ABSTRACT
The RNA-binding protein AUF1 regulates post-transcriptional gene expression by affecting the steady state and translation levels of numerous target RNAs. Remodeling of RNA structures by the largest isoform AUF1 p45 was recently demonstrated in the context of replicating RNA viruses, and involves two RNA remodeling activities, i.e. an RNA chaperone and an RNA annealing activity. AUF1 contains two non-identical RNA recognition motifs (RRM) and one RGG/RG motif located in the C-terminus. In order to determine the functional significance of each motif to AUF1’s RNA-binding and remodeling activities we performed a comprehensive mutagenesis study and characterized the wildtype AUF1, and several variants thereof. We demonstrate that each motif contributes to efficient RNA binding and remodeling by AUF1 indicating a tight cooperation of the RRMs and the RGG/RG motif. Interestingly, the data identify two distinct roles for the arginine residues of the RGG/RG motif for each RNA remodeling activity. First, arginine-mediated stacking interactions promote AUF1’s helix-destabilizing RNA chaperone activity. Second, the electropositive character of the arginine residues is the major driving force for the RNA annealing activity. Thus, we provide the first evidence that arginine residues of an RGG/RG motif contribute to the mechanism of RNA annealing and RNA chaperoning.
KEYWORDS: AUF1, RRM, RGG motif, RNA chaperone, RNA annealer, RNA remodeling, flavivirus, host factor
Introduction
AU-rich element (ARE) binding factor 1 (AUF1, also known as hnRNPD) is a family of four isoforms (p37, p40, p42 and p45) generated by alternative splicing of a common pre-mRNA resulting in isoform-specific sequences encoded by exons 2 and 7 [1]. AUF1’s canonical roles in cellular RNA metabolism include the regulation of stability and/or translation of mRNA targets based on recognition of AU-rich sequences within mRNA 3ʹ untranslated regions (3ʹUTR) [2,3]. In addition to its strong affinity to AU-rich sequences, PAR-CLIP analysis recently revealed a strong preference of AUF1 for U-/GU-rich sequences in mRNAs and non-coding RNAs [4]. A newly discovered role of the protein indicates a functional connection between AUF1 and Argonaute (AGO) proteins, the catalytic components of the RNA-induced silencing complex (RISC) [5]. AUF1 was found to promote loading of miRNA let-7b onto AGO2 which in turn triggered target mRNA decay [6].
All AUF1 isoforms share two tandem, non-identical RNA recognition motifs (RRM) containing canonical RNP-1 and RNP-2 sequence motifs. The RRM protein domain is the most abundant RNA-binding domain and is able to recognize a large number of different RNA sequences and shapes [7]. Conserved phenylalanine residues located in RNP-1 and RNP-2 at positions 100 and 142 within RRM1 and 185 and 227 within RRM2 have been implicated in RNA binding through stacking interactions [8–10]. Within its C-terminus AUF1 harbors an arginine-glycine-rich region (RGG/RG motif), which is, in the case of isoform p42 and p45, interrupted by the exon 7-encoded sequence (Figure 1(a)). The RGG/RG motif is an evolutionarily conserved sequence involved in mediating nucleic acid binding and protein interactions, functions that are often regulated by arginine methylation [11].
Figure 1.
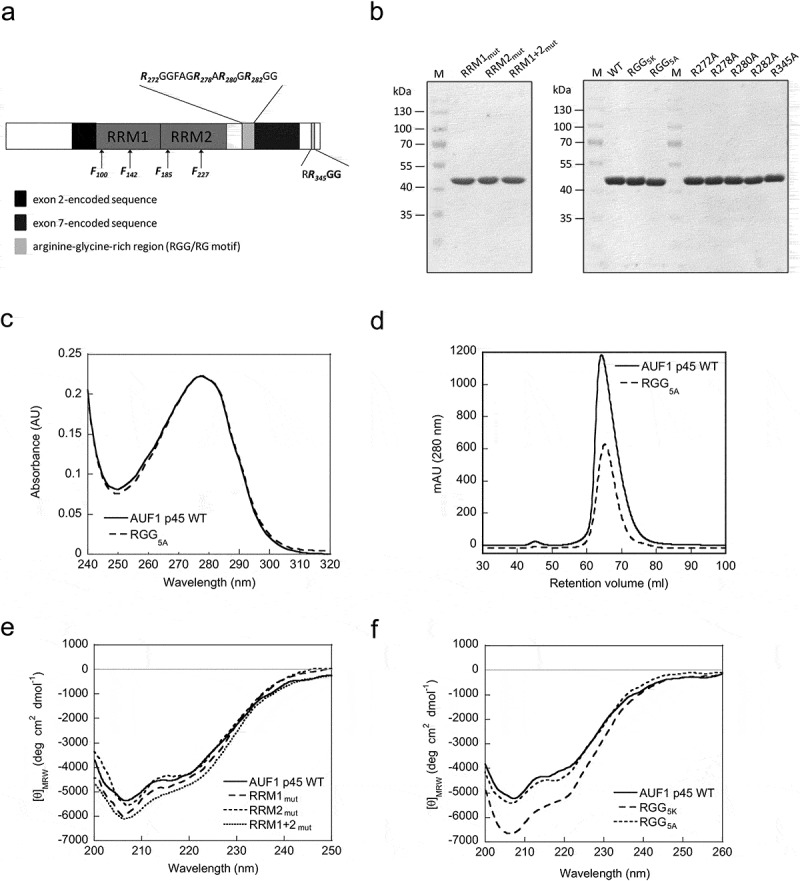
Characterization of substitution variants of AUF1 p45. (a) Organization of the AUF1 isoform p45. The RNA recognition motifs (RRM) are indicated as well as isoform-specific sequences encoded by exon 2 (black) and exon 7 (grey). The RGG/RG motif is depicted in light grey and the amino acid sequence is highlighted. The amino acid positions that were mutated in the variants used in this study are indicated. (b) AUF1 p45 and its substitution variants were produced in, and purified from, E. coli. About 4 µg of protein were analyzed on a Coomassie-stained SDS-gel in parallel with a molecular weight marker (M). (c) UV absorption spectra of wildtype AUF1 p45 and variant RGG5A in storage buffer. (d) Elution profiles of wildtype AUF1 p45 and variant RGG5A fractionated through a Superdex 75 gel filtration column (HiLoad 16/60). (e) Far-UV circular dichroism (CD) spectra of AUF1 p45 and of the indicated substitution variants were recorded. The acquired data were normalized to mean residue weight (MRW) ellipticities. (f) Same as in (e).
Folding and structural rearrangements of RNA molecules in order to achieve their functionally active conformation is often assisted by proteins with RNA remodeling activities, which can be classified into three types: RNA chaperones, RNA annealers and RNA helicases [12]. RNA chaperones and RNA annealers promote structural rearrangements of RNA molecules in an ATP-independent manner. RNA helicases mostly catalyze ATP-driven RNA unwinding, although ATP concentration is an important regulator of RNA helicase-guided remodeling of RNA structures [13–15]. Protein-guided remodeling of RNA structures was demonstrated in a wide variety of processes of RNA metabolism, such as ribosome assembly [16], pre-tRNA folding [17], spliceosome assembly [18,19], mRNA translation and turnover rates [20,21], rearrangement of cellular and viral riboswitches [22], reverse transcription of retroviruses [23] and RNA interference [24].
AUF1 was previously shown to act as a host factor that supports the genome replication of Flaviviridae family members, including the important human pathogens West Nile virus (WNV), Dengue virus (DENV) and Zika virus (ZIKV) [25–27]. Cyclization of the single-stranded flaviviral RNA genome is a prerequisite for the initiation of RNA synthesis and hence virus amplification [28]. Flavivirus genome cyclization involves the rearrangement of RNA secondary structures within the 5ʹ- and 3ʹ-end of the viral RNA and subsequent interactions of a set of complementary cyclization sequences (schematically depicted in Figure 3(a), reviewed in [29–31]. The largest AUF1 isoform p45 was shown to promote the cyclization of the flaviviral genome by two RNA remodeling activities, i.e. a helix-destabilizing RNA chaperone activity and an RNA annealing activity by which AUF1 p45 accelerates the hybridization of the cyclization sequences [25–27]. Furthermore, AUF1 p45 was shown to be consistently modified by arginine methylation in mammalian cells. The methylated variant turned out to be more efficient at stimulating the RNA chaperone activity whereas the RNA annealing activity of AUF1 p45 remained unaffected [26].
