ABSTRACT
Autophagy is a conserved self-degradation mechanism that governs a large array of cellular processes in filamentous fungi. Filamentous insect and nematode mycopthogens function in the natural control of host populations and have been widely applied for biological control of insect and nematode pests. Entomopathogenic and nematophagous fungi have conserved “core” autophagy machineries that are analogous to those found in yeast but also feature several proteins involved in specific aspects of the autophagic pathways. Here, we review the functions of autophagy in protecting fungal cells from starvation and stress cues and sustaining cell differentiation, asexual development and virulence. An emphasis is placed upon the regulatory mechanisms involved in autophagic and non-autophagic roles of some autophagy-related genes. Methods used for monitoring conserved or specific autophagic events in fungal pathogens are also discussed.
KEYWORDS: Entomopathogenic fungi, nematophagous fungi, autophagic events, asexual development, stress response, virulence, host-pathogen interactions
Introduction
Autophagy is an evolutionally conserved degradation process much beyond a simply starvation-responsive process considered previously in eukaryotic cells [1]. This self-degradation cellular process has been intensively studied in model yeast and occurs selectively or nonselectively in the form of microautophagy or macroautophagy. Microautophagy takes place via direct uptake of cytoplasm or organelles surrounded by invaginated vacuolar membranes. Nonselective macroautophagy involves random engulfment of cytoplasm and organelles by autophagsomes that appear in the vacuoles containing the contents to be degraded and recycled, contrasting to selective macroautophagy that enables to degrade specific organelles, such as mitochondria, peroxisomes and ribosomes, for removal of redundant or impaired organelles [2]. Autophagy is mediated by the cytoplasm-to-vacuole targeting (Cvt) pathway that is responsible for specific sorting of proteins to vacuoles [3]. Despite conserved features, autophagic proteins are functionally differentiated among fungi [4]. Filamentous fungi are highly divergent in morphology and lifestyle [5], and hence the roles of autophagic processes in their adaptation to host and environment may differ from one lineage to another [6].
Filamentous entomopathogenic and nematophagous fungi play important roles in the natural control of host populations and have been widely applied for biological control of pest insects and nematodes [7,8]. As classic insect mycopathogens, Beauveria bassiana and Metarhizium spp. are a large source of global mycoinsecticides and mycoacricides as alternatives to chemical pesticides [9,10]. Fungal conidia adhere to insect cuticle, where they geminate to infect host through cuticular penetration for entry into host hemocoel [11]. The success of fungal infection is followed by transition of penetrating hyphae into hyphal bodies (namely unicellular blastospores), a process called dimorphic transition that facilitates intrahemocoel proliferation of fungal cells by yeast-like budding until host mummification to death [12–14]. Upon host death, hyphal bodies become septate hyphae that penetrate the host cuticle again for outgrowth and ultimate conidiation on cadaver surfaces for a new infection cycle [15,16]. Nematophagous fungi can be divided into nematode-trapping, egg-parasitic, endoparasitic and toxin-producing groups [17]. The mycelia of the first two groups can form traps to capture namatodes and invade host eggs by the actions of mechanical forces and extracellular hydrolytic enzymes [18], respectively. The infection cycle of an entomopathogenic or nematophagous fungus comprises a wide array of cellular processes and events that are closely linked to autophagy [19,20]. This mini-review aims to update the understanding of autophagic events that are genetically regulated in insect and nematode mycopathgens and associated with their phenotypes crucial for biological control potential, including vegetative growth, cell differentiation, asexual or sexual development, host infection and virulence.
Overview of autophagy-related proteins in insect and nematode mycopathogens
The yeasts Saccharomyces cerevisiae, Komagataella pastoris (formerly Pichia pastoris) and K. phaffii are model species used in autophagic studies. Up to 42 genes have been found encoding autophagy-related proteins (ATGs) and mostly characterized in the yeasts, as summarized in Table 1. Among those, 18 are considered as core genes indispensable for autophagic processes while other 24 are involved into the induction of specific autophagic pathway or selective autophagy [21–23]. The core ATG genes are obligatory for all autophagy-related processes and fall into five functional groups, including ATG1 kinase complex (A1C), membrane recruiting system (MRS), phosphoinositide 3-kinase complex (PI3KC), ubiquitin-like conjugation system (ULCS), and degradation and transportation system (DTS). As illustrated in Figure 1, autophagic behavior is induced by A1C and PI3KC complexes through formation of preautophagosomal structures, followed by vesicle formation and expansion that rely upon ULCS during autophagosome maturation, hydrolyzation and recycling of all engulfed proteins and organelles by DTS in vacuoles [24], and a requirement of MRS for phagophore membrane expansion and vesicle completion [23].
Table 1.
Autophagy-related proteins (ATGs) found in the NCBI protein databases of yeasts, fungal entomopathogens and nematophagous fungi.
