Abstract
Study question
The primary objective was to assess whether germline selection events play a role during the inheritance of pathogenic mitochondrial DNA (mtDNA) variants and to provide possible underlying mechanisms for this selection.
Summary answer
We conclude that inheritance of mtDNA is mutation-specific and governed by a combination of random genetic drift and negative and/or positive selection.
What is known already
mtDNA inherits maternally through a genetic bottleneck, but the underlying mechanisms are largely unknown. Although random genetic drift is recognized as an important mechanism, selection mechanisms are thought to play a role as well.
Study design, size, duration
We determined the mtDNA mutation loads in 160 available oocytes, zygotes, and blastomeres of five carriers of the m.3243A>G mutation, one carrier of the m.8993T>G mutation, and one carrier of the m.14487T>C mutation.
Participants/materials, setting, methods
Mutation loads were determined in preimplantation genetic diagnosis (PGD) samples using PCR assays and analysed mathematically to test for random sampling effects. In addition, a meta-analysis has been performed on mutation load transmission data in the literature to confirm the results of the PGD samples.
Main results and the role of chance
By applying the Kimura distribution, which assumes random mechanisms, we found that mtDNA segregations patterns could be explained by variable bottleneck sizes among all our carriers (moment estimates ranging from 10 to 145). Marked differences in the bottleneck size would determine the probability that a carrier produces offspring with mutations markedly different than her own. We investigated whether bottleneck sizes might also be influenced by non-random mechanisms. We noted a consistent absence of high mutation loads in all our m.3243A>G carriers, indicating non-random events. To test this, we fitted a standard and a truncated Kimura distribution to the m.3243A>G segregation data. A Kimura distribution truncated at 76.5% heteroplasmy has a significant better fit (P-value = 0.005) than the standard Kimura distribution. For the m.8993T>G mutation, we suspect a skewed mutation load distribution in the offspring. To test this hypothesis, we performed a meta-analysis on published blood mutation levels of offspring-mother (O-M) transmission for the m.3243A>G and m.8993T>G mutations. This analysis revealed indeed some evidence that the O-M ratios for the m.8993T>G mutation are different from zero (P-value < 0.001), while for the m.3243A>G mutation there was little evidence that the O-M ratios are non-zero. Lastly, for the m.14487T>G mutation, where the whole range of mutation loads was represented, we found no indications for selective events during its transmission.
Large scale data
All data is included in the results section of this manuscript.
Limitations, reason for caution
The availability of human material for the mutations is scarce, requiring additional samples to confirm our findings.
Wider implications of the findings
Our data show that non-random mechanisms are involved during mtDNA segregation. We aimed to provide the mechanisms underlying these selection events. One explanation for selection against high m.3243A>G mutation loads could be as previously reported a pronounced OXPHOS deficiency at high mutation loads, which prohibits oogenesis (e.g. progression through meiosis). No maximum mutation loads of the m.8993T>G mutation seem to exist, as the OXPHOS deficiency is less severe, even at levels close to 100%. In contrast, high mutation loads seem to be favoured, probably because they lead to an increased mitochondrial membrane potential (MMP), a hallmark on which healthy mitochondria are being selected. This hypothesis could provide a possible explanation for the skewed segregation pattern observed. Our findings are corroborated by the segregation pattern of the m.14487T>C mutation, which does not affect OXPHOS and MMP significantly, and its transmission is therefore predominantly determined by random genetic drift. Our conclusion is that mutation-specific selection mechanisms occur during mtDNA inheritance, which has implications for preimplantation genetic diagnosis and mitochondrial replacement therapy.
Study funding/competing interest(s)
This work has been funded by GROW-School of Oncology and Developmental Biology. The authors declare no competing interests.
Keywords: mtDNA segregation, genetic drift, purifying selection, preimplantation genetic diagnosis, heteroplasmy
Introduction
Mutations in the mitochondrial DNA (mtDNA) are an important source of maternally inherited multi-systemic mitochondrial disease and over 250 pathogenic mtDNA mutations have been reported (Hellebrekers, et al., 2012). These pathogenic mtDNA mutations are often heteroplasmic, which is a co-existence of wild-type and mutated mtDNA genomes. The mtDNA mutation load (or heteroplasmy level) correlates with the severity of the disease (Wilson, et al., 2016). During its maternal transmission, the mtDNA passes through a bottleneck (Otten, et al., 2015), which is a restriction in the number of mtDNA molecules inherited (Cree, et al., 2008, Hauswirth, et al., 1982), followed by a strong amplification of these founder molecules. The mtDNA bottleneck is thought to be responsible for the large shifts in heteroplasmy levels observed between mother and child (Hauswirth, et al., 1982), leading to highly variable mutations loads in oocytes from mtDNA mutation carriers with an unpredictable recurrence risk. The mechanisms underlying this process are largely unknown. A central process determining the distribution of mtDNA heteroplasmy is thought to be random genetic drift (Brown, et al., 2001, Jenuth, et al., 1996). However, some reports describe differences in segregation patterns between mtDNA mutations (Howell, et al., 1992), which, apart from genetic drift, could also be explained by selection events (Meiklejohn, et al., 2007, Stewart, et al., 2008) during mtDNA transmission. Although recently statistical evidence has been provided for the existence of purifying selection for mtDNA mutations in human oocytes (De Fanti, et al., 2017), additional evidence is desirable to further resolve the contribution of genetic drift and selection to mtDNA inheritance, as well as to identify the nature of the selection events.
Our main objective was to explain the differences in inheritance pattern of mtDNA mutations. Therefore, we determined the mutation load of three mtDNA mutations (m.3243A>G, m.8993T>G and m.14487T>C) in available oocytes, zygotes and embryos from multiple preimplantation genetic diagnosis (PGD) cycles. In the case of genetic drift, the inheritance can be modelled mathematically using the Kimura distribution (Wonnapinij, et al., 2008), which describes the probability distribution of heteroplasmy values in the offspring as a function of the amount of mutation carried over by the mother, and the bottleneck size. We applied the Kimura distribution to estimate individual effective bottleneck sizes for every female carrier of mtDNA mutations and found that individual bottleneck sizes greatly varied among different mutations. However, our observations were unlikely to be explained completely by genetic drift only and our data support the existence of mutation-specific selection factors during oogenesis. Based on both our observations and literature data, we propose two selective mechanisms, occurring during oogenesis, which in combination with genetic drift provide a plausible explanation of mutation-specific bottleneck sizes, as we report here and as has been documented before (Li, et al., 2016, Wilson, et al., 2016). Our results are important for understanding mtDNA inheritance and predicting the recurrence risk for carriers of mtDNA mutations. Mutation-specific selection mechanisms have also implications for PGD and mitochondrial replacement therapy (MRT), as for some mutations even a small amount of mutated mtDNA (Mitalipov, et al., 2014) might reach significant levels due to selection.
Materials and methods
The study was performed with informed consent of the participants on rest material of routine clinical procedures, approved by the local ethical committee.
Patients and PGD procedure
Five female carriers (Table 1) of the m.3243A>G mutation, one carrier of the m.8993T>G mutation and one carrier of the m.14487T>C mutation were counselled for PGD. Verbal and written information on IVF and intracytoplasmic sperm injection (ICSI), single cell procedures, success rates of the procedures, risk of misdiagnosis, and the limited experience on the worldwide application of PGD for mitochondrial disorders was supplied to all carriers and their partners. Before the treatment, informed consent was given by both partners for both the procedure as well as for re-analysis of non-transferred and non-frozen oocytes and embryos. The IVF-ICSI-PGD and cryopreservation procedures have been performed as described before (Sallevelt, et al., 2013).
