Abstract
While alcohol use disorder (AUD) is a highly heritable psychiatric disease, efforts to elucidate that heritability by examining genetic variation (e.g., single nucleotide polymorphisms) have been insufficient to fully account for familial AUD risk. Perhaps not coincidently, there has been a burgeoning interest in novel non-genomic mechanisms of inheritance (i.e., epigenetics) that are shaped in the male or female germ cells by significant lifetime experiences such as exposure to chronic stress, malnutrition, or drugs of abuse. While many epidemiological and preclinical studies have long pointed to a role for the parental preconception environment in offspring behavior, over the last decade many studies have implicated a causal relationship between the environmentally-sensitive sperm epigenome and intergenerational phenotypes. This critical review will detail the heritable effects of alcohol and the potential role for epigenetics.
Keywords: alcohol, stress, epigenetic inheritance, germline
INTRODUCTION
Alcohol use disorder (AUD) is a mental health disorder with an astounding lifetime prevalence of 29.1 % (Grant et al., 2016), exerting massive societal costs that account for ~1% of the gross national product in high- to middle-income countries (Rehm et al., 2009). As risk for AUD is increased several fold by a family history of alcoholism (Cotton, 1979, Kendler et al., 2015, Morean et al., 2009, Worobec et al., 1990), much of the research to date has focused on elucidating the genetic underpinnings of AUD pathology. However, there is a growing appreciation that epigenetic mechanisms, which govern gene expression without a change in the underlying DNA sequence, are critical at fertilization to drive gene expression patterns and development at the earliest stages of embryonic growth. Unlike the inflexible genome, epigenetic mechanisms are sensitive to environmental perturbations. Indeed, many recent studies have found that alcohol has diverse epigenetic effects in somatic and germinal tissue (Chastain and Sarkar, 2017) which play a causal role in the developmental and adult features of subsequent generations (Finegersh et al., 2015).
Given the long-held belief that only genetic information is passed through the germline, the potential for preconception alcohol exposure to affect offspring health and development has been understudied. Still, there is evidence dating as far back as one hundred years (Stockard and Papanicolaou, 1918) directly implicating parental preconception alcohol exposure as a driver of offspring development. Particularly with the discovery that environmentally-sensitive epigenetic mechanisms in the germline are causally related to cross-generational phenotypes, such studies have now attracted major scientific consideration and public enthusiasm (Yehuda et al., 2018). This intrigue is warranted as the potential for parental preconception alcohol exposure to affect offspring behavior such as alcohol drinking has radical implications for how we appreciate the mechanistic role of family history in AUD risk.
The goal of this critical review is to summarize work examining the effects of preconception alcohol exposure on offspring heath and behavior. While gestational alcohol exposure has wide-ranging effects on child development and behavior, this review will primarily focus on such studies where preconception alcohol exposure impacts subsequent generations (i.e., the effects are transmitted through the germline) (Govorko et al., 2012, Nizhnikov et al., 2016). Notably, as preconception maternal exposures may have lasting effects on in utero conditions and the quality of maternal care (requiring embryo transfer and/or cross-fostering to elucidate the germline vs somatic origin of offspring phenotypes), the majority of studies examining cross-generational effects of preconception alcohol exposure have utilized male subjects. Therefore, this review will focus primarily on findings following paternal exposure, although maternal preconception studies are considered. Overall, this review will emphasize the causal role of alcohol in the transmission of complex behavioral phenotypes across generations, such as alcohol drinking, and how these effects are likely attributable to reprogramming of epigenetic mechanisms in the germline.
THE MISSING HERITABILITY OF ALCOHOL USE DISORDER
Individual risk of developing most psychiatric disorders, including AUD, is widely conceptualized as the product of gene × environment interactions (Meaney, 2017). That is, both heredity (i.e., familial factors presumed to be primarily genetic) and experiences throughout the lifetime (i.e., environmental factors) are considered to best define at risk populations for disease. For the clinical pathologist, the genetic component of the risk equation has been especially intriguing, given the implications for biomarker discovery and targeted disease treatment and prevention. Collectively, twin and adoption studies estimate that AUD is ~50% heritable (Prescott and Kendler, 1999, Young-Wolff et al., 2011, Ystrom et al., 2011). Thus, to examine the genetic component of AUD, investigators throughout the world have widely employed genome wide association studies (GWAS) to survey diverse populations for genetic marks -- commonly single nucleotide polymorphisms (SNPs) -- that predict alcohol-related phenotypic variation (e.g., AUD diagnosis).
The most well-recognized studies linking genetic variants and AUD risk, implicate genes directly involved in alcohol metabolism -- alcohol dehydrogenase (ADH) and acetaldehyde dehydrogenase (ALDH) enzymes (Bierut et al., 2012, Birley et al., 2008, Higuchi et al., 1995, Li et al., 2012). Identified SNPs on ADH1B and ALDH2 are associated with impaired clearance of acetaldehyde, the toxic byproduct of alcohol metabolism, and reduced AUD risk (Cederbaum, 2012), suggesting a function for alcohol metabolism-related genetic variants in AUD heritability. Nevertheless, although these risk variants are common in Asian populations, they are rarely identified in populations of central European origin (Bach et al., 2017). In addition, such reproducible and robust findings have been the few exceptions in an otherwise conflicted field of study. The results of many GWAS efforts often fail to replicate across studies (Treutlein and Rietschel, 2011, Bierut et al., 2010). Moreover, none of the SNPs identified are estimated to account for more than 0.1% of AUD heritability (Heath et al., 2011). Such outcomes are not unique to AUD. For example, Crohn’s disease is estimated to have 80% heritability, but the additive effect of all SNPs associated with the disease account for only ~20% of that estimate (Park et al., 2010). This common discrepancy between estimated heritability of disease predicted by familial vs genetic variation, is referred to as the “missing” heritability problem.
There are numerous factors that may help explain missing AUD heritability. For example, the innumerable variables that comprise the environmental component of the G x E interaction (e.g., cultural diversity, drug availability in addiction, early-life stress, environmental toxins, microbiota), as well as gender and age all severely compound the challenge of replication across heterogeneous sample populations (Ober and Vercelli, 2011). Moreover, psychiatric disorders feature complex symptomology comprised of multiple “intermediate” phenotypes (e.g., alcohol-induced body sway) that further challenge efforts to discover a unifying genetic signature for AUD (Blanco-Gomez et al., 2016). Finally, in addition to all of these plausible explanations for missing heritability, there is growing evidence to suggest that a portion of the heritability of complex phenotypes may result from parental preconception experience.
HERITABILITY OF THE PRECONCEPTION ENVIRONMENT
The genetic theory of evolution has been the unifying standard for modern biology. Harmonious with Charles Darwin’s theory of natural selection, genetic theory posits that, for a given environment, success or fitness of the species will be determined by selective pressure for phenotypic traits that are genomic in origin (Orr, 2005). By this principle, the ancestral or parental environment has no targeted mechanism to influence the phenotype in subsequent generations. However, even preceding Darwin’s On the Origin of Species, the French biologist Jean-Baptiste Lamarck described a more flexible theory of heredity, positing that the ancestral environment is the primary driver of phenotypic variation in posterity (Lamarck, 1802). While long-rejected in favor of a singular genetic-basis for heredity, over the past twenty years, a surfeit of clinical and preclinical evidence has reignited interest in Lamarckian theory and a potential complimentary role for the parental environment in heredity (Skinner, 2015).
Nongenomic inheritance of a given phenotype is described as intergenerational if it is imparted from father (i.e., the F0 generation) to offspring (i.e., the F1 generation). If the given phenotype is observed in the F2 or subsequent generations, it is described as transgenerational. In the context of germline inheritance, the transgenerational terminology indicates that the given phenotype was transferred through germ cells never harbored by the exposed F0 generation. Thus, for females, as the F0 female will harbor both the F1 fetus and the primordial germ cells from which the F2 generation will spawn, transgenerational effects are reserved for the F3 generation and beyond. This review will focus largely on mechanisms of intergenerational germline inheritance, though transgenerational effects will also be discussed.
Studies examining effects of ancestral environmental exposures require careful monitoring of subjects across multiple generations and are therefore difficult to conduct on human populations. Nonetheless, some of the earliest human studies reporting multigenerational effects stem from the 1944–1945 Dutch Famine Cohort. These studies revealed that men exposed to famine during prenatal development were more likely to have offspring with increased body weight and more adiposity in adulthood (Veenendaal et al., 2013, Painter et al., 2008). Other early study focused on the remote Överkalix population of northern Sweden which maintained cross-generational historical records of harvest and food supply throughout the 20th century. The Överkalix study found that the food supply of paternal grandparents was inversely related to male and female longevity (Bygren et al., 2001, Pembrey et al., 2006). Additionally, fathers that smoked prior to puberty were more likely to have sons with increased body-mass-index (Pembrey et al., 2006). More recent findings from Rachel Yehuda’s group demonstrate intergenerational effects of parental stress. Both paternal and maternal post-traumatic stress disorder (PTSD) --from traumatic episodes preceding conception-- were associated with reduced basal cortisol levels and greater dexamethasone-suppression of cortisol in adult offspring (Lehrner et al., 2014, Yehuda et al., 2007). In another study, Yehuda et al. found that offspring of women that survived the Holocaust prior to conception were at increased risk for the development of PTSD, depression, and anxiety disorders (Yehuda et al., 2008). Furthermore, a new study found that male descendants of civil war prisoners of war exhibited increased mortality (Costa et al., 2018). Finally, a family history of alcoholism is associated with altered inheritance of a unique epigenetic signature at two putative oncogenes (Hill et al., 2017).
