Abstract
Pseudoenzymes have been identified across a diverse range of enzyme classes and fulfill important cellular functions. Examples of pseudoenzymes exist within ubiquitin conjugating and deubiquitinase protein families. Here we characterize FAM105A/OTULINL, the only putative pseudodeubiquitinase (DUB) of the ovarian tumour protease (OTU domain) family in humans. The crystal structure of FAM105A revealed that the OTU domain possesses structural deficiencies in both active site and substrate-binding infrastructure predicted to impair normal DUB function. We confirmed the absence of catalytic function against all ubiquitin linkages and an inability of FAM105A to bind ubiquitin compared to catalytically active FAM105B/OTULIN. FAM105A co-localized with KDEL markers and Lamin B1 at the endoplasmic reticulum (ER) and nuclear envelope, respectively. Accordingly, the FAM105A interactome exhibited significant enrichment in proteins localized to the ER/outer nuclear, Golgi and vesicular membranes. In light of undetectable deubiquitinase activity, we posit that FAM105A/OTULINL functions through its ability to mediate protein-protein interactions.
Graphical Abstract
eTOC Blurb
FAM105A is an OTU-class pseudo-deubiquitinase with a disrupted catalytic triad and undetectable cleavage activity for any diubiquitin linkage. Surface conservation predicts that FAM105A has evolved an adaptor function unrelated to a direct interaction with ubiquitin.
INTRODUCTION
Post-translational attachment of ubiquitin (Ub) to a target protein is mediated by a three-enzyme E1/E2/E3 cascade. In brief, the carboxy-terminus of Ub Gly76 is activated in an ATP-dependent manner by the E1 and transferred to the E2 to form a reactive E2~Ub thioester intermediate. Final transfer of activated Ub to a free amino group on a target protein is mediated by the action of an E3, either through a direct or indirect mechanism. Ub itself can be modified with Ub on one of seven lysine side chains (K6, K11, K27, K29, K33, K48 and K63) or its free amino-terminus of methionine (M1) through iterative reaction cycles (Komander and Rape, 2012), to generate eight different types of ubiquitin polymers (or “chains”).
Different Ub linkage types can impact distinct biology through the action of specific binding effectors. For example, K48 linked chains mediate the destruction of their target proteins through recognition by the RPN10 and RPN13 subunits of the proteasome (Hamazaki et al., 2015). M1 linked chains modulate inflammation signalling through recognition by the UBAN domain of NEMO (Fennell et al., 2018; Rahighi et al., 2009), and K63 linked chains mediate DNA damage signalling via recognition by the UDM domain of RNF168 (Takahashi et al., 2018). Ub chains can be cleaved by deubiquitinases (DUBs), a group of proteolytic enzymes comprising at least seven subfamilies (Abdul Rehman et al., 2016; Coleman and Huang, 2018; Nijman et al., 2005), each differentiated by a distinct catalytic domain structure. DUB subfamilies include the JAMM/MPN+ family of metalloproteases and the more prevalent UCH, Josephin, USPs, MINDY, ZUFSP and OTU families of cysteine proteases.
OTU domain DUBs are evolutionarily conserved from yeast to human with examples also encoded by viruses. In contrast to the USP family of DUBs, which characteristically display poor chain linkage discriminating abilities (Faesen et al., 2011), many OTU domain DUBs display exquisite specificity for one or a small subset of Ub linkage types (Mevissen et al., 2013). Notable members with single linkage-type discriminating abilities include OTUD4 and OTUB1, which specifically cleave K48-linked Ub chains, OTULIN, which cleaves M1-linked Ub chains, and Cezanne and Cezanne2, which uniquely cleave K11-linked chains.
OTU DUBs discriminate between different chain types through the ability of the OTU domain to recognize the globular fold of both ubiquitin moieties on either side of the intra Ub isopeptide bond cleaved (in some cases with the assistance of flanking motifs). By convention, the distal Ub, which contributes its C-terminus to the di-Ub linkage, engages the S1 binding pocket of the OTU domain while the proximal Ub, which contributes a lysine side chain or Met1 amino group, engages the S1’ binding pocket.
Approximately 10% of all DUBs are predicted to be incompetent for ubiquitin chain cleavage due to apparent deficiencies in their catalytic infrastructure (Walden et al., 2018). The question of how these pseudoDUBs exert a biological function if not through the cleavage of ubiquitin chains remains an enigma. This question extends broadly to pseudoenzymes in general (Murphy et al., 2017). In the case of the eukaryotic protein kinase superfamily, useful insight into pseudoenzyme function has been gleaned by comparison of the pseudokinase in question to its most closely related protein kinase that retains catalytic function. Exemplars include the kinase / pseudokinase pairs RAF and KSR, IRE1 and RNase L, and HER4 and ERBB3 (aka HER3)(Byrne et al., 2017).
FAM105A is a predicted pseudoenzyme member of the OTU family of DUBs. Remarkably little is known about its cellular and biochemical function, although a potential role in insulin secretion has recently been proposed (Taneera et al., 2015). FAM105A is most similar in sequence to OTULIN (aka FAM105B/GUMBY), the highly M1-specific DUB (Keusekotten et al., 2013; Rivkin et al., 2013). M1 Ub chains (aka linear Ub chains) generated by the multi-subunit E3 LUBAC (linear ubiquitin assembly complex) comprised of HOIP, HOIL and SHARPIN (Kirisako et al., 2006) are notable for their functional roles in inflammation and innate immunity (Rittinger and Ikeda, 2017). In particular, linear Ub chains assembled on NEMO and RIPK1 potentiate NFκB signalling and the enzymatic activity of OTULIN was shown to counteract this effect (Keusekotten et al., 2013; Rivkin et al., 2013). In addition to its catalytic function, the involvement of OTULIN in NFκB signalling is dependent on its ability to bind directly to HOIP through a conserved motif N-terminal to the catalytic OTU domain (Elliott et al., 2014; Schaeffer et al., 2014)
To investigate the function of the predicted pseudoDUB FAM105A, we characterized its structure and function in vitro and in cells drawing on comparisons to the known activities of OTULIN. Using X-ray crystallography, we confirmed that FAM105A belongs to the OTU subfamily of DUBs and possesses structural deficiencies in both catalytic and substrate-binding infrastructure predicted to impair normal DUB function. We confirm an absence of catalytic function against all di-Ub linkage types, and an inability to bind mono Ub and linear diUb at all concentrations tested. Further analysis also revealed no compelling evidence for FAM105A binding to the ubiquitin-like (UBL) molecules UFM1, ATG8, FAT10, ISG15, SUMO1 and SUMO2. Although most similar in structure to OTULIN, we also showed that FAM105A plays no discernable role in LUBAC dependent NFκB signalling. A unique subcellular localization pattern for FAM105A relative to OTULIN and a distinct BioID interactome profile suggested a role for FAM105A in an ER membrane related process.
RESULTS
Sequence and structure analysis of FAM105A
FAM105A is present in mammals, reptiles, amphibia and birds with closest sequence similarity to the human OTU family member OTULIN with 32.6% amino acid identity overall (Walden et al., 2018). OTULIN appears to predate FAM105A as it is present in a larger range of organisms including fish and marine invertebrates/tunicates such as Ciona. FAM105A is located in close proximity to OTULIN on the chromosomes of all species examined in which both genes are identifiable (from human to Xenopus). These findings led us to hypothesize that FAM105A may have evolved from OTULIN and that the two gene products might share related or overlapping biochemical and biological functions.
FAM105A and OTULIN proteins display similar domain architectures consisting of a predicted unstructured N-terminal region followed by an OTU domain (Figure 1A). The N-terminal regions of FAM105A and OTULIN appear unrelated in sequence (Figure 1B); while OTULIN possesses a conserved HOIP interaction motif (aka PUB interacting motif (PIM) - residues 53 to 57) that facilitates association with the multi-subunit E3 LUBAC and the ability to modulate linear ubiquitin signaling (Elliott et al., 2014; Schaeffer et al., 2014), FAM105A possesses a conserved predicted membrane localization motif (PMLM) encompassing residues 43 to 62 (43-MAKGFVMLAVSFLVAAICYF-62) as assessed by the SPLIT-4 server (Juretic et al., 2002). The PMLM of FAM105A shares similarity to short sequences in a diverse set of proteins including transporters, channels and permeases and appeared to generally correspond to hydrophobic α-helical secondary structure elements. The OTU domains of FAM105A and OTULIN display greatest amino acid sequence similarity to each other (41% identical and 63% similar) and low similarity to the catalytic domain of the next closest related OTU family DUB OTUB1 with only 11.8% and 16% percent similarity and identity respectively.
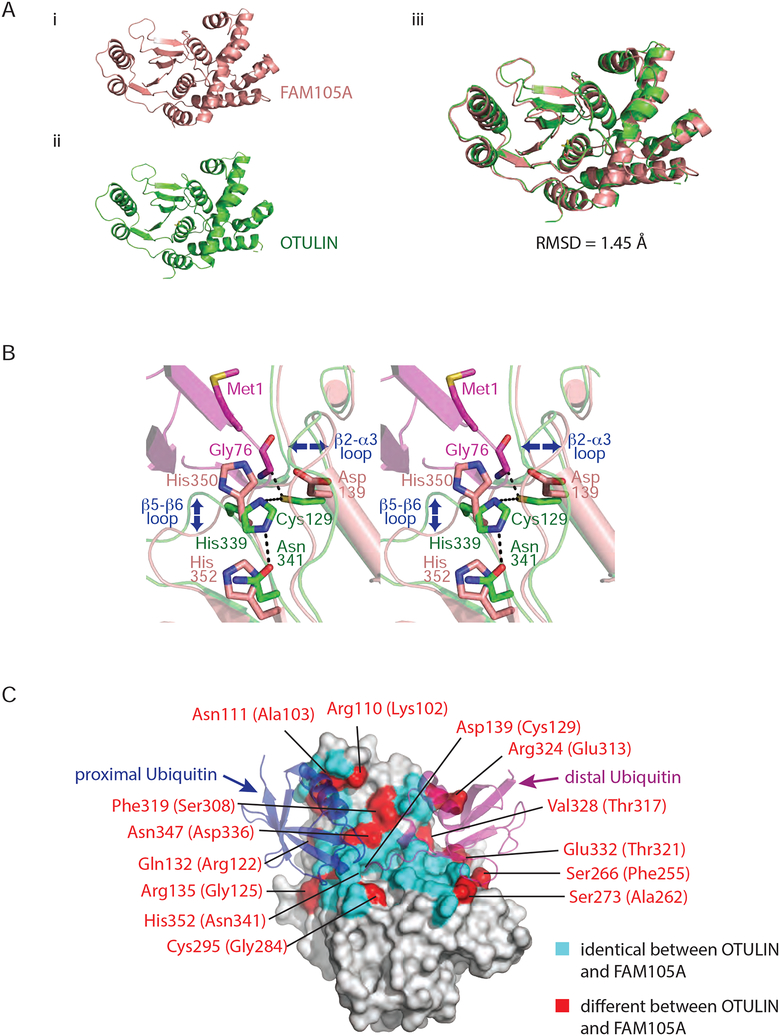
Figure 1. Domain architecture of FAM105A and OTULIN.
