Summary
Proteasomal shuttle factor UBQLN2 is recruited to stress granules and undergoes liquid-liquid phase separation (LLPS) into protein-containing droplets. Mutations to UBQLN2 have recently been shown to cause dominant x-linked inheritance of ALS and ALS/dementia. Interestingly, most of these UBQLN2 mutations reside in its proline-rich (Pxx) region, an important modulator of LLPS. Here, we demonstrated that ALS-linked Pxx mutations differentially affect UBQLN2 LLPS, depending on both amino acid substitution and sequence position. Using size-exclusion chromatography, analytical ultracentrifugation, microscopy, and NMR spectroscopy, we determined that those Pxx mutants that enhanced UBQLN2 oligomerization decreased saturation concentrations needed for LLPS and promoted solid-like and viscoelastic morphological changes to UBQLN2 liquid assemblies. Ubiquitin disassembled all LLPS-induced mutant UBQLN2 aggregates. We postulate that the changes in physical properties caused by ALS-linked Pxx mutations modify UBQLN2 behavior in vivo, possibly contributing to aberrant stress granule morphology and dynamics, leading to formation of inclusions, pathological characteristics of ALS.
Keywords: Liquid-liquid phase separation, ubiquilin-2, ubiquitin, ALS, viscoelasticity, proline-rich, self-assembly, oligomerization, aggregation
Graphical Abstract
eTOC Blurb
UBQLN2, part of protein quality control machinery in cells, phase separates under physiological conditions. In this issue of Structure, Dao, Martyniak et al. show that a subset of ALS-linked mutations in UBQLN2 disrupt phase separation, promote oligomerization, and change the material properties of UBQLN2 droplets in vitro.
Introduction
Liquid-liquid phase separation (LLPS) is proposed to drive the formation of dynamic, membraneless organelles, including stress granules. Stress granules contain various RNA-binding proteins, such as TDP-43, FUS, hnRNPA1/2, and TIA-1, all of whom undergo LLPS in vitro (Conicella et al., 2016; Lin et al., 2015; Mackenzie et al., 2017; Molliex et al., 2015; Patel et al., 2015; Ryan et al., 2018). Disease mutations in these proteins disrupt LLPS by facilitating a liquid-to-solid transition or the formation of aggregates that resemble cytoplasmic inclusions, pathological hallmark of neurodegenerative diseases such as amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD), and multisystem proteinopathy (Lin et al., 2015; Mackenzie et al., 2017; Molliex et al., 2015; Patel et al., 2015). These observations suggest that disease mutations alter LLPS properties of stress granules. In turn, stress granule dynamics are disrupted, leading to accumulation of persistent stress granules, which may mature into pathological inclusions (Molliex et al., 2015).
Important to stress granule maintenance are protein quality control (PQC) components such as the VCP/p97/CDC48 segregase and HSP70 chaperone proteins, among others (Alberti and Carra, 2018; Buchan et al., 2013; Jain et al., 2016; Walters and Parker, 2015). These PQC components clear misfolded proteins from and maintain liquidity of stress granules (Alberti and Carra, 2018; Mateju et al., 2017). We recently showed that the proteasomal shuttle factor UBQLN2, a PQC member of the ubiquitin-proteasome system and autophagy protein homeostasis mechanisms, is recruited to stress granules, and also undergoes LLPS in vitro (Dao et al., 2018). Moreover, LLPS behavior is critical for UBQLN2 to form stress-induced puncta inside cells (Dao et al., 2018). Through its STI1, UBA and UBL domains, UBQLN2 interacts with stress granule components HSP70, ubiquitin (Ub), and the proteasome, respectively (Kwon et al., 2007; Turakhiya et al., 2018). UBQLN2 also contains a unique C-terminal proline-rich (Pxx) region, a domain not present in the other four UBQLN paralogs (UBQLN1, UBQLN3, UBQLN4, UBQLNL). Most disease-associated mutations of UBQLN2 reside in the Pxx region, and disrupt Ub-mediated proteasome mechanisms and HSP70 chaperone processes (Chang and Monteiro, 2015; Deng et al., 2011; Hjerpe et al., 2016). These Pxx mutations are known to cause dominantly inherited X-linked ALS and FTD, with ALS patients carrying characteristic pathological inclusions in degenerated motor neurons (Deng et al., 2011; Le et al., 2016). Recent mutant UBQLN2 ALS mouse models also recapitulate ALS-like disease hallmarks such as reduced motor neuron function and/or motor neuron death (Le et al., 2016). However, the molecular mechanisms underlying how UBQLN2 Pxx mutations promote ALS disease states are not understood.