* Found in the NCBI protein databases of four yeasts (Hp: Hansenula polymorpha DL1; Kp: Komagataella pastoris NRRL Y-1603; Kph: K. phaffii GS115; Sc: S. cerevisiae S288C), 12 entomopathogenic fungi (Aal: Aschersonia aleyrodis RCEF 2490; Aap: Ascosphaera apis ARSEF 7405; Bba: Beauveria bassiana ARSEF 2860; Bbr: B. brongniartii RCEF 3172; Cm: Cordyceps militaris CM01; If: Isaria fumosorosea ARSEF 2679; Ll: Lecanicillium lecanii RCEF 1005; Mac: Metarhizium acridum CQMa102; Man: M. anisopliae BRIP 53293; Mr: M. robertsii ARSEF 23; Nr, Nomuraea rileyi RCEF 4871; Sp: Sporothrix insectorum RCEF 264), and two nematophagous fungi (Aol: A. oligospora ATCC 24927; Hm: Hirsutella minnesotensis 3608). Red items are the ATGs that have been characterized in insect and nematode mycopathogens.
Figure 1.
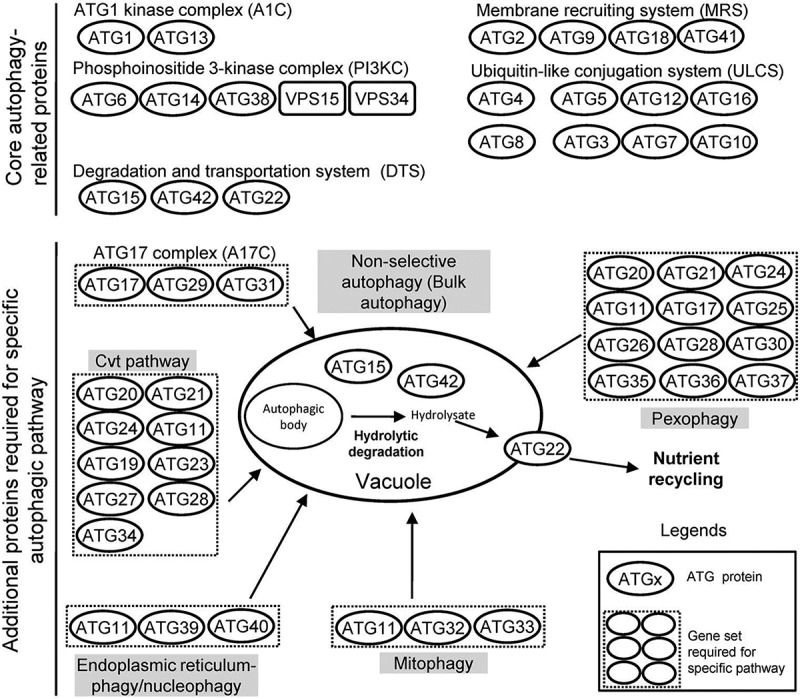
Autophagy-related (ATG) genes functioning in unicellular fungi. Forty-two ATG genes plus Vps14 and Vps34 involved in autophagy pathway of yeast species are sorted into two groups, of which one works in the “core” autophagy machinery and another participates in various specific pathways [24].
In insect and nematode mycopathogens, the ATG genes involved in different autophagic processes are not always identical with the yeast counterparts, and only those associated with A1C complex are completely conserved in fungi (Table 1). For instance, such mycopathogens lack not only ATG41 that interacts with ATG9 and participates in yeast autophagosome biogenesis [22] but also ATG14 and ATG38 that are associated with the yeast PI3KC required for vesicle formation and maturation [23]. ULCS is required for ATG8 activation and involved in two conjugation pathways. One pathway consists of the protease ATG4, the E1-like enzyme ATG7 and the E2-like enzyme ATG3 while another pathway comprises ATG7 and the E2-like enzyme ATG10 [25]. Interestingly, the yeast ATG10 homolog exists in some filamentous fungi [26] but is absent in Ascosphaera apis [27], suggesting less conserved structure or too low sequence identity for ATG10 to be located in the honeybee mycopathogen by BLAST search. ATG22 is a permease that uniquely transports degraded products from vacuole to cytosol in most yeast species [24,26]. In contrast, most of insect and nematode mycopathogens have more transporters homologous to the yeast ATG22. B. bassiana even possesses four ATG22 homologs, of which two (EJP69073 and EJP65688) are transcriptionally expressed during cell proliferation in host hemocoel [28] and another (EJP65315) is involved in fungal pathogenicity, which was reduced by its insertional mutagenesis [29]. This suggests a possibility for some filamentous entomopathogens to have evolved a strategy of utilizing multiple autophagy-related transporters at different stages of infection cycle.
