Abstract
Genotoxicity is thought to be the cause of many cancers. Genotoxicity due to environmental toxins may be partly responsible for the dramatic increase in the incidence of papillary thyroid cancer over the past two decades. Here, we present a fully automatable assay platform that directly quantifies the phosphorylation of nuclear histone gamma H2AX (γH2AX), a specific cellular marker for DNA double strand breaks (DSBs) via immunohistochemistry and laser scanning cytometry. It multiplexes γH2AX with total cell number measured as propidium iodide and calculates the percentage of cells with DSBs. Validation of this assay using NTHY-ori-3–1 human thyroid cells and etoposide showed that it was an excellent choice for high throughput applications. We used the assay to test the genotoxic effects of the EPA Toxcast Phase 1 pesticide library of 309 compounds. Compounds were evaluated in dose response and the DSB was quantified. We found that 19 pesticides induce DSB in vitro, highlighting a need to further assess these pesticides for their long-term oncogenic effects on the thyroid gland.
Keywords: DNA, double-strand breaks, pesticides, thyroid cells, thyroid cancer, γH2AX, high throughput screening, HTS, genotoxicity, toxicity profiling
INTRODUCTION
The genotoxic potential of new compounds, such as novel drugs and industrial chemicals, is of central interest. Many assays exist to assess this issue based on various indirect measurements of genotoxicity, such as reporter gene assays or assays based on recombination events. We have designed an assay that quantifies the cellular response to DNA double strand breaks (DSBs) caused by small molecules via quantification of H2AX phosphorylation in a cellular assay in high throughput.
DNA DSBs cause severe damage to cells and may lead to cancer through rearrangements in chromosomes [Khanna and Jackson, 2001; Mills et al., 2003]. The phosphorylation of histone H2AX at serine 139 located in its carboxy-terminal tail is central to the DNA repair process [Gallmeier et al., 2005; Huen and Chen, 2010]. It is one of the earliest events after DNA DSB occurrence and is a necessary anchor point for DNA repair proteins. This “gamma” phosphorylation of H2AX is accepted as a specific and direct indicator for the presence of DNA DSBs [Kuo and Yang, 2008; Ohnishi et al., 2009; Sharma et al., 2012]. This marker is especially pertinent as genotoxicity is often mediated through DNA DSB, and DSB occurrence is central to oncogenesis because mutations and/or re-arrangements in the genome caused by DSB may activate proto-oncogenes. DNA DSBs themselves have been found to be induced by genotoxic compounds or ionizing radiation [De Bont and van Larebeke, 2004; Mladenov and Iliakis, 2011]. In a healthy cell, DNA DSBs lead to downstream activation of repair processes [Burdak-Rothkamm and Prise, 2009]. The two main pathways for repair of DSBs are non-homologous end-joining and homologous recombination. Non-homologous end-joining is the main pathway by which cells repair damage from ionizing radiation because it does not require a template for repair and involves limited processing of the damaged ends prior to re-ligation of the DSB. This process is more likely to result in rearrangements leading to oncogenic mutations than repair by homologous recombination. Moreover, H2AX is also phosphorylated after exposure to alkylating agents and the formation of DNA adducts which may also be oncogenic [Staszewski et al., 2008].
The incidence rate of thyroid cancer has been increasing sharply since the mid-1990s. It is now the fastest-increasing cancer in both men and women [Siegel et al., 2012]. Among women, the age-adjusted incidence (cases per 100,000) of differentiated thyroid cancer increased from 6.4 per 100,000 in 1988 to 14.9 per 100,000 in 2005 [Chen et al., 2009]. Among men, the age-adjusted incidence of well-differentiated thyroid cancer increased from 2.5 per 100,000 in 1988 to 5.1 per 100,000 in 2005. Other studies confirmed the finding of a rising incidence of thyroid cancer [Aschebrook-Kilfoy et al., 2011; Enewold et al., 2009; Lim et al., 2017]. In an analysis of the SEER cancer database, the increasing incidence of thyroid cancer was found to have started around 1995 [Chen et al., 2009; Enewold et al., 2009]. Papillary cancer that now constitutes over 85% of thyroid cancers is responsible for the largest part of the increase. Although some of this increase may be attributed to ascertainment bias due to imaging studies of the thyroid gland followed by biopsy of malignant thyroid nodules [Davies and Welch, 2006], environmental causes have not been excluded. A possible scenario is that exposure to environmental toxins may be at least partly responsible for the increased incidence of papillary thyroid cancer [Chen et al., 2009].
