Abstract
Recent technical advances have enabled transcriptomics experiments at an unprecedented scale, and single-cell profiles from neural tissue are accumulating rapidly There has been considerable effort to use these profiles to understand cell diversity, primarily through unsupervised clustering and differential expression analysis. However, current practices to validate these findings vary. In this review, we describe recent efforts to evaluate clusters from single-cell RNA-sequencing data, and provide a framework for considering current evidence and practices in terms of their capacity to establish principles of cell biology. Single-cell RNA-sequencing has already transformed neuroscience. By facilitating detailed comparative and genetic perturbation analyses, it may provide the tools to uncover fundamental mechanisms of neural diversity throughout the tree of life.
Introduction
Single-cell RNA-seq (scRNA-seq) experiments use transcriptional profiling to characterize cells, sometimes grouping them into inferred cell types based on profile similarity [1]. Recent technical advances have increased the scale of scRNA-seq, making it feasible to profile thousands of cells in a single assay [2]. Already, studies of the nervous system span multiple species, anatomical regions, and developmental stages (e.g., [3-8]), and large scale efforts from consortia such as the Human Cell Atlas [9], the Brain Initiative Cell Census Network [10], and the CeNGEN project [11] only promise to increase the amount of available data. Many of these studies are focused on the same task, namely the classification of cells into types. However, because of the high degree of noise [12], the use of ad hoc methodology [13], and the lack of any unifying theory about what constitutes a cell type [14] it is important to assess how well scRNA-seq expression profiles replicate as an initial step toward their validation.
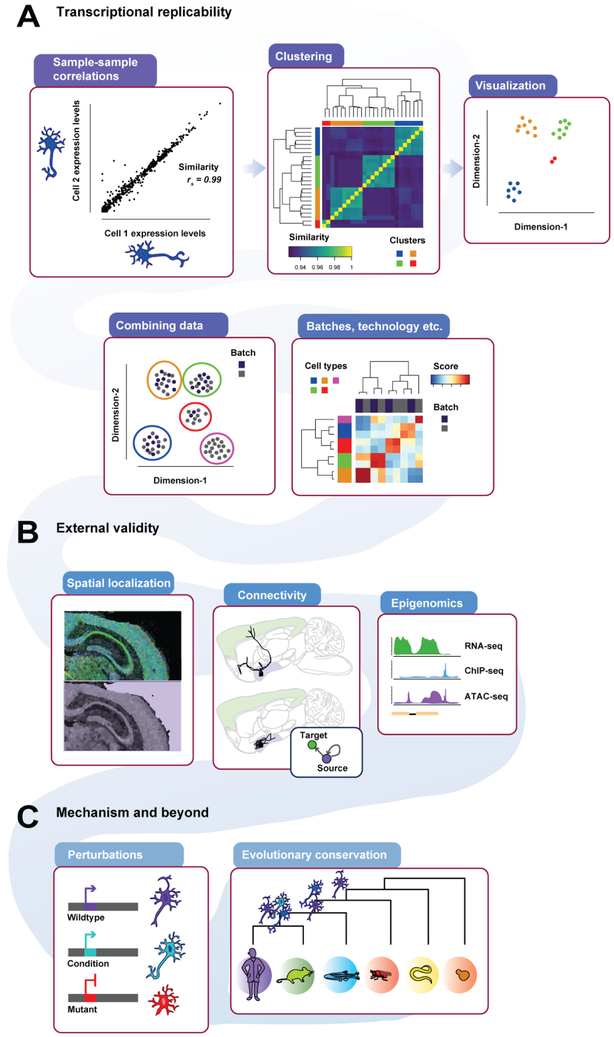
Indeed, the validation of single-cell clusters remains in its infancy, and in this review we aim to provide a framework for evaluating scRNA-seq clusters based on current practices in the field (see schematic, Figure 1). We note that this framework operates at the level of clusters, and we do not explicitly comment on the existence of specific cell types or subtypes, as definitions have yet to be established. In our view, evidence for scRNA-seq clusters can be divided into three levels, each of increasing consequence for establishing principles of cell biology: (i) replicability of expression levels, (ii) generalization to orthogonal data, and (iii) mechanistic validity. Assessments at each of these levels confirm robustness to particular classes of variation, discussed in further detail in the following.
Figure 1 -. ScRNA-seq replicability from transcriptional profile to external validity to mechanism and beyond.
Expression profiles can be compared (A-top left) to characterize replicability or clustered (A-top middle) to find groups of similar expression profiles, also visible as groupings of cells (A-top right) when the data is summarized via dimension reduction. New datasets should show the same clusters (A-bottom left), which can then be assessed for similarity (A-bottom right). External validity may be partially established by examining spatial localization of markers (B-left), connectivity (B-middle) or epigenomic profiles (B-right). Studies of genetic perturbation (C-left) and conservation (C-right) provide insight into the molecular and evolutionary mechanisms that drive expression diversity.