(A) FAM105A possesses an N-terminal predicted membrane localization motif (PMLM) and a C-terminal OTU domain. OTULIN possesses an N-terminal PUB interacting motif (PIM) and a C-terminal OTU domain. (B) Multiple sequence alignment of FAM105A and OTULIN from human, mouse, finch and Xenopus. Secondary structure elements are shown for FAM105A. Numbering refers to the human FAM105A and OTULIN sequences. Conserved residues are highlighted in grey. Residues comprising the linear diUb binding surface of OTULIN are highlighted in cyan in both OTULIN and FAM105A orthologues and red where divergent in FAM105A. Catalytic triad residues are indicated by red circles.
To facilitate functional and structural comparisons to OTULIN, we attempted to express FAM105A in bacteria. While full-length FAM105A was poorly expressed in soluble form, deletion of the N-terminal region containing the PMLM allowed for expression and purification of the OTU domain (residues 87–356) in high yield. The OTU domain of FAM105A readily crystallized enabling a structure determination at 2.1 Å resolution by molecular replacement using OTULIN (PDB ID = 4KSJ) as a search model (see Supplementary Table 1 for structure determination statistics and Supplementary Figure 1 for a representative electron density map). As predicted, the crystal structure of FAM105A displayed a canonical OTU fold (Figure 2A). Superposition of FAM105A and OTULIN structures revealed similarity over the entire OTU domain (RMSD of 1.45Å over 247 ordered Cα atoms Figure 2A). However, the two structures differed in two respects that suggested FAM105A is compromised for ubiquitin cleavage and ubiquitin binding functions.
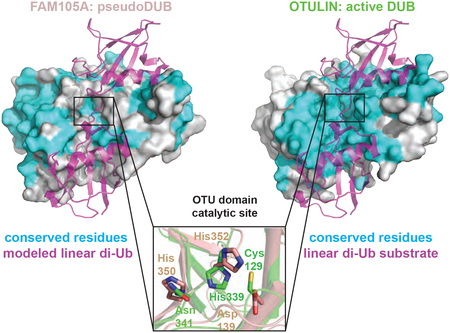
Figure 2. Structure of the OTU domain of FAM105A.
(A) Ribbon representations of FAM105A (top left) and OTULIN (bottom left). The two domains overlay (right panel) with a root mean square deviation (RMSD) of 1.45 Å for all Cα positions. (B) Zoom in view of the active site regions of FAM105A (pink) and OTULIN (green) bound to its cognate substrate linear di-Ub (purple). The catalytic triad of OTULIN composed of Cys129, His339, Asn341 superimpose most closely on the triad residues (Asp139, His350, His352) of FAM105A. Blue arrows indicate deviations in Cα-backbone positions (C) Surface representation of FAM105A modeled in complex with linear diUb. OTU domain residues identical between FAM105A and OTULIN are colored cyan with divergent residues colored red and labeled as indicated.
First, the catalytic triad of FAM105A appeared defective with no compensating features apparent in close vicinity of the active site (Figure 2B). While the active site in OTULIN conforms to a canonical structure and composition displayed by other OTU family members, two positions of the triad are substituted in FAM105A. In OTULIN, the catalytic triad consists of Cys129, His339 and Asn341. In its productive orientation bound to a linear diUb substrate, the His339 side chain functions to deprotonate the Cys129 side chain to generate a reactive nucleophile that can attack the carbonyl carbon of the target substrate peptide bond. This generates a substrate fragment linked to the enzyme through a thioester bond and a substrate fragment with a free amino group. The free enzyme is then regenerated by hydrolysis of the thioester intermediate coupled to the deprotonation of His339. In this capacity, the Asn341 side chain functions to stabilize the conformation of the His339 side chain while increasing its electronegativity. In the structure of FAM105A, the corresponding catalytic triad positions are occupied by Asp139, His350 and His352, respectively. While precedents for conservative substitutions of essential active site residues have been described for other classes of proteases (e.g. replacement of the serine position in the serine protease-like active site of Tobacco Etch Virus protease with cysteine (PDB 1LVM (Phan et al., 2002)) and substitution of the Asp-Asp catalytic diad in Aspartyl proteases with a His-Asp diad in the Plasmodium falciparum HAP protease (PDB:3QVC, 3QVI; (Bhaumik et al., 2009)), a more radical substitution of the catalytic cysteine residue by aspartic acid has not been demonstrated in the broader cysteine protease family. Thus, we reasoned that FAM105A lacks the capacity to cleave peptide bonds.
Second, the projected substrate-binding surfaces of FAM105A, while conserved in some respects with OTULIN, display critical substitutions that are predicted to perturb binding to the preferred linear diUb substrate of OTULIN. Specifically, 14 of 42 residues overall on the Ub substrate-binding site of OTULIN (PDB: 3ZNZ, 4KSK), are not conserved in FAM105A including 8 of 21 residues on the projected S1 (proximal Ub binding) binding surface and 6 of 21 residues on the projected S1’ (distal Ub) binding surface (Figure 2C). Among the residue differences between FAM105A and OTULIN in the S1-proximal Ub pocket, the bulkier side chain of Phe319 (compared to Ser308 in OTULIN) alters the conformation of the β5-β6 loop including two of the corresponding catalytic triad residues (His350, His352) (Figure 2B). An additional FAM105A substitution of Arg135 (Gly125 in OTULIN) located within the β2-α3 loop region exhibits altered Cα backbone positioning and generates a major steric clash with a modeled proximal ubiquitin (Figure 2B). Within the distal S1’ (distal Ub binding) pocket of FAM105A, Arg324 (Glu313 in OTULIN) and Ser273 (Ala262 in OTULIN) substitutions are predicted to cause major clashes with the distal ubiquitin (Figure 2C). While the substitutions on the S1 and S1’ surfaces suggested that FAM105A is unlikely to bind to linear diUb substrate in a manner predictable by OTULIN, it did not rule out the possibility that FAM105A could bind another Ub chain type or another conjugated form of Ub, or to a Ub-like molecule, as considerable variation of residues on the ubiquitin binding surfaces are observed in other OTU family DUBs.
Functional characterization of FAM105A in vitro
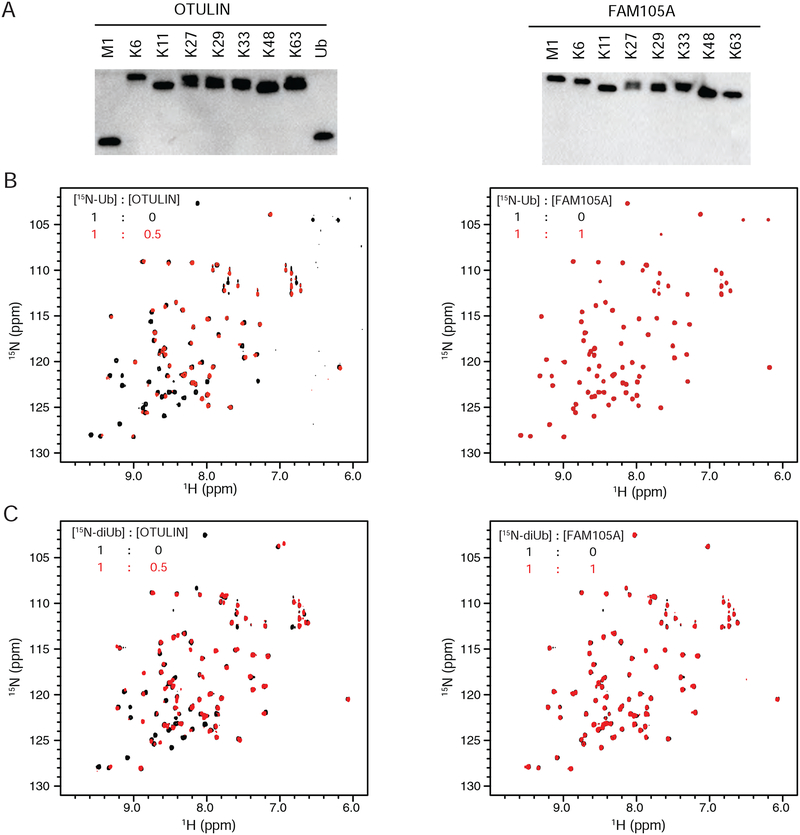
To probe the functional significance of our structural observations, we characterized the ability of FAM105A to cleave the eight different ubiquitin chain types. Consistent with a deficiency in the catalytic triad, FAM105A was not able to cleave any of the diUb chain substrates. In contrast, OTULIN displayed the expected preference for linear Ub chains (Figure 3A). Furthermore, a FAM105A double mutant was generated in which the canonical catalytic active site triad observed in OTULIN was restored. FAM105A D139C/H352N was similarly incompetent for cleavage of all diUb linkages and the model substrate AMC-Ub in contrast to the highly active DUB USP2 (Supplementary Figure 7).
Figure 3. In vitro cleavage and binding analysis of FAM105A against various ubiquitin substrates.
(A) Cleavage activity analysis of OTULIN (left) and FAM105A (right) against the indicated diUb chain substrates. (B) Superpositions of the 1H,15N-HSQC spectra of 15N-Ub in isolation and in 1:0.5 molar ratio with OTULINC129S (left panel) or in 1:1 molar ratio with FAM105A (right panel). (C) Superpositions of the 1H,15N-HSQC spectra of 15N-linear diUb in isolation and in 1:0.5 molar ratio with OTULINC129S (left panel) or in 1:1 molar ratio with FAM105A (right panel).
Next we used nuclear magnetic resonance spectroscopy (NMR) to assess the interaction of FAM105A with 15N-Ub and 15N-linear diUb in heteronuclear single quantum coherence (HSQC) experiments. In striking contrast to OTULIN, which causes pronounced chemical shift perturbations and peak intensity changes to the HSQC spectra of both 15N-Ub and 15N-linear diUb reflective of binding interactions (Figure 3B left panel, Figure 3C left panel respectively), FAM105A caused no detectable changes (Figure 3B left panel, Figure 3C left panel respectively). As the HSQC analysis is highly sensitive to even weak binding events, we conclude that FAM105A is unlikely to bind to linear di-Ub or any other conjugated form of Ub as some manifestation of binding to mono Ub would have been expected in this regard (Rivkin et al., 2013).