We previously identified the Pxx region as an important modulator of UBQLN2 LLPS (Dao et al., 2018). Since disruption of LLPS is thought to lead to the formation of inclusions and Pxx mutations in UBQLN2 cause inclusions in ALS patients, we hypothesized that these mutations significantly alter UBQLN2 LLPS behavior. In this work, we systematically examined the LLPS properties of eleven ALS-linked Pxx mutations in a C-terminal construct of UBQLN2 via a comprehensive suite of biochemical and biophysical experiments including fluorescence microscopy, spectrophotometric assays, size-exclusion chromatography, analytical ultracentrifugation and NMR spectroscopy. Interestingly, we found that many, but not all, of these mutations decreased the saturation concentrations needed for LLPS compared to WT UBQLN2, promoted liquid-to-solid transitions, and/or induced the formation of amorphous aggregates over time. We determined that the type and location of amino acid substitution modulate the oligomerization state of UBQLN2, and in turn, its phase separation.
Results
We expressed and purified eleven UBQLN2 PXX mutants, including T487I, A488T, P497H, P497L, P497S, P500S, P506A, P506S, P506T, P509S, and P525S (Deng et al., 2011; Fahed et al., 2014; Gellera et al., 2013; Teyssou et al., 2017; Vengoechea et al., 2013; Williams et al., 2012) (Figure 1A). For this study, we expressed these Pxx mutants in the UBQLN2 450–624 background, which is the minimal UBQLN2 construct that undergoes phase separation, and exhibits similar LLPS properties as full-length UBQLN2 (Dao et al., 2018). UBQLN2 450–624 is easy to express and purify in milligram quantities from E. coli, and is more amenable for NMR spectroscopy permitting atomic-level detail on protein structure and dynamics (Dao et al., 2018). We purified tagless versions of these proteins by using UBQLN2’s LLPS properties to isolate UBQLN2 from E. coli lysate (Dao et al., 2018) (See STAR Methods).
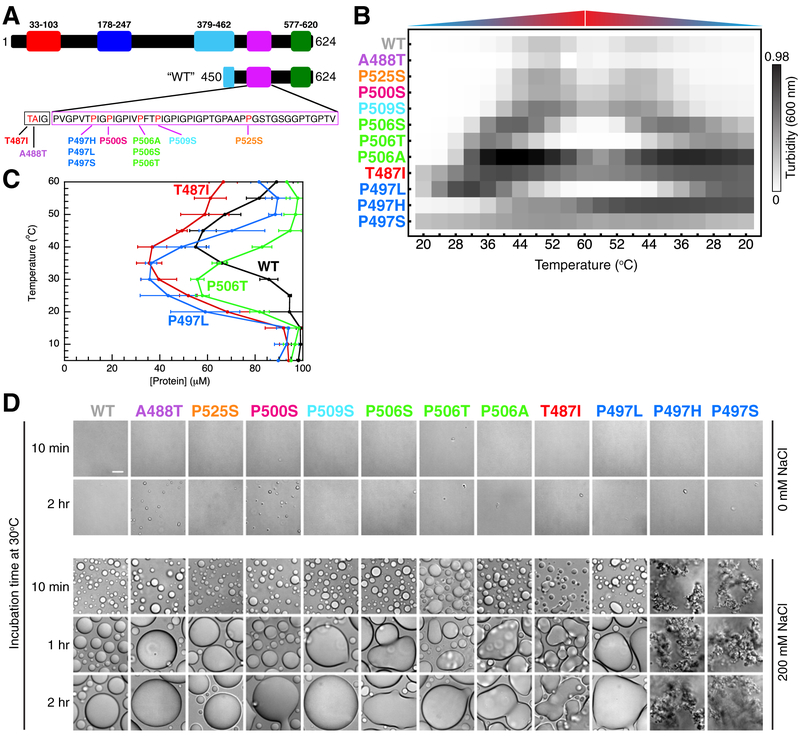
Figure 1. ALS-linked mutations disrupt UBQLN2 LLPS and alter droplet morphology.