Despite these intriguing studies demonstrating effects of parental lifetime exposures on offspring health and raising the possibility of epigenetic inheritance manifesting in the human population, they cannot rule out a genetic contribution based on potential pre-existing genetic variation in the sample population. Additionally, maternal in utero effects and parental investment in child development top a long list of potential confounding variables, making a germline-specific mechanism extremely difficult to discern. Thus, to test the hypotheses inspired by cross-generational epidemiological data, preclinical investigators have utilized isogenic rodent strains under controlled laboratory conditions to directly test the effects of various paternal preconception exposures to a vast range of environmental insults on an equally expansive number of biological and behavioral measures in offspring. Most prominently featuring studies pertaining to dietary change, chronic stress and various drugs of abuse, many paternal preconception exposures with rodents have now been found to directly impart complex physiological and behavioral phenotypes to offspring (for excellent reviews, see: Rando and Simmons, 2015, Chan et al., 2017b, Goldberg and Gould, 2018). For instance, obese fathers confer deficits in glucose metabolism to offspring (Chen et al., 2016a, Cropley et al., 2016, de Castro Barbosa et al., 2016, Fullston et al., 2013). Furthermore, numerous paternal chronic stress paradigms reshape physiological and behavioral stress vulnerability across generations (Rodgers et al., 2013, Dietz et al., 2011, Gapp et al., 2014, Short et al., 2016) and paternal cocaine exposure alters cocaine preference and hippocampal-dependent memory in male offspring (Vassoler et al., 2013, Le et al., 2017, Wimmer et al., 2017). Other paternal conditions with cross-generational effects in rodents include exposures to environmental toxicants such as bisphenol A (Fan et al., 2018) as well as exposures to beneficial conditions such as enriched housing (Yeshurun et al., 2017) or free access to a running wheel (Short et al., 2017). As rodent sires are not involved in offspring gestation or rearing in these studies, the contribution of the father to offspring development is likely restricted to the germline. While factors such as sire “fitness” can influence maternal care for offspring through social interaction during breeding (Mashoodh et al., 2012), several paternal exposure studies have partially or fully validated the germline origin of intergenerational effects using in vitro fertilization experiments (Dias and Ressler, 2014, Chen et al., 2016a, Sharma et al., 2016, Dietz et al., 2011, Huypens et al., 2016).
HERITABILITY OF PRECONCEPTION ALCOHOL EXPOSURE
To date, there have been over forty published paternal preconception alcohol exposure studies in rodents (for review, see: Finegersh et al., 2015), most varying in species, route of administration, and duration. Despite these differences, some intergenerational (F1) effects have been consistent such as low fetal and birth weight (Bielawski et al., 2002, Ledig et al., 1998, Chang et al., 2017, Chang et al., 2019), altered organ weights (Abel, 1993b, Ledig et al., 1998, Lee et al., 2013, Chang et al., 2017), and increased number of runts (Bielawski et al., 2002, Bielawski and Abel, 1997). Additional phenotypes reported include increased cortical thickness (Jamerson et al., 2004), reduced testosterone (Abel and Lee, 1988), and altered neurotransmitter levels (Nelson et al., 1988). Furthermore, several intergenerational behavioral alterations have been reported including reduced spatiotemporal learning (Wozniak et al., 1991), increased anxiety- and impulsivity-like phenotypes (Kim et al., 2014, Liang et al., 2014), and increased sensitivity to amphetamine-induced hyperlocomotion (Abel, 1993a). Overall, these preclinical studies affirm that paternal alcohol exposure can confer fetal alcohol syndrome-like defects, molecular and physiological alterations, and behavioral phenotypes to offspring.
Most of the above referenced studies were conducted from the perspective of the paternal contribution to fetal alcohol syndrome-like phenotypes. Surprisingly, until recently, no published studies had examined the effect of paternal alcohol exposure on offspring alcohol drinking or alcohol sensitivity-related phenotypes. In 2014, Finegersh and Homanics started to fill this gap in the literature, by examining a battery of alcohol-related behaviors in the offspring of sires that were exposed to chronic alcohol exposure. They discovered that C57BL/6J (B6) adult male mice exposed intermittently to alcohol vapor over five weeks (average blood alcohol concentration ~160 mg/dL following each 8 hour exposure) sired hybrid B6 × Strain 129 adult male offspring with increased sensitivity to the anxiolytic effects of a low dose alcohol injection and decreased alcohol drinking preference and consumption (Finegersh and Homanics, 2014). Those key findings were later replicated with mice on a pure B6 background (Rompala et al., 2017). Using a chronic alcohol liquid diet exposure, another study reported a bidirectional effect of paternal alcohol exposure on conditioned place preference (CPP) for alcohol in male offspring depending on the paternal alcohol exposure dosage (i.e., increased CPP at a low dose (0.5 g/kg), decreased CPP at a high dose (1.5 mg/kg)) (Ceccanti et al., 2016). Another study examining cross-generational alcohol drinking behaviors found that paternal binge-alcohol exposure over eight days increased alcohol intake by intraoral infusion in postnatal offspring (Hollander et al., 2018). Finally, female mice exposed to alcohol during postnatal development exhibited reduced sensitivity to alcohol-induced loss of righting reflex and passed this phenotype to male and female offspring (Popoola et al., 2017). Intriguingly, in three of these studies, the cross-generational alcohol-related phenotypes coincided with altered BDNF signaling in reward-related brain regions (Ceccanti et al., 2016, Finegersh and Homanics, 2014, Rompala et al., 2017). In addition, a recent study found that like alcohol, paternal cocaine reduced cocaine seeking behavior in male offspring and the phenotype was reversible with a BDNF antagonist (Vassoler et al., 2013). This suggests that the BDNF locus may be an attractive target for examining mechanistic effects of paternal alcohol exposure on offspring neurobiology across reward-related brain regions. Taken together, these novel studies suggest that paternal preconception alcohol exposure is a heritable factor capable of driving alcohol-related phenotypes in the next generation.
Given the complexity of factors driving alcohol drinking in humans relative to rodents, more work needs to be done with rodent models to more fully characterize how paternal preconception alcohol affects the offspring alcohol drinking phenotype. Illustrating this point, while it was initially reported that chronic paternal cocaine self-administration reduced cocaine preference in male offspring, another study found that sires selected for high cocaine motivation imparted increased cocaine preference to offspring (Le et al., 2017). Moreover, paternal cocaine exposure had divergent effects on offspring cocaine preference depending on whether the cocaine exposure was voluntarily (self-administered) or forced (Le et al., 2017). Thus, the results from the handful of studies to date are insufficient and more cross-generational alcohol exposure paradigms need to be evaluated to better understand their translational potential. Likewise, as no studies to date have employed next generation sequencing techniques (e.g., RNA-seq, ATAC-seq) to examine the effects of paternal alcohol on offspring brain, such experiments are needed for an unbiased characterization of transcriptomic and epigenetic mechanisms potentially underlying intergenerational alcohol-related behaviors.
CROSS-GENERATIONAL RELATIONSHIP BETWEEN ALCOHOL AND STRESS
Many studies have characterized the positive feedback loop propelling the relationship between stress and alcohol (i.e., stress promotes chronic alcohol abuse and chronic alcohol abuse enhances vulnerability to subsequent stressors) (see Koob et al., 2014 and Becker et al., 2011 for excellent reviews). Now, several studies indicate a unique relationship between alcohol and stress stretching across generations. For instance, alcohol exposure paradigms such as parental adolescent injections (Przybycien-Szymanska et al., 2014), maternal preconception gavage (Jabbar et al., 2016) and paternal exposure during prenatal development (Govorko et al., 2012) were found to alter hypothalamic-pituitary-adrenal (HPA) axis function in offspring. These studies are notable in light of the extensive literature showing that a family history of alcoholism is associated with impaired HPA axis function (Stephens and Wand, 2012).
One recent investigation carried out by our lab found that male offspring of sires exposed to preconception alcohol exhibit blunted stress responsivity (Rompala et al., 2016). Interestingly, this blunted HPA axis phenotype in the F1 generation has also been observed in response to paternal preconception chronic stress exposure (Rodgers et al., 2013). Similarly, we have found that both paternal preconception alcohol and paternal preconception stress reduce alcohol drinking selectively in males (Rompala et al., 2018b, Finegersh and Homanics, 2014), thus suggesting that paternal alcohol and stress act similarly to influence the alcohol drinking and stress phenotypes of the next generation. Indeed, alcohol acts directly on the HPA axis, acutely increasing corticosterone/cortisol levels in rodents (Rivier, 2014) and humans (Valimaki et al., 1984, Mendelson and Stein, 1966), indicating a likely shared mechanism of the paternal alcohol and stress exposures. Notably, glucocorticoid receptors are expressed throughout the male reproductive tract (Silva et al., 2010, Schultz et al., 1993) and may be a shared somatic mechanism to influence epigenetic factors in the germline (Chan et al., 2018).
GERMLINE EPIGENETIC MECHANISMS
As many of the studies demonstrating an intergenerational effect of alcohol exposure were carried out with males which do not contribute to offspring fetal and postnatal development, the effects are likely transmitted through the germline. This section will review the heritable mechanisms implicated in non-genomic inheritance of complex behavior phenotypes such as alcohol drinking behavior.
Epigenetic processes are broadly defined as the molecular factors that drive gene expression while not altering the primary DNA nucleotide sequence. Among the most well-understood epigenetic mechanisms are cytosine methylation, chromatin modifications, and noncoding RNAs. Methylation of DNA at a cytosine nucleotide that is followed by a guanine nucleotide (CpG) within gene promoters is associated with transcriptional silencing by local recruitment of heterochromatic proteins that impede transcription factor binding (Illingworth et al., 2008). Post-translational chromatin modifications regulate the affinity of positively-charged histone proteins for the phosphate-rich DNA, in turn influencing gene expression by determining whether the chromatin is “open” or “closed” to transcription (Smith and Shilatifard, 2010). Finally, noncoding RNAs function by regulating diverse transcriptional and translational processes, often targeting messenger RNAs with sequence homology (Cech and Steitz, 2014). Collectively, epigenetic mechanisms are the primary regulators of transcription and underlie the cellular diversity able to emerge from a single genome. This section will focus primarily on the unique epigenomic state of the male germline (see Figure 1 for overview of sperm epigenome).
Figure 1. Mechanisms of the sperm epigenome.
