ABSTRACT
Staphylococcus argenteus and Staphylococcus schweitzeri are newly identified species of the S. aureus-related complex. S. argenteus, as occurring globally and showing significant prevalence and comparable infection and morbidity rates compared to S. aureus, is becoming clinically important. Whole genome sequencing has revealed the presence of several virulence genes but the molecular mechanisms of S. argenteus infection and virulence are largely unknown. Here, we studied the effect of a previously characterized clinical S. argenteus isolate on human cells in vitro. The clinical isolate, together with the S. argenteus type strain MSHR1132T and the S. schweitzeri type strain FSA084T, had a cytotoxic effect on the cells, which showed necrotic cell death after a few hours of treatment. The protein causing the cytotoxic effect was purified and identified by mass spectrometry as alpha-hemolysin, Hla, which is awell-known pore-forming toxin in S.aureus. The cytotoxic effect could be blocked with an antibody against Hla. S.argenteus showed 12–15 fold higher expression levels of hla at the RNA level and 4–6 fold higher expression levels at the protein level compared to S.aureus. The higher expression levels of hla were supported by higher RNA levels of the regulatory factors sarA and saeR. Also, the RNAIII component of the accessory gene regulator (agr) quorum sensing system was 8,000–10,000 fold higher in the S.argenteus isolates compared to S.aureus. This is the first study on the effect of S.argenteus on ahuman cell line and strengthens the idea of significant virulence of S.argenteus.
KEYWORDS: Staphylococcus argenteus, Staphylococcus schweitzeri, Staphylococcus aureus, virulence, toxin, alpha-hemolysin, hla, agr, quorum sensing
Introduction
Staphylococcus argenteus was first described in 2009 as part of the Staphylococcus aureus clonal complex (CC) 75 [1], but was formally named in 2015 and together with Staphylococcus schweitzeri forms the S. aureus-related complex, including also CC2250, CC1223, CC2854, and CC2198 [2,3]. S. argenteus is globally distributed and has been isolated from both humans and animals [2–6], while S. schweitzeri has not been associated with human infections, and has only been isolated from non-human primates [2,7]. Depending on the sample collection, the proportion of S. argenteus within the S. aureus complex varies from 0.16% to 0.35% in some studies [4,8] to 10% in others [6]. The differences in prevalence may be due to methodological issues as screening methods are mostly still based on S. aureus, and more S. argenteus-optimized diagnostics might lead to a higher number of positive isolates.
S. argenteus is mostly connected to skin and soft tissue infections and has only rarely caused invasive disease [6,9,10]. At first, S. argenteus was thought to be less virulent than S. aureus, due to the lack of the yellow pigment staphyloxantin [11], which confers resistance against oxidative stress and neutrophil killing [12]. Indeed, S. argenteus was later shown to be more susceptible to oxidative stress and neutrophil killing in vitro than S. aureus and had reduced virulence in murine sepsis and skin infection models [2,3,13], although this was not connected to the deficient staphyloxantin expression [9]. However, as compared to S. aureus, similar [3,6] or even higher [14] human mortality rates have been reported for S. argenteus.
S. argenteus and S. schweitzeri have been shown to carry several toxin genes, such as staphylococcal enterotoxins sea, sec, and sed, hemolysins hla and hlb and Panton-Valentine leucocidin (pvl) [3,7,15-18], although some studies have indicated a lower prevalence of these genes compared to S. aureus [3,17]. Using S. aureus-specific PCR primers, the majority of S. argenteus isolates were first shown to be PVL-negative [3]. However, whole genome sequencing (WGS) later revealed both a nucleotide sequence similarity of only approximately 75% between S. argenteus and S. aureus in the pvl gene, and actually much higher prevalence of pvl among S. argenteus isolates [18]. Alpha-hemolysin (Hla) was the first staphylococcal exotoxin to be identified and belongs to the group of membrane-damaging toxins (reviewed in [19]), with a well-studied function and role in S. aureus virulence. Hla is secreted in a water-soluble form and binds to human cells via its target cell receptor ADAM10 resulting in the formation of heptamer pores in cell membranes. Low levels of Hla result in cell permeabilization, which induces apoptosis both intrinsically through the mitochondrial pathway including caspases 9 and 3 [20], but also extrinsically through TNF-α upregulation and caspase 8 activation [21]. High levels of Hla can also trigger necrosis due to a rapid Ca2+ influx [22–24]. Hla, and several other secreted toxins, are regulated by a complex network of factors including SarA, SarT, SaeR, and the quorum sensing accessory gene regulator (agr) system, which is active in late logarithmic to stationary phase of growth [25]. For hla, the regulatory RNAIII molecule of the agr system specifically plays an important role by activating both hla transcription and translation [26,27].
We previously characterized a clinical S. argenteus isolate (RK308) and, using WGS, were able to identify several toxin genes in the genome [18]. To further investigate the molecular mechanisms of S. argenteus virulence, we here studied the effect of this particular isolate on human cells in vitro and could show that, as opposed to S. aureus, S. argenteus was cytotoxic due to high levels of the secreted toxin Hla.
