Pathogens sense and respond to several small molecules within the GI tract to modulate expression of their virulence repertoire. Indole is a signaling molecule produced by the gut microbiota. Here we show that indole concentrations are higher in the lumen, where the microbiota is present, than in the intestinal tissue. The enteric pathogens EHEC and C. rodentium sense indole to downregulate expression of their virulence genes, as a read-out of the luminal compartment. We also identified the bacterial membrane-bound HK CpxA as an indole sensor. This regulation ensures that EHEC and C. rodentium express their virulence genes only at the epithelial lining, which is the niche they colonize.
KEYWORDS: Citrobacter rodentium, CpxA, indole, enterohemorrhagic E. coli (EHEC), locus of enterocyte effacement (LEE), microbiota
ABSTRACT
Microbial establishment within the gastrointestinal (GI) tract requires surveillance of the gut biogeography. The gut microbiota coordinates behaviors by sensing host- or microbiota-derived signals. Here we show for the first time that microbiota-derived indole is highly prevalent in the lumen compared to the intestinal tissue. This difference in indole concentration plays a key role in modulating virulence gene expression of the enteric pathogens enterohemorrhagic Escherichia coli (EHEC) and Citrobacter rodentium. Indole decreases expression of genes within the locus of enterocyte effacement (LEE) pathogenicity island, which is essential for these pathogens to form attaching and effacing (AE) lesions on enterocytes. We synthetically altered the concentration of indole in the GI tracts of mice by employing mice treated with antibiotics to deplete the microbiota and reconstituted with indole-producing commensal Bacteroides thetaiotaomicron (B. theta) or a B. theta ΔtnaA mutant (does not produce indole) or by engineering an indole-producing C. rodentium strain. This allowed us to assess the role of self-produced versus microbiota-produced indole, and the results show that decreased indole concentrations promote bacterial pathogenesis, while increased levels of indole decrease bacterial virulence gene expression. Moreover, we identified the bacterial membrane-bound histidine sensor kinase (HK) CpxA as an indole sensor. Enteric pathogens sense a gradient of indole concentrations in the gut to probe different niches and successfully establish an infection.
INTRODUCTION
There is a plethora of signals present in the human gut that mediate host-microbiota communication to maintain a homeostatic gastrointestinal (GI) environment (1, 2). The colon contains tryptophan derivatives such as indole, which is a microbiota-derived signaling molecule (3). Indole is also known to be absorbed by host cells and helps strengthen the integrity of the intestinal barrier and is regarded as a beneficial chemical cue within microbe-host interactions (4). Indole is synthesized by tryptophanase, the enzyme that catalyzes L-tryptophan conversion to indole (3) and is encoded by the tnaA gene. Both Escherichia coli (the predominant component of the Gammaproteobacteria phylum in the intestine) and Bacteroides thetaiotaomicron (B. theta) (one of the prominent species in the Bacteroidetes phylum [5]) have a tnaA gene (BT_1492 is the B. theta homolog [5]) and produce indole. The concentration of indole present in the human colon is not known. However, commensal and pathogenic strains of E. coli have been shown to produce approximately 500 μM indole in cultures (4), and the concentration of indole in human stools has been detected to be between 250 and 1,000 μM (6, 7). The continuous production of indole by the microbiota in the lumen and its absorption by the host cells suggest that a gradient concentration of indole exists in the intestine.
Enteric pathogens exploit intestinal chemistry to program virulence gene expression, leading to successful colonization of the GI tract (1). EHEC colonizes the human colon leading to outbreaks of bloody diarrhea and hemolytic-uremic syndrome (HUS) worldwide (8). EHEC virulence determinants include the production of the potent Shiga toxin that causes HUS and the genes necessary for the attaching and effacing (AE) lesion formation on enterocytes. AE lesion formation requires genes contained within the locus of enterocyte effacement (LEE) pathogenicity island (PI) (9). The LEE region contains five major operons, LEE1 to LEE5 (10) (Fig. 1A), which encode a type III secretion system (T3SS) (11), an adhesin (intimin) (12) and its receptor (Tir) (13), and effector proteins (14). T3SSs are molecular syringes that translocate bacterial effectors into the host cells, leading to changes in signaling and actin remodeling, culminating in the formation of AE lesions and contributing to overall EHEC pathogenesis (15). The ler gene (within the LEE1 operon) encodes the master regulator of the LEE genes (10) (Fig. 1A). Transcription of ler is regulated by multiple bacterium- and host-derived signals (16). This exquisite regulation is necessary because deregulation of the LEE-encoded T3SS in EHEC constitutes an energy burden that hampers this pathogen’s ability to successfully compete with the enteric microbiota for a colonization niche (17, 18).
FIG 1.
An indole concentration gradient is important to regulate virulence gene expression. (A, top) Schematic of the LEE pathogenicity island (LEE1 to LEE5 operons) and the representative virulence genes used in qRT-PCR experiments. (Bottom) Cartoon depicts the type III secretion system (encoded by the LEE) used by EHEC to inject bacterial effectors into the host cell. (B) Schematic representation for the indole concentration gradient in the human gut. Indole is produced by the microbiota in the lumen, where its concentration is higher. It is absorbed by epithelial cells, with its concentration decreasing at the epithelial lining. (C and D) Mass spectrometry measurement of indole and tryptophan (Trp) concentrations from colon tissues (CT) and colon content (CC) of C3H/HeJ mice before infection (C) and day 4 postinfection (D). Two-tailed unpaired t test was used to perform statistical analysis (n = 3 or 4 mice per group). Error bars represent standard errors of means (SEM). Values that are significantly different (P < 0.05) are indicated by a bar and asterisk. Values that are not significantly different (ns) are indicated. (E) Scheme of the EHEC tna operon depicting indole production resulting from tryptophan metabolism. (F) qRT-PCR for the expression of select virulence genes under increasing indole concentrations ranging from 1 to 250 μM. P values were determined by using one-way ANOVA followed by Bonferroni’s multiple-comparison test. Error bars represent standard deviations (SD). *, P < 0.05; **, P < 0.01. Experiments were performed in anaerobic conditions using low-glucose DMEM, and samples were harvested in late log phase. Data are representative of at least three independent experiments with three biological replicates and three technical replicates. Fold change were calculated relative to rpoA as an internal control. See also Fig. S1 in the supplemental material. (G) qRT-PCR for the expression of select virulence genes grown with 500 μM indole. Statistics were performed using unpaired t test followed by multiple comparison by Bonferroni-Dunn method. ***, P < 0.001. Experiments were performed under anaerobic conditions using low-glucose DMEM, and samples were harvested in late log phase. Data are representative of at least three independent experiments with three biological replicates and three technical replicates. Fold change were calculated relative to rpoA as an internal control. See also Fig. S1. (H) qRT-PCR analysis for the expression of virulence genes with various tryptophan concentrations. Statistics were calculated using ANOVA followed by Bonferroni’s multiple-comparison test. (F, G, and H) Experiments were performed in anaerobic conditions using low-glucose DMEM, and samples were harvested in late log phase. Data are representative of at least three independent experiments with three biological replicates and three technical replicates. Fold change were calculated relative to rpoA as an internal control. See also Fig. S1.
High expression of virulence genes is observed in late logarithmic phase. Download FIG S1, PDF file, 0.3 MB (310.3KB, pdf) .
