Why some but not all individuals infected with Mycobacterium tuberculosis develop disease is poorly understood. Previous studies have revealed an important influence of the microbiota on host resistance to infection with a number of different disease agents. Here, we investigated the possible role of the individual’s microbiome in impacting the outcome of M. tuberculosis infection in rhesus monkeys experimentally exposed to this important human pathogen. Although M. tuberculosis infection itself caused only minor alterations in the composition of the gut microbiota in these animals, we observed a significant correlation between an individual monkey’s microbiome and the severity of pulmonary disease. More importantly, this correlation between microbiota structure and disease outcome was evident even prior to infection. Taken together, our findings suggest that the composition of the microbiome may be a useful predictor of tuberculosis progression in infected individuals either directly because of the microbiome’s direct influence on host resistance or indirectly because of its association with other host factors that have this influence. This calls for exploration of the potential of the microbiota composition as a predictive biomarker through carefully designed prospective studies.
KEYWORDS: microbiome, nonhuman primate, tuberculosis
ABSTRACT
The factors that determine host susceptibility to tuberculosis (TB) are poorly defined. The microbiota has been identified as a key influence on the nutritional, metabolic, and immunological status of the host, although its role in the pathogenesis of TB is currently unclear. Here, we investigated the influence of Mycobacterium tuberculosis exposure on the microbiome and conversely the impact of the intestinal microbiome on the outcome of M. tuberculosis exposure in a rhesus macaque model of tuberculosis. Animals were infected with different strains and doses of M. tuberculosis in three independent experiments, resulting in a range of disease severities. The compositions of the microbiotas were then assessed using a combination of 16S rRNA and metagenomic sequencing in fecal samples collected pre- and postinfection. Clustering analyses of the microbiota compositions revealed that alterations in the microbiome after M. tuberculosis infection were of much lower magnitude than the variability seen between individual monkeys. However, the microbiomes of macaques that developed severe disease were noticeably distinct from those of the animals with less severe disease as well as from each other. In particular, the bacterial families Lachnospiraceae and Clostridiaceae were enriched in monkeys that were more susceptible to infection, while numbers of Streptococcaceae were decreased. These findings in infected nonhuman primates reveal that certain baseline microbiome communities may strongly associate with the development of severe tuberculosis following infection and can be more important disease correlates than alterations to the microbiota following M. tuberculosis infection itself.
OBSERVATION
Tuberculosis (TB) is the leading cause of death due to a single infectious agent (1). The WHO estimates a third of the world’s population to be latently infected with Mycobacterium tuberculosis. Nevertheless, only a small percentage of those individuals exposed to M. tuberculosis develop active disease during their lifetime. Furthermore, some exposed individuals appear to be able to clear the bacilli before the establishment of an adaptive host immune response (2). The factors that determine this broad spectrum of M. tuberculosis infection outcome remain poorly defined. One important candidate is the host intestinal microbiome, which in a wide range of previous studies has been shown to influence host resistance to a variety of different infectious and inflammatory diseases both in the gut and at extraintestinal sites (3–5).
Previous studies by our group and others have revealed effects of M. tuberculosis infection and treatment on the microbiota in both mouse models and humans (6, 7). Nevertheless, none of this work has directly investigated possible associations of the microbiome with outcome of TB exposure. Laboratory mice, while important experimental models, do not present with the same TB disease spectrum observed in humans (8) and present little interindividual variation in their intestinal microbiomes (9). Even in patients, longitudinal before and after M. tuberculosis infection sampling is not possible without sampling a large population over a long duration. To circumvent these issues, we employed a nonhuman primate rhesus macaque model to examine the possible association of the microbiome with TB disease outcome. While rhesus macaques are highly susceptible to M. tuberculosis and fail to develop latent infection under most experimental settings, their variable disease progression resembles that seen in humans (8, 10, 11). Additionally, their microbiotas display a high level of interindividual variability comparable to that observed in clinical sampling (12).