Figure 3.
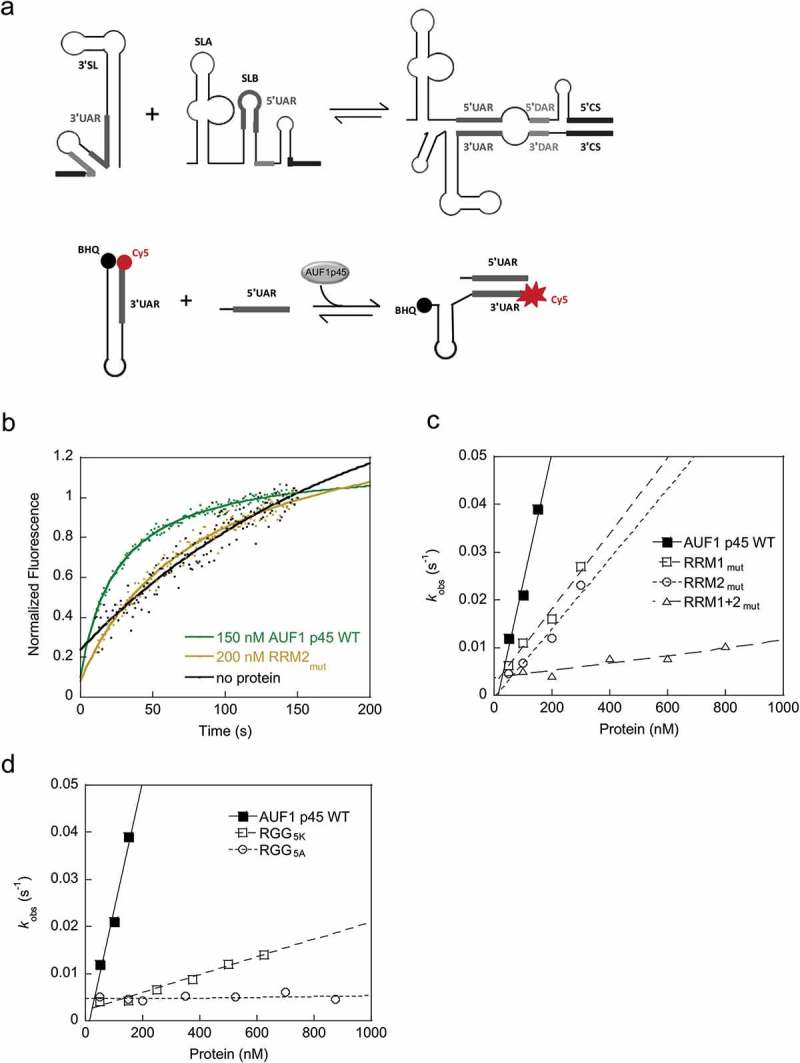
The RRMs as well as the RGG/RG motif contribute to the RNA chaperone activity of AUF1 p45. (a) (Top) Scheme of the structural rearrangement of the 5ʹ and 3ʹ termini, specifically of the 5ʹUAR and 3ʹUAR elements, during cyclization of the DENV RNA genome. (Bottom) Scheme of the fluorescence-based 3’SLtrunc-5ʹUAR interaction assay to detect AUF1 p45-mediated conformational rearrangement of DENV RNA by de-quenching of Cy5. (b) Example kinetic traces with Cy5 and BHQ (black hole quencher) labeled 3’SLtrunc incubated with the indicated concentrations of AUF1 p45 or variant RRM2mut. Following the addition of 5ʹUAR RNA the fluorescence signals were measured, plotted as a function of time, and fitted according to a first-order reaction (no protein; Equation (1) or second-order reaction (in the presence of protein; Equation (2). (c) and (d) The observed rate constants kobs (s−1) that were measured for the RNA-RNA interaction reaction in the presence of AUF1 p45 or variants thereof were plotted as a function of the protein concentration.
To what extent the RRMs and the RGG/RG motif of AUF1 within the full-length protein contribute to its RNA binding, as well as RNA remodeling activities, has not been investigated yet. Here, we deciphered the role of the RRMs and the RGG/RG motif of AUF1 p45 by an extensive mutagenesis approach. We report that these RNA-binding motifs are specific determinants of AUF1 p45 that contribute differently to its RNA-binding, RNA chaperone and RNA annealing activities. We propose that the arginine patch of the RGG/RG motif is, on the one hand, required for AUF1’s RNA chaperone activity through stacking interactions with nucleobases in order to destabilize helices. On the other hand, it supports AUF1’s RNA annealing activity by overcoming electrostatic barriers prior to helix formation, providing the first evidence for the essential role of arginine residues of an RGG/RG motif in RNA remodeling. Finally, free energy calculations revealed that the arginine residues of the RGG/RG motif contribute additively to RNA-binding, which in turn determines the RNA annealing activity.
Results
Preparation and characterization of AUF1 p45 variants
An amino acid exchange mutagenesis study was carried out to determine how the RRMs and the RGG/RG motif contribute to AUF1’s RNA-binding and restructuring activities. The first set of protein variants contained amino acid substitutions in the RRMs at positions that have been implicated to be involved in RNA binding (Figure 1(a)) [8–10]. We generated protein variants by substituting phenylalanine residues F100 and F142 each to alanine to impair the function of RRM1 (referred to as RRM1mut). In order to impair RRM2 we substituted residues F185 and F227 each to isoleucine (referred to as RRM2mut), because the respective alanine substitution variant was prone to oligomerization (data not shown). Furthermore, we substituted all the aforementioned phenylalanine residues to either alanine (RRM1) or isoleucine (RRM2) in order to impair both RRMs (referred to as RRM1 + 2mut). The second set of protein variants contained arginine to either lysine or alanine substitutions within the RGG/RG motif at positions that were previously shown to be modified by methylation [26]. The five methylatable arginine residues within the RGG/RG motif (Figure 1(a)) were exchanged to either lysine (referred to as RGG5K), in order to preserve the charge of the protein, or to alanine (referred to as RGG5A) to examine the role of the positive charge of the arginine residues. Additionally, we generated single-amino acid substitution-variants, where only one methylatable arginine residue at a time was exchanged to an alanine. The protein variants were purified from E. coli using the same protocol as for the wildtype (WT) AUF1 p45. Using this approach, we obtained homogeneous preparations of the variants that had the same quality as the WT protein, i.e., they were free of impurities (Figure 1(b)) and, an important aspect when studying protein-RNA interactions, free of contaminating nucleic acids. The UV absorption spectra for wildtype and variants revealed 280 nm/260 nm ratios of 1.8 (Figure 1(c) and data not shown). These corresponded closely to the theoretical value of 1.6, indicating the absence of co-purifying nucleic acids.
The monomeric state of AUF1 p45 was demonstrated previously by analytical ultracentrifugation [25]. The final step of the protein purification protocol, size exclusion chromatography, revealed elution volumes for the variants very similar to the WT (Figure 1(d) and data not shown), strongly indicating all variants to be monomeric.
Since the exchange of amino acids might be detrimental to the overall structure of the protein, we analyzed the structural integrity of the protein variants by far-UV circular dichroism (CD). The overall CD signal of the WT AUF1 p45 is rather low emphasizing the high content of disordered regions, especially at the N- and C-termini [26] (Figure 1(e)). Importantly, the far-UV CD spectra of the RRM variants as well as of the AUF1 p45 5A variant, closely resembled the characteristics of the WT AUF1 p45 demonstrating the structural integrity of the variants (Figure 1(e,f)). The chirality change of the RGG/RG variant RGG5K compared to the WT AUF1 p45 might indicate a conformational change of the protein (Figure 1(f)). Nevertheless, the general characteristics of the WT spectrum were maintained indicating a similar secondary structure and confirmed the structural integrity of this variant.
The RRM motifs determine specificity and contribute differently to AUF1’s RNA-binding activity
In order to decipher specific functions associated with the RNA-binding motifs of AUF1 p45, we conducted fluorescence equilibrium measurements using short, fluorescently-labeled RNA molecules. We sought to compare the binding efficiency of the WT and its variants to a 16 nt-long AU/GU-rich, single-stranded RNA and to a randomly composed single-stranded RNA of the same length (Figure 2(a)). For three reasons, the AU/GU-rich RNA should be a bona fide target RNA of AUF1: i) AUF1 shows specificity for AU/GU-rich sequences [4], ii) the AU/GU-rich RNA is part of the AU-rich region of the West Nile virus genome, which was previously identified as a high-affinity binding site for AUF1 p45 [25] and iii) the AU/GU-rich RNA contains two UUAG motifs, which are specifically recognized by AUF1 [32]. RNA binding was analyzed according to a single-site binding model as previously described for AUF1 [6]. As expected, AUF1 p45 WT displayed a higher affinity (100-fold) for the AU/GU-rich RNA (KD = 1.6 nM) than for the random RNA (KD = 157 nM) (Figure 2(b,c); Table 1).