To investigate the possibility that divergence of the projected ubiquitin binding surfaces on FAM105A from OTULIN might reflect a change of binding preference to a Ub-like molecule, we employed a thermal shift binding assay. In this assay, the binding of a ligand to a target of interest is detected by a shift in the thermal denaturation temperature (Tm) as measured by a change in fluorescence of a hydrophobic reporter dye (Kopec and Schneider, 2011) that selectively recognizes the unfolded state of the target. Consistent with our NMR HSQC results, increasing concentrations of mono Ub and linear diUb caused a progressive stabilization of the Tm of OTULIN (Supplementary Figure 2A,B left and right respectively) but no detectable change to the Tm of FAM105A (Supplementary Figure 2C,D left and right respectively). Having established the functionality of the thermal shift assay, we next tested the effect of the Ub-like proteins UFM1, ATG8, FAT10, ISG15, SUMO1 and SUMO2 on the Tm of FAM105A (Supplementary Figure 3A). We observed no significant shift in the Tm of FAM105A even at the highest concentrations of each Ubl ligand tested (400μM) (Supplementary Figure 3C–I). The Tm values for each Ubl were either beyond the temperature range tested (i.e. not observed) or at values far greater than that of FAM105A and thus would not complicate interpretation of the thermal shift profiles (Supplementary Figure 3B). The interaction of SUMO2 with UBC9/UBE2I (Supplementary Figure 3J) further validated this screening approach. We conclude that FAM105A lacks an ability to bind conjugated forms of Ub or the Ub-like molecules examined.
In the absence of detectable binding of Ub or Ub-like molecules to FAM105A, we questioned whether FAM105A uses its projected Ub binding surface for an undetermined function. If true, we reasoned that the surface residues in question would be conserved across FAM105A orthologues. Indeed, conservation analysis revealed that residues that comprise the linear Ub binding surface of OTULIN are almost invariant across functional orthologues (Supplementary Figures 5, 6B). This proved not to be the case for FAM105A (Supplementary Figures 4, 6A), suggesting that FAM105A does not employ the particular surfaces in question to carry out an important but undetermined function. Interestingly however, other surfaces of the FAM105A OTU domain appear highly conserved. As residues in the corresponding positions of OTULIN are not conserved across its orthologues, this suggests that whatever function FAM105A performs with these conserved surfaces has no inferable function from OTULIN.
Role of FAM105A in LUBAC signalling
Despite the above biochemical and biophysical findings, we reasoned that previously uncharacterized functions might exist for FAM105A that overlap with OTULIN and that these functions would be more easily discerned in a cellular context. While no data on FAM105A protein tissue distribution was available, FAM105A mRNA is expressed at moderate to high levels in lung, gastrointestinal tract, prostate, seminal gland and bone marrow (Uhlen et al., 2015).
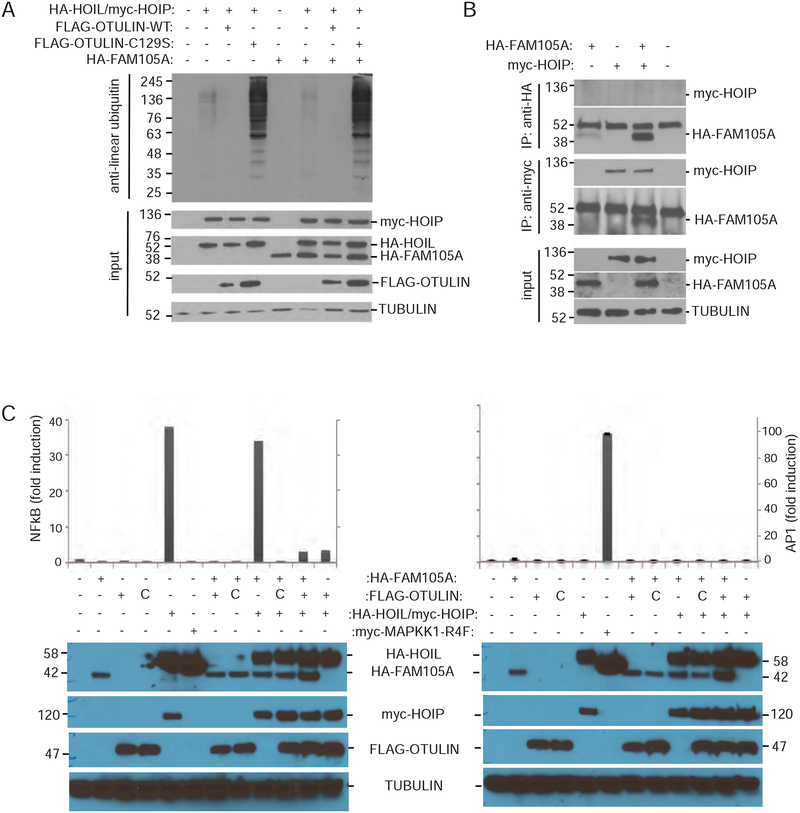
The overexpression of OTULIN in HEK293T cells gives rise to marked reduction of detectable linear ubiquitin chains in cells when overexpressed with LUBAC subunits. Consistent with FAM105A lacking the ability to cleave linear Ub chains in vitro, overexpression of FAM105A, in contrast to OTULIN, did not affect cellular levels of linear Ub when co-expressed with HOIP/HOIL (Figure 4A). To test whether FAM105A might interact with HOIP, as OTULIN does, we performed immunoprecipitation assays in HEK293T cells overexpressing Myc-HOIP and HA-FAM105A. While Myc-HOIP could recover HA-FAM105A in these assays, immunoprecipitation of HA-FAM105A did not recover HOIP (Figure 4B) suggesting that the interaction between these proteins is not considerable. In addition, the over expression of OTULIN in HEK293T cells causes a marked reversal of LUBAC-dependent NFκB signalling, one of the most studied terminal effectors of linear chain production by LUBAC. Unlike OTULIN, co-transfection of FAM105A with LUBAC subunits and an NFκB luciferase reporter did not affect LUBAC dependent induction of NFκB activity (Figure 4C). In these experiments, the general transcriptional activity assessed by an AP1-luciferase reporter was not affected by LUBAC in the presence or absence of either OTULIN or FAM105A. Overall these cellular results, which are consistent with the above in vitro results, led us to conclude that despite sharing a common evolutionary origin with OTULIN, FAM105A must play a functional role distinct from that of OTULIN.
Figure 4. FAM105A does not counteract LUBAC activity.
(A) Immunoblot using anti-Met1Ub antibody detected decreased levels of Met1Ub-conjugated proteins in HEK293T cells upon co-expression of FLAG-OTULIN, but not FLAG-OTULINC129S or FAM105A with HAHOIL and myc-HOIP. (B) Immunoprecipitation with anti-HA antibody (IP:anti-HA) of HA-tagged FAM105A (HA-FAM105A) does not recover myc-tagged HOIP (myc-HOIP). In immunoprecipitation with anti-myc antibody (IP: anti-myc), however, myc-HOIP recovered HA-FAM105A. Input amounts and Tubulin levels are shown. (C) Co-expression of HAFAM105A did not affect activity of FLAG-OTULIN in a luciferase reporter assay. FLAG-OTULIN represses myc-HOIP/HA-HOIL dependent activation of an NFκB-dependent luciferase reporter. Co-transfection of HA-FAM105A does not impact myc-HOIP/HA-HOIL dependent activation or FLAG-OTULIN dependent repression of this luciferase reporter. General transcriptional activity of an AP1 luciferase reporter was unaffected. HA-MAPKK1-R4F acts as a positive control for the AP1 luciferase assay.
Analysis of FAM105A subcellular localization
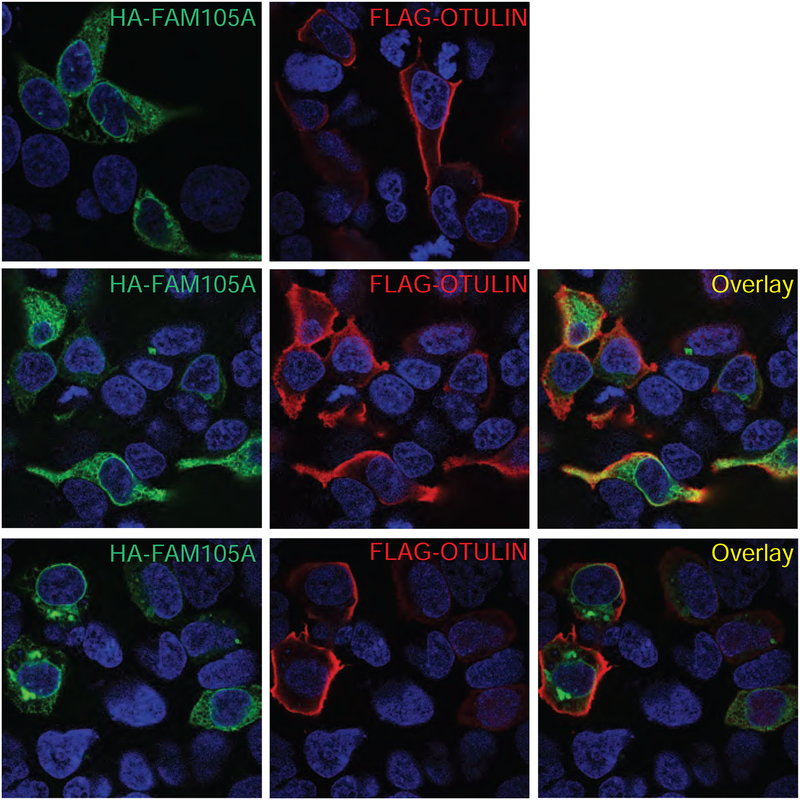
The conspicuous presence of a twenty residue stretch of conserved hydrophobic residues corresponding to a putative PMLM in the N-terminal region of FAM105A led us to question whether FAM105A localizes to membrane structures. To address this question, we epitope tagged FAM105A with an N-terminal HA tag and, for comparison purposes, OTULIN with an N-terminal FLAG tag. Transient transfection of each construct individually in Neuro2A cells followed by immunofluorescence analysis revealed distinct patterns of subcellular localization (Figure 5). Specifically FAM105A localized to the cytoplasm, nuclear envelope and ER membrane-like structures whereas OTULIN localized at or near the plasma membrane and in the cytoplasm (Figure 5A,B). Co-expression of FAM105A and OTULIN corroborated that the individual subcellular localization patterns of each protein are distinct and the expression of each protein doesn’t affect the localization of the other (Figure 5C,D).
Figure 5. FAM105A and OTULIN show differential subcellular localization.