Materials and methods
Bacterial isolates and cultivation
The S. argenteus clinical isolate RK308 has been described previously [18]. The S. argenteus type strain MSHR1132T (isolated from a human sample [11]), and the S. schweitzeri type strain FSA084T (isolated from a red-tailed monkey in Gabon, Africa [7]) have been described previously [2] and were purchased from DSMZ, Braunschweig, Germany. The S. warneri type strain CCUG 7325T, the S. haemolyticus type strain CCUG 7323T and the S. epidermidis type strain CCUG 18000AT were purchased from CCUG, Gothenburg, Sweden. The S. aureus type strain 1800T and the MRSA reference strain CCUG35601 were also included for comparison. Bacteria were resuscitated from frozen stocks at −70°C onto blood agar and subsequently grown overnight in Müller-Hinton broth at 37°C.
Cell lines and culturing
The HeLa human cervical cancer and the HT29 human colon cancer cell lines were maintained in RPMI 1640 media (Swedish National Veterinary Institute, Uppsala, Sweden) supplemented with 2 mM glutamine (Gibco by life technologies, Carlsbad, California, US), 10% fetal bovine serum (FBS, Gibco by life technologies), 100 U/ml Penicillin and 100 μg/ml Streptomycin (PeSt, Swedish National Veterinary Institute) and grown at 37°C and in 5% CO2.
Cell infection and cytotoxicity assay
For infection experiments, bacteria from overnight cultures were centrifuged at 8000xg for 5 min, diluted in cell culture media and added to cells at a MOI of 100. For the cytotoxicity assays, the bacterial broth from overnight cultures was instead collected and sterile filtrated through a 0.22 μm filter. Filtrated broth (10–100 μl) was added to cells grown in RPMI 1640 media (Swedish National Veterinary Institute) supplemented with 2 mM glutamine (Gibco by life technologies) and 10% fetal bovine serum (FBS, Gibco by life technologies) and incubated at 37°C and in 5% CO2. Un-inoculated broth was used as mock. For neutralization, the anti-Hla antibody (8B7, Abcam ab190467, Abcam, Cambridge, UK) was added at a concentration of 4 μg/ml at the same time as the broth. At 6 h, cells were photographed through the microscope lens.
MTT assay
The tetrazolium dye MTT (Thiazolyl Blue Tetrazolium Bromide; Sigma-Aldrich (Merck), Darmstadt, Germany) was added directly to cell cultures after 6-h incubation to a final concentration of 200 μg/ml. Cells were incubated for an additional 1–2 h until a sufficient amount of dye had precipitated. Precipitates in the cell culture media were dissolved in equal amounts of 0.04 M HCl in isopropanol and measured spectrophotometrically at 570 nm. One well was set up without cells and used as blank in the spectrophotometer. The viability of the cells was calculated as the inverted absorbance value and compared with cells treated with un-inoculated bacterial broth, which was set to 100%.
SDS-PAGE and western blot
Bacterial broth was concentrated 10x in PBS on Pierce Protein Concentrator columns (3K MWCO, Thermo Fisher Scientific, Waltham, USA) and run on 4–12% SDS-PAGE gels. Gels were either stained with Coomassie Brilliant Blue or blotted to nitrocellulose filters for western blot. The western blot was blocked with 2% milk in TBS+0.05% Tween and detected with a mouse anti-Hla primary antibody (8B7, ab190467; Abcam, Cambridge, UK) diluted 1:1000 and a goat anti-mouse-HRP secondary antibody (A28177; Thermo Fisher Scientific) diluted 1:2000, both in TBS+0.05% Tween. Bands were visualized using the 1-Step Ultra TMB-Blotting Solution (Thermo Fisher Scientific). The blot was scanned and bands quantified in Photoshop using integrated density measurements.
Mass spectrometry analysis
Selected bands were excised from the Coomassie Brilliant Blue-stained SDS-PAGE for mass spectrometry analyses. The samples were in-gel digested with trypsin according to a standard protocol and analyzed by LC-Orbitrap MS/MS at the MS Facility, SciLifeLab, Uppsala University. Resulting peptide sequences were matched against all translated ORFs in the sequenced RK308 genome to identify the corresponding full-length ORFs, which were then blasted against Staphylococcus proteins to identify the proteins and genes.
Genomics
All included isolates are previously whole genome sequenced. Sequences for isolates included in cytotoxicity assays and RNA expression analyses are available online in the GenBank database (http://www.ncbi.nlm.nih.gov/Genbank/index.html) under the following accession numbers: S. argenteus RK308, LSFQ00000000; S. argenteus MSHR1132T, FR821777; S. schweitzeri FSA084T, CCEL00000000; S. aureus 1800T (DSM20231), AMYL00000000 and MRSA CCUG 35601, REGS00000000. For a complete list of all 116 sequenced S. argenteus isolates used to confirm the presence of the hla gene, please refer to [18]. Sequence alignments and phylogenetic analyses were done in CLC Main Workbench (Qiagen, Hilden, Germany) using standard program settings.
