Abstract
Bioconjugation of siRNAs with chemical moieties is an effective strategy to improve the stability and cellular uptake of siRNAs. However, chemical conjugations of siRNAs are always challenging because of siRNAs’ extremely poor stability. Therefore, a new strategy to attach a chemical moiety to siRNA without chemical reaction is highly needed. Peptide nucleic acids (PNAs) are DNA analogues in which the phosphate ribose ring in the backbone is replaced with a polyamide. Compared to DNA, PNA has a higher affinity for complementary DNA and better chemical stability. We, therefore, employed PNAs as a complementary linker to attach chemical moieties to siRNAs by annealing. The objective of this study is to develop an easy but efficient strategy to noncovalently attach chemical moieties to siRNAs without chemical modification of the siRNAs. We identified a PNA complementary sequence for hybridizing with siRNAs. Also, we compared the stability and silencing effects of different siRNA–PNA chimeras, which were annealed at different termini of the siRNA. siRNAs with a PNA annealed to the 3′ end of the sense strand exhibited enhanced stability in the serum and maintained a good silencing effect. The siRNA–PNA chimera was then employed in two delivery systems to deliver the PCBP2 siRNA, a potential antifibrotic siRNA, to hepatic stellate cells. In both systems, the chimera demonstrated high cellular uptake and silencing activity. The results suggested that the siRNA–PNA chimera is an easy and efficient approach to attach targeting ligands or chemical moieties to siRNAs without chemical modification of the siRNA. This new technology will greatly reduce the difficulty and cost in conjugating chemical moieties to siRNAs.
Keywords: PNA, siRNA, chimera, bioconjugation, targeting moiety, peptide, nanocomplex
Graphical Abstract
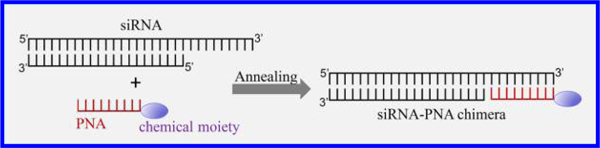
INTRODUCTION
RNA interference (RNAi), an endogenous mechanism employed by many species to regulate gene expression, has received extensive interest from academia and industry since its discovery.1–3 Small interfering RNAs (siRNAs) are a class of double-stranded RNAs comprising 21–25 nucleotides, which can precisely degrade complementary mRNA through the RNAi pathway. siRNAs have numerous distinct advantages, including high specificity, high potency, and easy accessibility to various targets. A number of siRNA therapeutics are evaluated in clinical trials, and the recent success of Alnylam’s siRNA therapeutics (Patisiran) in a phase III trial may pave the way for the approval of the first-ever siRNA drug. Despite its promising therapeutic potential, it still faces several challenges, including poor stability of siRNAs and the lack of efficiently targeted delivery systems. Various nonviral carriers, such as liposomes, lipids, and polymers, have been extensively studied to overcome these challenging barriers and have shown promising results in preclinical studies. However, potential cytotoxicity and undesired immune response associated with the use of these materials, particularly cationic agents, limit their clinical applications. In contrast, direct conjugation of various moieties, such as lipids, peptides, polymers, or proteins, to siRNAs represents an alternative strategy to deliver siRNAs into target cells. These bioconjugates have exhibited improved pharmacokinetic performance in vivo and enhanced delivery efficiency to target tissues without compromising the gene silencing activity.4,5 Besides, siRNA can be conjugated to a chemical moiety and then incorporated in a nanoscale delivery system.6,7 However, because siRNAs are susceptible to degradation by various factors, the stability of siRNA is always a major concern in the chemical conjugation process. Moreover, in most cases, the method used to purify conjugated siRNA is complicated and usually results in a significant loss of the product.
Peptide nucleic acids (PNAs) are oligonucleotide analogues that contain normal DNA bases, but the phosphodiester backbone is substituted with a polyamide structure consisting of repeating N-(2-aminoethyl) glycine units.8 Due to this change, PNAs attain several unique advantages over DNA or RNA when used as therapeutics, including high affinities for complementary DNA and RNA, resistance to nucleases and proteases, and high stability in a biological environment.9 Since the introduction of PNA two decades ago, the applications of PNA have been extended from antisense and antigene agents to biomolecular tools, molecular probes, and biosensors. PNAs have been adopted as biosupramolecular tags for programmable assemblies and reactions.10 Moreover, PNAs have been used as adapters to attach ligands or synthetic peptides, such as nuclear localization signals and transferrins, to plasmids, resulting in enhanced nuclear localization and targeting efficacy.11,12 In this study, we employed PNA as a complementary linker to noncovalently attach chemical moieties to siRNAs by annealing. The objective of this study is to identify a relatively easier but more efficient strategy to link chemical moieties to siRNAs and improve their stability while maintaining their silencing activity.
MATERIALS AND METHODS
Materials.
Luciferase siRNA, PCPB2 siRNA (Sense strand 5′-GUCAGUGUGGCUCUCUUAC-3′), scrambled siRNA, and Lipofectamine 2000 were purchased from Invitrogen (Carlsbad, CA). Cell culture reagents were ordered from Mediatech, Inc. (Manassas, VA). PNAs and PNA conjugates were ordered from PNA Bio Inc. (Newbury Park, CA).
Cell Culture.
HeLa cells were obtained from American Type Culture Collection (Manassas, VA). HSC-T6 cells were kindly provided by Dr. Scott L. Friedman (Mount Sinai School of Medicine, New York University). Both HeLa and HSC-T6 cells were cultured in DMEM medium supplemented with 10% FBS, penicillin (100 unit/mL), and streptomycin (100 μg/mL).
Annealing of siRNA Duplex with PNA.
Equal amounts (100 μM) of sense and antisense strands of the siRNA were mixed in the presence of annealing buffer. The mixture was then incubated at 95 °C for 3 min and slowly cooled to room temperature. The formed siRNA duplex was then annealed with the PNA solution at a 1:1 molar ratio in TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0). The annealed siRNA–PNA chimera was confirmed by electrophoresis on a 20% native polyacrylamide gel and visualized by staining with GelRed.
Serum Stability of the siRNA–PNA Chimera.
siRNAs or siRNA–PNA chimeras were incubated with 50% rat serum at 37 °C for a series of time intervals. After incubation, the samples were collected, and the stability of siRNA–PNA chimeras was evaluated by electrophoresis on a 20% native polyacrylamide gel and visualized by staining with GelRed. Intensities of the gel bands were quantitated with ImageJ.
Luciferase Assay.
The firefly luciferase plasmid, renilla luciferase plasmid, and siRNA were cotransfected using Lipofectamine 2000. Cotransfection of a reporter plasmid with its correspondent siRNA is a standard protocol to screen the silencing activity of numerous siRNAs at protein level in a short period of time.13,14 Briefly, HeLa cells were seeded in a 96-well plate at a density of 1.0 × 104 cells/well in complete DMEM medium 12 h before the transfection. For each well, 60 ng of the firefly luciferase plasmid and 6 ng of the renilla luciferase plasmid were diluted with 12.5 μL of Opti-MEM I reduced serum medium, and 0.13 μL of Lipofectamine 2000 was diluted with 12.5 μL of Opti-MEM I medium. The Lipofectamine 2000 and plasmid dilutions were then mixed and incubated at room temperature for 25 min before transfection.