Copyright © 2019 Kumar and Sperandio.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In this study, we show that self- and microbiota-derived indole decreases EHEC and Citrobacter rodentium LEE gene expression in vitro and during murine infection. We map the indole signaling cascade through RNA-Seq and identify the histidine sensor kinase CpxA as an indole sensor. Moreover, we also show that a gradient of indole exists within the mammalian GI tract. Indole is more prevalent in the lumen, where it is synthesized by the microbiota, and is decreased at the epithelium surface, where it is absorbed by host cells.
RESULTS
Exogenous indole decreases EHEC virulence gene expression.
Indole is synthesized from the amino acid tryptophan by the bacterial TnaA enzyme. Its levels within the mammalian intestine are not known. It has been shown that E. coli growing in vitro produces 500 μM indole (4) and that the concentration of indole in human stools is estimated to be between 250 and 1,000 μM (6, 7). Indole is produced by the microbiota that reside in the lumen and is absorbed by intestinal epithelial cells, suggesting that higher indole concentrations would be present in the lumen (4) (Fig. 1B). Congruent with this hypothesis, the concentration of indole is significantly higher in the lumen of the colon than in colonic tissues, while tryptophan concentrations remain unchanged in these compartments during either uninfected or C. rodentium-infected conditions (Fig. 1C and D).
EHEC has a tryptophanase (tnaA) gene and can produce its own indole (Fig. 1E). To assess the role of the indole gradient in regulating the expression of virulence genes, a mutant of EHEC that lacks tnaA and cannot produce its own indole was constructed. Treatment of ΔtnaA EHEC with increasing concentration of indole decreases the expression of virulence genes (LEE genes espA, espB, and eae) (Fig. 1F and G; see also Fig. S1A and B in the supplemental material). Importantly, there is no effect of its precursor molecule tryptophan on the expression of virulence genes (Fig. 1H). These data emphasize that indole specifically acts as a signaling molecule.
Endogenous and exogenous sources of indole decrease EHEC virulence gene expression.
Wild-type (WT) EHEC produces indole because it harbors tnaA. Congruent with indole decreasing LEE gene expression (Fig. 1F), LEE-encoded (espA, tir, and eae) as well as non-LEE-encoded Shiga toxin (stx2a) expression is increased in ΔtnaA EHEC, and this phenotype can be complemented (Fig. 2A and B). It is noteworthy that the increase in LEE (espA and eae genes) and stx2a expression in ΔtnaA is more pronounced in late logarithmic growth phase (Fig. S1C to E), which is when tnaA expression is increased (Fig. S1F). The LEE is required for AE lesion formation. AE lesions are characterized by remodeling of the epithelial cell cytoskeleton forming a pedestal-like structure that cups EHEC (8). In accordance with the LEE expression phenotype, AE lesion formation is enhanced in the ΔtnaA mutant strain compared to WT and complemented strains (Fig. 2C). Exogenous addition of 500 μM indole to both WT and ΔtnaA EHEC decreased the expression of virulence genes, as well as AE lesion formation. Although exogenous indole dampened virulence-related phenotypes, it had no effect on bacterial growth or viability (Fig. 1G and 2D and Fig. S2A to D). These findings are congruent with previous literature that reported that only high indole concentrations (5 mM), which are 15-fold higher than the physiological concentration measured in E. coli, have toxic effects on growth (19). More importantly, exogenously added indole (500 μM) to ΔtnaA EHEC rescues the WT EHEC phenotype (Fig. 2E and F). This combined with the fact that the ΔtnaA EHEC has no growth defect compared to WT EHEC (Fig. S1C) indicates that indole-dependent LEE regulation is not due to a metabolic defect caused by deletion of tnaA, but to indole signaling.
FIG 2.
Indole decreases the expression of virulence genes. (A) qRT-PCR analysis to compare the expression of select virulence genes from WT EHEC, ΔtnaA EHEC, ptna (tna gene on a plasmid). Experiments were performed under anaerobic conditions using low-glucose DMEM, and samples were harvested in late log phase. Data are representative of at least three independent experiments with three biological replicates and three technical replicates. Fold change was calculated relative to rpoA as an internal control. Statistical analysis was performed using one-way ANOVA, followed by Bonferroni’s multiple-comparison test. ***, P < 0.001. (B) Western blot analysis of secreted protein EspB. EHEC (WT, ΔtnaA, or ptna) were grown in low-glucose DMEM under anaerobic conditions. Cells were harvested in late logarithmic phase at the same OD600 (same number of bacterial cells), and supernatants were concentrated for secreted proteins. BSA is used as a loading control to ensure no variability in the concentration step. αEspB, anti-EspB antibody. (C) Fluorescein actin staining analysis. HeLa cells were infected with WT EHEC, ΔtnaA EHEC, or ptna EHEC. At 5 h postinfection, cells were washed and stained with FITC-phalloidin to visualize actin (green) and propidium iodide to stain for bacteria and nuclei (red). Pedestals were visualized as green puncta (white arrows). Pedestals were enumerated for each field, with each field containing approximately 20 cells. The number of pedestals per infected cell was quantified (n = 3). Error bars represent standard deviations. **, P < 0.01 by one-way ANOVA. (D) qPCR for assessing the effect of addition of 500 μM indole on virulence gene expression of WT EHEC grown anaerobically. Statistical analysis was performed using unpaired t test, followed by multiple comparison by Bonferroni-Dunn method. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (E) qRT-PCR expression analysis of select virulence genes from WT EHEC, ΔtnaA EHEC, and ΔtnaA EHEC with 500 μM exogenously added indole. Bacterial cells were grown under anaerobic conditions. Statistical significance was calculated using one-way ANOVA, followed by Bonferroni’s multiple-comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (F) Western blot comparison of the secreted protein EspB from WT and ΔtnaA EHEC grown anaerobically in the presence or absence of 500 μM indole. BSA was used as a loading control. All qRT-PCR data are representative of at least three independent experiments with three biological replicates and three technical replicates. Fold change was calculated relative to an internal control rpoA. Error bars represent standard deviations. See also Fig. S1 and S2.
High indole concentration decreases EHEC virulence without affecting its growth. Download FIG S2, PDF file, 0.5 MB (521.5KB, pdf) .
Copyright © 2019 Kumar and Sperandio.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Indole moonlights as a metabolite and a signaling molecule, and as an extracellular signal, it has been reported to require minimal concentrations of 500 μM to be active (3). We measured the indole and tryptophan concentrations within bacterial cells and in supernatants in our in vitro experiments. Exogenous addition of 500 μM indole to ΔtnaA EHEC led to the recovery of a small fraction of indole from the cellular fraction, which was comparable to the amount of indole present in the cellular fraction of WT EHEC (Fig. 3A). Indole has been reported to freely diffuse across membranes (20). Our data suggest that either a high concentration of exogenous indole is necessary to cross the bacterial membrane or that once indole enters the cell, it is quickly shuffled in as a metabolite, which could account for the decrease in its cellular concentration. WT EHEC can metabolize tryptophan; therefore, there is less tryptophan in the cellular fraction of WT EHEC than in ΔtnaA EHEC. However, less tryptophan was obtained from ΔtnaA EHEC treated with 500 μM indole, because the addition of indole decreases expression of the tryptophan synthetic pathway (Fig. 3B). Measurements of the indole concentrations from supernatants of WT or ΔtnaA EHEC in the presence or absence of 500 μM indole suggest that WT EHEC converts most of the tryptophan to indole, while there was no change in the concentrations of tryptophan in the ΔtnaA EHEC (Fig. 3C and D).
FIG 3.