i. Replicability of expression levels
ScRNA-sequencing has become a convenient method for characterizing heterogeneity in biological systems, yet there are many sources of both technical and biological variation that can influence whether an experimental result is replicable. Technical variation may result from differences in library preparation protocols (e.g., 5’, 3’ or full-length mRNA capture, the inclusion of unique molecular identifiers, linear or PCR-based amplification); differences in sequencing depth; or differences in sample handling (e.g., dissociation procedures may affect cell viability, the probability of doublets, or the expression of immediate early genes). Biological variation is expected between cells, between individuals, and as a result of experimental selection procedures, such as sorting cells based on marker expression, or isolating RNA from nuclei. When comparing across datasets, further interpretive variation exists from the use of different clustering methods to group cells within each dataset Importantly, because scRNA-seq requires destructive sampling of cells, standard experimental design approaches for disentangling technical and biological sources of variability are not applicable. In the following we discuss the evaluation of scRNA-seq results across sources of technical and biological variation, and how these evaluations can provide insight into the replicability of inferred cell groupings.
i.i. Comparing expression profiles
The simplest way to compare two expression profiles is to plot one against another in a sample-sample scatterplot, and to calculate the Pearson or Spearman rank correlation coefficient In these plots, each point represents a gene and its position is determined by the gene’s expression level in each of the samples. In general, correlations between RNA-seq samples are high even when the samples are quite distinct biologically [15], because genes have fairly consistent expression levels. These correlations are typically very significant (e.g., p<10−10) because there are many data points (genes). Correlation is a fairly crude measure of sample replicability, and in bulk RNA-seq there have been efforts to encourage the use of visualizations and quality control (QC) measures that are more sensitive to distributional changes, such as Bland-Altman plots [16]. However, as long as both positive and negative cases are considered, sample-sample scatterplots can be useful for exploratory QC.
One thoughtful implementation of sample-sample comparison looks to recapitulate bulk expression profiles from merged single cells (e.g., [17,18]). Here, the biological signal is held constant (cell identity) to assess different technical approaches (bulk or single cell RNA-seq). Average expression profiles of groups of ten or more cells are often observed to be quite consistent with bulk profiles, even when their individual signals are much more variable (e.g., [18,19]). The observed cell-to-cell variability in scRNA-seq data is sometimes used as evidence for the existence of cell subtypes, and it is exploited by clustering algorithms to group cells into putatively biologically meaningful classes, even though it is also consistent with technical noise [20]. In spite of this limitation, clustering analyses have proven irresistible in the single-cell field, with studies routinely reporting that biologically relevant tissue heterogeneity has been ‘revealed’ by scRNA-seq [5,17-19,21]. We discuss the evaluation of these clusters below.
i.ii. Evaluating clusters
The assessment of scRNA-seq clusters follows two main strategies. The first strategy is to use multiple clustering pipelines to analyze a given data set, or to re-cluster data subsets, and determine how well the new clusters align with the original results (e.g., [19,22]). Here, almost all technical and biological sources of variation within the data are held constant In general, replicability analyses can only provide insight into the type of variation that is being sampled from. Because this strategy tests for in silico replication, it can only speak to the robustness of the results with respect to the parameter choices used for analysis. Consequently, results of these analyses are often good, likely because methods are only lightly calibrated to the data to begin with [23,24].
The second strategy is to perform additional experimental replicates and see whether clusters contain samples from all replicates. More formally, this is an assessment of whether the identified cell clusters replicate across batches, assuming the biological makeup of the replicate experiments can be held constant In scRNA-seq the most pernicious batch effects appear to arise during the preparation of sequencing libraries from cellular RNA [25,26], similar to what was previously observed for bulk RNA-seq [27]. In practice, this means that all cells prepared in a single 10x Chromium well, or on a single Fluidigm C1 chip, will tend to have higher within-batch correlations than across-batch, possibly due to ambient RNA [28] or other biochemical factors (reagents, temperature, etc.). This has prompted the invention of ‘cell hashing’ techniques that use barcoded antibodies to label samples so that they can be pooled in a given reaction (e.g., tumor and normal) [29]. While useful, increased sample multiplexing does not provide a measure of the robustness of results to sources of technical variation. To address this, one successful approach has been to generate multiple scRNA-sequencing libraries from biological replicates, and then validate clusters based on their reproducibility. This approach prevents the over-interpretation of clusters attributable to technical variation, such as differences in the fraction of the transcriptome that is assayed [30]. A prominent example comes from the landmark Drop-seq paper, where the authors aimed to transcriptionally characterize the retina [31]. Here, over forty thousand cells were profiled across seven batches, where each batch contained pooled retinal tissue from four to six mice. The authors identified thirty-nine clusters, and noted that all but one sampled proportionately from all batches. The cluster that failed to replicate more broadly appeared to contain non-retinal cell markers, suggesting a potential dissection artifact yielding cell type ascertainment bias. This experimental design and analysis strategy successfully separates replicable clusters from likely technical artifacts.
