Abstract
Mitochondrial DNA (mtDNA) mutations cause inherited diseases and are implicated in the pathogenesis of common late-onset disorders, but how they arise is not clear1,2. Here we show mtDNA mutations are present in primordial germ cells (PGCs) within healthy female human embryos. Isolated PGCs which have a profound reduction in mtDNA content, with discrete mitochondria containing ~5 mtDNA molecules. Single cell deep mtDNA sequencing of in vivo human female PGCs showed rare variants reaching higher heteroplasmy levels in later PGCs, consistent with the observed genetic bottleneck. We also saw the signature of selection against non-synonymous protein-coding, tRNA gene and D-loop variants, concomitant with a progressive upregulation of genes involving mtDNA replication and transcription, and linked to a transition from glycolytic to oxidative metabolism. The associated metabolic shift would expose deleterious mutations to selection during early germ cell development, preventing the relentless accumulation of mtDNA mutations in the human population predicted by Muller’s ratchet. Mutations escaping this mechanism will show massive shifts in heteroplasmy levels within one human generation, explaining the extreme phenotypic variation seen in human pedigrees with inherited mtDNA disorders.
The human mitochondrial genome (mtDNA) is a 16.5 Kb molecule of double stranded DNA that encodes 37 genes essential for the synthesis of 13 polypeptide subunits of the respiratory chain, the final common pathway of oxidative metabolism1. MtDNA is inherited exclusively down the maternal line, and the amount present within human cells is tightly regulated. Diploid cells typically contain 1-10,000 mtDNA molecules partitioned between a network of fusing and budding mitochondria. Exposed to a potent source of oxygen free radicals and lacking protective histone proteins, mtDNA is particularly vulnerable to mutation, creating a mixed population of wild-type and mutated mtDNA within a cell (heteroplasmy)2. MtDNA mutations accumulate in healthy humans during life, particularly in non-dividing (post mitotic) cells including neurons, skeletal and cardiac muscle. Higher levels have been observed in organs affected by common late onset human disorders including Alzheimer’s disease and Parkinson’s disease. Although the overall tissue mutation burden is low, very high levels within single cells lead to bioenergetic failure and ultimately cause cell death1. These findings led to the proposal that mtDNA mutations contribute to the pathogenesis of several common human diseases, and also play a part in the ageing process itself3, but the origin of these mutations has not been unclear.
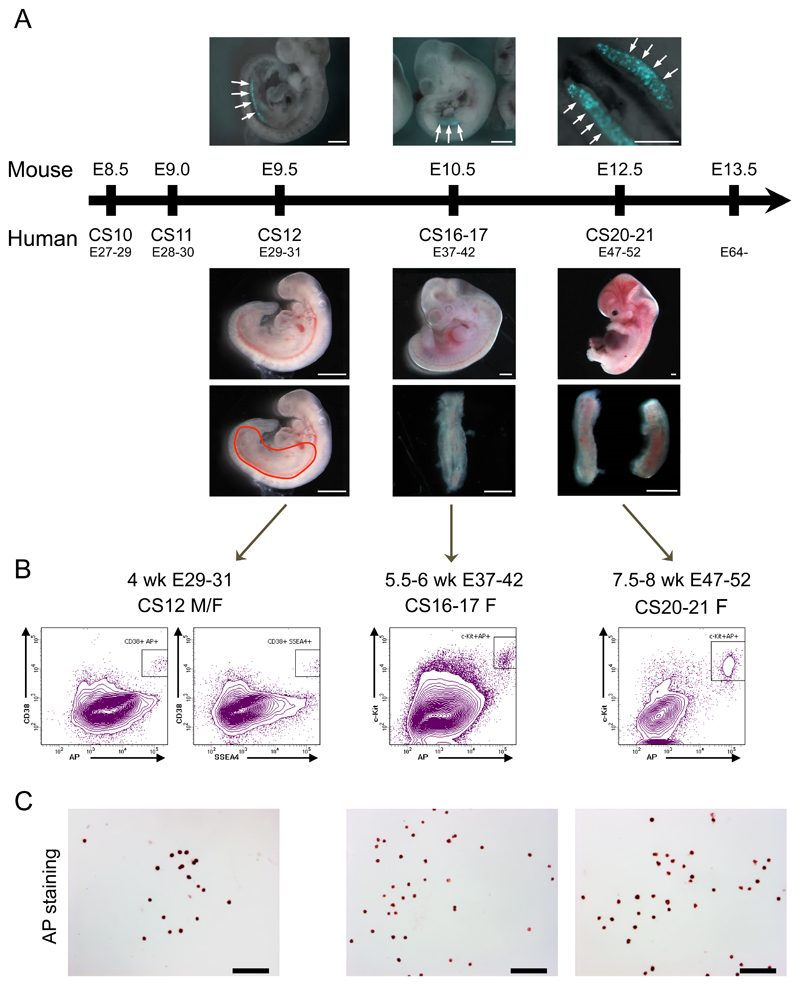
Age-associated mtDNA mutations were assumed to have arisen in somatic tissues during life, but recent massively parallel deep sequencing of maternal relatives raised the possibility that low-level mtDNA heteroplasmic point mutations may be inherited down the maternal line4. To determine whether this is the case, we developed a protocol to isolate pure primordial germ cells (PGCs) from first trimester karyotypically normal human female embryos (Fig.1). Anatomical regions containing migrating PGCs were dissected from fresh 4-week embryos, and genital ridges were isolated from 5 - 8 week embryos. Enzymatic dissociation formed single cell suspensions, which were then labelled with fluorochrome-conjugated antibodies against putative human germ cell surface markers, and sorted using fluorescence-activated cell sorting (FACS). Tissue non-specific alkaline phosphatase (TNAP) histochemistry5, VASA immunocytochemistry6 (Supplementary Fig.1), and germ-cell specific transcripts (Supplementary Figs.1&2)7, were used to determine the proportion of PGCs after FACS sorting at appropriate developmental stages. Highly purified (>97%) PGCs were isolated in 4 week (Carnegie stage, CS12) embryos using a combination of TNAP, stage-specific embryonic antigen 4 (SSEA4) and the PGC-associated cell-surface marker CD38; and for weeks 5-8 (CS16/17 – CS20/21) we used TNAP and c-KIT antibodies (Fig.1b).
Figure 1. Isolation of a pure population of human primordial germ cells (PGCs).
(a) Human and mouse developmental time points used in this study. Upper panel - Prdm1 (Blimp1)-mVenus and Dppa3 (Stella)-eCFP [BVSC] mouse embryos (E9.5 and E10.5) and gonads (E12.5) 17. Arrows show the fluorescent mouse PGCs subsequently isolated by fluorescent sorting. Upper panel scale bars from left to right: 0.5 mm, 1 mm, 0.5 mm. Lower panel – fresh human female embryos showing lower core-body dissected area (red line drawing) for 4 Wk (Carnegie stages, CS, 12) embryos (left), and dissected female embryonic gonads for 5.5 - 6 Wk (CS16/17) and 8 Wk (CS20/21) embryos (middle & right) Lower panel scale bar = 1 mm.
(b) Isolation of human PGCs by fluorescent sorting using: for week 4 (CS12) dissected embryos - a triple positive combination for TNAP, stage-specific embryonic antigen 4 (SSEA4) and CD38, and for weeks 5-8 (CS16/17 – CS20/21) gonadal tissue double positive for TNAP and c-KIT. Boxes in the FACS plots show the sorted cell population.
(c) Purity of isolated hPGCs was validated by alkaline phosphatase activity-staining showing >97% positive cells. Scale bar = 100 μm.
See also Fig.4a-c and Supplementary Figs. 1&2. Observations for b & c were independently replicated three times at each developmental stage.