Filamentous fungal ATG proteins involved in specific autophagy pathway exhibit a low degree of conservation [26]. During nonselective macroautophagy induced by starvation, A1C associates with the ATG17 complex (A17C) consisting of ATG17, ATG29 and ATG31 in S. cerevisiae [30]. Of those, ATG31 seems to exist only in S. cerevisiae since its homolog is absent in filamentous and other yeast species, such as Hansenula polymorpha, K. pastoris and K. phaffii. This implicates that a novel mechanism might exist in bulk autophagy of the species other than the budding yeast. In addition, selective autophagy required for cellular homeostasis includes mitophagy, pexophagy, ribophagy, reticulophagy and the Cvt pathway [31]. In selective degradation processes, cargo must be recognized by a receptor and forms a cargo-receptor complex (CRC). ATG11 acts as an essential scaffold protein that mediates the CRC interaction with the core proteins essential for autophagosome formation [32] and is highly conserved in yeasts and filamentous fungi [26]. ATG11 acts as a conserved adaptor which interacts with specific receptors in various pathways [23]. In the budding yeast, aminopeptidase I (Ape1) is translocated into vacuoles via the Cvt pathway, and ATG19 functions as a receptor between Ape1 and ATG11 [23]. In spite of weakly conserved ATG19-B proteins in some yeasts [26], ATG19 is absent in insect and nematode mycopathogens (Table 1), in which it remains unknown whether the Cvt pathway exists and what protein acts as the receptor if it exists. In pexophagy, ATG30 and ATG36 function as the receptor in K. pastoris and S. cerevisiae, respectively [33,34], but both of them are absent in insect and nematode mycopathogens. ATG32, a protein associated with mitochondrial membrane, acts as a receptor and mediates selective degradation of mitochondria (mitophagy) [35]. ATG39 is anchored in perinuclear endoplasmic reticulum (ER) for initiation of reticulophagy and nucleophagy. ATG40 is localized to cortical and cytoplasmic ER for mediation of specific ER degradation [36]. However, such receptors acting in the selective autophagic processes of yeasts lack homologs in insect and nematode mycopathogens. ATG41 existing only in S. cerevisiae has been found to interact with ATG9 and play a role in autophagasome formation [22]. As a newly characterized vacuolar serine carboxypeptidase, ATG42 (Ybr139w) is required for normal vacuole function and the terminal steps of autophagy in S. cerevisiae [21] and exist in all examined yeasts and insect/nematode mycopathognes (Table 1), suggesting its highly conserved role in fungi. In B. bassiana, selective autophagy is evidently associated with cellular stress response, development and virulence. Loss-of-function mutation of ATG11 in B. bassiana has been shown to not completely block autophagic process in vacuoles but to abolish pexophagy and mitophagy during growth in vitro and in vivo [37] although actual receptors involved in the processes remain unclear.
Overall, many of yeast ATG homologs, particularly those receptors, are distinct or absent in the genomic databases of insect and nematode mycopathogens [7,38–40]. This is likely attributable to their essential or nonessential roles in fungal adaptation to hosts and habitats and/or extremely low identities of their sequences to the counterparts in the model yeasts.
Monitoring autophagic events in insect and nematode mycopathogens
Interest in unveiling the prominent role of autophagy in the life cycles of insect and nematode mycopathogens is increasing in the postgenomic era. Effective methods have been explored to monitor autophagic events in these mycopathogens. The acidophilic dye monodansyl cadaverine (MDC) has been used as an indicator of acidic autophagosomes to examine whether the stained structures accumulate in the vacuoles of ΔATG mutants in B. bassiana [41] or at the early stage of mycelial trap formation in the nematode-trapping fungus Arthrobotrys oligospora [20]. Due to a high affinity to acid environment, however, MDC is not suitable for the detection of autophagesomes when other acid vesicles exist [42].
Highly conserved ATG8 is localized on the membrane of autophagosomes to be translocated into vacuoles and considered as a molecular marker for autophagic tracking due to its essentiality for preautophagomal structure formation and autophagosome maturation in eukaryotic cells [43,44]. Fluorescence protein-tagged ATG8 fusion proteins have been successfully used to monitor autophagic events in germlings, hyphae, aerial conidia, submerged blastospores and in vivo hyphal bodies of B. bassiana [20] and in the appressoria formed at the initial stage of infection by Metarhizium robertsii, another important insect mycopathogen [45].
Transmission electron microscopy (TEM) is the most effective method that allows for observation of various autophagic structures, such as phagophores, autophagosomes and autophagic bodies [4]. This method has been employed to unveil the absence/presence of autophagic bodies under autophagy-inducing conditions [20] or during asexual development in the absence of important genes in B. bassiana [13]. In M. robertsii, autophagic bodies in the vacuoles of hyphal cells stressed by starvation are also well visualized via TEM [45]. Recently, dual RNA-seq analysis has been adopted to reveal all possible ATG genes that are expressed during B. bassiana propagation within host hemocoel [28]. This suggests that the molecular detection method is highly effective to monitor the activities of all ATG genes in the in vivo sample of small size.