Table 1. Age and mutations loads in different tissues of carriers for different mtDNA mutations.
| Carrier | Age, first cycle (yrs) | Blood mut-% | Hair mut-% | Urine mut-% | Muscle Mut-% | Avrg mut-% |
|---|---|---|---|---|---|---|
| m.3243A>G #1 | 36 | 13 | 26 | 55 | ND | 31 |
| m.3243A>G #2 | 30 | 25 | 10 | ND | 28 | 21 |
| m.3243A>G #3 | 28 | 27 | 29 | 59 | ND | 37 |
| m.3243A>G #4 | 32 | 19 | 26 | 38 | ND | 28 |
| m.3243A>G #5 | 31 | 16 | 29 | 28 | ND | 24 |
| m.8993T>G | 30 | 4 | 3 | 5 | ND | 4 |
| m.14487T>C | 40 | 34 | ND | 65 | 55 | 51 |
Mutation load analysis
Primers used for PCR amplification conditions and digestion controls for the m.3243A>G and m.8993T>G mutations were as reported before (Monnot, et al., 2011, Sallevelt, et al., 2013), while for the m.14487T>C mutation, these were: ND6_199F & ND6_199F (6-FAM) (ATACTCTTTCACCCACAGCAC), ND6_340R (ATAGTTTTTTTAATTTATTTAGGGCA), SpikeTspRI-F (vic, AGCTGGTAAAAAGAGGCCTAAC) and SpikeTspRI-R (GCGCCGAATAATAGGTATAGTG) and digestion with TspRI. Samples were analysed by capillary electrophoresis on an ABI Prism 3730 Genetic Analyser followed by GeneScan analysis. To calculate the mutation load, the area of the mutation peak was divided by the sum of the peak area of the wild-type and mutation peak. Validation experiment of our mutation load calculations using a Shewart chart has been performed as described before (Sallevelt, et al., 2013).
For cleavage stage embryos from which multiple blastomeres were available, the average was calculated and reported as the mutation load for this embryo in order to exclude that a single embryo is repeatedly reported. In all cases, the mutation loads in individual blastomeres from a single embryo were not substantially different, consistent with previous findings (Sallevelt, et al., 2013). Furthermore, mutation loads in blood, hair, urine and/or muscle was established, using the same procedures (Table 1).
Moment estimation of the effective bottleneck size
We estimated the effective bottleneck size (Neff) for a carrier, assuming genetic drift, by using the equation Neff = -t / ln (b), where t is the number of mtDNA generations, which has generally been interpreted as 15 cell divisions during embryogenesis. As the average heteroplasmy level in the dataset (p0) is taken as a moment estimator of the amount of mutation carried over from the mother, this is used to determine the bottleneck parameter (b) using equation 6 in reference (Wonnapinij, et al., 2008).
Maximum likelihood estimation for the standard and the truncated Kimura distribution
All presented models were performed using the freely available program R (Ihaka, 1996) and the publicly available library ‘hypergeo’. The standard Kimura distribution (as presented in equations 2-4 of (Wonnapinij, et al., 2008)) was implemented in R. The truncated distribution was obtained by computing the standard distribution and normalized by the cumulative distribution function up to the threshold. The maximum likelihood estimates were then obtained by optimizing the model so that the Kimura curves fit best through the data. To this end, we fitted equation 5 from reference (Wonnapinij, et al., 2008) to the data and obtained maximum likelihood estimated for the Kimura distribution. The inference criterion used for comparing the models was their ability to predict the observed data, i.e. models were compared directly through their minimized minus log-likelihood. When the numbers of parameters in models differed, they were penalized by adding the number of estimated parameters, a form of the Akaike information criterion (AIC) (Akaike, 1998). For both models, the bottleneck 95% confidence intervals were obtained by computing likelihood profiles.
Meta-analysis of published heteroplasmy data (m.3243A>G & m.8993T>G)
Blood heteroplasmy data was collected from literature for the m.3243A>G (n=111) and the m.8993T>G (n=118) mutation (Table S1). The blood heteroplasmy levels for the m.3243A>G mutation were age-corrected to account for the exponential loss of mutated mtDNA in blood leukocytes by using an equation: H(corr) =H(t) eSt, where H(corr) is the corrected heteroplasmy value, H(t) is the heteroplasmy measured in blood at age t in years and S is 0.020 ± 0.003 per year, a parameter previously defined (Rajasimha, et al., 2008). Patients with mutation load(s) <15% were excluded, as this improved the correlation between blood and muscle heteroplasmy (data not shown). For both mutations, the offspring heteroplasmy value was divided by the maternal heteroplasmy level (O-M ratio). The data was transformed to a logarithmic scale and a Gaussian curve was fitted to the frequency histograms. To test whether the data departed from a Gaussian distribution with a mean equal to 0 (no selection bias), we used a two-tailed t-test. Results were considered significant if the P-value was <0.05.
Results
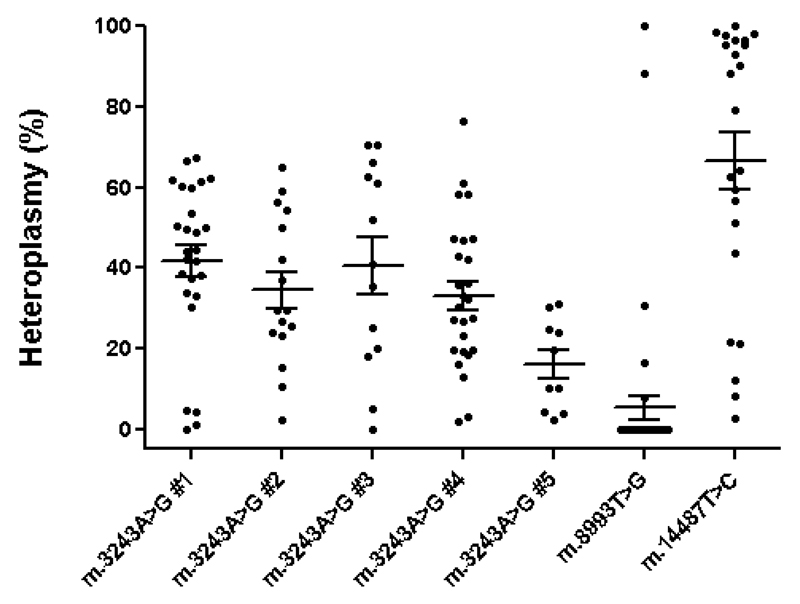
We studied the inheritance of three mtDNA mutations in five carriers of the m.3243A>G mutations, one carrier of the m.8993T>G mutation and one carrier of the 14487T>G mutation for whom PGD was performed in our clinic. We measured the mtDNA mutation loads of female carriers in various tissues of the carrier (Table 1) and in available oocytes, zygotes and blastomeres. We constructed mutation load distributions for each individual carrier (Figure 1). Based on the mutation load distribution and assuming random genetic drift, we computed the moment estimates of the Kimura probability distribution to obtain the effective (or mathematical) bottleneck size (Neff) (Table 2), as previously described (Wonnapinij, et al., 2008). The obtained Neff values are highly variable, being highest for the m.3243A>G carrier with the lowest heteroplasmy level (carrier #5: ~145), whereas the Neff for the m.8993T>G and the m.14487T>C carriers are consistently smaller, ~10 and ~22 respectively. Thus, a clear difference exists between these two carriers and the m.3243A>G carriers. For the m.3243A>G mutation, the reported values are close to the estimated effective bottleneck size of 108 in a female carrier of the mutation of which 82 single primary oocytes have been analysed (Wonnapinij, et al., 2008), as well as the estimated bottleneck size of 72, calculated for a carrier of which 35 preimplantation embryos have been analysed (Monnot, et al., 2011). Together, these results suggest that the observed mtDNA segregation patterns could be explained by a Kimura distribution, under the assumption that bottlenecks are variable and potentially mutation-specific, the latter arguing against random mechanism only.
Figure 1.
Distribution of the percentages of m.3243A>G, m.8993T>G and m.14487T>C mutation in surplus oocytes, blastomeres and zygotes from seven different carriers. When multiple blastomeres of a single cleavage stage embryo were analysed, the mutation load plotted for this embryo was the average of these blastomeres. Every dot thus represents the (average) mutation load of a single oocyte, embryo or zygote. Horizontal lines indicate average mutation loads with SEM.