For transfection, the siRNA or siRNA–PNA chimera was diluted with 12.5 μL of Opti-MEM I medium to a final concentration of 50 nM in the plate. Next, 0.2 μL of Lipofectamine 2000 was diluted with 12.5 μL of Opti-MEM I medium, and the complex was formed after two solutions were mixed. After the complex has formed, the cell culture media was removed from the seeded wells, and 50 μL of antibiotic-free Opti-MEM I medium was added into each well, followed by 25 μL of plasmid DNA and siRNA or the siRNA–PNA chimera. After a 6 h incubation at 37 °C in a CO2 incubator, the medium was replaced with 100 μL of DMEM supplemented with 10% FBS, penicillin (100 unit/mL), and streptomycin (100 μg/mL). 48 h after the transfection, firefly and Renilla luciferase activities were measured sequentially from a single sample using the dual-luciferase reporter (DLR) assay system. The relative firefly luciferase activity was calculated by normalizing results with Renilla luciferase activity.
Cellular Uptake Study Using Flow Cytometry.
Cells were seeded in 6-well plates at a density of 2.0 × 105 cells/well and incubated at 37 °C overnight. On the next day, the cells were washed with DPBS and incubated with the Alexa Fluor 647-labeled siRNA–PNA-peptide-431 for 6 h in Opti-MEM I reduced serum medium. After the incubation, the cells were incubated in DPBS containing 1 mg/mL of heparin at 37 °C for 15 min. Subsequently, the cells were washed with DPBS three times, trypsinized, and centrifuged to remove trypsin. Finally, the fluorescent signals in the cells were analyzed on a FACSCalibur flow cytometer.
Silencing Activity of siRNA–PNA-Peptide.
HSC-T6 cells were seeded in a 24-well plate at a density of 5 × 104 cells/well 12 h before transfection. PNA-peptide-431 was purchased from PNA Bio Inc. (Newbury Park, CA) using the solid-phase peptide synthesis method. The cells were then washed with DPBS and incubated with the siRNA–PNA-peptide-431 at a final concentration of 2 μM siRNA in Opti-MEM I reduced serum medium. After incubation for 24 h, the cells were harvested for RNA isolation. Silencing activity was evaluated by real-time RT-PCR as we described before.15 The primers used in this study were as follows: PCBP2, 5′-ACCAATAGCACAGCTGCCAGTAGA-3′ (forward primer) and 5′-AGTCTCCAACATGACCACGCAGAT-3′ (reverse primer), and 18s rRNA (as internal control), 5′-GTCTGTGATGCCCTTA-GATG-3′ (forward primer), and 5′-AGCTTATGACCCGCACT-TAC-3′ (reverse primer).
Silencing Activity of the PCBP2 siRNA Nanocomplex Containing siRNA–PNA–Bitoin.
Biotin-labeled PNA was purchased from PNA Bio Inc. (Newbury Park, CA) and annealed with PCBP2 siRNA containing the complementary sequence to form the siRNA–PNA–biotin chimera. Subsequently, siRNA–PNA–biotin chimera, biotin-labeled IGF2R-specific peptide-431, and neutravidin were mixed in a 3.9:0.1:1 molar ratio at room temperature for 10 min to form the siRNA/neutravidin/peptide complex, which was further condensed with protamine at a N/P ratio of 2.5:1 for 30 min to form the final PCBP2 siRNA nanocomplex as described.7 Protamine is a cationic peptide that has been widely used for nucleic acid delivery.16–18 Moreover, protamine has been approved as a heparin antagonist by the FDA. In a recent clinical study for heparin reversal, toxicities were not observed in 380 patients who received systematic administration of protamine at an average dose of 39.1 mg.19 HSC-T6 cells were transfected with the PCPB2 siRNA nanocomplex at a final concentration of 50 nM siRNA in Opti-MEM I reduced serum medium for 6 h at 37 °C. The cells were incubated in complete medium for 18 h and then harvested for RNA isolation, and the silencing activity at the mRNA level was evaluated as described above.
Western Blot.
HSC-T6 cells were transfected with siRNA–PNA–peptide-431 or siRNA nanocomplex for 24 and 6 h, respectively, at 37 °C, followed by incubation in a fresh DMEM medium supplemented with 10% FBS for another 24 or 42 h. Subsequently, the cells were lysed, and proteins were obtained as we described before.20 Twenty micrograms of the protein were loaded to a 12% SDS-PAGE gel and separated under 70 V for 2 h. The proteins were then transferred to a PVDF membrane, blocked with 5% nonfat milk, and probed with the primary anti-PCPB2 antibody.
Statistics Analysis.
Data were presented as the mean ± standard deviation (SD). The difference between any two groups was determined by ANOVA with Tukey’s post hoc test, and P < 0.05 was considered statistically significant.
RESULTS
Optimization of the Complementary Sequences for Hybridizing PNAs with siRNAs.
The objective of this study is to develop a PNA-based platform to noncovalently attach chemical moieties to siRNAs without comprising their silencing activity (Figure 1A). The first step is to identify a PNA sequence that can form a stable chimera with siRNA. We accordingly designed two PNA sequences: an 8-mer PNA with the sequence CACCACTC and a 9-mer PNA with the sequence CACCACCAC. The melting temperatures of the 8-mer PNA and 9-mer PNAs with complementary RNA are 43.7 and 54.2 °C, respectively. The PNAs were annealed to the 3′ end of the sense strand of the luciferase siRNA at room temperature for 30 min to form siRNA–PNA chimeras. The chimeras were then incubated at 37 °C in TE buffer for various time intervals to evaluate their stability at physiological temperature. If the 8-mer PNA was annealed to the siRNA, approximately half of the chimera dissociated at 37 °C after 2 h (Figure 1B). In contrast, the chimera of the siRNA with the 9-mer PNA displayed better stability, and most of the chimera was stable for up to 2 h at 37 °C (Figure 1C). We, therefore, use the 9-mer PNA for the following studies.
Figure 1.
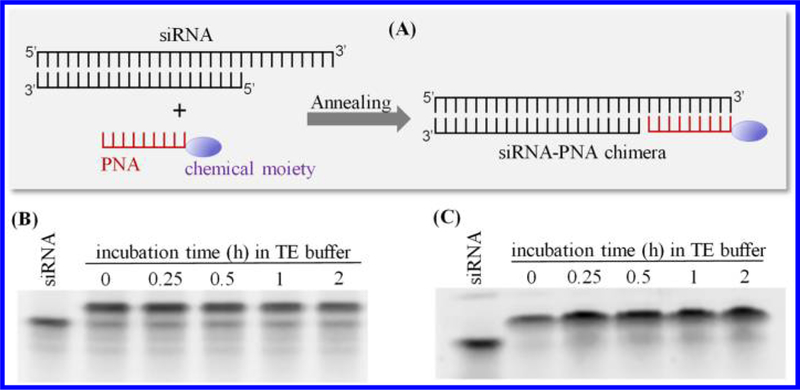
Scheme and stability of the siRNA–PNA chimera at physiological temperature. (A) Scheme of the siRNA–PNA chimera. siRNA duplex containing an extended sequence at the 3′ end of the sense strand was annealed with PNA at a 1:1 molar ratio. The chimeras were then incubated at 37 °C for a series of time intervals to evaluate their stability. (B) Stability of the chimera formed with the 8-mer PNA CACCACTC. (C) Stability of the chimera formed with the 9-mer PNA CACCACCAC.