Autophagic events associated with pest control potential of mycopathogens
The biological control potential of a fungal insect or nematode pathogen depends on not only the virulence or pathogenicity as an indicative ability to invade the host but also cell tolerance to environmental adversity and the asexual development that is critical for in vivo propagation of fungal cells and efficiency of in vitro mass-production [46]. Thus, the fungal potential against pest insects and nematodes is definitely an output of cellular functions and processes that are linked to autophagic events, as illustrated in Figure 2.
Figure 2.
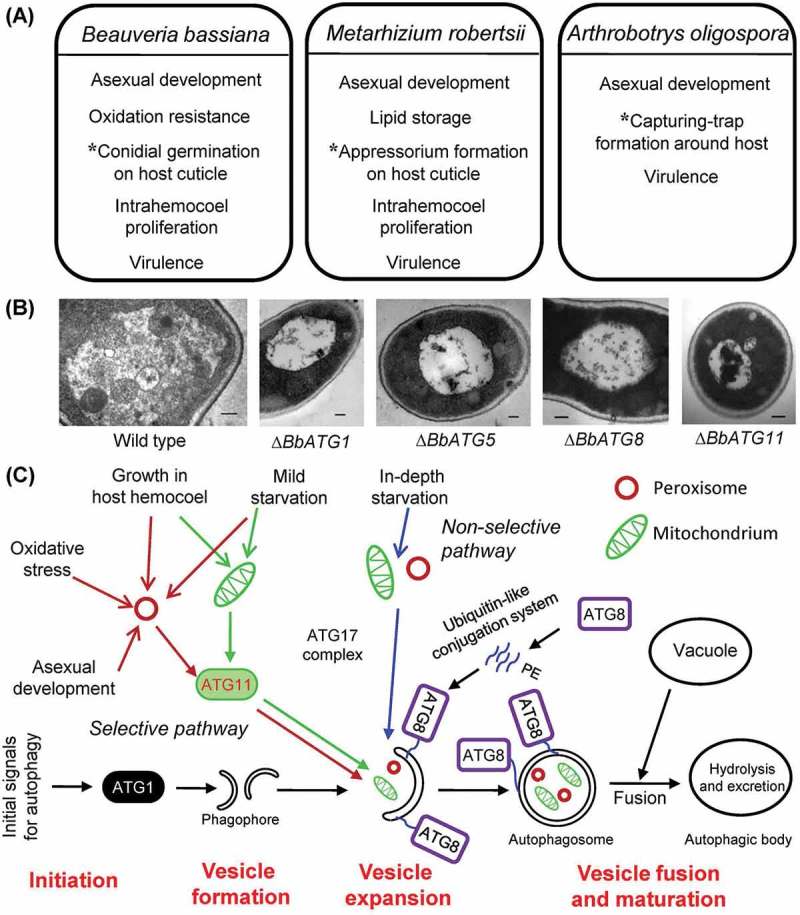
Overview of autophagic events in entomopathogenic and nematophagous fungi. (A) Divergent roles of autophagic events in sustaining the in vitro and in vivo cellular processes of B. bassiana, M. robertsii and A. oligospora, three representative mycopathogens that have evolved for adaptation to distinct host spectra and associated habitats and fall into different lineages. Autophagy mediates the asterisked process that is distinct for each of the fungal pathogens to penetrate through the host cuticle after conidial germination. (B) Transmission electronic microscopic images (scale bars: 0.2 μm) for intravacuolar autophagic events altered by singular deletions of ATG1, ATG5, ATG8 and ATG11 in B. bassiana. (C) Proposed model for autophagy pathways in B. bassiana, including starvation-induced or non-selective autophagy, selective autophagy and bulk autophagy. PE: phosphatidylethanolamine.
Autophagy in response to nutritional starvation/shift
During conidial germination on scant media or oligotrophic insect cuticle, B. bassiana may make use of autophagy to mobilize and recycle intracellular stored nutrients. This role is evidenced with abolishment of autophagic process in the absence of some ATG genes, such as ATG1, ATG5 and ATG8 [20,41]. The conidia of these ΔATG mutants germinated as well as the wild-type conidia on rich medium but suffered germination defects in response to nutritional starvation on water agar and host cuticle. Additionally, deletion of ATG11 in B. bassiana resulted in a block of selective autophagy during conidial germination on oligotrophic substrata and significant germination defects under nutrient deficient conditions [37]. Similarly, nematode surface is also a nutritionally poor substratum for nematode-trapping fungi [47], in which autophagic process is induced by amino acid starvation [19].