RNA preparation from bacteria and cDNA synthesis
Bacteria from overnight cultures were first treated with 4 μg/ml lysozyme in TE buffer for 1 h at 37°. Bacterial RNA was thereafter prepared using TRIreagent (Thermo Fisher Scientific) according to the manufacturer’s protocol. The RNA was DNase treated twice using dsDNase (Thermo Fisher Scientific) with phenol/chisam extraction and ethanol precipitation performed in between. The concentration of the RNA was determined using Nanodrop, and the integrity of the RNA was verified on a 1% agarose gel. A total of 100 ng of RNA was reverse transcribed (RT) using Maxima First Strand cDNA Synthesis Kit for qPCR (Thermo Fisher Scientific) according to the manufacturer’s protocol. A control reaction without RT was set up for each sample to rule out residual DNA. The cDNA synthesis reaction was diluted in water and 1/2000 was used for qPCR. At least three independent RNA preparations were used for qPCR.
Quantitative PCR
Real-time qPCR was performed in the Bio-Rad CFX96 Touch cycler (Bio-Rad, Hercules, USA) using the DyNAmo HS SYBR Green qPCR kit (Thermo Fisher Scientific). Primer sequences for 16S rRNA, hla and regulatory genes are available upon request. For expression analyses, expression levels of each gene were calculated as 2−CT, normalized to those of 16S rRNA and expressed as fold over that of the S. aureus 1800T isolate.
Statistical evaluation
Student’s t-test (unpaired, two-tailed, n = 3) was used to evaluate statistical differences between samples as described in figure legends where *** refers to p < 0.001; ** to p < 0.01 and * to p < 0.05.
Results
S. argenteus and S. schweitzeri are cytotoxic to human cancer cells
To assess the effect of S. argenteus on human cells, an in vitro infection model was used in which HeLa cervical cancer and HT29 colon cancer cells were infected with the S. argenteus RK308 isolate and the S. argenteus type strain MSHR1132T. For comparison, the S. schweitzeri type strain FSA084T, the S. aureus type strain 1800T, the MRSA reference strain CCUG 35601, the S. warneri type strain CCUG 7325T, the S. haemolyticus type strain CCUG 7323T and the S. epidermidis type strain CCUG 18000AT were also included in the in vitro infection analyses.
Both S. argenteus isolates and the S. schweitzeri isolate had a cytotoxic effect on HeLa and HT29 cells, visible already at approximately four hours after adding bacteria (data not shown). Epithelial cells showed clear signs of cell necrosis with swelling and membrane disruption, which finally led to cell lysis. To see if the effect was caused by a secreted protein, sterile filtered broth from overnight cultures was added to cell cultures. The same cytotoxic effect as seen with living S. argenteus and S. schweitzeri bacteria was detected (Figure 1(a)). However, none of the other Staphylococcus isolates tested showed a cytotoxic effect on the cells, neither with living bacteria nor with broth (Figure 1 and data not shown).
Figure 1.
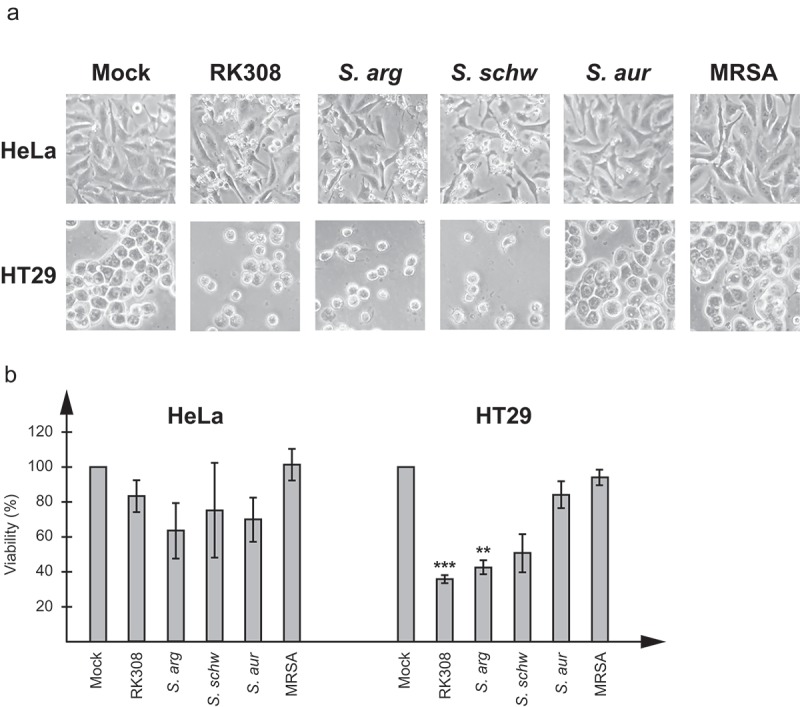
S. argenteus and S. schweitzeri isolates are cytotoxic to human cancer cells. Broth from overnight cultures of S. argenteus RK308 (RK308), S. argenteus MSHR1132T (S. arg), S. schweitzeri FSA084T (S. schw), S. aureus 1800T (S. aur) and MRSA CCUG 35601 (MRSA) was added to HeLa and HT29 cells for 6 h. Un-inoculated broth was used as mock. (a) Cells were photographed through the microscope lens. (b) Viability of cells from (a) calculated from the MTT assay compared to mock, which was set to 100%. Asterisks above bars show significant differences as compared to S. aureus 1800T.