In order to evaluate the genotoxic activity of pesticides on thyroid cells, we adapted γH2AX measurements to a high throughput method that quantifies the cellular DNA damage response directly. This high throughput platform is useful for screening small molecules such as drugs and industrial chemicals, including pesticides and other environmental toxins, for their ability to cause DNA DSB in thyroid cells via quantification of γH2AX occurrence through laser scanning cytometry as readout on cell-by-cell resolution. Our results identified several pesticides that were able to cause DNA DSB in this human thyroid cell line that had not been detected by other genotoxicity assays.
MATERIALS AND METHODS
Cell Lines
We used the NTHY-ori-3–1 cell line now available from Sigma-Aldiich, St. Louis, MO [Lemoine et al., 1989]. NTHY-ori-3–1 cells are maintained in RPMI 1640 media containing 2 mM glutamine and 10% fetal bovine serum plus penicillin-streptomycin (100 units/mL and 0.1 mg/mL respectively; Life Technologies, Carlsbad, CA).
Reagents
Etoposide (Accord Healthcare, Durham, NC) was utilized as a positive marker for double strand DNA breaks. DSBs were probed via phospho-histone H2AX (Ser139) (20E3) rabbit mAb (Cell Signaling Technology) followed by goat anti-rabbit IgG antibody conjugated to Alexa Fluor 488 (Life Technologies). The ToxCast Phase 1 library of 309 compounds was obtained from the US Environmental Protection Agency [Judson et al., 2010]. Supporting Information Table 1 provides a list of these compounds. All other reagents were obtained from Sigma-Aldrich.
Methods
Cells were grown to 60–80% confluence in a 75-cm2 flask. The cell pellet was released using 0.25% trypsin-EDTA in 10 mL RPMI media containing 10% FBS and 200 mM Glutamax. The cell density was counted with a hemocytometer. Then cells were seeded at 1500–2000 cells per well in 384 well plates (Greiner, Bio One, Monroe, NC, 781091) for each allotment via a multidrop reagent dispenser manifold (Thermo Scientific, Canoga Park, CA). The cells were allowed to adhere at 37°C, 5% CO2 overnight. The test compounds were added using a Biomek FX (Beckman Coulter, Brea, CA) liquid handler and incubated for 24 hr. All subsequent steps of washing with PBS and dispensing of reagents were performed with the ELx405 liquid handler (BioTek Instruments, Winooska, VT) and the multidrop reagent dispenser, respectively. After 24 hr, the cells were washed and fixed in 4% paraformaldehyde for 25 min at room temperature. The plate of cells was washed again and permeabilized with 1% Triton X-100 and incubated for 1 hr at room temperature. After washing, the plate was blocked with 3% bovine serum albumin for 60 min at ambient temperature. During the remaining steps, the plates were shielded from light. Blocking agent was washed from the plate. Then the phosho-H2AX antibody diluted to 1:225 in 3% BSA, 0.3% Triton X-100 in PBS was added, and the cells were incubated overnight at 4°C. Primary antibody was washed off and secondary antibody labeled with Alexa 488 dye (1:250 in same buffer as the primary antibody) was added to each well and incubated at room temperature for 1 hr. The secondary antibody was washed off, and a propidium iodide (PI) solution (100 μg/mL RNase, 5 μM PI, 0.3% Triton X-100) added and incubated for 20 min at room temperature. This γH2AX readout was multiplexed with PI for enumeration of the total cell number for normalization. The plates were washed three times on an ELX 405 plate washer between each staining step. The plates were read on an Acumen Explorer ex3 (TTP LabTech, Melburn, England) laser scanning cytometer. This reader allows for quantification of nuclear fluorescence on a cell-by-cell level indicating the presence of γH2AX in any given cell. Etoposide was used as a positive control and DMSO (the solvent for the test compounds) as the negative control. Solvent controls indicated DMSO compatibility well over 1%. We used a maximum of 1% DMSO in our experiments. Total cell count as a measure of toxicity was assessed via the number of nuclei that was quantified by PI stain.
