Abstract
Despite expanding knowledge regarding the role of astroglia in regulating neuronal function, little is known about regional or functional subgroups of brain astroglia and how they may interact with neurons. We use an astroglia-specific promoter fragment in transgenic mice to identify an anatomically defined subset of adult grey matter astroglia. Using transcriptomic and histological analyses, we generate a combinatorial profile for in vivo identification and characterization of this astroglia subpopulation. These astroglia are enriched in cortical layer V, express distinct molecular markers, including norrin and LGR6, with corresponding layer-specific neuronal ligands, are found in human cortex and modulate neuronal activity. Astrocytic norrin appears to regulate dendrites and spines and its loss, as occurs in Norrie disease, contributes to cortical dendritic spine loss. These studies provide evidence that human and rodent astroglia subtypes are regionally and functionally distinct, can regulate local neuronal dendrite and synaptic spine development and contribute to disease.
Introduction
Astroglia are the most abundant cell type in the central nervous system (CNS) and have essential roles in the development and homeostasis of the nervous system, maturation and maintenance of synapses, and regulation of neural transmission1. Disturbances of these essential roles may contribute to various CNS diseases, including amyotrophic lateral sclerosis (ALS) 1, 2. For more than 100 years, astroglia have been broadly defined into two morphologically described subgroups: protoplasmic astroglia, which are localized to gray matter, and fibrous astroglia, which are localized to white matter. However, recent years have witnessed a growing appreciation for potential astroglia diversity beyond simple morphology, with accumulating evidence suggesting the existence of functionally distinct astroglia subpopulations 1, 3–7. Current knowledge surrounding regional specialization of astroglia populations comes in part from insight provided by the positional identity of astroglia in the developing spinal cord, where defined anatomical locations also define different astroglia subpopulations 4–6. Each of these subpopulations displays a unique biological profile that allows the cell to maintain its physiological niche 8. In disease, altered function in regional astroglia can contribute to neurotoxic events, such as impaired glutamate uptake by perisynaptic astroglia in the context of neurodegeneration 9. However, very little is known about the molecular identities of different astroglia subgroups in the majority of CNS tissues and how these subgroups might regulate local neuronal function. This limited understanding of astroglia is due in part to the lack of RNA or protein markers to identify different subgroups in the adult CNS, making histological or functional studies of different populations nearly impossible. To begin to develop insight into possible astroglia subgroups, we explored the biology of an astroglia-specific targeted protein, the glutamate transporter EAAT2, known to be focally altered in some disorders 10.
All astroglia normally express EAAT2, also known as GLT1 11. Astroglial GLT1 is generally expressed uniformly throughout the CNS, although the levels of GLT1 are approximately 10-fold higher in the hippocampus and cortex relative to the spinal cord 12. Dramatic regional dysregulation of GLT1 expression has been observed in a variety of neurological and psychiatric disorders 11, 13, 14. For example, GLT1 is downregulated in a subset of astroglia in the ventral spinal cord and layers of motor cortex in human ALS patients as well as in rodent models of ALS 2. Although the biological basis of this focal dysregulation has historically focused on how neurons may focally regulate GLT1 expression 15, in our attempts to define the regulation of GLT1 expression within astroglia specifically, we discovered a cortical layer specific subpopulation with unique molecular/protein signature that define, in part, a functional role in regulating local neuronal dendrite and spine formation. We find that this novel astroglial pathway plays a fundamental role in the childhood neurological disorder, Norrie disease.
Results
Identification and Validation of 8.3-Astroglia: Glt1 Promoter Reporter Mouse Lines
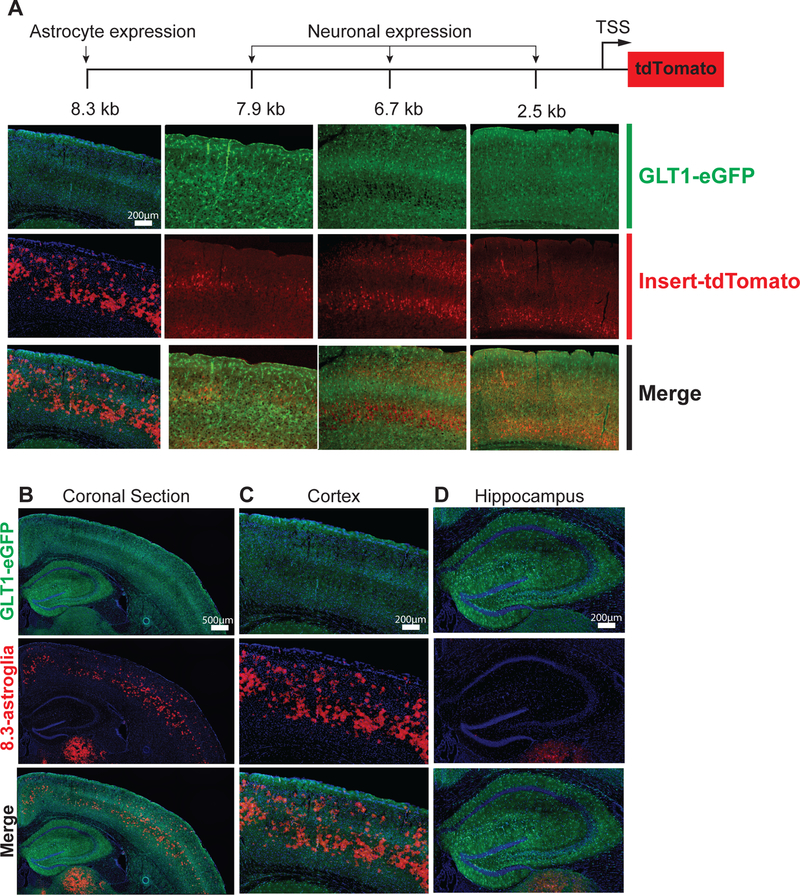
To examine Glt1 regulation within astroglia, we created multiple mouse lines using DNA inserts that contained fragments of the Glt1 promoter upstream of the Glt1 transcriptional start site. Downstream of the Gtl1 transcriptional start site, the insertion of a tdTomato reporter allowed us to visualize cell populations actively transcribing the transgenic constructs. The fragments of the Glt1 promoter were of various sizes, ranging from 2.5 kb to 8.3 kb upstream of the transcription start site (Fig. 1A, Fig S1A). These fragment sizes were chosen based on conserved genomic regions with high methylation between the mouse and human Glt1gene (human nomenclature, EAAT2) (Fig. S1A). Multiple founder lines expressing a promoter fragment ≤ 7.9 kb showed tdTomato expression that was restricted to neurons (Fig. 1A, Fig. S2A - B). Unexpectedly, founder lines with only a slightly larger insert size, 8.3 kb, showed tdTomato fluorescence limited to grey-matter astroglia (Fig. 1A–C, Fig. 2A–C, Fig. S3B). Notably, a specific and reproducible subpopulation of grey-matter astroglia expressed tdTomato in the cerebral cortex and several other regions unexplored in this study (Fig. 1B–D, Fig. S3A, Video S1). To compare this tdTomato-astroglia population (henceforth referred to as “8.3-astroglia”) with all other grey-matter cortical astroglia, we crossed 8.3-astroglia-labeled mice with a BAC-GLT1-eGFP mouse line (GLT1-eGFP), which labels all CNS astroglia with eGFP (Fig. 1A–D, Fig. S3B) 12. In the double transgenic mouse line (8.3-astroglia/GLT1-eGFP), all 8.3-astroglia were also eGFP-positive, indicating that these cells are indeed astroglia expressing GLT1 (Fig. 2A–C, Fig. S3B). In adult cortex grey matter, definable subsets of all eGFP-astroglia were also tdTomato-positive (Fig. 1B–C, Fig. S3B). Notably, 8.3-astroglia were completely absent from the hippocampus (Fig. 1D, Movie S1).