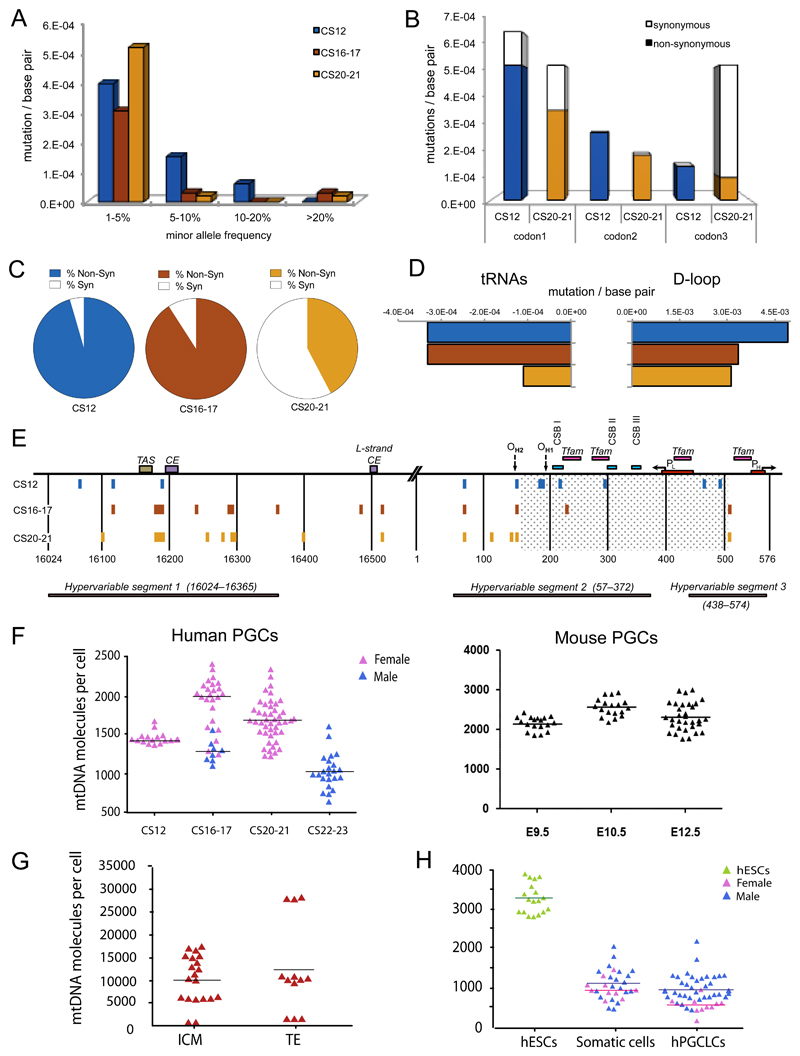
After cell lysis, mtDNA was amplified in overlapping 2Kb fragments to minimise potential contamination from nuclear-encoded mitochondrial insertions (NumtS), before >5000-fold depth re-sequencing. NumtS contamination was further minimized by filtering-out all variants with a minor allele frequency (MAF) <1%. Sequencing of a cloned mtDNA template revealed using the same protocol showed no variants present at ≥1% MAF (Supplementary Fig.3). However, we observed mtDNA single nucleotide variants (SNVs) at >1% heteroplasmy in PGCs isolated from all 12 embryos studied (Fig.2a), which did not map to known human NumtS8. This shows that low-level mtDNA heteroplasmy is present within the female germ line in healthy humans. The variants were predominantly nucleotide transitions, which are less prone to in vitro sequencing error and less likely to be due to oxidative damage9, and are consistent with errors during in vivo DNA synthesis by mtDNA polymerase ϒ or the deamination of cytidine and adenosine10.
Figure 2. Mitochondrial DNA variants in human primordial germ cells.
(a-e) Mitochondrial DNA (mtDNA) variants exceeding the assay detection threshold (minor allele frequency, MAF) of >1% during human primordial germ cell (PGC) development. Carnegie stage, CS12 = blue, CS16/17 = red, CS20/21 = orange. Data were derived from n=80 single cells sorted from two CS12 embryos, n=240 cells sorted from four CS16/17 embryos, and n=360 cells sorted from six CS20/21 embryos.
(a) Frequency distribution of mtDNA variants at sequential stages of germ line development.
(b) Codon positions affected by the coding region mutations (No codon bias for CS12, P = NS, significant codon bias for CS20/21, P = 0.005, Fisher’s exact text, 2-sided).
(c) Proportion of non-synonymous (shaded) variants and synonymous (open) (dN/dS, P = 0.02, Fisher’s exact text, 2-sided).
(d) Evidence of selection against mtDNA variants in tRNA genes (P=0.03, Fisher’s exact text, 2-sided) and the non-coding Displacement (D)-loop (P = 0.003) between CS12 and CS20/21.
(e) Position of the variants detected in the non-coding mtDNA D-loop region.
(f) MtDNA content of single in vivo PGCs from human (left, n = 117) and Blimp1-mVenus and Stella-eCFP [BVSC] mouse (right, n = 68) embryos. Each data point represents the mtDNA content in a single cell corresponding to the mean value from the independent qPCR measurements.
(g) MtDNA content of single human inner cell mass (ICM, n = 20), trophectoderm (TE, n = 12) cells from five embryos in vivo. Each data point represents the mtDNA content in a single cell corresponding to the mean value from the independent qPCR measurements.
(h) MtDNA content of single human embryonic stem cells (hESCs, n = 18), somatic cells (n = 30), and human primordial germ-cell like cells (hPGCLC, n = 48) isolated in vitro7,21.
Two independent PGCLC inductions were performed for both the male (WIS2) and female (H9) ESC lines, with cells analysed at day 5/6. Each data point represents the mtDNA content in a single cell corresponding to the mean value from the independent qPCR measurements. Horizontal lines in (f-h) represent the mean values.
We also observed a change in the mutation spectrum from CS12 to CS20/21 (Fig.2a). For protein coding genes, there was a decrease in the proportion of non-synonymous variants (non-synonymous/synonymous ratio, dN/dS, P=0.02) and a corresponding change in the codon bias (Fig.2b&c). We also saw a decrease in the mutation rate / base pair for tRNA genes (Fig.2d). These are the hallmarks of selection against potentially deleterious mtDNA variants. Similar patterns have been seen in mice transmitting mtDNA heteroplasmy11,12, but the timing and mechanism of the selection was not known. Based on our observations, this occurs at a very early stage in the developing human female germ line, during PGC specification and migration. We were surprised to see some evidence of selection against variants in the non-coding mtDNA D-loop (Fig.2d), but these did not appear to be evenly distributed. CS12 PGCs contained variants in conserved regions not present in CS20/21PGCs (Fig.2e), consistent with selection against variants in regions critically involved in mtDNA transcription and replication13.
Given that the majority of the mtDNA variants were present at low heteroplasmy levels (Fig.2a), below the conventional threshold required for a biochemical effect, it was not immediately apparent how the selection could occur. To explore the potential mechanism, we measured the absolute amount of mtDNA in the human PGCs at different stages of development in vivo, and then sequenced individual late-stage PGCs to look for evidence of variant segregation. First we studied human oocytes from three healthy donors, which contained a mean of 1.22 x106 mtDNA molecules (SD=189,742, n=3)14. This was in keeping with previous measurements15 and confirmed the assay reliability. The amount of mtDNA in human PGCs from 14 embryos in vivo was ~1000-fold less than in oocytes (CS12 mtDNA copies: median 1425, mean 1446, SD=77) (Fig.2f), consistent with a human genetic bottleneck when compared to oocyte mtDNA levels. The mtDNA levels in female and male PGCs were similar, as previously seen in mice16. Overall, these observations resembled measurements made in parallel in Blimp1-mVenus and Stella-eCFP [BVSC] mice17 (Fig.2f), where E9.5 PGCs in the mouse corresponds to CS12 in the human, and E12.5 in the mouse corresponds to CS20/21 PGCs in the human. This validates previous findings in mice with different nuclear genetic backgrounds16,18,19, and shows a similar mechanism in both humans and mice. However, the lower amount of mtDNA we observed in human PGCs (human/mouse: CS12/E9.5, P<0.0001; CS16-7/E10.5, P<0.0001; CS20-21/E12.5, P<0.0001) predicts a ‘tighter’ genetic bottleneck in humans than in mice, and thus the more rapid segregation of heteroplasmy. This is in accordance with observations in human and mouse pedigrees where more dramatic shifts in heteroplasmy are seen in humans (see Fig.4a in Ref20), consistent with a ‘tighter’ bottleneck in humans.
