, Gothenburg, Sweden) using a Tissue-Tek TEC 5 Tissue Embedding Console System (Sakura Finetek Co. Ltd., Tokyo, Japan). Fixation times varied with tissue size. Processing, fixation, dehydration, and embedding of older samples (1988–1999) involved a similar protocol. After processing, FFPE samples were stored at room temperature.
DNA extraction, quantity and quality assessment
Samples were sent to the Broad Institute of MIT and Harvard, Cambridge, MA, USA for DNA extraction and WGS. The DNA was extracted from the FFPE samples as cores, scrolls, or slide sections. DNA extraction was conducted on all samples with a column-based QIAamp DNA FFPE Tissue kit (Qiagen, Hilden, Germany). After deparaffinization and de-crosslinking, the samples were lysed with proteinase. Buffering conditions were adjusted for optimal DNA binding to the column. Lysed samples were added to the column and the DNA was selectively bound to the column membrane as a contaminant and passed through enzyme inhibitors in the washing steps. The DNA was eluted with Tris-EDTA (TE) buffer, quantified in triplicate with the Quant-iT PicoGreen DNA assay kit (Thermo Fisher Scientific, Waltham, MA, USA), and normalized to a minimum concentration of 2 ng/uL. Aliquots of ≥ 150 ng per sample were transferred for library preparation using the one-well protocol developed by the Broad Institute [19]. One library was provided and the typical median library insert size was 330 bp. Samples did not undergo post shearing clean-up because they were already highly degraded. The Kapa QC Q-ratios for Q129/Q41 base-pair target regions were calculated with a Kapa Library Quantification and QC kit (Roche Diagnostics, Risch-Rotkreuz, Switzerland) [20]. Samples lacking accurately calculated storage times were excluded.
Whole-genome sequencing
WGS was conducted on samples using the Illumina HiSeq X and Illumina HiSeq 2000 (Illumina, San Diego, CA, USA). It generated 150-bp paired-end reads and short-insert, paired-end reads (2 × 100 bp), respectively, to reach the 30x coverage target (where x = number of sequenced bases / total genome size). Binary Alignment Map (BAM) files were demultiplexed, aggregated, and aligned. All reads per sample with the same index were demultiplexed from the other samples in the same run. The samples were aggregated into one BAM file and aligned to the reference genome (GRCh37/Hg19). Recalibration, indel realignment, and duplicate marking were performed on the raw reads in the BAM files. The Picard Informatics Pipeline was used to aggregate all of the data from a particular sample into a single BAM file including all reads, all bases from all reads, and original- or vendor-assigned quality scores.
The BAM files were indexed with SAMtools [21]. GATK v. 3.5.0 (Broad Institute, Cambridge, MA, USA) was used for additional WGS sequencing quality metrics analyses and variant calling. GATK-HaplotypeCaller and GATK-GenotypeGVCF were used for variant calling. Joint genotype calling of single-nucleotide variants (SNVs) and indels (insertion or deletion) was performed according to GATK Best Practices [22–24]. Variant calling was run on each sample individually and Genomic Variant Calling Format (GVCF) files were generated. The GVCF files were combined into one Variant Call Format (VCF) file. All 85 samples were included in joint genotyping processing. GATK-VariantRecalibrator and GATK-ApplyRecalibration were used for variant filtering. GATK-SelectVariants was used to isolate individual samples which did not pass variant filtering. Additional and complementary sequencing quality metrics and coverage analyses were performed with GATK-CollectWgsMetrics and GATK-VariantEval. GATK-CollectWgsMetrics calculated the mean coverage using overlap and duplication values [25]. These analyses generated data on coverage, TiTv ratios, and the mean percentages of the genome at 20x, 10x, and 0x coverage. Phred scores were available from the original- or vendor-assigned quality scores. A Phred score of Q20 indicated a 1:100 chance of an incorrect base call and was used during variant filtering [26]. The TiTv ratio is the ratio of transitions (point mutation between purines A ↔ G or pyrimidines C ↔ T) to transversions (point mutations between different types of nucleotides: A ↔ C, A ↔ T, C ↔ G or G ↔ T) in the SNVs in a WGS file. Since there are more possible transversions, the theoretical ratio is 0.5. The most common mutation is a cytosine (C) to thymine (T) transition. The expected whole-genome sequencing TiTv ratio in humans is therefore 2.0–2.1 [27]. TiTv is relatively higher in exonic regions and comparatively lower in intronic regions. Only WGS data for chromosome 6 was used in this study. Storage times were calculated for all WGS samples.