Fig. 1. Generation of GLT1 promoter reporter mice employing increasing lengths of the DNA sequence upstream of the transcription start site.
Promoter reporter mice were generated by employing different lengths of the DNA sequences upstream of the transcriptional start site (TSS) of GLT1 followed by tdTomato leading to astroglia-specific tdTomato-expression. (A) Varying length promoter reporter fragments resulted in differential cell-specific tdTomato expression. A fragment of 8.3 kb was required for astroglia-restricted tdTomato expression, whereas fragments ≤ 7.9 kb resulted in neuronal tdTomato expression and no astroglial expression in adults. (B) Coronal section of the 8.3 kb-tdTomato expression. (C) Magnified image of motor cortex representing overlap between 8.3 kb-tdTomato and Glt1-eGFP astrocyte specific expression. (D) No 8.3 kb-tdTomato expression was detected in the hippocampus. N=3 different mice were imaged with analysis of 5 images per mouse.
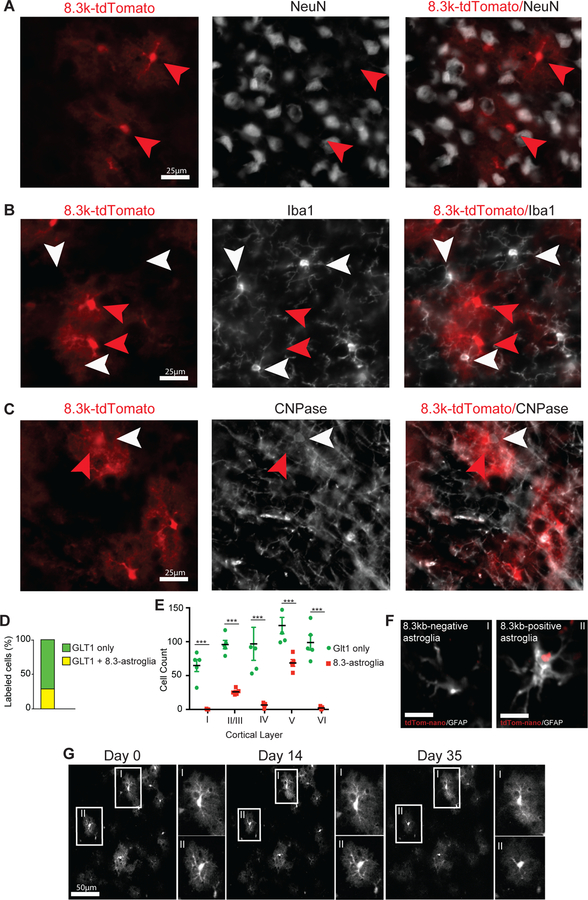
Fig. 2. 8.3 kb-tdTomato expression is static and limited to an astroglia subset in the cerebral motor cortex.
(A) 8.3 kb-tdTomato does not colocalize with neuronal marker, NeuN. (B) 8.3 kb-tdTomato does not colocalize with microglia marker, Iba1. (C) 8.3 kb-tdTomato does not colocalize with myelin and oligodendrocyte marker, CNPase. (D) 8.3-astroglia consist of about 25% of all grey matter astroglia in the motor cortex. (E) 8.3-astroglia are heavily enriched in cortical layers II/III and V. Glt1-eGFP only astroglia are equally distributed across all cortical layers. (F) 8.3 kb-tdTomato nanoparticles are expressed by a subset of GFAP-positive astroglia enriched in layers II/III and V of the mouse motor cortex (i,ii scale bars 15μM). (G) Cortical multiphoton in vivo imaging performed weekly for 5 weeks in adult mice tracking individual 8.3-astroglia (N = 5 mice, 100 cells). Astroglia cell counting was performed with sections imaged and subjected to validated automated cell counting of eGFP and tdTomato fluorescence cells. Statistics used to compare eGFP only to 8.3-astroglia per cortical layer include two-way ANOVA with Tukey post hoc analysis, error bars represent standard error of mean (SEM). p-value statistics used are as following * p< 0.05, ** p< 0.01, *** p< 0.001. For nanoparticle injections, n = 5 mice were intracortically injected and analyzed. For all experiments n = 5 mice were analyzed, 3–5 images per mouse. Red arrows indicate 8.3kb-astroglia and white arrows indicate non-8.3-astroglia cell.
The 8.3-astroglia consisted of approximately 28% of all cortical astroglia as evaluated by the number of 8.3-astroglia/GLT1-eGFP double-positive astroglia in the adult cortex (Fig. 2D, Fig. S3C). Layer V of the cortex was preferentially enriched with 8.3-astroglia compared to all other cortical layers, whereas the GLT1-eGFP single-positive astroglia were evenly distributed among the cortical layers as assessed by traditional 2D as well as 3D imaging approaches (Fig. 2E, Video S1).
To verify that these observations were not due to an artifact of genomic integration and to provide an alternative approach for studying this astroglia subset without the need for labeled transgenic mice, we introduced the 8.3 kb-tdTomato plasmid in vivo in wild-type mice using nanoparticles. Nanoparticles have the advantage of accepting large molecular constructs and have excellent in vivo tissue distribution properties compared to adeno-associated viruses or lentiviruses 16, 17. CMV-eGFP control and 8.3 kb-tdTomato plasmids were packaged into separate nanoparticles that were approximately the size of nanometer-sized exosomes as visualized by transmission electron microscopy (Fig. S3F–G). Following intracortical injection of CMV-eGFP control nanoparticles, eGFP fluorescence was distributed across the entire hemisphere and in all cell types (Fig. S3D). After intracortical injection of the 8.3 kb-tdTomato nanoparticles, however, tdTomato expression was limited to astroglia in the grey matter and specifically enriched in layers II/III and V, consistent with the 8.3-astroglia transgenic mouse (Fig. 2F, Fig. S3E). These data support the hypothesis that the 8.3 kb sequence upstream of the Glt1 transcriptional start site is utilized only by a subset of astroglia.