Caspase activity was assessed via Caspase-Glo 3/7 Assay System (Promega, Madison, WI; G8092) according to manufacturers’ protocol. NTHY-ori-3–1 cells were dissociated and seeded in the same fashion as above on white 384-well plates (Greiner Bio-One, 781080). After varying incubation durations with compound, luminescence was measured 1 hr post addition of Caspase-GLo 3/7 reagent on Analyst GT (Molecular Devices, Sunnyvale, CA).
Assay Execution
Each of the 309 compounds of the ToxCast Phase 1 library was tested in a quantitative high-throughput approach in 10 dilutions going from micromolar to nanomolar concentrations by successive twofold dilutions. Preliminary study indicated 24 hr as optimal time for incubation with the test compounds (data not shown). Compounds that caused dose-related DSB were retested in 3–6 additional assays using the NTHY-ori-3–1 human thyroid cell line for confirmation of the results. Using the Prism 5 software (Graphpad, La Jolla, CA), a dose response curve was constructed and the EC50 was calculated for the positive compounds. For time courses, the assay was executed as above but with a shorter exposure time as indicated in the individual experiments.
RESULTS
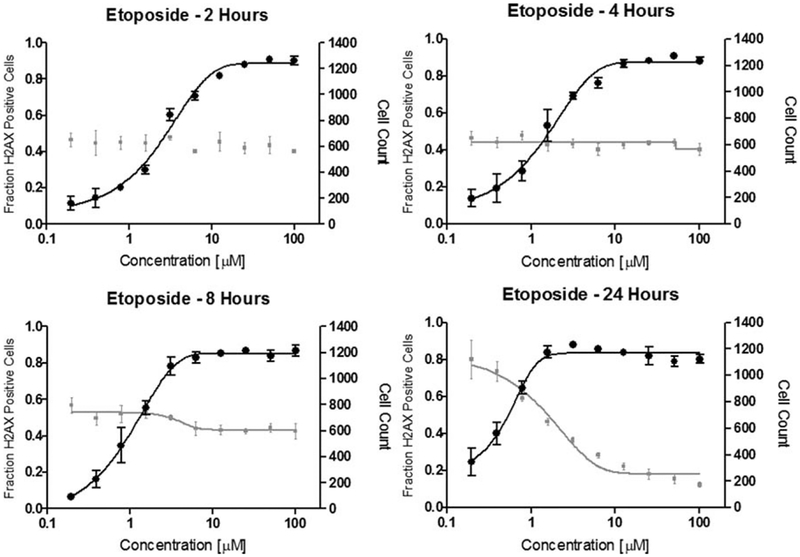
In a first step, we evaluated our test using etoposide as positive control in dose response and we also evaluated the dose response after 2, 4, 8, and 24 hr (Fig. 1) Etoposide is a topoisomerase II inhibitor that induces DNA DSBs and is used in the clinic for its anti-cancer activity. We found that the onset of the DNA damage marker γH2AX upon etoposide exposure was visible via immunohistology by eye in the image generated by the Acumen Explorer after 2 hr. This went hand in hand with a significant shift in γH2AX intensity as shown in the histogram plots (Fig. 2) and gave rise to a solid dose response curve. Moreover, the typical foci of the γH2AX lesions are visible upon examination with a confocal microscope (Fig. 3). Z′ statistics (14) yielded Z′ > 0.75 indicating an excellent assay with full compatibility for high throughput screening (HTS). (For comparison, a Z′ > 0.5 indicates a separation of positive and negative populations of more than 12 standard deviations.) The standard deviations observed were smaller than 10% for most data points. An interesting feature of this assay is its linear range over two orders of magnitude of the etoposide concentration (Fig. 1). Also, the γH2AX response is independent of cellular toxicity. For example, etoposide did not drastically reduce the total number of cells at 2, 4, or 8 hr of incubation.