Immunofluorescence of (A) HA-FAM105A (red), (B) FLAG-OTULIN (green), or (C and D) co-expressed HA-FAM105A and FLAG-OTULIN in Neuro2A cells.
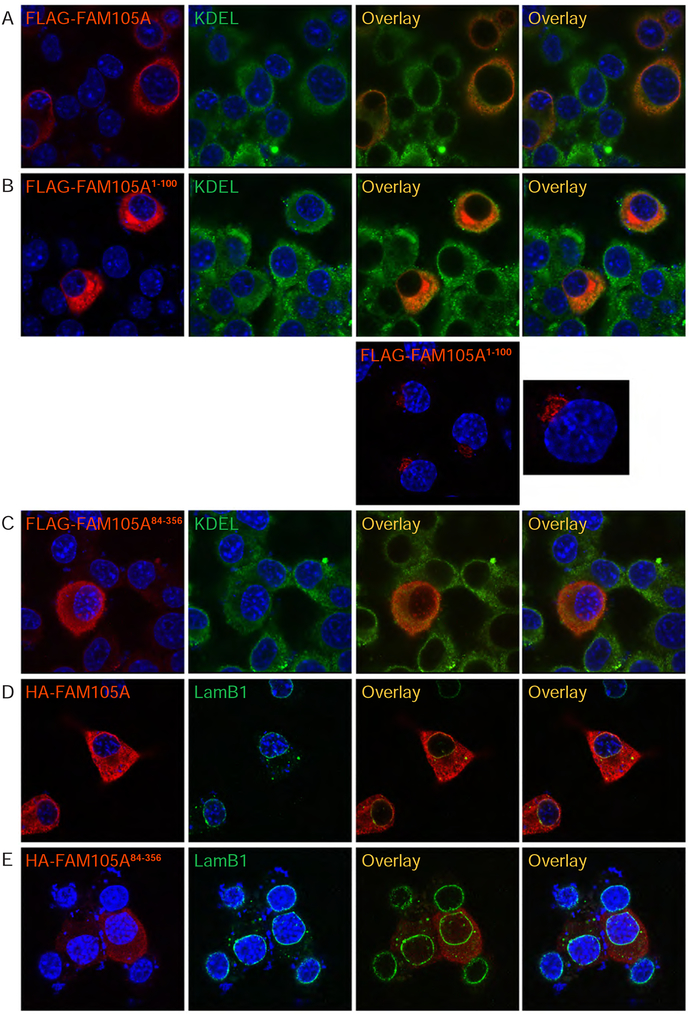
To determine if the N-terminal region of FAM105A containing the PMLM is responsible for the membrane localization pattern of the full-length protein, we epitope tagged the first 100 amino acids of FAM105A at the amino-terminus with a FLAG tag (FLAG-FAM105A1−100) and found that this protein recapitulated the localization observed for full-length FLAG-FAM105A protein to the ER as shown by co-localization with an antibody directed against the KDEL ER localization sequence (Figure 6, panels A vs B). We note that on occasion, FLAG-FAM105A1−100 also localized to Golgi-like structures (Figure 6B-lower panels), a behaviour that may reflect the instability of small protein fusions. When we deleted the first 83 amino-terminal amino acids of FAM105A, the resulting construct (FLAG-FAM105A84−356) comprising the OTU domain showed diffuse cytoplasmic and low level, at times punctate, nuclear distribution as well as loss of the normally sharp localization at the nuclear envelope (Figure 6C). Repeating a subset of experiments with HA tagged FAM105A constructs and an antibody against endogenous Lamin B1 (Figure 6 panels D vs E) gave similar results and confirmed localization of a fraction of FAM105A to the nuclear envelope with Lamin B1. These results are consistent with the dependence of FAM105A subcellular localization on its N-terminal region and likely on the PMLM. Together these studies indicated that FAM105A has evolved to occupy subcellular compartments distinct from those of OTULIN.
Figure 6. FAM105A amino-terminus directs its subcellular localization.
Immunofluorescence analysis in Neuro2A cells of FLAG-tagged (A) full-length FAM105A protein, (B) the FAM105A1−100 N-Terminus and (C) the FAM105A84−356 OTU domain. Coimmunostaining for endogenous KDEL (green) marked the endoplasmic reticulum. FLAG-FAM105A and FLAG-FAM105A1−100 largely colocalize with KDEL in the cytoplasm, but FLAG-FAM105A84−356 OTU-domain protein appears diffuse throughout the cytoplasm and in puncta within the nucleus. (D) HA-FAM105A, but not (E) the isolated OTU-domain, HA-FAM105A84−356, localizes to the nuclear envelope, which is marked by immunostaining for Lamin B1 (green). All FAM105A proteins are shown in red.
BioID analysis of FAM105A
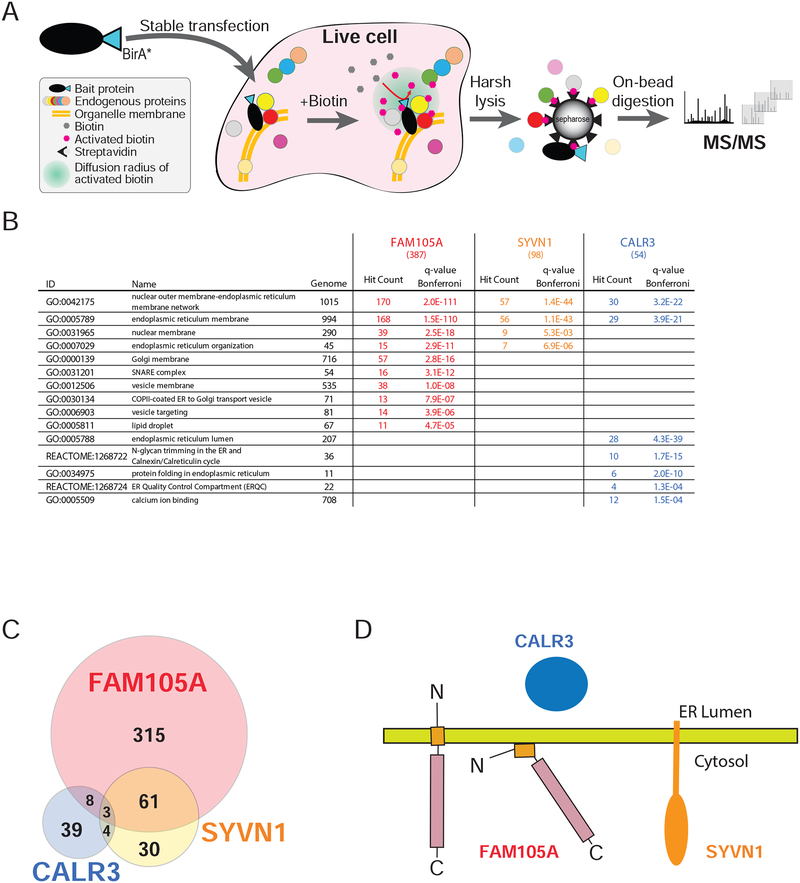
To better understand FAM105A localization, we identified proximal interacting protein partners using in vivo proximity-dependent biotinylation (BioID) (Roux et al., 2012). FAM105A was stably expressed with an in-frame C-terminal BirA-FLAG fusion tag in Flp-In T-REx 293 cells (Figure 7A), and biotinylated proteins identified using mass spectrometry (Coyaud et al., 2015). Consistent with the subcellular localization pattern, GO analysis of FAM105A interactors revealed significant enrichment for proteins localized to the ER/outer nuclear membrane (Figure 7B, Supplementary Tables 2,3). For comparative purposes, we also conducted BioID on: (i) the ER lumen chaperone protein calreticulin (CALR3) and; (ii) the ER membrane protein SYVN1/HRD1 (Figure 7B, Supplementary Tables 2,3). The FAM105A interactome displayed significantly higher similarity with HRD1 than with CALR3 (Figure 7C), consistent with either of two membrane topologies for FAM105A: a single pass transmembrane protein with the N-terminus in the ER lumen and C-terminus in the cytoplasm, or a peripherally associated membrane protein with both N terminus and the OTU domain in the cytosol (Figure 7D). In agreement with our lack of detection of roles for FAM105A in linear (de)ubiquitination, these analyses did not recover HOIP or any other known component of the linear (de)ubiquitination machinery. In contrast, the FAM105A interactome included a number of proteins previously linked to membrane contact sites (e.g. VAPA, VAPB, ESYT½, PDZD8, RAB3GAP½, VPS13A, INF2 and ACBD5), suggesting that FAM105A could play a role in ER-organelle communication. Additional study will be required to test this hypothesis.
Figure 7. Comparison of FAM105A, SYVN1 (an ER membrane protein), and CALR3 (an ER lumen protein) BioID interactomes.
(A) A C-terminally tagged FAM105A-BirA*FLAG protein (the “bait” polypeptide) was stably expressed in Flp-In T-REx 293 cells. Bait expression was induced with 1μM tetracycline, and 50μM biotin was added to the culture media for 24 hrs, allowing for in vivo biotin labelling of proximal interacting proteins. Cells were lysed and biotinylated polypeptides were captured on a streptavidin column. After washing, bound proteins were subjected to trypsin digestion and the resulting peptides identified using liquid-chromatography (LC) coupled in-line with tandem mass spectrometry (MS/MS). (B) Selected GO or Pathway categories significantly enriched (Bonferroni q-value<0.01) in the FAM105A, SYVN1, and CALR3 BioID interactomes (n.s.: not significant); Genome: number of human genes coding for proteins in the indicated category; Hit count: number of high confidence interactors for the indicated bait protein assigned to each category (total high confidence interactors indicated in brackets at the top of each column); q-value Bonferroni: significance value extracted from ToppGene Suite analysis; Grey font: not significant. (C) Venn diagram depicting the overlap between high confidence interacting partners identified in BioID analyses of FAM105A, SYVN1 and CALR3 (see Supplemental Table 2 and 3 for detailed data and enrichment analysis). (D) Models for FAM105A orientation at the ER membrane and cytosolic interface relative to SYVN1 and CALR3.
DISCUSSION
DUBs of the OTU family utilize an essential cysteine residue in their catalytic triad to cleave ubiquitin chains but often display unique preferences for different ubiquitin linkage types. FAM105A harbours an aspartate residue at this critical position of the triad suggesting it would lack the ability to cleave ubiquitin chains. The crystal structure of FAM105A87−356 confirmed its membership in the class of enzymes with an OTU-fold and revealed no unanticipated structural features that would compensate for its defective triad. Indeed, our tests showed that FAM105A does not cleave any of the 8 distinct di-Ub linkage types including M1 substrates that are preferred by its closest catalytically competent paralogue FAM105B/OTULIN. Given our findings that FAM105A is a pseudoDUB, we considered other roles that it may play to exert a biological function. Pseudoenzymes commonly function through one of four mechanisms (Murphy et al., 2017).