In order to quantify the cytotoxic effect, cell viability was determined by measurement of the cell lysis-induced increase of NAD(P)H-dependent metabolic activity in the cell culture media at 6 h using the MTT assay and compared with cells exposed to un-inoculated broth. For HT29 cells, 40% viability was seen for cells exposed to the broth from S. argenteus cultures, while S. schweitzeri FSA084T resulted in 50% viability. For HeLa cells, although the cytotoxic effect was clearly visible in the microscope, the measured NAD(P)H-dependent metabolic activity was not as high with neither S. argenteus isolates nor with S. schweitzeri FSA084T, and resulted in viability of about 60–80%.
Identification of the cytolysin by mass spectrometry analyses
To identify the cytotoxic protein, broths from overnight cultures used in cytotoxicity experiments were run on SDS-PAGE and stained with Coomassie Brilliant Blue (Figure 2(a)). Three strong bands, of approximately 45, 35, and 20 kDa, respectively, were specific for the RK308 isolate, and two of which were also present in the S. argenteus type strain MSHR1132T (Figure 2(a), lanes 2 and 4). These three bands (1, 2, and 3, Figure 2(a), lane 2) were excised from the gel and analyzed by mass spectrometry (MS). The results were matched against all translated ORFs in the RK308 sequence, and each full-length protein was then identified using BLASTp (Table S1).
Figure 2.
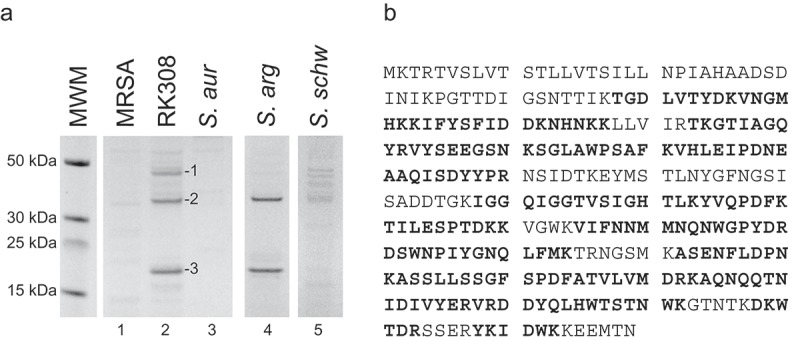
Identification of the cytolysin by MS analyses. (a) Coomassie brilliant blue stained SDS-PAGE of bacterial broth from S. argenteus RK308 (RK308), S. argenteus MSHR1132T (S. arg), S. schweitzeri FSA084T (S. schw), S. aureus 1800T (S. aur) and MRSA CCUG 35601 (MRSA) used in the cytotoxicity assay. Bands 1, 2 and 3 from the RK308 lane were excised from the gel for MS analyses. MWM, molecular weight marker. (b) Complete amino acid sequence of alpha-hemolysin, Hla, with peptides identified by MS shown in bold.
Band number 1 was identified as a cysteine protease and band number 3 as a DUF4888 domain-containing protein (S1 Table). As none of these proteins were likely to explain the cytotoxic property, they were not further investigated. Instead, band number 2, a protein of approximately 35 kDa and present in both S. argenteus and in the S. schweitzeri FSA084T isolates, was identified by BLASTp as an S. argenteus beta-channel forming cytolysin (Table S1), with 66% coverage in the MS analyses (Figure 2(b) and Table S1). Further BLAST searches on both the gene and protein sequences identified this cytolysin as Staphylococcus alpha-hemolysin (Hla).
In silico analyses confirmed the presence of the hla gene in all 116 S. argenteus isolates for which whole genome sequences were available (for a complete list of all sequenced S. argenteus isolates included, please refer to [18]). The amino acid sequences of the hla genes were almost identical within the S. argenteus group. The hla gene was also identified in S. schweitzeri FSA084T and S. aureus 1800T, while not detected in MRSA CCUG 35601, S. warneri CCUG 7325T, S. haemolyticus CCUG 7323T and S. epidermidis CCUG 18000AT used in the experiments.
The S. argenteus RK308 Hla protein sequence showed 90% and 89% similarity to S. schweitzeri FSA084T and S. aureus 1800T, respectively (Fig. S1A). The majority of amino acid residues, earlier shown to be important for binding S. aureus Hla to the cell membrane and pore formation [28], were conserved between the two S. argenteus, S. schweitzeri, and S. aureus isolates (Fig. S1A). However, phylogenetic analyses revealed a closer relationship between S. schweitzeri FSA084T and S. aureus 1800T while the two S. argenteus isolates were more distant (Fig. S1B).