Fig. 1.
A: Pseudo-image of the propidium iodide and γH2AX channel generated by laser scanning cytometry. The cells were permeabilized using Triton X100 and then subjected to staining. B: The resulting histogram in which cells showed an intensity of 200 RFUs and over were gated as positive. Even at a very low dose of 100 nM etoposide, enrichment of γH2AX positive cells can be clearly observed.
Fig. 2.
Dose response curve for DNA DSB caused by etoposide. Shown here is the fraction of nuclei showing γH2AX immunofluorescence—gated as 200 RFU and higher—divided by the total number of cells. Interestingly, the total number of cells does not change drastically although almost all cells turn γH2AX positive at high etoposide concentrations.
Fig. 3.
Confocal images of N-THY-ori-1 cells treated for 24 hr with either DMSO as vehicle, etoposide, or abamectin at 3.125 μM. Note the typical γH2AX foci located in the nuclei which are indicative of DNA DSB occurrence.
Next, we tested the ToxCast 1 library. Our results from the ToxCast library indicated that some compounds were toxic and some compounds were genotoxic. Specifically, the results showed that 19 compounds in the ToxCast 1 Library caused dose-related DNA DSB in the NTHY-ori-3–1 cells after incubation with the compound for 24 hr. Table I lists these compounds, their uses, and their lowest effective concentrations (LEC) in our assay. We used a three standard deviation cutoff above background to identify the LEC of the positive compounds. For a significant subset of the compounds, the LEC of γH2AX induction was in the single digit micromolar range which suggests significant genotoxic potential of these compounds (Table I). Representative plots of the γH2AX data for each of the compounds in Table I are shown in Supporting Information Figure 1.
TABLE I.
Pesticides That Caused DNA Double Strand Breaks and Brief Description of Principal Function
| Pesticide | Function | Lowest effective concentration (μM) | CAS number |
|---|---|---|---|
| Abamectin | insecticide, mitocide | 1.51 | 71751-41-2 |
| Amitraz | insecticide | 2.50 | 33089-61-1 |
| Captan | fungicide | 9.85 | 133-06-2 |
| Captafol | fungicide | 0.63 | 2425-06-1 |
| Difenoconazole | fungicide | 1.25 | 119446-68-3 |
| Diquat dibromide monohydrate | herbicide | 20.00 | 85-00-7/6385-62-2 |
| Fluazinam | fungicide | 10.00 | 79622-59-6 |
| 3-Iodo-2-propynyl-N-butylcarbamate | fungicide | 1.25 | 55406-53-6 |
| Maneb | fungicide | 40.00 | 12427-38-2 |
| Milbemectin | insecticide, acaricide | 19.50 | 51596-11-3/51596-10-2 |
| Naled | insecticide | 20.00 | 300-76-5 |
| Niclosamide | anti-helminthic | 1.25 | 50-65-7 |
| Prallethrin | insecticide | 4.98 | 23031-36-9 |
| Prodiamine | herbicide | 5.00 | 29091-21-2 |
| Propargite | insecticide | 10.00 | 2312-35-8 |
| Rotenone | insecticide, pesticide | 5.00 | 83-79-4 |
| Tebupirimfos | insecticide | 10.00 | 96182-53-5 |
| Tribufos | defoliant | 10.00 | 78-48-8 |
| Triclosan | fungicide, antibacterial | 20.40 | 3380-34-5 |
We also tested the ability of a subgroup of compounds to induce apoptosis as measured by caspase-3/7 activation as a function of time. What is of particular interest here is the fact that all compounds induced γH2AX after 2 hr. However, only some compounds are able to induce caspase-3/7 activation after just 2 hr (Fig. 4). For example, abamectin induced caspase-3/7 activation at 2 hr, captafol required 4 hr, and captan and etoposide required 8 hr for modest activation. Interestingly, captan showed bi-phasic behavior in terms of toxicity in this test. Similar bi-phasic behavior has been observed with other sulfur-containing compounds such as dithiothreitol (DTT) in testing toxicity based on oxidative stress [Held and Biaglow, 1994].