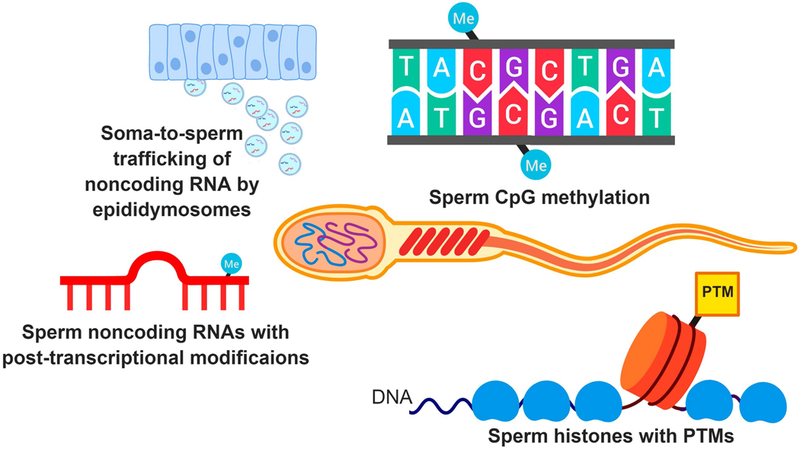
Sperm feature various epigenetic mechanisms, most notably DNA methylation, histone post-translational modifications (PTMs) and noncoding RNAs. The high levels of DNA methylation in sperm are primarily localized to cytosine phosphate guanine (CpG) dinucleotides. While many of the histones in sperm are exchanged for protamines, a small percentage of histones are maintained and contain PTMs. Finally, sperm contain diverse noncoding RNA types such as miRNAs and tDRs with accompanying post-transcriptional modifications. Many of the sperm noncoding RNAs are derived from extracellular vesicles secreted by epididymal somatic cells (i.e., epididymosomes).
Given the specialized function of the male germline in fertilization, sperm feature a unique nuclear structure. Most histones are replaced in the early stages of spermatogenesis with highly basic protamines that robustly neutralize the phosphate bond-rich DNA. The nucleus is further compressed by inter- and intra-molecular disulfide bonds, contributing to a condensed state at approximately 1/13 the volume of the oocyte nucleus (Martins and Krawetz, 2007). In this heavily neutralized state, and lacking major ribosomal machinery, mature sperm cells exhibit minimal transcriptional activity (Gur and Breitbart, 2006). Upon fertilization, most of the sperm genome is stripped of epigenetic marks to facilitate pluripotency in early embryo development (Feng et al., 2010). Thus, until recently, the male gametes were presumed to deliver little to no epigenetic memory to the fertilized oocyte. However, with the advancement of next generation sequencing technology allowing for enhanced resolution in methylation, chromatin modification, and noncoding RNA analysis, studies are beginning to characterize a unique, environmentally-responsive, and functional epigenetic landscape in sperm.
Indeed, not all genomic loci are stripped of DNA methylation at fertilization. For instance, large regions around intracisternal A particle (IAP) retrotransposons (Popp et al., 2010) and imprinting regions (Feng et al., 2010) are protected from global epigenetic reprogramming during embryogenesis. Additionally, only a small percentage of histones are retained in sperm (~1% in mouse, ~10% in humans) as the majority are exchanged for protamines during spermatogenesis to facilitate DNA condensation and stabilization (Bogliotti and Ross, 2012). Retained histones are concentrated at promoter regions of key genes for embryo development, thereby well-positioned to influence initial zygotic gene expression (Brykczynska et al., 2010, Hammoud et al., 2009). Moreover, adult exposure to the environmental toxin DDT or gestational exposure to vinclozolin lead to ectopic histone retention sites in sperm, showing that environmental exposures can influence germline histone displacement mechanisms (Skinner et al., 2018, Ben Maamar et al., 2018).
In addition to chromatin, sperm contain RNA, though only ~ 1% of that carried by somatic cells (Zhang et al., 2017). Due to the largely arrested transcriptional state of fully mature sperm, it was long assumed that this limited amount of RNA in sperm was purely comprised of degraded transcripts from the earlier transcriptionally-active stages of spermatogenesis in the testis. However, the discovery that sperm RNAs are delivered to the oocyte at fertilization raised the possibility that they could be functional epigenetic molecules in the early embryo (Ostermeier et al., 2004). Supporting this notion, subsequent deep sequencing studies have helped characterize a diverse and unique population of intact, functional noncoding RNA species in sperm. The majority of RNA species identified in sperm are “small” noncoding RNA (18–40 nucleotides), mainly microRNA (miRNA), transfer RNA-derived small RNAs (tDR), and piwi-interacting RNAs (piRNA) (Chen et al., 2016b). The miRNAs are the most well-studied small noncoding RNA with a putative function in mRNA silencing via binding to homologous “seed” regions of the 3’-UTR of target transcripts (Cai et al., 2009). The function of tDR is not well-understood in sperm though they have been implicated in various biological functions including transcriptional regulation (Chen et al., 2016a), miRNA-like mRNA silencing (Haussecker et al., 2010), and translational inhibition (Yamasaki et al., 2009). The piRNAs regulate expression of transposable elements during spermatogenesis and are the dominant noncoding RNA species in immature testis spermatozoa (Ernst et al., 2017). Finally, sperm also contain long noncoding RNAs (>200 nucleotides) involved in diverse transcriptional regulatory processes and additional novel RNA species will likely continue to be identified in the coming years. Overall, while the precise mechanism of these noncoding RNAs either in mature sperm, or upon delivery to the oocyte, remains to be determined, multiple studies have report an essential role for sperm RNA in early embryonic development (Yuan et al., 2016, Liu et al., 2012, Conine et al., 2018).
EFFECTS OF ENVIRONMENTAL INSULTS ON EPIGENETIC MECHANISMS IN SPERM
The earliest evidence of intergenerational/transgenerational phenotype associated with environmentally-induced epigenetic mechanisms in sperm showed that transient exposure of pregnant rats to endocrine disruptors (e.g., vinclozolin) imparts reduced fertility to males across multiple generations through the male germline (Anway et al., 2005, Nilsson and Skinner, 2015). This phenomenon was associated with altered DNA methylation patterns in sperm (Anway et al., 2005, Nilsson and Skinner, 2015) and those effects have more recently been found to expand to histone retention and altered small RNA content (Ben Maamar et al., 2018, Schuster et al., 2016).
Additional studies have found that the male germline is sensitive to environmentally-induced changes in DNA methylation well beyond prenatal development. For instance, adult-onset prediabetic conditions (Wei et al., 2014), obesity (Fullston et al., 2013), and cocaine-seeking motivation (Le et al., 2017) all modify the DNA methylome in sperm. One recent study reported similar changes in DNA methylation in sperm between tetrahydrocannabinol (THC)-treated rats and cannabis-smoking adults (Murphy et al., 2018). Other studies have reported effects of cocaine and stress on histone modifications in developing or mature sperm at specific-gene loci that coincided with altered expression of the same gene in adult offspring brain, suggesting cross-generational epigenetic memory (Vassoler et al., 2013, Dias and Ressler, 2014). Finally, various paternal preconception environmental exposures directly affect the noncoding RNA milieu in sperm (Rodgers et al., 2013, Fullston et al., 2013, Gapp et al., 2014, Chen et al., 2016a, Sharma et al., 2016, Short et al., 2017). In humans, alterations in sperm small noncoding RNAs have been associated with obesity (Donkin et al., 2016), early life stress (Dickson et al., 2018), and smoking history (Marczylo et al., 2012). Aside from sperm RNA abundance, there is increasing interest in post-transcriptional RNA modifications (i.e., the epitranscriptome) as some of these modifications on sperm RNA were found to be sensitive to paternal diet (Chen et al., 2016a) and deletion of the RNA methyltransferase gene DNMT2 is necessary for an established model of RNA-mediated inheritance of glucose intolerance from paternal high fat diet (Zhang et al., 2018). Interestingly, dissimilar to DNA methylation and histone modifications in sperm, which are hypothesized to be vulnerable to environmental exposures in the testis preceding or during spermatogenesis, the sperm RNA profile is dynamically shaped in the epididymis during final maturation (Nixon et al., 2015). Whether the sperm RNA profile is especially sensitive to environmental exposures during epididymal transit remains to be tested.
While experimental methods for loci-specific manipulation of DNA methylation or histone modifications in the germline have yet to be developed (although the relevant technology is progressing rapidly (Kungulovski and Jeltsch, 2016)), techniques have been established for directly testing the role of sperm RNA in established animal models of environmentally-induced intergenerational inheritance. Using 1-cell embryos, isolated sperm RNA can be directly injected into the fertilized oocyte to examine resultant progeny and determine if sperm RNA is sufficient to recapitulate the cross-generational phenotypes in question. To date, there have already been at least six studies using this method to draw a causal link between environmentally-responsive sperm RNAs and heritable phenotypes (see Table 1)(Rodgers et al., 2015, Gapp et al., 2014, Benito et al., 2018, Grandjean et al., 2015, Gapp et al., 2018, Chen et al., 2016a). Beginning in 2014, Isabella Mansuy and colleagues found that the intergenerational effects of maternal separation stress could be partially recapitulated in mice derived from one cell embryos from control, unstressed parents that were injected with total sperm RNA from stressed males (Gapp et al., 2014). Subsequently, Tracey Bale and colleagues discovered that the intergenerational effect of paternal chronic stress on stress responsivity in offspring could be recapitulated in mice derived from normal embryos injected with synthetic oligos for nine stress-enriched sperm miRNAs (Rodgers et al., 2015). Another study examining the intergenerational effects of paternal high fat diet found that sperm tDR could reproduce the diet-induced intergenerational effects on glucose tolerance in the next generation (Chen et al., 2016a). Finally, a similarly important role for sperm long noncoding RNAs was discovered in a model of postnatal trauma (Gapp and Bohacek, 2018). Collectively, this evidence supports a functional role for sperm-derived RNA in the early embryo and epigenetic inheritance. However, in each of these studies, the specific causal embryonic mechanism of the altered sperm RNA remains unclear. In Rodgers et al., stress-increased sperm miRNAs were found to reduce expression of canonical mRNA targets (Rodgers et al., 2015). In addition, one paternal diet-sensitive tRNA fragment (tRNA-Glu-CTC) was discovered to be essential for suppression of retrotransposon-dependent zygotic gene expression (Sharma et al., 2016). Given the pluripotent state of the early embryo, it is conceivable that transient sperm RNA-induced alterations in zygotic transcription may offset other mechanisms such as de novo DNA methylation to ultimately program a stable epigenetic state in the developing offspring (Greenberg et al., 2017).