Table 2. Average heteroplasmy levels and moment estimated bottleneck sizes for carriers with the m.3243A>G, m.8993T>G and m.14487T>C mutation using the Kimura distribution.
| MOMENT ESTIMATED KIMURA DISTRIBUTION | ||||
|---|---|---|---|---|
| carrier | n |
p0, heteroplasmy in samples (average ± SEM) |
b | Effective bottleneck size (Neff) (value [95% CI]) |
| m.3243A>G #1 | 26 | 0.42 ± 0.04 | 0.83 | 83.1 [36-116] |
| m.3243A>G #2 | 16 | 0.34 ± 0.05 | 0.84 | 86.5 [45-134] |
| m.3243A>G #3 | 13 | 0.41 ± 0.07 | 0.73 | 48.7 [23-93] |
| m.3243A>G #4 | 26 | 0.33 ± 0.04 | 0.85 | 92.4 [55-127] |
| m.3243A>G #5 | 10 | 0.16 ± 0.04 | 0.90 | 145.4 [67-349] |
| All m.3243A>G | 91 | 0.35 ± 0.02 | 0.82 | 73.6 [53-93] |
| m.8993T>G | 46 | 0.05 ± 0.03 | 0.21 | 9.6 [7-27] |
| m.14487T>C | 23 | 0.66 ± 0.07 | 0.50 | 21.7 [19-76] |
n, number of unique offspring (oocytes, embryos or zygotes) for every carrier
The m.8993T>G mutation carrier has the lowest estimated bottleneck size, while some of her offspring have mutation loads markedly different from the carrier (Figure 1 and Tables 1 and 2). This is much less the case for the m.3243A>G carrier, especially for carrier #5, where the offspring has mutation loads highly comparable to the carrier. We further investigated what apart from different bottleneck sizes could explain the different mutation distribution patterns. Strikingly, we noted an absence of high m.3243A>G mutation loads (>80%) in the offspring carriers (Figure 1). Especially for carrier #1 and #3, which has the highest average mutation loads (>30%, Tables 1 and 2), absence of high mutation loads is unexpected, as we did observe eight samples with mutation loads <20%, suggesting an asymmetric distribution. Absence of high mutation loads during inheritance of the m.3243A>G mutation supports previous observations (Brown, et al., 2001, Monnot, et al., 2011, Treff, et al., 2012).
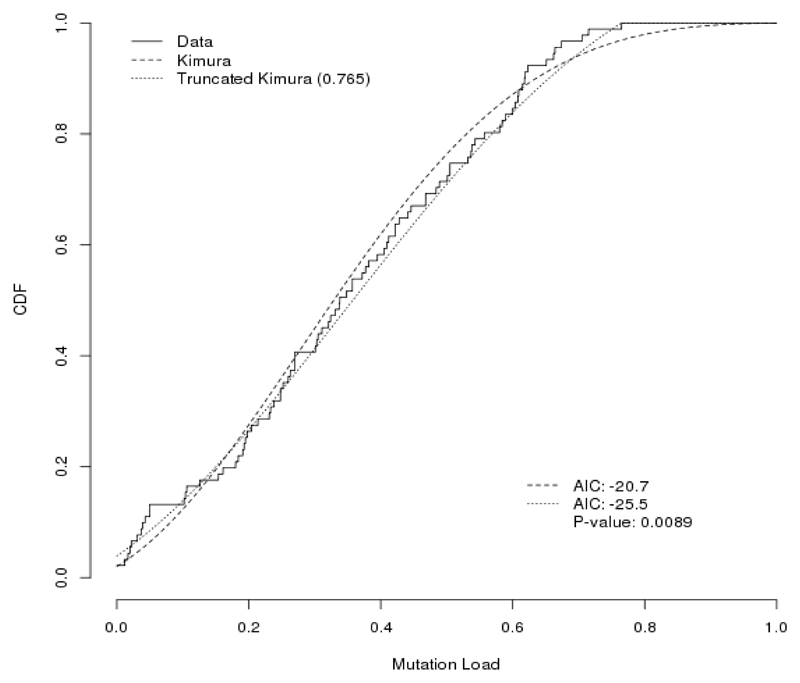
Potential mutation-specific bottleneck sizes and absence of high m.3243A>G, suggest non-random events are active during the inheritance. To test our hypothesis that high heteroplasmy levels are indeed consistently lacking during the inheritance of the m.3243A>G mutation, we tested an alternative model to explain its segregation. To this end, we computed the maximum likelihood ratios under H0, a fitted normal Kimura distribution, and H1, a fitted Kimura distribution that is truncated at the highest observed m.3243A>G mutation load observed (Table 3). The moment estimates provided in Table 2 are fair, as they are close to the actual Kimura maximum likelihood estimates presented in Table 3. Based on the AIC, the truncated model fits the data better than the standard Kimura model for the m.3243A>G carriers #1-3 and #5, as well as when all data for the five m.3243A>G carriers are combined (Table 3 and Figure 2). This implies that the truncated Kimura distribution is a better model for the inheritance data than the fitted normal Kimura distribution. This is a strong argument that there is indeed a (non-random) threshold effect for the m.3243A>G mutation around 80% heteroplasmy level during its inheritance.
Table 3. Maximum likelihood estimates (MLE) for the standard (H0) and truncated (H1) Kimura models. Bold indicated the hypothesis that is most likely based on the Akaike information criterion (AIC).
| carrier | Fit | p0 | b | Truncation (%) | Neff | AIC |
|---|---|---|---|---|---|---|
| [95% CI] | ||||||
| m.3243A>G #1 | H0 | 0.41 | 0.80 | 66.5 [50-86] | 2.0 | |
| H1 | 0.41 | 0.87 | 67.4 | 110.8 [90-151] | -11.4 | |
| m.3243A>G #2 | H0 | 0.34 | 0.85 | 94.4 [45-133] | -6.1 | |
| H1 | 0.34 | 0.88 | 65.1 | 113.3 [71-172] | -7.0 | |
| m.3243A>G #3 | H0 | 0.39 | 0.73 | 47.8 [23-68] | 8.7 | |
| H1 | 0.40 | 0.85 | 71.5 | 90.4 [67-127] | 3.3 | |
| m.3243A>G #4 | H0 | 0.33 | 0.86 | 97.0 [54-127] | -13.5 | |
| H1 | 0.33 | 0.86 | 76.5 | 102.3 [64-149] | -12.2 | |
| m.3243A>G #5 | H0 | 0.16 | 0.91 | 162.3 [64-352] | -13.2 | |
| H1 | 0.16 | 0.95 | 32.4 | 265.9 [180-447] | -14.5 | |
| All m.3243A>G | H0 | 0.35 | 0.81 | 71.2 [53-93] | -20.7 | |
| H1 | 0.35 | 0.84 | 76.5 | 83.3 [74-96] | -26.6 | |
| m.8993T>G | H0 | 0.05 | 0.36 | 14.5 [7-27] | 37.1 | |
| m.14487T>C | H0 | 0.64 | 0.68 | 39.0 [25-48] | 8.9 |
Figure 2.
Fit of the m.3243A>G transmission data. Shown are the actual data and the data fitted using the Kimura distribution and the Kimura distribution truncated at 76.5%. Chi-squared distribution with 1 degree of freedom indicated that the truncated Kimura model is a better fit (P-value 0.0089).
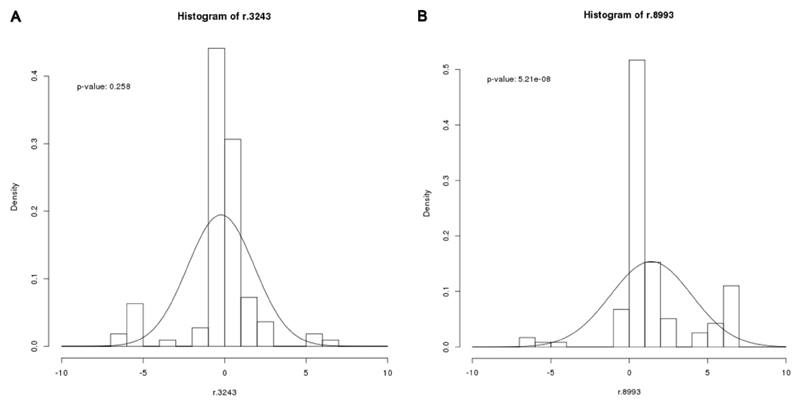
For the m.8993T>G carrier, the distribution in mutation loads in oocytes, zygotes and cleavage-stage embryos were fundamentally different from the m.3243A>G mutation and clearly non-Gaussian, showing a skewed segregation (Figure 1), which is in line to previous observations for this mutation (Blok, et al., 1997, Dahl, et al., 2000, Steffann, et al., 2006, Thorburn, et al., 2009, White, et al., 1999). To further study whether the inheritance of m.8993T>G is indeed skewed, we performed a meta-analysis on blood heteroplasmy levels of mother-child transmissions from published studies (Figure 3) and compared this with the m.3243A>G mutation. We generated O-M ratios by dividing the offspring heteroplasmy values (O) by those from the mother (M) and plotted a histogram of the log-values of these ratios for both mutations. For the m.3243A>G mutation, the average O-M ratio is centred close to zero (Figure 3A). The P-value (0.258) suggests that there is little evidence that the O-M ratios are non-zero. In contrast, for the m.8993T>G mutation (Figure 3B), the evidence that the O-M ratios are different from zero was statistically significant (P-value < 0.001).