Fig. 4.
A: Time resolved γH2AX dose-response graph for captan, captafol, abamectin, and etoposide with cell counts. Note that the induction of DNA damage has a relatively fast onset and is substantial after only 2 hr of treatment. B: Time resolved dose-response graph for caspase-3/7 activation.
DISCUSSION
Previously, the main tests for in vitro genotoxicity screening were a bacterial gene mutation test (Ames test), the mouse lymphoma thymidine kinase gene mutation assay (MLA test), micronucleus assay, comet assay, chromosomal aberration assay, the sister chromatid exchange assay, and GreenScreen. These tests have high false negative and/or positive rates [Kirkland et al., 2005]. This is due to the fact that they measure DNA damage indirectly as a single productive recombination in the genome of the host organism. Effectively, this detection of genotoxicity is analogous to detecting rare cellular events and thus makes these tests less sensitive and accurate. In testing the ToxCast 1 library by GreenScreen, only two of the 19 compounds listed in Table I were positive: 3-Iodo-2-propynyl-N-butylcarbamate and triclosan [Knight et al., 2009]. Tsamou et al. measured γH2AX in the human hepatoma cell line, HepG2, grown in 6-well plates with FACS as readout and reported that it was superior to the Ames and MLA tests [Tsamou et al., 2012].
Audebert et al. developed a 96-well plate assay for γH2AX to assess genotoxicity caused by polycyclic aromatic hydrocarbons in HepG2 and two colorectal adenocarcinoma cell lines [Audebert et al., 2010, 2012]. The Audebert laboratory also reported that their γH2AX assay, using HepG2 cells, gave consistent results in testing 61 compounds whose genotoxic potential had already been characterized [Khoury et al., 2013]. Graillot et al. evaluated the genotoxicity of seven mixtures of pesticides using their γH2AX assay in HepG2 cells [Graillot et al., 2012]. They reported that only the fungicides, fludioxonil and cyprodinil, were genotoxic alone; however, when these compounds were tested in a mixture that also included three other pesticides, genotoxicity occurred at much lower concentrations of the mixture, suggesting that the mixture may produce genotoxic metabolites.
Shah et al. recently reported a study of the effects of 967 chemicals from the ToxCast 1 and ToxCast 2 libraries using HepG2 cells; they developed a complex analysis that included 10 endpoints in order to determine a concentration-dependent tipping point of irreversible cellular damage; γH2AX, presumably induced by oxidative stress, was one endpoint [Shah et al., 2016]. They reported that 22.7% of the chemicals showed DNA damage (termed oxidative stress) based on γH2AX immunofluorescence, a much higher proportion of genotoxic compounds in the ToxCast 1 library than we found (6.1%) using the human thyroid cell in our assay; however, the vast majority of genotoxic compounds in their study were in the ToxCast 2 library that we did not test. Of the 19 compounds listed in Table I, only triclosan was positive in their study with an LEC of 42 (iM that was similar to that which we found. The differences between the results of the two studies may be attributable to the use of different cell types, as well as possible metabolism of the compounds by the hepatic cells.
