ABSTRACT
Nutrient iron sequestration is the most significant form of nutritional immunity and causes bacterial pathogens to evolve strategies of host iron scavenging. Cigarette smoking contains iron particulates altering lung and systemic iron homeostasis, which may enhance colonization in the lungs of patients suffering chronic obstructive pulmonary disease (COPD) by opportunistic pathogens such as nontypeable. NTHi is a heme auxotroph, and the NTHi genome contains multiple heme acquisition systems whose role in pulmonary infection requires a global understanding. In this study, we determined the relative contribution to NTHi airway infection of the four heme-acquisition systems HxuCBA, PE, SapABCDFZ, and HbpA-DppBCDF that are located at the bacterial outer membrane or the periplasm. Our computational studies provided plausible 3D models for HbpA, SapA, PE, and HxuA interactions with heme. Generation and characterization of single mutants in the hxuCBA, hpe, sapA, and hbpA genes provided evidence for participation in heme binding-storage and inter-bacterial donation. The hxuA, sapA, hbpA, and hpe genes showed differential expression and responded to heme. Moreover, HxuCBA, PE, SapABCDFZ, and HbpA-DppBCDF presented moonlighting properties related to resistance to antimicrobial peptides or glutathione import, together likely contributing to the NTHi-host airway interplay, as observed upon cultured airway epithelia and in vivo lung infection. The observed multi-functionality was shown to be system-specific, thus limiting redundancy. Together, we provide evidence for heme uptake systems as bacterial factors that act in a coordinated and multi-functional manner to subvert nutritional- and other sources of host innate immunity during NTHi airway infection.
KEYWORDS: Haemophilus influenzae, heme binding, iron nutritional immunity, protein moonlighting, respiratory infection
Introduction
The need of invading bacteria to acquire nutrient metals from their environments has caused vertebrate hosts to evolve to restrict their bioavailability. Nutrient iron sequestration is one mode of innate defence termed “iron nutritional immunity”, whereby iron levels are tightly controlled by host regulatory systems controlling its absorption, systemic transport, distribution, cellular uptake, and storage [1,2]. In response to this, pathogens evolve mechanisms of host iron piracy to scavenge this metal during infection [3].
Iron exists in the reduced ferrous (Fe2+) or oxidized ferric (Fe3+) forms. The Fe2+/Fe3+ redox potential makes it extremely versatile when incorporated into proteins as a catalytic center or electron carrier, and it is essential in numerous biological processes. Although abundant in nature, iron does not normally occur in its biologically active ferrous form. Under aerobic conditions, Fe2+ is unstable and generates Fe3+ and reactive oxygen species (ROS) via the Fenton reaction, the latter of which can damage lipids, DNA, and proteins [4]. Conversely, ferric iron is poorly water-soluble and requires specialized proteins to facilitate its mobilization and maintain intracellular reservoir (lactoferrin and transferrin for transport, ferritin for storage). However, the most abundant form of iron in vertebrates is bound within a porphyrin ring as ferriprotoporphyrin IX (heme) [5], most of which is bound to intracellular proteins (hemoglobin, myoglobin, cytochromes). Hemoglobin from lysed erythrocytes is bound by haptoglobin, whereas free heme is bound to hemopexin and, to a lesser extent, to albumin, under which circumstances it may become available to extracellular pathogens [6]. Many pathogens thus exploit host heme as a nutrient iron source [5,7,8].
In the lungs, iron is stored in ferritin, macrophages sequester heme from senescent erythrocytes, take and store iron from transferrin, and clear inhaled iron-containing particulate matter. Likewise, neutrophils release iron regulatory molecules helping sequester free iron [9–11]. Human lung iron levels range between 0.4–0.9 mg/g, and ~10 μg iron are exposed to lungs daily at a normal rate of breathing from the atmosphere [12]. Exposure to higher iron levels through inhalation of iron-containing particulate matter provides catalytically active iron resulting in tissue damage. Moreover, alveolar hemorrhaging observed in several lung disease conditions leads to increased iron levels. As a consequence, strong evidence for dysregulated iron homeostasis exists in major respiratory diseases including chronic obstructive pulmonary disease (COPD) [1,2,13–15]. COPD is a complex progressive condition characterized by chronic airway inflammation, associated with airway remodeling, alveolar destruction and persistent airflow obstruction [16]. Cigarette smoking, the single greatest risk factor for developing COPD [17], contains iron particulates inducing intracellular iron accumulation [13,14]. Elevated levels of iron, ferritin, and oxidative stress are found in the lungs of smokers, where alveolar macrophages increase iron loading [13,14,18–20]. Similarly, the COPD airways present altered levels of iron-binding proteins, increased iron loading by alveolar macrophages, and high rates of polymorphisms in genes associated with iron regulation [21–28]. Given that iron overload increases the virulence of numerous pathogens [29,30], dysregulation of iron homeostasis in the COPD lungs may also facilitate persistence by opportunistic pathogens in the lower airways [2,6,11].
Nontypeable Haemophilus influenzae (NTHi) is typically a commensal of the human upper airways, but also a common opportunistic pathogen in the lower airways of patients with COPD [31–35]. NTHi persistence within the COPD lungs contributes to airway inflammation that results in worsening of symptoms and promotes disease progression [33,36–38]. NTHi is incapable of synthesizing heme, but possesses a ferrochelatase that reversibly inserts iron into protoporphyrin IX (PPIX) to form heme [39,40]. Thus, it requires either iron in the presence of PPIX or exogenous heme to grow aerobically [41]. NTHi presents a variety of heme uptake-binding systems including TonB-dependent outer membrane systems that bind heme or hemoproteins (Hup [42], HemR [43], Hgps [44–50], HxuCBA [51–56]), and TonB-independent periplasmic proteins that bind heme and are linked to inner membrane ABC transporters (SapABCDFZ, HbpA-DppBCDF [57–62]). NTHi also stores and shares heme by using the outer membrane protein E (PE) [63] (Figure 1). Information on the contribution of heme uptake to NTHi respiratory infection is scattered [47,50,55,60,64], and a global understanding is lacking.
Figure 1.
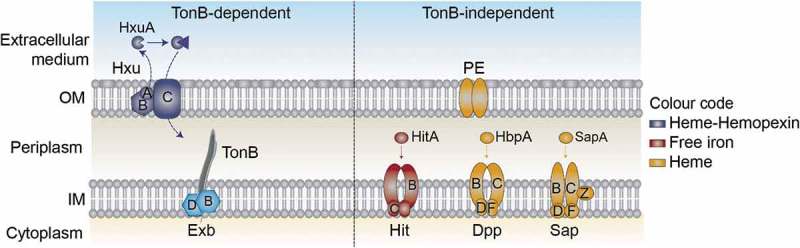
Schematic representation of NTHi heme uptake systems considered in this study. HxuCBA is a TonB-dependent system involved in heme-hemopexin binding. HxuC is a receptor and HxuAB is a two-partner secretion system. HxuA is exposed at the cell surface, and leads to heme release and its capture by HxuC. SapABCDFZ, HbpA-DppBCDF and PE are TonB-independent systems. SapA and HbpA are periplasmic proteins that bind heme and are linked to the inner membrane ABC transporters SapBCDFZ and DppBCDF, respectively. PE is an outer membrane protein that binds heme as a dimer. The HitABC free iron uptake system is also shown. HitA is a periplasmic protein that binds free iron linked to the HitBC transporter located at the inner membrane. Heme-iron sources are color-coded.
Existing evidence prompted us to hypothesize that pathological disruption of lung iron homeostasis and bacterial heme auxotrophy may have evolved NTHi toward the coordinated action of its repertoire of heme scavenging mechanisms, to evade nutritional immunity and facilitate the host-pathogen interplay. In this study, HxuCBA, PE, SapABCDFZ, and HbpA-DppBCDF were selected as representative H. influenzae heme binding systems at the outer membrane and within the periplasm (Figure 1). Systematic bacterial gene inactivation and phenotypic assessment revealed their relative contribution to NTHi airway infection, and their ability to bind heme was investigated in silico.
Materials and methods
Computational modeling
Docking of heme B to HbpA, SapA, PE, and HxuA was performed using the HADDOCK2.2 (High Ambiguity-Driven protein-protein DOCKing) server [65,66]. For HbpA and SapA, the I-TASSER server [67,68] was used to generate homology models based on the crystal structure of HbpA from Haemophilus parasuis in its ligand-bound form (PDB code 3M8U) [62]. For both proteins, the best scoring I-TASSER model was used for the docking experiment. To drive the docking, residues with 60% ligand binding probability as estimated by the COACH function within the I-TASSER suite [69,70] were included as part of an initial estimation of the location of the binding site (residues 61–64, 155, 280, 396, 398, 445–447, and 537 for HbpA; residues 54–55, 458–460, and 462 for SapA). In addition to full-length SapA, heme docking was also performed after removing residues 140–158. For PE, heme was docked to the crystal structure of the PE-dimer (3ZH5). Residues from both monomers within the predicted heme binding pocket (59, 65, 67, 94, 96, 98, 101, 103, and 107) were used to drive the docking. For HxuA, heme was docked to the HxuA crystal structure (PDB code 4RM6), and residues in a manually identified putative binding pocket (450, 452, 464, 516, 526, 556, 560, and 572) were used to drive the docking. Docking solutions were evaluated based on the HADDOCK score and Z-score (the number of standard deviations the HADDOCK score of a given solution is separated from the mean of all clusters). For HbpA, truncated SapA, and PE, a single statistically significant solution cluster was obtained (p < 0.05) whereas for HxuA, two equally good solutions were obtained. For full-length SapA, a second solution (cluster 4) with a non-significantly different HADDOCK score from the top solution was obtained. However, this was discarded due to the significantly higher Z-score (−0.2 compared to −1.7 for the top solution). Detailed docking statistics are given in Table S4. The top solution in each cluster was further analyzed in PyMOL (PyMOL Molecular Graphics System, version 2.0, Schrödinger, LLC), and LigPlot+ [71], which were also used to generate figures.
Bacterial strains, plasmids, and growth media
Strains and plasmids used in this study are described in Table 1 and Table S1. NTHi strains were grown for 16 h, at 37°C, 5% CO2, on chocolate agar (Biomérieux), Mueller Hinton-Fastidious agar (MH-F) (Biomérieux), Haemophilus Test Medium agar (HTM, Oxoid), or Brain-Heart Infusion agar (BHI, Oxoid). BHI was supplemented with 10 μg/ml nicotinamide adenine dinucleotide (NAD, factor V) (Sigma-Aldrich) (referred to as BHI-NAD or heme-deficient medium). Alternatively, HTM and BHI were supplemented with both NAD and 10 μg/ml heme (factor X) (Sigma-Aldrich) (referred to as sHTM and sBHI). Liquid cultures were grown in heme-deficient medium, sBHI or, alternatively, in a chemically defined minimal medium (CDMM) supplemented with 50 μM cystine (Sigma-Aldrich) or 50 μM glutathione disulfide (GSSG) (Sigma-Aldrich) (Table S2 [62,72,73]). GSSG (20 mM) was prepared in sterile water; 20 mM cystine was prepared in 1M HCl, filtered and maintained at 4°C until use. Erythromycin (Erm) at 11 μg/ml or spectinomycin (Spec) at 50 μg/ml was used when required. Escherichia coli was grown on Luria Bertani (LB) agar at 37°C, supplemented with ampicillin at 100 μg/ml, Erm at 150 μg/ml, or Spec at 50 μg/ml, when indicated.
Table 1.