Statistical analysis
Data are presented as means ± SD or medians and interquartile ranges (IQR). Categorical variables are percentages. WGS quality metrics were compared between groups by a two-sample t-test and by a Mann-Whitney U test for continuous variables. The associations among WGS quality metrics, storage time, and KAPA QC scores were evaluated by linear regression (including log transformed variables). KAPA QC, DNA yield, and storage time were factored into the multivariable regression analysis of the WGS quality metrics. P < 0.05 was deemed statistically significant. Data were processed in Stata/IC v. 15.1 (StataCorp LP, College Station, TX, USA).
Supporting information
(XLSX)
Acknowledgments
The authors thank Maegan V. Harden, Senior Project Scientist of Broad Institute of MIT and Harvard, for overseeing data pre-processing and for post-processing support. We also thank Cristina-Daria Ciornei Karlsson, biobank coordinator at the Medical Service of the Department of Medicine and Clinical Pathology at Lund Skane University Hospital for coordinating and managing the biobank samples.
Data Availability
Data cannot be shared publicly because of legal and ethical restrictions. However, all relevant data are within the manuscript and its Supporting Information files. Raw data is available to all interested researchers upon request. The instruction how to apply and criteria for access to confidential data is available by the Swedish Ethical Review Authority (http://etikprovning.se). Information on how to access samples in Swedish biobanks for research, method development or clinical trials is available by the Biobanking and molecular Resource Infrastructure of Sweden (http://BBMRI.se).
Funding Statement
This study was funded by the Swedish Heart-Lung Foundation, the Swedish Society of Medicine, a government grant for clinical research, Region Skane Research Funds, Donation Funds from Skane University Hospital, the Anna-Lisa and Sven Eric Lundgrens Foundation, and the Crafoord Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lund LH, Khush KK, Cherikh WS, Goldfarb S, Kucheryavaya AY, Levvey BJ, et al. The Registry of the International Society for Heart and Lung Transplantation: Thirty-fourth Adult Heart Transplantation Report—2017; Focus Theme: Allograft ischemic time. J Heart Lung Transplant. 2017;36(10):1037–1046. 10.1016/j.healun.2017.07.019 [DOI] [PubMed] [Google Scholar]
- 2.Crespo-Leiro MG, Alonso-Pulpon L, Vazquez de Prada JA, Almenar L, Arizon JM, Brossa V, et al. Malignancy after heart transplantation: incidence, prognosis and risk factors. Am J Transplant. 2008;8(5):1031–1039. 10.1111/j.1600-6143.2008.02196.x [DOI] [PubMed] [Google Scholar]
- 3.Lund LH, Edwards LB, Kucheryavaya AY, Dipchand AI, Benden C, Christie JD, et al. The Registry of the International Society for Heart and Lung Transplantation: Thirtieth Official Adult Heart Transplant Report—2013; focus theme: age. J Heart Lung Transplant. 2013;32(10):951–964. 10.1016/j.healun.2013.08.006 [DOI] [PubMed] [Google Scholar]
- 4.Carrick DM, Mehaffey MG, Sachs MC, Altekruse S, Camalier C, Chuaqui R, et al. Robustness of Next Generation Sequencing on Older Formalin-Fixed Paraffin-Embedded Tissue. PLoS One. 2015;10(7):e0127353 10.1371/journal.pone.0127353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baudhuin LM, Leduc C, Train LJ, Avula R, Kluge ML, Kotzer KE, et al. Technical Advances for the Clinical Genomic Evaluation of Sudden Cardiac Death: Verification of Next-Generation Sequencing Panels for Hereditary Cardiovascular Conditions Using Formalin-Fixed Paraffin-Embedded Tissues and Dried Blood Spots. Circ Cardiovasc Genet. 2017;10(6). [DOI] [PubMed] [Google Scholar]
- 6.Hedegaard J, Thorsen K, Lund MK, Hein AM, Hamilton-Dutoit SJ, Vang S, et al. Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PLoS One. 2014;9(5):e98187 10.1371/journal.pone.0098187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Allen EM, Wagle N, Stojanov P, Perrin DL, Cibulskis K, Marlow S, et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med. 2014;20(6):682–688. 10.1038/nm.3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hadd AG, Houghton J, Choudhary A, Sah S, Chen L, Marko AC, et al. Targeted, high-depth, next-generation sequencing of cancer genes in formalin-fixed, paraffin-embedded and fine-needle aspiration tumor specimens. J Mol Diagnostics. 2013;15(2):234–247. [DOI] [PubMed] [Google Scholar]