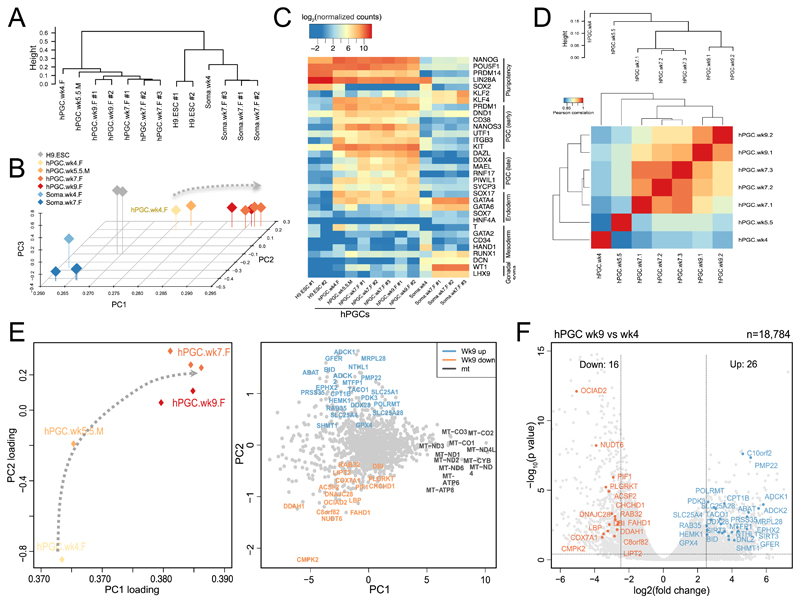
Figure 4. Transcriptome analysis of human primordial germ cells (PGCs).
Analysis of n = 13 transcriptomes.
(a) Unsupervised hierarchical clustering (UHC) of RNA-seq expression profiles in H9 Embryonic stem cells (ESCs), Wk4, Wk7, and Wk9 female (F) human PGCs and Wk5.5 male (M) hPGCs, and gonadal somatic cells (Soma) showing clustering of gene expression consistent with the appropriate developmental stage (see also Supplementary Fig.4).
(b) Principal component analysis (PCA) of RNA-seq gene expression in hPGC samples. Arrow line indicates developmental progression along PC1.
(c) Heat map showing expression of key PGC-associated transcripts (early and late) and of pluripotency, mesoderm, endoderm, and gonadal somatic (Soma) markers.
(d) Correlation of transcription profiles of human PGCs of various stages for nuclear-encoded mitochondrial transcripts predicted by MitoCarta2.0 (see Methods, Pearson’s correlation, two-sided) and mtDNA-encoded transcripts. Only genes with normalized reads counts with (log2 (normalized counts) > 2.5) were used for analysis. Expression levels were averaged over biological replicates (see 4A for the number of biological replicates in each group, total n=13). This shows a progressive change in transcriptome profiles for mitochondrial genes in a sequential manner during PGC development.
(e) Two-dimensional principal components analysis (PCA) of RNA-seq gene expression (PC2 against PC1) for mitochondrial nuclear-encoded and mtDNA-encoded genes (MitoCarta2.0) at the different stages of hPGCs (left panel), with a corresponding plot of the PC scores on PC2 and PC1 (right panel). Gene names are color-coded to illustrate up-regulated expression in hPGCs-wk9 (purple), and down-regulated expression in hPGs-wk9 (red) when compared with hPGCs-wk4 sample (see also Fig. 4f, Supplementary Fig.4&5 and Supplementary Table 4). Arrow line indicates potential mitochondrial-related-transcript progression for each developmental stage from migratory hPGC-wk4 to gonadal hPGC-wk9.
(f) Volcano plot showing differentially expressed genes (from MitoCarta2.0 list) between week 9 (n = 2 independent replicates) and week 4 (n = 1) hPGCs [log2(fold change)>2.5, p < 0.05, DEseq two-sided, see Methods]. See also Supplementary Fig.4&5 and Supplementary Table 4.
Note that the week 9 PGCs included in the RNAseq analysis were from a slightly later developmental stage than CS20/21, reflecting the clinical samples that were available.
It is not technically possible to isolate human PGCs from earlier stages because the cells are collected from clinical samples. However, to gain further insight we also measured mtDNA levels in single cells from both the human inner cell mass (ICM) and trophectoderm (TE) cells (Fig.2g). Although we saw a wide range of mtDNA levels in ICM cells, the mean level (9794, SD=5661) was >5-fold greater than in PGCs. We then studied embryonic stem cells (hESCs) and human PGC-like cells (hPGCLCs)7,21 which are derived from the embryonic epiblast in vitro, thus allowing us to estimate the amount of mtDNA at an intermediate stage of human germ cell development (Fig.2h). hESCs (mean=3264, SD=378) contained less mtDNA than ICM cells, and hPGCLCs (mean=896, SD=368) contained less mtDNA than hESCs (P=8.9x10-33). These findings are in keeping with a post implantation mtDNA bottleneck. The female hPGCLCs (mean=533, SD=203) contained significantly less mtDNA than male hPGCLCs (mean=991, SD=342, P=1.5x10-5), and less mtDNA than both male (mean=1102, SD=426, P=2.2x10-16) and female (mean=972, SD=248, P=2.2x10-16) human somatic cells were isolated from the same in vitro system but not expressing germ-cell markers. These findings are consistent with a more stringent mtDNA bottleneck in the human female germ line.