Neutralization of the cytotoxic effect by an anti-Hla antibody
To further confirm that the cytotoxic effect was actually caused by Hla, a commercially available antibody against S. aureus Hla was tested in neutralization assays on HT29 cells. As shown in Figure 3, the antibody efficiently blocked the cytotoxic effect of the broth added to cell cultures. This could be seen both in the microscope (Figure 3(a)) and as an increase in viability compared to cells only treated with Hla-containing broth (Figure 3(b)).
Figure 3.
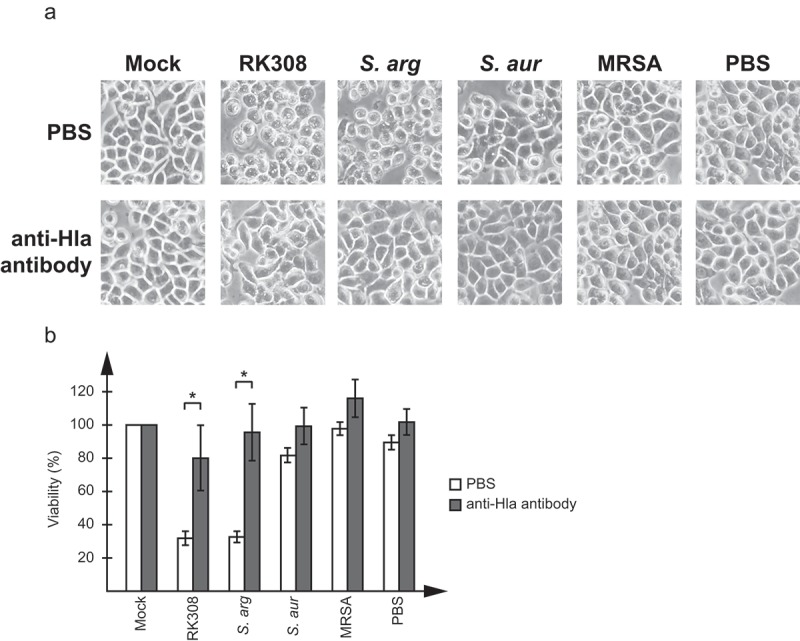
Neutralization of the cytotoxic effect by an anti-Hla antibody. Broth from overnight cultures of S. argenteus RK308 (RK308), S. argenteus MSHR1132T (S. arg), S. aureus 1800T (S. aur) and MRSA CCUG 35601 (MRSA) was added to HT29 cells for 6 h together with the anti-Hla antibody or PBS as indicated. Un-inoculated broth was used as mock. (a) Cells were photographed through the microscope lens. (b) Viability of cells from (a) calculated from the MTT assay compared to mock, which was set to 100%. Asterisks show significant differences between antibody- and PBS-treated cells.
S. argenteus expresses high levels of Hla
The presence of the hla gene was confirmed using PCR on genomic DNA. Figure 4(a) shows that hla was detected in both S. argenteus isolates, as well as in S. schweitzeri FSA084T and S. aureus 1800T, while not present in MRSA CCUG 35601, in accordance with the in silico analyses. The expression pattern of hla was thereafter analyzed at both RNA and protein levels, using RT-qPCR and western blotting, respectively. The S. argenteus isolates had 12–15 fold higher hla RNA levels compared to S. schweitzeri FSA084T and S. aureus 1800T (Figure 4(b)), resulting in 4–6 fold higher Hla protein levels (Figure 4(c)). Interestingly, S. schweitzeri FSA084T had lower hla RNA levels, in the same range as S. aureus 1800T, but equally high Hla protein levels as the S. argenteus isolates included (Figure. 4(b,c)).
Figure 4.
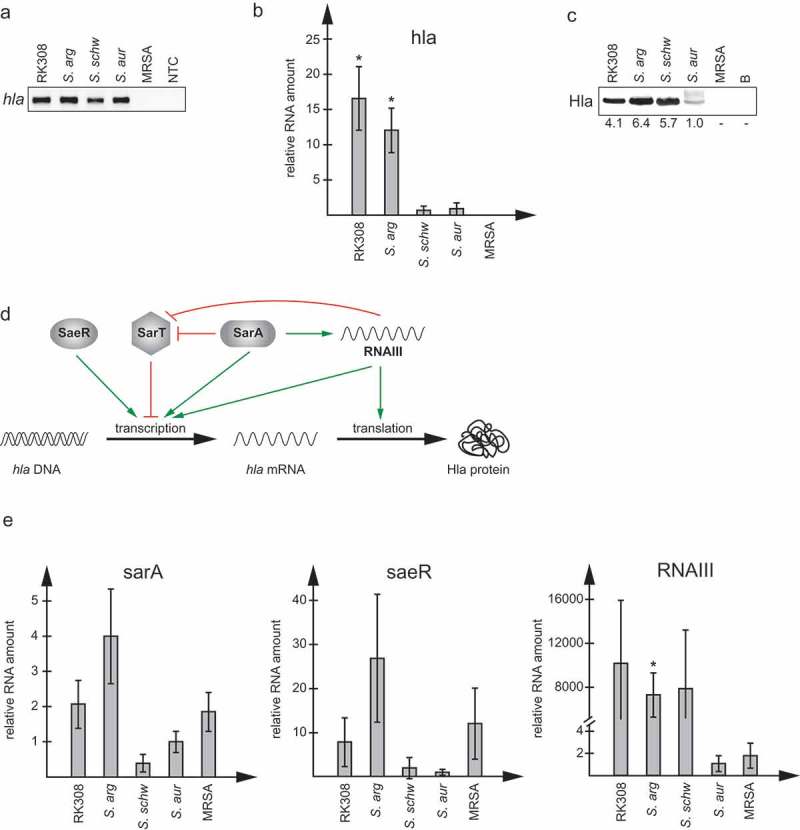
S. argenteus expresses high levels of alpha-hemolysin, hla. Expression analyses of hla and regulatory genes in S. argenteus RK308 (RK308), S. argenteus MSHR1132T (S. arg), S. schweitzeri FSA084T (S. schw), S. aureus 1800T (S. aur) and MRSA CCUG 35601 (MRSA). (a) Detection of the hla gene using PCR. NTC, no template control. (b) RNA levels of hla detected with RT-qPCR. RNA expression levels are normalized against 16SrRNA and expressed as fold compared to the S. aureus 1800T isolate. Asterisks above bars show significant differences as compared to S. aureus 1800T. (c) Hla protein levels in bacterial broth used in the cytotoxicity assay detected by western blot with an anti-Hla antibody. B, un-inoculated broth. Numbers below the blot show fold expression levels compared to S. aureus 1800T as quantified in Photoshop. (d) Schematic diagram showing the regulation of hla transcription and translation by regulatory factors. Adapted from [25]. (E) RNA levels of regulatory genes sarA, saeR, and RNAIII detected with RT-qPCR. RNA expression levels are normalized against 16SrRNA and expressed as fold compared to the S. aureus 1800T isolate. Asterisks above bars show significant differences as compared to S. aureus 1800T.