Intracellular and extracellular indole levels in EHEC. (A and B) Mass spectrometry measurement of indole and tryptophan obtained from the cellular fractions. WT EHEC, ΔtnaA EHEC, and ΔtnaA EHEC with 500 μM indole were grown anaerobically in low-glucose DMEM. After 5 h, cells were centrifuged to separate the cellular and supernatant fractions. (A) Addition of 500 μM indole to ΔtnaA EHEC rescued the WT levels of indole in the cellular fraction. (B) Tryptophan concentrations were measured as a control. The heat map to the right indicates the results of RNA sequencing analysis, revealing a decrease in the expression of tryptophan biosynthesis genes (trp operon) in the presence of indole. Statistics were performed using one-way ANOVA. (C) Mass spectrometry measurement of indole obtained from the supernatant fraction. Statistics were performed using unpaired t test. (D) Tryptophan concentration obtained from the supernatant fraction. Note that the concentration of tryptophan in DMEM is 78.431 μM. One-way ANOVA was performed to calculate statistical significance. Values that are significantly different are indicated by asterisks as follows: **, P < 0.01; ***, P < 0.001. n = 3 per group. Error bars indicate standard deviations (SD). See also Fig. S1.
CpxA is an indole sensor.
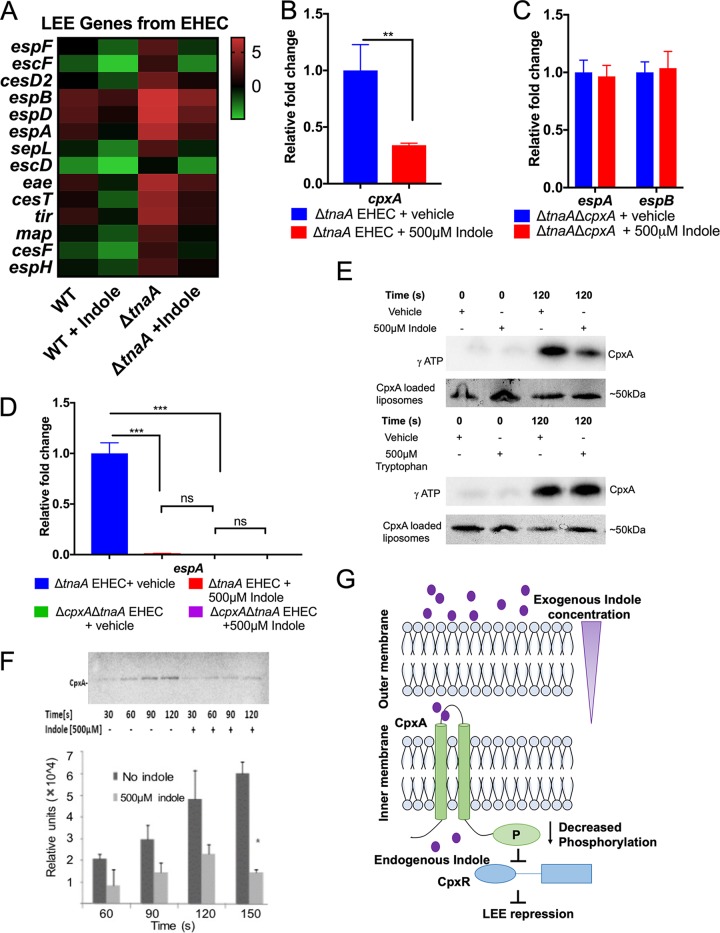
To probe the indole transcriptome, RNA-Seq was performed comparing gene expression from WT or ΔtnaA EHEC in the presence or absence of indole (GEO accession no. GSE119440). As expected, expression of the LEE genes decreased with the addition of indole (Fig. 4A). Previous studies reported sensing of small-molecule signals by bacterial membrane-bound histidine sensor kinases (HKs) (17, 21). Upon sensing a signal, HKs can either increase or decrease their autophosphorylation states, which lead to phosphorylation or dephosphorylation of response regulators (RRs) that are mostly transcription factors. Phosphorylation of RRs increases their ability to bind to DNA and regulate gene expression, while dephosphorylation inhibits their function. The cognate HK and RR pairs constitute a two-component signaling system (TCS) (22). Additionally, the signals tend to regulate expression of their sensor HKs (21). Among all the TCSs, the CpxA-CpxR system was the most significantly regulated by indole (Fig. S3A and B and Fig. S4A to C). The expression of cpxA was confirmed to be decreased with the addition of indole (Fig. 4B and Fig. S4A and B). The ΔtnaA ΔcpxA EHEC does not respond to indole to decrease transcription of the LEE (espA and espB), cpxR, and stx2a genes (Fig. 4C and D and Fig. S5A and B). Also, the ΔcpxA EHEC has decreased LEE expression and does not respond to indole (Fig. S5C and D). Additionally, autophosphorylation of CpxA reconstituted in liposomes is decreased in the presence of indole, and this phenotype is not responsive to tryptophan (Fig. 4E and F). CpxA is necessary for LEE gene activation in EHEC and virulence in C. rodentium (23, 24) (a murine pathogen extensively used as an infection model for EHEC [25], given that EHEC is strictly a human pathogen and cannot colonize mice). Therefore, CpxA senses indole as a signaling molecule to dephosphorylate itself, which can lead to dephosphorylation of CpxR, decreasing LEE gene expression (Fig. 4G). The CpxAR system is known to be activated by envelope perturbations; hence, at high toxic indole levels (2 mM), an E. coli cpxR mutant is responsive to indole, because of perturbations of membrane integrity (26).
FIG 4.
Indole regulates the virulence gene expression of EHEC through the histidine sensor kinase CpxA. (A) Heat map showing expression of LEE genes. WT EHEC and ΔtnaA EHEC in the presence or absence of 500 μM indole were grown until late logarithmic phase anaerobically. RNA sequencing was performed on the extracted RNA, and data analysis were conducted using ArrayStar. (B) qRT-PCR analysis on cpxA confirming that the presence of indole decreases its expression. △tnaA EHEC was grown anaerobically in the presence or absence of 500 μM indole. Statistical significance was calculated using unpaired t test. **, P < 0.05. (C) qRT-PCR showing that indole has no further effect on LEE4 (espA and espB) transcription in the absence of the CpxA sensor under anaerobic growth conditions. (D) qRT-PCR analysis comparing the expression of espA in the absence of cpxA with or without indole during anaerobic growth. Statistical analysis was performed using one-way ANOVA. In panels B to D, ΔtnaA EHEC was used as a background strain to avoid interference of endogenous indole. Fold change was calculated relative to rpoA as an internal control. All data are representative of two independent experiments with three biological and three technical replicates. Error bars represent standard deviations (SD). Values that are significantly different are indicated by asterisks as follows: **, P < 0.01; ***, P < 0.001. Values that are not significantly different (ns) are indicated. (E) Autophosphorylation of CpxA (loaded in liposomes) in the presence or absence of 500 μM (each) indole and tryptophan. Samples were run on 12% stain-free SDS-PAGE gel. CpxA loaded liposomes were visualized using Chemi Doc. Autoradiographs were obtained by using a phosphorimager. Data are representative of three independent experiments. (F) Time course for the autophosphorylation of CpxA (loaded in liposomes) in the presence or absence of indole. Autophosphorylation signal was quantified using Image Quant. Results were compared by unpaired t test, followed by multiple comparison by Bonferroni-Dunn method (n = 3). Error bars indicate SD. *, P < 0.05. (G) Schematic representation for indole signaling. High indole concentrations allow indole to cross the outer membrane of the enteric pathogens EHEC and C. rodentium. The histidine kinase CpxA in the inner membrane senses the presence of indole and decrease its phosphorylation leading to its decreased downstream activity and subsequent LEE repression. See also Fig. S4 and S5.