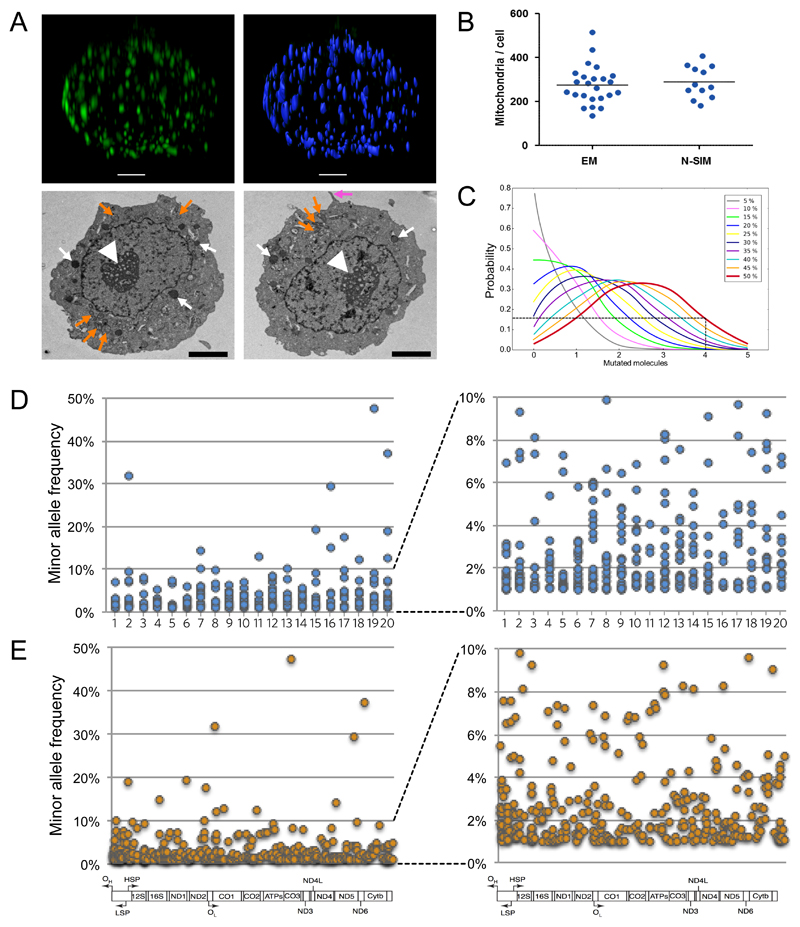
Returning to the PGCs isolated in vivo, next we analysed the sub-cellular distribution of mtDNA molecules within the developing human female germ line. Given the sparse and discrete mitochondrial content, we used the dissector method to measure the number of mitochondria in human PGCs on adjacent section electron micrographs (EM) (Fig.3a), validating our results with super-resolution whole cell imaging using Nikon Structured Illumination Microscopy in 3-dimensions (N-SIM), (Supplementary Movies 1 & 2, Fig.3b). The number of mitochondria did not change between CS12 and CS20-21, allowing us, to estimate the number of mtDNA per mitochondrion at ~5 per organelle in human PGCs. The reduction in mtDNA content after fertilization of the oocyte (Fig.2f), and associated intracellular compartmentalisation of mtDNA (Figs.3a&b), has important implications for the distribution of mtDNA heteroplasmy between individual organelles. Based on the assumption that mutated mtDNA molecules are randomly distributed between mitochondria, higher levels of heteroplasmy within a cell will be associated with a greater proportion of mitochondria containing very high levels (>80%) of a specific mutation (Fig.3c). For example, if the average level within a cell is 40-50% mutant, then 10-20% of the organelles will contain >80% mutant mtDNA, exceeding the threshold required to cause a biochemical defect for pathogenic mutations1,2. In this context, ~10-20% of the organelles would be dysfunctional, and potentially vulnerable to removal by mitochondrial quality control mechanisms. If correct, this mechanism would lead to an overall reduction in mutated mtDNA through selection at the organellar level. To determine whether this is plausible, we performed deep mtDNA sequencing on individual late-stage in vivo PGCs (Fig.3d&e). This showed much higher percentage levels of mtDNA heteroplasmy within individual germ cells, reaching up to 47% mutant. Thus, following the bottleneck, high levels of mtDNA mutations are found in single PGCs, and are predicted to reach very high levels in individual mitochondria (Fig.3c). Thus, the most likely explanation for the observed change in codon bias seen between CS12 and CS20-21 (Fig.2b), in the is the rapid segregation of variants above and below the assay detection threshold (1% MAF) due to the genetic bottleneck (Fig.2a&c), rather than de novo mutation events per se. In this context, even a subtle bias against deleterious variants would tend to suppress them below the detection threshold, whilst allowing non-functional variants to segregate freely to high levels (Fig.3d). Segregation of variants from <1% VAF detection threshold to higher levels could also explain why we saw a slight increase in non-synonymous variants in later-stage PGCs (Fig.2a)
Figure 3. Morphology and deep sequencing of single human primordial germ cells in vivo.
(a) Upper, N-SIM image of a human PGC labelled with anti-TOM20 (Supplementary movie 1) with mitochondria identified and counted by IMARIS (Supplementary movie 2). Scale bar = 2 μm. Lower, transmission electron micrographs (TEM) characteristic of human PGCs33. White fat arrow = nucleolus; white thin arrow = chromatin, electron dense glycogen particles, and lipid droplets; pink arrow = pseudopodia in the migratory stage (CS12) PGCs (right image). Round discrete mitochondria around the nucleus (orange arrows). Scale bar = 2 μm. Observations repeated on three independent occasions.
(b) Number of mitochondria in human primordial germ cells. Left (EM, n=24): mitochondrial count per cell using the dissector method and serial section EMs. Right (N-SIM, n=12): super-resolution microscopy. Each data point represents a measurement from a single PGC. EM: mean=275.9, median=272, SD 87.76. N-SIM: mean=288.8, median=272, SD=72.12. The horizontal bar represents the mean value.
(c) Probability of an individual mitochondrion containing mutated mtDNA molecules as a function of the overall heteroplasmy level within the cell. Binomial distributions assuming 5 molecules/mitochondrion that are randomly distributed between organelles. For cells containing 40-50% mutant mtDNA, many mitochondria are likely to contain 4 or more mutated molecules (>80%). If deleterious these high levels will potentially depolarise the cell triggering mitophagy, or inhibit mtDNA replication within that organelle, potentially leading to selection against mutations at the organellar level.
(d) Minor allele frequency (MAF) of mtDNA variants in 20 single human PGCs from three CS20/21 embryos detected by deep sequencing (left panel). Right panel shows the same data with the Y-axis scaled from 1-10% MAF. In both panels the detection threshold was set at MAF >1%.
(e) Position of the variants from single human PGCs (Fig. 4d) within the mitochondrial genome (left panel). Right panel shows the same data with the Y-axis scaled from 1-10% MAF. In both panels the detection threshold was set at MAF >1%.
To cast light on the possible mechanism of purifying selection, we analysed the transcriptome using RNA-seq on human PGCs from weeks 4 to 9. Unsupervised hierarchical clustering (UHC) of gene expression and principal component analysis (PCA) of RNA-seq data confirmed strong expression of germ-cell specific transcripts (Fig.4a-c). Importantly, we also observed a distinct transcript profile for the nuclear-encoded mitochondrial proteins (MitoCarta2.0)22 in the early stages of PGC development (Fig.4d). Principal component analysis (PCA) of both the nuclear-encoded and the mtDNA-encoded transcripts showed that the most dramatic changes were during the early stages of germ cell development, with mitochondrial genes forming a cluster distinct from late gonadal PGC genes, such as DAZL and DDX4 (Supplementary Fig.4). The mtDNA-encoded transcripts had the highest scores on PC1, accounting for 92% of the total variance (Fig.4e). This is consistent with the developmental progression of mtDNA gene expression during PGC development. In contrast, genes involved in the canonical glycolysis pathway had opposing trends on PC1 (Supplementary Fig.4&5). Although based solely on transcript analysis, these findings suggest that PGCs become less dependent on glycolytic metabolism and more dependent on oxidative metabolism as they develop. As a consequence, non-synonymous mtDNA mutations are progressively more likely to have phenotypic consequences during germ cell development. The proliferation of PGCs between CS12 and CS20/21 requires considerable mtDNA replication, inevitably introducing new mutations through ϒ polymerase errors. These new mutations are more likely to be synonymous (Fig.2) and have no functional consequence. The switch from glycolytic to oxidative metabolism provides an explanation for these findings, by exposing functional variants to the selective effects during this critical time period of PGC development.
Germ-line mtDNA selection at the organelle level has been proposed in mammals12,23 and Drosophila24,25, but it is not known how this occurs. One possibility is that mitochondria depolarise when the proportion of mutated respiratory chain subunits is high, activating intracellular surveillance systems mediated by Pink1 and Parkin, and leading to the selective removal of damaged organelles containing the mutated molecules by mitophagy26,27. An alternative mechanism involves impaired mtDNA replication in ATP-deficient organelles28, leading to the preferential turnover of organelles containing sufficient wild-type mtDNA; or, the transport of mitochondria may be compromised in respiratory deficient organelles, preventing their coalescence around the Balbiani body, which is a known site of intense mtDNA replication18. In our data, specific genes that were upregulated >2.5-fold (Fig.4f) included POLRMT, C10orf2 (Twinkle), SLC25A4 (ANT1), and MRPL28, which play a key role in mtDNA replication and gene expression (Supplementary Table 4). This was also reflected in the PCA, which showed a close correlation between mtDNA transcripts and nuclear genes involved in mtDNA transcription and translation (Supplementary Fig.4). Thus, mtDNA variants influencing mtDNA replication and expression will also be more likely to have an effect as PGCs develop, providing a possible explanation for the signature of selection we observed against tRNA and D-loop variants between CS12 and CS21 (Figs.2d&e).