Figure 2.
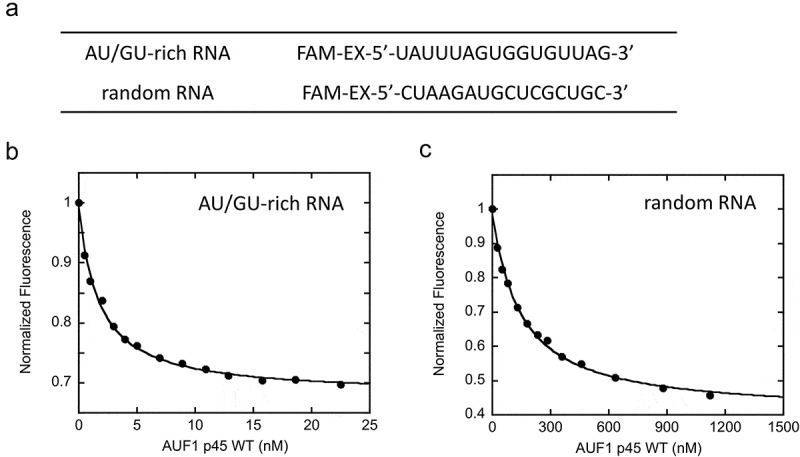
RNA binding of wildtype AUF1 p45. (a) FAM-EX-labeled RNA molecules that were applied for binding studies. (b) and (c) Representative binding isotherms with 0.2 nM AU/GU-rich RNA (b) or 25 nM random RNA (c) and increasing concentrations of AUF1 p45. The data were normalized to the initial intensity of the unbound FAM-EX-labeled RNA. The binding curves were fitted to a single-site binding model Equation (1) in materials and methods).
Table 1.
RNA binding parameters of AUF1 p45 and its variants.
| AU/GU-rich RNA |
random RNA |
|||||
|---|---|---|---|---|---|---|
| Protein | KD (nM)a | KD ratiob | ΔΔG (kJ/mol)c | KD (nM)a | KD ratiob | ΔΔG (kJ/mol)c |
| AUF1 p45 WT | 1.6 ± 0.5 | 1 | 0 | 157 ± 24 | 1 | 0 |
| AUF1 p45 RRM1mut | 25 ± 3.2 | 16 | 6.8 | 1260 ± 217 | 8 | 5.1 |
| AUF1 p45 RRM2mut | 24 ± 3.4 | 15 | 6.7 | 414 ± 49 | 2.6 | 2.4 |
| AUF1 p45 RRM1 + 2mut | 1040 ± 290 | 650 | 15.9 | 4170 ± 909 | 27 | 8.9 |
| AUF1 p45 RGG5K | 4.3 ± 1.0 | 2.7 | 2.4 | 314 ± 50 | 2 | 1.7 |
| AUF1 p45 RGG5A | 24 ± 3.1 | 15 | 6.7 | 2840 ± 191 | 18 | 7.1 |
aDissociation constants and standard deviations derived from at least three measurements.
bKD ratio was calculated to compare binding of wildtype (WT) vs protein variant.
cThe Gibbs free energy difference between a WT-RNA complex and a variant-RNA complex was calculated as follows: ΔΔG = -RT ln(KD WT/KD variant).
Interestingly, the RNA-binding capacity of the RRM variants depended on the RNA substrate used. Binding of the AU/GU-rich RNA by AUF1 p45 RRM1mut (KD = 25 nM) or AUF1 p45 RRM2mut (KD = 24 nM) was 15-fold less efficient than with the WT protein. When both RRMs (variant AUF1 p45 RRM1 + 2mut) were impaired, the binding efficiency was dramatically reduced by 650-fold (KD = 1044 nM; Table 1). This demonstrated that both RRMs contribute equally to binding of an AU/GU-rich RNA and that they can compensate for the loss of the other motif to a certain extent. On the other hand, the RRMs contributed differently to the binding of a randomly composed RNA of the same length. The AUF1 p45 variant RRM1mut showed a stronger reduction in affinity for a random RNA (KD = 1259 nM) than variant RRM2mut (KD = 414 nM) in comparison to the WT (8-fold vs. 2.6-fold; Table 1). This was taken to indicate that RRM2’s contribution to the binding of unspecific RNA is less pronounced than that of RRM1. Furthermore, these data revealed that both RRMs are necessary for high-affinity binding of RNAs confirming previous reports for AUF1 isoform p37 [33]. Another interesting observation was that the KD ratios differed depending on the RNA substrate used. In particular, the variant where both RRMs were impaired showed a 24-fold higher KD ratio for the AU/GU-rich RNA than that for the random RNA (650-fold vs 27-fold; Table 1). This indicates that the impairment of AUF1’s RRMs impacts the binding of specific RNAs more than that of an unspecific RNA. The mutagenesis of conserved phenylalanine residues within RRM1 and RRM2 were characterized by differences in the free-binding energy of 6.7–15.9 kJ/mol.
The RGG/RG motif of AUF1 p45 contributes to its RNA binding activity
We and others could show that five arginine residues within the RGG/RG motif of AUF1 are dimethylated in the cell [26,34]. To dissect the general role of these arginine residues in RNA binding we analyzed protein variants in which the five methylatable arginine residues were substituted by lysine or alanine (Figure 1(a)). The RNA-binding affinity of the arginine to lysine exchange variant AUF1 p45 RGG5K for both RNAs was only slightly reduced (AU/GU-rich RNA: KD = 4.3 nM; random RNA: KD = 314 nM; Table 1), indicating that this variant can compensate to some extent for the loss of arginine residues within the RGG/RG motif. In contrast, the binding of the arginine to alanine exchange variant AUF1 p45 RGG5A was more severely affected. The affinity for the AU/GU-rich RNA was reduced by 15-fold (KD = 24 nM), whereas the binding to the random RNA was similarly reduced (18-fold reduction, KD = 2840 nM) compared to the WT protein (Table 1). These data emphasize the importance of the positive charge of the arginine residues within the RGG/RG motif of AUF1 p45 for its RNA-binding activity. Thus, the RGG/RG motif of AUF1 p45 was revealed to be an additional key factor for the high-affinity recognition of AU/GU-rich sequences but also for the low-affinity binding of non-AU-rich sequences.
The RRM and RGG/RG motifs contribute differently to AUF1 p45’s RNA remodeling activities
In the next set of experiments, we performed two assays to study the RNA remodeling activities of AUF1 p45 and its variants. First, we applied an established fluorescence-based assay to mimic the rearrangement of the DENV RNA’s 3ʹ-terminal SL. To enable genome cyclization, the 3’SL must be denatured to allow 5ʹ-3ʹUAR hybridization (schematically depicted in Figure 3(a)). Using this assay, we previously showed that AUF1 p45 exhibits an RNA chaperone activity, which promotes the destabilization of flaviviral stem-loops involved in genome cyclization [25,27]. The assay applies a Cy5- and black-hole quencher (BHQ)-labeled RNA oligonucleotide (3’SLtrunc) corresponding to the lower part of the DENV 3’SL, including the 3ʹUAR sequence. The second RNA used in this assay is single-stranded and corresponds to a sequence of the DENV 5ʹUTR mainly comprising the 5ʹUAR cyclization sequence. Hybridization of both RNAs via the UAR elements requires a rearrangement of the stem structure, which dislocates the Cy5 fluorophore from the BHQ (Figure 3(a,b)) and can be measured time-dependently. The concentration-dependent efficiency of each variant’s RNA chaperone activity to support the 3’SLtrunc-5ʹUAR interaction was now analyzed by determining the rate constant for each reaction. The acceleration of the RNA-RNA interaction was most efficient with WT AUF1 p45 (Figure 3(b,c)). The single RRM-deficient variants RRM1mut and RRM2mut showed a moderate decrease in activity, whereas the double RRM-deficient variant RRM1 + 2mut displayed almost no ability to promote the 3’SLtrunc-5ʹUAR interaction (Figure 3(c)). Both RGG/RG variants of AUF1 p45 showed a drastic reduction in RNA chaperone activity, with the RGG5K variant showing activity only at very high protein concentrations, whereas the RGG5A variant showed no increase in the observed rate constants, even at the highest protein concentrations used (Figure 3(d)). These data imply that not only the positive charge but also a specific function of the arginine residues within the RGG/RG motif is required for the protein’s RNA chaperone activity.