Figure 3.
Frequency histograms of O-M values for m.3243A>G and m.8993T>G transmissions. Heteroplasmy data from blood for mother to child transmissions were compiled from published studies of the m.3243A>G (n=111) and the m.8993T>G mutation (n=118). The blood heteroplasmy for the m.3243A>G mutation was corrected for age to avoid potential bias. O-M values represent the ratio in heteroplasmy between generations (O=offspring, M=mother) of A) m.3243A>G and B) m.8993T>G Frequency. Histograms were plotted for both mutations, and a Gaussian curve fitted to each plot.
Lastly, the mutation distribution of the m.14487T>C mutation represents virtually all possible mutation loads (Figure 1) and we found no evident clues that mechanisms other than genetic drift are active during its inheritance.
Discussion
We analysed the mtDNA mutation load distribution in oocytes, zygotes and/or embryos of five carriers of three different mtDNA mutations. By assuming random mechanisms (genetic drift), the mtDNA segregations patterns could be explained by variable bottleneck sizes (Table 2). However, this may not equal the actual mtDNA copy number in the primordial germ cells (PGCs), when mechanisms other than genetic drift occur. Based on the previously reported equation (Wonnapinij, et al., 2008), there exists a (exponential/logarithmic) relation among the effective bottleneck size, the average mutation load, and the variance of mutation loads in the offspring. The formula implies that the maximum bottleneck size is achieved when there are as many wild type as mutated copies. More importantly, the formula also implies that the bottleneck sizes are inversely proportional to the variance. Thus, mutations with a smaller bottleneck size are more likely to produce offspring with a range of different mutation loads, even some very different from the mother. On the other hand, mutations with larger estimated bottleneck size are more likely to produce offspring with comparable mutation loads. These implications agree with our calculations. The m.8993T>G mutation carrier has the lowest calculated bottleneck size, and some of the offspring have more extreme mutation loads, which is much less the case for the m.3243A>G mutation.
Selective mechanisms during m.3243A>G transmission
The dependence on the mutation (Steffann, et al., 2015) implies that mechanisms other than genetic drift should be considered as well, in line with a recently reported purifying or filtering mechanism for pathogenic mtDNA variants, which occurred during the expulsion of the first and second polar body (De Fanti, et al., 2017). Our data support this. For the m.3243A>G mutation, estimated bottleneck sizes differed markedly from the other two mutations. Furthermore, high m.3243A>G mutation loads (>80%) were consistently lacking in all oocytes/zygotes/blastomeres (this paper, Brown, et al., 2001, Monnot, et al., 2011, Treff, et al., 2012). In case of genetic drift, one would expect a symmetrical mutation distribution. This would thus include mutations loads below 20% and above 80%, but the latter is not the case, not even for carriers with relatively high mutation loads. We fitted our data to either a standard Kimura distribution (H0) or a Kimura distribution that is truncated at the highest observed mutation load (H1, Table 3) and the truncated Kimura distribution was a better fit in four out of five carriers (Table 3). Strikingly, the truncated distribution was the best fit for carriers #1 and #3, which have the highest carrier loads, where one would have expected from a random distribution also higher mutation loads in their oocytes. A larger bottleneck size explains mutation loads closer to those of the carrier (Wonnapinij, et al., 2008), but cannot explain the absence of why only higher mutation loads. For all carriers together, a truncated distribution was also a better fit. This fits with reports that carriers of the >50% m.3243A>G mutation in blood tended to have offspring with same or lower heteroplasmy, which was less pronounced for mothers <50% (Uusimaa, et al., 2007). Taken together, these data support a negative selection above an m.3243A>G mutation load of ~80%, influencing the inheritance of the m.3243A>G mutation which is more marginal at lower levels. This was not observed for the other two mutations studied, implying a mutation-specific mechanism.
Preimplantation embryo survival is best served by a low level of metabolism, known as the “quiet embryo” hypothesis (Leese, 2002), with controversy on how “quiet” an optimal metabolism is (Conaghan, et al., 1993, Gardner, et al., 2013), as specific cellular metabolic processes appear to be important. For instance, PGCs depend on glycolysis (Ramalho-Santos, et al., 2009), whereas developing oocytes rely on OXPHOS for ATP production (Dumollard, et al., 2007, Van Blerkom, et al., 1995) to establish a minimal cytoplasmic steady-state ATP level in mature oocytes (Van Blerkom, et al., 1995, Zeng, et al., 2009) and early embryonic development (Dumollard, et al., 2004). A disturbed OXPHOS leads to a drop in ATP levels that interferes with oocyte maturation and preimplantation embryogenesis (Van Blerkom, 2011). The ATP demand can only be satisfied by a highly active mitochondrial OXPHOS (Dumollard, et al., 2004), as the glycolysis pathway is not active anymore (Shyh-Chang, et al., 2013). The m.3243A>G mutation affects the mitochondrial tRNA-LeuUUR (Goto, et al., 1990), resulting in defective assembly of complexes I, IV and V (Sasarman, et al., 2008) and, subsequently, a decrease in the ATP generating capacity. Our data suggest that >80% m.342A>G, the large ATP requirements during oogenesis cannot be met anymore, as the number of healthy energy-producing mitochondria/mtDNA becomes too low, resulting in failed oogenesis. This could already occur at an early stage, as high m.3243A>G mutation loads were absent in human primary (immature) oocytes (Brown, et al., 2001) of a single female carrier, although very high mutation loads in her offspring may also be unlikely due to the low mutation load in muscle (18%) and blood (7%). These observations are corroborated by an age-related decline of the m.3243A>G mutation load in blood over time, due to a loss of blood stem cells with high mutation levels (Rajasimha, et al., 2008). The figure of ~80% mutation load is in line with cybrids harbouring the m.3243A>G mutation (Desquiret-Dumas, et al., 2012, Picard, et al., 2014), where oxidative capacity started to decline at 60%, rapidly declined above 80% and was completely lost at 90% due to an almost complete lack of complex I, and severely reduced abundance of complex III and IV with a subsequent upregulation of the glycolysis pathway (Picard, et al., 2014). This has also been observed in muscle biopsies with high m.3243A>G mutation loads (Janssen, et al., 2008). Activating mtDNA replication has been suggested as an adaptive mechanism (Monnot, et al., 2013, Steffann, et al., 2015), leading to a larger number of mutant and wild-type mtDNA molecules, the latter rescuing the energy deficiency. This would also explain that the threshold can vary between 80% and 90%, most likely depending on the mtDNA copy number (Treff, et al., 2012), while 100% mutation loads were never observed.
The m.8993T>G mutation affects the ATPase6 subunit (Holt, et al., 1990), the terminal OXPHOS complex (complex V) (Nijtmans, et al., 2001), which generates ATP out of a proton gradient. As we observed very high (>95%) mutation loads in oocytes, zygotes and cleavage embryos of carriers, we concluded that no negative selection occured during oogenesis. Indeed, a study in lymphocytes showed that, despite high m.8993T>G mutation loads (80-95%), still 25-35% ATP synthesis capacity remained (Baracca, et al., 2007, Sgarbi, et al., 2006). Likewise, cybrids homoplasmic for the m.8993T>G mutation still displayed >25% residual ATP synthesis (Manfredi, et al., 1999). This implies that, even when homoplasmic for the m.8993T>G mutation, oxidative respiration is preserved to a moderate degree, which, in combination with a high mtDNA copy number in the oocyte (between 50,000 and 550,000 (Fragouli, et al., 2015, May-Panloup, et al., 2005)), produces sufficient ATP for normal oogenesis.