Table 1:
Studies where paternal exposure-induced alterations in sperm RNA coincide with intergenerational offspring phenotypes
| Paternal Exposure | Offspring Phonotype | Altered sperm noncoding RNA types and/or species | Citation | |
|---|---|---|---|---|
| DIET | low protein diet | impaired hepatic cholesterol biosynthesis | ↓ let7c, ↑ tRNA-Gly-GCC | Sharma et al. 2016 |
| offspring of obese fathers | glucose intolerance | ↑miR10a, ↓ tRNA-Glu-CTC | Cropley et al. 2016 | |
| high fat diet | glucose intolerance*, ↑ weight* | altered tDR, altered RNA modifications | Chen et al. 2016 | |
| obesity | ↑obesity, ↑insulin resistance | 11 altered miRNA | Fullston et al. 2013 | |
| western diet | ↑obesity* | ↑ miR19b | Grandjean et al. 2015 | |
| high fat diet | ↑weight, ↓glucose metabolism | ↑ let7c across both F0, F1 generations | de Castro Barbosa et al. 2016 | |
| obesity | ↑obesity, ↓fertility | altered long noncoding RNAs, mRNA | An et al. 2017 | |
| STRESS | chronic variable stress | ↓HPA responsivity* | 9 increased miRNAs | Rodgers et al. 2013 |
| maternal separation stress | ↓anxiety-like*, ↑depression-like behaviors* | several altered miRNA, piRNA, long noncoding RNA | Gapp et al. 2014, Gapp et al. 2018 | |
| chronic glucocorticoid treatment | ↑ anxiety-like behavior, ↓fear extinction | altered miRNAs | Short et al. 2016 | |
| chronic social instability | ↑anxiety-like behavior, ↓socialization | ↓ miR34c, ↓ miR449 | Saavenra et al. 2015, Dickson et al. 2018 | |
| ENRICHMENT | chronic voluntary exercise | ↓ anxiety-like behavior, ↓ fear reinstatement | altered miR19b, miR455, miR133a, tRNA-Gly-GCC | Short et al. 2016 |
| environmental enrichment | ↑ hippocampal-dependent learning* | ↑ miRNA 212/132 | Benito et al. 2018 | |
| TOXICANTS | ancestral vinclozolin exposure | ↓ fertility | piRNAs most affected | Ben Maamar et al. 2018, Schuster et al. 2016 |
| DTT exposure | obesity | piRNA, tRNA most affected | Skinner et al. 2015, Skinner et al. 2018 | |
| ALCOHOL | chronic intermittent alcohol | ↓alcohol drinking, ↑ alcohol sensitivity, ↓HPA responsivity | ↑miR10a, ↑ tRNA-Glu-CTC, two altered RNA modifications | Finegersh et al. 2014, Rompala et al. 2016, Rompala et al. 2018 |
Summary of rodent studies demonstrating heritable effects of paternal preconception exposures associated with epigenetic reprogramming of sperm noncoding RNA. Legend: mt-tRNA = mitochondrial tRNA.
= offspring phenotype can be recapitulated by zygotic injection of altered sperm RNA.
It does not appear that the effects of a direct environmental exposure on sperm RNA are maintained in the unexposed sperm of male offspring. Both paternal stress and high fat diet studies have reported inconsistent sperm RNA profiles between F0 and F1 generations (Gapp et al., 2014, Fullston et al., 2016). Conversely, alcohol exposure during gestation and adult cocaine addiction have been found to induce similar effects on gene promoter cytosine methylation in F0 and F1 sperm (Govorko et al., 2012, Le et al., 2017). How environmentally-induced changes in sperm methylation may escape reprogramming or undergo reestablishment in the germline after global demethylation remains unclear and an intriguing area for future study.
EFFECTS OF ALCOHOL ON EPIGNETIC MECHANISMS IN SPERM
Alcohol reduces sperm count (Rahimipour et al., 2013), circulating testosterone levels (Widenius et al., 1989, Kucheria et al., 1985), and overall fertility (Anderson et al., 1983) in rodents and similar effects on reproductive health have been reported in alcoholic men (reviewed in La Vignera et al., 2013). In addition to these deleterious effects of alcohol on reproductive health, numerous studies support a significant effect of alcohol on the epigenetic milieu in sperm.
DNA Methylation
As alcohol acts directly on DNA methylation machinery, reducing levels of the cytosine methyltransferase DNMT1 (Ponomarev et al., 2012) and S-Adenosyl-methionine (SAM) in somatic cells (Lu and Mato, 2005), many studies have investigated whether alcohol similarly affects DNA methylation in the male germline. Indeed, DNMT1 is reduced in sperm of animals undergoing intragastric chronic alcohol exposure (Bielawski et al., 2002). Moreover, several studies have found that DNA methylation at imprinted gene loci is reduced in sperm of chronic alcohol-treated mice (Knezovich and Ramsay, 2012, Finegersh and Homanics, 2014, Liang et al., 2014) as well as men with alcohol use disorder (Ouko et al., 2009). Recently, Finegersh and Homanics found that paternal preconception alcohol exposure reduced methylation of the BDNF promoter in sperm which coincided with increased BDNF in adult VTA of male offspring, thus implicating the BDNF locus in the cross-generational effects of alcohol (Finegersh and Homanics, 2014). In addition to these effects demonstrating vulnerability of the adult sperm methylome to alcohol, animals exposed to prenatal alcohol exhibited increased CpG methylation at the pro-opiomelanocortin (POMC) gene promoter in sperm that is associated with reduced POMC expression and suppression of its HPA-axis mediating function in offspring (Govorko et al., 2012). Not all results support methylation changes in response to ethanol, as one recent study found that paternal voluntary alcohol consumption, which altered cholesterol trafficking and hepatic fibrosis markers in offspring, had no effect on DNA methylation in sperm (Chang et al., 2017). In addition, despite a number of studies demonstrating effects of environmental perturbations on methylation in sperm, the effect sizes of significant changes in methylation at individual cytosines are often small (e.g., ~10–20 %) and thereby the significance for cross-generational inheritance is hard to discern, given the high penetrance observed in many preclinical paternal epigenetic inheritance models (Shea et al., 2015). Overall, emerging techniques that allow for loci-specific manipulation of epigenetic marks such as methylation will be critical for further examining the role of methylation in epigenetic inheritance.
Chromatin Modifications
While several studies have found effects of alcohol on histone post-transcriptional modifications in somatic tissue such as the liver or brain (see Berkel and Pandey, 2017 for an excellent review), only one study has directly examined the effect of alcohol on chromatin modifications in sperm. Chronic exposure to liquid alcohol (10% v/v) was found to impair chromatin condensation in mice (Rahimipour et al., 2013). Few studies to date have examined the effect of environmental insults on the chromatin profile with sequencing resolution in sperm, likely due to the minimal histone content retained following spermatogenesis. One recent study used novel next generation sequencing techniques to examine chromatin accessibility and conformation (e.g., ATAC-seq, Hi-C) in sperm, revealed a unique euchromatic landscape that was reflected in the developing embryo (Jung et al., 2017). However, another study found that studies such as Jung et al. using swim-up based sperm purification methods are likely to capture a significant fraction of infertile sperm characterized by incomplete histone to protamine transition (Yoshida et al., 2018). Nonetheless, as the unique sperm chromatin profile continues to be detailed with enhanced resolution, it will be essential to examine the effects of alcohol on these emerging epigenetic mechanisms.
Noncoding RNAs
To date, there has been one study examining the effects of alcohol exposure on small noncoding RNA in sperm. Chronic intermittent alcohol exposure, the same exposure found to reduce alcohol drinking and blunt stress responsivity in male offspring (Finegersh and Homanics, 2014, Rompala et al., 2016), was found to alter several miRNA and tDR species in sperm (Rompala et al., 2018a). Shared predicted mRNA targets of alcohol-responsive miRNAs were enriched for transcriptional regulators including some with defined roles in embryonic development. Prominently among the small RNAs increased in sperm by alcohol was miR10a, one of the few microRNAs found to be significantly enriched in the sperm relative to the oocyte,(Yang et al., 2016) and a putative post-transcriptional regulator of the alcohol-related gene BDNF (Jiajie et al., 2017). Among alcohol-responsive sperm tDR, tRNA-Glu-CTC was prominent as, along with tRNA-Gly-GCC, it is one of two extremely enriched small RNAs in mammalian sperm, together comprising more than half of the small RNA content (Peng et al., 2012). Supporting the importance of these sperm tDR, tRNA-Gly-GCC is necessary for controlling retrotransposon-regulated gene networks in the fertilized oocyte (Sharma et al., 2016). Moreover, in silico analysis revealed that tRNA-Glu-CTC is highly enriched for overall mRNA targets relative to other tDR species (Rompala et al., 2018a). Interestingly, both miR10a and tDR-Glu-CTC were prominently altered in F1 sons of obese fathers that impart glucose intolerance to the F2 male descendants (Cropley et al., 2016). Thus, interrogating the casual role of these and other alcohol-responsive small RNAs in intergenerational inheritance has great potential to unveil novel heritable mechanisms driving alcohol-related behavior.
Notably, the sperm RNA profile is shaped during epididymal transit via extracellular vesicles (i.e, epididymosomes) (Reilly et al., 2016, Sharma et al., 2016, Sharma et al., 2018). Indeed, the effects of alcohol on mature sperm were reflected in epididymosomes for some tDR species (Rompala et al., 2018a), suggesting a potential effect of alcohol on soma-to-germline RNA trafficking. Given the evidence implicating epididymosomes as the source of major RNA species in mature sperm, there is a fast-growing interest in characterizing gonadal and neuroendocrine regulation of epididymosome function. For instance, testosterone levels mediate epididymosomal protein trafficking to the sperm membrane (Suryawanshi et al., 2012) and emerging studies support such a role for glucocorticoids in RNA trafficking to sperm (Chan et al., 2018). Future studies should further investigate how adaptive changes in epididymal gene expression affect epididymosome RNA cargo sorting and delivery to sperm. One powerful tool for such experiments was recently introduced allowing for epididymal-synthesized RNAs to be tagged in vivo and then detected in sperm RNA using next generation sequencing (Sharma et al., 2018). This methodology can be used to examine the effects of various environmental perturbations directly on soma-to-germline RNA trafficking.