We next determined the stability of tdTomato fluorescence in this astroglia subset. Stable expression of tdTomato within a subset of astroglia throughout their lifespan, without the appearance of additional fluorescent signal from other cell populations, would support the use of tdTomato as a fluorescent marker to study the specific subset of cells utilizing the 8.3 kb promoter fragment. To address this possibility, we performed longitudinal multiphoton imaging of the motor cortex (up to 500 μm deep) of 8.3-astroglia-labeled transgenic mice (Suppl Video S2). We tracked individual 8.3-astroglia over a period of five weeks and saw no changes in fluorescence, nor did we observe the appearance of any additional tdTomato-expressing cells in the cortical regions studied (Fig. 2G, Suppl Video S2). The cell-specific tdTomato fluorescence was first observed during developmental astrogenesis (P5) and remained stable throughout adulthood (not shown). Finally, we assessed the longitudinal expression of tdTomato in vitro using isolated 8.3-astroglia from adult mouse cortex (P60-P90), finding that tdTomato fluorescence continued for up to two weeks prior to cell passaging (Fig. S4B - C). The continuous expression of tdTomato in 8.3-astroglia in vivo and in vitro supports the hypothesis that these cells represent a distinct subpopulation of astroglia and that this transgenic mouse model provides a tool to study this cell population in various contexts.
Molecular Properties of 8.3-Astroglia
We next assessed the unique molecular properties of the 8.3-astroglia by determining their transcriptomic profile. Adult mouse cortices (n = 3) were dissociated into single-cell suspensions and separated by FACS to collect three distinct populations: GLT1-eGFP only (grey-matter astroglia), 8.3-astroglia (grey-matter astroglia subset), and the negative-fluorescent cell population (all cells that are not grey-matter astroglia) (Fig. S4A) 18. During our FACS we noted that those astrocytes that were tdTomato positive displayed varying levels of tdTomato-fluorescence, however we were able to isolate only the high expressers which are continued to be referred to as 8.3-astroglia (Fig. S4A). Based on microarray analysis of these populations, each group displayed a unique RNA transcriptome (Fig. 3A). As expected, the two astroglia populations were both enriched in well-established astroglia markers such as Aldh1l1, Acsbg1, and Aqp4. (Fig. 3B) 19. However, when comparing 8.3-astroglia to GLT1-eGFP astroglia and the reporter-negative cell population, the 8.3-astroglia in the cortex displayed consistent and profound enrichment in several candidate markers (Fig. 3C–D, Table S1). Some of the highest expressing genes, that have also been shown to be involved in neurological disorders, include Kcnj10, Norrin, Olig2, Lgr6, and Fndc5 (Fig. 3C–D, Table S2); their expression was validated by quantitative PCR (Fig. 3D, data not shown).
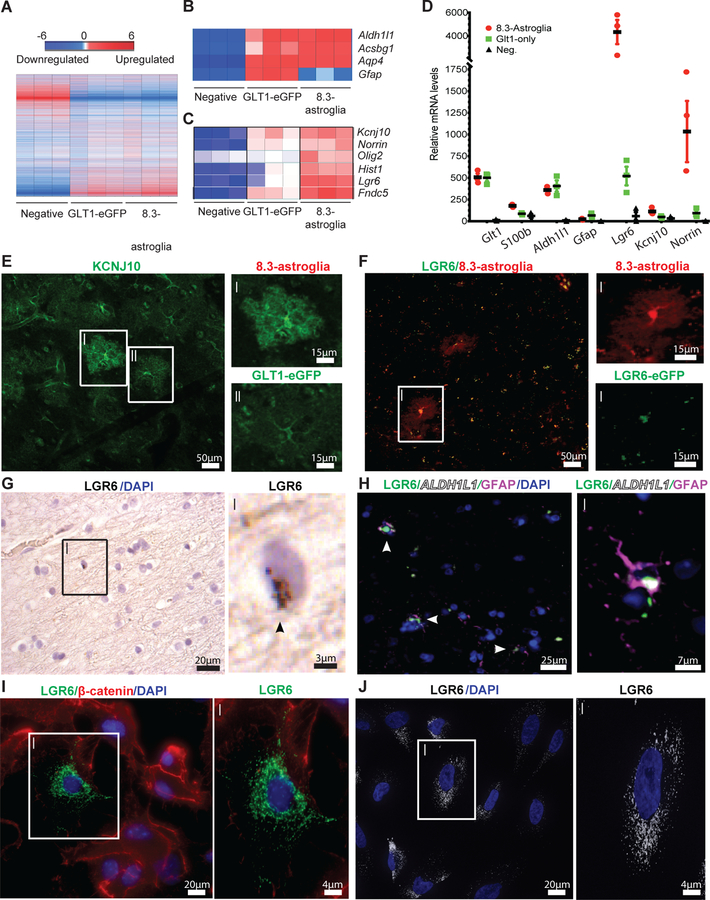
Fig. 3. Astroglial cell populations isolated from 8.3-astroglia mouse CNS and human cortex have unique transcriptomes as validated with immunohistochemistry (IHC) and RNA in situ hybridization.
(A) The cortex microarray heatmap displays unique genes differentially expressed between all rodent astroglia (GLT1-eGFP), 8.3 kb labeled astroglia (8.3-astroglia) and remaining non-labeled cells (negative). (B) Microarray expression of astroglia markers highly expressed in both rodent astroglia populations. (C) Selected markers highly enriched in 8.3-astroglia microarray data in the cortex. (D) qPCR validation of candidate markers from the microarray analyses in the cortex of the three cell populations. (E) KCNJ10 enhanced immunoreactivity in 8.3-astroglia compared to GLT1-eGFP astroglia in the mouse motor cortex (i,ii scale bars 15μm). (F) LGR6-GFP expression is restricted to 8.3-astroglia in the adult mouse cortex as evaluated in double transgenic LGR6-GFP-ires-CreERT2/8.3-astroglia mice (i,ii scale bars 15μm). (G) Analysis of Lgr6 mRNA expression in the human motor cortex in a subset of cells (i scale bar 2.5μm), arrow indicates RNA. (H) LGR6 protein in the human motor cortex colocalizes with GFAP and ALDH1L1 protein (i scale bar 7.5μm). (I) A subset of cultured primary mouse cortical astroglia are LGR6-positive (i scale bar 4μm). (J) A subset of cultured and matured human iPSC-astroglia are LGR6-immunoreactive (i scale bar 4μm). Three cell populations were isolated from the adult mouse whole cortex and subjected to FACS, microarray, and qPCR analyses from a total of three mice. For histological sections, n = five mice were imaged with a minimum of 3–5 images per mouse, representative images are shown. Human post-mortem tissue was evaluated in n = three non-neurological diseased cases with at least 3–5 images taken per case for both RNA ISH and immunohistochemistry. For mouse immunocytochemistry, n = five different cultures of mouse primary astroglia were generated and imaged at least 3–5 times per astroglia generation. For iPSC astroglia a minimum of three control human iPSC lines were generated. All images are representative of the total amount imaged. Statistics were performed by two-sided Student’s T-test of 8.3-astroglia vs negative cell population with represented values showing the mean and error bars representing SEM, *p<0.05, **p<0.01, ***p<0.001.
Hypothesizing that these enriched proteins could provide clues into the unique biological properties of these adult astroglia subpopulations and to allow us to create a combinatorial profile, we generated a list of markers enriched in single-positive GLT1-eGFP astroglia and double-positive astroglia populations and subjected these modified lists to software-based pathway analytics (Tables S1, S3, S4). We found that these astroglia populations were uniquely enriched in specific pathways, with enrichment of the Wnt/β-catenin signaling pathway in GLT1-eGFP astroglia and enrichment of the Sonic Hedgehog pathway in 8.3-astroglia (Fig. S4D). Both astroglia populations showed enrichment in pathways common to the function of all astroglia, such as pathways involved in fat synthesis. (Fig. S4D and Tables S2, S3) 20.