Bacterial strains used in this study.
| Strain | Description | Source |
|---|---|---|
| H. influenzae | ||
| NTHi375 | Wild-type, otitis media clinical isolate | [117] |
| NTHi375ΔhxuCBA-P682 | hxuCBA::spec, SpecR | This study |
| NTHi375ΔsapA-1665 | sapA::ermC::ermC, ErmR | This study |
| NTHi375ΔhbpA-P896 | hbpA::spec, SpecR | This study |
| NTHi375Δhpe-P897 | hpe::spec, SpecR | This study |
| RdKW20 | Laboratory strain, capsule-deficient serotype d | [97] |
| RdKW20ΔhxuCBA-P683 | hxuCBA::spec, SpecR | This study |
| RdKW20ΔsapA-P578 | sapA::ermC::ermC, ErmR | This study |
| RdKW20ΔhbpA-P885 | hbpA::spec, SpecR | This study |
| RdKW20ΔhitBC-P804 | hitBC::ermC, ErmR | This study |
| NTHi3655luxABCDE-P845 | Luminiscent strain derived from isolate NTHi3655. It contains a copy of the luxABCDE operon inserted in the genome. |
[63] |
| E. coli | ||
| TOP10 | Used for cloning assays | Thermofisher Scientific |
| SW102 | Derived from DY380, it contains a defective λ prophage with the recombination proteins exo, bet, and gam being controlled by the temperature-sensitive repressor cI857 |
[75] |
Generation of H. influenzae mutant strains
Plasmids, primers, and construct design are shown in Tables S1, S3, and Fig. S1A. Briefly, a DNA fragment containing each gene/operon and its respective flanking regions was PCR amplified with Phusion polymerase (ThermoScientific) using NTHi375 genomic DNA as template and primers gene+flanking region-F1 and gene+flanking region-R1, and cloned into pJET1.2/blunt (ThermoScientific) or pGEMT-easy (Promega), generating plasmids pJET1.2-hxuCBA, pGEMT-sapAB, pJET1.2-hbpA, and pJET1.2-hpe. To generate pJET1.2-hitABC, two DNA fragments of the hitABC operon were independently amplified using primers hitABC-F1/hitABC-R2 and hitABC-F2/hitABC-R1, respectively, KpnI digested and ligated, rendering a product with a 853 bp deletion of the hitB gene 3´ end and a 200 bp deletion of the hitC gene 5´ end, which was further amplified with primers hitABC-F1 and hitABC-R1 and cloned into pJET1.2 to generate pJET1.2-hitABC. An Erm-based disruption strategy was used to generate ΔsapA and ΔhitBC strains. pGEMT-sapAB was disrupted by inverse PCR with Phusion polymerase, using primers sapAB-F2 and sapAB-R2. A sapA gene internal fragment of 168 bp was replaced by a blunt-ended ErmC resistance cassette excised by SmaI digestion from pBSLerm [74], generating pGEMT-sapA::ermC, which was used as a template to amplify the sapA::ermC disruption cassette with primers sapAB-F1 and sapAB-R1. pJET1.2-hitABC was KpnI digested and used as cloning vector for a KpnI blunt-ended ErmC resistance gene excised by KpnI digestion from pBSLerm, generating pJET1.2-hitBC::ermC, which was used as a template to amplify the hitBC::ermC disruption cassette with primers hitABC-F1 and hitABC-R1. Alternatively, a Spec-based disruption strategy was used to generate ΔhxuCBA, ΔhbpA, and Δhpe strains. To do so, a Spec resistance gene was independently PCR amplified from pRSM2832 using gene-specific mutagenic primer pairs hxuCBA-F2/hxuCBA-R2, hbpA-F2/hbpA-R2, and hpe-F2/hpe-R2 [75]. E. coli SW102 cells were prepared for recombineering, separately co-electroporated with pJET1.2-hxuCBA, pJET1.2-hbpA or pJET1.2-hpe (Ampr) (50 ng), and with each gene-specific mutagenic cassette (Specr) (200 ng) [76]. Mutagenized clones containing pJET1.2-hxuCBA::spec, pJET1.2-hbpA::spec and pJET1.2-hpe::spec were selected on LB agar with Amp100 and Spec50. These plasmids were used as templates to amplify the hxuCBA::spec, hbpA::spec and hpe::spec disruption cassettes with primer pairs hxuCBA-F1/hxuCBA-R1, hbpA-F1/hbpA-R1, and hpe-F1/hpe-R1, respectively. Disruption cassettes were independently used to transform NTHi375 or Hi RdKW20 by using the MIV method [77]. Transformants were selected on sBHI agar with Erm at 11 μg/ml to obtain ΔsapA and ΔhitBC strains, and on sBHI agar with Spec at 50 μg/ml to obtain ΔhxuCBA, ΔhbpA, and Δhpe strains. All mutants were confirmed by PCR and sequencing. The sapABCDFZ and hbpA-HI0854/NTHI1022 are gene clusters containing 6 and 2 genes, respectively. Expression of the sapBCDFZ and HI0854/NTHI1022 genes in NTHi375ΔsapA and NTHi375ΔhbpA strains was tested by RT-PCR (see below), being comparable to that found in the wild type (WT) strain (Fig. S1B).
Bacterial growth
Strains grown on chocolate agar were used to generate bacterial suspensions normalized in PBS to OD600 = 1. Suspensions were spread on 20 ml BHI agar in the presence of sterile X + V factor disks (Oxoid), and grown during 20 h at 37°C. Growth diameter was measured on at least three independent occasions (n ≥ 3). Alternatively, normalized bacterial suspensions were diluted to OD600 = 0.05 in 2 ml sBHI, and 200 µl aliquots were transferred to individual wells in 96-well microtiter plates (Falcon). Plates were incubated with agitation at 37°C for 6 h in a Multiskan device (ThermoScientific), and OD636 was monitored every 15 min. Each growth curve was corrected to its blank values (sBHI). Independently, strains were grown on chocolate agar; biomass was inoculated in 7 ml sBHI and incubated for 8 h at 37°C with shaking (200 r.p.m.). Cultures were centrifuged (10 min, 4000 r.p.m.) and suspensions were normalized to OD600 = 1 in PBS. Then, 180 μl of CDMM supplemented with either 50 μM cystine or 50 μM GSSG were transferred to individual wells in 96-well microtiter plates (Sarstedt). Thereafter, 20 μl of the previously prepared bacterial suspensions were added to each well. Plates were incubated in a SpectraMAX 340 microplate reader at 37°C, and OD600 was recorded every 30 min for 8 h. In all cases, bacterial growth was monitored in triplicate at least three times (n ≥ 9).
Heme binding to the bacterial surface
As previously stated [63], strains were grown on chocolate agar, inoculated in 5 ml heme-deficient medium and grown for 8 h, at 37°C with shaking (200 r.p.m.). Next, 100 μl culture aliquots were inoculated into 20 ml sBHI and grown for 16 h under the same conditions. Normalized bacterial suspensions were collected by centrifugation (10 min, 4000 r.p.m.), washed three times with PBS, and resuspended in 5 ml PBS. For semi-quantitative determination of bacterial heme, 5 ml PBS suspensions were 2-fold serially diluted in PBS, 100 µl aliquots spotted on PVDF membranes, and heme was detected by an enhanced chemiluminescence (ECL) Western Blot detection kit (Pierce).
Co-culture of NTHi3655luxABCDE with heme donors
NTHi3655luxABCDE (recipient) was co-cultured with H. influenzae heme donors [63]. Donor strains were grown on chocolate agar, inoculated in 5 ml heme-deficient medium, and grown for 8 h with shaking (200 r.p.m.). Thereafter, 100 µl aliquots of these cultures were added to 20 ml sBHI and grown for 16 h under the same conditions. To generate heme starvation, NTHi3655luxABCDE was grown on chocolate agar, inoculated in 20 ml heme-deficient medium, and grown overnight with shaking (200 r.p.m.). All cultures were then recovered by centrifugation (10 min, 4000 r.p.m.), washed twice in BHI, and resuspended in 10 ml BHI. For co-cultures, appropriate volumes of each suspension (donor and recipient) containing identical number of colony forming units (CFU) were mixed in 5 ml heme-deficient medium generating a starting co-culture with OD600 = 0.1, to be incubated for 6 h. For recipient controls, similar NTHi3655luxABCDE cultures were prepared and grown in sBHI or heme-deficient medium (positive and negative controls, respectively). Luminescence was monitored every hour for 6 h in a 1,450 MicroBeta TriLux counter (PerkinElmer), using 100 µl bacterial culture aliquots and 96-well black plates (Uniplate®, Whatman). Experiments were performed in triplicates at ≥ 3 independent occassions (n ≥ 9).
Bacterial susceptibility to antimicrobials
Antibiotic minimal inhibitory concentrations (MICs) were determined by microdilution using commercial panels (STRHAE2; Sensititre), and interpreted by following the criteria of the Clinical Laboratory Standards Institute (CLSI) [78]. For polymyxin E (PxE), Etests (Biomérieux) on MH-F agar were performed: bacteria were grown on chocolate agar for 8 h, inoculated in 7 ml of heme-deficient medium or sBHI, and grown for 12 h at 37°C, 200 r.p.m. Cultures were centrifuged (10 min, 4000 r.p.m.), suspensions normalized to ~0.5 McFarland (equivalent to OD600 = 0.063) in saline solution, and used as inoculum on MH-F agar (n = 2). To monitor susceptibility to H2O2, bacteria grown on chocolate agar for 8 h were inoculated in 7 ml of sBHI, and grown for 12 h at 37°C, 200 r.p.m. Cultures were centrifuged (10 min, 4000 r.p.m.), suspensions were normalized to OD600 = 1 (~7×108 CFU/ml) in PBS and plated on sHTM agar on large Petri dishes (20 cm diameter; 80 ml sHTM agar/plate; 1.8 ml suspension/plate). H2O2 at 9.8 M (Sigma-Aldrich) was used as stock solution to be diluted in sterile distilled water. Sterile paper disks soaked on 10 μl H2O2 containing 25 or 50 μmol were located on the sHTM agar and incubated for 20 h at 37°C. Results are shown as diameter of bacterial growth inhibition (mm) around H2O2 disks (means ± SD). Assays were performed at ≥ 3 independent occassions (n ≥ 3).
RNA extraction and real-time quantitative PCR (RT-qPCR)
Two to 5 colonies of strains grown on chocolate agar were inoculated in 20 ml sBHI, grown for 11 h, diluted into 40 ml fresh sBHI to OD600 = 0.05, and grown to OD600 = 0.5–0.6 (named as conventional sBHI cultures). Alternatively, bacterial biomass was collected from chocolate agar, inoculated in 5 ml heme-deficient medium and grown for 8 h. Next, 100 μl aliquots were inoculated into 20 ml sBHI and grown overnight (37°C, 5% CO2, 200 r.p.m.). Overnight cultures were collected by centrifugation (10 min, 4000 r.p.m.), washed 3 times in PBS, resuspended in 5 ml PBS, diluted to OD600 = 0.05 in 40 ml fresh BHI-NAD or sBHI, and grown to OD600 = 0.4–0.5 [63]. Bacterial total RNA was isolated using TRIzol reagent (Invitrogen), and total RNA quality evaluated using RNA 6,000 Nano LabChips (Agilent 2,100 Bioanalyzer, Santa Clara, CA). All samples had intact 23S and 16S ribosomal RNA bands. Reverse transcription (RT) was performed using 1 µg of RNA by PrimerScript RT Reagent kit (Takara). PCR amplification was performed by using SYBR Premix Ex Taq II (Tli RNaseH Plus) (Takara). Fluorescence was analyzed with AriaMx Real-Time PCR System (Agilent Technologies). The comparative threshold cycle (Ct) method was used to obtain relative quantities of mRNAs that were normalized using 16S ribosomal RNA (16SrRNA) as an endogenous control. Primers were designed with Primer3 software (Table S3). All cultures were grown at least three times, and samples were processed with technical triplicates (n ≥ 3).
Cell culture and bacterial infection
A549 type II human pneumocytes (ATCC CCL-185) were maintained and seeded as described [79–86]. For infection, PBS-normalized bacterial suspensions (OD600 = 1) were prepared by using NTHi strains grown on chocolate agar. Alternatively, bacteria grown on chocolate agar were inoculated in 7 ml heme-deficient medium or sBHI, and grown for 12 h at 37°C, 200 r.p.m. Cultures were centrifuged (10 min, 4000 r.p.m.), suspensions normalized to OD600 = 1 in PBS, and then used as infecting inoculum. A multiplicity of infection (MOI) of ~100:1 was used. To monitor adhesion, cells were infected for 30 min. This assay does not completely exclude a possible internalization of some bacteria, and experimental conditions were previously set to mainly monitor adhesion [84]. For invasion testing, cells were incubated with bacteria for 2 h, washed 3 times with PBS, incubated for 1 h with RPMI 1640 medium containing 10% FCS, 10 mM Hepes, and 200 µg/ml gentamicin. After infection, cells were lysed with PBS-saponin 0.025%, 10-fold diluted and plated on sBHI agar. All infections were performed in triplicates at ≥ 3 independent occassions (n ≥ 9). Results are expressed as CFU/well.