Formation of the siRNA–PNA Chimera at Different Termini and a Comparison of Their Serum Stability.
The thermodynamic stability of the siRNAs’ termini determines their functionality. It is therefore critical to determine which terminus of siRNAs is the best site to form chimeras with PNAs. As described in Figure 2, the luciferase siRNA was annealed to the 9-mer PNA at four different termini (5′ and 3′ ends of the sense and antisense strands of the siRNA). Subsequently, all siRNA–PNA chimeras were incubated with 50% rat serum for a series of time intervals. The native siRNA was nearly completely degraded after a 4 h incubation at 37 °C. On the other hand, all siRNAs annealed with PNA exhibited enhanced serum stability. The chimeras in which PNA was annealed at the 3′ end of the sense (siRNA–PNA1) and 5′ end of the antisense (siRNA–PNA4) strands of siRNA exhibited better serum stability than the chimeras in which PNA was annealed at the 5′ end of the sense (siRNA–PNA2) and 3′ end of the antisense (siRNA–PNA3) strands of the siRNA.
Figure 2.
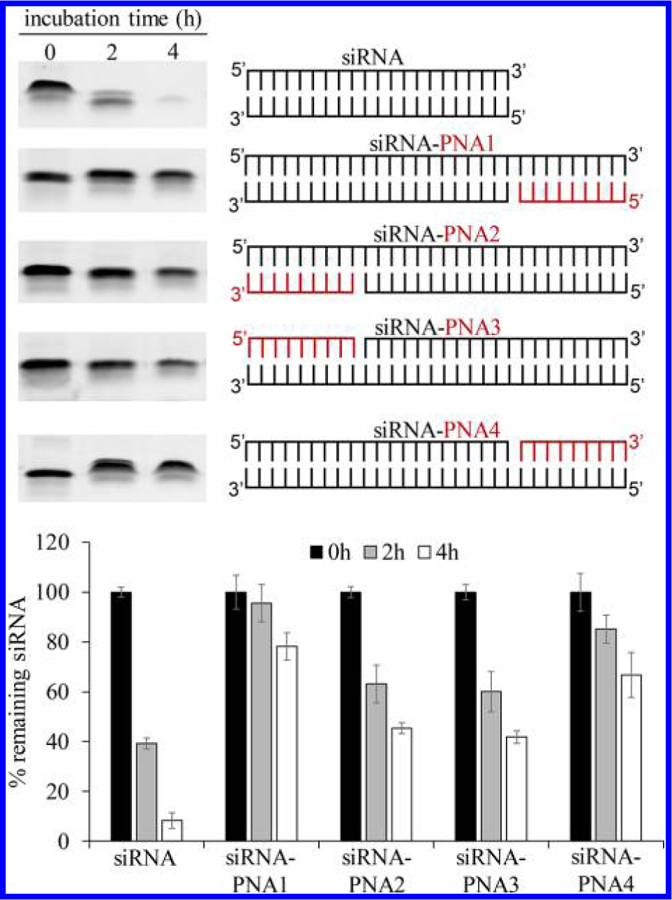
Serum stability of the siRNA–PNA chimeras annealed at different termini of the siRNA. The luciferase siRNA was annealed to the 9-mer PNA (CACCACCAC) at the 5′ and 3′ termini of the sense and antisense strands. The chimeras were incubated in 50% rat serum at 37 °C for a series of time points. Serum stability of the samples was evaluated by electrophoresis on a 20% native polyacrylamide gel and visualized by staining with GelRed. Intensities of the gel bands were quantitated with ImageJ. Results are presented as mean ± SD (n = 3 independent experiments).
Silencing Activity of the siRNA Annealed with PNA at Different Ends.
The silencing effects of the siRNAs annealed with PNA at different ends were evaluated. All luciferase siRNA–PNA chimeras and native luciferase siRNA were transfected into HeLa cells using Lipofectamine 2000. The silencing effect was detected using the Luciferase assay as described above. As shown in Figure 3, three of the siRNA–PNA chimeras showed reduced silencing activity compared to the native siRNA, except the siRNA annealed with PNA at the 3′ end of the sense strand. This siRNA–PNA chimera (siR–PNA1) retains the same silencing effect as the native siRNA. Therefore, PNA does not influence the silencing effect only when it is annealed to the 3′ end of the sense strand of the siRNA. This result also demonstrates that the annealed PNA in siR–PNA does not affect the unwinding of siRNA by the RNA-induced silencing complex (RISC) inside the cells.
Figure 3.
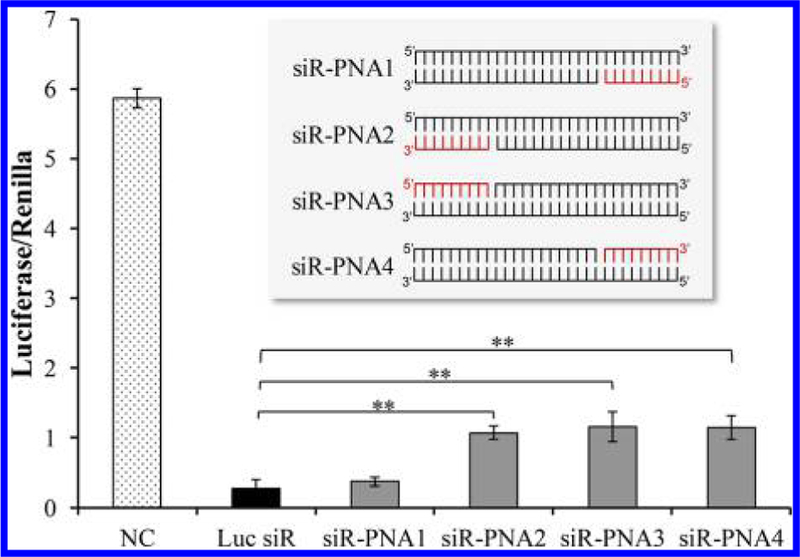
Silencing activity of the siRNA–PNA chimeras annealed at different termini of the siRNA. The luciferase siRNA was annealed to the 9-mer PNA (CACCACCAC) at 5′ and 3′ termini of the sense and antisense strands. Native siRNA or siRNA–PNA chimeras were cotransfected with the firefly luciferase plasmid and renilla luciferase plasmid using Lipofectamine 2000. A scrambled siRNA was used as the negative control. 48 h after transfection, the dual-luciferase reporter (DLR) Assay System was used to detect the relative luciferase expression level of each sample. Results are presented as mean ± SD (n = 3 independent experiments).
Silencing Activity and Serum Stability of the siRNA Annealed with Two PNAs at Both Ends.