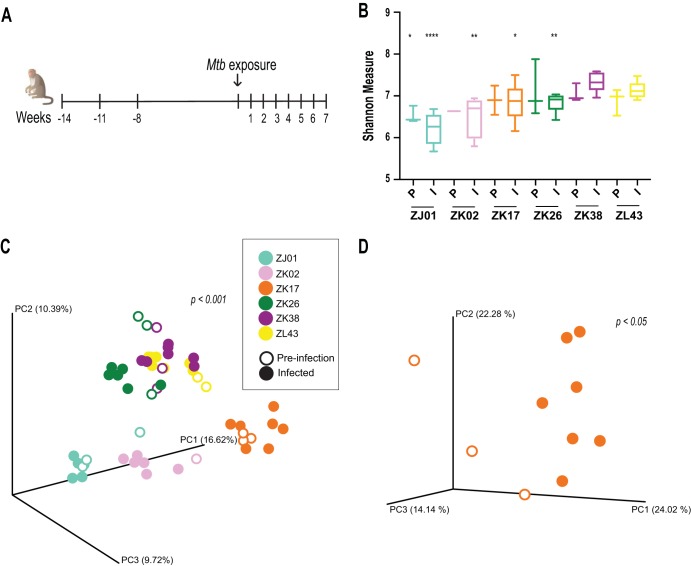
To address possible interactions between the rhesus macaque microbiome and TB disease, we analyzed the composition of the intestinal microbiota in fecal samples from monkeys in a published retrospective (13) as well as two newly performed infection studies involving different M. tuberculosis strains and intrabronchial inoculation doses. Fecal samples were collected at multiple time points pre- and postinfection (Fig. 1A), and the V4 region of the 16S rRNA gene was sequenced to determine the composition of the microbiota. In the first experiment analyzed involving six monkeys and pooling the time points from all animals, we failed to observe a significant difference in the alpha diversities before and after M. tuberculosis exposure or at different time points during infection (data not shown). However, we did observe major differences in alpha diversity between individual animals (Fig. 1B). Similarly, beta-diversity analyses using the Bray-Curtis dissimilarity index revealed that irrespective of infection status, the microbiota in the different fecal samples cluster by animal (Fig. 1C), indicating that the microbiome communities in the animals were different. When clustering analyses were performed for all of the time points from each individual animal, we observed a significant separation in the compositions of the pre- and postinfection microbiotas (Fig. 1D). Similar results were observed in the two additional independent infection experiments performed with two additional M. tuberculosis strains (see Fig. S1 in the supplemental material). Together, the above findings suggested that M. tuberculosis infection alters the intestinal microbiome in rhesus macaques, although in the absence of uninfected controls, the influence of age-related alterations cannot be ruled out. Regardless, the changes observed postinfection were of much lower magnitude than those associated with the interindividual variability of the animals involved.
FIG 1.
The interindividual variability in intestinal microbiota in rhesus macaques is greater than that induced by M. tuberculosis infection. (A) Six monkeys were exposed to <10 CFU of the Mycobacterium tuberculosis Erdman strain (Mtb) via intrabronchial instillation. Change in body weight was monitored over the course of infection as a measure of disease severity (13). Fecal samples were collected at the time points indicated (except for monkey ZK02, for which samples were collected 1 week prior to infection and for 6 weeks postinfection) to longitudinally monitor alterations in the intestinal microbiota. Background information on the animals is as follows (animal/gender [where F is female and M is male]/age in years): ZK02/F/4, ZK17/F/4.2, ZK26/F/4.2, ZK38/F/4.1, ZJ01/F/5.4, and ZL43/M/3.1. All animals were bred in Poolesville, MD, USA. Information about prior antibiotic exposure was not available. (B) Alpha-diversity estimates were calculated for each pre- and postinfection time point for each monkey using the Shannon index. The preinfection (P) and infected (I) time points were pooled and grouped by animal along the x axis. The box plot error bars indicate minimum and maximum values. Significance was then calculated between the preinfection time point of the monkey (ZK38) with the least severe disease (as determined from the PET/CT score and weight loss [13]) and the preinfection time point of the other animals. Significance between the pooled postinfection time points were calculated in a similar manner. Differences that were statistically significant are indicated (*, P < 0.05; **, P < 0.005; ****, P < 0.0001 [Student's t test]). (C) Beta-diversity clustering analyses of 16S sequence data from pre- and postinfection fecal samples of rhesus macaques were performed using the Bray-Curtis dissimilarity method, and the distances identified were visualized on a principal-component (PC) plot. Each circle represents a single time point, with the circles color coded by animal, as shown in the key. Open or closed circles indicate uninfected or infected status, respectively. Statistical testing of the Bray-Curtis distance between animals was performed using permutational multivariate analysis of variance (PERMANOVA) and was found to be significant (P < 0.001). (D) Clustering analysis of all time points for each animal was carried out independently to identify differences within pre- and postinfection microbiota. As an example, the clustering pattern for animal ZK17 (preinfection versus infected P < 0.05 [PERMANOVA]) is shown. The statistical significance of this comparison for the other animals was a P of <0.05 or lower (data not shown) except for ZK02, which was not tested due to the availability of only one preinfection time point.