LGR6 and Norrin expression by 8.3-Astroglia
We next sought to generate a panel of markers that could be used to uniquely identify 8.3-astroglia. Starting with candidates identified by our transcriptomic analyses, we assessed four genes that were highly enriched in 8.3-astroglia compared to both the GLT1-eGFP and negative-fluorescent cell populations: Kcnj10, Lgr6, Olig2, and Norrin (Fig. 3C–D, Table S2). In agreement with our RNA analyses, the cortical distribution of KCNJ10 was enriched in similar regions as the 8.3-astroglia (Fig. 3E, Fig. S5B–C). At higher magnification, 8.3-astroglia displayed a higher mean fluorescence intensity of KCNJ10 compared to GLT1-eGFP astroglia (Fig. 3E, Fig. S5B–C). To visualize Lgr6-expressing cells, we generated a double transgenic mouse model by crossing LGR6-GFP-ires-CreERT2 mice with the 8.3-astroglia mice 21. As expected, LGR6-eGFP localized to all 8.3-astroglia in the adult cortex, with varying degrees of fluorescence, of these double transgenic mice (Fig. 3F). The LGR6-EGFP fluorescence was only noted in 8.3-astroglia and no other cells. We also detected nuclear OLIG2 expression in 8.3-astroglia, consistent with the microarray data (Fig. S4E). We note that although OLIG2 is widely used to identify oligodendrocyte lineage cells in the CNS, it is also known to exist in some neuroprotective astroglia subpopulations 22, 23. In aggregate, the molecular identity and the unique CNS localization of the 8.3-astroglia are useful for elucidating the biological significance of this glial subgroup and allows for the studying of this subpopulation without the need of the transgenic mice.
To determine whether this astroglia subpopulation is also present in the human cortex, we used LGR6 expression as a surrogate marker for the 8.3-astroglia population. Using immunohistochemistry and RNA in situ hybridization on post-mortem adult human cortex tissue, we detected LGR6 expression in only a subset of astroglia in the human cortex, consistent with the results of our rodent studies (Fig. 3G–H, S5A). To validate the astroglia-specific expression of LGR6 in humans, we used immunohistochemistry to visualize astrocyte-specific marker ALDH1L1. LGR6 colocalized only with ALDH1L1-positive cells in the adult human cortex, providing further support that these cells are indeed a subset of human cortical astroglia (Fig. 3H, Fig. S5A).
LGR6-positive astroglia subsets could also be reliably identified in vitro in both pure mouse primary cortical cultures and in human induced pluripotent stem (hiPSCs) differentiated into astroglia (Fig. 3I–J). We also observed a higher colocalization of KCNJ10 and LGR6 in hiPSC-derived astroglia, supporting the results of our combinatorial profile (not shown). Thus, these studies establish a well-defined and geographically organized astroglia subpopulation in the adult human and rodent cortex and provide reliable markers to study their involvement in normal and diseased adult CNS physiology.
Functional Assessment of LGR6 in Astroglia
We next aimed to determine the functional significance of our observation that the receptor LGR6 is consistently enriched and labels this astroglia subpopulation. In addition, we focused on LGR6 because it showed the highest enrichment in our RNA analytics and has been widely shown to be astroglia-specific in the CNS 24–26. To address this question, we investigated the effects of addition of its ligand, R-spondin (RSPO1). RSPO1 has been shown to be neuron-specific in the adult cortex, but little is known about the downstream consequences of its interaction with LGR6 in the CNS 8. Consistent with published RNA in situ hybridization data, RSPO1 was highly enriched in areas of dense 8.3-astroglia in layer V (Fig. 4A, Fig. S6A–D) 27. Furthermore, we performed immunofluorescence for neuronal marker, NeuN, followed by RNA fluorescent in situ hybridization (FISH) for Rspo1 and noted that Rspo1 co-localized only to NeuN-positive neurons but only in a subset in the lower cortical layers (Fig. S6A–D). This provides additional support that Rspo1 is neuronal-specific in the adult motor cortex but that it is also limited to only a subset of neurons in cortical layer V.
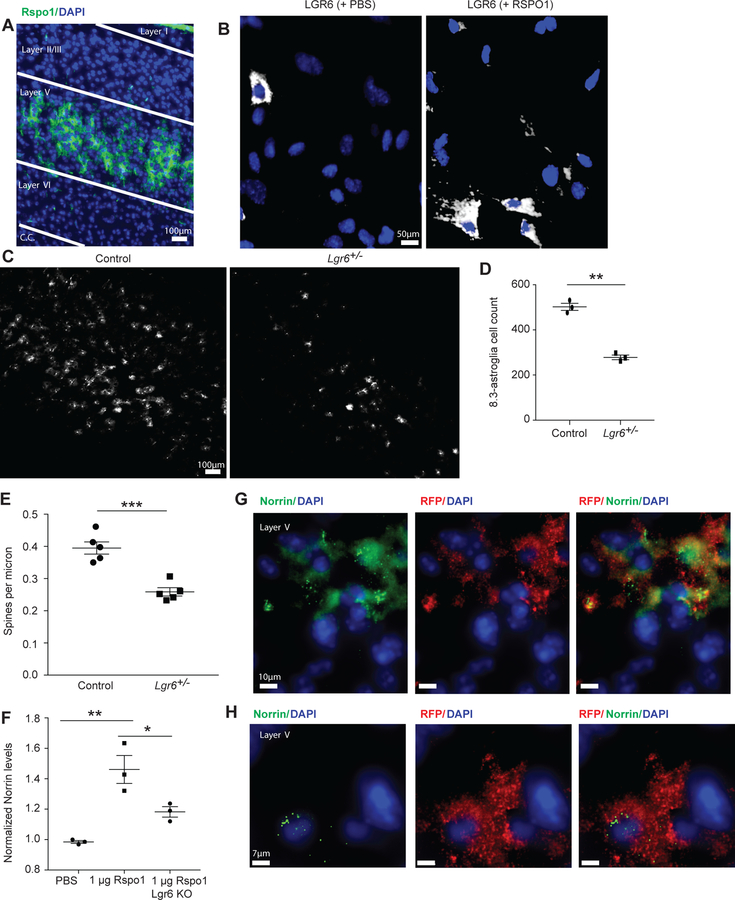
Fig. 4. Functional and dysfunctional properties of astroglia subpopulation by examination of LGR6 pathways in the cerebral motor cortex.