(A) Domain architecture map of UBQLN2 and the “WT” UBQLN2 450–624 construct used here. The Pxx sequence is shown with ALS-linked mutation sites highlighted in red. (B) Results from spectrophotometric turbidity assay as a function of temperature comparing LLPS of different UBQLN2 Pxx mutants using 25 μM protein in 20 mM NaPhosphate and 200 mM NaCl (pH 6.8). For P497S and P497H, unevenly distributed aggregates are observed at the end of each assay. Results were approximately organized in increasing turbidity. Data were averaged from n=6 experiments using proteins from two separate preps and three trials for each. (C) Phase diagrams for WT and mutant UBQLN2 450–624 constructed from saturation concentration measurements as a function of temperature. Measurements were obtained by using 100 μM protein in 20 mM NaPhosphate and 200 mM NaCl (pH 6.8). Error bars represent standard deviation over three trials. (D) DIC microscopy of UBQLN2 Pxx mutants over indicated times at 30°C using 100 μM protein in 20 mM NaPhosphate (pH 6.8) buffer in the absence of added NaCl (top panel) and presence of 200 mM NaCl (bottom panel). Scale bar = 10 μm.
ALS-linked mutants of UBQLN2 disrupt LLPS behavior
We used a spectrophotometric assay to screen for LLPS of UBQLN2 Pxx mutants over a temperature range between 20°C and 60°C (Figure 1B, Figure S1A,B). High and low A600 values correlate with UBQLN2 droplet formation and droplet clearance, respectively (Dao et al., 2018). For wild-type UBQLN2, protein solution became turbid with increasing temperature (Dao et al., 2018), but clarified slightly at high temperatures (approaching 55–60°C). Turbidity was reversible when temperature was decreased and the solution clarified at low temperatures (< 35°C). To investigate whether the UBQLN2 remained f olded over this temperature range, we used NMR spectroscopy to monitor the temperature dependence of UBQLN2 amide signals under both non-phase separating and phase-separating conditions (Figures S1C and S1D). In non-phase separating conditions, resonances for the UBA globular domain (residues 580–620) exhibited no peak intensity loss, suggesting that the protein remained folded between 25°C and 55°C. Backbone amide signals for the intrinsically disordered region (residues 450–580) broadened beyond detection with increasing temperature due to increased proton exchange rates with water and/or conformational exchange. In 200 mM NaCl, signal intensity for the UBA resonances decreased as UBQLN2 phase separated with increasing temperature. Importantly, UBA resonances were still detectable at 55°C and UB A chemical shifts were similar to the values obtained in the absence of added NaCl.
Next, we monitored turbidity of the eleven ALS-linked Pxx mutants in UBQLN2. We noted WT-like, reversible phase separation behavior for the A488T, P500S, P509S, and P525S mutants (top of Figure 1B). Strikingly, protein solutions of the remaining mutants (P506S, P506T, P506A, T487I, P497L, P497H, and P497S) showed increased turbidity at lower temperatures and remained turbid over a significantly larger temperature ranges than WT UBQLN2. Solutions of P506S, P506T and P497L mutants clarified completely at high temperatures (52–60°C). By contrast, T487I, P497H and P497S mutants remained turbid over the entire temperature range examined, and visible aggregates were present for P497H and P497S samples. We confirmed the reproducibility of these experiments by subjecting the same protein sample to multiple rounds of temperature-induced LLPS (Figure S1A).
Phase Diagrams of UBQLN2 Pxx Mutants
The turbidity assays suggested that some of the UBQLN2 Pxx mutants can be characterized by closed loop phase diagrams (Ruff et al., 2018) with both lower critical solution temperature (LCST) and upper critical solution temperature (UCST) phase transitions (Figure S1E). Between LCST and UCST, the system phase separates into a protein-dense phase (droplets) and protein-dilute phase (outside droplets). To quantify the effects of Pxx mutations on UBQLN2 LLPS, we measured the concentrations of the dilute phase (saturation concentrations, cs) as a function of temperature (Figure 1C). The cs values inform on the protein concentration beyond which protein partitions into droplets, and thus quantify the low-concentration arm of coexistence curves. We obtained cs by centrifugation of 100 μM samples (Figure 1C). We repeated these measurements using other concentrations and obtained similar trends across the mutants, but noted that the absolute cs increased with increased starting protein concentration. We speculate that centrifugation is not the optimal method to determine the cs values for UBQLN2. Indeed, centrifugation can disrupt phase boundaries and how the protein partitions between the dilute and dense phases (Posey et al., 2018). Our cs values corroborated the results of the turbidity assays in Figure 1B. For those mutants that exhibited altered LLPS properties compared to WT, cs values were considerably lower than WT at temperatures < 40 °C, suggesting these mutations significantly lower the concentrations necessary for LLPS. In addition, we observed that hydrophobic Pxx mutations T487I and P497L promoted LCST phase transitions at lower temperatures and concentrations similar to the effect of hydrophobic substitutions in elastin-like proteins (Urry et al., 1992). Closed loop phase diagrams are characteristic of nonpolar solutes in aqueous solutions (Moelbert and De Los Rios, 2003), in line with our previous suggestions that UBQLN2 LLPS is driven by hydrophobic interactions (Dao et al., 2018).