An extension of the second strategy is to assess cluster robustness across additional sources of biological or technical heterogeneity [32]. Comparisons between nuclear and whole-cell scRNA-seq, for example, have shown that cells of the same inferred type have broadly concordant expression profiles, with some explainable differences, such as the proportion of reads mapping to introns [33-35]. Comparisons may also be performed across published datasets, either to map cluster identity with respect to an external reference [36,37] or to determine the degree of evidence for putative subtypes, toward the goal of achieving field-wide consensus [38]. This approach has the advantage of sampling more broadly across all aspects of sample preparation and analysis, which are generally held constant within a laboratory. Cross-dataset analysis of expression clusters has a history in cancer, where independent groups aimed to verify cancer subtypes by comparing average profiles across studies [39]. Among single-cell studies, clusters derived from newly generated data are sometimes compared to previously reported clusters, treating prior results as post-hoc validation data (e.g., [40,41]). Cross-dataset comparisons of coarse-grained cell classes are often straightforward. As an example, Habib et al compared the clusters from their nuclear Drop-seq of mouse cortex and hippocampus to those characterized by the Allen Brain Atlas (ABA) using Smart-seq2 of the mouse cortex [19], finding positive correlations between cells of the same broad type (e.g., microglia) [42]. To map their results at finer resolution, the authors opted to take a machine learning approach, training a random forest classifier from their own GABAergic data to re-classify inhibitory clusters from the ABA, reporting the proportion of cells from each of the twenty-three ABA clusters to be associated with the eight Habib labels. While all of the clusters from the ABA were classified as one of the Habib labels, most Habib labels contained cells from multiple ABA clusters, suggesting differences in cluster resolution between the two datasets. Notably, three of the eight Habib clusters were not clearly associated with any particular ABA clusters. Although there are many thousands of cells in this comparison, the analysis here has an effective ‘n’ of two. In the absence of additional data, such as additional matched datasets (i.e., nuclear Drop-seq and Smart-seq2 of the same areas), or some other orthogonal validation to define ground-truth, it is impossible to say whether the observed failure of replicability is technical or biological. However, these results highlight the importance of critical evaluation of clustering results, and the difficulty of cross-dataset comparisons.
In general, three main issues plague cross-dataset analysis: heterogeneity of class structure between datasets (i.e., cell type proportions), including missing cell types; feature selection, typically of genes; and evaluation of dataset independence. Differences in dataset structure, including missing cell types, can have important consequences for cross-dataset comparison, particularly when model fitting (sometimes ‘manifold learning’) is performed with respect to within-dataset variability [43,44]. If two datasets are being merged, but they contain non-overlapping or only partially overlapping cell types, clusters that are quite distinct may incorrectly align to one another, generating clusters that contain a mixture of cell types. This is the primary failure mode of these approaches. Large class imbalances may also bear on the metrics used for evaluation [45]. Previous attempts to solve issues of feature selection and class structure between single-cell datasets have used machine learning approaches, as described above [22,42]. While these models are powerful and provide a deep summary of the data they are trained on, the primary challenges of machine learning – avoiding overfitting and improving interpretability [46] – are particularly problematic in the context of single-cell data, where artifacts may easily be confounded with biology. These issues also plague more recent approaches for data fusion in scRNA-seq [43,44,47-51]. In addition, establishing dataset independence is crucial for accurate interpretation of cross-study results, informing the degree to which a result may be considered validating. For example, we previously compared data from the ABA with that from Paul et al [52], and found almost perfect replicability of the Sst-Chodl interneuron profile [38]. However, we later learned that these two studies employed the same mouse line for their analyses [53], suggesting that this result is less surprising than it might appear. This is a clear example of non-independence, but there are likely to be cryptic dependencies between datasets due to their reliance on established marker genes, making this a difficult issue to solve.
Ultimately, while there are many complexities associated with cross-dataset analysis, we would emphasize that the outlook is bright Our work provided the first formal evaluation of single-cell RNA-seq data from the brain, indicating that approximately half of the inferred interneuron subtypes from three high profile publications were almost perfectly replicable [38]. This approach has since been adopted to characterize cell identity early in development [40,41], as well as across species [54], finding strong evidence of cluster replicability in each case. We anticipate that the availability of benchmark datasets from large consortia in combination with other comprehensive data resources [6,7], will soon allow for very detailed investigation of transcriptional variation within and across clusters, as well as the interaction of these profiles with other sources of biological and technical variability. We note that although approaches for joint analysis can often be quite complex, the reliance on mutual nearest neighbors is a recurrent theme [38,43,50]. Nearest neighbor or neighbor voting approaches have been shown to be useful across a wide range of machine learning tasks [55] and likely account for most of the performance of even very sophisticated approaches for joint modeling. Together, these data and analyses will enable the field to define consensus cluster profiles, which can then be assessed across additional modalities, and investigated for gene drivers, and evolutionary conservation, discussed in the following.
ii. Generalization to orthogonal data
As described above, scRNA-seq has been remarkably useful for identifying clusters that may represent cell types or subtypes. Yet historically, cell type definitions in neuroscience have been multimodal, requiring multiple strands of evidence to establish cell identity [56]. In light of this, it is common to assess the external validity of single-cell clustering results by performing experiments that can provide orthogonal information, such as imaging to assess spatial localization of marker genes [52], morphological reconstructions [19], retrograde or anterograde tracing [57], or electrophysiological recordings [58]. These data generally derive from different cells than those that were profiled with sequencing, though there are a number of examples of multimodal registration from the same cell (e.g., [59-61]). In the following, we discuss examples of three prominent types of orthogonal data used to provide evidence for neuronal clusters: spatial localization, connectivity, and epigenomic profiles.
ii.i. Spatial localization
A primary goal of scRNA-seq experiments is to discover novel marker genes that can distinguish between clusters of samples. Markers are useful because they can act as a “Rosetta stone”, enabling multimodal investigation of cell function through genetic targeting approaches, and they are typically identified through differential expression analysis. Often, the simplest follow-up experiment is to assess their spatial co-localization, particularly because established atlases can provide an initial filter on selected genes. While marker co-localization cannot definitively prove the existence of novel cell types, it provides validation that their co-expression is not a technical artifact of scRNA-seq. Certain spatial patterns known to reflect functional or developmental specification, such as layer-restricted distributions among neurons in the cortex, can also suggest shared lineages.