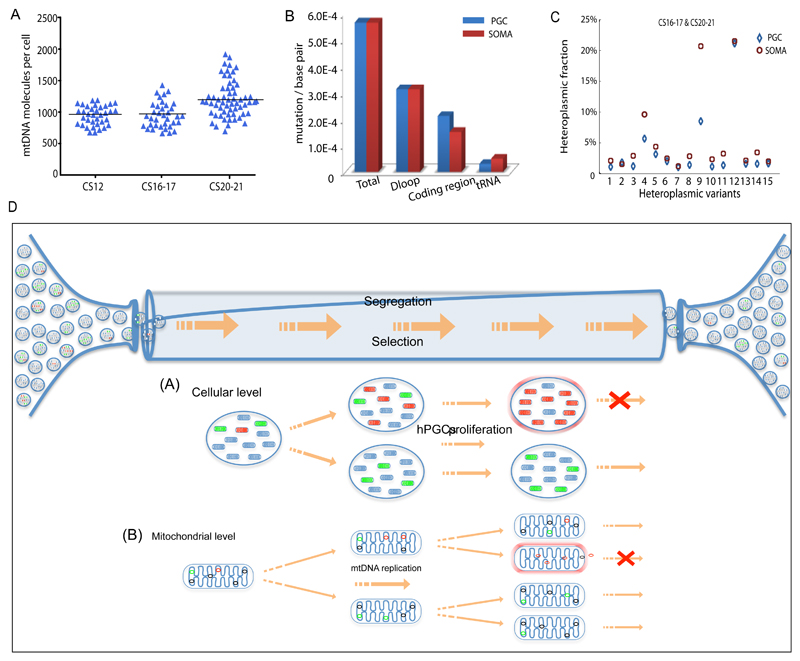
We also saw low levels of mtDNA somatic cells in vitro (Fig.2h) and in vivo (Fig.5a), in keeping with a somatic cell bottleneck, as previously observed in mice19. We also detected mtDNA variants in the somatic cells in vivo at similar levels to PGCs (Fig.5b), but the segregation pattern appeared to be different. Some of the mtDNA variants were detected in both the PGCs and the somatic cells from the same embryo, but by the later stages (n=15 shared variants, CS16-17 & CS20-21), the somatic cells contained higher heteroplasmy levels (P 0.034, Fig.5c). Although based on limited data, this suggests that a less stringent mtDNA bottleneck is present in somatic cells within human embryos, and that mtDNA variants are less constrained by selection in somatic tissues than in the germ line.
Figure 5. Mitochondrial DNA analysis of somatic cells in vivo & mechanisms of selection.
(a) MtDNA content of single human somatic cells in vivo at the same stage as the female primordial germ cells (PGCs) shown in Figs.1&2 (CS12 n=36, CS16-17 n=36; CS20-21 n=60). Each data point represents the mtDNA content in a single cell corresponding to the mean value from the independent qPCR measurements from 12 embryos. The horizontal bar represents the mean value.
(b) Mutation frequency for human female PGCs and corresponding somatic cells (soma) in developing human embryos (CS12-20/21) in vivo. Data derived from 12 embryos.
(c) Segregation of the same heteroplasmic mtDNA variants detected in somatic cells and PGCs from the same embryo at CS16-17 & CS20-21 (n = 15 variants) in vivo. Variants in the somatic cells had a higher heteroplasmy level than the PGCs (P=0.03, Fisher’s exact test, two-sided), which was not the case at CS12.
(d) In vivo PGCs contain ~ 300 discrete mitochondria which each containing 5 mtDNA molecules. In vitro data suggests that the amount of mtDNA may be even lower in female germ cells at an earlier stage. Some of the mtDNA molecules are mutated with either neutral (green) or deleterious (red) mutations. Between Carnegie Stage (CS) 12 and CS 20/21 the number of cells increases from ~3000 to ~80,000 accompanied by a massive proliferation of mtDNA molecules within the migrating PGCs. As the mitochondria replicate, the mutations will be copied. The random segregation of mtDNA molecules during mitochondrial fission will generate daughter mitochondria with either high or low levels of the original mutation. If the mutation is deleterious, then it will be selected against. This could be because it compromises ATP synthesis and activates surveillance systems that remove defective mitochondria, or because it compromises mtDNA replication, and thus prevents that particular mitochondrion from being copied. Both of these mechanisms can act in concert at the cellular level and at the mitochondrial level.
With rudimentary DNA repair mechanisms and negligible intermolecular recombination, uniparentally inherited genomes have limited means of removing de novo mutations. If left unchecked, the relentless accumulation of mtDNA mutations would eventually lead to ‘mutational meltdown’ through Muller’s ratchet, and extinction of the species29. The mtDNA genetic bottleneck has probably evolved to counteract this process, exposing potentially deleterious mutations to selective forces. Based on our findings, in humans, this occurs at a very early stage in germ-line development and is exquisitely sensitive (Fig.5d). However, if a mutation is sufficiently mild to escape germ-line selection, the same genetic bottleneck will have disastrous consequences. First observed in Holstein cows30, dramatic shifts in heteroplasmy levels have been observed within one generation. This has also been observed in families transmitting pathogenic mtDNA mutations, where a healthy mother harbouring a low level of heteroplasmy can have offspring with high levels of heteroplasmy and a severe multisystem disease. Pedigree analyses are consistent with a human mtDNA bottleneck31, but there has been no direct evidence in humans. Here we show that a human mtDNA genetic bottleneck exists, and is narrower than the bottleneck in mice16,18 (Fig.2f). Reducing the amount of mtDNA during this critical time of germ cell development will expose variants to selection acting at either the cellular, organellar or mtDNA level, because this coincides with germ cells expressing transcripts for oxidative phorphorylation as they proliferate and migrate to form the gonad. Harnessing the underlying cell biology could help prevent human mtDNA disease which currently affect ~1 in 11,000. Moreover, given the potential role of mtDNA mutations in common age-related disorders32, preventing their transmission may have a wider impact by promoting healthy ageing in future generations.