To understand the differences in hla RNA and protein levels between the included Staphylococcus isolates, we measured the expression levels of saeR, sarA and the RNAIII component of the agr quorum sensing system, the gene products of which are involved in regulation of hla expression at transcription and translation levels (Figure 4(d)). The transcriptional activators sarA and saeR showed 2–4 fold and 9–28 fold higher expression in S. argenteus isolates compared to S. aureus 1800T, respectively. RNAIII, which activates both hla transcription and translation, showed a remarkable 8,000–10,000 fold higher expression in S. argenteus and S. schweitzeri isolates compared to S. aureus.
Discussion
Since the discovery of S. argenteus in 2009, a lot of the research has focused on distribution in the population [2–6], prevalence rates [4,6,8], presence of virulence genes [3,7,15-18]and challenges in clinical diagnostics as compared to S. aureus, while information on S. schweizeri is more scarce. This is the first molecular study on the impact of S. argenteus and S. schweitzeri on human cells in vitro and describes a remarkable cytotoxic effect not seen with any other Staphylococcus species tested here. In this study, epithelial cells treated with S. argenteus and S. schweitzeri isolates showed clear signs of cell necrosis with swelling and finally cell lysis, probably caused by membrane disruption, which was further verified as an increase of NAD(P)H-dependent metabolic activity in the cell culture media, and hence reduced cell viability.
Using protein purification, mass spectrometry, and an antibody neutralization assay, we identified the cause of the cytotoxic effect as the secreted Hla toxin. The Hla toxin, which is a well-studied toxin in S. aureus, is known to be important for virulence, as it has been shown to directly damage host tissues and disable the immune system by lysing neutrophils [19]. In our study, the hla gene was detected in all sequenced S. argenteus isolates and was fairly conserved between S. argenteus, S. schweitzeri FSA084T and S. aureus 1800T. However, the toxin effect showed remarkable differences. As the majority of amino acid residues important for toxin function were conserved, the sequence variation itself was unlikely to explain the difference in the activity of the protein. We therefore decided to compare hla expression levels in our isolates. Measurements of hla RNA and protein levels in S. argenteus bacteria and broth confirmed high levels of both RNA and protein compared to S. aureus. Levels of hla RNA in S. schweitzeri were in the same range as for S. aureus while the secreted protein level was equally high as for S. argenteus. The high Hla protein levels in S. argenteus and S. schweitzeri are consistent with the observation that the cytotoxic effect seen was necrosis and not apoptosis, as Hla is known to cause apoptosis at low levels and necrosis at high levels [20–24]. However, it is possible that the cells treated with S. aureus broth, which did contain low levels of Hla according to the western blot, had started to show initial signs of apoptosis, although not yet visible in the microscope, or would eventually have developed apoptosis if treatment times had been extended.