To avoid relying on a small number of markers or on profiles from dissociated cells, recent work has aimed to assess transcription in intact tissue, either through highly multiplexed RNA fluorescence in situ hybridization (FISH) [62,63] or with in situ sequencing [64]. Here there are different limitations from those of scRNA-seq, related to microscopy and spatial constraints. For example, determining optimal parameters regarding probe design and optical density may be quite specific to the tissue to be assayed, which lengthens start-up time for the application of these protocols across biological contexts. Thus, while highly promising, these techniques have yet to be adopted widely.
ii.ii. Connectivity
Understanding the wiring diagram of the brain has been the major goal of systems neuroscience, and there have already been a number of interesting developments to link neuronal connectivity to transcription. In mouse, a handful of studies have used retrograde tracers or viral injections, sometimes in combination with transgenic approaches, to selectively target cells for scRNA-seq based on their projections to a single location [19,21,57,65,66]. An approach that assays connectivity more broadly by using molecular barcodes, MAPseq [67], has recently been combined with in situ sequencing of barcodes and FISH (BARseq) [68]. In initial analyses of BARseq data, projection profiles have been clustered and compared to their laminar distributions, showing that cells from the same location can have highly divergent connectivity profiles, and, notably, the authors failed to detect a layer 2/3 projection class that was predicted from gene expression [19]. Yet, it is difficult to determine what to expect when comparing clusters from projection profiles and from gene expression: one-to-one relationships may provide evidence that expression is associated with functional distinctions between samples, but they need not occur. Higher degrees of multiplexing, or the evaluation of newly defined cluster markers, will facilitate more comprehensive comparisons of projection mapping techniques to scRNA-seq data, helping to formalize and test hypotheses about the link between molecular and projection identity.
ii.iii. Epigenomics
Patterning of gene expression in different cell types is attributed to the ‘epigenome’ – all of the modifications to DNA that do not alter its sequence, but may be associated with differences in activity. New methods have made it possible to examine chromatin accessibility [69] and DNA methylation [70-72] in single cells, or even to assay expression and epigenetic profiles at once [73]. In a recent study of the human brain, single-nucleus chromatin accessibility and expression profiles were successfully combined to identify putative regulatory regions [74], though the authors noted that the epigenomic data could not resolve finer cell subtypes. Because epigenomic techniques are genome-wide rather than transcriptome-wide they provide very sparse data, which present a challenge. Yet these early results are promising: if epigenomic techniques can be used to map enhancers, they may contribute toward a mechanistic understanding of cell diversity, discussed next.
iii. Mechanistic validity: Perturbation and evolutionary conservation
In the previous section we described current practices to gain additional evidence for clusters identified by singlecell RNA-seq. These experiments are valuable because they tend to have different sources of noise than scRNA-seq, so their alignment to single-cell clusters suggests robustness to technical variation. However, because they are purely observational, they are necessarily limited in their capacity to provide causal information about the mechanisms that drive variation within and across cell clusters. Ultimately, the goal of scRNA-seq should not be to simply define replicable profiles across more and more data modalities, but rather to understand why they are replicable. Requiring this level of understanding clarifies the limitations of external validation through orthogonal data, and specifies what remains to be done: perturbation experiments. This is the gold-standard for experimental biology, but it has yet to be consistently applied as a test of the validity of scRNA-seq clusters, likely because mechanistic experiments can be difficult to design and execute. To our knowledge, there have been at least two studies that knocked down cluster-specific genes and validated their impact on cell fate. Both were performed in the planarian Schmidtea Mediterranea, and the approach was likely feasible because of the organism’s inherent regenerative capacity [75,76]. Eventually, with enough basic understanding of transcription factor and regulatory information, causal modeling may allow for in silico experimentation [77]. A more immediate solution may be found with the implementation of technologies that integrate pooled genetic perturbation screens with single-cell sequencing [78-80].