(A) Neuron-specific RSPO1 immunoreactive localization is restricted to cortical layer V in the mouse motor cortex (c.c. = corpus callosum). (B) RSPO1 treatment in vitro of primary mouse astroglia increases LGR6 immunoreactivity. (C) Lgr6-heterozygote mice display a reduction in the numbers of 8.3-astroglia. (D) Lgr6-heterozygote mice display a significant reduction in numbers of 8.3-astroglia, p-value 0.0003. (E) LGR6 alters neurite properties as Lgr6-heterozygote null mice have reduced spine density compared to control mice as assessed with the Golgi-Cox staining. (F) LGR6 receptor agonist RSPO1 affects astroglial Norrin. Norrin levels are increased following RSPO1 treatments on primary astroglia as assessed by ELISA. (G) Norrin mRNA colocalizes with 8.3-astroglia in the mouse motor cortex. (H) High magnification of colocalization of Norrin mRNA and tdTomato. N = five different mice with 3–5 coronal slices were imaged for IHC and RNA FISH. N = five mice were used for spine analysis with 5 neurons analyzed per mouse motor cortex. N = three different primary astroglia cultures were treated for 48 hours with RSPO1 and repeated 3–5 times for ELISA analysis (control vs Rspo1 p-value 0.0025, Rspo1 vs Lgr6-KO p-value 0.031). Statistics performed was two-sided Student’s T-test with values representing mean with SEM error bars, *p < 0.05, **p < 0.01, ***p < 0.001.
Next, we wanted to explore the astroglia response to RSPO1 in vitro. Treatment of primary astroglia cultures with increasing doses of RSPO1 resulted in significant enrichment in the overall numbers of LGR6-positive astroglia compared to PBS-treated control cultures (Fig. 4B, Fig. S7A–B). To determine whether this increase in LGR6-positive astroglia was due to proliferation, we stained astroglia with the proliferation marker Ki67, revealing a significant dose-response increase of Ki67-positive astroglia following RSPO1 treatment (Fig. S7A). This increase in proliferation was due to an increase of LGR6-positive astroglia (Fig. S7B). These in vitro findings suggest that in postnatal conditions, local neuronal release of RSPO1 could stimulate proliferation of astroglia, suggesting a specific interaction between a RSPO-positive neuron subpopulation and LGR6-positive 8.3-astroglia.
The stimulatory effect of LGR6 on proliferation of 8.3-astroglia in vitro suggested that loss of LGR6 could decrease the population of 8.3-astroglia. Indeed, we observed a dramatic loss of 8.3-astroglia in vivo in the heterozygote 8.3-astroglia/LGR6-GFP-ires-CreERT2 mice, which have 50% lower expression of LGR6 compared to wild-type mice; this decrease further correlated with a minimal but significant loss of cortical thickness (Fig. 4C–D, Fig. S7C). Since astroglia play a major role in neuronal synapse formation and elimination, we analyzed the overall density of spines on apical dendrites in layer V where 8.3-astroglia are highly enriched. We found a significant decrease in spine density in the heterozygote LGR6-eGFP/8.3-astroglia mouse model, correlating with the loss of 8.3-astroglia (Fig. 4E, Fig. S8A–B).
Next, we sought to determine which factor(s) released by 8.3-astroglia could be responsible for the deficits in dendritic spine density, focusing on secreted proteins that were highly enriched in 8.3-astroglia as identified by our transcriptomic analyses. To test for secretion of neuro-modulating proteins we performed stimulation of LGR6-positive astroglia with RSPO1 and used enzyme-linked immunosorbent assay (ELISA) to detect candidate secreted proteins and determine which proteins increased with RSPO1 treatment. One highly abundant protein not enriched in PBS control or LGR6-knockout primary astroglia was Norrin (Ndp, Norrie disease protein), a protein that is expressed in vivo by cortical astrocytes with strong colocalization with 8.3-astroglia in cortical layer V and has been shown to be astroglia-specific in the cortex (Fig. 4F–H) 24, 28.
Norrin Release by 8.3-Astroglia Regulates Dendritic Growth and Spine Formation
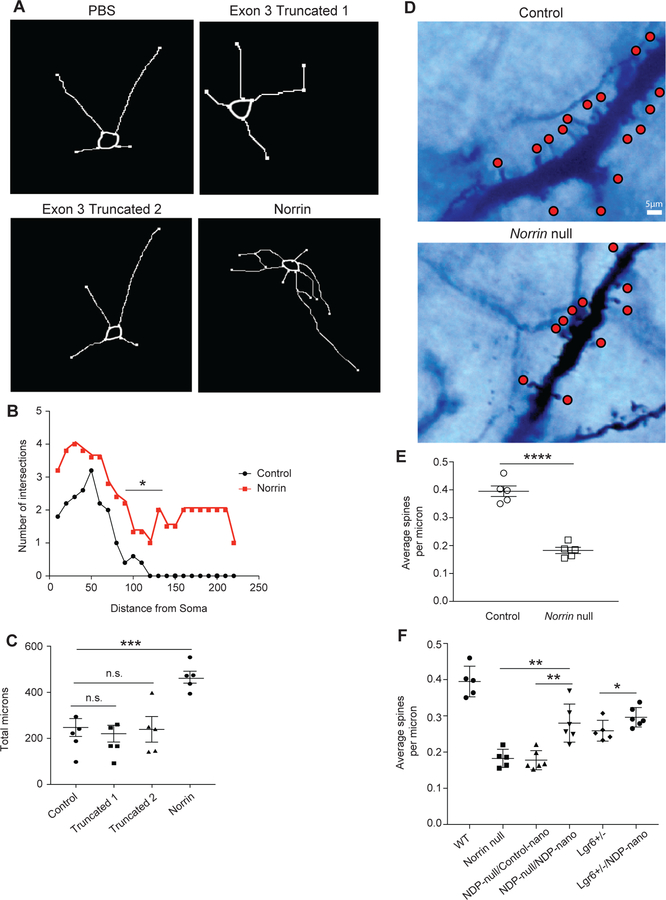
Norrin is known to be astroglia-specific in the adult cortex, where it binds to FZD4 and several other receptors such as LGR4 to activate the Wnt signaling pathway and thereby induce upregulation of neurotrophic growth factors, including Brain-Derived-Neurotrophic-Factor (BDNF) 29–31. Patients with Norrin mutations develop Norrie disease, a CNS and ocular disease that also has strong cognitive and behavioral deficits including mental retardation, psychosis, and early-onset dementia in many but not all patients 32, 33. Some patients with Norrie disease have a deletion of exon 2, resulting in the possibility of two truncated proteins from exon 3. To evaluate the effects of increased Norrin on cortical neurons, we treated primary mouse cortical neurons with recombinant Norrin, compared to the PBS vehicle control and the two Norrin truncated proteins from exon 3 (referred to as “Truncated 1” and “Truncated 2”) (Fig. 5A, Fig. S9A–D, Fig. S10A–D). We found that treatment with both truncated proteins did not affect dendritic arborization or length, however Norrin significantly affected dendritic arborization and increased dendritic length (Fig. 5B–C; data not shown). These findings show that the truncated Norrin proteins translated in Norrie disease patients is inefficient at effecting neuronal dendrites.
Fig. 5. Norrin is a neuro-modulating protein that affects cortical neuron dendritic morphology and spine density.