In a second approach, we investigated how efficiently the AUF1 p45 variants promote the annealing of single-stranded complementary RNAs. For this purpose, we applied a previously established FRET-based RNA annealing assay using the conserved flaviviral 5ʹ and 3ʹCS cyclization sequences, which are labeled with a Cy5 and a Cy3 fluorophore, respectively (Figure 4(a)) [26]. RNA-RNA annealing of the two fluorophore-labeled RNAs results in an increase of fluorescence, which can be monitored to determine the rate constants of the RNA complex formation.
Figure 4.
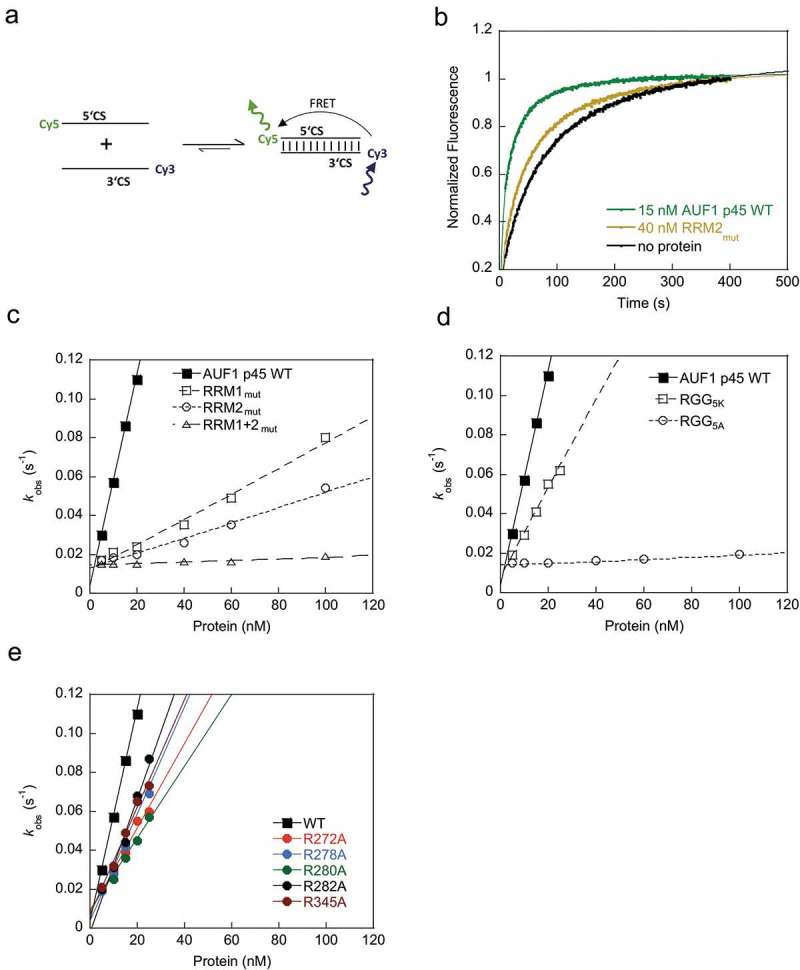
The RRMs as well as the RGG/RG motif contribute to the RNA annealing activity of AUF1 p45. (a) Scheme of the fluorescence-based 5ʹCS-3ʹCS RNA annealing assay to analyze the hybridization of the CS cyclization sequences. Annealing of the complementary 5′- and 3′CS RNAs that are fluorescently labeled with Cy5 and Cy3, respectively, leads to a detectable FRET signal. (b) Example kinetic traces of the RNA-RNA interaction in the presence of the indicated concentrations of AUF1 p45 or variant RRM2mut. Cy3-labeled 3′CS RNA was incubated with the respective protein, followed by the addition of Cy5-labeled 5′CS RNA. The fluorescence signals were analyzed according to a second-order reaction Equation (3). (c) – (e) The observed rate constants kobs (s−1) that were measured for the RNA-RNA interaction reaction in the presence of AUF1 p45 or variants thereof were plotted as a function of the protein concentration.
The WT protein accelerated the 5ʹ-3ʹCS RNA hybridization at low protein concentrations confirming its activity as an RNA annealer (Figure 4(b,c)). The single RRM-deficient variants RRM1mut and RRM2mut showed a clear decrease in activity, whereas the double RRM-deficient variant RRM1 + 2mut was completely unable to promote the 5ʹ-3ʹCS RNA annealing (Figure 4(c)). The AUF1 p45 RGG5K variant exhibited only a slight decrease in activity compared to the WT activity. In contrast, the RGG5A variant was severely affected in its ability to enhance the RNA annealing, showing almost no activity up to a protein concentration of 100 nM (Figure 4(d)). These data demonstrate that electrostatic interactions mediated by the RGG/RG motif play an essential role for AUF1’s RNA annealing activity.
The energetic contributions of the arginine residues of the RGG/RG motif to RNA binding and annealing are cumulative
In a final approach, we analyzed the energetic contribution of each arginine residue of the RGG/RG motif to the RNA annealing activity of AUF1 p45. For this purpose, we tested the RNA binding of the single arginine to alanine substitution variants R272A, R278A, R280A, R282A and R345A (Figure 1(b)) including the WT protein and the RGG5K and RGG5A variants to the annealing substrates 5ʹCS and 3ʹCS (each 14 nt long). The WT protein showed a moderate binding affinity to both annealing substrates (KD = approx. 100 nM; Table 2). The single substitution variants and the RGG5K variant differed in binding strength by approximately 2–2.5-fold, with the exception of variant R345A, which was less affected with a 1.4-fold decrease in binding affinity. The corresponding differences in binding free energy were in the range of 0.8 to 2.2 kJ/mol. As expected, the RGG5A substitution variant was the most detrimental with a 20-fold decrease in RNA-binding, which corresponded to a difference in binding free-energy of about 7 kJ/mol (Table 2).
Table 2.
RNA binding of AUF1 p45 and variants to flaviviral RNA annealing substrates.
| 5ʹCS |
3ʹCS |
|||||
|---|---|---|---|---|---|---|
| Protein | KD (nM)a | KD ratiob | ΔΔG (kJ/mol)c | KD (nM)a | KD ratiob | ΔΔG (kJ/mol)c |
| AUF1 p45 WT | 105 ± 27 | 1 | 0 | 101 ± 30 | 1 | 0 |
| AUF1 p45 RGG5K | 208 ± 19 | 2 | 1.7 | 198 ± 34 | 2 | 1.7 |
| AUF1 p45 RGG5A | 2157 ± 261 | 21 | 7.5 | 1813 ± 432 | 18 | 7.1 |
| AUF1 p45 R272A | 230 ± 18 | 2.2 | 1.9 | 201 ± 31 | 2 | 1.7 |
| AUF1 p45 R278A | 257 ± 51 | 2.5 | 2.2 | 242 ± 67 | 2.4 | 2.2 |
| AUF1 p45 R280A | 234 ± 56 | 2.2 | 1.9 | 228 ± 47 | 2.3 | 2 |
| AUF1 p45 R282A | 244 ± 60 | 2.3 | 2 | 234 ± 35 | 2.3 | 2 |
| AUF1 p45 R345A | 144 ± 5 | 1.4 | 0.8 | 139 ± 17 | 1.4 | 0.8 |
aDissociation constants and standard deviations derived from at least three measurements.
bKD ratio to compare binding of wildtype (WT) vs protein variant.
cThe binding free energy difference between a WT-RNA complex and a variant-RNA complex was calculated as follows: ΔΔG = -RT ln(KD WT/KD variant).
The results of the RNA annealing assay clearly showed that the exchange of each arginine residue to an alanine led to a slight decrease of RNA annealing activity of the corresponding variant as reflected by the observed rate constants Figure 4. These data demonstrate that the arginine residues of the RGG/RG motif individually contribute to the RNA annealing activity of AUF1 p45.