Secretion of IL-8
Bacteria grown on chocolate agar were collected with PBS, suspensions normalized to OD600 = 1 and used for 2 h infection with a MOI of ~100:1. Cells were washed three times with PBS, and incubated for 6 h in RPMI 1640 medium containing 10% FCS, 10 mM Hepes, and 100 µg/ml gentamicin. Supernatants were collected from the wells, cell debris was removed by centrifugation and samples were frozen at −80°C. IL-8 levels in supernatants were measured by ELISA (Abnova KA0115) with sensitivity <2 pg/ml. Infections were performed in duplicate and at least twice (n = 4). Results on IL-8 are expressed in pg/ml.
NTHi mouse lung infection
A mouse model of NTHi lung infection was used as described previously [86]. CD1 female mice were purchased from Charles River Laboratories (France), housed under pathogen-free conditions at the Institute of Agrobiotechnology facilities (registration number 365 ES/31-2016-000002-CR-SU-US), and used at 22–25 g (6–7 weeks). Animal handling and procedures were in accordance with the current European (Directive 2010/63/EU) and National (Real Decreto 53/2013) legislations, following the FELASA and ARRIVE guidelines, with the approval of Universidad Pública de Navarra (UPNa) Animal Experimentation Committee, and in accordance with the Helsinki declaration. Bacteria grown on chocolate agar were recovered in PBS generating normalized suspensions (~5 × 109 CFU/ml). Next, 20 µl suspensions (~1 × 108 CFU/mice) were placed at the entrance of the nostrils until complete inhalation, in mice previously anesthetized with ketamine-xylazine (3:1). At 24 or 48 h post-infection (hpi), mice were euthanized using cervical dislocation. Bronchoalveolar lavage fluid (BALF) samples were obtained by perfusion and collection of 0.7 ml of PBS, with help of a sterile 20G (1.1 mm diameter) VialonTM intravenous catheter (Becton-Dickinson) inserted into the trachea. Recovered BALF was serially 10-fold diluted in PBS, and plated in triplicate on sBHI agar to determine the number of viable bacteria. By following standardized published procedures [87], we considered that we could have a minimum of 3.3 CFU in 1 ml of sample without detecting bacteria (limit of detection <3–4 CFU/ml BALF), rendering log10 = 0.52. The left lung was individually weighed in sterile bags (Stomacher80, Seward Medical) and homogenized 1:10 (wt/vol) in PBS. Each homogenate was serially 10-fold diluted in PBS and plated in triplicate on sBHI agar to determine the number of CFU per lung. Infections were performed in groups of at least five mice per strain and time point (n ≥ 5).
Statistical analysis
In all cases, p < 0.05 value was considered statistically significant. Analyses were performed using Prism software, version 7 for Mac (GraphPad Software) statistical package, and are detailed in each figure legend.
Results
Structural basis of heme binding to the HbpA, SapA, PE, and HxuA proteins
First, we investigated the ability of HxuA, HbpA, PE, and SapA to bind heme in silico using the HADDOCK (High Ambiguity-Driven Protein DOCKing) server [65,66]. HxuA is part of the heme-hemopexin heme transport system which consists of the HxuC and the two-partner secretion system HxuBA. HxuA is exposed at the cell surface, and its interaction with hemopexin leads to heme release and its capture by HxuC [51–56]. PE is an outer membrane lipoprotein, the dimer of which has been proposed to bind heme [63]; and SapA and HbpA are Cluster C substrate-binding proteins (SBP) that locate at the periplasm and act in concert with the SapBCDFZ and DppBCDF inner membrane ABC transporters, respectively [57,59–62]. Docking results for all four proteins are summarized in Table S4.
Since there are no crystal structures of HbpA and SapA, homology models were constructed based on the crystal structure of HbpA from H. parasuis (HpHbpA) in its glutathione-bound form (PDB code 3M8U) [62] using I-TASSER [67,68]. The sequence identity for the structurally aligned region was 74% and 30% for HbpA and SapA, respectively. Despite the lower sequence identity for SapA, a sequence alignment with HbpA, HpHbpA and the structurally homologous dipeptide-binding protein DppA from E. coli (EcDppA) [88] confirmed high sequence similarity (Fig. S2), supporting HpHbpA as a good structural template for SapA as well as HbpA. Ligand binding site residues predicted by COACH [69,70], a ligand-binding prediction server integrated into I-TASSER suite, was used to drive the docking.
For HbpA, the highest scoring docking solution suggests heme binding at a cleft between the N-terminal and C-terminal domains (Figure 2(a–e)), similar to what is seen for binding of glutathione and dipeptide to HpHbpA and EcDppA respectively (Fig. S3A-B). In SapA, heme docks in a similar location but at a different angle due to the presence of a longer loop (residues 140–158, in orange in Figure 2(a,b)). This loop extension is not conserved in HbpA, HpHbpA and EcDppA, and thus seems to be a unique feature of SapA (Fig. S2). Since the position of this loop must be regarded as speculative due to its absence in the homology modeling template, we removed it from the SapA homology model and repeated the docking experiment. In the absence of the extended loop, the predicted heme binding mode is closer as to what is seen for HbpA, although the site is closer to the protein surface.
Figure 2.
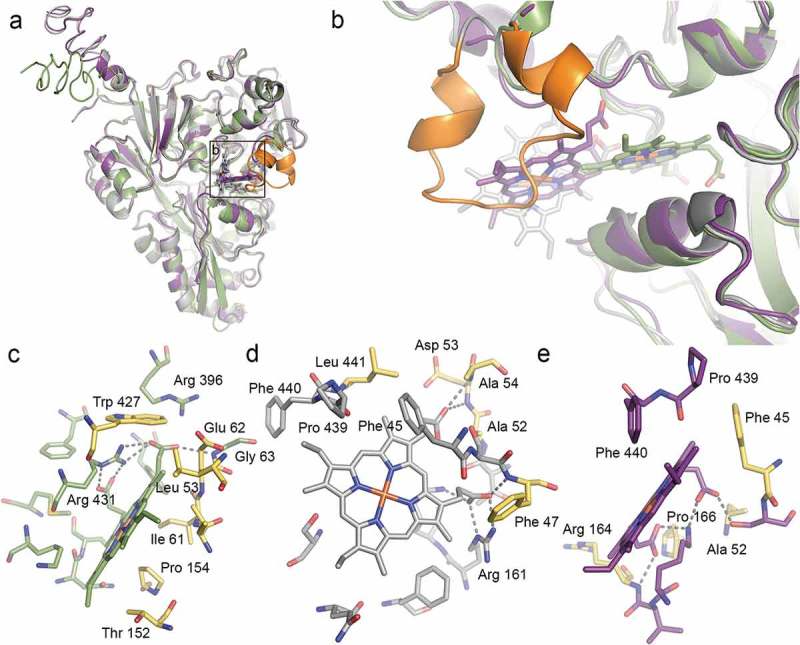
Structural prediction of heme binding to HbpA and SapA. HADDOCK was used to dock heme B to homology models of HbpA (green), full-length SapA (grey) and truncated SapA (purple) in which a non-conserved loop consisting of residues 140–158 has been removed. The truncated residues are colored orange in full-length SapA. (a) Overlay of the best scoring docking solutions for HbpA, SapA and truncated SapA. (b) Zoom-in on predicted heme binding sites showing how HbpA binds heme deeper into the ligand-binding pocket. In full-length SapA, the non-conserved loop (orange) prevents heme binding into the same pocket whereas in truncated SapA, the pocket is more accessible. (c-e) LigPlot+ analysis of the predicted heme binding site in HbpA (c), full-length SapA (d) and truncated SapA (e) with residues lining the binding sites in stick representation. Predicted hydrogen bonds are showed as dashed lines. Residues that are common for all three binding sites are colored yellow.
Analysis of the heme binding sites using LigPlot+ [71] shows predicted hydrogen bonds between heme propionates and positively charged side chains or main chain nitrogens in all three cases, as well as the presence of hydrophobic residues lining the pockets and providing van der Waals interaction with the rest of the heme group (Figure 2(c-e) and Fig. S3C-D). Despite the differences in heme binding mode, several residues are common for all three binding sites (colored yellow in Figure 2(c–e)).
For HbpA, the suggested site is supported by what has been previously predicted [89] and corresponds well to a very recent study where heme was docked to HpHbpA, OppA from H. influenzae (HiOppA) and NikA from E. coli (EcNikA) [90]. In the latter study, heme was suggested to bind in a site that was distinct but close to the site for other ligands (glutathione for HpHbpA, peptide for HiOppA, and butane-1,2,4-tricarboxylate-chelated nickel for EcNikA) and the ability of bind heme and other ligands simultaneously was demonstrated using surface plasmon resonance. Our docking results reveal a similar scenario, with heme binding close to- but not overlapping with the binding site for glutathione in HpHbpA (Fig. S3).
From the crystal structure of PE, it was suggested that heme could bind in a well-defined pocket formed at the PE dimer interface. Manual docking of heme into this pocket showed that it should be able to accommodate heme but no additional evaluation was performed [63,91]. Here we further explored heme binding to PE by docking heme into the predicted heme-binding pocket. Our results show that heme fits very well with predicted hydrogen bonds formed between heme propionates and polar residues within one of the PE monomers (Asn98, Asn101, and Thr105), as well as van der Waals interactions with hydrophobic residues on both sides of the pocket (Ile65 and Ile96) (Fig. S4A-C).
While HxuC reconstitution in E. coli previously showed its ability to act independently as a functional free heme receptor [53], direct heme binding to HxuA has not been demonstrated. Nevertheless, upon examination of the HxuA crystal structure (PDB code 4RM6) [92], we identified a pocket at the HxuA surface, close to the hemopexin binding site, that showed similarities with the predicted heme binding site in HbpA as indicated above, most notably the presence of several positively charged residues that may interact with heme propionates. The ability of this pocket to accommodate a heme molecule was investigated using HADDOCK, resulting in two solutions with similar docking scores (Table S4). As shown in Fig. S4D-E, heme fits well into this pocket, with the two solutions differing by the heme propionates pointing into or out of the pocket. In both solutions, several positively charged residues (Arg and/or Lys) are well positioned to interact with heme propionates via hydrogen bonds.
In summary, our computational studies provide plausible 3D models for how HbpA, SapA, PE, and HxuA interacts with heme.
Expression of NTHi heme binding protein encoding genes and their response to heme availability
H. influenzae genes encoding heme-binding proteins are transcribed during acute infection [93]. Multiple heme acquisition systems may counteract NTHi heme auxotrophy and human iron nutritional immunity, but could also lead to redundancy. Based on these assumptions, we asked if the systems under analysis could be inter-related to preserve bacterial fitness or prevent heme overloading toxicity, and if such relationship may occur at the gene expression level.
The NTHi375 strain was employed to independently generate hxuCBA, sapA, hbpA, and hpe mutants (gene accession numbers NF38_04090/NF38_04085/NF38_04080, NF38_08010/NF38_08015/NF38_08020/NF38_08025/NF38_08030/NF38_08035, NF38_00680, and NF38_04540) (Table 1). Initial characterization of the generated mutants rendered the following features: (i) growth in sBHI, with slightly longer lag phase for the hxuCBA, hbpA, and hpe mutants than for the WT strain (Fig. S5A). (ii) Aerobic growth on BHI agar around X + V factor disks comparable to that of the WT strain, except for ΔhxuCBA, whose growth showed a trend to be lower (NTHi375 WT, 138 ± 17 mm; NTHi375ΔhxuCBA, 122 ± 18 mm) (Fig. S5B). (iii) Anaerobic growth on BHI agar around V factor disks, and antibiotic susceptibility patterns (Table S5) similar to the WT strain. (iv) Inactivation of heme acquisition systems could modify bacterial iron levels and subsequently, iron reactivity with H2O2 via the Fenton reaction. To prove this phenomenon, inactivation of the sapA and hbpA genes increased growth inhibition around paper disks soaked with H2O2 25 and 50 μmol compared to the WT strain (Fig. S5C).
NTHi375 WT and mutant strains were used to determine the expression of the hxuA, sapA, hbpA, and hpe genes. First, conventional sBHI cultures were employed. The hbpA gene was highly expressed in all cases, with higher expression than that of the hxuA, sapA, and hpe genes. Inactivation of the hxuA, sapA, and hpe genes did not alter hbpA gene expression. However, expression of the hxuA, sapA, and hpe genes showed a consistent trend to be higher in the mutants than in the WT strain, being significant for the hxuA and hpe genes when comparing their expression in the WT and NTHi375ΔhbpA strains (Figure 3(a)).