Two additional infection experiments demonstrating high interindividual differences in the intestinal microbiomes of rhesus macaques before and after M. tuberculosis infection. (A, E) Outline of the experimental timeline of M. tuberculosis infection and fecal collection. Four monkeys in each experiment were exposed to 40 to 80 CFU of the M. tuberculosis Erdman-mCherry (A) or ∼18 CFU of the M. tuberculosis H37Rv-RFP (E) strain via intrabronchial instillation. Fecal samples were collected before and during the course of infection at the time points indicated. Preinfection fecal samples in panel E were collected prior to M. tuberculosis exposure. Background information on the animals is as follows (animal/gender/age in years): DF4A/M/3.8, DF8B/M/4.1, 0DH/F/4, DFN0/M/3.1, DFOX/M/3, DF1N/M/3, DF2H/M/3, and DFXA/M/3. All animals were from Morgan Island, SC, USA. Information about prior antibiotic exposure was not available. (B, F) Changes in body weight were monitored over the course of infection as a measure of disease severity. (C, G) Alpha-diversity estimates were calculated from the sequenced data of the one preinfection and all postinfection time points of each animal using the Shannon index. Preinfection (P) and infection (I) time points are grouped by animal along the x axis. (D, H) A beta-diversity clustering analysis of 16S rRNA sequence data from pre- and postinfection fecal samples of rhesus macaques was performed using the Bray-Curtis dissimilarity index and visualized here on a principal-component (PC) plot. Each circle represents a single time point, with the circles color coded by animal, as shown in the key. Open or closed circles indicate uninfected or infected status, respectively. Statistical testing of the Bray-Curtis distance between all of the animals in each experiment was performed using PERMANOVA and was found be significant (P < 0.001). Download FIG S1, PDF file, 0.6 MB (641.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
As reported previously, the rhesus macaques whose results are depicted in Fig. 1C presented with a wide range of disease severities, as determined by positron emission tomography-computed tomography (PET/CT) score and generalized weight loss (13). This enabled us to ask whether the extent of disease is associated with microbiota composition. To do so, we calculated the distance of the microbiome of each infected monkey relative to that of the healthiest macaque (ZK38) in the group and asked if that distance correlated with weight loss as a shared disease correlate. Indeed, there was a significant correlation between weight loss and the graphical distance from the time point-matched microbiome of the monkey with the least severe disease (Fig. 2A). Moreover, when clustering analysis was performed on the sequenced 16S rRNA data pooled from all three experiments described above, we observed that the animals that lost more weight grouped together and that the monkeys that were more resistant (i.e., failed to lose weight) formed a separate cluster (Fig. 2B). Of particular note, we observed this clustering pattern using just the preinfection microbiome points (Fig. S2). Employing Procrustes analysis, a tool which determines the statistical similarities of distribution patterns, no significant difference was observed in the clustering patterns between the pre- and postinfection microbiome time points (Fig. 2C). This finding suggests that it may be possible to predict the severity of TB disease progression from the composition of the baseline preinfection microbiome.