ALS-linked mutants alter droplet morphology and material states
We used differential interference contrast (DIC) microscopy to monitor droplet morphology over time (Figure 1D), as LLPS is not directly observed in the spectrophotometric assay. Under non-phase separating conditions (no added NaCl), no droplets were observed for any mutant over a 2-hour period; all solutions were clear. When LLPS was induced with 200 mM NaCl, droplets formed, but droplet morphology varied significantly depending on the type of amino acid substitution and site of mutation. The A488T, P500S, P509S, and P525S mutants all exhibited WT-like LLPS behavior, consistent with spectrophotometric turbidity assay results above. Droplets were round, and grew in size over time as a result of droplet fusion, consistent with liquid-like behavior. In contrast, the remaining Pxx mutants formed either amorphous droplets or aggregates. These characteristics were most apparent for P497L, P506A, T487I, P497H and P497S mutants. Notably, all three mutations at position 497 significantly altered droplet morphology, even producing aggregates that did not change over time, in the case of P497H or P497S. These aggregates were present even in a 2:1 mixture of WT and P497S proteins that were fluorescently labeled with Dylight-488 and 650 fluorophores, respectively (Figure S2A). To test the reversibilities of Pxx mutant assemblies, we added 1,6-hexanediol, a compound known to dissolve liquid assemblies, hydrogels, and some fibrillar assemblies (Kroschwald et al., 2015, 2017; Lin et al., 2016; Patel et al., 2007). In all cases, treatment with 5% 1,6-hexanediol cleared LLPS-induced solutions of mutant UBQLN2, including P497S and P497H aggregates. To determine whether these aggregates were filamentous in nature, we performed transmission electron microscopy (TEM) experiments for WT and P497S proteins. Most of the WT grid did not contain any visible species. Only with careful search did we find an area with amorphous aggregates (Figure S2B). In contrast, we observed short, filamentous assemblies throughout the P497S grid. These TEM data are consistent with recent TEM experiments for P506T protein (Sharkey et al., 2018). Therefore, mutant UBQLN2 may increase aggregation propensity and fibril formation.
To assess the material properties of mutant UBQLN2 droplets, we monitored fusion events (Figure 2A) and measured the characteristic relaxation times (Figure 2B) for WT and mutant droplets with samples that were incubated for 10 minutes under phase-separating conditions. Consistent with data above, droplets that exhibited WT-like behavior (A488T, P500S, P509S, and P525S) fused and returned to a round shape quickly at a similar rate as WT. In contrast, T487I fused droplets relaxed very slowly, resulting in amorphous droplets with apparent gel-like consistency. P497L, P506A, P506S, and P506T droplets relaxed at rates intermediate between WT and T487I droplets.
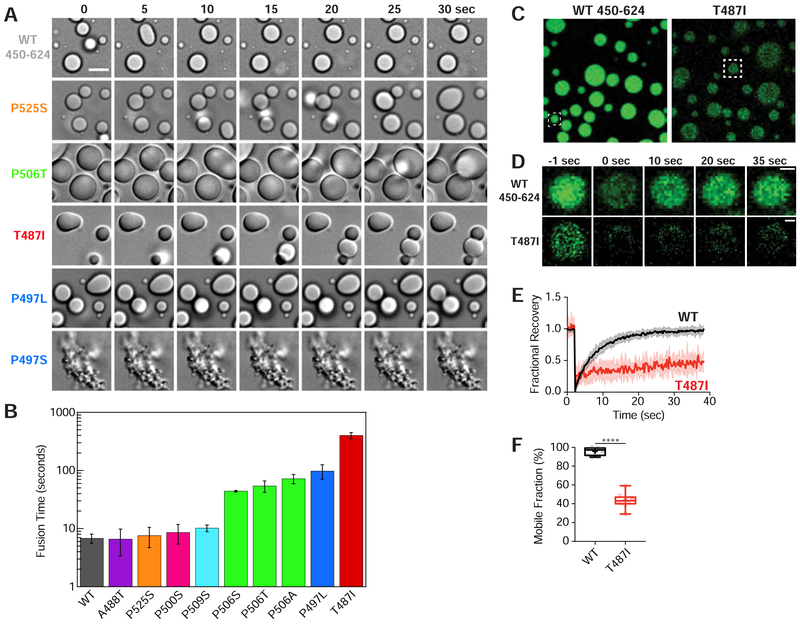
Figure 2. ALS-linked mutations promote viscoelastic behavior.