Figure 3.
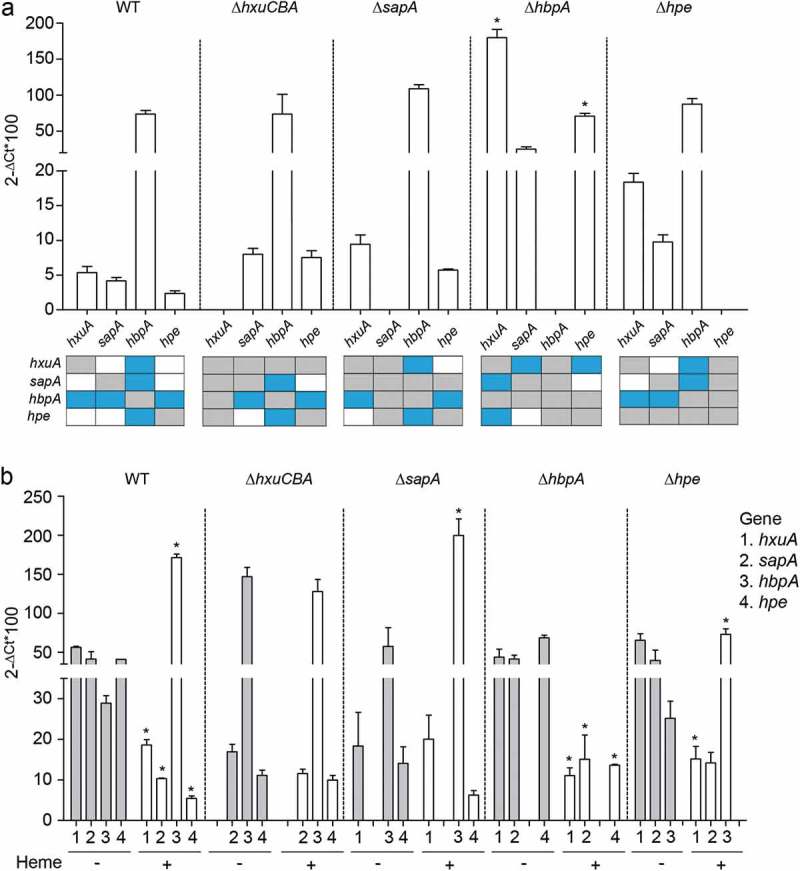
Expression of NTHi heme acquisition systems. (a) Expression of the hxuA, sapA, hbpA and hpe genes in NTHi375 WT, ΔhxuCBA, ΔsapA, ΔhbpA, and Δhpe strains grown in sBHI. Expression of the hbpA gene was higher than that of hxuA, sapA, and hpe (WT strain, higher expression of hbpA than that of hxuA (p < 0.05), sapA and hpe (p < 0.01); NTHi375ΔhxuCBA, higher expression of hbpA than that of sapA and hpe (p < 0.0005); NTHi375ΔsapA, higher expression of hbpA than that of hxuA and hpe (p < 0.0005); NTHi375Δhpe, higher expression of hbpA than that of hxuA (p < 0.05) and sapA (p < 0.0005)). Expression of the hxuA and hpe genes was higher in NTHi375ΔhbpA than in the WT strain (p < 0.0001 and p < 0.01, respectively). Bottom panel: schematic representation of each gene expression compared to each other, in the WT and mutant strains. Color code: blue, statistically significant differences; gray, not determined; white, no significant differences. Data are shown as mean ± SEM. Statistical comparisons of the means were performed with two-way ANOVA (Tukey´s multiple comparisons test). (b) Gene expression and heme sensing. In NTHi375 WT, ΔhbpA and Δhpe strains, hxuA, sapA, and hpe gene expression was higher in heme-restricted medium than in sBHI (WT: hxuA (p < 0.001), sapA (p < 0.01), hpe (p < 0.001); NTHi375ΔhbpA: hxuA (p < 0.01), sapA (p < 0.05), hpe (p < 0.0001); NTHi375Δhpe, hxuA, p < 0.05). In NTHi375 WT, ΔsapA and Δhpe strains, expression of hbpA was lower upon heme depletion (WT, p < 0.0001; NTHi375ΔsapA, (p < 0.001); NTHi375Δhpe, p < 0.05). Data are shown as mean ± SEM. Statistical comparisons of the means were performed with two-way ANOVA (Sidak´s multiple comparisons test).
Bacteria sense heme as means to alert pathogens upon contact with vertebrate tissues, thus modulating expression of systems involved in heme acquisition and metabolism [29,94]. Expression of genes encoding most heme uptake systems increases when iron or heme are restricted [95,96]. We next asked if gene expression responsive to heme availability occurs in a coordinated manner. NTHi375 WT and mutant strains were grown in heme-deficient medium or in sBHI for RNA purification and qRT-PCR analyses. In NTHi375 WT, ΔhbpA, and Δhpe strains, expression of the hxuA, sapA, and hpe (the latter in WT and ΔhbpA strains) genes was higher in heme-restricted medium than in sBHI. In NTHi375 WT, ΔsapA, and Δhpe strains, expression of the hbpA gene was lower upon heme depletion. Lastly, hxuCBA gene inactivation seemed to hamper the effect of heme availability on gene expression (Figure 3(b)).
Overall, these results support the notion that NTHi heme acquisition machinery may act in a coordinated manner, where HbpA seems to play a prominent role, likely followed by PE, and HxuCBA and SapABCDFZ participate in heme sensing, in turn having an impact on the expression of other uptake systems.
SapA, HbpA, and HxuA participate in heme donation to heme-starved NTHi bacterial cells
PE functions as a heme reservoir at the bacterial surface, which can be donated to other H. influenzae bacterial cells starved of heme [63]. We asked if this PE trait could be extended to the other heme-binding proteins. Monitoring heme donation as a means of luminescent emission requires co-culturing a recipient luminescent strain starved of heme and a donor strain cultured in the presence of heme [63]. Unexpectedly, NTHi375 was a poor heme donor, rendering very limited recipient growth (Fig. S6A); in contrast, H. influenzae reference strain RdKW20, which also contains the heme acquisition systems under study (hxuCBA, HI0262_HI0263_HI0264; sapABCDFZ, HI1638_HI1639_HI1640_HI1641_HI1642_HI1643; hbpA, HI0853) [97], donates heme at a significantly higher rate (Figure 4(a)). While comparing NTHi375 and RdKW20 donor strains, we found that the hbpA gene expression was significantly higher in the latter (Fig. S6B). This limitation by NTHi375 led us to perform gene inactivation in RdKW20 to analyze heme storage and inter-bacterial donation. The growth of the generated mutants in sBHI was comparable to that of the WT strain (Fig. S6C). We assessed the use of heme derived from SapA, HbpA, and HxuCBA by setting up co-cultures of NTHi3655luxABCDE starved of heme with RdKW20 WT or mutant strains cultured with heme. NTHi3655luxABCDE grew better when incubated with RdKW20 WT as compared to co-culture with RdKW20ΔsapA, RdKW20ΔhbpA, or RdKW20ΔhxuCBA mutants (Figure 4(a)). We next asked if heme interbacterial donation relates to its binding. To do so, we employed the free iron-binding system HitABC, in which HitA is a periplasmic protein that binds free iron linked to the HitBC transporter located at the inner membrane [98–101]. Inactivation of the hitABC system was performed in RdKW20 (HI0097_HI0098_HI0099). Growth of the generated mutant in sBHI was comparable to that of the WT strain (Fig. S6C). When co-culturing NTHi3655luxABCDE starved of heme with RdKW20ΔhitBC cultured with heme, NTHi3655luxABCDE growth was comparable to that obtained by co-culturing with the WT strain (Figure 4(a)), excluding participation of this free iron transport system in heme donation [98,99,101]. Lastly, deficiency in heme inter-bacterial donation may relate to lower heme binding at the bacterial surface. In fact, more heme was bound to WT than to the hxuCBA, sapA, and hbpA mutants. Unexpectedly, RdKW20ΔhitBC displayed a slight reduction in heme binding, but to a lower extent than that by the ΔhxuCBA, ΔsapA, and ΔhbpA strains (Figure 4(b)).
Figure 4.
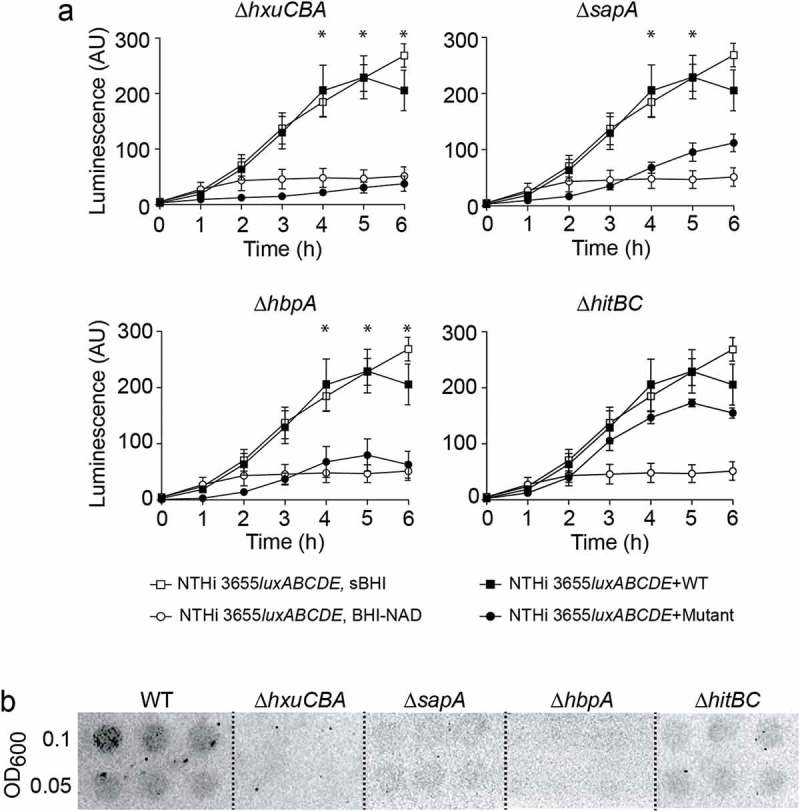
HxuCBA, SapA and HbpA participate in heme inter-bacterial donation. (a) Heme-starved NTHi 3655luxABCDE was used as a heme-recipient strain. Heme donation allowed recipient´s growth, measured as a means of luminescence. NTHi 3655luxABCDE co-culture with Hi RdKW20 WT rendered significant recipient growth, comparable to that of NTHi 3655luxABCDE grown in sBHI; in contrast, NTHi 3655luxABCDE co-culture with ΔhxuCBA, ΔsapA or ΔhbpA, did not promote recipient strain growth at the same extent (see growth time points 4, 5 and 6 h). HitABC does not contribute to surface-bound heme inter-bacterial donation. NTHi 3655luxABCDE co-culture with both Hi RdKW20 WT and ∆hitBC rendered significant recipient growth. White symbols, controls showing recipient strain growth in sBHI (square) or BHI-NAD (circle). Black symbols, recipient strain growth when co-cultured with WT (square) or mutant (circle) strains. *Indicates significant differences when comparing luminescence upon recipient co-culture with WT (square) or mutant (circle) strains. Data are shown as luminescence arbitrary units (AU) and represent mean ± SEM values. Statistical comparisons of the means were performed with two-way ANOVA (Bonferroni multiple comparisons test). (b) Semi-quantitative measure of heme binding on the surface of Hi RdKW20 WT and mutant strains. Normalized bacterial suspensions were 2-fold serially diluted (OD600 = 0.1 and 0.05) and spotted on a PVDF filter. ECL was used for heme detection. Blots are representative images of at least three independent experiments.
Overall, these results suggest that heme binding may work as a storage pool for heme to be shared between H. influenzae bacterial cells during shortage. This ability which was previously observed for the outer membrane lipoprotein PE [63] is now extended to HxuCBA, SapA, and HbpA.