Finally, to investigate the energetic contributions and nature of individual arginine-RNA interactions we analyzed the free energy change of the RNA annealing process upon mutation of AUF1. Here, we used the catalytic efficiency (kcat/KM) of each variant determined from the slope of the kobs vs protein concentration plot and calculated the free-energy changes. The data revealed that for each single substitution variant the total free energy is lowered by 2 kJ/mol demonstrating that the single substitution variants changed the RNA annealing activity only minimally Table 3. With the RGG5A variant a free-energy change of 11.6 kJ/mol was determined, which is close to the sum of the free-energy changes of the single substitution variants (10.1 kJ/mol). Interestingly, the RGG/RG single substitution variants showed similar free energy changes for RNA binding and annealing of about 2 kJ/mol. Thus, the arginine residues in the RGG/RG motif mainly contribute to the affinity of AUF1 p45 for the RNA in the initial complex formation.
Taken together, the RNA-binding affinities and free-energy changes of the RGG/RG variants closely correlate with the enzymatic activities of the RNA annealing assay. Moreover, each arginine residue of the RGG/RG motif is equally important for binding and annealing, suggesting an additive effect.
Discussion
Functional roles of the RRM motifs
In the context of the full-length protein, our study has confirmed the crucial role of conserved phenylalanine residues within the classical ß-sheet surface of AUF1’s RRMs for RNA binding [8,9]. The data further revealed a synergy between both RRMs within AUF1 because the free energy contribution to RNA binding of both RRMs together is greater than the sum of their individual contributions (Table 1). Although there is no structural information available yet for both AUF1 RRMs, we can speculate that the two independently folded RRMs are likely held in a fixed position, thus enabling the two RRMs to form an extended RNA-binding surface. Such a synergy of two tandem RRM domains forming a cleft with the RNA bound between the basic surfaces of the opposing ß sheets has been observed in the structures of several proteins, including hnRNPA1 [35–37], PABP [38], Sxl [39], Nucleolin [40], HuD [41] and Hrp1 [42]. In the case of hnRNPA1, Beusch and colleagues demonstrated that binding of both RRMs of hnRNPA1 to RNA is required to achieve optimal splicing repression [43]. In the case of the tandem RRMs of TDP-43, RNA-binding mediates extensive RRM-RRM interactions, which are crucial for the splicing activity of the protein [44].
Interestingly, we observed a general correlation between the RNA-binding and RNA remodeling activities of the RRM variants indicating that, at least with respect to the RRMs, RNA annealing simply depends on RNA-binding affinity, i.e. the binding energy is used for catalysis. This situation is different from hnRNPA1 where protein variants with mutated RNP-1 sub-motifs bound RNA almost as well as the WT protein but their RNA annealing activity was reduced [45].
Previous NMR studies with the isolated RRM1 of AUF1 suggested that non-specific interactions with RNA are mediated by the residues in the central region of the RNA-binding surface, while specific binding is mediated by those in the peripheral region [8]. Interestingly, the ratios of AUF1 RRM variants and wildtype KDs obtained here revealed differences depending on the RNA used. The loss of RNA-binding affinity of the RRM variants was much more pronounced when a high-affinity target RNA was used as opposed to an unspecific RNA (Table 1) indicating that the conserved phenylalanine residues within the RNP-1 and −2 sub-motifs also contribute to sequence-specific RNA recognition. One possible explanation for such an RNA discrimination mechanism involves different phenylalanine side-chain conformations at the RNA-binding surface of AUF1 that allow for high- or low-affinity binding, as was recently shown for RRM3 of CUG-binding protein 2 [46]. The role of the RGG/RG motif on the other hand can be attributed mainly to non-specific binding of RNA, since the ratios of the RGG/RG variants and wildtype KDs do not change with different RNAs used (Tables 1 and 2).
The calculated differences in binding free energies between wildtype and RRM variants revealed surprisingly low ΔΔG values compared to previous RRM-RNA binding data. For example the exchange of only one conserved phenylalanine residue in the isolated RRMs of U1A and Fox-1 showed ΔΔG values of 18–23 kJ/mol [47,48], whereas the combined exchange of two or four phenylalanine residues in RRM1 and 2 of AUF1 lowered the binding free-energy only by 7–16 kJ/mol (Table 1). This indicates that the free-energy differences are even more complex in the context of a full-length protein containing several RNA-binding motifs or that the stacking interactions of the conserved phenylalanine residues are not as powerful in RNA-binding by AUF1.
Functional roles of the RGG/RG motif
In the case of AUF1 isoform p37 it was shown that the two tandem RRM domains are necessary, but not sufficient, for high-affinity RNA-binding requiring additional contributions from N- and C-terminal domains to stabilize the resulting RNA-protein complex [33]. Another study revealed that the RNA footprint for maximal AUF1 binding is approximately 30 nt [49]. Because RRM domains bind an average of 4 nt [7], this implies N- and C-terminal regions to be crucially important by increasing the AUF1-RNA interaction network. Here, we provided evidence that the RGG/RG motif within the C-terminus of AUF1 p45 represents such a molecular determinant for high-affinity RNA binding. The interaction of arginine with RNA is mediated by hydrogen bonding, hydrophobic (stacking) interactions and electrostatic interactions. The observation that an arginine to lysine substitution variant largely retains the binding affinity to AU/GU-rich and unspecific RNAs (Table 1), highlights the importance of the positive charge of the arginine residues within the RGG/RG motif for AUF1 p45-RNA complex formation. One obvious explanation would be that the positively charged arginine residues stabilize the complex through electrostatic interactions with the negatively charged phosphates of the RNA. However, RNA-binding of the alanine substitution variant was only moderately affected, indicating an auxiliary role instead of an essential role for the RGG/RG motif. This picture changes completely when we analyzed the RNA remodeling activities of AUF1 p45. Since we observed a drastic loss of the RNA chaperone activity of the lysine substitution variant (Figure 3(d)), the arginine residues clearly confer a distinct property to the RGG/RG motif besides its positive charge. Arginine, in contrast to lysine, is capable of performing stacking interactions with RNA; hence, it seems reasonable to suggest that arginine-mediated stacking interactions with (flaviviral) stem-loop-RNAs contribute significantly to its helix-destabilizing RNA chaperone activity (see model in Figure 5). This idea is further supported by the recent observation that a methylated variant of AUF1 p45, in which the five arginine residues of the RGG/RG motif investigated here are dimethylated, showed a stronger RNA chaperone activity whereas the RNA annealing activity remained largely unaffected [26]. Through methylation the arginine becomes more hydrophobic and this may facilitate stacking with RNA bases. A similar situation where RNA remodeling activities are regulated by post-translational modifications has been observed for the RNA-binding proteins La and NF90. In the case of La a novel RNA chaperone domain was shown to be regulated by AKT phosphorylation [50]. NF90’s RNA annealing activity is mediated by an RGG motif within the protein’s C-terminus, which may be regulated by phosphorylation [22].
Figure 5.
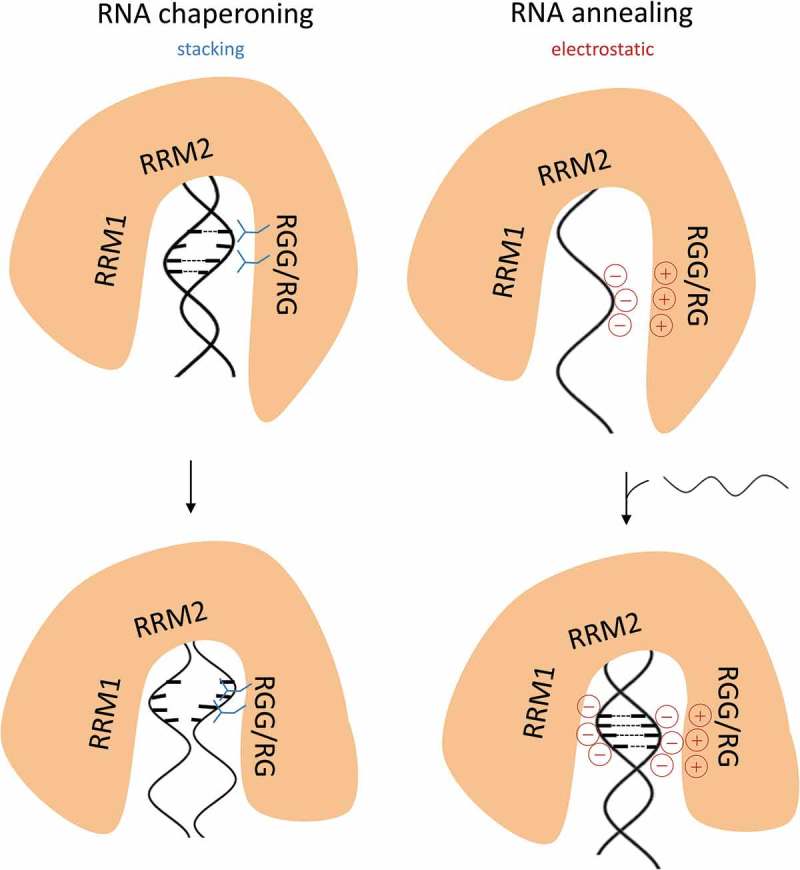
A model of AUF1’s RNA chaperone and RNA annealing activities mediated by its RGG/RG motif. Two distinct roles for the arginine residues of AUF1’s RGG/RG motif for each RNA remodeling activity are proposed. During RNA chaperoning, arginine-mediated stacking interactions promote destabilization of flexible RNA helices (left). During RNA annealing, the RGG/RG motif reduces the negative surface charge of the first RNA, which might result in the presentation of an annealing-competent conformation and facilitates subsequent annealing with the second RNA (right). The RNA remodeling activities of AUF1 might be accompanied by local folding of the intrinsically disordered C-terminus to provide energy for the RNA chaperoning/annealing process (indicated by a different shape of the protein).