FIG 2.
Microbiota clustering in rhesus macaques associates with disease severity both before and after M. tuberculosis infection. (A) The distance of the composition of the microbiota from each macaque from the corresponding time points of the animal with the least severe disease (ZK38) was quantified from the 3D space in Fig. 1C and plotted against percent weight change over the course of infection. The significance of the entire comparison (P < 0.01) was determined by regression analysis. (B) Clustering analysis of the 16S sequence data from all time points of the three independent experiments was performed using the Bray-Curtis dissimilarity index. Each circle/triangle represents one time point and is colored by animal, as indicated in the key. Animals that developed more-severe TB as determined by weight loss are represented with circles, and monkeys that presented with mild disease are depicted with triangles. The statistical significance of the Bray-Curtis distance between all animals with severe TB and those with mild TB was determined using PERMANOVA (P < 0.001). (C) Procrustes analysis was performed between the Bray-Curtis principal-component matrices generated from independent analyses of the preinfection and infection time points (Fig. S2) in order to test the congruence of the clustering pattern between the compositions of the microbiotas at every stage of the experiment. Each black line connects the preinfection (open circle/triangle) and infected (closed circle/triangle) time points of each animal, with the length of the line indicating the extent of change. The statistical significance of the congruence of the two distance matrices was determined using Monte-Carlo simulation to be a P of <0.001, which in this test indicates the absence of a difference in the clustering pattern. (D) LEfSe comparisons indicate differentially abundant families between mild and severe TB groups pre- and postinfection. Taxa that are relatively enriched in the monkeys with mild or severe TB before as well as after M. tuberculosis infection are indicted by an asterisk. Data are filtered for a P of <0.05 and a linear discriminant analysis (LDA) score of >2. (E) Metagenomic shotgun sequencing was performed on the fecal samples from the preinfection time points. The heatmap depicts unsupervised hierarchical clustering of the preinfection time points of animals colored by disease severity. Species-level taxa that were present at >500 parts per million (PPM) in at least 10% of the samples, with an adjusted P of <0.05 (Bonferroni correction), are displayed.
Clustering analysis of pre- and postinfection microbiotas of rhesus macaques. Beta-diversity clustering analysis was performed using the Bray-Curtis dissimilarity index independently at preinfection and infection time points from all three experiments. Each circle/triangle represents the microbiota composition of an animal averaged across all preinfection or infection time points, respectively. The principal coordinates generated from this analysis were utilized in the Procrustes analysis whose results are shown in Fig. 2B. Download FIG S2, PDF file, 0.4 MB (408.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
To further validate the association of microbiota composition with disease progression, we constructed a Dirichlet multinomial mixture model (14) of the pooled data from the three experiments to identify community types. The different samples were found to partition into two community types, with all time points from each monkey grouping into the same community (Fig. S3). Specifically, all animals that presented with mild disease partitioned into one community, and monkeys with severe disease grouped into the other community. Additionally, multivariate statistical analysis, in the absence of animal identification as a metadata variable, identified disease severity and not infection status or time point during the course of infection as the parameter that most significantly associates with microbiota composition (data not shown).
Microbiota community types associated with severe versus mild disease. The Dirichlet multinomial mixture model identified two microbiota community types among the 14 monkeys from all three experiments. Each animal/time point in the Bray-Curtis principal-component plot from Fig. 2B is colored by its microbiota community type. Community types 1 and 2 are indicated in red and blue, respectively. Download FIG S3, PDF file, 0.4 MB (432KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
We next asked which taxa are statistically distinct between the macaques with severe versus mild disease. A total of 15 or 36 taxa were significantly altered between the two groups before and after infection, respectively. Among those significantly increased in the animals with severe disease both pre- and postinfection were taxa belonging to the families Lachnospiraceae and Clostridiaceae 1, while members of the family Streptococcaceae and the Bacteroidales RF16 and Clostridiales vadin B660 groups were decreased in the same group (Fig. 2D). Members of the family Erysipelotrichaceae decreased and Ruminococcaceae increased in the severe-disease group following infection (Fig. 2D), with a number of taxa fluctuating in their abundances over the course of infection (Fig. S4).