The m.14487T>C mutation affects the ND6 subunit of complex I (Tarnopolsky, et al., 2013, Ugalde, et al., 2003). Its random distribution argues against negative selection and, indeed, biochemical investigations showed that, in skeletal muscle, complex I activity was only modestly decreased (Dermaut, et al., 2010, Gonzalo, et al., 2005). Selection on ATP production capacity has also been observed in the germline of an mtDNA mouse model (Fan, et al., 2008). A severe ND6 mutation, causing a premature termination of the protein and a subsequent inactivation of OXPHOS, was selectively eliminated during oogenesis within four generations, while a milder mutation in CO1, causing only 50% decrease in OXPHOS, was retained throughout multiple generations (Fan, et al., 2008).
In summary, our data indicate that cellular germline selection against mtDNA mutations depends on the degree that mutations affect the cellular OXPHOS-capacity of generating ATP in combination with the mtDNA copy number, but additional studies are required to provide further evidence. A study, in which the biochemical effects of several mtDNA mutations, including the m.3243A>G and m.8993T>G mutation, were studied in the same cybrid system (Pallotti, et al., 2004), revealed little difference between the two mutations, when homoplasmic, in terms of ATP production. However, low glucose concentrations were used, and only the oligomycin-sensitive ATP synthase activity was assessed in two extreme (WT and mutated homoplasmy) situations. Nevertheless, such observations indicate the limitations of using different cybrid models and that lack of uniform methods for assessing energy metabolism. Also, OXPHOS capacity may not be the only negative selection mechanism occurring. If mtDNA turnover is faster in oocytes that have high mutation loads (such as for the m.3243A>G mutation), the variance would increase (Johnston, et al., 2015). Furthermore, processes related to quality control and mitophagy may be involved as well (Tsukamoto, et al., 2008, Twig, et al., 2008), potentially causing the degradation of maternal (mutated) mtDNA or mitochondria during preimplantation. Whatever mechanism, selection against detrimental mtDNA mutations has been convincingly demonstrated in human (Li, et al., 2016), mouse (Stewart, et al., 2008) and drosophila (Hill, et al., 2014, Ma, et al., 2014).
Is there positive selection on the m.8993T>G mutation?
Genetic drift or selection on ATP production capacity are unlikely to explain the skewed pattern observed for the m.8993T>G mutation. Our data for this mutation is limited to a single carrier, but are confirmed by other studies showing that mutation loads for the m.8993T>G mutation are frequently skewed in oocytes/embryos:
-
-
Six out of seven oocytes from a m.8993T>G carrier had a mutant load of >95%, while the seventh was mutation-free (Blok, et al., 1997).
-
-
PGD for an asymptomatic carrier resulted in one embryo homoplasmic for the m.8993T>G mutation and two mutation-free embryos (Steffann, et al., 2006).
-
-
PGD in another m.8993T>G carrier showed skewing in 7 out of 10 embryos with a mutation load of 2.5% in one and >95% in six embryos (Thorburn, et al., 2009).
Prenatal diagnosis of the m.8993T>G mutation showed a skewed segregation in four cases with mutation loads >95% and in two cases no mutation (Bartley, et al., 1996, Ferlin, et al., 1997, Harding, et al., 1992, Steffann, et al., 2007, White, et al., 1999), whereas some intermediate (10-90%) mutation loads were reported as well (Steffann, et al., 2007). A large collection of pedigrees with a mutation at nucleotide 8993 (which can be either T>G or T>C) also showed that the mutation load distribution was predominantly skewed to the extremes (White, et al., 1999). The proportion of children with a high mutation load is increased, when the mutant load in the mother is high as well, particularly for the m.8993T>G mutation (Dahl, et al., 2000).
Finally, our meta-analysis of m.8993T>G mutation pedigrees showed an O-M ratio distribution different from zero (Figure 3), indicating that the offspring had a higher mutation load than their mother. In all studies, an ascertainment bias could have occurred, because the families were identified through an affected child with a very high mutation load. This bias was reduced by collecting our data through the mother, without the proband, similarly to the offspring analysis of the families of the meta-analysis, the PGD data (Steffann, et al., 2006, Thorburn, et al., 2009), and prenatal analysis (White, et al., 1999), but may not fully exclude the bias and impression of selection (Wilson, et al., 2016). In a previous study, preferential selection for both the m.3243A>G and the m.8993T>G mutation was suggested (Chinnery, et al., 2000), but the differences observed were likely due to a sampling effect. As we studied larger number of transmissions for both mutations, differences between the two mutations became more apparent. Moreover, in a comparable study, the m.3243A>G mutation is transmitted from mother to child in similar proportions, indicating a different transmission method than for m.8993T>G and mutation-specific mechanisms (ref).
The skewed segregation pattern for the m.8993T>G mutation indicated fixation at high mutation loads. In case of genetic drift, this could be explained by a very small number of mtDNA units being transmitted, estimated to be around 15. This would imply that the mtDNA sequence modulates the size of the genetic bottleneck (Wilson, et al., 2016). Alternatively, the consistent skewing could indicate positive selection for the m.8993T>G mutation, which, in turn, affects the number of segregating units being estimated. This might be explained by the effect the mutation has on the mitochondrial membrane potential (MMP). In metabolically active mitochondria, complex V is the major regulator of the MMP, which must be maintained to support ATP synthesis (Dumollard, et al., 2004). As a result, an increased MMP is necessary for oocyte maturation and for the competence of an embryo to develop (Ge, et al., 2012, Van Blerkom, 2011). Consequently, MMP as a sensor for normal healthy OXPHOS activity has become an important selection criterion during oogenesis, occurring at the level of the Balbiani bodies, which contain aggregates of highly active mitochondria with a high MMP (Fosslien, 2001, Zhang, et al., 2008, Zhou, et al., 2010) and which are preferentially transmitted to PGCs (Milani, 2015). In lymphocytes with >75% m.8993T>G mutation load or with a blocked complex V, the MMP was significantly increased compared to wild-type mitochondria (Baracca, et al., 2007), because the proton gradient could not be consumed by complex V. If comparable in oocytes, mitochondria with high m.8993T>G mutation loads are more likely to be recruited in Balbiani bodies and subsequently transmitted to the offspring, eventually leading to (almost) homoplasmy. This positive selection “competes” with genetic drift, resulting in the so far unexplained skewed segregation pattern, observed in oogenesis and early embryogenesis.
Selective advantage of the m.8993T>G mutation was also observed in somatic cell lines (Vergani, et al., 1999). Important to note is that the initial m.8993T>G heteroplasmy levels in this study were high (85 - 95%). This may corroborate another study, showing an increase of 0.75% per year in blood of an individual with high (79%) m.8993T>G mutation load whereas in an individual with low (7%) mutation load this percentage remained stable over 23 years (White, et al., 1999). This is the mean and this may vary at the single cell level. In contrast, for the m.8993T>C mutation, a random segregation pattern has been described, which can be explained by the fact that ATP production is relatively preserved and MMP is not significantly affected by the T>C substitution, as lymphocytes carrying >90% m.8993T>C mutation load have a MMP comparable to that in control lymphocytes (Baracca, et al., 2007). This is also the case for the m.3243A>G and m.14487T>C mutation, where no increasing effect on the MMP is expected.
In some cases, intermediate m.8993T>G mutation loads occur in PGD embryos (30-40% in three embryos (Thorburn, et al., 2009), 88% and 30% in two embryos (Sallevelt, et al., 2013)). Fixation might not have occurred, due to the potentially opposing mechanisms of genetic drift and positive selection, likely depending on the initial mutation load and possibly leading to cells without the mutation and with nearly 100% mutation after birth. Interestingly, three cases with somewhat larger differences between prenatal diagnostic (PND) samples and foetal tissues are in the higher mutant load range (Harding, et al., 1992, Steffann, et al., 2007). Although we think that positive selection on the MMP is a good explanation for the consistent skewing of the m.8993T>G mutation, additional (mechanistic) studies are needed to prove whether positive selection based on the MMP or a very small number of segregating units under random genetic drift is the most likely explanation.
Conclusion
Our data demonstrate that, during the bottleneck, genetic drift occurs together with mutation-specific negative and positive selection mechanisms, at the level of OXPHOS capacity (cellular) and MMP (organellar; Table 4). We expect that the selection processes are not per se a static event at one point in germline cell development, but may occur at different moments and may also affect adult stem cells (Rajasimha, et al., 2008). In any case, these mutation-specific events should be evaluated for each mtDNA mutation as the potential of preferential segregation of mutated mtDNA can have consequences for the threshold of transfer in PGD and the risk that mtDNA carry-over during mitochondrial replacement therapy (Hyslop, et al., 2016) can have clinical consequences.