On the other hand, in the case of the RNA annealing assay, the lysine substitution variant AUF1 p45 RGG5K can more efficiently compensate for the loss of the arginine residues (Figure 4(d)) indicating that the electropositive charge of arginine within the RGG/RG motif is the major driving force for the RNA annealing activity of the protein. Ionic bonds are formed between the RGG/RG motif of AUF1 and the first RNA, thus decreasing the net negative charge of the RNA, which might result in the presentation of an annealing-competent conformation (see model in Figure 5). This indicates that the electropositive RGG/RG motif promotes annealing, most likely by reducing the charge repulsion between the two annealing molecules. The requirement of a large electropositive patch for the activity of AUF1 confirms previous studies on the mechanism of protein-catalyzed RNA annealing [19,51–54]. The involvement of an RGG/RG motif in the process of RNA annealing also confirms previous data for the RNA annealers hnRNPA1 [55,56], FMRP [57] and NF90 [22]. However, with AUF1 we demonstrate for the first time that the electropositive character of the arginine residues within an RGG/RG motif is a crucial determinant for the protein’s RNA annealing activity.
The annealing activity of AUF1 p45 on RNA substrates should be considered as enzymatic reactions/catalytic reactions. To compare the WT and the variants we calculated the catalytic efficiencies (kcat/KM) which includes both the activation energies and the binding energies. We found that the free-energy changes between the WT and the arginine variants of AUF1 p45, which were calculated from the binding constants to RNA on the one hand, and the catalytic efficiencies of the RNA annealing assay on the other hand, closely correlate (Table 3). Thus, the higher annealing activity of the WT is characterized by an increase in RNA-protein-complex stability (enzyme-substrate complementarity) rather than by a rate advantage through lowering the activation energy barrier. Consequently, the energetic contributions of the positively charged guanidinium groups of the arginine residues of the RGG/RG motif of AUF1 p45 are related to the formation of the binary complex (RNA-AUF1 p45-complex similar to the ES complex), leaving the transition state unchanged.
Table 3.
Thermodynamic parameters of RGG/RG motif variants in RNA annealing.
| Protein | kcat/KM (106 s−1 M−1)a | ΔΔG≠ (kJ/mol)b |
|---|---|---|
| AUF1 p45 WT | 4.61 | 0 |
| AUF1 p45 RGG5K | 1.84 | 2.3 |
| AUF1 p45 RGG5A | 0.04 | 11.6 |
| AUF1 p45 R272A | 1.78 | 2.3 |
| AUF1 p45 R278A | 2.11 | 1.9 |
| AUF1 p45 R280A | 1.52 | 2.7 |
| AUF1 p45 R282A | 2.53 | 1.5 |
| AUF1 p45 R345A | 2.30 | 1.7 |
aThe catalytic efficiency kcat/KM was determined from the slope of the kobs values plotted as a function of the protein concentration.
bThe RNA annealing free energy difference ΔΔG≠ between the WT and the variants was calculated using Equation (7) (see materials and methods).
Proteins with RNA chaperone/annealing activities usually display a high content of intrinsically disordered regions (IDR). Based upon an entropy transfer model, IDRs of RNA chaperones undergo induced folding, i.e. they switch from an unstructured to a structured state upon binding of their target RNA [58]. Perhaps the arginine residues are functionally relevant in the induced folding of the intrinsically disordered C-terminus of AUF1 to provide energy for the RNA chaperoning/annealing process. For Tra2-ß1 it was shown that unstructured N- and C-terminal regions of its RRM interact with RNA and even participate in the specific recognition of sequences. Upon RNA binding these regions undergo a structural transition from a disordered to an ordered conformation [59]. Such RNA-induced positioning may also be the case for AUF1 where the C-terminal region of RRM2 includes the RGG/RG motif. Upon RNA binding the unstructured RGG/RG motif might fold and in this way stabilize the RNP-complex in order to achieve maximal binding activity (see model in Figure 5).
So far, the RNA remodeling activities of AUF1 were investigated in the context of flavivirus genomic RNA rearrangement and replication. Interestingly, recent studies linked AUF1 and the ARE-binding protein HuR to cellular processes involving RNA-RNA interactions, particularly miRNA binding to mRNAs targets, which may be influenced by mRNA target structure [60–62]. Moreover, the interdependence of local ARE structure and AUF1 binding was indicated to modulate mRNA decay activity, highlighting dynamic RNA conformations as an additional level to control gene expression [63–65]. Thus, unraveling the role of AUF1’s RNA restructuring activities during the post-transcriptional control of gene expression by trans-acting RNAs will be an important task for future investigations.
Materials and methods
Plasmids encoding SUMO-fusion proteins
The pETSUMO expression system was used for the expression of recombinant proteins in E. coli. The plasmid encoding AUF1 p45 is described elsewhere [25]. Plasmids encoding for amino acid substitution-variants of AUF1 p45 were generated by site-directed mutagenesis using the respective primer pairs (Supplementary Table S1).
Expression and purification of wildtype and variants of AUF1 p45 from E. coli
AUF1 p45 and its variants were purified from the soluble fraction of Escherichia coli BL21-CodonPlus® (DE3)-RP cells using nickel-agarose affinity chromatography (HisTrap™ HP, GE Healthcare) and, after cleavage with SUMO-protease, by heparin-sepharose affinity chromatography and gel-filtration (HiLoadTM 16/60 Superdex 200TM or Superdex 75TM, GE Healthcare). UV absorption spectra were measured using a JASCO V-550 spectrometer. The protein concentration was determined by measuring the absorbance at 280 nm using ϵ280 = 58,915 M−1 cm−1. All AUF1 p45 variants were stored at −80°C in 20 mM Tris/HCl, pH 7.6, 150 mM KCl, 1 mM Tris(2-carboxyethyl)phosphine (TCEP). To confirm the absence of co-purifying nucleic acids we determined the 280 nm/260 nm ratio of the purified protein and compared it with the theoretical 280 nm/260 nm ratio of AUF1 p45 using extinction coefficients for Trp, Tyr and Phe from the PhotochemCAD database [66].
Measurement of RNA-binding constants
The protein of interest was added to 5′-FAM-EX-labeled RNA (0.2–25 nM) in assay buffer (50 mM Hepes/NaOH, pH 8.0, 100 mM KCl, 5 mM MgCl2). Fluorescence changes were monitored using a Fluoromax-4 spectrofluorometer (Jobin Yvon) at 22°C. After attaining equilibrium, the signal amplitudes of the 5′-FAM-EX-labeled RNAs (Supplementary Table S2) were measured (excitation at 491 nm, emission at 515 nm) and corrected for the volume change. Fluorescence intensities relative to the starting fluorescence were plotted against the protein concentration. Fitting the binding isotherms to a single-site binding model according to Equation (1)[67,68] with KaleidaGraph (Synergy Software) yielded the KD values of the interaction of the protein and the labeled RNA.
| (1) |
ΔF - relative change in fluorescence intensity, γ – signal amplitude, b – total concentration of the RNA, c – total concentration of the protein, KD – dissociation constant.