In addition to small RNA abundance, two post-transcriptional RNA modifications were altered in sperm by chronic alcohol exposure (Rompala et al., 2018a). These modifications were specific to mitochondrial tRNAs and could affect expression of novel small mitochondrial RNA species (Ro et al., 2013) as well as sperm mitochondrial function. Notably, while Rompala et al. examined twenty-two RNA modifications, there have been over one hundred and fifty identified to date (Boccaletto et al., 2018). Moreover, Rompala et al. examined only the 30–40 nucleotide fraction of small RNAs, excluding all miRNA species (~21–23 nucleotides). Given that RNA modifications underlie small RNA stability (Chen et al., 2016a) and are essential to models of RNA-mediated inheritance (Kiani et al., 2013), future studies will need to fully characterize the effects of alcohol on the epitranscriptome.
Moving forward: linking alcohol-responsive germline epigenetics with alcohol drinking in offspring
Future studies will need to examine the causal role of alcohol-induced changes to the sperm epigenome on paternal alcohol-imparted offspring behavior. As novel genome editing tools are developed for applying locus-specific CpG methylation and histone modifications in the germline, the many cited effects of paternal environmental exposures and alcohol on the sperm chromatin will need to be tested directly. Conversely, there has been significant progress in examining the causal role of sperm RNA, as several studies examining the effects of paternal diet and paternal stress have directly tested whether injecting 1-cell embryos with altered paternal sperm RNA is sufficient to recapitulate intergenerational phenotypes in the resulting progeny. While this may not be a physiological manipulation (and does not establish necessity of the affected RNAs), it would be a critical step in establishing a role for sperm RNAs in driving the paternal effects. Studies targeting specific RNAs with mimics and/or inhibitors could also be useful, although RNA mimics were found to be sensitive to rapid degradation and lack potentially crucial post-transcriptional modifications (Chen et al., 2016a). Alternatively, given that extracellular vesicles are emerging as the likely mode of RNA biogenesis in epididymal sperm and that epididymosome RNAs can be trafficked to developing sperm in vitro (Sharma et al., 2018), the causal role of epididymosomal RNA in cross-generational inheritance remains to be studied. Finally, as novel candidate epigenetic marks emerge, clinical studies examining relevant population of subjects with alcohol abuse and AUD will be needed to define the translational relevance of current and forthcoming preclinical findings.
SUMMARY
Overall, the last decade has generated a wealth of evidence supporting inheritance driven by preconception environmental perturbations such as chronic alcohol exposure. While there has long been evidence for preconception alcohol exposures inducing fetal alcohol syndrome-like growth abnormalities, more recent studies have extended these findings to include intergenerational alcohol-related behaviors and the ability of alcohol to exert cross-generational effects similar to chronic stress. Finally, studies have begun to delineate genomic loci and RNAs in sperm that are sensitive to alcohol and associated with cross-generational effects (see Figure 2). All in all, these studies highlight the need to continue investigating the role of epigenetics to fully appreciate the heritability of AUD. Given the prevalence and societal impact of alcoholism, the effects of heavy alcohol exposure (or other stressors) on cross-generational alcohol drinking behavior have major implications for predicting familial risk for alcohol use disorder and development of novel disease prevention strategies.
Figure 2. Summary of evidence for paternal preconception alcohol exposure imparting alcohol-related behaviors to offspring.
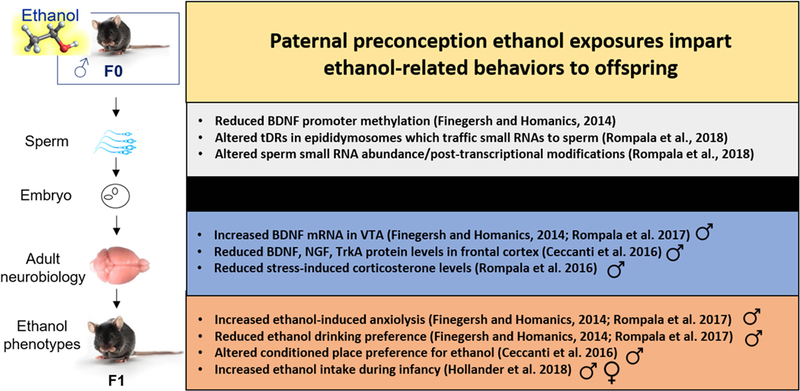
Studies investigating the intergenerational effects of paternal preconception alcohol have found effects of alcohol on the germline epigenome associated with alterations to neurobiology and alcohol-related behavior in adult male offspring.
Acknowledgments
Funding support: NIH/NIAAA AA010422, AA020889 and AA024670
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
REFERENCES
- Abel EL (1993a) Paternal alcohol exposure and hyperactivity in rat offspring: effects of amphetamine. Neurotoxicol Teratol 15:445–449. [DOI] [PubMed] [Google Scholar]
- Abel EL (1993b) Rat offspring sired by males treated with alcohol. Alcohol 10:237–242. [DOI] [PubMed] [Google Scholar]
- Abel EL, Lee JA (1988) Paternal alcohol exposure affects offspring behavior but not body or organ weights in mice. Alcohol Clin Exp Res 12:349–355. [DOI] [PubMed] [Google Scholar]
- Anderson RA Jr., Willis BR, Oswald C, Zaneveld LJ (1983) Ethanol-induced male infertility: impairment of spermatozoa. J Pharmacol Exp Ther 225:479–486. [PubMed] [Google Scholar]
- Anway MD, Cupp AS, Uzumcu M, Skinner MK (2005) Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 308:1466–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach P, Zois E, Vollstadt-Klein S, Kirsch M, Hoffmann S, Jorde A, Frank J, Charlet K, Treutlein J, Beck A, Heinz A, Walter H, Rietschel M, Kiefer F (2017) Association of the alcohol dehydrogenase gene polymorphism rs1789891 with gray matter brain volume, alcohol consumption, alcohol craving and relapse risk. Addict Biol [DOI] [PubMed]
- Ben Maamar M, Sadler-Riggleman I, Beck D, McBirney M, Nilsson E, Klukovich R, Xie Y, Tang C, Yan W, Skinner MK (2018) Alterations in sperm DNA methylation, non-coding RNA expression, and histone retention mediate vinclozolin-induced epigenetic transgenerational inheritance of disease. Environ Epigenet 4:dvy010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito E, Kerimoglu C, Ramachandran B, Pena-Centeno T, Jain G, Stilling RM, Islam MR, Capece V, Zhou Q, Edbauer D, Dean C, Fischer A (2018) RNA-Dependent Intergenerational Inheritance of Enhanced Synaptic Plasticity after Environmental Enrichment. Cell Rep 23:546–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkel TD, Pandey SC (2017) Emerging Role of Epigenetic Mechanisms in Alcohol Addiction. Alcohol Clin Exp Res 41:666–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielawski DM, Abel EL (1997) Acute treatment of paternal alcohol exposure produces malformations in offspring. Alcohol 14:397–401. [DOI] [PubMed] [Google Scholar]
- Bielawski DM, Zaher FM, Svinarich DM, Abel EL (2002) Paternal alcohol exposure affects sperm cytosine methyltransferase messenger RNA levels. Alcohol Clin Exp Res 26:347–351. [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, Hinrichs AL, Almasy L, Breslau N, Culverhouse RC, Dick DM, Edenberg HJ, Foroud T, Grucza RA, Hatsukami D, Hesselbrock V, Johnson EO, Kramer J, Krueger RF, Kuperman S, Lynskey M, Mann K, Neuman RJ, Nothen MM, Nurnberger JI Jr., Porjesz B, Ridinger M, Saccone NL, Saccone SF, Schuckit MA, Tischfield JA, Wang JC, Rietschel M, Goate AM, Rice JP, Gene EASC (2010) A genome-wide association study of alcohol dependence. Proc Natl Acad Sci U S A 107:5082–5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Goate AM, Breslau N, Johnson EO, Bertelsen S, Fox L, Agrawal A, Bucholz KK, Grucza R, Hesselbrock V, Kramer J, Kuperman S, Nurnberger J, Porjesz B, Saccone NL, Schuckit M, Tischfield J, Wang JC, Foroud T, Rice JP, Edenberg HJ (2012) ADH1B is associated with alcohol dependence and alcohol consumption in populations of European and African ancestry. Mol Psychiatry 17:445–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birley AJ, James MR, Dickson PA, Montgomery GW, Heath AC, Whitfield JB, Martin NG (2008) Association of the gastric alcohol dehydrogenase gene ADH7 with variation in alcohol metabolism. Hum Mol Genet 17:179–189. [DOI] [PubMed] [Google Scholar]
- Blanco-Gomez A, Castillo-Lluva S, Del Mar Saez-Freire M, Hontecillas-Prieto L, Mao JH, Castellanos-Martin A, Perez-Losada J (2016) Missing heritability of complex diseases: Enlightenment by genetic variants from intermediate phenotypes. Bioessays 38:664–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crecy-Lagard V, Ross R, Limbach PA, Kotter A, Helm M, Bujnicki JM (2018) MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 46:D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogliotti YS, Ross PJ (2012) Mechanisms of histone H3 lysine 27 trimethylation remodeling during early mammalian development. Epigenetics 7:976–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schubeler D, Stadler MB, Peters AHFM (2010) Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nature Structural & Molecular Biology 17:679–U647. [DOI] [PubMed] [Google Scholar]
- Bygren LO, Kaati G, Edvinsson S (2001) Longevity determined by paternal ancestors’ nutrition during their slow growth period. Acta Biotheor 49:53–59. [DOI] [PubMed] [Google Scholar]
- Cai Y, Yu X, Hu S, Yu J (2009) A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics 7:147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccanti M, Coccurello R, Carito V, Ciafre S, Ferraguti G, Giacovazzo G, Mancinelli R, Tirassa P, Chaldakov GN, Pascale E, Ceccanti M, Codazzo C, Fiore M (2016) Paternal alcohol exposure in mice alters brain NGF and BDNF and increases ethanol-elicited preference in male offspring. Addict Biol 21:776–787. [DOI] [PubMed] [Google Scholar]
- Cech TR, Steitz JA (2014) The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157:77–94. [DOI] [PubMed] [Google Scholar]
- Cederbaum AI (2012) Alcohol Metabolism. Clin Liver Dis 16:667–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J, Nugent B, Morrison K, Jasarevic E, Bhanu N, Garcia B, Bale T (2018) Epididymal glucocorticoid receptors promote intergenerational transmission of paternal stress. bioRxiv