- 9.Kerick M, Isau M, Timmermann B, Sultmann H, Herwig R, Krobitsch S, et al. Targeted high throughput sequencing in clinical cancer settings: formaldehyde fixed-paraffin embedded (FFPE) tumor tissues, input amount and tumor heterogeneity. BMC Med Genomics. 2011;4:68 10.1186/1755-8794-4-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ajay SS, Parker SC, Abaan HO, Fajardo KV, Margulies EH. Accurate and comprehensive sequencing of personal genomes. Genome Res. 2011;21(9):1498–1505. 10.1101/gr.123638.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meynert AM, Ansari M, FitzPatrick DR, Taylor MS. Variant detection sensitivity and biases in whole genome and exome sequencing. BMC Bioinformatics. 2014;15(1):247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Do H, Dobrovic A. Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin Chem. 2015;61(1):64–71. 10.1373/clinchem.2014.223040 [DOI] [PubMed] [Google Scholar]
- 13.Dallol A, Al-Ali W, Al-Shaibani A, Al-Mulla F. Analysis of DNA methylation in FFPE tissues using the MethyLight technology. Methods Mol Biol. 2011;724:191–204. 10.1007/978-1-61779-055-3_13 [DOI] [PubMed] [Google Scholar]
- 14.Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–474. 10.1038/nature26000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peters FS, Manintveld OC, Betjes MG, Baan CC, Boer K. Clinical potential of DNA methylation in organ transplantation. J Heart Lung Transplant. 2016;35(7):843–850. 10.1016/j.healun.2016.02.007 [DOI] [PubMed] [Google Scholar]
- 16.Bonnet E, Moutet M-LL, Baulard C, Bacq-Daian D, Sandron F, Mesrob L, et al. Performance comparison of three DNA extraction kits on human whole-exome data from formalin-fixed paraffin-embedded normal and tumor samples. PLoS One. 2018;13(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robbe P, Popitsch N, Knight SJLJL, Antoniou P, Becq J, He M, et al. Clinical whole-genome sequencing from routine formalin-fixed, paraffin-embedded specimens: pilot study for the 100,000 Genomes Project. Genet Med. 2018;20(10):1196–1205. 10.1038/gim.2017.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moran B, Das S, Smeets D, Peutman G, Klinger R, Fender B, et al. Assessment of concordance between fresh-frozen and formalin-fixed paraffin embedded tumor DNA methylation using a targeted sequencing approach. Oncotarget. 2017;8(29):48126–48137. 10.18632/oncotarget.18296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher S, Barry A, Abreu J, Minie B, Nolan J, Delorey TM, et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 2011;12(1):R1 10.1186/gb-2011-12-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.KapaBiosystems. KAPA Human Genomic DNA Quantification and QC Kit. 2016(October):1–12. [Google Scholar]
- 21.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aaron M, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Illumina. Sequencing Coverage Calculation Methods for Human Whole-Genome Sequencing. Ilumina. 2014:1–2.
- 26.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8(3):186–194. [PubMed] [Google Scholar]
- 27.Wang J, Raskin L, Samuels DC, Shyr Y, Guo Y. Genome measures used for quality control are dependent on gene function and ancestry. Bioinformatics. 2015;31(3):318–323. 10.1093/bioinformatics/btu668 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
Data cannot be shared publicly because of legal and ethical restrictions. However, all relevant data are within the manuscript and its Supporting Information files. Raw data is available to all interested researchers upon request. The instruction how to apply and criteria for access to confidential data is available by the Swedish Ethical Review Authority (http://etikprovning.se). Information on how to access samples in Swedish biobanks for research, method development or clinical trials is available by the Biobanking and molecular Resource Infrastructure of Sweden (http://BBMRI.se).