We also annealed two different 9-mer PNAs, ACACTCACG and CACCACCAC, to the 5′ and 3′ ends of the sense strand of the siRNA, respectively, to further increase the serum stability of the siRNA. The melting temperature of the PNA sequence ACACTCACG is 55.3 °C, which is similar to that of the PNA CACCACCAC. Two PNA sequences were incubated with the siRNA in TE buffer at 37 °C to allow the formation of the PNA–siRNA–PNA chimera. The formation of the chimera was confirmed by electrophoresis on a 20% native polyacrylamide gel and visualized by staining with GelRed. As illustrated in Figure 4, both PNA sequences were successfully annealed to the siRNA, and each PNA sequence specifically annealed to its complementary RNA sequence. For example, when the siRNA and the PNA (ACACTCACG) were annealed at a 1:2 molar ratio, the PNA only annealed to its complementary sequence at the 5′ end, but not 3′ end, of the sense strand, indicating precise annealing to its complementary sequence. Similar specificity was also observed when the PNA (CACCACCAC) was annealed to the 3′ end of the sense strand.
Figure 4.
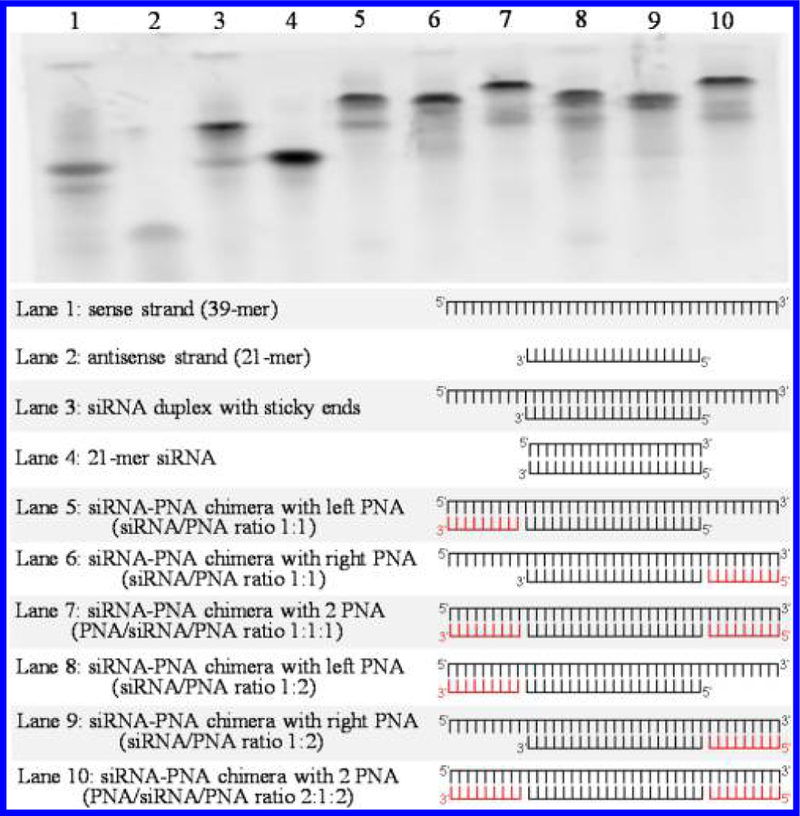
Formation of the siRNA–PNA chimeras containing one or two PNAs. Two 9-mer PNAs, ACACTCACG and CACCACCAC, were annealed to the 5′ and/or 3′ termini of the sense strand of the luciferase siRNA. The melting temperature of the PNA ACACTCACG is similar to that of the PNA CACCACCAC. Formation of the chimers was confirmed using a 20% native polyacrylamide gel (left PNA: ACACTCACG; right PNA: CACCACCAC).
However, silencing activity of the PNA–siRNA–PNA chimeras was compromised (Figure 5). We then inserted one and two nucleotides as a spacer between PNA and the antisense strand of the siRNA to minimize the effect of PNA on the thermodynamic stability of the siRNA. As Figure 6 showed, gene silencing activity of the PNA–siRNA–PNA chimera containing a single-nucleotide space was restored, suggesting an optimized distance between the PNA to the ends of the antisense strand is critical for its silencing activity. We then evaluated the serum stability of the siRNAs annealed with two PNAs. As illustrated in Figure 7, the siRNA duplex with sticky ends was immediately degraded into the 21-mer siRNA duplex in the serum, and the 21-mer siRNA was then degraded in a similar pattern as the native siRNA duplex in Figure 2. This is because single-strand RNA molecules are extremely unstable in the serum. In contrast, the PNA–siRNA–PNA chimera exhibited much better stability. However, the chimeras containing a gap between the PNA and the antisense strand were less stable in the serum. This result indicates that even a single nucleotide, which is not annealed to its complementary sequence, can be rapidly degraded in the serum. In conclusion, the best strategy for attaching PNA to siRNA is to anneal a single PNA to the 3′ end of the sense strand of siRNAs to achieve a good serum stability and high silencing activity.
Figure 5.
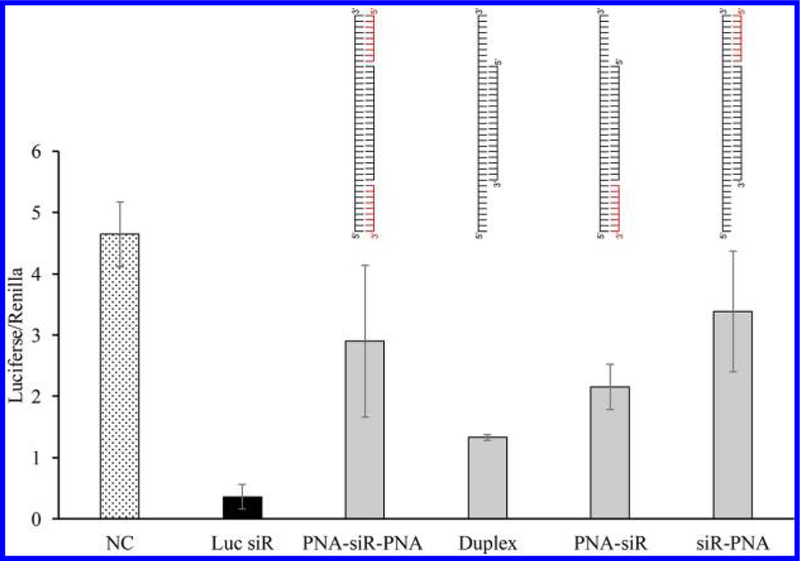
Silencing effect of the siRNA–PNA chimeras containing one or two PNAs. The native siRNA or siRNA–PNA chimeras were cotransfected with the firefly luciferase plasmid and renilla luciferase plasmid using Lipofectamine 2000. A scrambled siRNA was used as the negative control. A dual-luciferase reporter (DLR) assay system was used to detect the relative luciferase expression level of each sample. Results are presented as mean ± SD (n = 3 independent experiments).
Figure 6.