Taking this further, we suggest that cross-species analysis in combination with genetic perturbation is likely to provide the most nuanced view of factors driving cell identity. Though few cross-species analyses of neural singlecell data have been performed, most have focused on mouse and human expression [54,81]. The examination of a wider variety of species, and particularly non-model systems where genetic and environmental diversity can be assayed, will provide greater insight into the pathways that underlie cell specialization. In this direction, recent work examined the evolution of mammalian neocortex through comparative study of reptilian pallium, finding evidence that GABAergic interneurons are ancestral to both reptiles and mammals, indicating conservation of interneuron diversity [5]. An impressive investigation into the cell type repertoire of the cnidarian Nematostella vectensis found that neurons were enriched for genes originating at multiple evolutionary times, and that a subset of gene orthologs conserved in mouse, nematode (C. elegans) and Nematostella all showed neuronal expression specificity, though gene-gene co-expression relationships were distinct [8]. Taking inspiration from these studies, we envision that future comparative single-cell analyses will incorporate phylogeny, gene age, and natural history to an ever greater degree, allowing us to answer questions like: Did similar cell-types evolve independently? Over what interval did a given type arise? What pressures led to the diversification of cell types? Do all cell types have adaptive significance? Decreasing sequencing costs, and efficient gene targeting with CRISPR should enable comparative study of a wide variety of species so that we can move beyond phenomenology toward a more holistic understanding of cell phenotypes.
Conclusions
Currently, the meaning of “cell type” is richly debated, and thresholds for cluster assignment within any pipeline or acting on any single data set feel arbitrary and premature. We have deliberately avoided this debate in this review by focusing on current practices in single cell transcriptomics, and providing a framework for considering reports of replicability in the context of their explanatory capacity. Moving forward, we believe that meta-analysis across laboratories and species will become increasingly important Moreover, explicitly considering degree of conservation of cell phenotypes opens up the possibility of formalizing definitions of cell type. The analogy to species can be useful to consider: it is sometimes practical to treat species as discrete entities, and sometimes it is not Likewise, at some scales, cell types will be meaningful as discrete entities, but because their existence over evolutionary time is the product of incremental modification, at other scales, it may be more suitable to consider them along a continuum [82]. As comparative genomics and functional genetics join forces with single-cell analysis, not only will we begin to see what replicates, but we will also see what doesn’t. More importantly, we will know how and why the difference occurs.
Highlights.
Single-cell RNA-sequencing has been used to profile neural cells in many organisms.
Cell expression clusters are often treated as potential cell types or subtypes.
Cluster validation requires multiple strands of evidence.
Mechanistic and evolutionary studies may reveal principles of cell diversity.
Acknowledgments
We would like to thank Paul Pavlidis, Vilas Menon, Josh Huang, Shreejoy Tripathy, Tony Zador and Adam Kepecs for their helpful comments on earlier versions of this manuscript. We would also like to thank Sara Ballouz for drawing the schematic for this article. M.C. is supported by a postdoctoral fellowship from the National Institute of Mental Health (F32-MH114501) and a NARSAD Young Investigator grant from the Brain & Behavior Research Foundation (26140). J.G. is supported by grants from the National Institute of Mental Health (R01-MH113005-02) and from the National Library of Medicine (R01-LM012736-01). Any opinions, conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the Brain & Behavior Research Foundation or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
(* = special interest, ** = outstanding interest)
- *1.Wagner A, Regev A, Yosef N: Revealing the vectors of cellular identity with single-cell genomics. Nat Biotech 2016, 34:1145–1160.Insightful review article about the challenges of characterizing cell identity with current single-cell genomics technologies and computational methods.
- 2.Svensson V, Vento-Tormo R, Teichmann SA: Exponential scaling of single-cell RNA-seq in the past decade. Nature Protocols 2018, 13:599. [DOI] [PubMed] [Google Scholar]
- 3.Pandey S, Shekhar K, Regev A, Schier AF: Comprehensive Identification and Spatial Mapping of Habenular Neuronal Types Using Single-Cell RNA-Seq. Curr Biol 2018, 28:1052–1065.e1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenberg AB, Roco CM: Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. 2018, 360:176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tosches MA, Yamawaki TM: Evolution of pallium, hippocampus, and cortical cell types revealed by single-cell transcriptomics in reptiles. 2018, 360:881–888. [DOI] [PubMed] [Google Scholar]
- *6.Zeisel A, Hochgerner H, Lonnerberg P, Johnsson A, Memic F, van der Zwan J, Haring M, Braun E, Borm LE, La Manno G, et al. : Molecular Architecture of the Mouse Nervous System. Cell 2018, 174:999–1014.e1022.Heroic effort to apply single-cell RNA-sequencing across the adult mouse nervous system to characterize cell identity. Contemporaneous with Sauders et al 2018 (ref 7).
- *7.Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, Bien E, Baum M, Bortolin L, Wang S, et al. : Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell 2018, 174:1015–1030.e1016.Alongside Zeisel et al 2018 (ref 6), the first brain-wide canvas of adult mouse brain regions at single-cell level. The authors provide a nuanced description of cell identity based on independent component analysis.
- **8.Sebe-Pedros A, Saudemont B, Chomsky E, Plessier F, Mailhe MP, Renno J, Loe-Mie Y, Lifshitz A, Mukamel Z, Schmutz S, et al. : Cnidarian Cell Type Diversity and Regulation Revealed by Whole-Organism Single-Cell RNA-Seq. Cell 2018, 173:1520–1534.e1520.Tour-de-force of single-cell genomics. This paper combines expression profiling, epigenomics and evolutionary analysis to characterize whole-organism cell diversity in larval and adult Nematostella vectensis, showing conservation of neuronal gene expression but divergence of gene co-expression, between cnidaria and bilateria.