CpxA identified as an indole sensor. Download FIG S3, PDF file, 0.5 MB (570.5KB, pdf) .
Copyright © 2019 Kumar and Sperandio.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Indole signaling occurs through CpxA-CpxR node. Download FIG S4, PDF file, 0.6 MB (605.5KB, pdf) .
Copyright © 2019 Kumar and Sperandio.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A cpxA mutant is indole irresponsive and is attenuated for pathogenesis. Download FIG S5, PDF file, 0.2 MB (208.8KB, pdf) .
Copyright © 2019 Kumar and Sperandio.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Self-produced indole decreases C. rodentium virulence in mice.
Although C. rodentium is extensively used as a surrogate murine model for EHEC infections (27), it lacks the TnaA enzyme and cannot produce its own indole. To assess the effect of indole in vivo, the EHEC tna operon was inserted within the lacZ locus (lacZ does not contribute to C. rodentium murine infection [28]) of C. rodentium, which now produces indole (Fig. S6A and B). The murine microbiota was depleted as previously described (29, 30) to exclusively investigate the effect of self-produced indole in C. rodentium pathogenesis. Indole decreased the overall bacterial burden (Fig. 5A), mortality (Fig. 5B), and virulence gene expression (LEE genes espA and tir) (Fig. 5C). The levels of indole and tryptophan in vivo were measured by using mass spectrometry (Fig. 5E and F). This establishes that indole decreases the expression of virulence genes, as well as C. rodentium virulence in mice. However, indole-producing C. rodentium colonizes and causes mortality similar to that caused by WT C. rodentium in a microbiota-intact mouse (Fig. S6C and D). This is because similar amounts of indole were recovered from the stools of these mice, due to the high levels of indole being produced by the complex microbiota (Fig. S6E). These findings suggested that microbiota-derived or self-produced indole had similar effects in dictating C. rodentium infectivity. Indole signaling occurs through the CpxA HK (Fig. 4B to G and Fig. S5A to D). However, a C. rodentium cpxRA deletion strain is heavily attenuated and cannot colonize mice (24), as is a ΔcpxA strain (Fig. S5E and F), thereby limiting the ability to analyze indole signaling through the CpxRA system in vivo. Importantly, expression of cpxA and cpxR is decreased during murine infection with C. rodentium containing the tna operon (Fig. 5D). Hence, when enteric pathogens encounter high indole concentrations in the gut, indole is sensed by CpxA, which causes decreased expression of the LEE by modulating the expression and activity of CpxA (Fig. 4G).
FIG 5.
Indole limits C. rodentium colonization and pathogenesis in mice. (A) Colonization of C3H/HeJ mice with either WT C. rodentium or C. rodentium with the tna operon in microbiota-depleted mice. P value was determined by using Mann-Whitney U test. Each symbol represents the value for an individual mouse. Error bars indicate standard errors of means (SEM). (B) Survival analysis of mice infected with either WT C. rodentium or C. rodentium with the tna operon. A total of n = 15 mice per group were used for the study. Statistical significance was calculated using log rank (Mantel-Cox) test. ****, P < 0.0001. (C and D) Expression of bacterial virulence genes during murine infection. Stools from mice infected with either WT C. rodentium or C. rodentium with tna operon were collected, and RNA was extracted. qRT-PCR analysis was performed on select virulence genes espA and tir (C) and the indole sensor cpxA and its cognate response regulator cpxR (D). Fold change was determined relative to an internal control, rpoA. Each symbol represents the value for an individual mouse. Statistical significance was calculated using Mann Whitney U test. Error bars represent standard errors of means (SEM). Values that are significantly different are indicated by asterisks as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001. (E and F) Measurement of indole (E) or tryptophan (F) concentrations from fecal samples of mice infected with either WT C. rodentium or C. rodentium with the tna operon using mass spectrometry. N = 4 per group was used. Statistical analysis was performed using one-way ANOVA, followed by Bonferroni’s multiple-comparison test. Error bars indicate standard errors of means (SEM). **, P < 0.01; ns, not significant. See also Fig. S4, S5, and S6.
Self-produced indole or microbiota-derived indole dictates C. rodentium infectivity in a similar fashion. Download FIG S6, PDF file, 0.1 MB (114.6KB, pdf) .
Copyright © 2019 Kumar and Sperandio.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Microbiota-produced indole decreases C. rodentium virulence in mice.
Analysis of more than 1,000 metagenomes of American, European, and Chinese subjects showed that the tryptophanase genes are highly abundant and are present in one out of six microbial species (31). Within the human gut microbiota, tryptophanases are more prominent in Bacteroides species (31). Therefore, Bacteroides species occupying the anaerobic lumen are mainly responsible for production of the majority of the indole in the gut. To investigate the effect of microbiota-produced indole in C. rodentium murine infection, a Bacteroides thetaiotamicron (B. theta) ΔtnaA strain that cannot produce indole was constructed (Fig. S7A). The murine microbiota was depleted, and these animals were colonized with either WT or ΔtnaA B. theta (Fig. 6A). Microbiota-depleted and ΔtnaA B. theta-colonized mice had no indole detected in their stools, while mice colonized with WT B. theta had abundant levels of indole (Fig. S7B). Of note, both WT and ΔtnaA B. theta colonized mice to similar levels (Fig. S7C). The presence of indole-producing B. theta led to decreased C. rodentium burden and pathogenesis with prolonged survival of mice (Fig. 6B and C). C. rodentium LEE (espA, tir, eae, and escV) and stx2a gene expression was increased in mice colonized with ΔtnaA B. theta compared to mice colonized with the indole-producing WT B. theta (Fig. 6D). Hence, microbiota-produced indole in the lumen limits virulence of enteric pathogens. The luminal environment is anaerobic, while the epithelial surface is microaerophilic and becomes aerophilic during C. rodentium infection, leading to pathogen expansion because it is aerotolerant (32). Congruent with this scenario, EHEC LEE expression is increased during aerophilic conditions and decreases when oxygen tension is diminished, being barely detectable under anaerobiosis. Importantly, this decrease under anaerobiosis is dependent on indole, given that a ΔtnaA EHEC expresses high levels of the LEE proteins under anaerobiosis (Fig. 7A to C).
FIG 6.
Synthetically altering the microbiota to remodel gut indole concentration dictates C. rodentium infectivity. (A) Schematic representation of the microbiota remodeling and C. rodentium murine infection experiments. D, day; Bt, Bacteroides thetaiotaomicron. (B to D) C3H/HeJ mice precolonized with either WT B. thetaiotaomicron (B. theta) or ΔtnaA B. theta were infected with WT C. rodentium. (B) Mice were monitored for the C. rodentium colonization. Statistical significance was calculated using two-sided Mann-Whitney U test. Each symbol indicates the value for an individual mouse. (C) Survival of mice. P value was determined by using log rank (Mantel-Cox) test. A total of n = 15 mice per group was used. (D) Expression of select virulence genes. Statistical significance was calculated using unpaired t test, followed by multiple comparison using Bonferroni-Dunn method. Error bars represent standard errors of means (SEM). *, P < 0.05. See also Fig. S7.
FIG 7.