For a significant subset of the pesticides, the LEC of γH2AX induction was in the single digit micromolar range which points at significant genotoxic potential of these compounds (Table I). Of particular note are captan and captafol. Both compounds are thio-phthalimides substituted with chlorinated alkyl moieties. The difference is the size of the chloro-alky moiety which in captan’s case is a 1,1,2,2-tetrachloroethane moiety and in captafol’s case a trichloro-methane moiety. While chemically this can be expected to result in some differences in reactivity, the similarity of these compounds is obvious, and it can be expected that these compounds will have similar biological properties. Interestingly, the difference in the chloro-alkyl moiety results in a tenfold potency difference in terms of γH2AX induction, but both compounds are inducers of DNA damage. Historically, both compounds are suspected carcinogens; however, only captafol is no longer in use; captan is still a widely used pesticide [Giacinti et al., 2016]. The current maximally allowable residue of captan in strawberries is around 25 ppm, or 25 mg/L. For comparison, we observed an LEC of 9.85 μM for this pesticide in our system which equates to 2.7 ppm and is roughly one order of magnitude lower than the currently allowable residue. Interestingly, captan has been recently tied to an increased risk for myeloma [Presutti et al., 2016].
The use of pesticides designed to kill whole organisms such as insects (insecticides), fungi (fungicides), or weeds (herbicides) is still ubiquitous in agriculture [Lamberth et al., 2013] and the development of novel, safer alternatives will require us to rethink how these compounds are made, validated, and tested for their safety. This is highlighted by the fact that the mode of action through which these compounds might be inducing their genotoxic effects on thyroid cells is unclear. While the explanation of compound cytotoxicity that eventually goes hand in hand with apoptosis is tempting, the unusual kinetics of γ-H2AX and caspase-3/7 activation of some of these compounds preempts this conclusion. For example, in the case of captan, captafol, and abamectin, the γH2AX induction is visible after 2–4 hr and may or may not be accompanied by caspase-3/7 activation. These results point more toward a process that is analogous to the effects of TRAIL which can induce γH2AX within 1 hr of exposure to cells through a caspase-8 mediated signaling cascade that culminates in caspase-3/7 activation [Solier et al., 2009]. Interestingly, TRAIL leads to the generation of mutations in surviving cells and as such is “genotoxic.” Therefore, sublethal doses of toxicants can be expected—if they demonstrate genotoxic potential through γH2AX induction—to be genotoxic if they leave surviving cells that can carry the mutations forward [Lovric and Hawkins, 2010].
In conclusion, our results raise the possibility that some of these environmental toxins may have an etiologic role in the pathogenesis of thyroid cancer. The incidence rate of thyroid cancer has been increasing drastically in recent years and the results in this study suggest a potential explanation. The fact that it typically takes decades before the cancer becomes manifest makes the hypothesis difficult to prove or disprove. This puts particular emphasis on prevention through elimination of the use of genotoxic compounds. In addition, epidemiologic studies should be performed to evaluate the relationship of thyroid cancer to exposure to pesticides because there are not yet any definitive published epidemiologic studies that deal with this issue.
In summary, we implemented a high-throughput assay for measurement of DNA DSB in a human thyroid cell. We demonstrated the robustness of our novel assay using etoposide as tool compound. Screening of the ToxCast 1 Library of 309 pesticides with this assay revealed that 19 compounds caused dose-related DSB. The data provide a basis for further studies of environmental compounds as a possible cause of thyroid cancer.
Supplementary Material
ACKNOWLEDGMENT
Authors are grateful to Richard Judson and Kenneth Houck for providing the ToxCast 1 library for this study.The authors wish to acknowledge that the work was performed at the University of California Los Angeles (UCLA) Jonsson Comprehensive Cancer Center: Molecular Screening Shared Resource supported by the National Cancer Institute grant 5P30 CA016042 (to Kenneth Dorshkind) and also thank the UCLA Center for Medical Countermeasures Against Radiation for faciilities made possible by the National Institute of Allergy and Infectious Diseases grant U19AI067769–11 (William H. McBride, Joanne B. Weidhaas, John P. Chute).