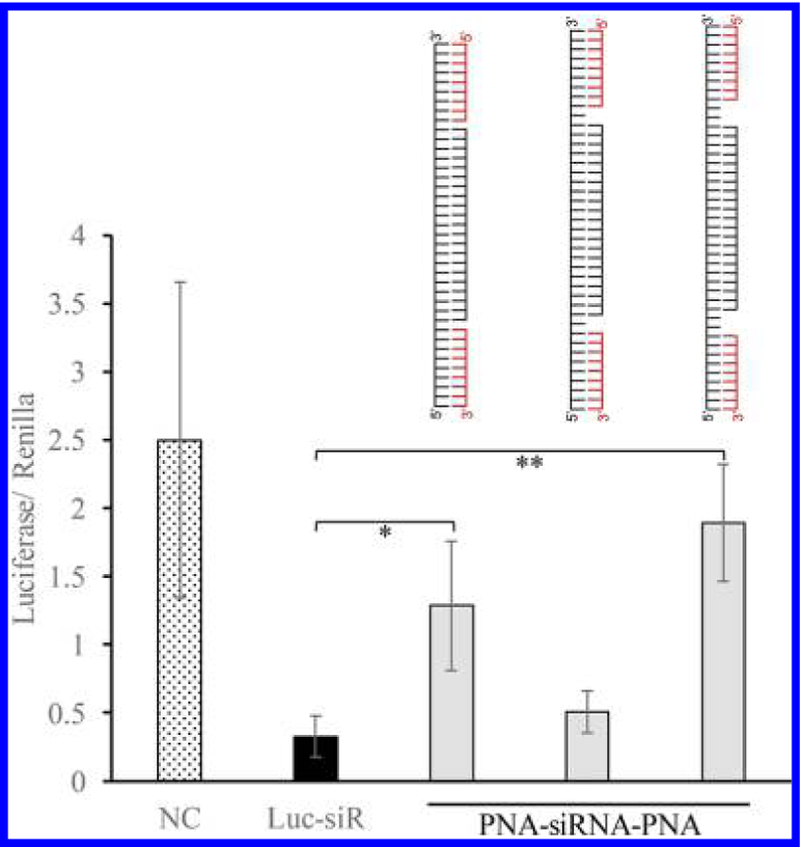
Silencing effect of the PNA–siRNA–PNA chimeras containing a space between the PNA and the antisense strand. The native siRNA or PNA–siRNA–PNA chimeras were cotransfected with the firefly luciferase plasmid and renilla luciferase plasmid using Lipofectamine 2000. A scrambled siRNA was used as the negative control. 48 h after transfection, a dual-luciferase reporter (DLR) assay system was used to detect the relative luciferase expression level of each sample. Results are presented as mean ± SD (n = 3 independent experiments).
Figure 7.
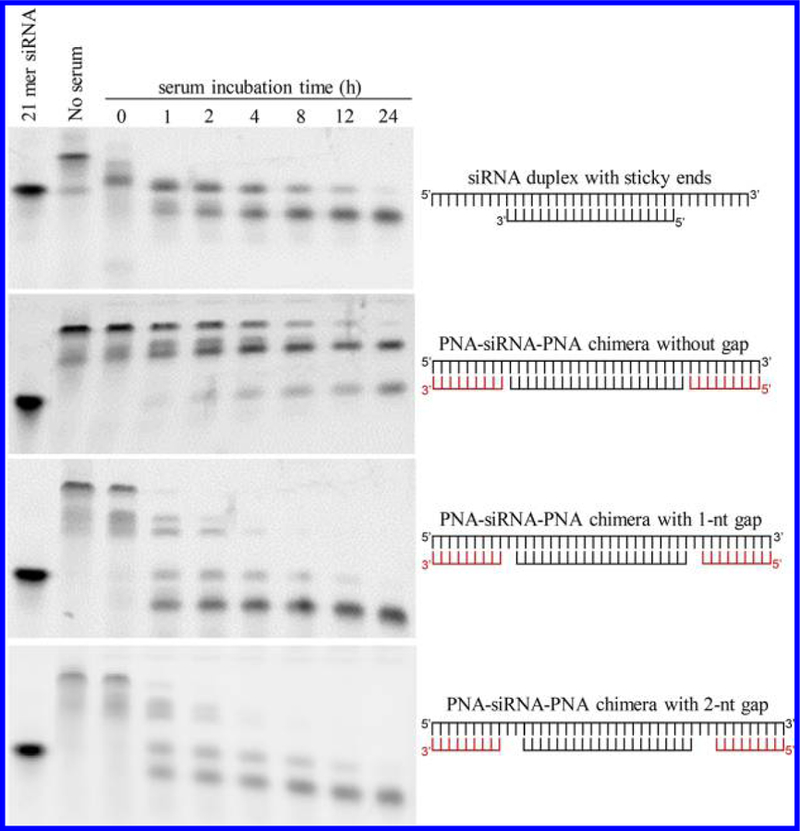
Serum stability of the PNA–siRNA–PNA chimeras containing a space between the PNA and the antisense strand. The PNA–siRNA–PNA chimeras were incubated in 50% rat serum at 37 °C for a series of time points. Serum stability of the samples was evaluated by electrophoresis on a 20% native polyacrylamide gel and visualized by staining with GelRed.
Application of the siRNA–PNA Platform in Attaching a Targeting Moiety to siRNA.
We next investigated whether the siRNA–PNA chimera platform can be used to attach a targeting ligand to siRNA and enhance its delivery to target cells. Herein, we employed an IGF2R-specific peptide ligand, peptide-431, which can be used to deliver therapeutic agents to hepatic stellate cells.21 Peptide-431 was linked to PNA with a spacer of five glycine residues to maintain the targeting capability of the peptide. As illustrated in Figure 8A, PNA–peptide-431 was annealed to the 3′ end of the sense strand of the PCPB2 siRNA, which was previously discovered by us as a promising siRNA for the treatment of liver fibrosis.20,22 The sense strand of the PCBP2 siRNA was labeled with Alexa Fluor 647, and cellular uptake of the siRNA–PNA–peptide-431 chimera was evaluated in a rat hepatic stellate cell line (HSC-T6) using flow cytometry. Native siRNA was used as a negative control. HSC-T6 cells took up significantly higher levels of the siRNA–PNA–peptide-431 chimera than the native siRNA, which only exhibited negligible cellular uptake (Figure 8B).
Figure 8.
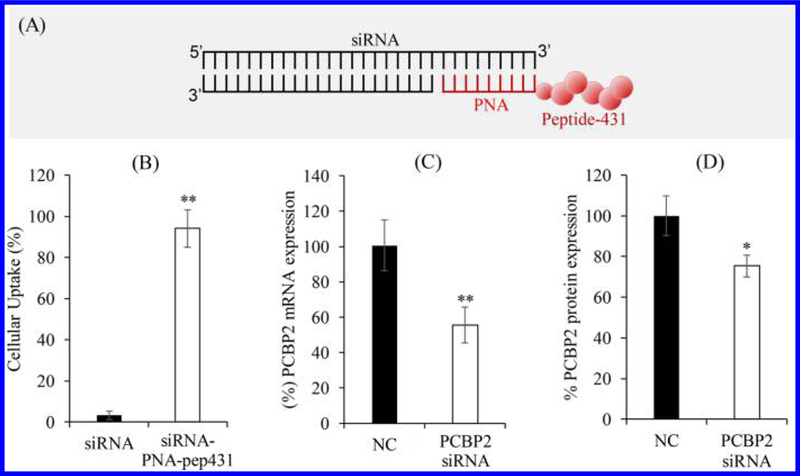
Attachment of the IGF2R-specific peptide-431 to PCBP2 siRNA using the siRNA–PNA platform. (A) Scheme of the PCBP2 siRNA–PNA–peptide-431 chimera. (B) Cellular uptake of free siRNA and the chimera in HSC-T6 cells. (C) Silencing activity of the chimera at the mRNA level of the PCBP2 gene. (D) Silencing activity of the chimera at the protein level of the PCBP2 gene.