- Chang RC, Skiles WM, Sarah SC, Wang H, Sutton GI, Bedi YS, Snyder M, Long CR, Golding MC (2017) DNA Methylation-Independent Growth Restriction and Altered Developmental Programming in a Mouse Model of Preconception Male Alcohol Exposure. Epigenetics: 12(10): 841–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang RC, Wang H, Bedi Y, Golding MC (2019) Preconception paternal alcohol exposure exerts sex-specific effects on offspring growth and long-term metabolic programming. Epigenetics Chromatin 12:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastain LG, Sarkar DK (2017) Alcohol effects on the epigenome in the germline: Role in the inheritance of alcohol-related pathology. Alcohol 60:53–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng GH, Peng H, Zhang X, Zhang Y, Qian J, Duan E, Zhai Q, Zhou Q (2016a) Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351:397–400. [DOI] [PubMed] [Google Scholar]
- Chen Q, Yan W, Duan E (2016b) Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nat Rev Genet 17:733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine CC, Sun F, Song L, Rivera-Perez JA, Rando OJ (2018) Small RNAs Gained during Epididymal Transit of Sperm Are Essential for Embryonic Development in Mice. Dev Cell 46:470–480 e473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa DL, Yetter N, DeSomer H (2018) Intergenerational transmission of paternal trauma among US Civil War ex-POWs. Proc Natl Acad Sci U S A 115:11215–11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton NS (1979) The familial incidence of alcoholism: a review. J Stud Alcohol 40:89–116. [DOI] [PubMed] [Google Scholar]
- Cropley JE, Eaton SA, Aiken A, Young PE, Giannoulatou E, Ho JW, Buckland ME, Keam SP, Hutvagner G, Humphreys DT, Langley KG, Henstridge DC, Martin DI, Febbraio MA, Suter CM (2016) Male-lineage transmission of an acquired metabolic phenotype induced by grand-paternal obesity. Mol Metab 5:699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro Barbosa T, Ingerslev LR, Alm PS, Versteyhe S, Massart J, Rasmussen M, Donkin I, Sjogren R, Mudry JM, Vetterli L, Gupta S, Krook A, Zierath JR, Barres R (2016) High-fat diet reprograms the epigenome of rat spermatozoa and transgenerationally affects metabolism of the offspring. Mol Metab 5:184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias BG, Ressler KJ (2014) Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat Neurosci 17:89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson DA, Paulus JK, Mensah V, Lem J, Saavedra-Rodriguez L, Gentry A, Pagidas K, Feig LA (2018) Reduced levels of miRNAs 449 and 34 in sperm of mice and men exposed to early life stress. Transl Psychiatry 8:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz DM, Laplant Q, Watts EL, Hodes GE, Russo SJ, Feng J, Oosting RS, Vialou V, Nestler EJ (2011) Paternal transmission of stress-induced pathologies. Biol Psychiatry 70:408–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkin I, Versteyhe S, Ingerslev LR, Qian K, Mechta M, Nordkap L, Mortensen B, Appel EV, Jorgensen N, Kristiansen VB, Hansen T, Workman CT, Zierath JR, Barres R (2016) Obesity and Bariatric Surgery Drive Epigenetic Variation of Spermatozoa in Humans. Cell Metab 23:369–378. [DOI] [PubMed] [Google Scholar]
- Ernst C, Odom DT, Kutter C (2017) The emergence of piRNAs against transposon invasion to preserve mammalian genome integrity. Nat Commun 8:1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Tian C, Liu Q, Zhen X, Zhang H, Zhou L, Li T, Zhang Y, Ding S, He D, Jin X, Liu J, Zhang B, Wu N, Manyande A, Zhu M (2018) Preconception paternal bisphenol A exposure induces sex-specific anxiety and depression behaviors in adult rats. PLoS One 13:e0192434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Jacobsen SE, Reik W (2010) Epigenetic reprogramming in plant and animal development. Science 330:622–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegersh Homanics GE (2014) Paternal alcohol exposure reduces alcohol drinking and increases behavioral sensitivity to alcohol selectively in male offspring. PLoS One 9:e99078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegersh A, Rompala GR, Martin DI, Homanics GE (2015) Drinking beyond a lifetime: New and emerging insights into paternal alcohol exposure on subsequent generations. Alcohol 49:461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullston T, Ohlsson-Teague EM, Print CG, Sandeman LY, Lane M (2016) Sperm microRNA Content Is Altered in a Mouse Model of Male Obesity, but the Same Suite of microRNAs Are Not Altered in Offspring’s Sperm. PLoS One 11:e0166076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullston T, Ohlsson Teague EM, Palmer NO, DeBlasio MJ, Mitchell M, Corbett M, Print CG, Owens JA, Lane M (2013) Paternal obesity initiates metabolic disturbances in two generations of mice with incomplete penetrance to the F2 generation and alters the transcriptional profile of testis and sperm microRNA content. FASEB J 27:4226–4243. [DOI] [PubMed] [Google Scholar]
- Gapp K, Bohacek J (2018) Epigenetic germline inheritance in mammals: looking to the past to understand the future. Genes Brain Behav 17:e12407. [DOI] [PubMed] [Google Scholar]
- Gapp K, Jawaid A, Sarkies P, Bohacek J, Pelczar P, Prados J, Farinelli L, Miska E, Mansuy IM (2014) Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci 17:667–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapp K, van Steenwyk G, Germain PL, Matsushima W, Rudolph KLM, Manuella F, Roszkowski M, Vernaz G, Ghosh T, Pelczar P, Mansuy IM, Miska EA (2018) Alterations in sperm long RNA contribute to the epigenetic inheritance of the effects of postnatal trauma. Mol Psychiatry [DOI] [PMC free article] [PubMed]
- Govorko D, Bekdash RA, Zhang C, Sarkar DK (2012) Male germline transmits fetal alcohol adverse effect on hypothalamic proopiomelanocortin gene across generations. Biol Psychiatry 72:378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean V, Fourre S, De Abreu DA, Derieppe MA, Remy JJ, Rassoulzadegan M (2015) RNA-mediated paternal heredity of diet-induced obesity and metabolic disorders. Sci Rep 5:18193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BF, Saha TD, Ruan WJ, Goldstein RB, Chou SP, Jung J, Zhang H, Smith SM, Pickering RP, Huang B, Hasin DS (2016) Epidemiology of DSM-5 Drug Use Disorder: Results From the National Epidemiologic Survey on Alcohol and Related Conditions-III. JAMA Psychiatry 73:39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg MV, Glaser J, Borsos M, Marjou FE, Walter M, Teissandier A, Bourc’his D (2017) Transient transcription in the early embryo sets an epigenetic state that programs postnatal growth. Nat Genet 49:110–118. [DOI] [PubMed] [Google Scholar]
- Gur Y, Breitbart H (2006) Mammalian sperm translate nuclear-encoded proteins by mitochondrial-type ribosomes. Genes Dev 20:411–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang HY, Purwar J, Carrell DT, Cairns BR (2009) Distinctive chromatin in human sperm packages genes for embryo development. Nature 460:473–U447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA (2010) Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 16:673–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Whitfield JB, Martin NG, Pergadia ML, Goate AM, Lind PA, McEvoy BP, Schrage AJ, Grant JD, Chou YL, Zhu R, Henders AK, Medland SE, Gordon SD, Nelson EC, Agrawal A, Nyholt DR, Bucholz KK, Madden PAF, Montgomery GW (2011) A Quantitative-Trait Genome-Wide Association Study of Alcoholism Risk in the Community: Findings and Implications. Biol Psychiat 70:513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi S, Matsushita S, Murayama M, Takagi S, Hayashida M (1995) Alcohol and aldehyde dehydrogenase polymorphisms and the risk for alcoholism. Am J Psychiatry 152:1219–1221. [DOI] [PubMed] [Google Scholar]
- Hill SY, Rompala G, Homanics GE, Zezza N (2017) Cross-generational effects of alcohol dependence in humans on HRAS and TP53 methylation in offspring. Epigenomics 9:1189–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander J, McNivens M, Pautassi RM, Nizhnikov ME (2018) Offspring of Male Rats Exposed to Binge Alcohol Exhibit Heightened Ethanol Intakeat Infancy and Alterations in T-Maze Performance. Alcohol [DOI] [PMC free article] [PubMed]
- Huypens P, Sass S, Wu M, Dyckhoff D, Tschop M, Theis F, Marschall S, Hrabe de Angelis M, Beckers J (2016) Epigenetic germline inheritance of diet-induced obesity and insulin resistance. Nat Genet 48:497–499. [DOI] [PubMed] [Google Scholar]
- Illingworth R, Kerr A, Desousa D, Jorgensen H, Ellis P, Stalker J, Jackson D, Clee C, Plumb R, Rogers J, Humphray S, Cox T, Langford C, Bird A (2008) A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol 6:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbar S, Chastain LG, Gangisetty O, Cabrera MA, Sochacki K, Sarkar DK (2016) Preconception Alcohol Increases Offspring Vulnerability to Stress. Neuropsychopharmacology 41:2782–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamerson PA, Wulser MJ, Kimler BF (2004) Neurobehavioral effects in rat pups whose sires were exposed to alcohol. Brain Res Dev Brain Res 149:103–111. [DOI] [PubMed] [Google Scholar]
- Jiajie T, Yanzhou Y, Hoi-Hung AC, Zi-Jiang C, Wai-Yee C (2017) Conserved miR-10 family represses proliferation and induces apoptosis in ovarian granulosa cells. Sci Rep 7:41304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung YH, Sauria MEG, Lyu X, Cheema MS, Ausio J, Taylor J, Corces VG (2017) Chromatin States in Mouse Sperm Correlate with Embryonic and Adult Regulatory Landscapes. Cell Rep 18:1366–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Edwards A, Myers J, Cho SB, Adkins A, Dick D (2015) The predictive power of family history measures of alcohol and drug problems and internalizing disorders in a college population. Am J Med Genet B Neuropsychiatr Genet 168B:337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiani J, Grandjean V, Liebers R, Tuorto F, Ghanbarian H, Lyko F, Cuzin F, Rassoulzadegan M (2013) RNA-mediated epigenetic heredity requires the cytosine methyltransferase Dnmt2. PLoS Genet 9:e1003498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P, Choi CS, Park JH, Joo SH, Kim SY, Ko HM, Kim KC, Jeon SJ, Park SH, Han SH, Ryu JH, Cheong JH, Han JY, Ko KN, Shin CY (2014) Chronic exposure to ethanol of male mice before mating produces attention deficit hyperactivity disorder-like phenotype along with epigenetic dysregulation of dopamine transporter expression in mouse offspring. J Neurosci Res 92:658–670. [DOI] [PubMed] [Google Scholar]