EHEC changes its virulence expression in different oxygenation conditions. (A to C) WT and △tnaA EHEC were grown under anaerobic (A), microaerophilic (B), or aerophilic (C) conditions. The cells were harvested in late log phase. The supernatants were collected, and the secreted protein fractions were probed using EspB antibody. BSA was used as a loading control. See also Fig. S1.
Bacteroides thetaiotaomicron (B. theta) murine colonization is not affected by tnaA. Download FIG S7, PDF file, 0.2 MB (173.3KB, pdf) .
Copyright © 2019 Kumar and Sperandio.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Enteric pathogens navigate the complex intestinal chemistry to find a suitable colonization niche. The microbiota plays an important part in shaping this chemistry (30, 33, 34) and has key interactions with enteric pathogens. These interactions are complex, with certain signals such as fucose decreasing EHEC LEE expression (17), while others such as succinate leads to increased pathogenesis (30). Both these signals are either made available or produced by a prominent member of the microbiota, B. theta. EHEC has a very low infectious dose, which is estimated to be 50 CFU (8), and colonizes the colon where the microbiota is very dense. In the colon, the major source for carbon is the mucus, which is decorated with sugars such as fucose. These sugars can be harvested by saccharolytic members of the microbiota, such as B. theta, and made available to other bacterial species that lack this capability (35). EHEC senses the fucose released from the mucus by B. theta to sense the lumen environment through the two-component system FusKR. In response to fucose, FusKR modulates EHEC’s metabolism to optimize its growth and decrease competition for carbon sources with commensal E. coli. It also decreases expression of the LEE genes that are not necessary in this gut compartment (17). As infection progresses, mucinases of EHEC and C. rodentium degrade the mucus layer (30). Importantly, because the main source of sugars in the colon is the mucus, this pathogen-induced obliteration of the mucus layer creates a nutrient-poor environment, which is gluconeogenic. Additionally, in this scenario, colonization of mice by B. theta profoundly changes the metabolic landscape of the murine gut, with organic acids such as succinate being enhanced (30, 36, 37). Succinate is then sensed by Cra to induce LEE expression at the epithelial lining (30, 38).
Indole is known to be produced by the microbiota and is present in the intestine. Indole has also been shown to be absorbed by the intestinal epithelial cells to improve barrier function (4). However, whether concentrations of indole vary between the lumen versus the vicinity of the epithelium was not established. Here we show that indole concentrations are indeed higher in the lumen compared to the epithelium (Fig. 1B to D). These data suggest that indole can also be used as a signal sensed by enteric pathogens to navigate the gut biogeography. Here we show that by sensing indole concentrations in the gut, EHEC and C. rodentium discriminate between the luminal environment, where expression of the T3SS is an unnecessary energy burden, from the epithelial surface, where T3SS expression is needed for host colonization. These findings establish a mechanism through which the microbiota confers resistance to pathogens. Moreover, they open the possibility that manipulation of indole concentrations in the GI tract by pre- or probiotics that produce indole can limit virulence of enteric pathogens.
MATERIALS AND METHODS
Strains, plasmids, and growth and culture conditions.
Strains and plasmids used in this study are listed in Table S1 in the supplemental material. WT enterohemorrhagic E. coli (EHEC) O157:H7 strain 86-24, Citrobacter rodentium (DBS770), and their isogenic mutants were routinely grown in LB overnight at 37°C. Bacteroides thetaiotaomicron (B. theta) VPI-5482 was routinely grown anaerobically overnight at 37°C in TYG medium. To express the type III secretion system (T3SS), low-glucose (1 g/liter) Dulbecco’s modified Eagle medium (DMEM) was used, as these conditions have been shown to induce the T3SS (38). Anaerobic growth was performed using either GasPak EZ anaerobe container system (Becton Dickinson) or Bactron EZ anaerobic chamber (Sheldon Manufacturing). For microaerophilic conditions, cultures were left standing in a 37°C incubator. For aerophilic conditions, cultures were grown under shaking conditions at 37°C and 250 rpm. Bacterial strains were grown in low-glucose DMEM, and the cultures were harvested in late logarithmic growth phase unless stated otherwise. HeLa cells were routinely cultured in high-glucose DMEM, defined as 4.5 g/liter glucose, DMEM, 10% FBS, and penicillin plus streptomycin plus glutamine (PSG) cocktail.
Bacterial strains. Download Table S1, PDF file, 0.3 MB (265.3KB, pdf) .
Copyright © 2019 Kumar and Sperandio.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Recombinant DNA techniques.
All primers for used for mutant and plasmid construction can be found in Table S2.
Oligonucleotides used in this study. Download Table S2, PDF file, 0.1 MB (78.9KB, pdf) .
Copyright © 2019 Kumar and Sperandio.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Construction of deletion mutants of EHEC and Citrobacter rodentium.
Isogenic mutants of EHEC 86-24 strain were created using the λ red recombination technique (39). Briefly, pKD4 was used to create the deletion PCR products. pKD46-expressing strain was used to perform the recombination and pCP20 to resolve the insertions. All mutant strains were confirmed by sequencing.
Construction of deletion mutant of B. theta.
The tnaA mutant was created in the B. thetaiotaomicron VPI-5482 Δtdk background, a gene encoding thymidine kinase as previously described (40). Briefly, the tnaA (BT1492) gene with ∼750-bp upstream and downstream sequence were ligated into a suicide vector, pExchange-tdk. The resulting construct was transformed into competent S17-1 λ pir. The knockout construct was conjugated in a B. theta Δtdk strain by growing them together. The integrant were selected on BHI-blood agar plates with 200 μg/ml gentamicin and 25 μg/ml erythromycin. The integrants were cultured in TYG medium overnight and then plated onto BHI-blood agar plates containing 200 μg/ml 5-fluoro-2-deoxyuridine (FUdR) to select for successful recombinants. The ΔtnaA B. theta deletions were screened by PCR and confirmed by sequencing.
Western blotting for secreted proteins.
From cultures grown in DMEM, secreted proteins were isolated as previously described (11). Ten micrograms of bovine serum albumin (BSA) was added to secreted protein samples as a loading control. Proteins were separated on a 5 to 20% SDS-polyacrylamide gel, transferred to a polyvinylidene fluoride (PVDF) membrane, and blocked with 3% milk in PBS containing 0.05% Tween (PBST). Membranes were probed with either anti-EspB or anti-EspA primary antibody, washed, and then incubated with a secondary rabbit antibody conjugated to streptavidin-horseradish peroxidase. Enhanced chemiluminescence (ECL) reagent was added, and the membranes were developed using the ChemiDoc touch imaging system (software 1.0.0.15) with Image Lab 5.2.1 software for image display.
RNA extraction and qRT-PCR.
Strains were grown in the presence or absence of indole until late log phase. RNA from three biological replicates was extracted using the RiboPure bacterial isolation kit according to the manufacturer’s protocols (Ambion). Quantitative reverse transcription-PCR (qRT-PCR) was performed as follows. Briefly, 2 μg of diluted extracted RNA was converted to cDNA with addition of superscript, random primers, DTT, and dNTPs. Validated primers (Table S2) and SYBR green were added to the cDNA, and the mix was run in Quantstudio 6 flex (Applied Biosystems). Data were collected using QuantStudio real-time PCR software v1.3, normalized to endogenous rpoA levels, and analyzed using the comparative cycle threshold (CT) method. For all the in vitro experiments, error bars indicate standard deviations (SD). A P value of less than 0.05 was considered significant.
Fluorescein actin staining assays.