Fluorescence-based RNA-RNA interaction assay
The assay was performed as described previously [27]. Briefly, AUF1 p45 wildtype (WT) or a variant thereof was added to 10 nM of 5ʹ-Cy5-labeled 3'SLtrunc DENV-RNA (purchased from IBA, Göttingen, Germany; Figure 3(a) and Supplementary Table S2) at different concentrations in assay buffer (50 mM Hepes/NaOH, pH 8.0, 100 mM KCl, 5 mM MgCl2). Then, 100 nM of non-labeled 5ʹUAR DENV-RNA was added and readings were taken for another 150 s. Changes in the fluorescence signals were monitored using a Fluoromax-4 spectrofluorometer (Jobin Yvon) at 22°C with the following parameters: excitation at 643 nm and emission at 667 nm. Fluorescence intensities relative to the starting fluorescence were plotted against time and fitted by KaleidaGraphTM (Synergy) to a first-order reaction when protein was omitted Equation (2) and a second-order reaction when protein was included Equation (3) yielding the corresponding rate constants kobs.
| (2) |
| (3) |
ΔF – total change of relative fluorescence amplitude, Foffset– fluorescence intensity at the start point of the reaction, Fmax - maximum signal amplitude, kobs – observed rate constant, t – time
FRET-based RNA annealing of complementary cyclization sequences
The assay was performed as described previously [26]. The fluorescent oligonucleotides (Figure 4(a), Supplementary Table S2) were purchased from IBA (Göttingen, Germany). Briefly, AUF1 p45 WT, or a variant thereof, was added at different concentrations to 10 nM of 5ʹ-Cy3-labeled 3ʹCS-RNA in assay buffer (50 mM Hepes/NaOH, pH 8.0, 100 mM KCl, 5 mM MgCl2). Then, 10 nM of 5ʹ-Cy5-labeled 5ʹCS-RNA was added and readings were taken for 400 s. Changes in the fluorescence signals were monitored using a Fluoromax-4 spectrofluorometer (Jobin Yvon, France) at 22°C. The Cy3 fluorophore was excited at 535 nm wavelength and readings were taken at the Cy5 emission wavelength 680 nm. Fluorescence intensities relative to the starting fluorescence were plotted against time and fitted by KaleidaGraphTM (Synergy software) to a second-order reaction Equation (3) (see above) yielding the corresponding rate constants kobs.
Calculation of free energy change of RNA annealing upon mutation of the arginine residues of the RGG/RG motif in AUF1 p45
Transition-state theory provides the relationship of the rate constant of a reaction and the free energy of activation.
| (4) |
| (5) |
where ΔG≠ is the free energy of activation, R is the universal gas constant, k is the rate constant of the reaction, kB is the Boltzmann constant, h is Planck’s constant, κ is the transmission factor and T is the absolute temperature.
The catalytic efficiency kcat/KM was determined from the slope of the plot kobs (second-order rate constant) in dependence on the protein concentration and used for the calculation of thermodynamic activation parameters [69,70]. The reaction of the wildtype is related to that of the respective variant in order to dissect its energetic contribution of the functional group by the term ΔΔG≠, which is the difference between the free energies of activation of the reaction catalysed by the wildtype (wt) and the variant (var).
| (6) |
Funding Statement
This work was supported by the Martin Luther University Halle-Wittenberg (TS, RPG, SF and S-EB), the Deutsche Forschungsgemeinschaft (DFG) under grant number BE1885/12-1 (awarded to SF and S-EB) and the DFG research training group (RTG) 1591 (awarded to AM and S-EB).
Acknowledgments
We thank Gary Sawers for critically reading the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Wagner BJ, DeMaria CT, Sun Y, et al. Structure and genomic organization of the human AUF1 gene: alternative pre-mRNA splicing generates four protein isoforms. Genomics. 1998;48:195–202. [DOI] [PubMed] [Google Scholar]
- [2].Moore AE, Chenette DM, Larkin LC, et al. Physiological networks and disease functions of RNA-binding protein AUF1. Wiley Interdiscip Rev RNA. 2014;5:549–564. [DOI] [PubMed] [Google Scholar]
- [3].White EJ, Matsangos AE, Wilson GM.. AUF1 regulation of coding and noncoding RNA. Wiley Interdiscip Rev RNA. 2017;8:e1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Yoon JH, De S, Srikantan S, et al PAR-CLIP analysis uncovers AUF1 impact on target RNA fate and genome integrity. Nat Commun. 2014;5:5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wu X, Chesoni S, Rondeau G, et al Combinatorial mRNA binding by AUF1 and Argonaute 2 controls decay of selected target mRNAs. Nucleic Acids Res. 2013;41:2644–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yoon JH, Jo MH, White EJ, et al AUF1 promotes let-7b loading on Argonaute 2. Genes Dev. 2015;29:1599–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Daubner GM, Clery A, Allain FH. RRM-RNA recognition: NMR or crystallography..and new findings. Curr Opin Struct Biol. 2013;23:100–108. [DOI] [PubMed] [Google Scholar]
- [8].Nagata T, Kurihara Y, Matsuda G, et al. Structure and interactions with RNA of the N-terminal UUAG-specific RNA-binding domain of hnRNP D0. J Mol Biol. 1999;287:221–237. [DOI] [PubMed] [Google Scholar]
- [9].Katahira M, Miyanoiri Y, Enokizono Y, et al. Structure of the C-terminal RNA-binding domain of hnRNP D0 (AUF1), its interactions with RNA and DNA, and change in backbone dynamics upon complex formation with DNA. J Mol Biol. 2001;311:973–988. [DOI] [PubMed] [Google Scholar]
- [10].Enokizono Y, Konishi Y, Nagata K, et al. Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D. J Biol Chem. 2005;280:18862–18870. [DOI] [PubMed] [Google Scholar]
- [11].Thandapani P, O‘Connor TR, Bailey TL, et al. Defining the RGG/RG motif. Mol Cell. 2013;50:613–623. [DOI] [PubMed] [Google Scholar]
- [12].Rajkowitsch L, Chen D, Stampfl S, et al. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–130. [DOI] [PubMed] [Google Scholar]
- [13].Yang Q, Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–13601. [DOI] [PubMed] [Google Scholar]
- [14].Kim Y, Myong S. RNA remodeling activity of DEAD box proteins tuned by protein concentration, RNA length, and ATP. Mol Cell. 2016;63:865–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gebhard LG, Kaufman SB, Gamarnik AV. Novel ATP-independent RNA annealing activity of the dengue virus NS3 helicase. PLoS One. 2012;7:e36244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kim H, Abeysirigunawarden SC, Chen K, et al. Protein-guided RNA dynamics during early ribosome assembly. Nature. 2014;506:334-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chakshusmathi G, Kim SD, Rubinson DA, et al. A La protein requirement for efficient pre-tRNA folding. EMBO J. 2003;22:6562–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Raghunathan PL, Guthrie C. A spliceosomal recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. Science. 1998;279:857–860. [DOI] [PubMed] [Google Scholar]
- [19].Didychuk AL, Montemayor EJ, Brow DA, et al. Structural requirements for protein-catalyzed annealing of U4 and U6 RNAs during di-snRNP assembly. Nucleic Acids Res. 2016;44:1398–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Santiago-Frangos A, Woodson SA. Hfq chaperone brings speed dating to bacterial sRNA. Wiley Interdiscip Rev RNA. 2018;9:e1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Schmidt T, Friedrich S, Golbik RP, et al. NF90-NF45 is a selective RNA chaperone that rearranges viral and cellular riboswitches: biochemical analysis of a virus host factor activity. Nucleic Acids Res. 2017;45:12441–12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Godet J, Boudier C, Humbert N, et al. Comparative nucleic acid chaperone properties of the nucleocapsid protein NCp7 and Tat protein of HIV-1. Virus Res. 2012;169:349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. [DOI] [PubMed] [Google Scholar]