Grant sponsor: U.S. Environmental Protection Agency
Footnotes
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- Aschebrook-Kilfoy B, Ward MH, Sabra MM, Devesa SS. 2011. Thyroid cancer incidence patterns in the united states by histologic type, 1992–2006. Thyroid 21:125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audebert M, Riu A, Jacques C, Hillenweck A, Jamin EL, Zalko D, Cravedi JP, et al. 2010. Use of the gammah2ax assay for assessing the genotoxicity of polycyclic aromatic hydrocarbons in human cell lines. Toxicol Lett 199:182–192. [DOI] [PubMed] [Google Scholar]
- Audebert M, Zeman F, Beaudoin R, Pery A, Cravedi JP. 2012. Comparative potency approach based on h2ax assay for estimating the genotoxicity of polycyclic aromatic hydrocarbons. Toxicol Appl Pharmacol 260:58–64. [DOI] [PubMed] [Google Scholar]
- Burdak-Rothkamm S, Prise KM. 2009. New molecular targets in radiotherapy: DNA damage signalling and repair in targeted and nontargeted cells. Eur J Pharmacol 625:151–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AY, Jemal A, Ward EM. 2009. Increasing incidence of differentiated thyroid cancer in the united states, 1988–2005. Cancer 115: 3801–3807. [DOI] [PubMed] [Google Scholar]
- Davies L, Welch HG. 2006. Increasing incidence of thyroid cancer in the united states, 1973–2002. JAMA 295:2164–2167. [DOI] [PubMed] [Google Scholar]
- De Bont R, van Larebeke N. 2004. Endogenous DNA damage in humans: A review of quantitative data. Mutagenesis 19:169–185. [DOI] [PubMed] [Google Scholar]
- Enewold L, Zhu K, Ron E, Marrogi AJ, Stojadinovic A, Peoples GE, Devesa SS. 2009. Rising thyroid cancer incidence in the united states by demographic and tumor characteristics, 1980–2005. Cancer Epidemiol Biomarkers Prev 18:784–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallmeier E, Winter JM, Cunningham SC, Kahn SR, Kern SE. 2005. Novel genotoxicity assays identify norethindrone to activate p53 and phosphorylate h2ax. Carcinogenesis 26:1811–1820. [DOI] [PubMed] [Google Scholar]
- Giacinti G, Raynaud C, Capblancq S, Simon V. 2016. Matrix-matching as an improvement strategy for the detection of pesticide residues. J Food Sci 81:T1342–T1350. [DOI] [PubMed] [Google Scholar]
- Graillot V, Takakura N, Hegarat LL, Fessard V, Audebert M, Cravedi JP. 2012. Genotoxicity of pesticide mixtures present in the diet of the french population. Environ Mol Mutagen 53:173–184. [DOI] [PubMed] [Google Scholar]
- Held KD, Biaglow JE. 1994. Mechanisms for the oxygen radicalmediated toxicity of various thiol-containing compounds in cultured mammalian cells. Radiat Res 139:15–23. [PubMed] [Google Scholar]
- Huen MS, Chen J. 2010. Assembly of checkpoint and repair machineries at DNA damage sites. Trends Biochem Sci 35:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson RS, Houck KA, Kavlock RJ, Knudsen TB, Martin MT, Mortensen HM, Reif DM, Rotroff DM, Shah I, Richard AM, et al. 2010. In vitro screening of environmental chemicals for targeted testing prioritization: The toxcast project. Environ Health Perspect 118:485–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna KK, Jackson SP. 2001. DNA double-strand breaks: Signaling, repair and the cancer connection. Nat Genet 27:247–254. [DOI] [PubMed] [Google Scholar]
- Khoury L, Zalko D, Audebert M. 2013. Validation of high-throughput genotoxicity assay screening using gammah2ax in-cell western assay on hepg2 cells. Environ Mol Mutagen 54:737–746. [DOI] [PubMed] [Google Scholar]