The high protein levels of Hla in S. argenteus and S. schweitzeri were supported by high expression levels of the regulatory factors sarA, saeR and RNAIII of the agr quorum sensing system. These factors have been shown to have direct effects on both transcription and translation of hla in S. aureus [25]. The saeR factor, which has a direct positive effect on hla transcription, was expressed to a 10–30 fold higher level in the S. argenteus isolates than in S. schweitzeri FSA084T and S. aureus 1800T, while sarA was expressed at 2–4 fold higher levels, respectively. However, SarA might be expected to have a larger impact on hla RNA and protein levels as it both activates hla transcription directly and also has an indirect effect on hla translation through stabilization of RNAIII. Moreover, SarA also seems to be able to regulate Hla protein levels by regulation of extracellular proteases as sarA mutants have been shown to express increased levels of extracellular proteases resulting in lower levels of Hla due to protease-mediated degradation [29]. However, the most striking difference in expression in this study was the 8,000–10,000 fold higher expression level of RNAIII in S. argenteus and S. schweitzeri compared to S. aureus. The agr system is known to be highly active in late-log to stationary phase of bacterial growth, which corresponds to the phase in which the bacterial broth was collected and used in the cytotoxicity assays in this study. There was no difference in sarA and saeR expression between S. schweitzeri and S. aureus but, S. schweitzeri had the same equally high expression levels of RNAIII as S. argenteus, which is likely the reason why S. schweitzeri also had such high Hla protein levels.
Hla has for a long time been the target of therapeutic approaches to limit the damage of S. aureus infection. This includes strategies such as specific antibodies or small chemical compounds to target not only Hla directly but also regulatory factors of the agr, sae, and sarA systems and the receptor ADAM10 [30], with the hope that this type of anti-virulence therapies might serve as a complement to antimicrobials. Given the high expression levels of Hla in S. argenteus and that many S. argenteus isolates are shown to be methicillin-resistant [8,18,31]these approaches might also be important to combat S. argenteus infections.
Of particular interest was the extremely high expression levels of RNAIII in the S. argenteus and S. schweitzeri isolates in this study. RNAIII is expressed from its own promoter (P3) in the agr locus, and further studies are needed to verify whether the other gene products of agr (agrA-D), expressed from a different promoter (P2) within the locus, are also increased in S. argenteus and S. schweitzeri, and whether this contributes to the general characteristics and virulence of these species.
To summarize, we present the first study looking at the effect of S. argenteus and S. schweitzeri on a human cell line. The findings showing high expression levels of Hla in S. argenteus and S. schweitzeri and resulting in cytolethal properties contributes to the understanding of S. argenteus virulence and support the earlier perceptions of S. argenteus being virulent and clinically relevant.
Funding Statement
This work was supported by ALF grants from the Uppsala University Hospital, Uppsala, Sweden.
Acknowledgments
The authors thank the Science for Life Laboratory Mass Spectrometry Based Proteomics Facility in Uppsala for the MS analyses.This work was supported by ALF grants from the Uppsala University Hospital, Uppsala, Sweden to HR. The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Data availability
All sequence data is available in the GenBank database (http://www.ncbi.nlm.nih.gov/Genbank/index.html) under the accession numbers provided above or in(18).
References
- [1].Ng JW, Holt DC, Lilliebridge RA, et al. Phylogenetically distinct Staphylococcus aureus lineage prevalent among indigenous communities in northern Australia. J Clin Microbiol. 2009;47(7):2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Tong SY, Schaumburg F, Ellington MJ, et al. Novel staphylococcal species that form part of a Staphylococcus aureus-related complex: the non-pigmented Staphylococcus argenteus sp. nov. and the non-human primate-associated Staphylococcus schweitzeri sp. nov. Int J Syst Evol Microbiol. 2015;65(Pt1):15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chantratita N, Wikraiphat C, Tandhavanant S, et al. Comparison of community-onset Staphylococcus argenteus and Staphylococcus aureus sepsis in Thailand: a prospective multicentre observational study. Clin Microbiol Infect. 2016;22(5):458e11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Argudin MA, Dodemont M, Vandendriessche S, et al. Low occurrence of the new species Staphylococcus argenteus in a Staphylococcus aureus collection of human isolates from Belgium. Eur J Clin Microbiol Infect Dis. 2016;35(6):1017–1022. [DOI] [PubMed] [Google Scholar]
- [5].Schuster D, Rickmeyer J, Gajdiss M, et al. Differentiation of Staphylococcus argenteus (formerly: Staphylococcus aureus clonal complex 75) by mass spectrometry from S. aureususing the first strain isolated from a wild African great ape. Int J Med Microbiol. 2017;307(1):57–63. [DOI] [PubMed] [Google Scholar]
- [6].Thaipadungpanit J, Amornchai P, Nickerson EK, et al. Clinical and molecular epidemiology of Staphylococcus argenteus infections in Thailand. J Clin Microbiol. 2015;53(3):1005–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Schaumburg F, Alabi AS, Kock R, et al. Highly divergent Staphylococcus aureus isolates from African non-human primates. Environ Microbiol Rep. 2012;4(1):141–146. [DOI] [PubMed] [Google Scholar]