(A) Norrin-treated mouse cortical neurons show increased length and branching. (B) Treatment with Norrin lead to a significant number of neurite changes including intersections and neurite branchings as revealed by Scholl analytics. (C) Neurite length is increased after Norrin treatment of cultured cortical mouse neurons. (D) Loss of Norrin in vivo leads to a significant defect in cortical neuron spine density. Layer V cortical neurons were identified in adult Norrin-null mouse motor cortex and dendritic spines were examined after Golgi-Cox staining. (E) Norrin-null mice have significantly reduced spine density compared to wild-type. (F) Intracortical nanoparticle injections containing 8.3 kb-Norrin significantly increase dendritic spine density in cortical layer V of the mouse motor cortex. For invitro studies (A,B,C) recombinant Norrin and it’s truncated analogs were applied to 10 different isolations of cortical neurons (n=5 mice) with a minimum of 20 neurons analyzed for each cohort for 48 hours. For spine density analysis, n = five P60-P90 mice were used using Golgi-Cox staining kit; 5–10 cortical neurons, located in the motor cortex cortical layer V, were counted and analyzed per genotype, representative images are shown. Statistics performed by two-sided Student’s T-test of comparing Norrin treated vs control (C,E,F), *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Values plotted represent the mean with SEM error bars.
Norrin has known effects on retinal biology, but its function in the cortex is unknown. To determine whether absence of Norrin would lead to neuronal deficits, we analyzed Norrin-null mice and quantified cortical neuron dendritic spine density. We found a significant loss of spines in cortical layer V in Norrin-null mice compared to their control littermates (Fig. 5D–E, Fig. S8A, C).
Next, we wanted to evaluate the effects of in vivo Norrin treatment on cortical neuronal dendritic spine density. To address this, we created a genetic plasmid with the astroglia subset specific promoter 8.3 kb followed by Norrin cDNA and packaged this plasmid into nanoparticles that were injected into the mouse motor cortex. Norrin secretion driven solely by the 8.3 kb promoter significantly increased dendritic spine density in the Norrin-null and LGR6-eGFP-ires-CreERT2 mice compared to the CMV-eGFP control nanoparticle injected Norrin-null and LGR6-eGFP-ires-CreERT2 mice (Fig. 5F). This provides evidence that Norrin production solely by 8.3-astroglia is sufficient to restore the dendritic abnormalities and suggested a role for this astroglial specific pathway in the regulation of neuronal spine and synaptic physiology.
Norrin Treatment effects the Electrophysiological Properties of Neurons
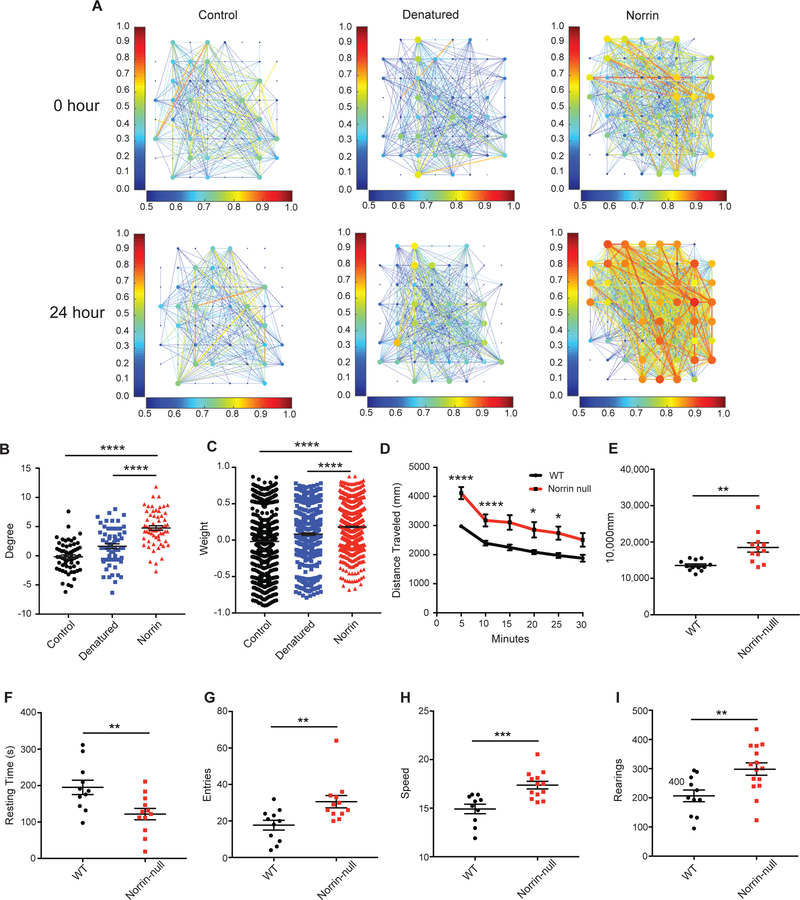
Next, we wanted to explore the effects of Norrin on the electrophysiology of cultured neurons. Using multi-electrode array (MEA) plates with rat cortical neurons and astrocytes, we monitored basal activity before and 24 hours after Norrin treatment (Fig. 6A, Fig S11A, S12A–F, S13A, and Videos 3 - 5). Spike and burst rates were normalized to pre-treatment activity to control for batch effects in baseline activity. There was a significant increase in the difference in both node degree and connection strength between electrodes, as well as a reorganization of spiking patterns wherein a greater percentage of detected spikes were organized into bursts (Fig. 6B–C, Fig. S12A–F). These findings demonstrate that Norrin could function to organize and strengthen cortical neuronal connectivity.
Fig. 6. Norrin alters the electrophysiological properties of cortical neurons and Norrin-null mice display neurobehavioral abnormalities.
(A) Norrin-treatment enhances cortical neuron connectivity and firing rate. (B) Norrin-treatment significantly increases the degree of neuronal firing. (C) Norrin-treatment significantly increases the weight of the neuronal firing strength. (D) Norrin-null mice are significantly more hyperactive than their wildtype littermate controls. (E) Total travel distance is significantly higher in norrin-null mice compared to their wildtype littermate controls. (F) Norrin-null have significantly decreased resting time compared than their wildtype littermate controls. (G) Norrin-null mice have significantly more arm entries than their wildtype littermate controls. (H) Norrin-null mice are significantly faster than their wildtype littermate controls. (I) Norrin-null mice have significantly more rearings than their wildtype littermate controls. MEA analyses were performed 3 times at each time point, each with three different treatments. For the open field assay and Y-Maze n = 15–20 mice were used per genotype. Statistics include 2-way repeated ANOVA, and two-sided Student T-test. For panels B-I, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Values plotted represent the mean with SEM error bars.
Norrin-null Mice Display Neurobehavioral Abnormalities
The Norrin-null mouse displayed a pronounced loss of dendritic spines in layer V of the cortex and effected both electrophysiology and dendrites in vitro, so we sought to determine if they also displayed altered behavior. To test this hypothesis, we subjected the Norrin-null mice and their wildtype littermates to an array of behavioral assays. Surprisingly, in the open field assay, Norrin-null mice were significantly more hyperactive than their wildtype littermates, as shown by increased exploration and decreased resting time (Fig. 6D–F). In the Y-maze assay, Norrin-null mice displayed increased combinations and arm entries, supportive of a hyperactive phenotype (Fig. 6G). Overall, the average speed of Norrin-null mice was significantly higher than the controls (Fig. 6H). When evaluating rearing, Norrin-null mice had significant more rearing behaviors than their wildtype littermates (Fig. 6I).). Furthermore, at this time period (P60 – P90), these mice did not display abnormalities in several assays such as elevated platform maze, fear conditioning, and rotarod (data not shown). These findings further support that Norrin-null mice have neurobehavioral abnormalities such as hyperactivity which may be the result of their cortical phenotype.