Longitudinal profiling of alterations in bacterial taxa in monkeys with mild versus severe TB disease. Average relative abundances of select bacterial families are plotted over the course of the three pooled experiments. Times are indicated in chronologic order and not by the exact time point of fecal collection. P, preinfection; I, infected. Download FIG S4, PDF file, 0.4 MB (471.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Finally, to investigate differences in the gene coding capacity of the microbiomes of animals that progressed to either severe or mild disease, we performed metagenomic shotgun sequencing on the fecal samples from the preinfection time points. In addition to corroborating the 16S rRNA gene data by showing an association between disease severity and the taxonomic structure of the microbiota (Fig. S5A, first panel), metagenomic sequencing also revealed a functional (taxonomically naive) difference between the predicted proteomes of microbiota samples as classified by seven different databases in the InterPro consortium (15) (Fig. S5A). Specifically, as also found in the 16S rRNA analysis, Roseburia intestinalis (family Lachnospiraceae), Succinivibrio dextrinosolvens, certain Ruminococcaceae, and Weissella (family Leuconostocaceae) were enriched and Streptococcus equinus (family Streptococcaceae) was decreased in some or all animals with severe disease (Fig. 2E). Although specific enzyme classes were found at different relative abundances between the two severity groups (Fig. S5B), the enzyme classes identified did not associate with a particular functional pathway.
Metagenomic shotgun sequencing and clustering analysis of the preinfection microbiome. (A) Principal-component plots depict the clustering pattern of preinfection microbiotas of animals in the mild- and severe-disease groups as determined from whole-genome shotgun sequencing. The principal-component analysis plot in the first panel is based on the relative abundances of taxa identified, and the remaining panels are based on the gene repertoire as determined by the various databases, as indicated above each panel. Statistical significance was calculated using PERMANOVA. (B) Unsupervised hierarchical clustering of the samples was performed based on their repertoire of enzyme-encoding genes. Only enzymes that were present at >50 parts per million in at least 15% of the samples at a P of <0.05 (Mann-Whitney test) were utilized for the clustering analysis and are displayed. Download FIG S5, PDF file, 0.6 MB (677.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
In a prior study employing cynomolgus macaques, modest changes in the pulmonary microbiome were detected following M. tuberculosis infection, which failed to correlate with the degree of lung inflammation observed (16). The present study focused on the intestinal microbiome of a different nonhuman primate species (rhesus macaques) and likewise showed only minor M. tuberculosis infection-induced changes in the microbiota. Instead, this work revealed a significant association between the composition of the gut microbiome and disease outcome as reflected in weight loss, which in this experimental model has been shown to reflect disease severity as determined by PET/CT score (13). Importantly, this association was evident at baseline before the animals encountered the infection, implicating the microbiome as a potential predictor of TB progression. Furthermore, this correlation evident in an initial experiment involving one M. tuberculosis strain was robustly maintained by the addition of data generated from two additional experiments employing less virulent bacterial strains. The functional significance of the observed microbiome/disease association is currently unclear. One straightforward hypothesis is that specific microbiota communities directly modify host responses involved in pathogenesis. However, an equally plausible explanation of the data is that these specific communities are instead indirect biomarkers of other host differences that themselves directly impact disease outcome. Since the mechanisms that underlie the heterogenous pathogenesis of M. tuberculosis infection in rhesus monkeys are poorly understood, it is impossible at present to distinguish between the above-described alternative hypotheses. Nevertheless, this report further highlights the need to investigate TB pathophysiology in more-relevant animal models, as recent mouse model studies of the TB microbiome interaction have revealed only a minimal role for the microbiome in host resistance to TB (17). Experiments involving large animal and human cohorts and assessing the innate as well as adaptive host resistance parameters previously linked to the microbiome are needed both to extend the association documented here and to identify possible mechanistic links between disease outcomes and the specific bacterial species associated with them.
Methods.