Table 4. Relative contribution of genetic drift and selection contribute to the segregation of three mtDNA mutations.
| Segregational mechanism | m.3243A>G | m.8993T>G | m.14487T>C |
|---|---|---|---|
| Genetic drift? | + | + | + |
Selection on OXPHOS function?
|
NEGATIVE At 90-100% OXPHOS assembly lost (CI) >80% respiratory defect ATP drops Severely reduced |
POSITIVE Only CV affected 100%: residual activity 20-30% Higher due to inefficient coupling |
NEUTRAL Stability and assembly CI affected OXPHOS capacity preserved No effect |
CI, complex I. CV, complex V.
Supplementary Material
Funding
We are grateful to GROW-School of Oncology and Developmental Biology for their fellowship to A.B.C.O. and H.J.M.S.
Footnotes
Author contributions
Conceptualization, A.B.C.O., S.C.E.H.S., and H.J.M.S.; Methodology, A.B.C.O., S.C.E.H.S. and P.J.C.; Formal Analysis, A.B.C.O., S.C.E.H.S., P.J.C., P.L., D.C.S. and H.J.M.S., Investigation, S.C.E.H.S., P.J.C., J.C.F.M.D., M.D. and S.S.; Resources, C.E.M.D., M.H., P.F.C. and H.J.M.S.; Writing – original draft, A.B.C.O., S.C.E.H.S., and H.J.M.S., Writing – Review & Editing, A.B.C.O., S.C.E.H.S., P.J.C., J.C.F.M.D., A.D.C.P., M.H., P.F.C., D.C.S. and H.J.M.S.; Visualization, A.B.C.O., P.J.C., P.L. and D.C.S.; Supervision, J.C.F.M.D., A.D.C.P., C.E.M.D. and H.J.M.S.; Funding Acquisition, H.J.M.S.
Conflict of Interest
None of the authors has any conflict of interest.
References
- Akaike H. Information Theory and an Extension of the Maximum Likelihood Principle. In: Parzen E, T K, Kitagawa G, editors. Selected Papers of Hirotugu Akaike. Springer; New York, NY: 1998. [Google Scholar]
- Baracca A, Sgarbi G, Mattiazzi M, Casalena G, Pagnotta E, Valentino ML, Moggio M, Lenaz G, Carelli V, Solaini G. Biochemical phenotypes associated with the mitochondrial ATP6 gene mutations at nt8993. Biochimica et biophysica acta. 2007;1767:913–919. doi: 10.1016/j.bbabio.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Bartley J, Senadheera D, Park P, Brar H, Wong LJ. Prenatal diagnosis of T8993G mitochondrial DNA point mutation in amniocytes by heteroplasmy detection. American journal of human genetics. 1996;59:A316. [Google Scholar]
- Blok RB, Gook DA, Thorburn DR, Dahl HH. Skewed segregation of the mtDNA nt 8993 (T-->G) mutation in human oocytes. American journal of human genetics. 1997;60:1495–1501. doi: 10.1086/515453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DT, Samuels DC, Michael EM, Turnbull DM, Chinnery PF. Random genetic drift determines the level of mutant mtDNA in human primary oocytes. American journal of human genetics. 2001;68:533–536. doi: 10.1086/318190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnery PF, Thorburn DR, Samuels DC, White SL, Dahl HM, Turnbull DM, Lightowlers RN, Howell N. The inheritance of mitochondrial DNA heteroplasmy: random drift, selection or both? Trends in genetics : TIG. 2000;16:500–505. doi: 10.1016/s0168-9525(00)02120-x. [DOI] [PubMed] [Google Scholar]
- Conaghan J, Handyside AH, Winston RM, Leese HJ. Effects of pyruvate and glucose on the development of human preimplantation embryos in vitro. Journal of reproduction and fertility. 1993;99:87–95. doi: 10.1530/jrf.0.0990087. [DOI] [PubMed] [Google Scholar]
- Cree LM, Samuels DC, de Sousa Lopes SC, Rajasimha HK, Wonnapinij P, Mann JR, Dahl HH, Chinnery PF. A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nature genetics. 2008;40:249–254. doi: 10.1038/ng.2007.63. [DOI] [PubMed] [Google Scholar]
- Dahl HH, Thorburn DR, White SL. Towards reliable prenatal diagnosis of mtDNA point mutations: studies of nt8993 mutations in oocytes, fetal tissues, children and adults. Human reproduction. 2000;15(Suppl 2):246–255. doi: 10.1093/humrep/15.suppl_2.246. [DOI] [PubMed] [Google Scholar]
- De Fanti S, Vicario S, Lang M, Simone D, Magli C, Luiselli D, Gianaroli L, Romeo G. Intra-individual purifying selection on mitochondrial DNA variants during human oogenesis. Human reproduction. 2017;32:1100–1107. doi: 10.1093/humrep/dex051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermaut B, Seneca S, Dom L, Smets K, Ceulemans L, Smet J, De Paepe B, Tousseyn S, Weckhuysen S, Gewillig M, et al. Progressive myoclonic epilepsy as an adult-onset manifestation of Leigh syndrome due to m.14487T>C. Journal of neurology, neurosurgery, and psychiatry. 2010;81:90–93. doi: 10.1136/jnnp.2008.157354. [DOI] [PubMed] [Google Scholar]
- Desquiret-Dumas V, Gueguen N, Barth M, Chevrollier A, Hancock S, Wallace DC, Amati-Bonneau P, Henrion D, Bonneau D, Reynier P, et al. Metabolically induced heteroplasmy shifting and l-arginine treatment reduce the energetic defect in a neuronal-like model of MELAS. Biochimica et biophysica acta. 2012;1822:1019–1029. doi: 10.1016/j.bbadis.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumollard R, Duchen M, Carroll J. The role of mitochondrial function in the oocyte and embryo. Current topics in developmental biology. 2007;77:21–49. doi: 10.1016/S0070-2153(06)77002-8. [DOI] [PubMed] [Google Scholar]
- Dumollard R, Marangos P, Fitzharris G, Swann K, Duchen M, Carroll J. Sperm-triggered [Ca2+] oscillations and Ca2+ homeostasis in the mouse egg have an absolute requirement for mitochondrial ATP production. Development. 2004;131:3057–3067. doi: 10.1242/dev.01181. [DOI] [PubMed] [Google Scholar]
- Fan W, Waymire KG, Narula N, Li P, Rocher C, Coskun PE, Vannan MA, Narula J, Macgregor GR, Wallace DC. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science. 2008;319:958–962. doi: 10.1126/science.1147786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlin T, Landrieu P, Rambaud C, Fernandez H, Dumoulin R, Rustin P, Mousson B. Segregation of the G8993 mutant mitochondrial DNA through generations and embryonic tissues in a family at risk of Leigh syndrome. The Journal of pediatrics. 1997;131:447–449. doi: 10.1016/s0022-3476(97)80074-1. [DOI] [PubMed] [Google Scholar]
- Fosslien E. Mitochondrial medicine--molecular pathology of defective oxidative phosphorylation. Annals of clinical and laboratory science. 2001;31:25–67. [PubMed] [Google Scholar]
- Fragouli E, Spath K, Alfarawati S, Kaper F, Craig A, Michel CE, Kokocinski F, Cohen J, Munne S, Wells D. Altered levels of mitochondrial DNA are associated with female age, aneuploidy, and provide an independent measure of embryonic implantation potential. PLoS genetics. 2015;11:e1005241. doi: 10.1371/journal.pgen.1005241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner DK, Wale PL. Analysis of metabolism to select viable human embryos for transfer. Fertility and sterility. 2013;99:1062–1072. doi: 10.1016/j.fertnstert.2012.12.004. [DOI] [PubMed] [Google Scholar]
- Ge H, Tollner TL, Hu Z, Dai M, Li X, Guan H, Shan D, Zhang X, Lv J, Huang C, et al. The importance of mitochondrial metabolic activity and mitochondrial DNA replication during oocyte maturation in vitro on oocyte quality and subsequent embryo developmental competence. Molecular reproduction and development. 2012;79:392–401. doi: 10.1002/mrd.22042. [DOI] [PubMed] [Google Scholar]
- Gonzalo R, Garcia-Arumi E, Llige D, Marti R, Solano A, Montoya J, Arenas J, Andreu AL. Free radicals-mediated damage in transmitochondrial cells harboring the T14487C mutation in the ND6 gene of mtDNA. FEBS letters. 2005;579:6909–6913. doi: 10.1016/j.febslet.2005.11.034. [DOI] [PubMed] [Google Scholar]