First, pseudoenzymes can act as negative regulators of active enzymes by competition for substrate binding. We showed that FAM105A has no detectable interaction with ubiquitin as assessed by NMR or with other UBLs (specifically UFM1, ATG8, FAT10, ISG15, SUMO1 or SUMO2) as assessed by thermal denaturation analysis. Thus, FAM105A is unlikely to act by competing for Ub or Ubl linked substrates.
Second, pseudoenzymes may interact with a catalytically active paralogue to function as an allosteric regulator. For example, the metalloprotease DUB BRCC36 forms a heterodimeric regulatory complex with the structurally related pseudoDUB KIAA0157/ABRAXAS (Zeqiraj et al., 2015). We showed that FAM105A did not impact the catalytic function of its closest paralogue OTULIN or interact with any other OTU family DUB as assessed by BioID. Furthermore, there are no precedents for OTU family DUBs functioning as higher-order oligomers thus it is unlikely that FAM105A functions as an allosteric activator of one or more OTU family DUBs.
Third, pseudoenzymes may act as a regulator of adjacent functional domains within the same polypeptide chain. The dual protein kinase ribonuclease RNase L exemplifies this mode of action, where its catalytically deficient protein kinase domain serves to regulate the catalytic output of its neighbouring ribonuclease domain (Huang et al., 2014). The simple architecture of FAM105A, which lacks any recognizable domain beyond its pseudoOTU domain is unlikely to function in this manner.
Finally, pseudoenzymes can gain altogether novel functions unrelated to those of their catalytically active paralogues. For example, the DUF1669 domain of FAM83 proteins comprises a pseudophospholipase D domain with no detectable enzymatic activity (Bozatzi and Sapkota, 2018). Instead, the pseudophospholipase D domains of FAM83 proteins have evolved a unique ability to bind casein kinase isoforms and anchor them to specific subcellular sites (Fulcher et al., 2018). Similarly, the pseudokinase VRK3 does not detectably bind ATP but functions as a negative regulator of MAP kinase signaling by interacting with VHR phosphatase and promoting dephosphorylation of ERKs (Kang and Kim, 2006; Scheeff et al., 2009). While a functional role for the OTU domain of FAM105A has yet to be identified, we note that the high evolutionary conservation of surface residues remote from the DUB active-site suggests a potential protein interaction function worthy of future investigation. In addition to the ER membrane localization properties mediated by the unique N-terminus, the plethora of ER proximal interactors identified by BioID provide further leads for understanding the biochemical and cellular function of this enigmatic pseudoenzyme.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Frank Sicheri (sicheri@lunenfeld.ca).
METHODS DETAILS
Protein expression for biochemistry and structural biology.
Human FAM105A comprising residues 87–356 was cloned from IMAGE clone (BC011524) into ProEx as an N-terminally His6-tagged protein. FAM105A was expressed in BL21 E. coli cells following an 18 hour induction with 0.25 mM IPTG at 18°C. The cell pellet was suspended in 20 mM HEPES pH 7.5, 400 mM NaCl and 5 mM Imidazole and lysed by passage through a homogenizer (Avestin Inc. Ottawa, CA). Following centrifugation, clarified supernatant was applied to a 5ml HiTrap Ni-chelation column (GE LifeScience Inc.). The column was washed with buffer containing 20 mM imidazole and eluted in a gradient to 300 mM imidazole. Fractions containing FAM105A were pooled and incubated overnight at 6°C with His-TEV protease to cleave the N-terminal His6-tag. Protein was dialyzed into 20 mM HEPES pH 7.5, 400 mM NaCl, 5 mM imidazole and flowed over a 1ml HiTrap Ni-chelation column to remove His-TEV and the cleaved His6-tag. A 120ml S75 size exclusion column was equilibrated in 20 mM HEPES pH7.0, 100 mM NaCl, 1 mM DTT and FAM105A eluted in a single peak. Protein was concentrated to 175 μM (6.0 mg/ml) and stored at −80°C. FAM105A was observed to slowly precipitate at protein concentrations greater than 200 μM and upon thawing previously frozen samples so in vitro experiments were performed with freshly purified protein.
OTULIN55−352 C129S, ubiquitin and linear diubiquitin were purified as described previously (Rivkin et al., 2013). His-tagged OTULIN and linear diubiquitin were each expressed in BL21 E.coli cells, purified using a HiTrap nickel chelting HP column (GE Healthcare) and eluted in 20 mM HEPES pH 7.5, 400 mM NaCl, 300 mM Imidazole. Following incubation with TEV protease and 2 mM DTT, protein was dialyzed in HiTrap loading buffer and flowed over a subtractive HiTrap nickel-chelating column. OTULIN or linear diubiquitin was concentrated for injection onto a 120 ml Superdex S75 (GE Healthcare) in 20 mM HEPES pH 7.5, 100 mM NaCl, 5 mM β-mercaptoethanol, concentrated to 10–25 mg/ml and stored frozen at −80°C. GST-tagged Ubiquitin ex pressed in BL21 E.coli cells was purified using glutathione Sepharose resin (GE Healthcare), washed extensively and incubated with TEV protease and 2 mM DTT. Cleaved ubiquitin was separated from His-tagged TEV by subtraction over a subtractive HiTrap nickel-chelating column and concentrated for injection onto a 120 ml Superdex S75 equilibrated in 20 mM HEPES pH 7.5, 100 mM NaCl, 5 mM β-mercaptoethanol. Ubiquitin was concentrated to 2.0 mM and stored frozen at −80°C.
The Ubl proteins, ATG81−117, UFM11−83, ISG151−157, SUMO11−107, SUMO21−93, FAT10-N8−82 and Fat10-C82−165 (Theng et al., 2014) were obtained and cloned into ProEx. Each protein was expressed in BL21 E.coli cells and purified by the same method as described above for linear diubiquitin. Purified Ubl proteins were concentrated to >800 μM and stored frozen at −80°C.
Protein crystallography.
Crystals of FAM105A87−356 were grown in hanging drops by mixing 1 μl protein solution containing 175 μM protein with 1 μl well solution (6% PEG8K, 0.1M Tris-HCl pH8.0, 150 mM NaCl) at 20°C. For cryoprotection, crystals were soaked in well solution supplemented with 20% glycerol. Diffraction data was collected from a single frozen crystal at 0.97919 Å wavelength on beamline NE-CAT 24-ID-C (APS, Chicago, Il) and processed with Xia2-XDS (Winter et al., 2013). Molecular replacement was performed using Phaser (McCoy et al., 2007) and the starting model of OTULIN (PDB code 4KSJ). Model building and refinement were performed using Coot (Emsley et al., 2010) and Phenix (Adams et al., 2011). Software used in this project was curated by SBGrid (Morin et al., 2013). The diffraction data and final refinement statistics are shown in Supplementary Table 1.
In vitro ubiquitin linkage cleavage assay.
Di-Ub chains were either purchased (Lifesensors: K11, Boston Biochem: K6, K27, K29, K33), expressed recombinantly (M1) or synthesized enzymatically (K48, K63) by methods previously described (Pickart and Raasi, 2005; Rivkin et al., 2013). 2 μM of FAM105A, 2 μM of OTULIN or 100 nM USP2 (Ernst et al., 2013) was incubated with 0.25–1 μg of each di-Ub chain type in 25 mM HEPES pH 7.0, 300 mM NaCl 1 mM DTT and 0.1 mg/ml BSA in 10 uL reactions for 18 hrs at 25°C. Reactions were terminated by addition of 2× Laemmli buffer, resolved on 20% SDS-PAGE gels and stained with Coomassie or transferred to PVDF membranes by semi-dry transfer methods. Membranes were probed first with Mouse monoclonal anti-Ub antibody (Covance MMS-257P) and subsequently with HRP linked-anti Mouse antibody. Images were taken by a Bio-Rad ChemiDoc MP Imaging System.
Ub-AMC Assay.
Ub-AMC (Boston Biochem) was diluted in reaction buffer (20 mM HEPES pH 7.0, 100 mM NaCl, 1 mM DTT). For each reaction 10 μl of Ub-AMC in a black 384-well low volume plate (Corning) was mixed with 10 μl of either: 50 nM USP2 (Ernst et al., 2013), 500 nM OTULIN, 500 nM FAM105A wt or FAM105A D139C/H352N at 30°C. The rate of AMC generation was measured using a Synergy Neo plate reader (BioTek). Fluorescent intensities were recorded following excitation at 345 nm and emission at 445 nm.
NMR Spectroscopy.
15N-ubiquitin and 15N-linear diubiquitin were expressed in E.coli using M9 medium supplemented with 15NH4Cl. Purification was performed as described for unlabelled proteins. NMR data were acquired at 25°C on a 600 MHz Bruker AVANCE III spectrometer equipped with a 1.7 mm TCI CryoProbe. 15N-ubiquitin or 15N-linear diubiquitin was used at 75 μM and 150 μM respectively for the collection of 1H, 15N-HSQC titration spectra with OTULIN55−352 C129S or FAM105A87–356. All NMR samples were prepared in 20 mM HEPES, pH 7.0, 100 mM NaCl, 1 mM DTT, and 5–7% D2O. All NMR spectra were processed using NMRPipe/NMRDraw (Delaglio et al., 1995) and further analyzed using NMRView (Johnson, 2004). Backbone resonance assignments for human ubiquitin and linear diubiquitin were reported previously (Vincendeau et al., 2016; Wang et al., 1995).
Thermal Stability Assay.
Differential scanning fluorimetry experiments were performed in duplicate. FAM105A87−356 or OTULIN55−352 C129S at 25 μM was incubated in 20 mM HEPES pH 7.0, 50 mM NaCl, 1 mM DTT with 10X SYPRO orange (ThermoFisher) and a titration of Ub or various Ubl proteins in duplicate. Samples in a final volume of 20 μl were incubated in 384-well white microplates (cat# 04729749001, Roche) and heated from 25°C to 85°C at a rate of 1.0°C per minute in a LightCycler 480 II (Roche). F luorescence intensity was plotted and the protein melting temperature (Tm) was determined from first derivative plots of the thermal melt plot using instrument software.
Co-Immunoprecipitation.