We then evaluated the silencing activity of the siRNA–PNA–peptide-431 chimera in HSC-T6 cells. As shown in Figure 8C, compared to scrambled siRNA, the siRNA–PNA–peptide-431 chimera silenced the PCBP2 at the mRNA level for 40%. Similar silencing activity at the protein level was also observed by Western blot analysis (Figure 8D).
Application of the siRNA–PNA Platform in the Fabrication of the siRNA Nanocomplex.
Having shown that the siRNA–PNA chimera strategy can be used to noncovalently link a targeting ligand to siRNA to enhance the delivery efficacy of the siRNA, we also investigated whether this strategy can be used to fabricate siRNA into nanocomplexes. In our previous work, we have developed a neutravidin-based siRNA nanocomplex to deliver siRNA to hepatic stellate cells. The nanocomplex protects siRNA from degradation and exhibits high silencing activity.6,15 Moreover, a whole blood assay was conducted to evaluate the effect of neutravidin nanocomplex on the expression of inflammatory cytokines. The results showed that the neutravidin nanocomplex did not induce the expressions of TNFα and IL-6 after 48 h incubation. The nanocomplex only slightly induces the expression of IFNγ after 48 h incubation.6
Although the neutravidin siRNA nanocomplex demonstrated high silencing activity with good safety, the siRNA was chemically conjugated to biotin, which limits the application of the siRNA nanocomplex in animal studies because of the difficulty and low yield of the chemical conjugation. Here, we conjugated biotin to PNA and then annealed the PNA–biotin to the PCBP2 siRNA to form the siRNA–PNA–biotin chimera, which was then mixed with neutravidin, biotin–peptide-431, and protamine to form the siRNA nanocomplex (Figure 9A). The siRNA nanocomplex was incubated with HSC-T6 cells and exhibited nearly 70% silencing activity at the mRNA level (Figure 9B). Similar silencing activity at the protein level was also observed in Western blot analysis (Figure 9C). The silencing activity is comparable to the siRNA nanocomplex using biotin-conjugated siRNA in our previous reports,6,7 suggesting that the siRNA–PNA–biotin has the same capability as the biotin-conjugated siRNA to form the siRNA nanocomplex and subsequently silence the target gene.
Figure 9.
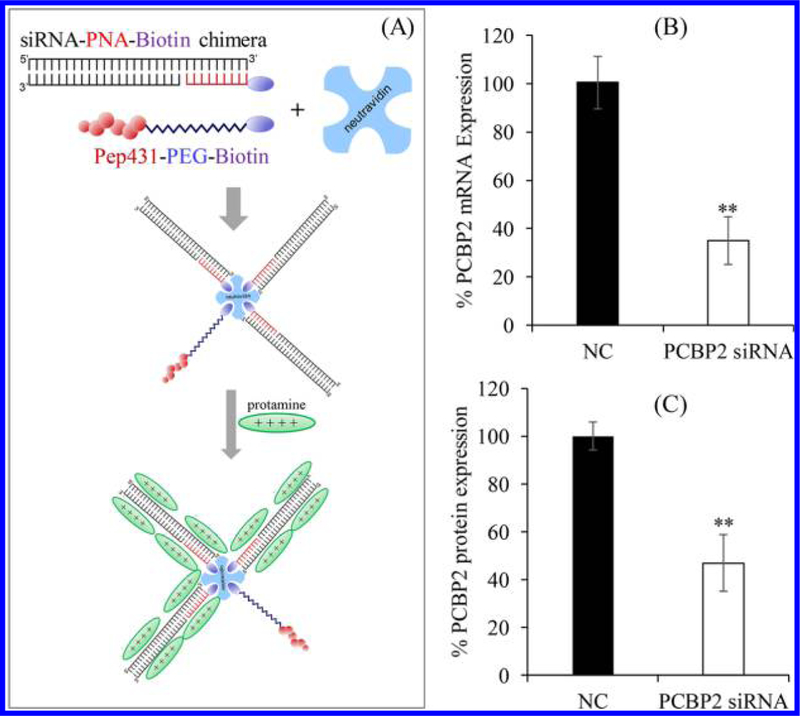
Fabrication of the neutravidin-based siRNA nanocomplex using the siRNA–PNA–biotin chimera. (A) Scheme of the neutravidin-based siRNA nanocomplex containing the siRNA–PNA–biotin chimera. (B) Silencing activity of the nanocomplex at the mRNA level of the PCBP2 gene. (C) Silencing activity of the nanocomplex at the protein level of the PCBP2 gene.
DISCUSSION
Since its discovery in 1998, the RNA interference has been widely used to regulate the expression of specific genes. siRNAs are one of the most widely used RNAi tools for the short-term downregulation of gene expression by degrading the mRNA. Over the past two decades, tremendous efforts have focused on developing novel siRNA delivery systems for therapeutic application. Among all approaches, nonviral delivery systems that use cationic lipids, polymers, or peptides to form nanoscale complexes with anionic siRNA have been widely studied. However, the significant toxicity of these cationic materials both in vitro and in vivo cannot be ignored.23 An alternative strategy that conjugates the siRNA to a targeting ligand, such as an antibody, peptide, or small molecule, can directly deliver the siRNA to the target tissue, thus increasing the selectivity and reducing the toxicity of cationic materials.7,24–26
siRNAs are mainly conjugated to targeting ligands using disulfide bonds and thioester bonds as the cleavable linker to release native siRNA inside cells. However, because of the poor stability of RNA, chemical conjugation of siRNAs is always complicated with a very low yield, thus limiting their application in animal studies. Therefore, there is a great need to develop a new method to noncovalently link targeting ligands to siRNAs without chemically modifying siRNA. In our study, we employed PNA as a complementary linker to form the siRNA–PNA chimera by annealing. Targeting ligands or other chemical moieties can be chemically conjugated to PNA, which is more stable than siRNAs in chemical reactions. Subsequently, the PNA–ligand conjugate is simply annealed to the siRNA duplex to form the siRNA–PNA chimera. Also, when the siRNA–PNA–chimera is unwound, the PNA–ligand will be detached to release the native siRNA to exert its silencing activity.