Moonlighting features of H. influenzaeheme-binding systems
Besides heme binding, SapA is required for NTHi resistance to antimicrobial peptides (AMP) and potassium acquisition, HbpA is essential for glutathione import, and PE binds extracellular matrix proteins [62,91,102–106]. To get further insight on the multi-functionality of the heme acquisition systems under study, we tested their contribution to NTHi resistance to polymyxin E (PxE) as a representative AMP. Inactivation of the hxuCBA, sapA, and hpe genes rendered reduced MIC for this AMP; conversely, hbpA gene inactivation increased PxE MIC. Mutant strains showed comparable relative phenotypes when grown in heme-deficient medium or in sBHI (Figure 5(a)). Moreover, we tested their contribution to glutathione import, a vital L-cysteine-containing tripeptide mediating protection against oxidative, xenobiotic, and metal iron stresses [107,108]. H. influenzae is a cysteine and glutathione auxotroph, and imports both molecules. Exogenous cysteine and glutathione, and their oxidized forms cystine and glutathione disulfide (GSSG), sustain H. influenzae growth comparably [73]. NTHi375 WT and mutant strains were inoculated in CDMM supplemented with cystine or GSSG. CDMM supplemented with cystine sustained bacterial growth in all cases. In contrast, CDMM supplemented with GSSG did not sustain NTHi375ΔhbpA growth. NTHi375ΔsapA growth was lower than that of the WT strain, but comparable in CDMM supplemented with cystine or GSSG (Figure 5(b)).
Figure 5.
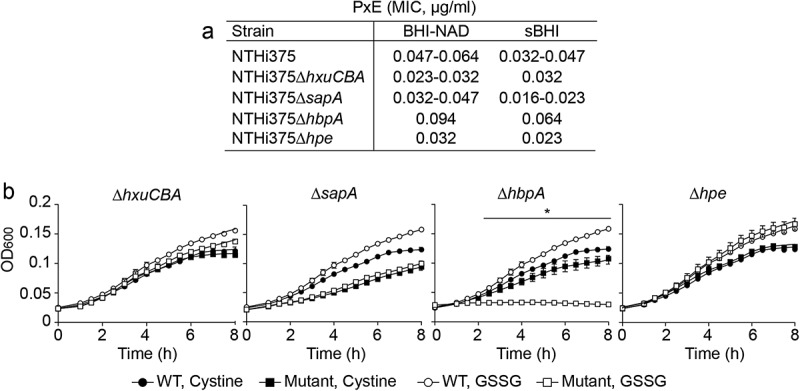
Moonlighting features of NTHi heme acquisition systems. (a) HxuCBA, SapABCDFZ and PE are involved in NTHi resistance to PxE. MIC for PxE was shown to be lower for ΔhxuCBA, ΔsapA and Δhpe mutants than for the WT strain. Inactivation of the hbpA gene rendered higher MIC for PxE. Bacterial inocula were prepared on heme-deficient or sBHI media. (b) HbpA, but not HxuCBA, SapABCDFZ or PE, participates in glutathione import by NTHi. Bacteria were grown in CDMM supplemented with cystine or GSSG. CDMM+cystine sustained bacterial growth in all cases. NTHi375ΔhbpA growth was lower in CDMM+GSSG than in CDMM+cystine (at 2.5 h, p < 0.05; 3 h, p < 0.001; from 3.5 to 8 h p < 0.0001). Data are shown as mean ± SEM. Statistical comparisons of the means were performed using two-way ANOVA (Tukey´s multiple comparisons test).
In conclusion, besides their common role in heme binding, HxuCBA, SapA, and PE also account for AMP resistance, and HbpA accounts for gluthatione import.
NTHi heme acquisition systems contribute to host airways-pathogen interplay
Production of dedicated heme scavenging machinery may also relate to successful bacterial-host interplay [1,6]. We asked whether inactivation of the systems under analysis alters NTHi interplay with A549 type II human pneumocytes. Epithelial invasion by NTHi375ΔhxuCBA, NTHi375ΔsapA, NTHi375ΔhbpA, and NTHi375Δhpe was lower than that observed for the WT strain (Figure 6(a)). All strains stimulated epithelial IL-8 secretion, but such inflammatory response was lower for NTHi375ΔhxuCBA, NTHi375ΔsapA, and NTHi375Δhpe than for the WT strain (Fig. S7A). We next assessed if heme availability modulates the NTHi-host cell interplay by growing NTHi375 WT and mutant strains in heme-deficient medium or sBHI before infection. Epithelial invasion for the mutants was lower than for the WT strain in all cases. Moreover, BHI-NAD rendered lower invasion when compared to that of bacteria grown in sBHI. However, while comparing invasion by BHI-NAD to that of sBHI grown bacteria, the ratio of increased invasion upon growth in sBHI was higher for the mutants (for NTHi375ΔhxuCBA, 17.5-fold; for NTHi375ΔsapA, 42-fold; for NTHi375ΔhbpA, 22.3-fold; for NTHi375Δhpe, 18.5-fold) than for the WT strain, 4.5-fold (Figure 6(b)). At a lower extent, similar observations were obtained when assessing epithelial adhesion (Fig. S7B). Last, we used an in vivo mouse respiratory infection model system [79–81,85,86]. At 24 hpi, NTHi375ΔhxuCBA lung and BALF bacterial numbers were lower than those recovered for the WT strain (Figure 6(c), left panels). At 48 hpi, the four tested mutants rendered numbers lower than those of the WT strain in both lung and BALF samples (Figure 6(c), right panels).
Figure 6.
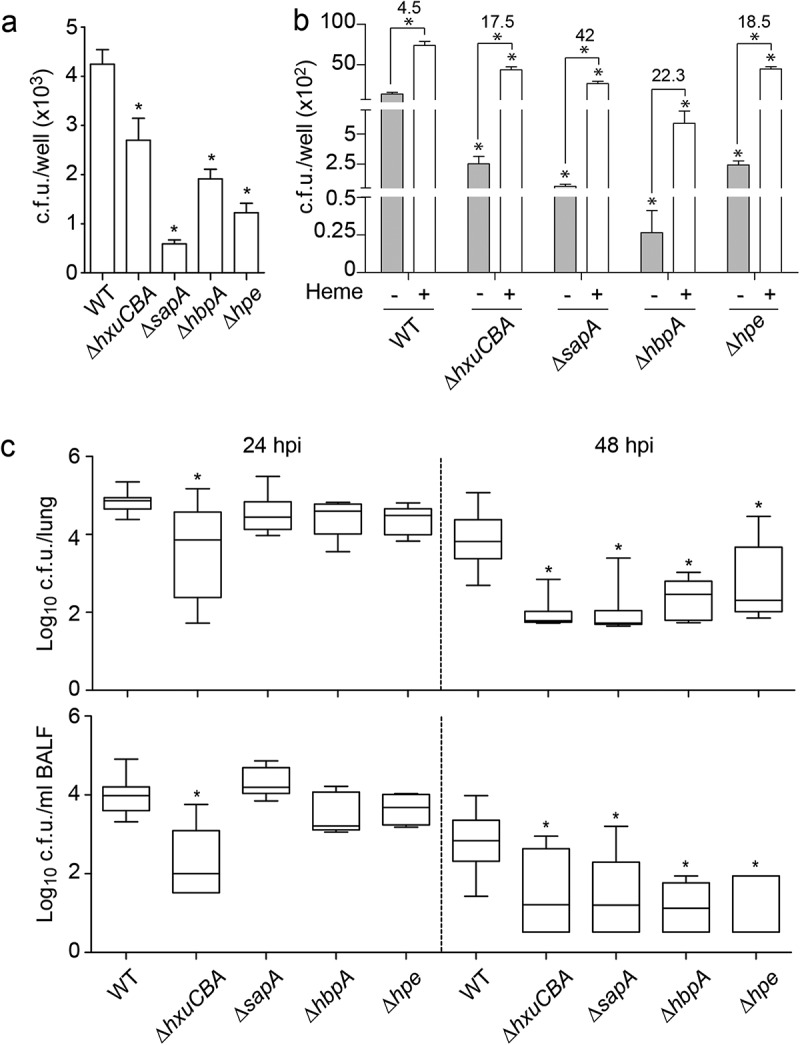
NTHi heme acquisition systems contribute to the host airway-pathogen interplay. (a) Effect of heme uptake inactivation in NTHi interplay with cultured cells. A549 cells were used to quantify invasion by NTHi375 WT, ΔhxuCBA, ΔsapA, ΔhbpA and Δhpe strains. Gene inactivation rendered significantly lower entry into A549 cells than that shown by the WT strain (ΔhxuCBA, p < 0.05; ΔsapA and Δhpe, p < 0.0001; ΔhbpA, p < 0.001). Mean ± SEM values are shown, statistical comparisons of the means were performed using one-way ANOVA (Dunnett’s multiple comparison test). (b) Inactivation of heme-binding systems lowers NTHi interaction with airway epithelial cells in a heme-dependent manner. BHI-NAD (gray bars) and sBHI (white bars) led to decreased A549 invasion rates for the mutants, compared to that of the WT strain (p < 0.0001). Bacterial growth in sBHI increased invasion rates, compared to BHI-NAD (for WT, ΔhxuCBA, ΔsapA and Δhpe, p < 0.0001). Top numbers indicate invasion fold increase for each strain upon growth in sBHI compared to BHI-NAD. Mean ± SEM values are shown, statistical comparisons of the means were performed using two-way ANOVA (Sidak’s multiple comparison test). (c) Effect of heme uptake inactivation in NTHi pulmonary infection. CD1 mice were intranasally infected with WT and mutant strains, euthanized at 24 and 48 hpi, and bacterial loads quantified. Results are reported as log10 CFU/lung and log10 CFU/ml BALF in upper and lower panels, respectively, and represented as box plot graphs (lines inside boxes represent median values). Statistical comparisons were performed using one-way ANOVA (Dunnett’s multiple comparison test). In the lungs, NTHi375ΔhxuCBA showed significantly lower loads at 24 hpi (ΔhxuCBA, p < 0.0005), and all mutants showed significantly lower loads at 48 hpi (ΔhxuCBA, ΔsapA, p < 0.0001; ΔhbpA, p < 0.001; Δhpe, p < 0.05) than those shown by the WT strain. In BALF samples, NTHi375ΔhxuCBA showed significantly lower loads at 24 hpi (p < 0.0001), and all mutants showed significantly lower loads at 48 hpi (ΔhxuCBA, p < 0.05; Δhpe, p < 0.01; ΔsapA and ΔhbpA p < 0.005) than those shown by the WT strain.
In summary, these results support that, under the conditions tested, inactivation of the HxuCBA, SapABCDFZ, HbpA, and PE systems impairs airway infection by NTHi, and that bacterial heme binding may facilitate its interplay with host epithelial cells.
Discussion
Microorganisms have evolved or acquired a variety of specialized iron uptake systems to overcome its environmental limitation and take up iron from extracellular sources, including during infection [109]. For many Gram negative bacteria (including uropathogenic E. coli, Pseudomonas aeruginosa, Neisseria meningitidis, Acinetobacter baumannii), Gram positive species (Staphylococcus aureus), and fungal pathogens (Candida albicans, Cryptococcus neoformans), heme provides the potential for a rich source of iron [110]. Here, we tackled the H. influenzae case, a human host-restricted opportunistic pathogen colonizing the lower airways of patients who undergo iron homeostasis unbalancing (i.e. COPD), and that requires exogenous heme for aerobic respiration, lacks siderophore production, and presents a panel of heme acquisition systems. In this study, differential contribution of HxuCBA, SapA, HbpA, and PE systems to NTHi respiratory infection was evaluated.
The proteins HxuA, SapA, HbpA, and PE share the ability to bind heme. We provide evidence for heme binding to SapA and HbpA Cluster C substrate-binding proteins (SBP) as well as information on the shared predicted structural basis of such binding. In fact, one striking feature of the Cluster C family is that members play dual roles in nutrient uptake, importing their canonical substrates and heme. In NTHi, four Cluster C proteins with capability to bind heme have been identified: HbpA, SapA, and OppA, which bind hydrophobic oligopeptides, and NTHI0310 with an unknown canonical substrate specificity. These proteins present overlapping functionality and multisubstrate specificity [90]. This may not be exclusive to NTHi given that the E. coli Cluster C nickel SBP NikA binds heme and plays a role as a heme chaperone in the periplasm [111]. Regarding HxuA, it should be stressed that our structural analyses may support heme binding, together with the experimental evidence presented in Figure 4 whereas previous data suggests heme binding to HxuC, when expressed together with the NTHi TonB complex in an E. coli heterologous host [53]. The mode of heme binding to PE has been predicted previously [63], and our docking analysis supports this and gives further details about the residues involved.