Effectene transfection reagent (Qiagen) was used to transfect human HEK293T cells. Cells were lysed 24–48 h post-transfection in 50 mM Tris pH 7.4, 100 mM EDTA, 150 mM NaCl and 0.5% Triton X-100 and protease inhibitor cocktail (Complete mini EDTA free, Roche). Lysates were pre-cleared with 10 μl of protein A/G plus agarose beads (Santa Cruz) for 30 minutes with rocking at 4°. Aliquots containing 400 μg total protein were incubated for 1 hour with 1 μg Flag antibody (Mouse M2, Sigma), 2 μg myc antibody (Mouse, Santa Cruz), 1 μg GFP antibody (Mouse, Roche) or 2 μg HA antibody (Rat, High Affinity, Roche). 20 μl of Protein A/G plus agarose bead slurry was added, incubated for 4 hours at 4° with rocking and washed thrice with 20 mM HEPES pH7.5, 500 mM NaCl, 10% glycerol, 0.5% Triton X-100, 1.5 mM MgCl2, 10 mM NaPO4 pH 7.5 and protease inhibitor cocktail. Samples were analyzed by immunoblot.
Analysis of linear ubiquitinated proteins.
Analyses of linear ubiquitinated proteins by immunoblot were performed as described previously (Rivkin et al., 2013). 293T cells were washed twice with PBS and lysed in 8 M urea, 50 mM Tris pH 7.5, 25 mM NaCl, 2 mM EDTA, 2 mM N-ethylmaleimide and complete mini protease inhibitors (Roche) and were disrupted by passing through an 18-gauge needle and analyzed by immunoblot with an anti-Met1Ub antibody (Genentech).
NFκB and AP1 activation assays.
NFκB dependent transcription was assayed in HEK293T cells co-transfected with various expression constructs, with the normalizing transfection efficiency control PRL vector and with either a reporter containing 6 NFκB binding sites or a control plasmid with 6 AP1 binding sites that drive firefly luciferase expression using Effectene (Qiagen), as described in Rivkin et al. (Rivkin et al., 2013). Dual luciferase assays (Promega) were performed 20–24hrs post-transection. Values were normalized relative to FLAG-pcDNA3.1. At least three independent experiments were performed with samples in triplicate for each set of constructs. pMCL-HA-MAPKK1-R4F [delta(31–51)/S218/S222D] was a gift from Natalie Ahn (Mansour et al., 1994) (Addgene plasmid # 40810; http:/n2t.net/addgene:40810).
Immunofluorescence.
HEK293T or Neuro2A cells were fixed with 4% paraformaldehyde, permeabilized with 0.3% Triton X-100/PBS, incubated for 1–3 hrs in histo-buffer (5%BSA/5% normal Donkey serum/5% normal Goat serum/0.1%Triton X-100/PBS), and then incubated with primary antibodies in histo-buffer overnight. After three washes in 0.1% Triton X-100/PBS, samples were incubated with secondary antibodies in histo-buffer for 1hr at room temperature, washed and mounted with Vectashield mounting media with 4’, 6-Diamidino-2-phenylindole dihydrochloride (DAPI) (Vector Laboratories) for visualization. Images were acquired using 60× or 100× oil immersion objective lens (Nikon D-eclipse C1 confocal microscope system) and analyzed with Adobe Photoshop.
Antibodies used were: mouse anti-Flag M2 (Clone F1804 Sigma, 1:1,000), rat anti-HA (1:250), mouse anti-KDEL (Abcam; [MAC 256] ab50601; 1:300) and rat anti-Lamin B1 (Abcam ab16048; 1:10,000), human anti-linear ubiquitin (Genentech, 1 mg/ml) antibodies. Secondary antibodies were all used at 1:500 dilutions and were: donkey anti-Rat Cy3, donkey anti-mouse Cy3 (both from Jackson Immunoresearch), donkey anti-mouse AlexaFluor 488, donkey anti-rat AlexaFluor 488 (both from Invitrogen, Molecular Probes).
Stable cell lines for BioID analysis.
BioID (Roux et al., 2012) was carried out as previously described (Gupta et al., 2015). In brief, full length human FAM105A, CALR3 and SYVN1 coding sequences (from clones BC011524, BC014595 and BC030530) were amplified by PCR, and cloned into our pcDNA5 FRT/TO BirA*FLAG (C-terminal epitope tag for FAM105A and SYVN1) or pcDNA5 FRT/TO FLAGBirA* (N-terminal epitope tag for CALR3) expression vectors. Using the Flp-In system (Invitrogen), 293 T-REx Flp-In cells stably expressing FlagBirA* alone or FlagBirA*/BirA*Flag fusions were generated. After selection (DMEM + 10% FBS + 200 μg/ml Hygromycin B), 5 × 150 cm2 plates (per biological replicate) of sub-confluent (60%) cells were incubated for 24 hr in complete media supplemented with 1 μg/ml tetracycline (Sigma), 50 μM biotin (BioShop). Cells were collected and pelleted (2,000 rpm, 3 min), the pellet was washed twice with PBS, and dried pellets were snap frozen. Two biological replicates were prepared for each bait protein.
Biotin-streptavidin affinity purification.
Cell pellets were resuspended in 10 mL of lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 0.1% SDS, 1:500 protease inhibitor cocktail (Sigma-Aldrich), 1:1,000 turbonuclease), incubated on an end-over-end rotator at 4°C for 1 h r, briefly sonicated to disrupt any visible aggregates, then centrifuged at 45,000 g for 30 min at 4°C. The supernatant was transferred to a fresh 15 ml conical tube, 30 μl of packed, pre-equilibrated streptavidin-sepharose beads (GE Healthcare) were added, and the mixture incubated for 3 hr at 4°C with end-over-end rotation. Beads were pelleted by centrifugation at 2,000 rpm for 2 min and transferred with 1 ml of lysis buffer to a fresh Eppendorf tube. Beads were washed once with 1 ml lysis buffer and twice with 1 ml of 50 mM ammonium bicarbonate pH 8.3 (ammbic). Beads were transferred in ammbic to a fresh tube, and washed two more times with 1 ml ammbic. Tryptic digestion was performed by incubating the beads with 1 μg MS grade TPCK trypsin (Promega) dissolved in 200 μl of 50 mM ammbic overnight at 37°C. The following morning, an additional 0.5 μg trypsin was added, and the beads incubated for 2 hr at 37°C. Beads were pelleted and the supernatant transferred to a fresh Eppendorf tube. Beads were washed twice with 150 μl of 50 mM ammbic, and washes were pooled with the eluate. The sample was lyophilized and resuspended in buffer A (0.1% formic acid). 1/5th of the sample was analyzed per MS run. Two MS analyses (i.e. two technical replicates) were conducted for each biological replicate.
Mass spectrometry analysis.
High performance liquid chromatography was conducted either on a Thermo Q-Exactive HF quadrupole-Orbitrap (QEHF; FAM105A and associated control runs) or on a hybrid LTQ-Orbitrap Velos mass spectrometer (Velos; SYVN1 and CALR3 and associated control runs). Samples analyzed on the QEHF were loaded on a 2 cm pre-column (Acclaim PepMap 20 mm × 75 μm inner diameter (ID)) and 50 cm analytical column (Acclaim PepMap, 500 mm × 75 μm diameter; C18; 2 μm; 100 Å, Thermo Fisher Scientific), and eluted along a 120 min reversed-phase buffer gradient at 225 nl/min via a Proxeon EASY-nLC 1000 pump. A parent ion scan was performed using a resolving power of 60,000, then up to the twenty most intense peaks were selected for MS/MS (minimum ion count of 1,000 for activation) using higher energy collision induced dissociation (HCD) fragmentation. Dynamic exclusion was activated such that MS/MS of the same m/z (within a range of 10 ppm; exclusion list size 500) were excluded from analysis for 5 sec. CALR3 and SYVN1 were analyzed on a Velos mass spectrometer: Analytical columns (75-μm inner diameter) and pre-columns (150-μm inner diameter) were made in-house from fused silica capillary tubing (InnovaQuartz) and packed with 100 Å C18–coated silica particles (Magic, Michrom Bioresources). Peptides were subjected to liquid chromatography (LC)-electrospray ionization-tandem mass spectrometry, using a similar reversed-phase buffer gradient running at 250 nl/min on a Proxeon EASY-nLC pump. A parent ion scan was performed in the Orbitrap using a resolving power of 60,000, then up to the twenty most intense peaks were selected for MS/MS (minimum ion count of 1,000 for activation), using standard collision induced dissociation fragmentation. Fragment ions were detected in the LTQ. Dynamic exclusion was activated such that MS/MS of the same m/z (within a range of 15 ppm; exclusion list size 500) detected twice within 15 s were excluded from analysis for 30 s.
For protein identification, Thermo .RAW files were converted to the .mzXML format using Proteowizard (Kessner et al., 2008), then searched using X!Tandem (Craig and Beavis, 2004) and Comet (Eng et al., 2013) against the Human RefSeq Version 45 database (containing 36113 entries). Search parameters specified a parent ion mass tolerance of 10 ppm (15ppm for Velos data), and an MS/MS fragment ion tolerance of 0.4 Da, with up to 2 missed cleavages allowed for trypsin. Variable modifications of +16@M and W, +32@M and W, +42@N-terminus, and +1@N and Q were allowed. Proteins identified with an iProphet cut-off of 0.9 (corresponding to ≤1% FDR) and at least two unique peptides were analyzed with SAINT Express v.3.6. Each set of data, consisting of two biological replicates each analyzed with two technical replicates, was compared to 20 control runs (analyzed on the QEHF for FAM105A, and on the Velos for SYVN1 and CALR3; 20 runs from cells expressing the FlagBirA* epitope tag only) collapsed to the two highest spectral counts for each prey, and high confidence interactors were defined as those with BFDR≤0.01. All raw mass spectrometry files have been deposited at the MassIVE archive (massive.ucsd.edu), ID MSV000082315.
Mass Spectrometry Data analysis.
A high confidence interactor list for FAM105A is presented in Supplemental Table 2. Enrichment analysis was conducted using the ToppGene Suite (Chen et al., 2009). Selected categories for each of the the three bait proteins are highlighted in Figure 7B, and complete analysis is included in Supplemental Table 3.
QUANTIFICATION AND STATISTICAL ANALYSIS
DSF assays were performed in duplicate for each Ubl titration or n=4 for ubiquitin and linear diubiquitin experiments. NFκB and AP1 activation assays were performed with triplicate samples for each set of constructs. Two biological replicates were prepared for each BioID bait protein. Statistical analysis of mass spectrometry results are included in Supplementary Table 3.
DATA AND SOFTWARE AVAILABILITY
Data Resources.
The FAM105A87−356 coordinates and structure factors have been deposited in the Protein Data Bank under accession code 6DRM. The FAM105A BioID data has been deposited in the raw mass spectrometry data archive at MassIVE: MSV000082315.
Supplementary Material
BioID list of FAM105A high confidence hits.