A plenty of fatty acids and lipids are used as carbon sources by insect mycopathogens during their infection to host through cuticular penetration [48]. Upon entry into the host hemocoel, fungal cells need metabolize hemolymph-rich trehalose and other carbohydrates and convert them to glucose for use in intrahemocoel propagation [49]. Pexophagy was first found in the cells of K. pastoris grown in an oleic acid-based medium and then shifted into a glucose-based medium [50]. In B. bassiana, both pexophagy and mitophagy are evidently involved in cell response to carbon shift [37].
Autophagy in response to oxidative stress
Autophagy plays an important role in scavenging damaged organelles and proteins in the response of mammal cells to oxidative stress [51]. In B. bassiana, ATG1 and ATG8 are functionally different in antioxidant response since total activity of superoxide dismutases (SODs) decreased by 50–70% in absence of ATG8 but was not affected in absence of ATG1 [20]. This contrasts to increased resistance to oxidative stress in the absence of either ATG1 or ATG8 in Aspergillus niger [52]. Similarly, loss-of-function mutations of ATG1 and ATG8 in S. cerevisiae resulted in enhanced SOD activities [53]. Apparently, ATG1 and ATG8 could be independent of each other in the response of B. basssiana to oxidative stress, and mechanistically different from their homologs in the antioxidant response of both mentioned fungi.
Reactive oxygen species (ROS) causing oxidative damage to mitochondria can be scavenged by selective autophagy for mitochondrial homeostasis [54]. Interestingly, ATG11 has been confirmed to function in antioxidant response of B. bassiana through pexophagy rather than mitophagy [37] although a mechanism underlying the ATG11-induced pexophagy under oxidative stress remains unclear.
Autophagy during cell differentiation and development
Autophagy has been widely linked to cell differentiation in many filamentous fungi [55]. In B. bassiana, normal autophagy is required for asexual development and morphogenesis. For instance, deletion of ATG1, ATG5 or ATG8 has been shown to greatly reduce the yields of aerial conidia as infective propagules or submerged blastospores as an index of in vivo dimorphic transition rate [20,41]. Autophagy is also involved in the conidiation of M. robertsii [45] or in the formation of mycelial nematode traps by A. oligospora [19].
Moreover, some ATG genes may regulate conidiation via different pathways. For example, a conidial protein (BbCP15) is required for conidiation due to the role of its acting as a downstream target of ATG1 instead of ATG8 in B. bassiana [20]. ATG5 is linked to conidial morphology in B. bassiana due to conidial size enlarged in absence of ATG5 [41]. These findings indicate distinct roles for some ATG genes in the cell differentiation and development that are associated with the in vitro and in vivo life cycles of insect and nematode mycopathogens.
Autophagy associated with host infection and fungal virulence
Fungal virulence is a pleiotropic phenotype linked to an array of cellular processes and events. In M. robertsii, ATG8 is essential for the formation of appressoria that initiate cuticular penetration in the course of host infection [45]. Deletion of ATG1, ATG5 or ATG8 resulted in blocked autophagy and attenuated virulence in B. bassiana [20,41], M. robertsii [45] or A. oligospora [19]. These studies demonstrate important impacts of autophagy on the virulence of insect and nematode mycopahtogens but are somewhat different from an absolute requirement of autophagy for the pathogenesis of Magnaporthe grisea, a phytopathogenic fungus [56]. The limited ATG genes characterized to date indicate a close linkage of normal autophagy with the virulence of entomopathogenic and nematophagous fungi. Therefore, different lineages of insect and nematode mycopathogens are ideal models for exploring diverse mechanisms involved in autophagic linkage to fungal virulence.
Regulatory network of autophagy in insect and nematode mycopathogens
In eukaryotes, autophagy is a precisely regulated self-degrading process. The target of rapamycin (TOR) pathway is considered to be a main regulator of autophagy and can inhibit autophagy via phosphorylation of ATG13, a regulator of ATG1 complex [57]. The TOR kinase is inactivated by sensing signals from upstream pathways, followed by formation of autophagy-inducing complex [2]. In B. bassiana, the ATG1 kinase may induce autophagy in response to starving cues [20]. Transcriptional networks learned from some insect and nematode mycopathogens also play important roles in autophagic processes. In Sordaria macrospora (a filamentous ascomycete), a bZIP transcription factor required for vegetative growth and fruiting-body development represses transcriptional expression of ATG4 and ATG8 [58]. Fungus-nematode interaction induces the autophagy of A. oligospora by amino acid starvation in a manner absolutely depending on transcriptional regulation of GCN4 which activates a set of genes required for amino acid biosynthesis [19]. G-protein receptor 3 is required for transcription of ATG1 and ATG2 during the in vitro blastospore formation of B. bassiana [59], suggesting an involvement of the G-protein pathway in signal transduction during autophagy. An in vivo transcriptomic analysis has uncovered that all ATG genes are expressed during B. bassiana propagation in host hemocoel, including ATG4, ATG8 and ATG10 regulated by alternative splicing [28]. In addition, two core eisosome proteins (Pil1A and Pil1B) simultaneously localized at the periphery of hyphal cells have been shown to play opposite roles in the autophagic regulation of B. bassiana, as unveiled by blocked autophagy in absence of Pil1B, restored autophagy in absence of Pil1A and opposite changes in transcript levels of many ATG genes in the mutant strains [13]. These studies indicate a complicated autophagy-regulatory network that remains poorly understood in insect and nematode mycopathogens.