Apart from the requirement for aerobic respiration, bound heme may also be stored at the bacterial surface or donated to neighboring heme-starved bacterial cells, by contribution of these systems. SapA and HbpA are predicted periplasmic proteins, and may need to be coupled to other currently unknown bacterial systems for effective heme uptake and donation. Moreover, we should not exclude that deficiency in heme donation by the sapA and hbpA mutants could also be an indirect effect. We acknowledge that using RdKW20 mutants to assess heme inter-bacterial donation may be a potential limitation given our use of the NTHi375 strain in the entire study, however, that it should be noted that RdKW20 rendered a useful tool to generate this specific information. Expression of the hxuA, sapA, and hpe genes was comparable between NTHi375 and RdKW20 WT strains, and was in parallel to expression to the same level of the hitB, tonB, tbpA, and dppB heme-iron uptake related genes; only hbpA gene expression was higher in RdKW20 than in NTHi375 strain, which could have an effect on the observed differences between strain backgrounds (Fig. S6B). Although the precise reason for limited heme inter-bacterial donation by NTHi375 is unknown, we provide several evidences suggesting heme binding at this bacterial surface: heme availability increased alveolar epithelial infection by NTHi375, and also increased NTHi375 susceptibility to PxE (more clearly shown for WT, sapA, hxuCBA, and hpe mutant strains) supporting the notion of AMP and heme competition for binding to SapA [58], HxuA or PE.
When tackling a possible relationship among heme uptake systems at the gene expression level, a prominent role for HbpA was observed, based on its high expression and on the effect of its inactivation on the expression of the hxuA, sapA, and hpe genes, suggesting HbpA contribution to down-regulating the activity of other heme piracy systems. However, HbpA does not seem to participate in heme sensing. In fact, in NTHi375 WT, ΔhbpA, and Δhpe strains, expression of the hxuA, sapA, and hpe genes was higher in heme-restricted than in sBHI medium, supporting the notion that the expression of those genes is up-regulated upon heme restriction in a HbpA- and PE-independent manner. Conversely, in NTHi375 WT, ΔsapA, and Δhpe strains, expression of the hbpA gene was lower upon heme depletion, suggesting hbpA gene expression being down-regulated upon heme restriction in a SapABCDFZ- and PE-independent manner. For unknown reasons, we noticed that expression of the hxuA, sapA, and hpe genes in NTHi375ΔhbpA was higher when conventionally grown in sBHI (Figure 3(a)) than in sBHI while assessing the effect of heme availability (Figure 3(b)). We acknowledge that BHI is a medium with non-defined composition that could render reproducibility issues, and emphasize the importance of preserving the use of exactly the same product in each experimental design, as performed in this study. Although to a lesser extent for HxuCBA, SapABCDFZ, and PE, heme acquisition interlinks may contribute to regulating the expression of the different systems, altogether compatible with the notion of a coordinated heme uptake network preventing toxicity by heme excess.
Notably, the heme uptake systems studied present moonlighting features (summarized in Figure 7). We highlight a relationship between NTHi heme acquisition and AMPs. In addition to the already reported involvement of SapABCDFZ in resistance to AMPs [102,103,112], HxuCBA and PE also seem to subvert these elements of the human soluble innate immunity. This was not the case for NTHi375ΔhbpA, which could be linked to the observed higher expression levels of the hxuA, sapA, and hpe genes upon hbpA gene inactivation. Monitoring AMP resistance was motivated by an earlier study where we conducted a genomic analysis of three NTHi clonal isolates serially collected from a bronchiectasis patient, which showed increasing AMP resistance and carried amino acid substitutions in SapABCDFZ and HxuCBA [113]. Based on that and current observations, molecular crosstalk between NTHi heme acquisition and AMP resistance will be addressed in further studies. We also asked if glutathione import could add to protein multi-functionality. This was only the case for HbpA, in line with previous observations [62].
Figure 7.
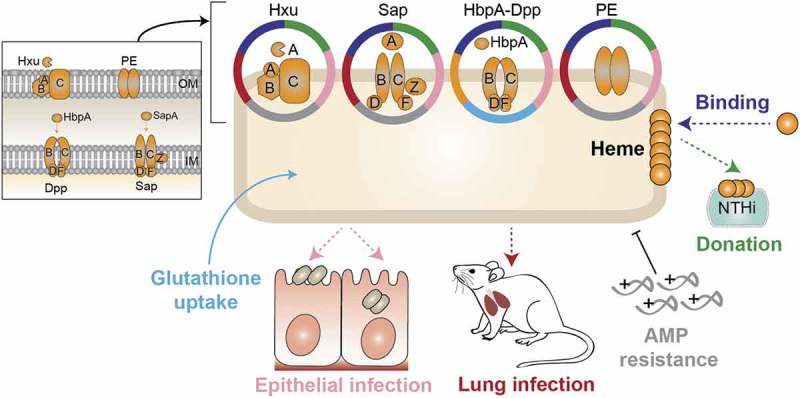
Schematic model showing moonlighting patterns by HxuCBA, SapABCDFZ, HbpA-DppBCDF and PE heme acquisition systems.
The use of cultured airway epithelia and murine pulmonary infection model systems revealed a significant involvement for heme binding systems in the bacterial-host interplay. Data obtained for NTHi375Δhpe support the role of PE as an adhesin [105,106]. In contrast, the sapA gene inactivation has been shown to increase bacterial invasiveness [104], which could relate to the use of non-polarized (this study) versus polarized epithelial cells grown at an air-liquid interface [104]. On the other hand, increased invasion by the WT NTHi strain when pre-grown in sBHI, and heme rescue of hxuCBA, sapA, hbpA, and hpe mutant deficiencies upon epithelial infection, could relate to pathogen subversion of host cell heme receptors such as heme uptake proteins HCP1 and HRG1 by using bound heme as a ligand. Our results somehow differ from those shown by transient iron deprived bacteria developing intracellular bacterial communities [64,114], which could relate to the use of different host cell lines, bacterial strains, or growth conditions. A role for HbpA and HxuCBA in causing bacteremia has been shown in a rat model of invasive disease [55,60], and for SapABCDFZ in a chinchilla model of otitis media (SapA, SapF [103,112,115]) and a mouse model of lung infection (SapF [116]). By using a mouse model of lung infection, we expanded previous observations on HbpA, HxuCBA and SapABCDFZ, and provided new roles for the PE heme-acquisition system in vivo.
In conclusion, the “iron-loving” bacterial species H. influenzae has evolved redundant mechanisms to obtain heme from the human host, and several of them present specific moonlighting properties. We also began elucidating the complex regulatory networks used by NTHi to adapt to the host airways. In general terms, pharmacological modification of host heme-iron metabolism and/or acquisition may hold a key to develop novel strategies to treat or prevent infection. Iron chelators with improved pharmacokinetic properties may become important antimicrobials, and heme-iron transporters and acquisition pathways attractive targets for the development of new treatments to fight infections in an era of increasing burden of antibiotic resistance.
Funding Statement
I.R.A. was funded by a PhD studentship from Universidad Pública de Navarra, Spain; S.M. is funded by a postdoctoral contract from CIBERES. This work has been funded by grants from MINECO SAF2012-31166 and SAF2015-66520-R, from Health Department, Regional Govern from Navarra, Spain, reference 03/2016, and from SEPAR 31/2015 to J.G. CIBER is an initiative from Instituto de Salud Carlos III (ISCIII), Madrid, Spain. This work was also supported by grants from Foundations of Anna and Edwin Berger (KR), the Swedish Medical Research Council (KR: grant number K2015-57X-03163-43-4, www.vr.se), the Cancer Foundation at the University Hospital in Malmö (KR), the Royal Physiographical Society (Forssman’s Foundation) (TAJ), Skåne County Council’s research and development foundation (KR), the Heart Lung Foundation (KR: grant number 20150697, www.hjart-lungfonden.se). The FP7 WeNMR (project# 261572), H2020 West-Life (project# 675858) and the EOSC-hub (project# 777536) European e-Infrastructure projects are acknowledged for the use of their web portals, which make use of the EGI infrastructure with the dedicated support of CESNET-MetaCloud, INFN-PADOVA, NCG-INGRID-PT, TW-NCHC, SURFsara and NIKHEF, and the additional support of the national GRID Initiatives of Belgium, France, Italy, Germany, the Netherlands, Poland, Portugal, Spain, UK, Taiwan and the US Open Science Grid.
Acknowledgments
We thank Lucía Caballero and Drs. Pau Martí-Lliteras and Javier Moleres for technical support, Dr. José Leiva (CUN, Spain) for helpful discussion, and Dr. Auxiliadora Prieto (CIB-CSIC, Spain) for allowing access to a Multiskan device.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental Material
Supplemental data for this article can be accessed here.