Methods
Isolation of single cells
Ethically approved human embryonic tissue samples were provided by the Human Developmental Biology Resource (HDBR), Newcastle University, UK. For CS12 embryos, the lower section (see Fig. 1) was dissected in PBS from intact healthy embryos from healthy donors. For CS16 to CS21 embryos genital ridges from individual embryos were dissected in PBS and the surrounding mesonephric tissue was removed. The dissected tissue was dissociated to single cell suspension with 350 μl in the case of CS12 and 200 and 250 μl in the case of CS16/7 and CS20/1 respectively of 5U/ml Liberase DL Research Grade (Roche, Basel, Switzerland) at 37 °C for 20-35 minutes (depending on the stage-size of the tissue). The dissociated sample was then washed with 750-800μl of cold Flow Cytometry Staining Buffer (FCSB) (R&D Systems, Minneapolis, USA) and the solution was passed through a 50 μm cell strainer. 20μl were removed for cell count using Tali® Image Cytometer (Life Technologies), before the cell suspension was centrifuged at 300 x g for 5 minutes at 4°C. Cells were suspended in cold FCSB and incubated with 5 μl of Brilliant Violet 421-conjugated anti-human CD38 (Biolegend, 303526, San Diego, USA), 20 μl of Alexa Fluor 647-conjugated SSEA-4 (BD Pharmingen, 560796) and 5 μl of Alexa Fluor 488-conjugated anti-alkaline phosphatase (BD Pharmingen, 561495) in the case of CS12 embryonic tissue, and 5 μl of Brilliant Violet 421-conjugated anti-human CD117 (c-Kit) (Biolegend, 313216, San Diego, USA), and 5 μl of Alexa Fluor 488-conjugated anti-alkaline phosphatase (BD Pharmingen, 561495) in the case of CS16/7 and CS20/1 gonadal tissue for 30 min at 4°C. Cells were the washed with 1 ml of FCSB and centrifuged at 300 x g for 5 minutes at 4°C. Cells were suspended in fresh cold FACB and sorted using FACS Aria II (BD Biosciences, San Jose, USA). To determine the right antibody combination for isolation of pure human PGCs different cell populations were sorted onto Polysine™ Microscope Adhesion Slides (Thermo Fisher Scientific, Inc, USA) and left to settle in a humidified chamber at room temperature (RT) for 20-30 min before fixed with 4% PFA for 5 min at RT. Purity for each sorted population was tested with alkaline phosphatase staining using Leukocyte Alkaline Phosphatase Kit (Sigma-Aldrich, St. Louis, USA). Greater than 97% pure samples were used for downstream analysis. Initially, gender was determined by PCR as described in 34 and then verified by karyotype analysis. Somatic cells were isolated at the same time using the same protocols, and inner cell mass and trophectoderm cells were manually dissected from in vitro fertilized human Day 6 embryos as described35. In vitro human embryonic stem cells (hESCs), human primordial germ cell-like cells (hPGCLCs), and somatic cells were cultured, induced and isolated as described previously7,21. Two independent PGCLC inductions were performed for both the male (WIS2) and female (H9) ESC lines. Single cells were isolated by FACS using the NANOS3-tdTomato knock-in reporter and the cell surface marker for tissue non-specific alkaline phosphatase (TNAP) in the WIS line, and TNAP/CD38 double positivity in the h9 line. The somatic were isolated during the same FACS sort and were negative for these markers.
The work with the human Day 6 embryos was done under licence from the UK Human Fertilisation and Embryology Authority (research licence numbers: R0075 and R0133) and also has local ethical approval (UK National Health Service Research Ethics Committee Reference: 06/Q0702/90). Informed consent was obtained from all subjects and the experiments conformed to the principles set out in the WMA Declaration of Helsinki and the NIH Belmont Report. No financial inducements are offered for donation.
Measuring mtDNA content
Lysis of PGCs
PGCs collected in 96-well plates in 40 or 80 cells per well (Life Technologies, Paisley, UK) were lysed for 16 hours in 50 mM Tris-HCl, pH 8.5, with 0.5% Tween 20 and 200 ng/ml proteinase K, at 55°C, followed by heat inactivation at 95°C for 10 minutes.
Lysis of single PGC
PGCs collected in 96-well plates in single cell per well (Life Technologies, Paisley, UK) were lysed for 30 minutes in 4 μl of 50 mM Tris-HCl, pH 8.5, with 0.5% Tween 20 and 200 ng/ml proteinase K, at 37°C, followed by heat inactivation at 80°C for 15 minutes.
Quantification of mtDNA copy number
Quantitative real-time PCR (qPCR) was performed on a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA). Human mitochondrial DNA copy number was calculated by absolute quantification using a singleplex Taqman assay targeting the mitochondrial MT-ND1 gene (Supplementary Table 1a). Mouse mitochondrial DNA copy number was calculated by absolute quantification using a singleplex Taqman assay targeting the mitochondrial MT-ND5 gene (Supplementary Table 1b). Standard curves using PCR-generated templates were used for absolute quantification. Samples and standards were measured in triplicate.
Somatic cells, human inner cell mass, trophectoderm, human embryonic stem cells, human primordial germ cell-like cells, and in vitro somatic cells
MtDNA levels were measured using the same protocols as the PGCs.
Immunocytochemistry
PGCs were sorted on Polysine™ Microscope Adhesion Slides (Thermo Fisher Scientific, Inc, USA) and left to settle in a humidified chamber at room temperature (RT) for 20-30 min before fixation with 4% PFA for 10 min at RT. Washed three times with PBS, 0.1% Tween-20, (5 min each wash) and incubated 20 min with PBS, 0.3% Triton-X buffer, and washed three times with PBS, 0.1% Tween-20 (5 min each wash). Cells were subsequently blocked for 1hr with PBS, 2.5% BSA, 0.1% Tween-20 and 5% normal goat serum at RT followed by staining with DDX4 (VASA) mouse monoclonal IgG1 Antibody (Selleckchem, Houston, USA) diluted 1:200 in the blocking solution at 4°C overnight. The slides were then washed three times with PBS, 0.1% Tween-20 (5 min each wash) and incubated with Alexa fluorophore conjugated secondary antibody (Molecular Probes) 1:500 diluted in PBS, 0.1% Tween-20 for 1 hr at room temperature in the dark, washed three times in PBS, 0.1% Tween-20 (5 min each wash) and once in PBS for 5 min. Finally, the glass bottom dishes were mounted in ProLong® Gold Antifade Mountant with DAPI (Molecular Probes) and imaged using a Zeiss Axio Imager Z1/Apotome Microscope.
Electron microscopy
PGCs collected in 1.5 ml microcentrifuge tubes containing 2.5% EM rade gluteraldehyde (TAAB Laboratories Equipment Ltd, Aldermaston, UK) in Sorenson’s phosphate buffer 0.2 M pH 7.4 (Electron Microscopy Sciences, Hatfield, UK) and stored at 4°C O/N. The cells were enrobed in low-melting point agarose (4%) to form a small block. After secondary fixation in 2% osmium tetroxide (Agar Scientific, Stansted, Essex, UK), the cells were dehydrated in graded acetone, embedded in epoxy resin (TAAB) and polymerised at 60°C. Semi-thin sections (0.5 μm) were taken from the resin block and stained with toluidine blue to identify the first cells. As soon as cells were found in the block, ultrathin serial sections (70 nm) were taken and collected on copper washer grids (Gilder Grids, Grantham, Lincolnshire, UK). Serial sections were taken for approximately 150 μm to ensure that there would be at least 6 whole cells sectioned. The grids were stained with uranyl acetate and lead citrate (Leica UK, Milton Keynes) and viewed on a Philips CM100 TEM (FEI, Eindhoven). Cells that were completely sectioned were imaged from each sample and mitochondria counted using the disector technique 36. Pairs of images 140 nm apart were taken at several levels through each cell. Each pair is comprised of a ‘reference’ and ‘look-up’ section. The number of mitochondria that are hit by the reference section but not by the look-up section is counted. To increase efficiency, the roles of the reference and look-up sections can be reversed. If the volume of the disector is known then the number density Nv can be obtained. The volume of the disector equals the area of cell profile multiplied by the distance between the paired sections. Multiplying number density by cell volume gives mitochondrial number.
Super-resolution microscopy
PGCs collected in 35 mm diameter glass bottom dishes, (Glass No 1.5, 0.170 ± 0.005 mm, High Tolerance Coverslip) (MatTek Corporation, Ashland, USA) and left to settle in a humidified chamber at room temperature (RT) for 20-30 min before fixed with 4% PFA for 10 min at RT. Dishes were washed three times with TBS (pH 7.6), 0.1% Tween-20 (5 min each wash) and then permeabilized for 10 min using TBS, 0.1% sodium citrate, 0.2% Triton-X buffer, before washed three times with TBS, 0.1% Tween-20 (5 min each wash). Cells were subsequently blocked for 1hr with TBS, 0.3M glycine, 2.5% BSA, 5% normal goat serum, 0.1% Tween-20 buffer at RT followed by staining with TOM20 mouse monoclonal IgG2a Antibody (F-10), sc-17764 (Santa Cruz Biotechnology, Inc., Texas, USA) diluted 1:1000 in the blocking solution at 4°C overnight. The glass bottom dishes were then washed three times with TBS, 0.1% Tween-20 (5 min each wash) and incubated with Alexa fluorophore conjugated secondary antibody (Molecular Probes) 1:1000 diluted in TBS, 0.1% Tween-20 for 1 hr at room temperature in the dark, washed three times in PBS, 0.1% Tween-20 (5 min each wash) and once in TBS for 5 min. Finally, the glass bottom dishes were mounted in ProLong® Gold Antifade Mountant with DAPI (Molecular Probes) and imaged using a Nikon Structured Illumination Microscope (N-SIM).