- Knezovich JG, Ramsay M (2012) The effect of preconception paternal alcohol exposure on epigenetic remodeling of the h19 and rasgrf1 imprinting control regions in mouse offspring. Front Genet 3:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucheria K, Saxena R, Mohan D (1985) Semen analysis in alcohol dependence syndrome. Andrologia 17:558–563. [DOI] [PubMed] [Google Scholar]
- Kungulovski G, Jeltsch A (2016) Epigenome Editing: State of the Art, Concepts, and Perspectives. Trends Genet 32:101–113. [DOI] [PubMed] [Google Scholar]
- La Vignera S, Condorelli RA, Balercia G, Vicari E, Calogero AE (2013) Does alcohol have any effect on male reproductive function? A review of literature. Asian J Androl 15:221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarck J (1802) Recherches sur l’organisation des corps vivans Paris: Chez L’auteur, Maillard. [Google Scholar]
- Le QM, Yan B, Yu XC, Li YQ, Song HK, Zhu HW, Hou WQ, Ma DGL, Wu FZ, Zhou YQ, Ma L (2017) Drug-seeking motivation level in male rats determines offspring susceptibility or resistance to cocaine-seeking behaviour. Nat Commun 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledig M, Misslin R, Vogel E, Holownia A, Copin JC, Tholey G (1998) Paternal alcohol exposure: developmental and behavioral effects on the offspring of rats. Neuropharmacology 37:57–66. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Ryu JS, Choi NY, Park YS, Kim YI, Han DW, Ko K, Shin CY, Hwang HS, Kang KS, Ko K (2013) Transgenerational effects of paternal alcohol exposure in mouse offspring. Anim Cells Syst 17:429–434. [Google Scholar]
- Lehrner A, Bierer LM, Passarelli V, Pratchett LC, Flory JD, Bader HN, Harris IR, Bedi A, Daskalakis NP, Makotkine I, Yehuda R (2014) Maternal PTSD associates with greater glucocorticoid sensitivity in offspring of Holocaust survivors. Psychoneuroendocrinology 40:213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhao H, Gelernter J (2012) Strong protective effect of the aldehyde dehydrogenase gene (ALDH2) 504lys (*2) allele against alcoholism and alcohol-induced medical diseases in Asians. Hum Genet 131:725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Diao L, Liu J, Jiang N, Zhang J, Wang H, Zhou W, Huang G, Ma D (2014) Paternal ethanol exposure and behavioral abnormities in offspring: associated alterations in imprinted gene methylation. Neuropharmacology 81:126–133. [DOI] [PubMed] [Google Scholar]
- Liu WM, Pang RT, Chiu PC, Wong BP, Lao K, Lee KF, Yeung WS (2012) Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proc Natl Acad Sci U S A 109:490–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SC, Mato JM (2005) Role of methionine adenosyltransferase and S-adenosylmethionine in alcohol-associated liver cancer. Alcohol 35:227–234. [DOI] [PubMed] [Google Scholar]
- Marczylo EL, Amoako AA, Konje JC, Gant TW, Marczylo TH (2012) Smoking induces differential miRNA expression in human spermatozoa: a potential transgenerational epigenetic concern? Epigenetics 7:432–439. [DOI] [PubMed] [Google Scholar]
- Martins RP, Krawetz SA (2007) Nuclear organization of the protamine locus. Soc Reprod Fertil Suppl 64:1–12. [DOI] [PubMed] [Google Scholar]
- Mashoodh R, Franks B, Curley JP, Champagne FA (2012) Paternal social enrichment effects on maternal behavior and offspring growth. Proc Natl Acad Sci U S A 109 Suppl 2:17232–17238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaney MJ (2017) Epigenetics and the Biology of Gene × Environment Interactions, in Gene-Environment Transactions in Developmental Psychopathology: The Role in Intervention Research, Gene-Environment Transactions in Developmental Psychopathology: The Role in Intervention Research (TOLAN PH, LEVENTHAL BL eds), pp 59–94, Springer International Publishing, Cham. [Google Scholar]
- Mendelson JH, Stein S (1966) Serum Cortisol Levels in Alcoholic and Nonalcoholic Subjects During Experimentally Induced Ethanol Intoxication. Psychosom Med 28:616–626. [DOI] [PubMed] [Google Scholar]
- Morean ME, Corbin WR, Sinha R, O’Malley SS (2009) Parental history of anxiety and alcohol-use disorders and alcohol expectancies as predictors of alcohol-related problems. J Stud Alcohol Drugs 70:227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SK, Itchon-Ramos N, Visco Z, Huang Z, Grenier C, Schrott R, Acharya K, Boudreau MH, Price TM, Raburn DJ, Corcoran DL, Lucas JE, Mitchell JT, McClernon FJ, Cauley M, Hall BJ, Levin ED, Kollins SH (2018) Cannabinoid exposure and altered DNA methylation in rat and human sperm. Epigenetics [DOI] [PMC free article] [PubMed]
- Nelson BK, Brightwell WS, MacKenzie-Taylor DR, Burg JR, Massari VJ (1988) Neurochemical, but not behavioral, deviations in the offspring of rats following prenatal or paternal inhalation exposure to ethanol. Neurotoxicol Teratol 10:15–22. [DOI] [PubMed] [Google Scholar]
- Nilsson EE, Skinner MK (2015) Environmentally Induced Epigenetic Transgenerational Inheritance of Reproductive Disease. Biol Reprod 93:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon B, Stanger SJ, Mihalas BP, Reilly JN, Anderson AL, Tyagi S, Holt JE, McLaughlin EA (2015) The microRNA signature of mouse spermatozoa is substantially modified during epididymal maturation. Biol Reprod 93:91. [DOI] [PubMed] [Google Scholar]
- Nizhnikov ME, Popoola DO, Cameron NM (2016) Transgenerational Transmission of the Effect of Gestational Ethanol Exposure on Ethanol Use-Related Behavior. Alcohol Clin Exp Res 40:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober C, Vercelli D (2011) Gene-environment interactions in human disease: nuisance or opportunity? Trends Genet 27:107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA (2005) The genetic theory of adaptation: a brief history. Nat Rev Genet 6:119–127. [DOI] [PubMed] [Google Scholar]
- Ostermeier GC, Miller D, Huntriss JD, Diamond MP, Krawetz SA (2004) Reproductive biology: delivering spermatozoan RNA to the oocyte. Nature 429:154. [DOI] [PubMed] [Google Scholar]
- Ouko LA, Shantikumar K, Knezovich J, Haycock P, Schnugh DJ, Ramsay M (2009) Effect of alcohol consumption on CpG methylation in the differentially methylated regions of H19 and IG-DMR in male gametes: implications for fetal alcohol spectrum disorders. Alcohol Clin Exp Res 33:1615–1627. [DOI] [PubMed] [Google Scholar]
- Painter RC, Osmond C, Gluckman P, Hanson M, Phillips DI, Roseboom TJ (2008) Transgenerational effects of prenatal exposure to the Dutch famine on neonatal adiposity and health in later life. BJOG 115:1243–1249. [DOI] [PubMed] [Google Scholar]
- Park JH, Wacholder S, Gail MH, Peters U, Jacobs KB, Chanock SJ, Chatterjee N (2010) Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet 42:570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pembrey ME, Bygren LO, Kaati G, Edvinsson S, Northstone K, Sjostrom M, Golding J, Team AS (2006) Sex-specific, male-line transgenerational responses in humans. Eur J Hum Genet 14:159–166. [DOI] [PubMed] [Google Scholar]
- Peng H, Shi J, Zhang Y, Zhang H, Liao S, Li W, Lei L, Han C, Ning L, Cao Y, Zhou Q, Chen Q, Duan E (2012) A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res 22:1609–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD (2012) Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci 32:1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popoola DO, Nizhnikov ME, Cameron NM (2017) Strain-specific programming of prenatal ethanol exposure across generations. Alcohol 60:191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W (2010) Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 463:1101–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott CA, Kendler KS (1999) Genetic and environmental contributions to alcohol abuse and dependence in a population-based sample of male twins. Am J Psychiatry 156:34–40. [DOI] [PubMed] [Google Scholar]
- Przybycien-Szymanska MM, Rao YS, Prins SA, Pak TR (2014) Parental binge alcohol abuse alters F1 generation hypothalamic gene expression in the absence of direct fetal alcohol exposure. PLoS One 9:e89320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimipour M, Talebi AR, Anvari M, Sarcheshmeh AA, Omidi M (2013) Effects of different doses of ethanol on sperm parameters, chromatin structure and apoptosis in adult mice. Eur J Obstet Gynecol Reprod Biol 170:423–428. [DOI] [PubMed] [Google Scholar]
- Rehm J, Mathers C, Popova S, Thavorncharoensap M, Teerawattananon Y, Patra J (2009) Alcohol and Global Health 1 Global burden of disease and injury and economic cost attributable to alcohol use and alcohol-use disorders. Lancet 373:2223–2233. [DOI] [PubMed] [Google Scholar]
- Reilly JN, McLaughlin EA, Stanger SJ, Anderson AL, Hutcheon K, Church K, Mihalas BP, Tyagi S, Holt JE, Eamens AL, Nixon B (2016) Characterisation of mouse epididymosomes reveals a complex profile of microRNAs and a potential mechanism for modification of the sperm epigenome. Sci Rep 6:31794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivier C (2014) Role of hypothalamic corticotropin-releasing factor in mediating alcohol-induced activation of the rat hypothalamic-pituitary-adrenal axis. Front Neuroendocrinol 35:221–233. [DOI] [PubMed] [Google Scholar]