- Goto Y, Nonaka I, Horai S. A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1990;348:651–653. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- Harding AE, Holt IJ, Sweeney MG, Brockington M, Davis MB. Prenatal diagnosis of mitochondrial DNA8993 T----G disease. American journal of human genetics. 1992;50:629–633. [PMC free article] [PubMed] [Google Scholar]
- Hauswirth WW, Laipis PJ. Mitochondrial DNA polymorphism in a maternal lineage of Holstein cows. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:4686–4690. doi: 10.1073/pnas.79.15.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellebrekers DM, Wolfe R, Hendrickx AT, de Coo IF, de Die CE, Geraedts JP, Chinnery PF, Smeets HJ. PGD and heteroplasmic mitochondrial DNA point mutations: a systematic review estimating the chance of healthy offspring. Human reproduction update. 2012;18:341–349. doi: 10.1093/humupd/dms008. [DOI] [PubMed] [Google Scholar]
- Hill JH, Chen Z, Xu H. Selective propagation of functional mitochondrial DNA during oogenesis restricts the transmission of a deleterious mitochondrial variant. Nature genetics. 2014;46:389–392. doi: 10.1038/ng.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt IJ, Harding AE, Petty RK, Morgan-Hughes JA. A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. American journal of human genetics. 1990;46:428–433. [PMC free article] [PubMed] [Google Scholar]
- Howell N, Halvorson S, Kubacka I, McCullough DA, Bindoff LA, Turnbull DM. Mitochondrial gene segregation in mammals: is the bottleneck always narrow? Human genetics. 1992;90:117–120. doi: 10.1007/BF00210753. [DOI] [PubMed] [Google Scholar]
- Hyslop LA, Blakeley P, Craven L, Richardson J, Fogarty NM, Fragouli E, Lamb M, Wamaitha SE, Prathalingam N, Zhang Q, et al. Towards clinical application of pronuclear transfer to prevent mitochondrial DNA disease. Nature. 2016;534:383–386. doi: 10.1038/nature18303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R, G R. R: A Language for Data Analysis and Graphics. Journal of Computational and Graphical Statistics. 1996;5:299–314. [Google Scholar]
- Janssen AJ, Schuelke M, Smeitink JA, Trijbels FJ, Sengers RC, Lucke B, Wintjes LT, Morava E, van Engelen BG, Smits BW, et al. Muscle 3243A-->G mutation load and capacity of the mitochondrial energy-generating system. Annals of neurology. 2008;63:473–481. doi: 10.1002/ana.21328. [DOI] [PubMed] [Google Scholar]
- Jenuth JP, Peterson AC, Fu K, Shoubridge EA. Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nature genetics. 1996;14:146–151. doi: 10.1038/ng1096-146. [DOI] [PubMed] [Google Scholar]
- Johnston IG, Burgstaller JP, Havlicek V, Kolbe T, Rulicke T, Brem G, Poulton J, Jones NS. Stochastic modelling, Bayesian inference, and new in vivo measurements elucidate the debated mtDNA bottleneck mechanism. eLife. 2015;4:e07464. doi: 10.7554/eLife.07464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leese HJ. Quiet please, do not disturb: a hypothesis of embryo metabolism and viability. BioEssays : news and reviews in molecular, cellular and developmental biology. 2002;24:845–849. doi: 10.1002/bies.10137. [DOI] [PubMed] [Google Scholar]
- Li M, Rothwell R, Vermaat M, Wachsmuth M, Schroder R, Laros JF, van Oven M, de Bakker PI, Bovenberg JA, van Duijn CM, et al. Transmission of human mtDNA heteroplasmy in the Genome of the Netherlands families: support for a variable-size bottleneck. Genome research. 2016 doi: 10.1101/gr.203216.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Xu H, O'Farrell PH. Transmission of mitochondrial mutations and action of purifying selection in Drosophila melanogaster. Nature genetics. 2014;46:393–397. doi: 10.1038/ng.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredi G, Gupta N, Vazquez-Memije ME, Sadlock JE, Spinazzola A, De Vivo DC, Schon EA. Oligomycin induces a decrease in the cellular content of a pathogenic mutation in the human mitochondrial ATPase 6 gene. The Journal of biological chemistry. 1999;274:9386–9391. doi: 10.1074/jbc.274.14.9386. [DOI] [PubMed] [Google Scholar]
- May-Panloup P, Chretien MF, Jacques C, Vasseur C, Malthiery Y, Reynier P. Low oocyte mitochondrial DNA content in ovarian insufficiency. Human reproduction. 2005;20:593–597. doi: 10.1093/humrep/deh667. [DOI] [PubMed] [Google Scholar]
- Meiklejohn CD, Montooth KL, Rand DM. Positive and negative selection on the mitochondrial genome. Trends in genetics : TIG. 2007;23:259–263. doi: 10.1016/j.tig.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Milani L. Mitochondrial membrane potential: a trait involved in organelle inheritance? Biol Lett. 2015;11 doi: 10.1098/rsbl.2015.0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitalipov S, Wolf DP. Clinical and ethical implications of mitochondrial gene transfer. Trends Endocrinol Metab. 2014;25:5–7. doi: 10.1016/j.tem.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnot S, Gigarel N, Samuels DC, Burlet P, Hesters L, Frydman N, Frydman R, Kerbrat V, Funalot B, Martinovic J, et al. Segregation of mtDNA throughout human embryofetal development: m.3243A>G as a model system. Human mutation. 2011;32:116–125. doi: 10.1002/humu.21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnot S, Samuels DC, Hesters L, Frydman N, Gigarel N, Burlet P, Kerbrat V, Lamazou F, Frydman R, Benachi A, et al. Mutation dependance of the mitochondrial DNA copy number in the first stages of human embryogenesis. Human molecular genetics. 2013;22:1867–1872. doi: 10.1093/hmg/ddt040. [DOI] [PubMed] [Google Scholar]
- Nijtmans LG, Henderson NS, Attardi G, Holt IJ. Impaired ATP synthase assembly associated with a mutation in the human ATP synthase subunit 6 gene. The Journal of biological chemistry. 2001;276:6755–6762. doi: 10.1074/jbc.M008114200. [DOI] [PubMed] [Google Scholar]
- Otten AB, Smeets HJ. Evolutionary defined role of the mitochondrial DNA in fertility, disease and ageing. Human reproduction update. 2015 doi: 10.1093/humupd/dmv024. [DOI] [PubMed] [Google Scholar]
- Pallotti F, Baracca A, Hernandez-Rosa E, Walker WF, Solaini G, Lenaz G, Melzi D'Eril GV, Dimauro S, Schon EA, Davidson MM. Biochemical analysis of respiratory function in cybrid cell lines harbouring mitochondrial DNA mutations. The Biochemical journal. 2004;384:287–293. doi: 10.1042/BJ20040561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard M, Zhang J, Hancock S, Derbeneva O, Golhar R, Golik P, O'Hearn S, Levy S, Potluri P, Lvova M, et al. Progressive increase in mtDNA 3243A>G heteroplasmy causes abrupt transcriptional reprogramming. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E4033–4042. doi: 10.1073/pnas.1414028111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasimha HK, Chinnery PF, Samuels DC. Selection against pathogenic mtDNA mutations in a stem cell population leads to the loss of the 3243A-->G mutation in blood. American journal of human genetics. 2008;82:333–343. doi: 10.1016/j.ajhg.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho-Santos J, Varum S, Amaral S, Mota PC, Sousa AP, Amaral A. Mitochondrial functionality in reproduction: from gonads and gametes to embryos and embryonic stem cells. Human reproduction update. 2009;15:553–572. doi: 10.1093/humupd/dmp016. [DOI] [PubMed] [Google Scholar]
- Sallevelt SC, Dreesen JC, Drusedau M, Spierts S, Coonen E, van Tienen FH, van Golde RJ, de Coo IF, Geraedts JP, de Die-Smulders CE, et al. Preimplantation genetic diagnosis in mitochondrial DNA disorders: challenge and success. Journal of medical genetics. 2013;50:125–132. doi: 10.1136/jmedgenet-2012-101172. [DOI] [PubMed] [Google Scholar]