Fluorescein actin staining (FAS) assays were performed as described previously (41). Briefly, HeLa cells were grown overnight to about 80% confluence at 37°C and 5% CO2 on coverslips in wells containing high-glucose (4.5 g/liter) DMEM. Prior to infection, fresh DMEM lacking antibiotics replaced overnight medium. To infect HeLa cells, late-log-phase bacterial cultures with equal CFU grown in low-glucose DMEM anaerobically for 5 h were used. HeLa cells were infected with an MOI of 100. After 5 h, the coverslips were washed, fixed, and permeabilized. The samples were treated with fluorescein isothiocyanate (FITC)-labeled phalloidin to visualize actin accumulation and propidium iodide to visualize bacterial DNA and HeLa nuclei. The coverslips were then mounted on slides and imaged with a confocal microscope (Zeiss LSM 880 confocal/multiphoton) at 60× resolution. The number of pedestals per HeLa cell was quantified. Replicate coverslips from multiple experiments were quantified, and statistical analyses were performed using Student’s unpaired t test, followed by multiple comparison by Bonferroni-Dunn method wherever required. A P value of less than 0.05 was considered significant.
RNA sequencing library preparation and analysis.
Briefly, RNAs extracted from three biological replicates were used to perform RNA sequencing experiments. Sequencing was run at the University of Texas (UT) Southwestern Medical Center Next Generation Sequencing (NGS) core. RNA libraries were prepared using Illumina ScriptSeq Complete kit (Bacteria). RNA libraries were run on an Illumina HiSeq 2500 sequencer with SE-50. To analyze the data, DNASTAR Lasergene software was used. Reads were mapped to the Escherichia coli O157:H7 strain sakai genome (NCBI txid 386585). Data were analyzed using ArrayStar, and all experiments were normalized by reads assigned per kilobase of target per million mapped reads (RPKM). Statistical significance was calculated using Student’s t test followed by FDR (Benjamini Hochberg) correction. A P value of less than 0.05 was considered significant. All experiments were normalized by reads assigned per kilobase of target per million mapped reads (RPKM).
Membrane protein purification.
E. coli (BL21 DE3) bacteria harboring pET21a-CpxA (His tagged) were grown until stationary phase (optical density [OD] of ∼0.7 to 0.8). Cultures were induced with 0.1 mM IPTG and grown further at 18°C overnight. The bacterial cultures were spun down at 10,000 rpm and 4°C for 30 min, and the pellets were stored at −80°C until further use. The bacterial pellets were resuspended in lysis buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 2% lauryldimethylamine N-oxide [LDAO], 10% glycerol), and protease inhibitor cocktail (PIC) was added. The lysates were sonicated for 30 s (on) and 30 s off for 8 min at 85% amplitude, followed by centrifugation at 10,000 rpm and 4°C for 30 min. The supernatants were filtered using 0.22-μm filter tube. Ni-NTA beads (500 μl) were added to 40 ml of lysate and kept on a shaker for up to 2 h at 4°C to allow protein binding to the Ni beads. The bound lysates were passed through 5-ml disposable columns twice. The column was washed with 5 column volumes (CV) of wash buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 30 mM imidazole). Proteins were eluted from the column by adding elution buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 250 mM imidazole). The eluted proteins were concentrated using Amicon Ultra 15 centrifugal filter units (10 kDa), and excess imidazole was removed by adding 50 mM Tris (pH 8.0) and 150 mM NaCl as a dialyzing buffer. The protein concentration was estimated using Bradford assay and were immediately loaded on liposomes for autophosphorylation assays.
Autophosphorylation assays.
The autophosphorylation experiments were performed as previously described (21). Liposomes were reconstituted as described previously (21). Briefly, 50 mg of E. coli phospholipids (Avanti Polar Lipids; 20 mg/ml in chloroform) were evaporated and then dissolved into 5 ml phosphate buffer containing 80 mg N-octyl-β-D-glucopyranoside. The solution was dialyzed overnight against phosphate buffer. The resulting liposome suspension underwent freeze/thawing in liquid N2. The liposomes were stored at −80°C until further use. The liposomes (1 ml) were then destabilized by the addition of 5.8 mg dodecylmaltoside, and CpxA-His was added in the ratio of 40:1, stirring at room temperature for 10 min. Biobeads (58 mg) were then added to remove the detergent, and the resulting solution was allowed to incubate at 4°C overnight. The supernatant was then incubated with fresh Biobeads (58 mg) for another hour. The resulting liposomes containing reconstituted CpxA-His was used for autophosphorylation experiments. Ten microliters of the CpxA-His liposome was adjusted to 10 mM MgCl2,1 mM DTT, 500 μM indole, 500 μM tryptophan or no signal, freeze/thawed rapidly in liquid N2, and kept at room temperature for 1 h. [γ-32P]ATP (0.25 μl) was added to each reaction mixture. At each time point, 2 μl of 5× SDS loading buffer was added to stop the reaction. The samples were run on 12% SDS-PAGE according to standard procedures and visualized via a phosphorimager (Typhoon FLA 9500; GE).
Indole colorimetric test.
Portions (50 μl) of bacterial cultures were added with 50 μl of either p-dimethylaminocinnamaldehyde (DMACA) indole reagent droppers (Becton Dickinson) or with BBL indole reagent droppers (Becton Dickinson). The presence of indole was detected by change in color. The presence of indole was observed by formation of green coloration on treatment with DMACA indole reagent and pink color with BBL indole reagent dropper.
Murine infections.
Three- to 4-week-old female C3H/HeJ mice were purchased from The Jackson Laboratory and housed in a specific-pathogen-free facility at the UT Southwestern Medical Center. All experiments were performed under IACUC-approved protocols.
Microbiota depletion method.
At 3 to 4 weeks of age, female C3H/HeJ mice were orally administered a combination of four antibiotics, ampicillin, neomycin, metronidazole, and vancomycin (Sigma-Aldrich), via oral gavage for 2 days to deplete gut microbiota (5 mg of each antibiotic per mouse per day). Fecal pellets were collected before and after antibiotic treatment to confirm depletion of the gut microbiota. Feces were resuspended in PBS at 1 g/ml and plated on brain heart infusion (BHI)-blood agar plates containing no antibiotics. Colony counts were performed after 48-h incubation at 37°C under both aerobic and anaerobic conditions.
B. theta reconstitution experiment.
Following antibiotic treatment, half of the mice were administered 3 × 109 CFU WT B. theta, while the remainder of the mice were given 3 × 109 CFU ΔtnaA B. theta via oral gavage. Twenty-four hours later, mice were mock infected with PBS or orally infected with 1 × 107 CFU strain DBS770. The experiments were performed twice with a total of 15 mice per group.
Infection with C. rodentium with the tna operon.
C. rodentium with tna operon was generated by inserting the tna operon from EHEC in the lacZ locus of the WT C. rodentium (DBS770) strain. Microbiota-depleted mice were infected with 1 × 107 CFU of either WT DBS770 or DBS770 with the tna operon. On the indicated days, feces were collected for colonization, qRT-PCR, and survival analysis. The experiments were performed twice with a total of 15 mice per group.
Colony enumeration.
The feces from infected mice were collected and placed in microcentrifuge tubes on ice. Feces were resuspended in PBS, normalized to feces weight, and plated on plates with the appropriate media and antibiotics. The colonies were counted, and the statistical comparison between groups were performed by unpaired Mann-Whitney U test.
Tissue collection for mass spectrometry, RNA isolation, and qRT-PCR.