All experimental procedures were in compliance with protocols approved by the NIAID Animal Care and Use Committee. Fecal samples were collected and processed as described previously (18). The V4 region of the 16S rRNA gene was amplified and sequenced as previously described (18, 19). The sequence data were processed and analyzed using the QIIME2/DADA2 (20, 21) pipeline, and the operational taxonomic units (OTUs) were classified using the SILVA database (22). For alpha- and beta-diversity analyses, samples were rarefied to 23,000 reads/sample. The week 7 time point of monkey ZK26 was not included in the analyses due to insufficient numbers of reads. Procrustes analysis was performed on the pre- and postinfection beta-diversity clustering pattern using their principal-component distances to determine the congruence of the two shapes. The Dirichlet multinomial mixture model implemented in mothur (23) was used to identify community types, while LEfSe (24) was employed to identify differentially abundant taxa. Whole-genome shotgun sequencing was performed according to the Nextera DNA Flex protocol using the Illumina NextSeq 500 platform and the metagenomic data analyzed as previously described (25). Briefly, reads were first filtered for quality and removal of host DNA sequences, after which they were de novo assembled and the contigs were annotated ab initio. Taxonomic classification was performed by means of a k-mer spectrum analysis using custom databases built from NCBI genome entries. The predicted proteome from the contigs of each sample was characterized using InterProScan (15).
Data availability.
Sequence data are available in the NCBI Short Read Archive (SRA) database under BioProject ID PRJNA541010 (https://trace.ncbi.nlm.nih.gov/Traces/study/?acc=SRP194962).
ACKNOWLEDGMENTS
This work was supported in whole or part by the Intramural Research Programs of the NIAID and the NCI, NIH.
We are grateful to members of the NIAID Building 33 Animal Facility staff for their help with fecal sample collection. We also acknowledge the NIH HPC Biowulf cluster, NIAID Bioinformatics and Computational Biosciences Branch and NIH library for providing computational resources and Bruno Andrade for helpful discussions.
We declare that we have no conflicts of interest.
Footnotes
Citation Namasivayam S, Kauffman KD, McCulloch JA, Yuan W, Thovarai V, Mittereder LR, Trinchieri G, Barber DL, Sher A. 2019. Correlation between disease severity and the intestinal microbiome in Mycobacterium tuberculosis-infected rhesus macaques. mBio 10:e01018-19. https://doi.org/10.1128/mBio.01018-19.
Contributor Information
Stefan H. E. Kaufmann, Max Planck Institute for Infection Biology.
Olivier Neyrolles, CNRS - University of Toulouse.
Hardy Kornfeld, University of Massachusetts Medical School.
Cecilia Lindstam Arlehamn, La Jolla Institute for Allergy & Immunology.
REFERENCES
- 1.WHO. 2018. Global tuberculosis report 2018. WHO, Geneva, Switzerland. [Google Scholar]
- 2.Simmons JD, Stein CM, Seshadri C, Campo M, Alter G, Fortune S, Schurr E, Wallis RS, Churchyard G, Mayanja-Kizza H, Boom WH, Hawn TR. 2018. Immunological mechanisms of human resistance to persistent Mycobacterium tuberculosis infection. Nat Rev Immunol 18:575–589. doi: 10.1038/s41577-018-0025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamada N, Seo SU, Chen GY, Nunez G. 2013. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol 13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 4.Honda K, Littman DR. 2012. The microbiome in infectious disease and inflammation. Annu Rev Immunol 30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rooks MG, Garrett WS. 2016. Gut microbiota, metabolites and host immunity. Nat Rev Immunol 16:341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Namasivayam S, Sher A, Glickman MS, Wipperman MF. 2018. The microbiome and tuberculosis: early evidence for cross talk. mBio 9:e01420-18. doi: 10.1128/mBio.01420-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong BY, Maulen NP, Adami AJ, Granados H, Balcells ME, Cervantes J. 2016. Microbiome changes during tuberculosis and antituberculous therapy. Clin Microbiol Rev 29:915–926. doi: 10.1128/CMR.00096-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Garra A, Redford PS, McNab FW, Bloom CI, Wilkinson RJ, Berry MP. 2013. The immune response in tuberculosis. Annu Rev Immunol 31:475–527. doi: 10.1146/annurev-immunol-032712-095939. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen TLA, Vieira-Silva S, Liston A, Raes J. 2015. How informative is the mouse for human gut microbiota research? Dis Model Mech 8:1–16. doi: 10.1242/dmm.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharpe S, White A, Gleeson F, McIntyre A, Smyth D, Clark S, Sarfas C, Laddy D, Rayner E, Hall G, Williams A, Dennis M. 2016. Ultra low dose aerosol challenge with Mycobacterium tuberculosis leads to divergent outcomes in rhesus and cynomolgus macaques. Tuberculosis (Edinb) 96:1–12. doi: 10.1016/j.tube.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Maiello P, DiFazio RM, Cadena AM, Rodgers MA, Lin PL, Scanga CA, Flynn JL. 2018. Rhesus macaques are more susceptible to progressive tuberculosis than cynomolgus macaques: a quantitative comparison. Infect Immun 86:e00505-17. doi: 10.1128/IAI.00505-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yasuda K, Oh K, Ren B, Tickle TL, Franzosa EA, Wachtman LM, Miller AD, Westmoreland SV, Mansfield KG, Vallender EJ, Miller GM, Rowlett JK, Gevers D, Huttenhower C, Morgan XC. 2015. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe 17:385–391. doi: 10.1016/j.chom.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kauffman KD, Sallin MA, Sakai S, Kamenyeva O, Kabat J, Weiner D, Sutphin M, Schimel D, Via L, Barry CE, Wilder-Kofie T, Moore I, Moore R, Barber DL. 2018. Defective positioning in granulomas but not lung-homing limits CD4 T-cell interactions with Mycobacterium tuberculosis-infected macrophages in rhesus macaques. Mucosal Immunol 11:462–473. doi: 10.1038/mi.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmes I, Harris K, Quince C. 2012. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS One 7:e30126. doi: 10.1371/journal.pone.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, Bridge A, Brown SD, Chang HY, El-Gebali S, Fraser MI, Gough J, Haft DR, Huang H, Letunic I, Lopez R, Luciani A, Madeira F, Marchler-Bauer A, Mi H, Natale DA, Necci M, Nuka G, Orengo C, Pandurangan AP, Paysan-Lafosse T, Pesseat S, Potter SC, Qureshi MA, Rawlings ND, Redaschi N, Richardson LJ, Rivoire C, Salazar GA, Sangrador-Vegas A, Sigrist CJA, Sillitoe I, Sutton GG, Thanki N, Thomas PD, Tosatto SCE, Yong SY, Finn RD. 2019. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res 47:D351–D360. doi: 10.1093/nar/gky1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cadena AM, Ma Y, Ding T, Bryant M, Maiello P, Geber A, Lin PL, Flynn JL, Ghedin E. 2018. Profiling the airway in the macaque model of tuberculosis reveals variable microbial dysbiosis and alteration of community structure. Microbiome 6:180. doi: 10.1186/s40168-018-0560-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dumas A, Corral D, Colom A, Levillain F, Peixoto A, Hudrisier D, Poquet Y, Neyrolles O. 2018. The host microbiota contributes to early protection against lung colonization by Mycobacterium tuberculosis. Front Immunol 9:2656. doi: 10.3389/fimmu.2018.02656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Namasivayam S, Maiga M, Yuan W, Thovarai V, Costa DL, Mittereder LR, Wipperman MF, Glickman MS, Dzutsev A, Trinchieri G, Sher A. 2017. Longitudinal profiling reveals a persistent intestinal dysbiosis triggered by conventional anti-tuberculosis therapy. Microbiome 5:71. doi: 10.1186/s40168-017-0286-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108Suppl 1:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, Hickman HD, McCulloch JA, Badger JH, Ajami NJ, Trinchieri G, Pardo-Manuel de Villena F, Yewdell JW, Rehermann B. 2017. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell 171:1015–1028. doi: 10.1016/j.cell.