Discussion
The majority of evidence for astroglia heterogeneity has arisen from developmental studies in the spinal cord or found as a pathological consequence in neurological disorders 6, 11, 34. To date, there is little data that molecularly and/or physiologically defines the presence of astroglia subsets in the adult nervous system that function to interact and/or regulate regional neuronal populations. Furthermore, no reliable tools exist to accurately and robustly identify these subpopulations in normal and disease states. With the generation of the 8.3-astroglia mouse line, we have discovered and are now able to easily study a specific astroglia subset in the cortex of adult mice. This astroglia subset, 8.3-astroglia, has distinct cortical patterning and molecular profiles compared to other grey-matter astroglia, and can be robustly identified using unique markers such as OLIG2, NORRIN, KCNJ10 and LGR6 in rodent and human CNS, as well as in primary mouse and hiPSC-astroglia cultures.
Our extensive profiling data has allowed us to generate a protein/RNA profile to molecularly identify 8.3-astroglia. LGR6 colocalizes with 8.3-astroglia in the mouse and human cortex, along with subsets of in vitro astroglia from rodents and in hiPSC-derived astroglia. LGR6 contributes to Wnt signaling, one of the pathways enriched in grey-matter astroglia, and its ligand, RSPO1, is specifically released by pyramidal neurons 8, 21, 35. In the cortex of adult mice, we demonstrate that RSPO1 is restricted to a neuronal subset in layer V, the areas highly enriched in LGR6-positive 8.3-astroglia. LGR6 and Norrin have both been shown to be highly enriched and limited to astroglia 24, 26. The correlation of RSPO1, LGR6, Norrin, and 8.3-astroglia cortical patterning strongly suggests that there may be a crosstalk between neurons and astroglia, where extrinsic signaling, including the possible release of RSP01 from neuronal subtypes could influence astroglia subtype-specific responses (Fig. S14). In support of this model, RSPO1 is largely restricted to layer 5 of the cortex. It is well known that neurons influence astroglial physiology, for example via astroglia expression of glutamate transporters EAAT2 36. Future studies should address the positional identity of specific astroglia and neuron populations where neuronal diversity may influence astroglia diversity and vice versa.
The unique anatomical localization of 8.3-astroglia and their enrichment in LGR6 and Norrin suggest that they may be involved in glial-based pathogenesis of Norrie disease. We document a selective loss of spine density in both the LGR6 heterozygote and Norrin-null mice in cortical layer V, corresponding to the location of 8.3-astroglia and Norrin expression. This is particularly interesting as some Norrie disease patients with genetic deletion of exon 2 of the Norrin gene developed epilepsy. Epileptic patients have been document to have reduced spine density and altered dendritic arbor 37. Unfortunately, it is not possible to analyze spine density on Norrie disease patients as there is no available post-mortem brain tissue.
Norrin’s effect on cortical neuron dendrites is particularly interesting. We show that treatment with Norrin on neurons leads to increases in dendritic length and arborization. However, treatment with the two truncated Norrin proteins found in Norrie disease patients with deletion of exon 2 exhibited no effect on normal neuronal dendrites. There is a loss of spines in a widespread of disorders. In an ALS mouse model, there is a significant loss of spines in pyramidal layer V neurons in the motor cortex 38. This is similar to the pathology in Alzheimer’s disease, where dendritic spine loss is also involved 39. In transcriptomic data of Alzheimer’s disease (AD) mice, Norrin is significantly downregulated in astrocytes 40. There is also an observed loss of Norrin levels in the hippocampus in AD. Finally, mental retardation (another documented phenotype in some Norrie disease patients) reduced spine density is also observed 39.
Norrin could serve as a potential therapeutic in neurological disorders with a spine density pathology. This is not the first report of astroglia regulating synapse maturation and receptors. Recently, the ability of astroglia to regulate synaptic GluA2 AMPA receptors and drive synapse maturation via Chordin-like 1 was documented 41. Now, we show that using the 8.3 kb specific promoter to drive the expression of Norrin in the mouse cortex is sufficient at restoring and improving cortical spine density. This data shows that the sole release of Norrin strictly by 8.3-astroglia could serve as a therapeutic to modulate neuronal spines and synapses. Additionally, the usage of a ubiquitous promoter would hypothetically elevate Norrin levels even more and could lead to a more dramatic increase in spine density, alterations to dendritic arborization and length, and the electrophysiology of cortical neurons. In fact, the multielectrode array studies allowed us to test the effects of Norrin on the electrophysiological properties of neurons and Norrin treatment led to improved neuronal connectivity and strength supporting a possible role for this astroglia subpopulation in regulating neuronal connectivity and a candidate therapy.
To assess the clinical phenotype of these mutant mice, we performed a wide array of behavioral assays on the Norrin-null mice. We found that Norrin-null mice displayed abnormal behavior in the open field assay and Y-maze. Overall the Norrin-null mice were hyperactive, which is similar to what has been shown in BDNF mutant mice and it has been shown that Norrin can drive the expression of BDNF and other neurotrophic proteins 30. These neurobehavioral abnormalities might be expected in light of the cortical pathology in the Norrin-null mice.
Lastly, the focal localization of the cortical layer of 8.3-astroglia is quite intriguing. It has been shown recently that in the cortex, different layers exhibit different populations of astroglia, when based on their morphology 42. We now show that the 8.3-astroglia subset are the dominant population of layer V astroglia. It is well established that different cortical layers exhibit different neuronal populations. Our findings provide further evidence that suggest a layer-specific cross-talk between regional subsets of neurons and astroglia potentially indicating a localized neuron-glial functional specification. Future studies could explore this relationship, especially in disorders where a subset of cells is affected (i.e. ALS motor neurons) as well as in paradigms of cortical synaptic plasticity.
Taken together, these findings expand the growing understanding of functional astroglial heterogeneity by uncovering and defining a unique subset of astroglia in the CNS and has led to new tools to manipulate this astroglia subset in efforts to ultimately discover novel therapeutic avenues for neurological disorders affected by this and other astroglia subsets. This has also led to advances in understanding Norrie disease, which we now define as also involving astroglia and the contributions that astroglia may play in its pathophysiology. These studies set a foundation to build upon for understanding astroglial subset, their powerful role in neuronal spine biology and unique neuronal glial pairing in normal and pathological states.
Supplementary Material
Acknowledgements
We thank the Johns Hopkins Deep Sequencing and Microarray Core and C. Conover Talbot Jr. for insight on microarray, and analyses in Partek and Spotfire software suites, Dr. Hao Zhang for assistance with FACS at the Johns Hopkins University School of Public Health FACS Center, the Johns Hopkins Medicine Microscopy Core for use of the Zeiss LSM 700, Dr. Jeremy Nathans for the Norrin mice, Dr. Lyle Ostrow for postmortem tissue, and members of the Rothstein lab for helpful discussions. This work was funded by grants from the National Science Foundation Graduate Fellowship Research Program (S.J.M.), R01NS092067 (J.D.R., M.B.R.), and NIH S10 OD016374 (J.D.R.).