- Ro S, Ma HY, Park C, Ortogero N, Song R, Hennig GW, Zheng H, Lin YM, Moro L, Hsieh JT, Yan W (2013) The mitochondrial genome encodes abundant small noncoding RNAs. Cell Res 23:759–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers AB, Morgan CP, Bronson SL, Revello S, Bale TL (2013) Paternal stress exposure alters sperm microRNA content and reprograms offspring HPA stress axis regulation. J Neurosci 33:9003–9012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers AB, Morgan CP, Leu NA, Bale TL (2015) Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proc Natl Acad Sci U S A 112:13699–13704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompala GR, Finegersh A, Homanics GE (2016) Paternal preconception ethanol exposure blunts hypothalamic-pituitary-adrenal axis responsivity and stress-induced excessive fluid intake in male mice. Alcohol 53:19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompala GR, Finegersh A, Slater M, Homanics GE (2017) Paternal preconception alcohol exposure imparts intergenerational alcohol-related behaviors to male offspring on a pure C57BL/6J background. Alcohol 60:169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompala GR, Mounier A, Wolfe CM, Lin Q, Lefterov I, Homanics GE (2018a) Heavy Chronic Intermittent Ethanol Exposure Alters Small Noncoding RNAs in Mouse Sperm and Epididymosomes. Front Genet 9:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompala GR, Simons A, Kihle B, Homanics GE (2018b) Paternal Preconception Chronic Variable Stress Confers Attenuated Ethanol Drinking Behavior Selectively to Male Offspring in a Pre-Stress Environment Dependent Manner. Front Behav Neurosci 12:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz R, Isola J, Parvinen M, Honkaniemi J, Wikstrom AC, Gustafsson JA, Pelto-Huikko M (1993) Localization of the glucocorticoid receptor in testis and accessory sexual organs of male rat. Mol Cell Endocrinol 95:115–120. [DOI] [PubMed] [Google Scholar]
- Schuster A, Skinner MK, Yan W (2016) Ancestral vinclozolin exposure alters the epigenetic transgenerational inheritance of sperm small noncoding RNAs. Environ Epigenet 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F, Song L, Carone BR, Ricci EP, Li XZ, Fauquier L, Moore MJ, Sullivan R, Mello CC, Garber M, Rando OJ (2016) Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351:391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U, Sun F, Conine CC, Reichholf B, Kukreja S, Herzog VA, Ameres SL, Rando OJ (2018) Small RNAs Are Trafficked from the Epididymis to Developing Mammalian Sperm. Dev Cell 46:481–494 e486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea JM, Serra RW, Carone BR, Shulha HP, Kucukural A, Ziller MJ, Vallaster MP, Gu H, Tapper AR, Gardner PD, Meissner A, Garber M, Rando OJ (2015) Genetic and Epigenetic Variation, but Not Diet, Shape the Sperm Methylome. Dev Cell 35:750–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short AK, Fennell KA, Perreau VM, Fox A, O’Bryan MK, Kim JH, Bredy TW, Pang TY, Hannan AJ (2016) Elevated paternal glucocorticoid exposure alters the small noncoding RNA profile in sperm and modifies anxiety and depressive phenotypes in the offspring. Transl Psychiatry 6:e837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short AK, Yeshurun S, Powell R, Perreau VM, Fox A, Kim JH, Pang TY, Hannan AJ (2017) Exercise alters mouse sperm small noncoding RNAs and induces a transgenerational modification of male offspring conditioned fear and anxiety. Transl Psychiatry 7:e1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva EJ, Queiroz DB, Honda L, Avellar MC (2010) Glucocorticoid receptor in the rat epididymis: expression, cellular distribution and regulation by steroid hormones. Mol Cell Endocrinol 325:64–77. [DOI] [PubMed] [Google Scholar]
- Skinner MK (2015) Environmental Epigenetics and a Unified Theory of the Molecular Aspects of Evolution: A Neo-Lamarckian Concept that Facilitates Neo-Darwinian Evolution. Genome Biol Evol 7:1296–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Ben Maamar M, Sadler-Riggleman I, Beck D, Nilsson E, McBirney M, Klukovich R, Xie Y, Tang C, Yan W (2018) Alterations in sperm DNA methylation, non-coding RNA and histone retention associate with DDT-induced epigenetic transgenerational inheritance of disease. Epigenetics Chromatin 11:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Shilatifard A (2010) The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol Cell 40:689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens MA, Wand G (2012) Stress and the HPA axis: role of glucocorticoids in alcohol dependence. Alcohol Res 34:468–483. [PMC free article] [PubMed] [Google Scholar]
- Stockard CR, Papanicolaou GN (1918) Further studies on the modification of the germ‐cells in mammals: The effect of alcohol on treated guinea‐pigs and their descendants. Journal of Experimental Zoology 26:119–126. [Google Scholar]
- Suryawanshi AR, Khan SA, Joshi CS, Khole VV (2012) Epididymosome-mediated acquisition of MMSDH, an androgen-dependent and developmentally regulated epididymal sperm protein. J Androl 33:963–974. [DOI] [PubMed] [Google Scholar]
- Treutlein J, Rietschel M (2011) Genome-wide association studies of alcohol dependence and substance use disorders. Curr Psychiatry Rep 13:147–155. [DOI] [PubMed] [Google Scholar]
- Valimaki MJ, Harkonen M, Eriksson CJ, Ylikahri RH (1984) Sex hormones and adrenocortical steroids in men acutely intoxicated with ethanol. Alcohol 1:89–93. [DOI] [PubMed] [Google Scholar]
- Vassoler FM, White SL, Schmidt HD, Sadri-Vakili G, Pierce RC (2013) Epigenetic inheritance of a cocaine-resistance phenotype. Nat Neurosci 16:42–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenendaal MV, Painter RC, de Rooij SR, Bossuyt PM, van der Post JA, Gluckman PD, Hanson MA, Roseboom TJ (2013) Transgenerational effects of prenatal exposure to the 1944–45 Dutch famine. BJOG 120:548–553. [DOI] [PubMed] [Google Scholar]
- Wei Y, Yang CR, Wei YP, Zhao ZA, Hou Y, Schatten H, Sun QY (2014) Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proc Natl Acad Sci U S A 111:1873–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widenius TV, Eriksson CJ, Ylikahri RH, Harkonen M (1989) Inhibition of testosterone synthesis by ethanol: role of luteinizing hormone. Alcohol 6:241–244. [DOI] [PubMed] [Google Scholar]
- Wimmer ME, Briand LA, Fant B, Guercio LA, Arreola AC, Schmidt HD, Sidoli S, Han Y, Garcia BA, Pierce RC (2017) Paternal cocaine taking elicits epigenetic remodeling and memory deficits in male progeny. Mol Psychiatry 22:1653. [DOI] [PubMed] [Google Scholar]
- Worobec TG, Turner WM, O’Farrell TJ, Cutter HS, Bayog RD, Tsuang MT (1990) Alcohol use by alcoholics with and without a history of parental alcoholism. Alcohol Clin Exp Res 14:887–892. [DOI] [PubMed] [Google Scholar]
- Wozniak DF, Cicero TJ, Kettinger L 3rd, Meyer ER (1991) Paternal alcohol consumption in the rat impairs spatial learning performance in male offspring. Psychopharmacology (Berl) 105:289–302. [DOI] [PubMed] [Google Scholar]
- Yamasaki S, Ivanov P, Hu GF, Anderson P (2009) Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 185:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Lin J, Liu M, Li R, Tian B, Zhang X, Xu B, Liu M, Zhang X, Li Y, Shi H, Wu L (2016) Highly sensitive sequencing reveals dynamic modifications and activities of small RNAs in mouse oocytes and early embryos. Sci Adv 2:e1501482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehuda R, Bell A, Bierer LM, Schmeidler J (2008) Maternal, not paternal, PTSD is related to increased risk for PTSD in offspring of Holocaust survivors. J Psychiatr Res 42:1104–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehuda R, Lehrner A, Bierer LM (2018) The public reception of putative epigenetic mechanisms in the transgenerational effects of trauma. Environ Epigenet 4:dvy018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehuda R, Teicher MH, Seckl JR, Grossman RA, Morris A, Bierer LM (2007) Parental posttraumatic stress disorder as a vulnerability factor for low cortisol trait in offspring of holocaust survivors. Arch Gen Psychiatry 64:1040–1048. [DOI] [PubMed] [Google Scholar]
- Yeshurun S, Short AK, Bredy TW, Pang TY, Hannan AJ (2017) Paternal environmental enrichment transgenerationally alters affective behavioral and neuroendocrine phenotypes. Psychoneuroendocrinology 77:225–235. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Muratani M, Araki H, Miura F, Suzuki T, Dohmae N, Katou Y, Shirahige K, Ito T, Ishii S (2018) Mapping of histone-binding sites in histone replacement-completed spermatozoa. Nat Commun 9:3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young-Wolff KC, Enoch MA, Prescott CA (2011) The influence of gene-environment interactions on alcohol consumption and alcohol use disorders: A comprehensive review. Clin Psychol Rev 31:800–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ystrom E, Reichborn-Kjennerud T, Aggen SH, Kendler KS (2011) Alcohol Dependence in Men: Reliability and Heritability. Alcoholism-Clinical and Experimental Research 35:1716–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Schuster A, Tang C, Yu T, Ortogero N, Bao J, Zheng H, Yan W (2016) Sperm-borne miRNAs and endo-siRNAs are important for fertilization and preimplantation embryonic development. Development 143:635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Gao F, Fu J, Zhang P, Wang Y, Zeng X (2017) Systematic identification and characterization of long non-coding RNAs in mouse mature sperm. PLoS One 12:e0173402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang X, Shi J, Tuorto F, Li X, Liu Y, Liebers R, Zhang L, Qu Y, Qian J, Pahima M, Liu Y, Yan M, Cao Z, Lei X, Cao Y, Peng H, Liu S, Wang Y, Zheng H, Woolsey R, Quilici D, Zhai Q, Li L, Zhou T, Yan W, Lyko F, Zhang Y, Zhou Q, Duan E, Chen Q (2018) Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat Cell Biol 20:535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]