Concluding remarks
Autophagic events exert comprehensive effects on the in vitro and in vivo life cycles of entomopathogenic and nematophagous fungi, in which many ATG genes remain to be functionally explored. The previous studies restricted to several conserved ATG genes have unveiled that their roles in autophagic events are not necessarily similar to those learned from model yeasts or phytopathogenic fungi. We speculate that insect and nematode mycopathogens could have evolved a distinct autophagy-regulatory network that warrants their adaptation to entomopathogenic or nematophagous lifestyle, which could have originated from different evolution histories. In classic insect mycopathogens, for instance, the Beauveria/Cordyceps lineage is considered to have evolved insect pathogenicity 130 million years earlier than the Metarhizium lineage from plant affinity or pathogenicity [38–40,60]. Perhaps for this reason, host spectra differ greatly between B. bassiana and Metarhizium spp [10]. So do their genetic backgrounds required for adaptation to different host spectra and associated habitats. Due to their high potential for use in pest control programs, it is necessary to functionally characterize the ATG family genes of the representative lineages, elucidate contributions of autophagic events to their potential against pest insects and nematodes, and explore possible mechanisms underlying the events. Future emphasis is expectedly placed upon distinct roles of some ATG genes in sustaining biological control potential of insect and nematode mycopathogens. The new knowledge will facilitate development and application of fungal formulations against target pests.
Funding Statement
This work was supported by funding from the National Natural Science Foundation of China (Grant No.: 31670144) and the Ministry of Science and Technology of the People’s Republic of China (Grant No.: 2017YFD0201202).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Ryter SW, Cloonan SM, Choi AMK.. Autophagy: a critical regulator of cellular metabolism and homeostasis. Mol Cells. 2013;36(1):7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Reggiori F, Klionsky DJ. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics. 2013;194(2):341–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lynch-Day M, Klionsky DJ. The Cvt pathway as a model for selective autophagy. FEBS Lett. 2010;584(7):1359–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Pollack JK, Harris SD, Marten MR. Autophagy in filamentous fungi. Fungal Genet Biol. 2009;46(1):1–8. [DOI] [PubMed] [Google Scholar]
- [5].Klein DA, Paschke MV. Filamentous fungi: the indeterminate lifestyle and microbial ecology. Microb Ecol. 2004;47(3):224–235. [DOI] [PubMed] [Google Scholar]
- [6].Voigt O, Pöggeler S. Self-eating to grow and kill: autophagy in filamentous ascomycetes. Appl Microbiol Biotechnol. 2013;97(21):9279–9290. [DOI] [PubMed] [Google Scholar]
- [7].Yang J, Wang L, Ji X, et al. Genomic and proteomic analyses of the fungus Arthrobotrys oligospora provide insights into nematode-trap formation. PLoS Pathog. 2011;7(9):e1002179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wang CS, Wang SB. Insect pathogenic fungi: genomics, molecular interactions, and genetic improvements. Annu Rev Entomol. 2017;62:73–90. [DOI] [PubMed] [Google Scholar]
- [9].de Faria MR, Wraight SP. Mycoinsecticides and Mycoacaricides: a comprehensive list with worldwide coverage and international classification of formulation types. Biol Control. 2007;43(3):237256. [Google Scholar]
- [10].Wang CS, Feng MG. Advances in fundamental and applied studies in China of fungal biocontrol agents for use against arthropod pests. Biol Control. 2014;68:129–135. [Google Scholar]
- [11].Ortiz-Urquiza A, Keyhani NO. Action on the surface: entomopathogenic fungi versus the insect cuticle. Insects. 2013;4(3):357374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wang J, Ying SH, Hu Y, et al. Mas5, a homologue of bacterial DnaJ, is indispensable for the host infection and environmental adaptation of a filamentous fungal insect pathogen. Environ Microbiol. 2016;18(3):10371047. [DOI] [PubMed] [Google Scholar]
- [13].Zhang LB, Tang L, Ying SH, et al. Two eisosome proteins play opposite roles in autophagic control and sustain cell integrity, function and pathogenicity in Beauveria bassiana. Environ Microbiol. 2017;19(5):2037–2052. [DOI] [PubMed] [Google Scholar]
- [14].Tong SM, Zhang AX, Guo CT, et al. Daylight length-dependent translocation of VIVID photoreceptor in cells and its essential role in conidiation and virulence of Beauveria bassiana. Environ Microbiol. 2018;20(1):169–185. [DOI] [PubMed] [Google Scholar]