Complete BioID analysis of FAM105A, SYVN1 and CALR3.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Ub (P4D1) | Covance | MMS-257P |
| Anti-FLAG (Mouse M2) | Sigma | F1804 |
| Anti-Myc (mouse 9E10) | Santa Cruz | Sc-40 |
| Anti-GFP (mouse) | Roche | 11814460001 |
| Anti-HA (rat - high affinity) | Roche | 1186742300 |
| Anti-Metl Ub (1E3) | V. Dixit (Genentech) | N/A |
| Anti-KDEL (mouse MAC 256) | Abcam | Ab50601 |
| Anti-Lamin B1 (rat) | Abcam | Ab16048 |
| Anti-Cy3 rat (donkey) | Jackson Immunoresearch | 712-165-150 |
| Anti-Cy3 mouse (donkey) | Jackson Immunoresearch | 715-165-150 |
| Anti-mouse AlexaFluor 488 (donkey) | Invitrogen | A-21202 |
| Anti-rat AlexaFluor 488 (donkey) | Invitrogen | A-21208 |
| Bacterial and Virus Strains | ||
| BL21(DE3) chemically competent E.coli | Thermo Fisher | C600003 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Linear diUbiquitin | Boston Biochem. | UC-700 |
| K6 diUbiquitin | Life Sensors | SI0602 |
| K11 diUbiquitin | Boston Biochem. | UC-40 |
| K27 diUbiquitin | Boston Biochem. | UC-61B |
| K29 diUbiquitin | Boston Biochem. | UC-81 |
| K33 diUbiquitin | Boston Biochem. | UC-101 |
| K48 diUbiquitin | Boston Biochem. | UC-200 |
| K63 diUbiquitin | Boston Biochem. | UC-300 |
| Ub-AMC | Boston Biochem. | U-550 |
| Ubiquitin | Rivkin et.al, 2013 | N/A |
| Sypro Orange | Thermo Fisher | S6650 |
| 15N-Ammonium Chloride | Cambridge Isotope | NLM-467–50 |
| Effectene transfection reagent | Qiagen | 301425 |
| Protease inhibitor - Complete mini EDTA free | Sigma-Aldrich | 11836170001 |
| Protein A/G plus agarose beads | Santa Cruz | SC-2003 |
| Paraformaldehyde | Sigma-Aldrich | P6148 |
| 4’,6-Diamidino-2-phenylindole dihydrochloride (DAPI) | Vector Laboratories | H-1200–10 |
| D-Biotin | Biobasic | BB0078 |
| Mammalian protease inhibitor cocktail | Sigma-Aldrich | P8340 |
| Streptavidin immobilized to Sepharose | GE | 17-5113-01 |
| Sequencing grade modified trypsin | Promega | V5113 |
| T etracycline | Sigma-Aldrich | T7660 |
| Turbonuclease | Sigma-Aldrich | T4330–50KU |
| TPCK trypsin | Promega | V-5111 |
| USP2 | Ernst et al., 2012 | N/A |
| Critical Commercial Assays | ||
| Protein Thermal Melt Assay | Roche | LightCycler 480 |
| Multiwell microplates | Roche | 04 729 749 001 |
| Dual luciferase assays | Promega | E1910 |
| Deposited Data | ||
| Crystal structure of FAM105A 87–356 | This paper | PDB: 6DRM |
| Raw mass spectrometry data archive | This paper | MassIVE: MSV000082315 |
| Crystal structure of OTULIN 79–352 | Rivkin et al., 2013 | PDB: 4KSJ |
| Crystal structure of OTULIN and M1-diUb | Rivkin et al., 2013 | PDB: 4KSK |
| Crystal structure of OTULIN and M1-diUb | Keusekotten et al., 2014 | PDB: 3ZNZ |
| Experimental Models: Cell Lines | ||
| Neuro2A | Rivkin et al., 2013 | N/A |
| HEK293T | Rivkin et al., 2013 | N/A |
| Flp-In T-REx 293 | ThermoFisher | R78007 |
| Oligonucleotides | ||
| FAM105A-N87: cgggatccaacctcagtgtggaggcagag | Millipore Sigma | N/A |
| FAM105A-C356: ccgctcgagttaaaagactggaatgtggtagtggcg | Millipore Sigma | N/A |
| D139C: cgacaagtaaggagagataactatTGT gctctcagatcagtgttatttcag | Millipore Sigma | N/A |
| H352N: ctgaccgagaacgaccgccactacAACattccagtcttttaactcgaggc | Millipore Sigma | N/A |
| SUMO 1-N1: gcggatccatgtctgaccaggaggcaaaacc | Millipore Sigma | N/A |
| SUMO 1-C97: ccgctcgagttaaccccccgtttgttcctg | Millipore Sigma | N/A |
| SUMO2-N1: cgggatccatggccgacgaaaagcccaaggaagg | Millipore Sigma | N/A |
| SUMO2-C93: ccgctcgagttaacctcccgtctgttggaacac | Millipore Sigma | N/A |
| ATG8-N1: gcggatccatgaagttccagtacaaggaggac | Millipore Sigma | N/A |
| ATG8-C116: ccgctcgagttacccatagacactctcatcactgtaggc | Millipore Sigma | N/A |
| ISG15-N1: cgggatccatgggctgggacctgacggtg | Millipore Sigma | N/A |
| ISG15-C157: ggcctcgagctagcctccccgcaggcgcag | Millipore Sigma | N/A |
| FAT 10-N8: cgggatccctctgtgtgcatgtccgttccgag | Millipore Sigma | N/A |
| FAT 10-C82: ccgctcgagttacttcaccactttcagggtaaggtg | Millipore Sigma | N/A |
| FAT10-N82: cgggatccaagcccagtgatgaggagctgccc | Millipore Sigma | N/A |
| FAT 10-C165: ccgctcgagtcaccctccaatgcaataacatgccag | Millipore Sigma | N/A |
| Ufm1-N1: cgggatccatgtcgaaggtttcctttaagatcacgc | Millipore Sigma | N/A |
| Ufm1-C83: ccgctcgagttatccaacacgatctctaggaataatccgc | Millipore Sigma | N/A |
| FAM105A-WT-F: aggcgcgccaatggcggcgacaaggagccccacg | Millipore Sigma | N/A |
| FAM105A-M84-F: aggcgcgccattcaaaaggaacctcagtgtggag | Millipore Sigma | N/A |
| FAM105A-C100X-R: cactcgagtcaacaataactgagtaaatcaacctc | Millipore Sigma | N/A |
| FAM105A-WT-R: cactcgaggctaaaagactggaatgtggtagtggcggtc | Millipore Sigma | N/A |
| GW-FAM105A-C100-R: ggggaccactttgtacaagaaagctgggtaacaataactgagtaaatcaacctc | Millipore Sigma | N/A |
| GW-FAM105A-F1: ggggacaagtttgtacaaaaaagcaggctggagacatggcggcgacaaggagccccacg | Millipore Sigma | N/A |
| GW-FAM105A-R1: ggggaccactttgtacaagaaagctgggtaaaagactggaatgtggtagtggcggtc | Millipore Sigma | N/A |
| Recombinant DNA | ||
| ProEx FAM105A - human 87–356 | This paper | BC011524 |
| ProEx AT G8 - human 1–116 | This paper | BC009309 |
| ProEx ISG15 - human 1–157 | This paper | DQ892714 |
| ProEx FAT10 - human 8–82, 82–165 | This paper | BC012472 |
| ProEx SUMO1 - human 1–97 | This paper | BC053528 |
| ProEx SUMO2 - human 1–93 | This paper | BC008450 |
| ProEx UFM1 - human 1–83 | This paper | BC005193 |
| pGEX-Ub | Rivkin et.al, 2013 | N/A |
| PRoEx-linear diUb | Rivkin et.al, 2013 | N/A |
| HA-FAM105A 1–356 | This paper | BC011524 |
| HA-FAM105A 84–356 | This paper | BC011524 |
| FLAG-FAM105A 1–356 | This paper | BC011524 |
| FLAG-FAM105A 1–100 | This paper | BC011524 |
| FLAG-FAM105A 84–356 | This paper | BC011524 |
| FLAG-OTULIN | Rivkin et al, 2013 | N/A |
| FLAG-OTULIN C93S | Rivkin et al, 2013 | N/A |
| HA-HOIL | Rivkin et al, 2013 | N/A |
| Myc-HOIP | Rivkin et al, 2013 | N/A |
| Myc-MAPKK 1-R4F | Mansour et al, 1994 | http:/n2t.net/addgene:40810 |
| pcDNA5 FRT/TO FAM105A | This paper | BC011524 |
| pcDNA5 FRT/TO CALR3 | This paper | BC014595 |
| pcDNA5 FRT/TO SYVN1 | This paper | BC030530 |
| Software and Algorithms | ||
| Adobe Photoshop | Adobe | CS6 |
| Xia2 | Winter et al., 2013 | 0.5.542 |
| XDS | Kabsch et al., 2010 | 20170923 |
| Phaser | McCoy et al., 2007 | 2.5.2 |
| PHENIX | Adams et al., 2011 | 1.13 |
| COOT | Emsley et al., 2004 | 0.8.9.1 |
| NMRPipe/NMRDraw | Delagio et al., 1995 | 20180515 |
| NMRView | Johnson et al., 2004 | 8.0.3 |
| Light Cycler Protein Melting software | Roche | N/A |
| Excel | MicroSoft | 16.16.5 |
| ProHits | Guomin et al., 2010 | 5.0.2 |
| Proteowizard | Kessner et al., 2008 | v.3 |
| X!Tandem | Craig and Beavis, 2004 | Alanine 2017.02.01 |
| Comet | Eng et al., 2013 | 2018011 |
| SAINTexpress | Teo et al., 2014 | 3.6.1 |
| ToppGene | Chen et al., 2009 | https://toppgene.cchmc.org/ |
Highlights:
FAM105A contains an OTU domain most closely related to OTULIN.
No deubiquitinase activity was detected for FAM105A.
FAM105A does not detectably interact with ubiquitin or UBLs.
FAM105A localizes to the endoplasmic reticulum via an N-terminal sequence element.
ACKNOWLEDGEMENTS
This work was supported by CIHR Operating grant 342269 and NSERC Discovery grant RGPIN-2015–05475 to S.P.C., CIHR Foundation grant FDN143301 to A-C.G., CIHR Project grant PJT156093 to B.R. and a CIHR Foundation grant FDN143277 to F.S.