References
- [1].Ali MK, Kim RY, Karim R, et al. Role of iron in the pathogenesis of respiratory disease. Int J Biochem Cell Biol. 2017. July;88:181–195. PubMed PMID: 28495571. [DOI] [PubMed] [Google Scholar]
- [2].Ghio AJ. Disruption of iron homeostasis and lung disease. Biochim Biophys Acta. 2009. July;1790(7):731–739. PubMed PMID: 19100311. [DOI] [PubMed] [Google Scholar]
- [3].Barber MF, Elde NC. Buried treasure: evolutionary perspectives on microbial iron piracy. Trends Genet. 2015. November;31(11):627–636. PubMed PMID: 26431675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Krewulak KD, Vogel HJ. Structural biology of bacterial iron uptake. Biochim Biophys Acta. 2008. September;1778(9):1781–1804. PubMed PMID: 17916327. [DOI] [PubMed] [Google Scholar]
- [5].Choby JE, Skaar EP. Heme synthesis and acquisition in bacterial pathogens. J Mol Biol. 2016 August28;428(17):3408–3428. PubMed PMID: 270192980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cassat JE, Skaar EP. Iron in infection and immunity. Cell Host Microbe. 2013 May15;13(5):509–519. PubMed PMID: 23684303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hood MI, Skaar EP. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol. 2012 July16;10(8):525–537. PubMed PMID: 22796883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Runyen-Janecky LJ. Role and regulation of heme iron acquisition in gram-negative pathogens. Front Cell Infect Microbiol. 2013;3:55 PubMed PMID: 24116354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gammella E, Buratti P, Cairo G, et al. Macrophages: central regulators of iron balance. Metallomics. 2014. August;6(8):1336–1345. PubMed PMID: 24905850. [DOI] [PubMed] [Google Scholar]
- [10].Ganz T. Macrophages and systemic iron homeostasis. PubMed PMID: 22441209 J Innate Immun. 2012;4(5–6):446–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ghio AJ, Turi JL, Yang F, et al. Iron homeostasis in the lung. Biol Res. 2006;39(1):67–77. PubMed PMID: 16629166. [DOI] [PubMed] [Google Scholar]
- [12].Cloonan SM, Mumby S, Adcock IM, et al. The “iron”-y of iron overload and iron deficiency in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2017 November1;196(9):1103–1112. PubMed PMID: 28410559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ghio AJ, Hilborn ED, Stonehuerner JG, et al. Particulate matter in cigarette smoke alters iron homeostasis to produce a biological effect. Am J Respir Crit Care Med. 2008 December1;178(11):1130–1138. PubMed PMID: 18723436. [DOI] [PubMed] [Google Scholar]
- [14].Ghio AJ, Stonehuerner JG, Richards JH, et al. Iron homeostasis and oxidative stress in idiopathic pulmonary alveolar proteinosis: a case-control study. Respir Res. 2008 January23;9:10 PubMed PMID: 18215276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gozzelino R, Arosio P. Iron homeostasis in health and disease. Int J Mol Sci. 2016 January20;17(1):130 PubMed PMID: 26805813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Berg K, Wright JL. The pathology of chronic obstructive pulmonary disease: progress in the 20th and 21st centuries. Arch Pathol Lab Med. 2016. December;140(12):1423–1428. PubMed PMID: 27922768. [DOI] [PubMed] [Google Scholar]
- [17].Wong J, Magun BE, Wood LJ. Lung inflammation caused by inhaled toxicants: a review. Int J Chron Obstruct Pulmon Dis. 2016;11:1391–1401. PubMed PMID: 27382275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Nelson ME, O’Brien-Ladner AR, Wesselius LJ. Regional variation in iron and iron-binding proteins within the lungs of smokers. Am J Respir Crit Care Med. 1996. April;153(4 Pt 1):1353–1358. PubMed PMID: 8616566. [DOI] [PubMed] [Google Scholar]
- [19].O’Brien-Ladner AR, Nelson SR, Murphy WJ, et al. Iron is a regulatory component of human IL-1b production. Support for regional variability in the lung. Am J Respir Cell Mol Biol. 2000. July;23(1):112–119. PubMed PMID: 10873160. [DOI] [PubMed] [Google Scholar]
- [20].Thompson AB, Bohling T, Heires A, et al. Lower respiratory tract iron burden is increased in association with cigarette smoking. J Lab Clin Med. 1991. June;117(6):493–499. PubMed PMID: 2045717. [PubMed] [Google Scholar]
- [21].Chappell SL, Daly L, Lotya J, et al. The role of IREB2 and transforming growth factor b-1 genetic variants in COPD: a replication case-control study. BMC Med Genet. 2011 February14;12:24 PubMed PMID: 21320324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Cloonan SM, Glass K, Laucho-Contreras ME, et al. Mitochondrial iron chelation ameliorates cigarette smoke-induced bronchitis and emphysema in mice. Nat Med. 2016. February;22(2):163–174. PubMed PMID: 26752519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].DeMeo DL, Mariani T, Bhattacharya S, et al. Integration of genomic and genetic approaches implicates IREB2 as a COPD susceptibility gene. Am J Hum Genet. 2009. October;85(4):493–502. PubMed PMID: 19800047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Eagan TM, Damas JK, Ueland T, et al. Neutrophil gelatinase-associated lipocalin: a biomarker in COPD. Chest. 2010. October;138(4):888–895. PubMed PMID: 20495108. [DOI] [PubMed] [Google Scholar]
- [25].Engstrom G, Segelstorm N, Ekberg-Aronsson M, et al. Plasma markers of inflammation and incidence of hospitalisations for COPD: results from a population-based cohort study. Thorax. 2009. March;64(3):211–215. PubMed PMID: 18988660. [DOI] [PubMed] [Google Scholar]
- [26].Philippot Q, Deslee G, Adair-Kirk TL, et al. Increased iron sequestration in alveolar macrophages in chronic obstructive pulmonary disease. PLoS One. 2014;9(5):e96285 PubMed PMID: 24789352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pillai SG, Ge D, Zhu G, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009. March;5(3):e1000421 PubMed PMID: 19300482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tandara L, Grubisic TZ, Ivan G, et al. Systemic inflammation up-regulates serum hepcidin in exacerbations and stabile chronic obstructive pulmonary disease. Clin Biochem. 2015. December;48(18):1252–1257. PubMed PMID: 26164540. [DOI] [PubMed] [Google Scholar]
- [29].Skaar EP. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 2010 August12;6(8):e1000949 PubMed PMID: 20711357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Soares MP, Weiss G. The iron age of host-microbe interactions. EMBO Rep. 2015. November;16(11):1482–1500. PubMed PMID: 26474900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ahearn CP, Gallo MC, Murphy TF. Insights on persistent airway infection by non-typeable Haemophilus influenzae in chronic obstructive pulmonary disease. Pathog Dis. 2017 June1;75(4). PubMed PMID: 28449098 DOI: 10.1093/femspd/ftx042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bandi V, Apicella MA, Mason E, et al. Nontypeable Haemophilus influenzae in the lower respiratory tract of patients with chronic bronchitis. Am J Respir Crit Care Med. 2001 December1;164(11):2114–2119. PubMed PMID: 11739144. [DOI] [PubMed] [Google Scholar]
- [33].Finney LJ, Ritchie A, Pollard E, et al. Lower airway colonization and inflammatory response in COPD: a focus on Haemophilus influenzae. Int J Chron Obstruct Pulmon Dis. 2014;9:1119–1132. PubMed PMID: 25342897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sethi S, Evans N, Grant BJ, et al. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N Engl J Med. 2002 August15;347(7):465–471. PubMed PMID: 12181400. [DOI] [PubMed] [Google Scholar]
- [35].Sethi S, Wrona C, Eschberger K, et al. Inflammatory profile of new bacterial strain exacerbations of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2008 March1;177(5):491–497. PubMed PMID: 18079493. [DOI] [PubMed] [Google Scholar]
- [36].Anzueto A. Impact of exacerbations on COPD. Eur Respir Rev. 2010. June;19(116):113–118. PubMed PMID: 20956179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Desai H, Eschberger K, Wrona C, et al. Bacterial colonization increases daily symptoms in patients with chronic obstructive pulmonary disease. Ann Am Thorac Soc. 2014. March;11(3):303–309. PubMed PMID: 24423399. [DOI] [PubMed] [Google Scholar]
- [38].Murphy TF, Brauer AL, Schiffmacher AT, et al. Persistent colonization by Haemophilus influenzae in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2004 August1;170(3):266–272. PubMed PMID: 15117742. [DOI] [PubMed] [Google Scholar]
- [39].Loeb MR. Ferrochelatase activity and protoporphyrin IX utilization in Haemophilus influenzae. J Bacteriol. 1995. June;177(12):3613–3615. PubMed PMID: 7768877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Schlor S, Herbert M, Rodenburg M, et al. Characterization of ferrochelatase (hemH) mutations in Haemophilus influenzae. Infect Immun. 2000. May;68(5):3007–3009. PubMed PMID: 10769004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Granick S, Gilder H. The porphyrin requirements of Haemophilus influenzae and some functions of the vinyl and propionic acid side chains of heme. J Gen Physiol. 1946. September;30:1–13. PubMed PMID: 20997746. [PMC free article] [PubMed] [Google Scholar]
- [42].Morton DJ, Smith A, Ren Z, et al. Identification of a haem-utilization protein (Hup) in Haemophilus influenzae. Microbiology. 2004. December;150(Pt 12):3923–3933. PubMed PMID: 15583146. [DOI] [PubMed] [Google Scholar]
- [43].LaCross NC, Marrs CF, Gilsdorf JR. Otitis media associated polymorphisms in the hemin receptor HemR of nontypeable Haemophilus influenzae. Infect Genet Evol. 2014. August;26:47–57. PubMed PMID: 24820341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Jin H, Ren Z, Pozsgay JM, et al. Cloning of a DNA fragment encoding a heme-repressible hemoglobin-binding outer membrane protein from Haemophilus influenzae. Infect Immun. 1996. August;64(8):3134–3141. PubMed PMID: 8757844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Jin H, Ren Z, Whitby PW, et al. Characterization of hgpA, a gene encoding a haemoglobin/haemoglobin-haptoglobin-binding protein of Haemophilus influenzae. Microbiology. 1999. April;145(Pt 4):905–914. PubMed PMID: 10220170. [DOI] [PubMed] [Google Scholar]
- [46].Maciver I, Latimer JL, Liem HH, et al. Identification of an outer membrane protein involved in utilization of hemoglobin-haptoglobin complexes by nontypeable Haemophilus influenzae. Infect Immun. 1996. September;64(9):3703–3712. PubMed PMID: 8751920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Morton DJ, Bakaletz LO, Jurcisek JA, et al. Reduced severity of middle ear infection caused by nontypeable Haemophilus influenzae lacking the hemoglobin/hemoglobin-haptoglobin binding proteins (Hgp) in a chinchilla model of otitis media. Microb Pathog. 2004. January;36(1):25–33. PubMed PMID: 14643637. [DOI] [PubMed] [Google Scholar]
- [48].Morton DJ, Whitby PW, Jin H, et al. Effect of multiple mutations in the hemoglobin- and hemoglobin-haptoglobin-binding proteins, HgpA, HgpB, and HgpC, of Haemophilus influenzae type b. Infect Immun. 1999. June;67(6):2729–2739. PubMed PMID: 10338475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ren Z, Jin H, Morton DJ, et al. hgpB, a gene encoding a second Haemophilus influenzae hemoglobin- and hemoglobin-haptoglobin-binding protein. Infect Immun. 1998. October;66(10):4733–4741. PubMed PMID: 9746572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Seale TW, Morton DJ, Whitby PW, et al. Complex role of hemoglobin and hemoglobin-haptoglobin binding proteins in Haemophilus influenzae virulence in the infant rat model of invasive infection. Infect Immun. 2006. November;74(11):6213–6225. PubMed PMID: 16966415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cope LD, Love RP, Guinn SE, et al. Involvement of HxuC outer membrane protein in utilization of hemoglobin by Haemophilus influenzae. Infect Immun. 2001. April;69(4):2353–2363. PubMed PMID: 11254593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Cope LD, Thomas SE, Hrkal Z, et al. Binding of heme-hemopexin complexes by soluble HxuA protein allows utilization of this complexed heme by Haemophilus influenzae. Infect Immun. 1998. September;66(9):4511–4516. PubMed PMID: 9712810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Fournier C, Smith A, Delepelaire P. Haem release from haemopexin by HxuA allows Haemophilus influenzae to escape host nutritional immunity. Mol Microbiol. 2011. April;80(1):133–148. PubMed PMID: 21276097. [DOI] [PubMed] [Google Scholar]
- [54].Hanson MS, Pelzel SE, Latimer J, et al. Identification of a genetic locus of Haemophilus influenzae type b necessary for the binding and utilization of heme bound to human hemopexin. Proc Natl Acad Sci U S A. 1992 March01;89(5):1973–1977. PubMed PMID: 1542695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Morton DJ, Seale TW, Madore LL, et al. The haem-haemopexin utilization gene cluster (hxuCBA) as a virulence factor of Haemophilus influenzae. Microbiology. 2007. January;153(Pt 1):215–224. PubMed PMID: 17185550. [DOI] [PubMed] [Google Scholar]
- [56].Wong JC, Patel R, Kendall D, et al. Affinity, conservation, and surface exposure of hemopexin-binding proteins in Haemophilus influenzae. Infect Immun. 1995. June;63(6):2327–2333. PubMed PMID: 7768617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hanson MS, Slaughter C, Hansen EJ. The hbpA gene of Haemophilus influenzae type b encodes a heme-binding lipoprotein conserved among heme-dependent Haemophilus species. Infect Immun. 1992. June;60(6):2257–2266. PubMed PMID: 1339409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Mason KM, Raffel FK, Ray WC, et al. Heme utilization by nontypeable Haemophilus influenzae is essential and dependent on Sap transporter function. J Bacteriol. 2011. May;193(10):2527–2535. PubMed PMID: 21441512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Morton DJ, Madore LL, Smith A, et al. The heme-binding lipoprotein (HbpA) of Haemophilus influenzae: role in heme utilization. FEMS Microbiol Lett. 2005 December15;253(2):193–199. PubMed PMID: 16289530. [DOI] [PubMed] [Google Scholar]