- [15].He PH, Dong WX, Chu XL, et al. The cellular proteome is affected by a gelsolin (BbGEL1) during morphological transitions in aerobic surface versus liquid growth in the entomopathogenic fungus Beauveria bassiana. Environ Microbiol. 2016;18(11):4153–4169. [DOI] [PubMed] [Google Scholar]
- [16].Cai Q, Wang JJ, Fu B, et al. Gcn5-dependent histone H3 acetylation and gene activity is required for the asexual development and virulence of Beauveria bassiana. Environ Microbiol. 2018;20(4):1484–1497. [DOI] [PubMed] [Google Scholar]
- [17].Liu X, Xiang M, Che Y. The living strategy of nematophagous fungi. Mycoscience. 2009;50(1):20–25. [Google Scholar]
- [18].Li J, Zou C, Xu J, et al. Molecular mechanisms of nematode-nematophagous microbe interactions: basis for biological control of plant-parasitic nematodes. Annu Rev Phytopathol. 2015;53:67–95. [DOI] [PubMed] [Google Scholar]
- [19].Chen YL, Gao Y, Zhang KQ, et al. Autophagy is required for trap formation in the nematode-trapping fungus Arthrobotrys oligospora. Env Microbiol Rep. 2013;5(4):511–517. [DOI] [PubMed] [Google Scholar]
- [20].Ying SH, Liu J, Chu XL, et al. The autophagy-related genes BbATG1 and BbATG8 have different functions in differentiation, stress resistance and virulence of mycopathogen Beauveria bassiana. Sci Rep. 2016;6:26376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Parzych KR, Ariosa A, Mari M, et al. A newly characterized vacuolar serine carboxypeptidase, Atg42/Ybr139w, is required for normal vacuole function and the terminal steps of autophagy in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2018;29(9):1089–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yao Z, Delorme-Axford E, Backues SK, et al. Atg41/Icy2 regulates autophagosome formation. Autophagy. 2015;11(12):2288–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Farré J-C, Subramani S. Mechanistic insights into selective autophagy pathways: lessons from yeast. Nat Rev Mol Cell Biol. 2016;17(9):537–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yang Z, Huang J, Geng J, et al. Atg22 Recycles amino acids to link the degradative and recycling functions of autophagy. Mol Biol Cell. 2006;17(12):5094–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Reumann S, Voitsekhovskaja O, Lillo C. From signal transduction to autophagy of plant cell organelles: lessons from yeast and mammals and plant-specific features. Protoplasma. 2010;247(3–4):233–256. [DOI] [PubMed] [Google Scholar]
- [26].Meijer WH, van der Klei IJ, Veenhuis M, et al. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy. 2007;3(2):106–116. [DOI] [PubMed] [Google Scholar]
- [27].Cornman RS, Bennett AK, Murray KD, et al. Transcriptome analysis of the honey bee fungal pathogen, Ascosphaera apis: implications for host pathogenesis. BMC Genomics. 2012;13:285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Dong WX, Ding JL, Gao Y, et al. Transcriptomic insights into the alternative splicing-mediated adaptation of the entomopathogenic fungus Beauveria bassiana to host niches: autophagy-related gene 8 as an example. Environ Microbiol. 2017;19(10):4126–4139. [DOI] [PubMed] [Google Scholar]
- [29].Kim S, Lee SJ, Nai YS, et al. Characterization of T-DNA insertion mutants with decreased virulence in the entomopathogenic fungus Beauveria bassiana JEF-007. Appl Microbiol Biotechnol. 2016;100(20):8889–8900. [DOI] [PubMed] [Google Scholar]
- [30].Kawamata T, Kamada Y, Kabeya Y, et al. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell. 2008;19(5):2039–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kraft C, Reggiori F, Peter M. Selective types of autophagy in yeast. BBA-Mol Cell Res. 2009;1793(9):404–1412. [DOI] [PubMed] [Google Scholar]
- [32].Suzuki K. Selective autophagy in budding yeast. Cell Death Differ. 2013;20(1):43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Farré JC, Manjithaya R, Mathewson RD, et al. PpAtg30 tags peroxisomes for turnoer by selective autophagy. Dev Cell. 2008;14(3):365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Motley AM, Nuttall JM, Hettema EH. Pex-3anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J. 2012;31(13):2852–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kondo-Okamoto N, Noda NN, Suzuki SW, et al. Autophagy-related protein 32 acts as autophagic degron and directly initiate mitophagy. J Biol Chem. 2012;287(13):10631–10638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mochida K, Oikawa Y, Kimura Y, et al. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. 2015;522(7556):359–362. [DOI] [PubMed] [Google Scholar]
- [37].Ding JL, Peng YJ, Chu XL, et al. Autophagy-related gene BbATG11 is indispensable for pexophagy and mitophagy, and contributes to stress response, conidiation and virulence in the insect mycopathogen Beauveria bassiana. Environ Microbiol. 2018. DOI: 10.1111/1462-2920.14329 [DOI] [PubMed] [Google Scholar]