- Kirkland D, Aardema M, Henderson L, Muller L. 2005. Evaluation of the ability of a battery of three in vitro genotoxicity tests to discriminate rodent carcinogens and non-carcinogens i. Sensitivity, specificity and relative predictivity. Mutat Res 584:1–256. [DOI] [PubMed] [Google Scholar]
- Knight AW, Little S, Houck K, Dix D, Judson R, Richard A, McCarroll N, Akerman G, Yang C, Birrell C, et al. 2009. Evalvation of high-throughput genotoxicty assays used in profiling the US EPA Toxcast chemicals. Regul Toxicol Pharmacol 55:188–199. [DOI] [PubMed] [Google Scholar]
- Kuo LJ, Yang LX. 2008. Gamma-h2ax—A novel biomarker for DNA double-strand breaks. In Vivo 22:305–309. [PubMed] [Google Scholar]
- Lamberth C, Jeanmart S, Luksch T, Plant A. 2013. Current challenges and trends in the discovery of agrochemicals. Science 341:742–746. [DOI] [PubMed] [Google Scholar]
- Lemoine NR, Mayall ES, Jones T, Sheer D, McDermid S, Kendall-Taylor P, Wynford-Thomas D. 1989. Characterisation of human thyroid epithelial cells immortalised in vitro by simian virus 40 DNA transfection. Br J Cancer 60:897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H, Devesa SS, Sosa JA, Check D, Kitahara CM. 2017. Trends in thyroid cancer incidence and mortality in the United States, 1974–2013. JAMA 317:1338–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovric MM, Hawkins CJ. 2010. Trail treatment provokes mutations in surviving cells. Oncogene 29:5048–5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills KD, Ferguson DO, Alt FW. 2003. The role of DNA breaks in genomic instability and tumorigenesis. Immunol Rev 194:77–95. [DOI] [PubMed] [Google Scholar]
- Mladenov E, Iliakis G. 2011. Induction and repair of DNA double strand breaks: The increasing spectrum of non-homologous end joining pathways. Mutat Res 711:61–72. [DOI] [PubMed] [Google Scholar]
- Ohnishi T, Mori E, Takahashi A. 2009. DNA double-strand breaks: Their production, recognition, and repair in eukaryotes. Mutat Res 669:8–12. [DOI] [PubMed] [Google Scholar]
- Presutti R, Harris SA, Kachuri L, Spinelli JJ, Pahwa M, Blair A, et al. 2016. Pesticide exposures and the risk of multiple myeloma in men: An analysis of the north american pooled project. Int J Cancer 139:1703–1714. [DOI] [PubMed] [Google Scholar]
- Shah I, Setzer RW, Jack J, Houck KA, Judson RS, Knudsen TB, Liu J, Martin MT, Reif DM, Richard AM, et al. 2016. Using toxcast data to reconstruct dynamic cell state trajectories and estimate toxicological points of departure. Environ Health Perspect 124: 910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Singh K, Almasan A. 2012. Histone h2ax phosphorylation: A marker for DNA damage. Methods Mol Biol 920:613–626. [DOI] [PubMed] [Google Scholar]
- Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, et al. 2012. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin 62:220–241. [DOI] [PubMed] [Google Scholar]
- Solier S, Sordet O, Kohn KW, Pommier Y. 2009. Death receptor-induced activation of the chk2- and histone h2ax-associated DNA damage response pathways. Mol Cell Biol 29:68–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staszewski O, Nikolova T, Kaina B. 2008. Kinetics of gamma-h2ax focus formation upon treatment of cells with uv light and alkylating agents. Environ Mol Mutagen 49:734–740. [DOI] [PubMed] [Google Scholar]
- Tsamou M, Jennen DG, Claessen SM, Magkoufopoulou C, Kleinjans JC, van Delft JH. 2012. Performance of in vitro gammah2ax assay in hepg2 cells to predict in vivo genotoxicity. Mutagenesis 27:645–652. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.