- 9.Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, Bodenmiller B, Campbell P, Carninci P, Clatworthy M, et al. : The Human Cell Atlas. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ecker JR, Geschwind DH, Kriegstein AR, Ngai J, Osten P, Polioudakis D, Regev A, Sestan N, Wickersham IR, Zeng H: The BRAIN Initiative Cell Census Consortium: Lessons Learned toward Generating a Comprehensive Brain Cell Atlas. Neuron 2017, 96:542–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammarlund M, Hobert O, Miller DM 3rd, Sestan N: The CeNGEN Project: The Complete Gene Expression Map of an Entire Nervous System. Neuron 2018, 99:430–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brennecke P, Anders S, Kim JK, Kolodziejczyk AA, Zhang X, Proserpio V, Baying B, Benes V, Teichmann SA, Marioni JC, et al. : Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods 2013, 10:1093–1095. [DOI] [PubMed] [Google Scholar]
- 13.Zappia L, Phipson B, Oshlack A: Exploring the single-cell RNA-seq analysis landscape with the scRNA-tools database. PLOS Computational Biology 2018, 14:e1006245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *14.What Is Your Conceptual Definition of "Cell Type" in the Context of a Mature Organism? : Cell Syst 2017, 4:255–259.A diverse group of scientists answer this timely and thought-provoking question, revealing the absence of field-wide consensus.
- 15.Ballouz S, Gillis J: AuPairWise: A Method to Estimate RNA-Seq Replicability through Co-expression. PLoS Comput Biol 2016, 12:e1004868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teng M, Love MI, Davis CA, Djebali S, Dobin A, Graveley BR, Li S, Mason CE, Olson S, Pervouchine D, et al. : A benchmark for RNA-seq quantification pipelines. Genome Biology 2016, 17:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, Marques S, Munguba H, He L, Betsholtz C, et al. : Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 2015, 347:1138–1142. [DOI] [PubMed] [Google Scholar]
- 18.Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, Wildberg A, Gao D, Fung HL, Chen S, et al. : Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 2016, 352:1586–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, et al. : Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci 2016, 19:335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA: The technology and biology of single-cell RNA sequencing. Mol Cell 2015, 58:610–620. [DOI] [PubMed] [Google Scholar]
- 21.Hockley JRF, Taylor TS, Callejo G, Wilbrey AL, Gutteridge A, Bach K, Winchester WJ, Bulmer DC, McMurray G, Smith ESJ: Single-cell RNAseq reveals seven classes of colonic sensory neuron. Gut 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **22.Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, et al. : Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell 2016, 166:1308–1323.e1330.Exemplary high-throughput single-cell RNA-sequencing experimental design, analysis and validation. This paper builds from Macosko et al 2015 (ref 27) to further characterize bipolar neuron subtypes of the retina using both high- and low-throughput single-cell approaches, as well as morphological and functional validation.
- 23.Freytag S, Tian L, Lönnstedt I, Ng M, Bahlo M: Comparison of clustering tools in R for medium-sized 10x Genomics single-cell RNA-sequencing data [version 1; referees: 1 approved, 2 approved with reservations]. F1000Research 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duò A, Robinson M, Soneson C: A systematic performance evaluation of clustering methods for single-cell RNA-seq data [version 2; referees: 2 approved]. F1000Research 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *25.Tung P-Y, Blischak JD, Hsiao CJ, Knowles DA, Burnett JE, Pritchard JK, Gilad Y: Batch effects and the effective design of single-cell gene expression studies. Scientific Reports 2017, 7:39921.An important investigation of the sources of technical variation in single-cell RNA-sequencing data.
- 26.Natarajan KN, Miao Z, Jiang M, Huang X, Zhou H, Xie J, Wang C, Qin S, Zhao Z, Wu L, et al. : Comparative analysis of sequencing technologies platforms for single-cell transcriptomics. bioRxiv 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Consortium SM-I: A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nat Biotechnol 2014, 32:903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young MD, Behjati S: SoupX removes ambient RNA contamination from droplet based single cell RNA sequencing data. bioRxiv 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, Smibert P: Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 2017, 14:865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *30.Hicks SC, Townes FW, Teng M, Irizarry RA: Missing data and technical variability in single-cell RNA-sequencing experiments. Biostatistics 2017.This was the first paper to demonstrate the influence of systemic batch effects and technical variability on single-cell RNA-sequencing results. First available on bioRxiv in 2015.