- [8].Hansen TA, Bartels MD, Hogh SV, et al. Whole genome sequencing of Danish Staphylococcus argenteus reveals a genetically diverse collection with clear separation from Staphylococcus aureus. Front Microbiol. 2017;8:1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tong SYC, Sharma-Kuinkel BK, Thaden JT, et al. Virulence of endemic nonpigmented Northern Australian Staphylococcus aureus clone (clonal complex 75, S-argenteus) is not augmented by staphyloxanthin. J Infect Dis. 2013;208(3):520–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Rigaill J, Grattard F, Grange S, et al. Community-acquired Staphylococcus argenteus sequence type 2250 bone and joint infection, France, 2017. Emerg Infect Dis. 2018;24(10):1958–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Holt DC, Holden MT, Tong SY, et al. A very early-branching Staphylococcus aureus lineage lacking the carotenoid pigment staphyloxanthin. Genome Biol Evol. 2011;3:881–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Liu GY, Essex A, Buchanan JT, et al. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J Exp Med. 2005;202(2):209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dupieux C, Blonde R, Bouchiat C, et al. Community-acquired infections due to Staphylococcus argenteus lineage isolates harbouring the Panton-Valentine leucocidin, France, 2014. Euro Surveill. 2015;20(23):21154. [DOI] [PubMed] [Google Scholar]
- [14].Chen SY, Lee H, Wang XM, et al. High mortality impact of Staphylococcus argenteus on patients with community-onset staphylococcal bacteraemia. Int J Antimicrob Agents. 2018;52(6):747–753. [DOI] [PubMed] [Google Scholar]
- [15].Indrawattana N, Pumipuntu N, Suriyakhun N, et al. Staphylococcus argenteus from rabbits in Thailand. Microbiologyopen. 2018;8(4):e00665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wakabayashi Y, Umeda K, Yonogi S, et al. Staphylococcal food poisoning caused by Staphylococcus argenteus harboring staphylococcal enterotoxin genes. Int J Food Microbiol. 2018;265:23–29. [DOI] [PubMed] [Google Scholar]
- [17].Kaden R, Engstrand L, Rautelin H, et al. Which methods are appropriate for the detection of Staphylococcus argenteus and is it worthwhile to distinguish S. argenteus from S. aureus? Infect Drug Resist. 2018;11:2335–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhang DF, Zhi XY, Zhang J, et al. Preliminary comparative genomics revealed pathogenic potential and international spread of Staphylococcus argenteus. BMC Genomics. 2017;18(1):808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Berube BJ, Bubeck Wardenburg J.. Staphylococcus aureus alpha-toxin: nearly a century of intrigue. Toxins (Basel). 2013;5(6):1140–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bantel H, Sinha B, Domschke W, et al. alpha-Toxin is a mediator of Staphylococcus aureus-induced cell death and activates caspases via the intrinsic death pathway independently of death receptor signaling. J Cell Biol. 2001;155(4):637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Haslinger B, Strangfeld K, Peters G, et al. Staphylococcus aureus alpha-toxin induces apoptosis in peripheral blood mononuclear cells: role of endogenous tumour necrosis factor-alpha and the mitochondrial death pathway. Cell Microbiol. 2003;5(10):729–741. [DOI] [PubMed] [Google Scholar]
- [22].Essmann F, Bantel H, Totzke G, et al. Staphylococcus aureus alpha-toxin-induced cell death: predominant necrosis despite apoptotic caspase activation. Cell Death Differ. 2003;10(11):1260–1272. [DOI] [PubMed] [Google Scholar]
- [23].Prince LR, Graham KJ, Connolly J, et al. Staphylococcus aureus induces eosinophil cell death mediated by alpha-hemolysin. PLoS One. 2012;7(2):e31506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Eichstaedt S, Gabler K, Below S, et al. Effects of Staphylococcus aureus-hemolysin A on calcium signalling in immortalized human airway epithelial cells. Cell Calcium. 2009;45(2):165–176. [DOI] [PubMed] [Google Scholar]
- [25].Bronner S, Monteil H, Prevost G. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol Rev. 2004;28(2):183–200. [DOI] [PubMed] [Google Scholar]
- [26].Morfeldt E, Taylor D, von Gabain A, et al. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995;14(18):4569–4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Novick RP, Ross HF, Projan SJ, et al. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. Embo J. 1993;12(10):3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Walker B, Bayley H. Key residues for membrane binding, oligomerization, and pore forming activity of staphylococcal alpha-hemolysin identified by cysteine scanning mutagenesis and targeted chemical modification. J Biol Chem. 1995;270(39):23065–23071. [DOI] [PubMed] [Google Scholar]
- [29].Zielinska AK, Beenken KE, Joo HS, et al. Defining the strain-dependent impact of the Staphylococcal accessory regulator (sarA) on the alpha-toxin phenotype of Staphylococcus aureus. J Bacteriol. 2011;193(12):2948–2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kong C, Neoh HM, Nathan S. Targeting Staphylococcus aureus toxins: a potential form of anti-virulence therapy. Toxins (Basel). 2016;8(3):72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Tang Hallback E, Karami N, Adlerberth I, et al. Methicillin-resistant Staphylococcus argenteus misidentified as methicillin-resistant Staphylococcus aureus emerging in western Sweden. J Med Microbiol. 2018;67:968–971. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data is available in the GenBank database (http://www.ncbi.nlm.nih.gov/Genbank/index.html) under the accession numbers provided above or in(18).