Image segmentation analysis for measuring mitochondria content
Three-dimensional reconstruction of super-resolution z-stack images was rendered using NIS-Elements software (Nikon). Mitochondria content count, and visualization of PGCs was performed using Imaris software (Bitplane, Belfast, UK).
Ultra-deep sequencing and variant analysis
Details of collected samples are given in Supplementary Table 2. PGCs and somatic cells were sorted and lysed in 96-well plates as described above and mtDNA was amplified using PrimeSTAR GXL DNA polymerase (error rate=0.00108%, Takara Bio, Saint-Germain-en-Laye, France) in nine overlapping fragments (Supplementary Table 1c) grouped cells or in two overlapping fragments (Supplementary Table 1d) for the single cells. The specificity of mtDNA amplification was assessed using DNA extracted from Rho0 cell lines previously shown to be devoid of mtDNA 4. There was no template amplification after >40 PCR cycles confirming that the primers only amplified mtDNA and not nuclear-mitochondrial sequences (NUMTs). No PCR products were generated with any of the primers. Amplified products were assessed by gel electrophoresis, against DNA+ve and DNA-ve controls. Each amplicon was individually purified using Agencourt AMPure XP beads (Beckman-Coulter, USA), quantified using a Qubit 2.0 fluorimeter (Life Technologies, Paisley, UK) and amplicons from the same sample were pooled. Libraries were prepared for Next Generation Sequencing (NGS) using the Illumina Nextera XT DNA sample preparation kit (Illumina, CA, USA), pooled amplicons were ‘tagmented’, amplified, cleaned, normalised and pooled in equimolar concentrations for library preparation. The library was sequenced using MiSeq Reagent Kit v3.0 (Illumina, CA, USA) in paired-end, 250 bp reads. Post-run FASTQ files were analysed using an in-house developed bioinformatic pipeline. Reads were aligned to the rCRS (NC_012920) using BWA v0.7.4 invoking –mem 37. Aligned reads were sorted and indexed using Samtools v0.1.8 38 duplicate reads were removed using Picard v1.85 (http://broadinstitute.github.io/picard/). Variant calling was performed in tandem using VarScan v2.3.1 39 (minimum depth=1500, supporting reads=10, base-quality (BQ)=>30, mapping quality (MQ)=>20 and variant threshold=1.0%). Variants were annotated using ANNOVAR v529 40. Heteroplasmic variants are defined as >1% minor allele frequency. Our previous work using this method showed that variants present at >1% heteroplasmy were highly likely to have been present in the original DNA sample, and unlikely to be artefact. In-house Perl scripts were used to extract base/read quality data and coverage data. In addition, low quality variants, present in low complexity regions, were not included in the analysis. Sequencing a cloned mtDNA template with the same protocol showed minimal background sequencing error (Supplementary Fig.3)
RNA-Seq Library Preparation
PGCs were sorted into 100 μl of extraction buffer of PicoPure RNA Isolation Kit (Applied Biosystems) and RNA was extracted following the manufacturer’s protocol. The Ovation RNA-Seq System V2 (Nugen) was used for reverse transcription and amplification of total RNA (0.5 to 2 ng) into cDNA. Fragments of 250 bp were generated by sonication of amplified cDNA using the Covaris S220 Focused-ultrasonicators. Subsequently, using Ovation Rapid DR Multiplex System (Nugen) and 500 ng of fragmented cDNA an RNASeq library was prepared. Quantification of the library was done by qPCR using KAPA Library Quantification Kit (Kapa Biosystems) using QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems). Libraries were sequenced by HiSeq 2000/2500 sequencing system (Illumina) with single-end 50 bp read length. Every 4 indexed libraries were multiplexed to one lane of a flowcell, resulting in >40 millions single end reads per sample.
RNA-Seq Analysis
After removal of adaptor sequence and low-quality reads, RNA-seq reads were mapped to the human reference genome (UCSC GRCh37/hg19) using TopHat2 v2.0.13 (http://ccb.jhu.edu/software/tophat) guided by ENSEMBL gene models. The annotations of transcripts were based on Ensembl (Release 74) considering protein coding, long-noncoding RNA and processed transcripts. Using featureCounts the raw counts per transcripts were obtained. The counts for nuclear-encoded transcripts were normalized, and statistical tests for differential expression of transcripts were performed by using the R Bioconductor DESeq package (www.bioconductor.org). To account for the profound reduction in mtDNA content, mitochondrial transcript counts were normalized by the set of DESeq-scaling factors obtained for nuclear-encoded transcripts before evaluating their differential abundance. Further normalization of expression-normalized transcript counts was done by transcript length (per kB). Hierarchical clustering plots were generated with the R ‘hclust’ functions using the Ward’s method. Principal components were computed by singular value decomposition with the R ‘prcomp’ functioned on scaled DESeq-normalized expression values. Only the 80% most highly expressed transcripts were used for clustering and principal component analysis. By selecting the most equivalent time points in human PGC development, mitochondrial genes that were significantly differentially expressed (log2FC > 2.5 or < -2.5 and p value < 0.05) between hPGCs (Wk 7 or 9) and hPGCs (Wk4) were compiled and highlighted in the plots of the principal component (PC) scores for each gene (Fig. 5e). Human mitochondrial related genes were adopted from MitoCarta2.0 inventory.
Quantitative Reverse Transcription PCR (RT-qPCR)
Total RNA from sorted PGCs or somatic cells was extracted by PicoPure RNA Isolation Kit (Applied Biosystems). cDNA was synthesized and amplified using Ovation RNA-Seq System V2 (Nugen). RT-qPCR reactions were set up with SYBR Green JumpStart Taq ReadyMix (Sigma) and 200 nM of forward and reverse primers (Table S5). RT-qPCR was then performed on QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems) at 95°C for 10 minutes, 40 cycles of 95°C for 15 seconds and 60°C for 1 minute, followed by a melt curve stage. Mean threshold cycles were determined from 2-3 technical replicates. Relative expression levels were calculated using the comparative Ct method, by normalization to GAPDH and relative to CS12 human PGCs or soma (Supplementary Figs.1&2).
Supplementary Material
Supplementary Information is available in the online version of the paper.
Acknowledgements
PFC is a Wellcome Trust Senior Fellow in Clinical Science (101876/Z/13/Z), and a UK NIHR Senior Investigator, who receives support from the Medical Research Council Mitochondrial Biology Unit (MC_UP_1501/2), the Medical Research Council (UK) Centre for Translational Muscle Disease research (G0601943), EU FP7 TIRCON, and the National Institute for Health Research (NIHR) Biomedical Research Centre based at Cambridge University Hospitals NHS Foundation Trust and the University of Cambridge. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. WCCT is supported by a Croucher Foundation studentship, and MAS by a Wellcome Investigator Award.
Footnotes
Data availability
Deep-sequencing (mtDNA-seq, and RNA–seq) data that support the findings of this study are available in the Supplementary Information or have been deposited in the Gene Expression Omnibus (GEO). Previously published RNA-seq data that were re-analysed here are available under accession codes SRX857044, SRX857046, SRX901022, SRX901024, SRX901025, SRX901026, SRX901027, SRX901028, SRX901029, SRX901030, and RX901031. The new RNA-seq data have the following accession codes: hPGC female week 4 GSM2060944 & Somatic cells female week 4 GSM2060945. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Author Contributions
V.I.F developed methods and isolated the in vivo human and mouse PGCs, and performed the microscopy; S.D. performed RNA-Seq bioinformatic analysis and contributed to the manuscript; A.P. and W.W.C.T. carried out the real-time PCR assays; W.W.C.T. performed the RNA-seq experiments and N.A. derived and isolated the hESCs, hPGCLCs, and in vitro somatic cells; A.C. and L.N. isolated the human inner cell mass and trophectoderm cells, overseen by D.I. and Y.K. J.C. carried out the library preparation and deep sequencing; M.C. helped with the human tissue dissection; H.S. helped with the super-resolution microscopy; B.P. preformed the technical validation of the deep sequencing protocol; M.S. provided the BVSC mouse; G.H. advised on the NGS data analysis; M.A.S. supervised the RNA-Seq experiments and real-time PCR expression assays and advised on the project; P.F.C. supervised the project, designed experiments, analysed the data, and wrote the paper. All authors contributed to the manuscript.