Footnotes
Competing Financial Interests.
A patent has been filed on the use of norrin (S.J.M. and J.D.R). The reminaing authors declare no competing financial interests.
REFERENCES
- 1.Zhang Y & Barres BA Astrocyte heterogeneity: an underappreciated topic in neurobiology. Curr Opin Neurobiol 20, 588–594 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Bruijn LI, et al. ALS-linked SOD1 mutant G85R mediates damage to astrocytes and promotes rapidly progressive disease with SOD1-containing inclusions. Neuron 18, 327–338 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Molofsky AV, et al. Astrocyte-encoded positional cues maintain sensorimotor circuit integrity. Nature 509, 189–194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsai HH, et al. Regional astrocyte allocation regulates CNS synaptogenesis and repair. Science 337, 358–362 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oberheim NA, Goldman SA & Nedergaard M Heterogeneity of astrocytic form and function. Methods Mol Biol 814, 23–45 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaboub LS & Deneen B Developmental origins of astrocyte heterogeneity: the final frontier of CNS development. Developmental neuroscience 34, 379–388 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller SJ Astrocyte Heterogeneity in the Adult Central Nervous System. Frontiers in Cellular Neuroscience 12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeisel A, et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Rothstein JD, Van Kammen M, Levey AI, Martin LJ and Kuncl RW Selective loss of glial glutamate transporter GLT-1 in amyotrophic lateral sclerosis. Ann Neurol 38, 73–84 (1995). [DOI] [PubMed] [Google Scholar]
- 10.Rothstein JD, et al. Glutamate transporter subtypes: role in excitotoxicity and amyotrophic lateral sclerosis. Ann.Neurol 36, 282 (1994). [Google Scholar]
- 11.Rothstein JD, Van Kammen M, Levey AI, Martin LJ & Kuncl RW Selective loss of glial glutamate transporter GLT-1 in amyotrophic lateral sclerosis. Ann. Neurol 38, 73–84 (1995). [DOI] [PubMed] [Google Scholar]
- 12.Regan MR, et al. Variations in Promoter Activity Reveal a Differential Expression and Physiology of Glutamate Transporters by Glia in the Developing and Mature CNS. J. Neurosci 27, 6607–6619 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka K, et al. Epilepsy and exacerbation of brain injury in mice lacking the glutamate transporter GLT-1. Science 276, 1699–1702 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Higashimori H, et al. Astroglial FMRP-dependent translational down-regulation of mGluR5 underlies glutamate transporter GLT1 dysregulation in the fragile X mouse. Hum Mol Genet 22, 2041–2054 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swanson RA, et al. Neuronal Regulation of Glutamate Transporter Subtype Expression in Astrocytes. J. Neurosci 17, 932–940 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellis BL, Hirsch ML, Porter SN, Samulski RJ & Porteus MH Zinc-finger nuclease-mediated gene correction using single AAV vector transduction and enhancement by Food and Drug Administration-approved drugs. Gene Ther 20, 35–42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parr-Brownlie LC, et al. Lentiviral vectors as tools to understand central nervous system biology in mammalian model organisms. Front Mol Neurosci 8, 14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foo LC Purification of astrocytes from transgenic rodents by fluorescence-activated cell sorting. Cold Spring Harb. Protoc 2013, 551–560 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci 28, 264–278 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nieweg K, Schaller H & Pfrieger FW Marked differences in cholesterol synthesis between neurons and glial cells from postnatal rats. J. Neurochem 109, 125–134 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Snippert HJ, et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science 327, 1385–1389 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Jiang P, et al. hESC-derived Olig2+ progenitors generate a subtype of astroglia with protective effects against ischaemic brain injury. Nat. Commun 4, 2196 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tatsumi K, et al. Olig2-Lineage Astrocytes: A Distinct Subtype of Astrocytes That Differs from GFAP Astrocytes. Front Neuroanat 12, 8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hrvatin S, et al. Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat Neurosci 21, 120–129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quake SR, Wyss-Coray T & Darmanis S Transcriptomic characterization of 20 organs and tissues from mouse at single cell resolution creates a Tabula Muris. Nature (2018). [Google Scholar]
- 26.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34, 11929–11947 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li JY, et al. LGR4 and its ligands, R-spondin 1 and R-spondin 3, regulate food intake in the hypothalamus of male rats. Endocrinology 155, 429–440 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye X, Smallwood P & Nathans J Expression of the Norrie disease gene (Ndp) in developing and adult mouse eye, ear, and brain. Gene Expr Patterns 11, 151–155 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Braunger BM, The TER different functions of Norrin. Retinal Degenerative Diseases 723, 679–683 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Seitz R, Hackl S, Seibuchner T, Tamm ER & Ohlmann A Norrin mediates neuroprotective effects on retinal ganglion cells via activation of the Wnt/beta-catenin signaling pathway and the induction of neuroprotective growth factors in Muller cells. J Neurosci 30, 5998–6010 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng C, et al. Multi-functional norrin is a ligand for the LGR4 receptor. J Cell Sci 126, 2060–2068 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ott S, Patel RJ, Appukuttan B, Wang X & Stout JT A novel mutation in the Norrie disease gene. Journal of American Association for Pediatric Ophthalmology and Strabismus 4, 125–126 (2000). [DOI] [PubMed] [Google Scholar]
- 33.M, W. Norrie’s disease. Birth defects orginal article series 7, 117–124 (1971). [PubMed] [Google Scholar]
- 34.Tong X, et al. Astrocyte Kir4.1 ion channel deficits contribute to neuronal dysfunction in Huntington’s disease model mice. Nat. Neurosci 17, 694–703 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao XH & Nguyen H Epidermal expression of Lgr6 is dependent on nerve endings and Schwann cells. Exp Dermatol 23, 195–198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Y, et al. Presynaptic regulation of astroglial excitatory neurotransmitter transporter GLT1. Neuron 61, 880–894 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swann JW, A.-N.S. Spine Loss and Other Dendritic Abnormalities in Epilepsy. Hippocampus 10, 617–625 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Fogarty MJ, Noakes PG & Bellingham MC Motor cortex layer V pyramidal neurons exhibit dendritic regression, spine loss, and increased synaptic excitation in the presymptomatic hSOD1(G93A) mouse model of amyotrophic lateral sclerosis. J Neurosci 35, 643–647 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fiala JC, S.J., Harris KM Dendritic Spine Pathology: Cause or Consequence of Neurological Disorders? Brain Research Reviews 39, 29–54 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Orre M, et al. Isolation of glia from Alzheimer’s mice reveals inflammation and dysfunction. Neurobiol Aging 35, 2746–2760 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Blanco-Suarez E, Liu TF, Kopelevich A & Allen NJ Astrocyte-Secreted Chordin-like 1 Drives Synapse Maturation and Limits Plasticity by Increasing Synaptic GluA2 AMPA Receptors. Neuron 100, 1116–1132 e1113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lanjakornsiripan D, et al. Layer-specific morphological and molecular differences in neocortical astrocytes and their dependence on neuronal layers. Nat Commun 9, 1623 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.