At the indicated time points, mice were euthanized, and the colon tissue and content were collected. The tissue was washed in PBS twice to remove any residual fecal content. The content and tissues were snap-frozen in liquid nitrogen. Mass spectrometry analysis was performed to estimate the amount of indole and tryptophan in those fractions. The feces from infected mice were collected in tubes. The tubes were snap-frozen in liquid nitrogen and stored in −80°C until use. RNA was isolated from individual mouse fecal pellets using RNeasy Power Microbiome kit (Qiagen) per the manufacturer’s instructions. Monocolonization of B. theta exacerbates C. rodentium infectivity (30), causing mice to be very sick with very little fecal content obtained from each mouse. In such cases, fecal samples from three or four mice in the same cage were combined and RNA was extracted from the combined material. A total of 10 mice per group were used to obtain RNA from the experiment. qRT-PCR was performed as described earlier using Quantstudio 6 flex (Applied Biosystems). rpoA was used as an internal control for Citrobacter rodentium. Significance was determined either by unpaired t test or Mann-Whitney U test as stated in figure legends.
Mass spectrometry analysis of tryptophan and indole.
Tissue and content/feces weights were homogenized in a threefold volume of methanol (3 × weight of tissue in grams = volume methanol in milliliters; total homogenate volume [in milliliters] = 4 × weight of tissue). Homogenates were made using a BeadBug microtube homogenizer run for 2 min at 2,800 rpm and BeadBug-prefilled tubes with 3.0-mm zirconium beads (catalog no. Z763802; Sigma). Standards were made by spiking 100 μl of control BM homogenate with various known concentrations of indole and processed like samples. Tryptophan standards used BM homogenate diluted 1:5,000 in methanol and were spiked with various known concentrations of tryptophan. The tissue homogenate samples were incubated at room temperature for 10 min. Samples were spun at 13,000 for 5 min. The supernatant (100 μl) was combined 50:50 with ddH2O containing 0.2% formic acid and 100 ng/ml 13Trp IS and spun again, The supernatant was transferred to an HPLC vial w/insert and analyzed by HPLC/MS. The Qtrap 6500 and analytical conditions follow: buffer A was ddH2O plus 0.1% formic acid; buffer B was MeOH plus 0.1% formic acid; column, Agilent C18 XDB column with 5-μm packing and 50- by 4.6-mm size. The gradient conditions were as follows: 0 to 1.0 min, 0% buffer B; 1 to 1.5 min, gradient to 100% buffer B; 1.5 to 3.5 min, 100% buffer B; 3.5 to 4.0 min, gradient to 0% buffer B; 4 to 5 min, 0% buffer B. The ion source/gas parameters follow: CUR = 30, CAD = 9, IS = 5500, TEM = 200, GS1 = 20, GS2 = 10. Indole transition, 118.0 to 91.0; tryptophan transition, 205.09 to 188; 13 Trp IS transition: 216.043 to 155.2. Samples were injected twice, once at 20 μl with the indole standard curve and again at 5 μl with the tryptophan standard curve. This was done instead of making dilutions.
Quantification and statistical analysis.
For all in vitro experiments (qRT-PCR and FAS) and mass spectrometry data, a Student’s unpaired t test was used when comparing two groups and a one-way ANOVA was used when comparing more than two groups. Multiple-comparison correction using Bonferroni-Dunn method was used wherever required. For experiments with mice, the nonparametric Mann-Whitney U test was used to determine the statistical significance. Statistics for survival analysis was calculated using log rank (Mantel-Cox) test. A P value of <0.05 was considered significant. Detailed information on the numbers of biological samples and animals used can be found in the figure legends.
Data and software availability.
RNA-Seq data can be accessed using accession number (GSE119440) at the NCBI GEO database.
ACKNOWLEDGMENTS
We thank Sebastian Winter for critically reviewing the manuscript. We thank Noelle Williams and Jessica Kilgore from the UT Southwestern Pharmacology Core facility for her help with the indole measurements.
This study was supported by NIH grants AI053067, AI05135, AI077613, and AI114511 to V.S.
A.K. conceived the studies, performed experiments, analyzed data, and wrote the paper. V.S. supervised all experiments, analyzed data, and wrote the paper.
Footnotes
Citation Kumar A, Sperandio V. 2019. Indole signaling at the host-microbiota-pathogen interface. mBio 10:e01031-19. https://doi.org/10.1128/mBio.01031-19.
REFERENCES
- 1.Baumler AJ, Sperandio V. 2016. Interactions between the microbiota and pathogenic bacteria in the gut. Nature 535:85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cameron EA, Sperandio V. 2015. Frenemies: signaling and nutritional integration in pathogen-microbiota-host interactions. Cell Host Microbe 18:275–284. doi: 10.1016/j.chom.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang D, Ding X, Rather PN. 2001. Indole can act as an extracellular signal in Escherichia coli. J Bacteriol 183:4210–4216. doi: 10.1128/JB.183.14.4210-4216.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bansal T, Alaniz RC, Wood TK, Jayaraman A. 2010. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc Natl Acad Sci U S A 107:228–233. doi: 10.1073/pnas.0906112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 6.Karlin DA, Mastromarino AJ, Jones RD, Stroehlein JR, Lorentz O. 1985. Fecal skatole and indole and breath methane and hydrogen in patients with large bowel polyps or cancer. J Cancer Res Clin Oncol 109:135–141. doi: 10.1007/BF00391888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuccato E, Venturi M, Di Leo G, Colombo L, Bertolo C, Doldi SB, Mussini E. 1993. Role of bile acids and metabolic activity of colonic bacteria in increased risk of colon cancer after cholecystectomy. Dig Dis Sci 38:514–519. doi: 10.1007/BF01316508. [DOI] [PubMed] [Google Scholar]
- 8.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 9.McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci U S A 92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mellies JL, Elliott SJ, Sperandio V, Donnenberg MS, Kaper JB. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol Microbiol 33:296–306. doi: 10.1046/j.1365-2958.1999.01473.x. [DOI] [PubMed] [Google Scholar]
- 11.Jarvis KG, Giron JA, Jerse AE, McDaniel TK, Donnenberg MS, Kaper JB. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc Natl Acad Sci U S A 92:7996–8000. doi: 10.1073/pnas.92.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jerse AE, Yu J, Tall BD, Kaper JB. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci U S A 87:7839–7843. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kenny B, DeVinney R, Stein M, Reinscheid DJ, Frey EA, Finlay BB. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511–520. doi: 10.1016/S0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 14.Elliott SJ, Wainwright LA, McDaniel TK, Jarvis KG, Deng YK, Lai LC, McNamara BP, Donnenberg MS, Kaper JB. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol Microbiol 28:1–4. [DOI] [PubMed] [Google Scholar]
- 15.Stevens MP, Frankel GM. 2014. The locus of enterocyte effacement and associated virulence factors of enterohemorrhagic Escherichia coli. Microbiol Spectr 2:EHEC-0007-2013. doi: 10.1128/microbiolspec.EHEC-0007-2013. [DOI] [PubMed] [Google Scholar]
- 16.Mellies JL, Lorenzen E. 2014. Enterohemorrhagic Escherichia coli virulence gene regulation. Microbiol Spectr 2:EHEC-0004-2013. doi: 10.1128/microbiolspec.EHEC-0004-2013. [DOI] [PubMed] [Google Scholar]