NMR spectrometers were funded by the Canada Foundation for Innovation and the NMR Core Facility was supported by the Princess Margaret Cancer Foundation. We thank Dr. Geneviève Seabrook for technical expertise and access to the NMR Core Facility. Northeastern Collaborative Access Team beamlines used for diffraction studies are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P41 GM103403). The Pilatus 6M detector on 24-ID-C beam line is funded by a NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02–06CH11357.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information includes seven figures and three tables that can be found with this article online.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abdul Rehman SA, Kristariyanto YA, Choi SY, Nkosi PJ, Weidlich S, Labib K, Hofmann K, and Kulathu Y (2016). MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes. Mol Cell 63, 146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Echols N, Headd JJ, Hung LW, Jain S, Kapral GJ, Grosse Kunstleve RW, et al. (2011). The Phenix software for automated determination of macromolecular structures. Methods 55, 94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik P, Xiao H, Parr CL, Kiso Y, Gustchina A, Yada RY, and Wlodawer A (2009). Crystal structures of the histo-aspartic protease (HAP) from Plasmodium falciparum. J Mol Biol 388, 520–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozatzi P, and Sapkota GP (2018). The FAM83 family of proteins: from pseudo-PLDs to anchors for CK1 isoforms. Biochem Soc Trans 46, 761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne DP, Foulkes DM, and Eyers PA (2017). Pseudokinases: update on their functions and evaluation as new drug targets. Future Med Chem 9, 245–265. [DOI] [PubMed] [Google Scholar]
- Chen J, Bardes EE, Aronow BJ, and Jegga AG (2009). ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37, W305–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman KE, and Huang TT (2018). In a Class of Its Own: A New Family of Deubiquitinases Promotes Genome Stability. Mol Cell 70, 1–3. [DOI] [PubMed] [Google Scholar]
- Coyaud E, Mis M, Laurent EM, Dunham WH, Couzens AL, Robitaille M, Gingras AC, Angers S, and Raught B (2015). BioID-based Identification of Skp Cullin F-box (SCF)beta-TrCP½ E3 Ligase Substrates. Mol Cell Proteomics 14, 1781–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R, and Beavis RC (2004). TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, and Bax A (1995). NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Elliott PR, Nielsen SV, Marco-Casanova P, Fiil BK, Keusekotten K, Mailand N, Freund SM, Gyrd-Hansen M, and Komander D (2014). Molecular basis and regulation of OTULIN-LUBAC interaction. Mol Cell 54, 335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, Jahan TA, and Hoopmann MR (2013). Comet: an open-source MS/MS sequence database search tool. Proteomics 13, 22–24. [DOI] [PubMed] [Google Scholar]
- Ernst A, Avvakumov G, Tong J, Fan Y, Zhao Y, Alberts P, Persaud A, Walker JR, Neculai AM, Neculai D, et al. (2013). A strategy for modulation of enzymes in the ubiquitin system. Science 339, 590–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faesen AC, Luna-Vargas MP, Geurink PP, Clerici M, Merkx R, van Dijk WJ, Hameed DS, El Oualid F, Ovaa H, and Sixma TK (2011). The differential modulation of USP activity by internal regulatory domains, interactors and eight ubiquitin chain types. Chem Biol 18, 1550–1561. [DOI] [PubMed] [Google Scholar]
- Fennell LM, Rahighi S, and Ikeda F (2018). Linear ubiquitin chain-binding domains. FEBS J 285, 2746–2761. [DOI] [PubMed] [Google Scholar]
- Fulcher LJ, Bozatzi P, Tachie-Menson T, Wu KZL, Cummins TD, Bufton JC, Pinkas DM, Dunbar K, Shrestha S, Wood NT, et al. (2018). The DUF1669 domain of FAM83 family proteins anchor casein kinase 1 isoforms. Sci Signal 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta GD, Coyaud E, Goncalves J, Mojarad BA, Liu Y, Wu Q, Gheiratmand L, Comartin D, Tkach JM, Cheung SW, et al. (2015). A Dynamic Protein Interaction Landscape of the Human Centrosome-Cilium Interface. Cell 163, 1484–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamazaki J, Hirayama S, and Murata S (2015). Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis. PLoS Genet 11, e1005401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Zeqiraj E, Dong B, Jha BK, Duffy NM, Orlicky S, Thevakumaran N, Talukdar M, Pillon MC, Ceccarelli DF, et al. (2014). Dimeric structure of pseudokinase RNase L bound to 2–5A reveals a basis for interferon-induced antiviral activity. Mol Cell 53, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA (2004). Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol 278, 313–352. [DOI] [PubMed] [Google Scholar]
- Juretic D, Zoranic L, and Zucic D (2002). Basic charge clusters and predictions of membrane protein topology. J Chem Inf Comput Sci 42, 620–632. [DOI] [PubMed] [Google Scholar]
- Kang TH, and Kim KT (2006). Negative regulation of ERK activity by VRK3-mediated activation of VHR phosphatase. Nat Cell Biol 8, 863–869. [DOI] [PubMed] [Google Scholar]
- Kessner D, Chambers M, Burke R, Agus D, and Mallick P (2008). ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24, 2534–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keusekotten K, Elliott PR, Glockner L, Fiil BK, Damgaard RB, Kulathu Y, Wauer T, Hospenthal MK, Gyrd-Hansen M, Krappmann D, et al. (2013). OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell 153, 1312–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, and Iwai K (2006). A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J 25, 4877–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, and Rape M (2012). The ubiquitin code. Annu Rev Biochem 81, 203–229. [DOI] [PubMed] [Google Scholar]
- Kopec J, and Schneider G (2011). Comparison of fluorescence and light scattering based methods to assess formation and stability of protein-protein complexes. J Struct Biol 175, 216–223. [DOI] [PubMed] [Google Scholar]
- Mansour SJ, Matten WT, Hermann AS, Candia JM, Rong S, Fukasawa K, Vande Woude GF, and Ahn NG (1994). Transformation of mammalian cells by constitutively active MAP kinase kinase. Science 265, 966–970. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J Appl Crystallogr 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevissen TE, Hospenthal MK, Geurink PP, Elliott PR, Akutsu M, Arnaudo N, Ekkebus R, Kulathu Y, Wauer T, El Oualid F, et al. (2013). OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell 154, 169–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin A, Eisenbraun B, Key J, Sanschagrin PC, Timony MA, Ottaviano M, and Sliz P (2013). Collaboration gets the most out of software. Elife 2, e01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JM, Mace PD, and Eyers PA (2017). Live and let die: insights into pseudoenzyme mechanisms from structure. Curr Opin Struct Biol 47, 95–104. [DOI] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, and Bernards R (2005). A genomic and functional inventory of deubiquitinating enzymes. Cell 123, 773–786. [DOI] [PubMed] [Google Scholar]
- Phan J, Zdanov A, Evdokimov AG, Tropea JE, Peters HK 3rd, Kapust RB, Li M, Wlodawer A, and Waugh DS (2002). Structural basis for the substrate specificity of tobacco etch virus protease. J Biol Chem 277, 50564–50572. [DOI] [PubMed] [Google Scholar]
- Pickart CM, and Raasi S (2005). Controlled synthesis of polyubiquitin chains. Methods Enzymol 399, 21–36. [DOI] [PubMed] [Google Scholar]
- Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, et al. (2009). Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell 136, 1098–1109. [DOI] [PubMed] [Google Scholar]
- Rittinger K, and Ikeda F (2017). Linear ubiquitin chains: enzymes, mechanisms and biology. Open Biol 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivkin E, Almeida SM, Ceccarelli DF, Juang YC, MacLean TA, Srikumar T, Huang H, Dunham WH, Fukumura R, Xie G, et al. (2013). The linear ubiquitin-specific deubiquitinase gumby regulates angiogenesis. Nature 498, 318–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Raida M, and Burke B (2012). A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196, 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer V, Akutsu M, Olma MH, Gomes LC, Kawasaki M, and Dikic I (2014). Binding of OTULIN to the PUB domain of HOIP controls NF-kappaB signaling. Mol Cell 54, 349–361. [DOI] [PubMed] [Google Scholar]
- Scheeff ED, Eswaran J, Bunkoczi G, Knapp S, and Manning G (2009). Structure of the pseudokinase VRK3 reveals a degraded catalytic site, a highly conserved kinase fold, and a putative regulatory binding site. Structure 17, 128–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi TS, Hirade Y, Toma A, Sato Y, Yamagata A, Goto-Ito S, Tomita A, Nakada S, and Fukai S (2018). Structural insights into two distinct binding modules for Lys63-linked polyubiquitin chains in RNF168. Nat Commun 9, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneera J, Fadista J, Ahlqvist E, Atac D, Ottosson-Laakso E, Wollheim CB, and Groop L (2015). Identification of novel genes for glucose metabolism based upon expression pattern in human islets and effect on insulin secretion and glycemia. Hum Mol Genet 24, 1945–1955. [DOI] [PubMed] [Google Scholar]
- Theng SS, Wang W, Mah WC, Chan C, Zhuo J, Gao Y, Qin H, Lim L, Chong SS, Song J, et al. (2014). Disruption of FAT10-MAD2 binding inhibits tumor progression. Proc Natl Acad Sci U S A 111, E5282–5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, et al. (2015). Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- Vincendeau M, Hadian K, Messias AC, Brenke JK, Halander J, Griesbach R, Greczmiel U, Bertossi A, Stehle R, Nagel D, et al. (2016). Inhibition of Canonical NF-kappaB Signaling by a Small Molecule Targeting NEMO-Ubiquitin Interaction. Sci Rep 6, 18934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden M, Masandi SK, Pawlowski K, and Zeqiraj E (2018). Pseudo-DUBs as allosteric activators and molecular scaffolds of protein complexes. Biochem Soc Trans 46, 453–466. [DOI] [PubMed] [Google Scholar]
- Wang AC, Grzesiek S, Tschudin R, Lodi PJ, and Bax A (1995). Sequential backbone assignment of isotopically enriched proteins in D2O by deuterium-decoupled HA(CA)N and HA(CACO)N. J Biomol NMR 5, 376–382. [DOI] [PubMed] [Google Scholar]
- Winter G, Lobley CM, and Prince SM (2013). Decision making in xia2. Acta Crystallogr D Biol Crystallogr 69, 1260–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeqiraj E, Tian L, Piggott CA, Pillon MC, Duffy NM, Ceccarelli DF, Keszei AF, Lorenzen K, Kurinov I, Orlicky S, et al. (2015). Higher-Order Assembly of BRCC36-KIAA0157 Is Required for DUB Activity and Biological Function. Mol Cell 59, 970–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
BioID list of FAM105A high confidence hits.
Complete BioID analysis of FAM105A, SYVN1 and CALR3.
Data Availability Statement
Data Resources.
The FAM105A87−356 coordinates and structure factors have been deposited in the Protein Data Bank under accession code 6DRM. The FAM105A BioID data has been deposited in the raw mass spectrometry data archive at MassIVE: MSV000082315.