Author Information
The authors declare no competing financial interests.
“Reprints and permissions information is available at www.nature.com/reprints”.
References
- 1.Vafai SB, Mootha VK. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491:374–383. doi: 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- 2.Stewart JB, Chinnery PF. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nature Reviews Genetics. 2015;16:530–542. doi: 10.1038/nrg3966. [DOI] [PubMed] [Google Scholar]
- 3.Wallace DC. Colloquium paper: bioenergetics, the origins of complexity, and the ascent of man. Proc Natl Acad Sci U S A. 2011;107(Suppl 2):8947–8953. doi: 10.1073/pnas.0914635107. 0914635107 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Payne BA, et al. Universal heteroplasmy of human mitochondrial DNA. Hum Mol Genet. 2013;22:384–390. doi: 10.1093/hmg/dds435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanley NA, et al. SRY, SOX9, and DAX1 expression patterns during human sex determination and gonadal development. Mech Dev. 2000;91:403–407. doi: 10.1016/s0925-4773(99)00307-x. [DOI] [PubMed] [Google Scholar]
- 6.Anderson RA, Fulton N, Cowan G, Coutts S, Saunders PT. Conserved and divergent patterns of expression of DAZL, VASA and OCT4 in the germ cells of the human fetal ovary and testis. BMC developmental biology. 2007;7:136. doi: 10.1186/1471-213X-7-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irie N, et al. SOX17 is a critical specifier of human primordial germ cell fate. Cell. 2015;160:253–268. doi: 10.1016/j.cell.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dayama G, Emery SB, Kidd JM, Mills RE. The genomic landscape of polymorphic human nuclear mitochondrial insertions. Nucleic Acids Res. 2014;42:12640–12649. doi: 10.1093/nar/gku1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kennedy SR, Salk JJ, Schmitt MW, Loeb LA. Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PLoS Genet. 2013;9:e1003794. doi: 10.1371/journal.pgen.1003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Copeland WC, Longley MJ. Mitochondrial genome maintenance in health and disease. DNA Repair (Amst) 2014;19:190–198. doi: 10.1016/j.dnarep.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart JB, Freyer C, Elson JL, Larsson NG. Purifying selection of mtDNA and its implications for understanding evolution and mitochondrial disease. Nat Rev Genet. 2008;9:657–662. doi: 10.1038/nrg2396. nrg2396 [pii] [DOI] [PubMed] [Google Scholar]
- 12.Freyer C, et al. Variation in germline mtDNA heteroplasmy is determined prenatally but modified during subsequent transmission. Nat Genet. 2012;44:1282–1285. doi: 10.1038/ng.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 14.Pyle A, et al. Extreme-Depth Re-sequencing of Mitochondrial DNA Finds No Evidence of Paternal Transmission in Humans. PLoS Genet. 2015;11:e1005040. doi: 10.1371/journal.pgen.1005040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Craven L, et al. Pronuclear transfer in human embryos to prevent transmission of mitochondrial DNA disease. Nature. 2010;465:82–85. doi: 10.1038/nature08958. nature08958 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cree LM, et al. A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat Genet. 2008;40:249–254. doi: 10.1038/ng.2007.63. ng.2007.63 [pii] [DOI] [PubMed] [Google Scholar]
- 17.Ohinata Y, Sano M, Shigeta M, Yamanaka K, Saitou M. A comprehensive, non-invasive visualization of primordial germ cell development in mice by the Prdm1-mVenus and Dppa3-ECFP double transgenic reporter. Reproduction. 2008;136:503–514. doi: 10.1530/REP-08-0053. [DOI] [PubMed] [Google Scholar]
- 18.Wai T, Teoli D, Shoubridge EA. The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat Genet. 2008;40:1484–1488. doi: 10.1038/ng.258. ng.258 [pii] [DOI] [PubMed] [Google Scholar]
- 19.Cao L, et al. The mitochondrial bottleneck occurs without reduction of mtDNA content in female mouse germ cells. Nat Genet. 2007;39:386–390. doi: 10.1038/ng1970. [DOI] [PubMed] [Google Scholar]
- 20.Wonnapinij P, Chinnery PF, Samuels DC. Previous estimates of mitochondrial DNA mutation level variance did not account for sampling error: comparing the mtDNA genetic bottleneck in mice and humans. Am J Hum Genet. 2010;86:540–550. doi: 10.1016/j.ajhg.2010.02.023. S0002-9297(10)00139-4 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobayashi T, et al. Principles of early human development and germ cell program from conserved model systems. Nature. 2017;546:416–420. doi: 10.1038/nature22812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44:D1251–1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stewart JB, et al. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol. 2008;6:e10. doi: 10.1371/journal.pbio.0060010. 07-PLBI-RA-2319 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma H, Xu H, O'Farrell PH. Transmission of mitochondrial mutations and action of purifying selection in Drosophila melanogaster. Nat Genet. 2014;46:393–397. doi: 10.1038/ng.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill JH, Chen Z, Xu H. Selective propagation of functional mitochondrial DNA during oogenesis restricts the transmission of a deleterious mitochondrial variant. Nat Genet. 2014;46:389–392. doi: 10.1038/ng.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, Dorn GW., 2nd PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narendra DP, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukuoh A, et al. Screen for mitochondrial DNA copy number maintenance genes reveals essential role for ATP synthase. Molecular systems biology. 2014;10:734. doi: 10.15252/msb.20145117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muller HJ. The relation of recombination to mutational advance. Mutat Res. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 30.Upholt WB, Dawid IB. Mapping of mitochondrial DNA of individual sheep and goats: rapid evolution in the D loop region. Cell. 1977;11:571–583. doi: 10.1016/0092-8674(77)90075-7. [DOI] [PubMed] [Google Scholar]
- 31.Chinnery PF, et al. The inheritance of mtDNA heteroplasmy: random drift, selection or both? Trends Genet. 2000;16:500–505. doi: 10.1016/s0168-9525(00)02120-x. [DOI] [PubMed] [Google Scholar]
- 32.Keogh M, Chinnery PF. Hereditary mtDNA heteroplasmy: a baseline for aging? Cell metabolism. 2013;18:463–464. doi: 10.1016/j.cmet.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 33.Funkuda T. Ultrastructure of primordial germ cells in human embryo. Virchows Arch B Cell Pathol. 1976;20:85–89. [PubMed] [Google Scholar]
- 34.Villesen P, Fredsted T. Fast and non-invasive PCR sexing of primates: apes, Old World monkeys, New World monkeys and Strepsirrhines. BMC Ecol. 2006;6:8. doi: 10.1186/1472-6785-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noli L, Capalbo A, Ogilvie C, Khalaf Y, Ilic D. Discordant Growth of Monozygotic Twins Starts at the Blastocyst Stage: A Case Study. Stem cell reports. 2015;5:946–953. doi: 10.1016/j.stemcr.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sterio DC. The unbiased estimation of number and sizes of arbitrary particles using the disector. J Microsc. 1984;134:127–136. doi: 10.1111/j.1365-2818.1984.tb02501.x. [DOI] [PubMed] [Google Scholar]
- 37.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koboldt DC, et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.