- 17.Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V. 2012. Fucose sensing regulates bacterial intestinal colonization. Nature 492:113–117. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC, Nunez G. 2012. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garbe TR, Kobayashi M, Yukawa H. 2000. Indole-inducible proteins in bacteria suggest membrane and oxidant toxicity. Arch Microbiol 173:78–82. doi: 10.1007/s002030050012. [DOI] [PubMed] [Google Scholar]
- 20.Pinero-Fernandez S, Chimerel C, Keyser UF, Summers DK. 2011. Indole transport across Escherichia coli membranes. J Bacteriol 193:1793–1798. doi: 10.1128/JB.01477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. 2006. The QseC sensor kinase: a bacterial adrenergic receptor. Proc Natl Acad Sci U S A 103:10420–10425. doi: 10.1073/pnas.0604343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung K, Fried L, Behr S, Heermann R. 2012. Histidine kinases and response regulators in networks. Curr Opin Microbiol 15:118–124. doi: 10.1016/j.mib.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 23.De la Cruz MA, Morgan JK, Ares MA, Yáñez-Santos JA, Riordan JT, Girón JA. 2016. The two-component system CpxRA negatively regulates the locus of enterocyte effacement of enterohemorrhagic Escherichia coli involving sigma(32) and Lon protease. Front Cell Infect Microbiol 6:11. doi: 10.3389/fcimb.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Thomassin JL, Giannakopoulou N, Zhu L, Gross J, Salmon K, Leclerc JM, Daigle F, Le Moual H, Gruenheid S. 2015. The CpxRA two-component system is essential for Citrobacter rodentium virulence. Infect Immun 83:1919–1928. doi: 10.1128/IAI.00194-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mallick EM, McBee ME, Vanguri VK, Melton-Celsa AR, Schlieper K, Karalius BJ, O’Brien AD, Butterton JR, Leong JM, Schauer DB. 2012. A novel murine infection model for Shiga toxin-producing Escherichia coli. J Clin Invest 122:4012–4024. doi: 10.1172/JCI62746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raffa RG, Raivio TL. 2002. A third envelope stress signal transduction pathway in Escherichia coli. Mol Microbiol 45:1599–1611. doi: 10.1046/j.1365-2958.2002.03112.x. [DOI] [PubMed] [Google Scholar]
- 27.Luperchio SA, Schauer DB. 2001. Molecular pathogenesis of Citrobacter rodentium and transmissible murine colonic hyperplasia. Microbes Infect 3:333–340. doi: 10.1016/S1286-4579(01)01387-9. [DOI] [PubMed] [Google Scholar]
- 28.Mundy R, Pickard D, Wilson RK, Simmons CP, Dougan G, Frankel G. 2003. Identification of a novel type IV pilus gene cluster required for gastrointestinal colonization of Citrobacter rodentium. Mol Microbiol 48:795–809. doi: 10.1046/j.1365-2958.2003.03470.x. [DOI] [PubMed] [Google Scholar]
- 29.Kuss SK, Best GT, Etheredge CA, Pruijssers AJ, Frierson JM, Hooper LV, Dermody TS, Pfeiffer JK. 2011. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science 334:249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Curtis MM, Hu Z, Klimko C, Narayanan S, Deberardinis R, Sperandio V. 2014. The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe 16:759–769. doi: 10.1016/j.chom.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Devlin AS, Marcobal A, Dodd D, Nayfach S, Plummer N, Meyer T, Pollard KS, Sonnenburg JL, Fischbach MA. 2016. Modulation of a circulating uremic solute via rational genetic manipulation of the gut microbiota. Cell Host Microbe 20:709–715. doi: 10.1016/j.chom.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez CA, Miller BM, Rivera-Chávez F, Velazquez EM, Byndloss MX, Chávez-Arroyo A, Lokken KL, Tsolis RM, Winter SE, Bäumler AJ. 2016. Virulence factors enhance Citrobacter rodentium expansion through aerobic respiration. Science 353:1249–1253. doi: 10.1126/science.aag3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Antunes LC, Andersen SK, Menendez A, Arena ET, Han J, Ferreira RB, Borchers CH, Finlay BB. 2011. Metabolomics reveals phospholipids as important nutrient sources during Salmonella growth in bile in vitro and in vivo. J Bacteriol 193:4719–4725. doi: 10.1128/JB.05132-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu W, Winter MG, Byndloss MX, Spiga L, Duerkop BA, Hughes ER, Büttner L, de Lima Romão E, Behrendt CL, Lopez CA, Sifuentes-Dominguez L, Huff-Hardy K, Wilson RP, Gillis CC, Tükel Ç, Koh AY, Burstein E, Hooper LV, Bäumler AJ, Winter SE. 2018. Precision editing of the gut microbiota ameliorates colitis. Nature 553:208–211. doi: 10.1038/nature25172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marcobal A, Southwick AM, Earle KA, Sonnenburg JL. 2013. A refined palate: bacterial consumption of host glycans in the gut. Glycobiology 23:1038–1046. doi: 10.1093/glycob/cwt040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macy JM, Ljungdahl LG, Gottschalk G. 1978. Pathway of succinate and propionate formation in Bacteroides fragilis. J Bacteriol 134:84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferreyra JA, Wu KJ, Hryckowian AJ, Bouley DM, Weimer BC, Sonnenburg JL. 2014. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe 16:770–777. doi: 10.1016/j.chom.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Njoroge JW, Nguyen Y, Curtis MM, Moreira CG, Sperandio V. 2012. Virulence meets metabolism: Cra and KdpE gene regulation in enterohemorrhagic Escherichia coli. mBio 3:e00280-12. doi: 10.1128/mBio.00280-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pudlo NA, Urs K, Kumar SS, German JB, Mills DA, Martens EC. 2015. Symbiotic human gut bacteria with variable metabolic priorities for host mucosal glycans. mBio 6:e01282-15. doi: 10.1128/mBio.01282-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knutton S, Baldwin T, Williams PH, McNeish AS. 1989. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun 57:1290–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
High expression of virulence genes is observed in late logarithmic phase. Download FIG S1, PDF file, 0.3 MB (310.3KB, pdf) .
Copyright © 2019 Kumar and Sperandio.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
High indole concentration decreases EHEC virulence without affecting its growth. Download FIG S2, PDF file, 0.5 MB (521.5KB, pdf) .
Copyright © 2019 Kumar and Sperandio.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
CpxA identified as an indole sensor. Download FIG S3, PDF file, 0.5 MB (570.5KB, pdf) .
Copyright © 2019 Kumar and Sperandio.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Indole signaling occurs through CpxA-CpxR node. Download FIG S4, PDF file, 0.6 MB (605.5KB, pdf) .
Copyright © 2019 Kumar and Sperandio.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A cpxA mutant is indole irresponsive and is attenuated for pathogenesis. Download FIG S5, PDF file, 0.2 MB (208.8KB, pdf) .
Copyright © 2019 Kumar and Sperandio.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Self-produced indole or microbiota-derived indole dictates C. rodentium infectivity in a similar fashion. Download FIG S6, PDF file, 0.1 MB (114.6KB, pdf) .
Copyright © 2019 Kumar and Sperandio.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacteroides thetaiotaomicron (B. theta) murine colonization is not affected by tnaA. Download FIG S7, PDF file, 0.2 MB (173.3KB, pdf) .
Copyright © 2019 Kumar and Sperandio.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial strains. Download Table S1, PDF file, 0.3 MB (265.3KB, pdf) .
Copyright © 2019 Kumar and Sperandio.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Oligonucleotides used in this study. Download Table S2, PDF file, 0.1 MB (78.9KB, pdf) .
Copyright © 2019 Kumar and Sperandio.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.