PNAs are oligonucleotide analogues in which the negatively charged sugar–phosphate backbone is replaced with neutral N-(2-aminoethyl) glycine units.8,27 Due to this neutral backbone, PNA anneals to its complementary DNA or RNA sequence without electrostatic repulsion. As a result, PNA has a higher affinity and selectivity for its complementary sequence and a relatively higher Tm compared to DNA or RNA.28 Since PNA was first described, it has been widely used as antisense and antigene agents, microarrays, biosensors, imaging probes, fluorescence in situ hybridization (FISH), and PCR clamping.29–32 In this study, a PNA is annealed to an siRNA to generate the siRNA–PNA chimera. Our results indicated that Tm is critical in forming a stable siRNA–PNA chimera. The Tm must be sufficiently high to maintain the thermal stability of the chimera at physiological temperature. On the other hand, the Tm must not be too high to prevent the siRNA from unwinding and incorporating into the RISC, which would significantly reduce the silencing activity of the siRNA. Ursula Giesen et al. developed a formula to calculate the Tm for a PNA/DNA hybrid that exhibited a good predictive ability.33 Two PNA sequences, an 8-mer PNA CACCACTC and a 9-mer PNA CACCACCAC, were annealed to the siRNA duplex, and the Tm of the siRNA–PNA chimera was calculated using the reported formula. Tm of the siRNA–PNA(8-mer) chimera is 43.7 °C, whereas the Tm of the siRNA–PNA (9-mer) chimera is 54.2 °C. Accordingly, the 9-mer PNA forms a more stable chimera with the siRNA (Figure 1) and therefore is selected as the complementary linker for this study.
An siRNA duplex has four potential sites for conjugation: the 5′ and 3′ ends of the sense and antisense strands. Czauderna et al. evaluated the silencing activities of siRNAs when the 5′ and 3′ ends of the sense and antisense strands were modified. Modifying the 5′ end of the antisense strand dramatically reduces the siRNA silencing activity, but no negative effects are observed when other three termini were modified.34 Because the 5′ end of the antisense strand must be phosphorylated by the RISC for recognition, it is usually not modified.35 In our study, the PNA was annealed to four possible sites on the siRNA: the 5′ and 3′ ends of the sense and antisense strands of the siRNA duplex. Accordingly, 5′ or 3′ ends of the antisense or sense strands of the siRNA duplex were extended by a 9-mer complementary RNA sequence for annealing. As shown in Figure 3, the siR–PNA4 chimera contains an extended RNA sequence at the 5′ end of the antisense strand and therefore exhibits reduced silencing efficacy. This is in accordance with others’ reports indicating that the 5′ end of the antisense is essential for the activity of siRNA.34,35 The siR–PNA3 chimera also shows diminished activity because its antisense strand was extended at the 3′ end, which affects its complementation with its target mRNA. In addition, the siR–PNA2 chimera containing an extended RNA sequence at the 5′ end of the sense strand also exhibits a diminished silencing activity. Only the siR–PNA1 chimera with an extended RNA sequence at the 3′ end of the sense strand maintains the original activity of the siRNA. This could be because annealing of PNA to the 5′ end of the sense strand affects unwinding of the siRNA duplex inside the cells. As a result, the 3′ end of the sense strand is the best annealing position for PNA to form a stable siRNA–PNA chimera without comprising silencing effect.
Another advantage of using PNA in the siRNA chimera is its resistance to nuclease and protease degradation, which improves the serum stability of the siRNA–PNA chimera. For example, the siRNA–PNA chimera exhibits enhanced serum stability in Figure 2. In addition, the serum stability of the siRNA is further improved when it is annealed to two PNA sequences (Figure 7). However, silencing activity of the PNA–siRNA–PNA chimera was compromised. It is postulated that two PNAs form a quite stable duplex with the sense strand, which limits the unwinding of the antisense strand from the sense strand. As a result, the chimeras containing only one PNA is the optimized strategy to noncovalently link a chemical moiety to siRNA without affecting its silencing activity.
CONCLUSION
In summary, we developed a new method to noncovalently attach chemical moieties to siRNAs using a 9-mer PNA as a complementary linker. Compared to native siRNA, the siRNA–PNA chimera exhibits improved serum stability and maintains the silencing activity. We also demonstrated that this chimera platform can be used to construct a peptide–siRNA conjugate and an siRNA nanocomplex without chemical modification of the siRNA. The constructed siRNA agents were successfully delivered to hepatic stellate cells and silenced its target gene. This new technology will greatly reduce the difficulty and cost in conjugating chemical moieties to siRNAs. This is particularly true for animal studies using siRNA, in which a large quantity of siRNA is always needed.
ACKNOWLEDGMENTS
This work is supported by awards (2R01AA021510 and 1R01GM121798) from the National Institutes of Health. Kun Cheng, PhD, was also supported by an American Cancer Society–Lee National Denim Day Research Scholar Grant (RSG-15-132-01-CDD).
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Elbashir SM; Harborth J; Lendeckel W; Yalcin A; Weber K; Tuschl T Duplexes of 21-nucleotide RNAs Mediate RNA Interference in Cultured Mammalian Cells. Nature 2001, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- (2).Fire A; Xu S; Montgomery MK; Kostas SA; Driver SE; Mello CC Potent and Specific Genetic Interference by Double-stranded RNA in Caenorhabditis Elegans. Nature 1998, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- (3).Kennerdell JR; Carthew RW Use of DsRNA-mediated Genetic Interference to Demonstrate that Frizzled and Frizzled 2 Act in the Wingless Pathway. Cell 1998, 95, 1017–1026. [DOI] [PubMed] [Google Scholar]
- (4).Nishina K; Unno T; Uno Y; Kubodera T; Kanouchi T; Mizusawa H; Yokota T Efficient in vivo Delivery of siRNA to the Liver by Conjugation of Alpha-tocopherol. Mol. Ther 2008, 16, 734–740. [DOI] [PubMed] [Google Scholar]
- (5).Qin B, Jin W, Cheng K Bioconjugation of siRNA for Site-Specific Delivery. In Advanced Delivery and Therapeutic Applications of RNAi; Cheng K, Mahato R, Eds.; Wiley, 2013. [Google Scholar]