- [25].Friedrich S, Schmidt T, Geissler R, et al. AUF1 p45 promotes West Nile virus replication by an RNA chaperone activity that supports cyclization of the viral genome. J Virol. 2014;88:11586–11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Friedrich S, Schmidt T, Schierhorn A, et al. Arginine methylation enhances the RNA chaperone activity of the West Nile virus host factor AUF1 p45. RNA. 2016;22:1574–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Friedrich S, Engelmann S, Schmidt T, et al. The host factor AUF1 p45 supports flavivirus propagation by triggering the RNA switch required for viral genome cyclization. J Virol. 2018;92:e01647–01617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Filomatori CV, Lodeiro MF, Alvarez DE, et al. A 5‘ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 2006;20:2238–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Fernandez-Sanles A, Rios-Marco P, Romero-Lopez C, et al. Functional information stored in the conserved structural RNA domains of flavivirus genomes. Front Microbiol. 2017;8:546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Selisko B, Wang C, Harris E, et al. Regulation of Flavivirus RNA synthesis and replication. Curr Opin Virol. 2014;9:74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gebhard LG, Filomatori CV, Gamarnik AV. Functional RNA elements in the dengue virus genome. Viruses. 2011;3:1739–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kajita Y, Nakayama J, Aizawa M, et al. The UUAG-specific RNA binding protein, heterogeneous nuclear ribonucleoprotein D0. Common modular structure and binding properties of the 2xRBD-Gly family. J Biol Chem. 1995;270:22167–22175. [DOI] [PubMed] [Google Scholar]
- [33].DeMaria CT, Sun Y, Long L, et al. Structural determinants in AUF1 required for high affinity binding to A + U-rich elements. J Biol Chem. 1997;272:27635–27643. [DOI] [PubMed] [Google Scholar]
- [34].Fellows A, Deng B, Mierke D, et al. Peptides modeled on the RGG domain of AUF1/hnRNP-D regulate 3‘ UTR-dependent gene expression. Int Immunopharmacol. 2013;17:132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Xu RM, Jokhan L, Cheng X, et al. Crystal structure of human UP1, the domain of hnRNP A1 that contains two RNA-recognition motifs. Structure. 1997;5:559–570. [DOI] [PubMed] [Google Scholar]
- [36].Shamoo Y, Krueger U, Rice LM, et al. Crystal structure of the two RNA binding domains of human hnRNP A1 at 1.75 A resolution. Nat Struct Biol. 1997;4:215–222. [DOI] [PubMed] [Google Scholar]
- [37].Ding J, Hayashi MK, Zhang Y, et al. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999;13:1102–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Deo RC, Bonanno JB, Sonenberg N, et al. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. [DOI] [PubMed] [Google Scholar]
- [39].Handa N, Nureki O, Kurimoto K, et al. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature. 1999;398:579–585. [DOI] [PubMed] [Google Scholar]
- [40].Allain FH, Bouvet P, Dieckmann T, et al. Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin. EMBO J. 2000;19:6870–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wang X, Tanaka Hall TM. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat Struct Biol. 2001;8:141–145. [DOI] [PubMed] [Google Scholar]
- [42].Perez-Canadillas JM. Grabbing the message: structural basis of mRNA 3‘UTR recognition by Hrp1. EMBO J. 2006;25:3167–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Beusch I, Barraud P, Moursy A, et al. Tandem hnRNP A1 RNA recognition motifs act in concert to repress the splicing of survival motor neuron exon 7. Elife. 2017;6:e25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lukavsky PJ, Daujotyte D, Tollervey JR, et al. Molecular basis of UG-rich RNA recognition by the human splicing factor TDP-43. Nat Struct Mol Biol. 2013;20:1443–1449. [DOI] [PubMed] [Google Scholar]
- [45].Mayeda A, Munroe SH, Caceres JF, et al. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 1994;13:5483–5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Diarra Dit Konte N, Krepl M, Damberger FF, et al. Aromatic side-chain conformational switch on the surface of the RNA Recognition Motif enables RNA discrimination. Nat Commun. 2017;8:654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Nolan SJ, Shiels JC, Tuite JB, et al. Recognition of an essential adenine at a protein−RNA interface: comparison of the contributions of hydrogen bonds and a stacking interaction. J Am Chem Soc. 1999;121:8951–8952. [Google Scholar]
- [48].Auweter SD, Fasan R, Reymond L, et al. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J. 2006;25:163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zucconi BE, Ballin JD, Brewer BY, et al. Alternatively expressed domains of AU-rich element RNA-binding protein 1 (AUF1) regulate RNA-binding affinity, RNA-induced protein oligomerization, and the local conformation of bound RNA ligands. J Biol Chem. 2010;285:39127–39139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kuehnert J, Sommer G, Zierk AW, et al. Novel RNA chaperone domain of RNA-binding protein La is regulated by AKT phosphorylation. Nucleic Acids Res. 2015;43:581–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Muller UF, Goringer HU. Mechanism of the gBP21-mediated RNA/RNA annealing reaction: matchmaking and charge reduction. Nucleic Acids Res. 2002;30:447–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Doetsch M, Furtig B, Gstrein T, et al. The RNA annealing mechanism of the HIV-1 Tat peptide: conversion of the RNA into an annealing-competent conformation. Nucleic Acids Res. 2011;39:4405–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Panja S, Schu DJ, Woodson SA. Conserved arginines on the rim of Hfq catalyze base pair formation and exchange. Nucleic Acids Res. 2013;41:7536–7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zheng A, Panja S, Woodson SA. Arginine patch predicts the RNA annealing activity of hfq from gram-negative and gram-positive bacteria. J Mol Biol. 2016;428:2259–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kumar A, Wilson SH. Studies of the strand-annealing activity of mammalian hnRNP complex protein A1. Biochemistry. 1990;29:10717–10722. [DOI] [PubMed] [Google Scholar]
- [56].Munroe SH, Dong XF. Heterogeneous nuclear ribonucleoprotein A1 catalyzes RNA.RNA annealing. Proc Natl Acad Sci U S A. 1992;89:895–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Gabus C, Mazroui R, Tremblay S, et al. The fragile X mental retardation protein has nucleic acid chaperone properties. Nucleic Acids Res. 2004;32:2129–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18:1169–1175. [DOI] [PubMed] [Google Scholar]
- [59].Clery A, Jayne S, Benderska N, et al. Molecular basis of purine-rich RNA recognition by the human SR-like protein Tra2-beta1. Nat Struct Mol Biol. 2011;18:443–450. [DOI] [PubMed] [Google Scholar]
- [60].Chang N, Yi J, Guo G, et al. HuR uses AUF1 as a cofactor to promote p16INK4 mRNA decay. Mol Cell Biol. 2010;30:3875–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Hsiao KY, Wu MH, Chang N, et al. Coordination of AUF1 and miR-148a destabilizes DNA methyltransferase 1 mRNA under hypoxia in endometriosis. Mol Hum Reprod. 2015;21:894–904. [DOI] [PubMed] [Google Scholar]
- [62].Fotinos A, Fritz DT, Lisica S, et al. Competing repressive factors control bone morphogenetic protein 2 (BMP2) in mesenchymal cells. J Cell Biochem. 2015;117:439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wilson GM, Sutphen K, Chuang K, et al. Folding of A+U-rich RNA elements modulates AUF1 binding. Potential roles in regulation of mRNA turnover. J Biol Chem. 2001;276:8695–8704. [DOI] [PubMed] [Google Scholar]
- [64].Wilson GM, Sutphen K, Moutafis M, et al. Structural remodeling of an A + U-rich RNA element by cation or AUF1 binding. J Biol Chem. 2001;276:38400–38409. [DOI] [PubMed] [Google Scholar]
- [65].Fialcowitz EJ, Brewer BY, Keenan BP, et al. A hairpin-like structure within an AU-rich mRNA-destabilizing element regulates trans-factor binding selectivity and mRNA decay kinetics. J Biol Chem. 2005;280:22406–22417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Taniguchi M, Du H, Lindsey JS. PhotochemCAD 3: diverse modules for photophysical calculations with multiple spectral databases. Photochem Photobiol. 2018;94:277–289. [DOI] [PubMed] [Google Scholar]
- [67].Schmidt T, Knick P, Lilie H, et al. Coordinated action of two double-stranded RNA binding motifs and an RGG motif enables nuclear factor 90 to flexibly target different RNA substrates. Biochemistry. 2016;55:948–959. [DOI] [PubMed] [Google Scholar]
- [68].Schmidt T, Knick P, Lilie H, et al. The properties of the RNA-binding protein NF90 are considerably modulated by complex formation with NF45. Biochem J. 2017;474:259–280. [DOI] [PubMed] [Google Scholar]
- [69].Wilkinson AJ, Fersht AR, Blow DM, et al. Site-directed mutagenesis as a probe of enzyme structure and catalysis: tyrosyl-tRNA synthetase cysteine-35 to glycine-35 mutation. Biochemistry. 1983;22:3581–3586. [DOI] [PubMed] [Google Scholar]
- [70].Fersht AR. Structure and mechanism in protein science. 1st ed. New York: Freeman; 1998. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