- Sasarman F, Antonicka H, Shoubridge EA. The A3243G tRNALeu(UUR) MELAS mutation causes amino acid misincorporation and a combined respiratory chain assembly defect partially suppressed by overexpression of EFTu and EFG2. Human molecular genetics. 2008;17:3697–3707. doi: 10.1093/hmg/ddn265. [DOI] [PubMed] [Google Scholar]
- Sgarbi G, Baracca A, Lenaz G, Valentino LM, Carelli V, Solaini G. Inefficient coupling between proton transport and ATP synthesis may be the pathogenic mechanism for NARP and Leigh syndrome resulting from the T8993G mutation in mtDNA. The Biochemical journal. 2006;395:493–500. doi: 10.1042/BJ20051748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Daley GQ, Cantley LC. Stem cell metabolism in tissue development and aging. Development. 2013;140:2535–2547. doi: 10.1242/dev.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffann J, Frydman N, Gigarel N, Burlet P, Ray PF, Fanchin R, Feyereisen E, Kerbrat V, Tachdjian G, Bonnefont JP, et al. Analysis of mtDNA variant segregation during early human embryonic development: a tool for successful NARP preimplantation diagnosis. Journal of medical genetics. 2006;43:244–247. doi: 10.1136/jmg.2005.032326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffann J, Gigarel N, Corcos J, Bonniere M, Encha-Razavi F, Sinico M, Prevot S, Dumez Y, Yamgnane A, Frydman R, et al. Stability of the m.8993T->G mtDNA mutation load during human embryofetal development has implications for the feasibility of prenatal diagnosis in NARP syndrome. Journal of medical genetics. 2007;44:664–669. doi: 10.1136/jmg.2006.048553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffann J, Monnot S, Bonnefont JP. mtDNA mutations variously impact mtDNA maintenance throughout the human embryofetal development. Clin Genet. 2015;88:416–424. doi: 10.1111/cge.12557. [DOI] [PubMed] [Google Scholar]
- Stewart JB, Freyer C, Elson JL, Wredenberg A, Cansu Z, Trifunovic A, Larsson NG. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS biology. 2008;6:e10. doi: 10.1371/journal.pbio.0060010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarnopolsky M, Meaney B, Robinson B, Sheldon K, Boles RG. Severe infantile leigh syndrome associated with a rare mitochondrial ND6 mutation, m.14487T>C. American journal of medical genetics Part A. 2013;161A:2020–2023. doi: 10.1002/ajmg.a.36000. [DOI] [PubMed] [Google Scholar]
- Thorburn DR, Wilton L, Stock-Myer S. Healthy baby girl born following PGD for the mitochondrial DNA mutation m.8993T>G. Mitochondrion. 2009;10:222. [Google Scholar]
- Treff NR, Campos J, Tao X, Levy B, Ferry KM, Scott RT., Jr Blastocyst preimplantation genetic diagnosis (PGD) of a mitochondrial DNA disorder. Fertility and sterility. 2012;98:1236–1240. doi: 10.1016/j.fertnstert.2012.07.1119. [DOI] [PubMed] [Google Scholar]
- Tsukamoto S, Kuma A, Mizushima N. The role of autophagy during the oocyte-to-embryo transition. Autophagy. 2008;4:1076–1078. doi: 10.4161/auto.7065. [DOI] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. The EMBO journal. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugalde C, Triepels RH, Coenen MJ, van den Heuvel LP, Smeets R, Uusimaa J, Briones P, Campistol J, Majamaa K, Smeitink JA, et al. Impaired complex I assembly in a Leigh syndrome patient with a novel missense mutation in the ND6 gene. Annals of neurology. 2003;54:665–669. doi: 10.1002/ana.10734. [DOI] [PubMed] [Google Scholar]
- Uusimaa J, Moilanen JS, Vainionpaa L, Tapanainen P, Lindholm P, Nuutinen M, Lopponen T, Maki-Torkko E, Rantala H, Majamaa K. Prevalence, segregation, and phenotype of the mitochondrial DNA 3243A>G mutation in children. Annals of neurology. 2007;62:278–287. doi: 10.1002/ana.21196. [DOI] [PubMed] [Google Scholar]
- Van Blerkom J. Mitochondrial function in the human oocyte and embryo and their role in developmental competence. Mitochondrion. 2011;11:797–813. doi: 10.1016/j.mito.2010.09.012. [DOI] [PubMed] [Google Scholar]
- Van Blerkom J, Davis PW, Lee J. ATP content of human oocytes and developmental potential and outcome after in-vitro fertilization and embryo transfer. Human reproduction. 1995;10:415–424. doi: 10.1093/oxfordjournals.humrep.a135954. [DOI] [PubMed] [Google Scholar]
- Vergani L, Rossi R, Brierley CH, Hanna M, Holt IJ. Introduction of heteroplasmic mitochondrial DNA (mtDNA) from a patient with NARP into two human rho degrees cell lines is associated either with selection and maintenance of NARP mutant mtDNA or failure to maintain mtDNA. Human molecular genetics. 1999;8:1751–1755. doi: 10.1093/hmg/8.9.1751. [DOI] [PubMed] [Google Scholar]
- Wai T, Ao A, Zhang X, Cyr D, Dufort D, Shoubridge EA. The role of mitochondrial DNA copy number in mammalian fertility. Biology of reproduction. 2010;83:52–62. doi: 10.1095/biolreprod.109.080887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SL, Collins VR, Wolfe R, Cleary MA, Shanske S, DiMauro S, Dahl HH, Thorburn DR. Genetic counseling and prenatal diagnosis for the mitochondrial DNA mutations at nucleotide 8993. American journal of human genetics. 1999;65:474–482. doi: 10.1086/302488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SL, Shanske S, Biros I, Warwick L, Dahl HM, Thorburn DR, Di Mauro S. Two cases of prenatal analysis for the pathogenic T to G substitution at nucleotide 8993 in mitochondrial DNA. Prenatal diagnosis. 1999;19:1165–1168. [PubMed] [Google Scholar]
- White SL, Shanske S, McGill JJ, Mountain H, Geraghty MT, DiMauro S, Dahl HH, Thorburn DR. Mitochondrial DNA mutations at nucleotide 8993 show a lack of tissue- or age-related variation. Journal of inherited metabolic disease. 1999;22:899–914. doi: 10.1023/a:1005639407166. [DOI] [PubMed] [Google Scholar]
- Wilson IJ, Carling PJ, Alston CL, Floros VI, Pyle A, Hudson G, Sallevelt SC, Lamperti C, Carelli V, Bindoff LA, et al. Mitochondrial DNA sequence characteristics modulate the size of the genetic bottleneck. Human molecular genetics. 2016;25:1031–1041. doi: 10.1093/hmg/ddv626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonnapinij P, Chinnery PF, Samuels DC. The distribution of mitochondrial DNA heteroplasmy due to random genetic drift. American journal of human genetics. 2008;83:582–593. doi: 10.1016/j.ajhg.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng HT, Yeung WS, Cheung MP, Ho PC, Lee CK, Zhuang GL, Liang XY, O WS. In vitro-matured rat oocytes have low mitochondrial deoxyribonucleic acid and adenosine triphosphate contents and have abnormal mitochondrial redistribution. Fertility and sterility. 2009;91:900–907. doi: 10.1016/j.fertnstert.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Zhang YZ, Ouyang YC, Hou Y, Schatten H, Chen DY, Sun QY. Mitochondrial behavior during oogenesis in zebrafish: a confocal microscopy analysis. Development, growth & differentiation. 2008;50:189–201. doi: 10.1111/j.1440-169X.2008.00988.x. [DOI] [PubMed] [Google Scholar]
- Zhou RR, Wang B, Wang J, Schatten H, Zhang YZ. Is the mitochondrial cloud the selection machinery for preferentially transmitting wild-type mtDNA between generations? Rewinding Muller's ratchet efficiently. Current genetics. 2010;56:101–107. doi: 10.1007/s00294-010-0291-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.