- (6).Jain A; Barve A; Zhao Z; Jin W; Cheng K Comparison of Avidin, Neutravidin, and Streptavidin as Nanocarriers for Efficient siRNA Delivery. Mol. Pharmaceutics 2017, 14, 1517–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Zhao Z; Li Y; Jain A; Chen Z; Liu H; Jin W; Cheng K Development of a Peptide-modified siRNA Nanocomplex for Hepatic Stellate Cells. Nanomedicine 2018, 14, 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Nielsen PE; Egholm M; Berg RH; Buchardt O Sequence-selective Recognition of DNA by Strand Displacement with A Thymine-substituted Polyamide. Science 1991, 254, 1497–1500. [DOI] [PubMed] [Google Scholar]
- (9).Dean DA Peptide Nucleic Acids: Versatile Tools for Gene Therapy Strategies. Adv. Drug Delivery Rev 2000, 44, 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Barluenga S; Winssinger N PNA as a Biosupramolecular Tag for Programmable Assemblies and Reactions. Acc. Chem. Res 2015, 48, 1319–1331. [DOI] [PubMed] [Google Scholar]
- (11).Branden LJ; Mohamed AJ; Smith CI A Peptide Nucleic Acid-nuclear Localization Signal Fusion that Mediates Nuclear Transport of DNA. Nat. Biotechnol 1999, 17, 784–787. [DOI] [PubMed] [Google Scholar]
- (12).Liang KW; Hoffman EP; Huang L Targeted Delivery of Plasmid DNA to Myogenic Cells via Transferrin-conjugated Peptide Nucleic Acid. Mol. Ther 2000, 1, 236–243. [DOI] [PubMed] [Google Scholar]
- (13).Gitlin L; Karelsky S; Andino R Short Interfering RNA Confers Intracellular Antiviral Immunity in Human Cells. Nature 2002, 418, 430–434. [DOI] [PubMed] [Google Scholar]
- (14).Yu JY; DeRuiter SL; Turner DL RNA Interference by Expression of Short-Interfering RNAs and Hairpin RNAs in Mammalian Cells. Proc. Natl. Acad. Sci. U. S. A 2002, 99, 6047–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Shukla RS; Tai W; Mahato R; Jin W; Cheng K Development of Streptavidin-based Nanocomplex for siRNA Delivery. Mol. Pharmaceutics 2013, 10, 4534–4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Nakamura T; Yamada K; Fujiwara Y; Sato Y; Harashima H Reducing the Cytotoxicity of Lipid Nanoparticles Associated with a Fusogenic Cationic Lipid in a Natural Killer Cell Line by Introducing a Polycation-Based siRNA Core. Mol. Pharmaceutics 2018, 15, 2142–2150. [DOI] [PubMed] [Google Scholar]
- (17).Ding J; Liang T; Min Q; Jiang L; Zhu JJ ″Stealth and Fully-Laden″ Drug Carriers: Self-Assembled Nanogels Encapsulated with Epigallocatechin Gallate and siRNA for Drug-Resistant Breast Cancer Therapy. ACS Appl. Mater. Interfaces 2018, 10, 9938–9948. [DOI] [PubMed] [Google Scholar]
- (18).Rengaswamy V; Zimmer D; Suss R; Rossler J RGD liposome-protamine-siRNA (LPR) nanoparticles targeting PAX3-FOXO1 for alveolar rhabdomyosarcoma therapy. J. Controlled Release 2016, 235, 319–327. [DOI] [PubMed] [Google Scholar]
- (19).Gurses KM; Kocyigit D; Yalcin MU; Evranos B; Yorgun H; Sahiner ML; Kaya EB; Oto MA; Ozer N; Aytemir K Safety and efficacy outcomes of protamine administration for heparin reversal following cryoballoon-based pulmonary vein isolation. J. Interv Card Electrophysiol 2015, 43, 161–167. [DOI] [PubMed] [Google Scholar]
- (20).Liu H; Chen Z; Jin W; Barve A; Wan Y-JY; Cheng K Silencing of α-Complex Protein-2 Reverses Alcohol- and Cytokine-Induced Fibrogenesis in Hepatic Stellate Cells. Liver Research 2017, 1, 70–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Chen Z; Jin W; Liu H; Zhao Z; Cheng K Discovery of Peptide Ligands for Hepatic Stellate Cells Using Phage Display. Mol. Pharmaceutics 2015, 12, 2180–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Shukla RS; Qin B; Wan YJ; Cheng K PCBP2 siRNA Reverses the Alcohol-induced Pro-fibrogenic Effects in Hepatic Stellate Cells. Pharm. Res 2011, 28, 3058–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Lv H; Zhang S; Wang B; Cui S; Yan J Toxicity of Cationic Lipids and Cationic Polymers in Gene Delivery. J. Controlled Release 2006, 114, 100–109. [DOI] [PubMed] [Google Scholar]
- (24).Hassler MR; Turanov AA; Alterman JF; Haraszti RA; Coles AH; Osborn MF; Echeverria D; Nikan M; Salomon WE; Roux L; Godinho B; Davis SM; Morrissey DV; Zamore PD; Karumanchi SA; Moore MJ; Aronin N; Khvorova A Comparison of Partially and Fully Chemically-modified siRNA in Conjugate-mediated Delivery in vivo. Nucleic Acids Res 2018, 46, 2185–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Alam MR; Ming X; Fisher M; Lackey JG; Rajeev KG; Manoharan M; Juliano RL Multivalent Cyclic RGD Conjugates for Targeted Delivery of Small Interfering RNA. Bioconjugate Chem 2011, 22, 1673–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Zhou J; Lazar D; Li H; Xia X; Satheesan S; Charlins P; O’Mealy D; Akkina R; Saayman S; Weinberg MS; Rossi JJ; Morris KV Receptor-Targeted Aptamer-siRNA Conjugate-Directed Transcriptional Regulation of HIV-1. Theranostics 2018, 8, 1575–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Corey DR Peptide Nucleic Acids: Expanding the Scope of Nucleic Acid Recognition. Trends Biotechnol 1997, 15, 224–229. [DOI] [PubMed] [Google Scholar]
- (28).Egholm M; Buchardt O; Christensen L; Behrens C; Freier SM; Driver DA; Berg RH; Kim SK; Norden B; Nielsen PE PNA Hybridizes to Complementary Oligonucleotides Obeying the Watson-Crick Hydrogen-bonding Rules. Nature 1993, 365, 566–568. [DOI] [PubMed] [Google Scholar]
- (29).Nagai Y; Miyazawa H; Huqun; Tanaka T; Udagawa K; Kato M; Fukuyama S; Yokote A; Kobayashi K; Kanazawa M; Hagiwara K Genetic Heterogeneity of the Epidermal Growth Factor Receptor in Non-small Cell Lung Cancer Cell Lines Revealed by A Rapid and Sensitive Detection System, the Peptide Nucleic Acid-locked Nucleic Acid PCR Clamp. Cancer Res 2005, 65, 7276–7282. [DOI] [PubMed] [Google Scholar]
- (30).Brandt O; Hoheisel JD Peptide Nucleic Acids on Microarrays and Other Biosensors. Trends Biotechnol 2004, 22, 617–622. [DOI] [PubMed] [Google Scholar]
- (31).Marciniak RA; Cavazos D; Montellano R; Chen Q; Guarente L; Johnson FB A Novel Telomere Structure in A Human Alternative Lengthening of Telomeres Cell Line. Cancer Res 2005, 65, 2730–2737. [DOI] [PubMed] [Google Scholar]
- (32).Ivanova GD; Fabani MM; Arzumanov AA; Abes R; Yin H; Lebleu B; Wood M; Gait MJ PNA-peptide Conjugates as Intracellular Gene Control Agents. Nucleic Acids Symp. Ser. (Oxf) 2008, 52, 31–32. [DOI] [PubMed] [Google Scholar]
- (33).Giesen U; Kleider W; Berding C; Geiger A; Orum H; Nielsen PE A Formula for Thermal Stability (Tm) Prediction of PNA/DNA Duplexes. Nucleic Acids Res 1998, 26, 5004–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Czauderna F; Fechtner M; Dames S; Aygun H; Klippel A; Pronk GJ; Giese K; Kaufmann J Structural Variations and Stabilising Modifications of Synthetic siRNAs in Mammalian Cells. Nucleic Acids Res 2003, 31, 2705–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Ma JB; Yuan YR; Meister G; Pei Y; Tuschl T; Patel DJ Structural Basis for 5′-end-specific Recognition of Guide RNA by the A. Fulgidus Piwi Protein. Nature 2005, 434, 666–670. [DOI] [PMC free article] [PubMed] [Google Scholar]