- 31.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. : Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015, 161:1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ziegenhain C, Vieth B, Parekh S, Reinius B, Guillaumet-Adkins A, Smets M, Leonhardt H, Heyn H, Hellmann I, Enard W: Comparative Analysis of Single-Cell RNA Sequencing Methods. Molecular Cell 2017, 65:631–643.e634. [DOI] [PubMed] [Google Scholar]
- 33.Habib N, Li Y, Heidenreich M, Swiech L, Avraham-Davidi I, Trombetta JJ, Hession C, Zhang F, Regev A: Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science 2016, 353:925–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bakken TE, Hodge RD, Miller JM, Yao Z, Nguyen TN, Aevermann B, Barkan E, Bertagnolli D, Casper T, Dee N, et al. : Equivalent high-resolution identification of neuronal cell types with single-nucleus and single-cell RNA-sequencing. bioRxiv 2018. [Google Scholar]
- 35.Lake BB, Codeluppi S, Yung YC, Gao D, Chun J, Kharchenko PV, Linnarsson S, Zhang K: A comparative strategy for single-nucleus and single-cell transcriptomes confirms accuracy in predicted cell-type expression from nuclear RNA. Scientific Reports 2017, 7:6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin C, Jain S, Kim H, Bar-Joseph Z: Using neural networks for reducing the dimensions of single-cell RNA-Seq data. Nucleic Acids Research 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiselev VY, Yiu A, Hemberg M: scmap: projection of single-cell RNA-seq data across data sets. Nature Methods 2018. [DOI] [PubMed] [Google Scholar]
- *38.Crow M, Paul A, Ballouz S, Huang ZJ, Gillis J: Characterizing the replicability of cell types defined by single cell RNA-sequencing data using MetaNeighbor. Nature Communications 2018, 9:884.This was the first paper to assess the robustness of single-cell clusters across diverse datasets and gene sets, showing that independently identified mouse cortical interneuron subtypes had highly replicable transcriptional profiles.
- 39.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. : Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A 2003, 100:8418–8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalish BT, Cheadle L, Hrvatin S, Nagy MA, Rivera S, Crow M, Gillis J, Kirchner R, Greenberg ME: Single-cell transcriptomics of the developing lateral geniculate nucleus reveals insights into circuit assembly and refinement. Proceedings of the National Academy of Sciences 2018:201717871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mi D, Li Z, Lim L, Li M, Moissidis M, Yang Y, Gao T, Hu TX, Pratt T, Price DJ, et al. : Early emergence of cortical interneuron diversity in the mouse embryo. Science 2018, 360:81–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Habib N, Avraham-Davidi I, Basu A, Burks T, Shekhar K, Hofree M, Choudhury SR, Aguet F: Massively parallel single-nucleus RNA-seq with DroNc-seq. 2017, 14:955–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haghverdi L, Lun ATL, Morgan MD, Marioni JC: Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R: Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saito T, Rehmsmeier M: The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PloSone 2015, 10:e0118432–e0118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ching T, Himmelstein DS, Beaulieu-Jones BK, Kalinin AA, Do BT, Way GP, Ferrero E, Agapow P-M, Zietz M, Hoffman MM, et al. : Opportunities and obstacles for deep learning in biology and medicine. Journal of The Royal Society Interface 2018, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hie BL, Bryson B, Berger B: Panoramic stitching of heterogeneous single-cell transcriptomic data. bioRxiv 2018. [Google Scholar]
- 48.Alquicira-Hernandez J, Nguyen Q, Powell JE: scPred: Single cell prediction using singular value decomposition and machine learning classification. bioRxiv 2018. [Google Scholar]
- 49.Welch J, Kozareva V, Ferreira A, Vanderburg C, Martin C, Macosko E: Integrative inference of brain cell similarities and differences from single-cell genomics. bioRxiv 2018. [Google Scholar]
- 50.Barkas N, Petukhov V, Nikolaeva D, Lozinsky Y, Demharter S, Khodosevich K, Kharchenko PV: Wiring together large single-cell RNA-seq sample collections. bioRxiv 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Stoeckius M, Smibert P, Satija R: Comprehensive integration of single cell data. bioRxiv 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paul A, Crow M, Raudales R, He M, Gillis J, Huang ZJ: Transcriptional architecture of synaptic communication delineates GABAergic neuron identity. Cell 2017, 171:522–539. e520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He M, Tucciarone J, Lee S, Nigro MJ, Kim Y, Levine JM, Kelly SM, Krugikov I, Wu P, Chen Y, et al. : Strategies and Tools for Combinatorial Targeting of GABAergic Neurons in Mouse Cerebral Cortex. Neuron 2016, 91:1228–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **54.Hodge RD, Bakken TE, Miller JA, Smith KA, Barkan ER, Graybuck LT, Close JL, Long B, Penn O, Yao Z, et al. : Conserved cell types with divergent features between human and mouse cortex. bioRxiv 2018.This preprint from the Allen Brain Institute describes similarities and differences between cells from the adult human and mouse middle temporal gyrus, cataloguing conservation of multiple cell types, as well as variation in their proportions, laminar distributions, gene expression and morphology between species.
- 55.Caruana R, Niculescu-Mizil A: An empirical comparison of supervised learning algorithms. In Proceedings of the 23rd international conference on Machine learning: ACM: 2006:161–168. [Google Scholar]
- 56.Ascoli GA, Alonso-Nanclares L, Anderson SA, Barrionuevo G, Benavides-Piccione R, Burkhalter A, Buzsaki G, Cauli B, Defelipe J, Fairen A, et al. : Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat Rev Neurosci 2008, 9:557–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *57.Tasic B, Yao Z, Smith KA, Graybuck L, Nguyen TN, Bertagnolli D, Goldy J, Garren E, Economo MN, Viswanathan S, et al. : Shared and distinct transcriptomic cell types across neocortical areas. bioRxiv 2017.An in-depth investigation of adult mouse neocortical cell diversity from researchers at the Allen Brain Institute.