- [38].Gao Q, Jin K, Ying SH, et al. Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genet. 2011;7(1):e1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zheng P, Xia Y, Xiao G, et al. Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional Chinese medicine. Genome Biol. 2011;12(1):R116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Xiao G, Ying SH, Zheng P, et al. Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci Rep. 2012;2:483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhang L, Wang J, Xie XQ, et al. The autophagy gene BbATG5, involved in the formation of the autophagosome, contributes to cell differentiation and growth but is dispensable for pathogenesis in the entomopathogenic fungus Beauveria bassiana. Microbiol-SGM. 2013;159:243–252. [DOI] [PubMed] [Google Scholar]
- [42].Manafo DB, Colombo MI. A novel assay to study autophagy: regulation of autophagosome vacuole size by amino acid deprivation. J Cell Sci. 2001;114(20):3619–3629. [DOI] [PubMed] [Google Scholar]
- [43].Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3(3):181–206. [DOI] [PubMed] [Google Scholar]
- [44].Klionsky DJ, Abdalla FC, Abeliovich H, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12(1):1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Duan Z, Chen Y, Huang W, et al. Linkage of autophagy to fungal development, lipid storage and virulence in Metarhizium robertsii. Autophagy. 2013;9(4):538–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Zhang LB, Feng MG. Antioxidant enzymes and their contributions to biological control potential of fungal insect pathogens. Appl Microbiol Biotechnol. 2018;102(12):4995–5004. [DOI] [PubMed] [Google Scholar]
- [47].Schmidt AR, Dorfelt H, Perrichot V. Palaeoanellus dimorphus gen. et sp. nov. (Deuteromycotina): a Cretaceous predatory fungus. Am J Bot. 2008;95(10):1328–1334. [DOI] [PubMed] [Google Scholar]
- [48].Gołębiowski M, Boguś MI, Paszkiewicz M, et al. The composition of the cuticular and internal free fatty acids and alcohols from Lucilia sericata males and females. Lipids. 2012;47(6):613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lu YX, Zhang Q, Xu WH. Global metabolomic analyses of the hemolymph and brain during the initiation, maintenance, and termination of pupal diapause in the cotton bollworm, Helicoverpa armigera. PLoS One. 2014;9(6):e99948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Gould SJ, McCollum D, Spong AP, et al. Development of the yeast Pichia pastoris as a model organism for a genetic and molecular analysis of peroxisome assembly. Yeast. 1992;8(8):613–628. [DOI] [PubMed] [Google Scholar]
- [51].Kiffin R, Bandyopadhyay U, Cuervo AM. Oxidative stress and autophagy. Antioxid Redox Signal. 2006;8(1–2):152–162. [DOI] [PubMed] [Google Scholar]
- [52].Nitsche BM, Burggraaf-van Welzen AM, Lamers G, et al. Autophagy promotes survival in aging submerged cultures of the filamentous fungus Aspergillus niger. Appl Microbiol Biotechnol. 2013;97(18):8205–8218. [DOI] [PubMed] [Google Scholar]
- [53].Zhang Y, Qi H, Taylor R, et al. The role of autophagy in mitochondria maintenance: characterization of mitochondrial functions in autophagy deficient S. cerevisiae strains. Autophagy. 2007;3(4):337–346. [DOI] [PubMed] [Google Scholar]
- [54].Ashrafi G, Schwarz TL. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013;20(1):31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Deng YZ, Qu Z, Naqvi NI. Role of macroautophagy in nutrient homeostasis during fungal development and pathogenesis. Cells. 2012;1(3):449–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kershaw MJ, Talbot NJ. Genome-wide functional analysis reveals that infection-associated fungal autophagy is necessary for rice blast disease. Proc Natl Acad Sci USA. 2009;106(37):15967–15972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Russell RC, Yuan HX, Guan KL. Autophagy regulation by nutrient signaling. Cell Res. 2014;24(1):42–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Voigt O, Herzog B, Jakobshagen A, et al. bZIP transcription factor SmJLB1 regulates autophagy-related genes Smatg8 and Smatg4 and is required for fruiting-body development and vegetative growth in Sordaria macrospora. Fungal Genet Biol. 2013;61(1):50–60. [DOI] [PubMed] [Google Scholar]
- [59].Ying SH, Feng MG, Keyhani NO. A carbon responsive G-protein coupled receptor modulates broad developmental and genetic networks in the entomopathogenic fungus, Beauveria bassiana. Environ Microbiol. 2013;15(11):2902–2921. [DOI] [PubMed] [Google Scholar]
- [60].Shang Y, Xiao G, Zheng P, et al. Divergent and convergent evolution of fungal pathogenicity. Genome Biol Evol. 2016;8(5):1374–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]