- [60].Morton DJ, Seale TW, Bakaletz LO, et al. The heme-binding protein (HbpA) of Haemophilus influenzae as a virulence determinant. Int J Med Microbiol. 2009. November;299(7):479–488. PubMed PMID: 19451029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Morton DJ, Seale TW, Vanwagoner TM, et al. The dppBCDF gene cluster of Haemophilus influenzae: role in heme utilization. BMC Res Notes. 2009 August24;2:166 PubMed PMID: 19703293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Vergauwen B, Elegheert J, Dansercoer A, et al. Glutathione import in Haemophilus influenzae Rd is primed by the periplasmic heme-binding protein HbpA. Proc Natl Acad Sci U S A. 2010 July27;107(30):13270–13275. PubMed PMID: 20628015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Al Jubair T, Singh B, Fleury C, et al. Haemophilus influenzae stores and distributes hemin by using protein E. Int J Med Microbiol. 2014. July;304(5–6):662–668. PubMed PMID: 24863527. [DOI] [PubMed] [Google Scholar]
- [64].Szelestey BR, Heimlich DR, Raffel FK, et al. Haemophilus responses to nutritional immunity: epigenetic and morphological contribution to biofilm architecture, invasion, persistence and disease severity. PLoS Pathog. 2013;9(10):e1003709 PubMed PMID: 24130500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].van Zundert GCP, Rodrigues J, Trellet M, et al. The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J Mol Biol. 2016 February22;428(4):720–725. PubMed PMID: 26410586. [DOI] [PubMed] [Google Scholar]
- [66].Wassenaar TA, Van Dijk M, Loureiro-Ferreira N, et al. WeNMR: Structural biology on the grid. J Grid Comput. 2012;10(4):743–767. [Google Scholar]
- [67].Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010. April;5(4):725–738. PubMed PMID: 20360767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008 January23;9:40 PubMed PMID: 18215316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Yang J, Roy A, Zhang Y. Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics. 2013 October15;29(20):2588–2595. PubMed PMID: 23975762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Yang J, Roy A, Zhang Y. BioLiP: a semi-manually curated database for biologically relevant ligand-protein interactions. Nucleic Acids Res. 2013. January;41(Database issue):D1096–103. PubMed PMID: 23087378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011 October24;51(10):2778–2786. PubMed PMID: 21919503. [DOI] [PubMed] [Google Scholar]
- [72].Herriott RM, Meyer EY, Vogt M, et al. Defined medium for growth of Haemophilus influenzae. J Bacteriol. 1970. February;101(2):513–516. PubMed PMID: 5308770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Vergauwen B, Pauwels F, Vaneechoutte M, et al. Exogenous glutathione completes the defense against oxidative stress in Haemophilus influenzae. J Bacteriol. 2003. March;185(5):1572–1581. PubMed PMID: 12591874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Allen S, Zaleski A, Johnston JW, et al. Novel sialic acid transporter of Haemophilus influenzae. Infect Immun. 2005. September;73(9):5291–5300. PubMed PMID: 16113244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Tracy E, Ye F, Baker BD, et al. Construction of non-polar mutants in Haemophilus influenzae using FLP recombinase technology. BMC Mol Biol.2008 November11;9:101 PubMed PMID: 19014437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Sinha S, Mell JC, Redfield RJ. Seventeen Sxy-dependent cyclic AMP receptor protein site-regulated genes are needed for natural transformation in Haemophilus influenzae. J Bacteriol. 2012. October;194(19):5245–5254. PubMed PMID: 22821979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Herriott RM, Meyer EM, Vogt M. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J Bacteriol. 1970. February;101(2):517–524. PubMed PMID: 5308771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Clinical & Laboratory Standards Institute. CSLI Guidelines. Performance Standards for Antimicrobial Susceptibility Testing: 27th Edition. CLSI M100-S27 2017. [Google Scholar]
- [79].Euba B, Lopez-Lopez N, Rodriguez-Arce I, et al. Resveratrol therapeutics combines both antimicrobial and immunomodulatory properties against respiratory infection by nontypeable Haemophilus influenzae. Sci Rep. 2017 October16;7(1):12860 PubMed PMID: 29038519 DOI: 10.1038/s41598-017-13034-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Euba B, Moleres J, Segura V, et al. Genome expression profiling-based identification and administration efficacy of host-directed antimicrobial drugs against respiratory infection by nontypeable Haemophilus influenzae. Antimicrob Agents Chemother. 2015. December;59(12):7581–7592. PubMed PMID: 26416856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Euba B, Moleres J, Viadas C, et al. Relationship between azithromycin susceptibility and administration efficacy for nontypeable Haemophilus influenzae respiratory infection. Antimicrob Agents Chemother. 2015. May;59(5):2700–2712. PubMed PMID: 25712355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Euba B, Moleres J, Viadas C, et al. Relative contribution of P5 and Hap surface proteins to nontypeable Haemophilus influenzae interplay with the host upper and lower airways. PLoS One. 2015;10(4):e0123154 PubMed PMID: 25894755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Lopez-Gomez A, Cano V, Moranta D, et al. Host cell kinases, α5 and β1 integrins, and Rac1 signalling on the microtubule cytoskeleton are important for non-typeable Haemophilus influenzae invasion of respiratory epithelial cells. Microbiology. 2012. September;158(Pt 9):2384–2398. PubMed PMID: 22723286. [DOI] [PubMed] [Google Scholar]
- [84].Morey P, Cano V, Marti-Lliteras P, et al. Evidence for a non-replicative intracellular stage of nontypeable Haemophilus influenzae in epithelial cells. Microbiology. 2011. January;157(Pt 1):234–250. PubMed PMID: 20929955. [DOI] [PubMed] [Google Scholar]
- [85].Morey P, Viadas C, Euba B, et al. Relative contributions of lipooligosaccharide inner and outer core modifications to nontypeable Haemophilus influenzae pathogenesis. Infect Immun. 2013. November;81(11):4100–4111. PubMed PMID: 23980106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Rodriguez-Arce I, Marti S, Euba B, et al. Inactivation of the thymidylate synthase thyA in non-typeable Haemophilus influenzae modulates antibiotic resistance and has a strong impact on its interplay with the host airways. Front Cell Infect Microbiol. 2017;7:266 PubMed PMID: 28676846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Grillo MJ, Blasco JM, Gorvel JP, et al. What have we learned from brucellosis in the mouse model? Vet Res.2012 April13;43:29 PubMed PMID: 22500859; PubMed Central PMCID: PMCPMC3410789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Dunten P, Mowbray SL. Crystal structure of the dipeptide binding protein from Escherichia coli involved in active transport and chemotaxis. Protein Sci. 1995. November;4(11):2327–2334. PubMed PMID: 8563629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Dunten P, Mowbray SL. Modeling of the structure of the Haemophilus influenzae heme-binding protein suggests a mode of heme interaction. Protein Sci. 1995. November;4(11):2335–2340. PubMed PMID: 8563630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Tanaka KJ, Pinkett HW. Oligopeptide-binding protein from nontypeable Haemophilus influenzae has ligand-specific sites to accommodate peptides and heme in the binding pocket. J Biol Chem.2019 January18;294(3):1070–1082. PubMed PMID: 30455346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Singh B, Al-Jubair T, Morgelin M, et al. The unique structure of Haemophilus influenzae protein E reveals multiple binding sites for host factors. Infect Immun. 2013. March;81(3):801–814. PubMed PMID: 23275089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Zambolin S, Clantin B, Chami M, et al. Structural basis for haem piracy from host haemopexin by Haemophilus influenzae. Nat Commun.2016 May18;7:11590 PubMed PMID: 27188378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Whitby PW, Sim KE, Morton DJ, et al. Transcription of genes encoding iron and heme acquisition proteins of Haemophilus influenzae during acute otitis media. Infect Immun. 1997. November;65(11):4696–4700. PubMed PMID: 9353052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Stauff DL, Skaar EP. The heme sensor system of Staphylococcus aureus. Contrib Microbiol. 2009;16:120–135. PubMed PMID: 19494582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Whitby PW, Seale TW, VanWagoner TM, et al. The iron/heme regulated genes of Haemophilus influenzae: comparative transcriptional profiling as a tool to define the species core modulon. BMC Genomics.2009 January07;10:6 PubMed PMID: 19128474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Whitby PW, Vanwagoner TM, Seale TW, et al. Transcriptional profile of Haemophilus influenzae: effects of iron and heme. J Bacteriol. 2006. August;188(15):5640–5645. PubMed PMID: 16855256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Fleischmann RD, Adams MD, White O, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science.1995 July28;269(5223):496–512. PubMed PMID: 7542800. [DOI] [PubMed] [Google Scholar]
- [98].Adhikari P, Kirby SD, Nowalk AJ, et al. Biochemical characterization of a Haemophilus influenzae periplasmic iron transport operon. J Biol Chem.1995 October20;270(42):25142–25149. PubMed PMID: 7559648. [DOI] [PubMed] [Google Scholar]
- [99].Anderson DS, Adhikari P, Nowalk AJ, et al. The hFbpABC transporter from Haemophilus influenzae functions as a binding-protein-dependent ABC transporter with high specificity and affinity for ferric iron. J Bacteriol. 2004. September;186(18):6220–6229. PubMed PMID: 15342592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Anderson DS, Adhikari P, Weaver KD, et al. The Haemophilus influenzae hFbpABC Fe3+ transporter: analysis of the membrane permease and development of a gallium-based screen for mutants. J Bacteriol. 2007. July;189(14):5130–5141. PubMed PMID: 17496104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Sanders JD, Cope LD, Hansen EJ. Identification of a locus involved in the utilization of iron by Haemophilus influenzae. Infect Immun. 1994. October;62(10):4515–4525. PubMed PMID: 7927717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Mason KM, Bruggeman ME, Munson RS, et al. The non-typeable Haemophilus influenzae Sap transporter provides a mechanism of antimicrobial peptide resistance and SapD-dependent potassium acquisition. Mol Microbiol. 2006. December;62(5):1357–1372. PubMed PMID: 17064364. [DOI] [PubMed] [Google Scholar]
- [103].Mason KM, Munson RS Jr., Bakaletz LO. A mutation in the sap operon attenuates survival of nontypeable Haemophilus influenzae in a chinchilla model of otitis media. Infect Immun. 2005. January;73(1):599–608. PubMed PMID: 15618200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Raffel FK, Szelestey BR, Beatty WL, et al. The Haemophilus influenzae Sap transporter mediates bacterium-epithelial cell homeostasis. Infect Immun. 2013. January;81(1):43–54. PubMed PMID: 23071138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Ronander E, Brant M, Eriksson E, et al. Nontypeable Haemophilus influenzae adhesin protein E: characterization and biological activity. J Infect Dis.2009 February15;199(4):522–531. PubMed PMID: 19125675. [DOI] [PubMed] [Google Scholar]
- [106].Ronander E, Brant M, Janson H, et al. Identification of a novel Haemophilus influenzae protein important for adhesion to epithelial cells. Microbes Infect. 2008. January;10(1):87–96. PubMed PMID: 18069033. [DOI] [PubMed] [Google Scholar]
- [107].Morris D, Khurasany M, Nguyen T, et al. Glutathione and infection. Biochim Biophys Acta. 2013. May;1830(5):3329–3349. PubMed PMID: 23089304. [DOI] [PubMed] [Google Scholar]
- [108].Sporer AJ, Kahl LJ, Price-Whelan A, et al. Redox-based regulation of bacterial development and behavior. Annu Rev Biochem.2017 June20;86:777–797. PubMed PMID: 28654321. [DOI] [PubMed] [Google Scholar]
- [109].Huang W, Wilks A. Extracellular heme uptake and the challenge of bacterial cell membranes. Annu Rev Biochem.2017 June20;86:799–823. PubMed PMID: 28426241. [DOI] [PubMed] [Google Scholar]
- [110].Carver PL. The battle for iron between humans and microbes. PubMed PMID: 28730969 Curr Med Chem. 2018;25(1):85–96. [DOI] [PubMed] [Google Scholar]
- [111].Shepherd M, Heath MD, Poole RK. NikA binds heme: a new role for an Escherichia coli periplasmic nickel-binding protein. Biochemistry.2007 May1;46(17):5030–5037. PubMed PMID: 17411076. [DOI] [PubMed] [Google Scholar]
- [112].Shelton CL, Raffel FK, Beatty WL, et al. Sap transporter mediated import and subsequent degradation of antimicrobial peptides in Haemophilus. PLoS Pathog. 2011. November;7(11):e1002360 PubMed PMID: 22072973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Garmendia J, Viadas C, Calatayud L, et al. Characterization of nontypeable Haemophilus influenzae isolates recovered from adult patients with underlying chronic lung disease reveals genotypic and phenotypic traits associated with persistent infection. PLoS One. 2014;9(5):e97020 PubMed PMID: 24824990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Hardison RLHD, Harrison A, Beatty WL, et al. Transient nutrient deprivation promotes macropinocytosis-dependent intracellular bacterial community development. mSphere. 2018;3(5):e00286–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Vogel AR, Szelestey BR, Raffel FK, et al. SapF-mediated heme-iron utilization enhances persistence and coordinates biofilm architecture of Haemophilus. Front Cell Infect Microbiol. 2012;2:42 PubMed PMID: 22919633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Gawronski JD, Wong SM, Giannoukos G, et al. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci U S A.2009 September22;106(38):16422–16427. PubMed PMID: 19805314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Hood DW, Makepeace K, Deadman ME, et al. Sialic acid in the lipopolysaccharide of Haemophilus influenzae: strain distribution, influence on serum resistance and structural characterization. Mol Microbiol. 1999. August;33(4):679–692. PubMed PMID: 10447878. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