- 58.Fuzik J, Zeisel A, Mate Z, Calvigioni D, Yanagawa Y, Szabo G, Linnarsson S, Harkany T: Integration of electrophysiological recordings with single-cell RNA-seq data identifies neuronal subtypes. Nat Biotechnol 2016, 34:175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cadwell CR, Palasantza A, Jiang X, Berens P, Deng Q, Yilmaz M, Reimer J, Shen S, Bethge M, Tolias KF, et al. : Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat Biotechnol 2016, 34:199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Foldy C, Darmanis S, Aoto J, Malenka RC, Quake SR, Sudhof TC: Single-cell RNAseq reveals cell adhesion molecule profiles in electrophysiologically defined neurons. Proc Natl Acad Sci U S A 2016, 113:E5222–5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Angermueller C, Clark SJ, Lee HJ, Macaulay IC, Teng MJ, Hu TX, Krueger F, Smallwood SA, Ponting CP, Voet T, et al. : Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nature Methods 2016, 13:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Codeluppi S, Borm LE, Zeisel A, La Manno G, van Lunteren JA, Svensson CI, Linnarsson S: Spatial organization of the somatosensory cortex revealed by cyclic smFISH. bioRxiv 2018. [DOI] [PubMed] [Google Scholar]
- 63.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X: Spatially resolved, highly multiplexed RNA profiling in single cells. Science 2015, 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Ferrante TC, Terry R, Turczyk BM, Yang JL, Lee HS, Aach J, et al. : Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nature Protocols 2015, 10:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chevée M, De Jong Robertson J, Cannon GH, Brown SP, Goff LA: Variation in Activity State, Axonal Projection, and Position Define the Transcriptional Identity of Individual Neocortical Projection Neurons. Cell reports 2018, 22:441–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chung S, Weber F, Zhong P, Tan CL, Nguyen TN, Beier KT, Hörmann N, Chang W-C, Zhang Z, Do JP, et al. : Identification of preoptic sleep neurons using retrograde labelling and gene profiling. Nature 2017, 545:477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kebschull JM, Garcia da Silva P, Reid AP, Peikon ID, Albeanu DF, Zador AM: High-Throughput Mapping of Single-Neuron Projections by Sequencing of Barcoded RNA. Neuron 2016, 91:975–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen X, Kebschull JM, Zhan H, Sun Y-C, Zador AM: High-throughput mapping of long-range neuronal projection using in situ sequencing. bioRxiv 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ: Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 2015, 523:486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guo H, Zhu P, Wu X, Li X, Wen L, Tang F: Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome research 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Farlik M, Sheffield Nathan C, Nuzzo A, Datlinger P, Schönegger A, Klughammer J, Bock C: Single-Cell DNA Methylome Sequencing and Bioinformatic Inference of Epigenomic Cell-State Dynamics. Cell Reports 2015, 10:1386–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luo C, Keown CL, Kurihara L, Zhou J, He Y, Li J, Castanon R, Lucero J, Nery JR, Sandoval JP: Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science 2017, 357:600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cao J, Cusanovich DA, Ramani V, Aghamirzaie D, Pliner HA, Hill AJ, Daza RM, McFaline-Figueroa JL, Packer JS, Christiansen L, et al. : Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *74.Lake BB, Chen S, Sos BC, Fan J, Kaeser GE, Yung YC, Duong TE, Gao D, Chun J, Kharchenko PV: Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nature biotechnology 2018, 36:70.The first comparison of single-cell transcriptional and epigenomic profiles from adult human brain.
- 75.Plass M, Solana J, Wolf FA, Ayoub S, Misios A, Glažar P, Obermayer B, Theis FJ, Kocks C, Rajewsky N: Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science 2018. [DOI] [PubMed] [Google Scholar]
- 76.Fincher CT, Wurtzel O, de Hoog T, Kravarik KM, Reddien PW: Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bareinboim E, Pearl J: Causal inference and the data-fusion problem. Proceedings of the National Academy of Sciences 2016, 113:7345–7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dixit A, Parnas O, Li B, Chen J, Fulco CP, Jerby-Arnon L, Marjanovic ND, Dionne D, Burks T, Raychowdhury R, et al. : Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 2016, 167:1853–1866.e1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jaitin DA, Weiner A, Yofe I, Lara-Astiaso D, Keren-Shaul H, David E, Salame TM, Tanay A, van Oudenaarden A, Amit I: Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell 2016, 167:1883–1896.e1815. [DOI] [PubMed] [Google Scholar]
- 80.Datlinger P, Rendeiro AF, Schmidl C, Krausgruber T, Traxler P, Klughammer J, Schuster LC, Kuchler A, Alpar D, Bock C: Pooled CRISPR screening with single-cell transcriptome readout. Nature Methods 2017, 14:297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.La Manno G, Gyllborg D, Codeluppi S, Nishimura K, Salto C, Zeisel A, Borm LE, Stott SRW, Toledo EM, Villaescusa JC, et al. : Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell 2016, 167:566–580.e519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cembrowski MS, Menon V: Continuous Variation within Cell Types of the Nervous System. Trends in Neurosciences 2018, 41:337–348. [DOI] [PubMed] [Google Scholar]