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Two additional infection experiments demonstrating high interindividual differences in the intestinal microbiomes of rhesus macaques before and after M. tuberculosis infection. (A, E) Outline of the experimental timeline of M. tuberculosis infection and fecal collection. Four monkeys in each experiment were exposed to 40 to 80 CFU of the M. tuberculosis Erdman-mCherry (A) or ∼18 CFU of the M. tuberculosis H37Rv-RFP (E) strain via intrabronchial instillation. Fecal samples were collected before and during the course of infection at the time points indicated. Preinfection fecal samples in panel E were collected prior to M. tuberculosis exposure. Background information on the animals is as follows (animal/gender/age in years): DF4A/M/3.8, DF8B/M/4.1, 0DH/F/4, DFN0/M/3.1, DFOX/M/3, DF1N/M/3, DF2H/M/3, and DFXA/M/3. All animals were from Morgan Island, SC, USA. Information about prior antibiotic exposure was not available. (B, F) Changes in body weight were monitored over the course of infection as a measure of disease severity. (C, G) Alpha-diversity estimates were calculated from the sequenced data of the one preinfection and all postinfection time points of each animal using the Shannon index. Preinfection (P) and infection (I) time points are grouped by animal along the x axis. (D, H) A beta-diversity clustering analysis of 16S rRNA sequence data from pre- and postinfection fecal samples of rhesus macaques was performed using the Bray-Curtis dissimilarity index and visualized here on a principal-component (PC) plot. Each circle represents a single time point, with the circles color coded by animal, as shown in the key. Open or closed circles indicate uninfected or infected status, respectively. Statistical testing of the Bray-Curtis distance between all of the animals in each experiment was performed using PERMANOVA and was found be significant (P < 0.001). Download FIG S1, PDF file, 0.6 MB (641.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Clustering analysis of pre- and postinfection microbiotas of rhesus macaques. Beta-diversity clustering analysis was performed using the Bray-Curtis dissimilarity index independently at preinfection and infection time points from all three experiments. Each circle/triangle represents the microbiota composition of an animal averaged across all preinfection or infection time points, respectively. The principal coordinates generated from this analysis were utilized in the Procrustes analysis whose results are shown in Fig. 2B. Download FIG S2, PDF file, 0.4 MB (408.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Microbiota community types associated with severe versus mild disease. The Dirichlet multinomial mixture model identified two microbiota community types among the 14 monkeys from all three experiments. Each animal/time point in the Bray-Curtis principal-component plot from Fig. 2B is colored by its microbiota community type. Community types 1 and 2 are indicated in red and blue, respectively. Download FIG S3, PDF file, 0.4 MB (432KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Longitudinal profiling of alterations in bacterial taxa in monkeys with mild versus severe TB disease. Average relative abundances of select bacterial families are plotted over the course of the three pooled experiments. Times are indicated in chronologic order and not by the exact time point of fecal collection. P, preinfection; I, infected. Download FIG S4, PDF file, 0.4 MB (471.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Metagenomic shotgun sequencing and clustering analysis of the preinfection microbiome. (A) Principal-component plots depict the clustering pattern of preinfection microbiotas of animals in the mild- and severe-disease groups as determined from whole-genome shotgun sequencing. The principal-component analysis plot in the first panel is based on the relative abundances of taxa identified, and the remaining panels are based on the gene repertoire as determined by the various databases, as indicated above each panel. Statistical significance was calculated using PERMANOVA. (B) Unsupervised hierarchical clustering of the samples was performed based on their repertoire of enzyme-encoding genes. Only enzymes that were present at >50 parts per million in at least 15% of the samples at a P of <0.05 (Mann-Whitney test) were utilized for the clustering analysis and are displayed. Download FIG S5, PDF file, 0.6 MB (677.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
Sequence data are available in the NCBI Short Read Archive (SRA) database under BioProject ID PRJNA541010 (https://trace.ncbi.nlm.nih.gov/Traces/study/?acc=SRP194962).