(A) Snapshots of droplet fusion over a 30 second window, indicating differences in droplet fusion kinetics. Droplets were imaged 10 minutes after incubating 100 μM protein at 30°C in buffer containing 20 mM NaPhosphate and 200 mM NaCl (pH 6.8). Arrowheads denote example droplet fusions. Scale bar = 5 μm. (B) Average characteristic relaxation times for WT and mutant droplet fusion. Error bars represent the standard deviation (SD) over nine droplets. (C) Representative snapshots of droplets used in FRAP experiments. Boxed are droplets whose FRAP events are shown in (D). (D) FRAP of UBQLN2 droplets. Representative fluorescence images of photobleaching experiment. Scale bar = 1 μm. (E) Normalized fluorescence intensity over time with thick lines indicating averages over 10 droplets. Error bars represent the SD. (F) Box and whisker plot representing minimum and maximum values. Cross represents the mean. Student’s unpaired T-test, p < 0.0001.
To examine the liquidity of UBQLN2 droplets, we performed fluorescence recovery after photobleaching (FRAP) experiments. Fast and complete fluorescence recovery rates suggest that the droplets maintain liquid-like behavior with rapid exchange of protein in and out, whereas slow recovery rates suggest that protein mobility is significantly impaired. For these experiments, we photobleached a portion of 2 mm droplets for WT and T487I mutant. For WT UBQLN2, droplet fluorescence signal quickly and fully recovered with a mobile fraction of 95.9 ± 1.3% (Figure 2C–2F). By contrast, T487I droplet fluorescence only partly recovered (43.6 ± 2.4%) after photobleaching (Figure 2C–2F). These results were entirely consistent with droplet fusion data above. Together, these data suggested that single point mutations in the Pxx region can drastically alter UBQLN2 LLPS and change the material properties of LLPS-induced droplets. Importantly, mutant UBQLN2 droplets are not irreversible aggregates, as they can be dissolved by 1,6-hexanediol and undergo reversible phase separation.
Disease-linked mutants promote UBQLN2 oligomerization
Previously, we determined that UBQLN2 self-association was a prerequisite for LLPS behavior (Dao et al., 2018), consistent with other systems that phase separate in vitro and in vivo (Bouchard et al., 2018; Marzahn et al., 2016; Mitrea et al., 2018). We hypothesized that those disease-linked UBQLN2 Pxx mutants that phase-separated at lower temperatures than WT and exhibited amorphous droplet morphology were more prone to oligomerize. We monitored self-association using size-exclusion chromatography (SEC) and sedimentation velocity analytical ultracentrifugation (SV-AUC) under non-phase separating conditions (no added NaCl).
SEC data for WT UBQLN2 show evidence for protein self-association as protein concentration is increased between 10 mM and 500 mM, as previously demonstrated (Figure 3A) (Dao et al., 2018). To identify the oligomeric species present in solution, we performed SV-AUC experiments under identical experimental conditions as SEC (Figure 3B, Figure S3B). WT UBQLN2 450–624 was monomeric at 10 μM and 100 μM concentrations (experimental sedimentation coefficient of 1.7S). As protein concentration was increased towards 500 μM, additional peaks at 3.4S appeared (Figure 3C), likely indicating a mixture consisting of dimer, trimer or tetramer species in equilibrium with monomers. To determine the mechanism by which UBQLN2 450–624 oligomerizes, we determined the average molar masses of the sedimenting species by employing a two-dimensional size-and-shape analysis (c(s,fr)) of the SV-AUC data at each protein concentration (Brown and Schuck, 2006). Transformation of each c(s,fr) distribution into a sedimentation coefficient-molar mass distribution (c(s,M)) revealed that UBQLN2 is largely monomeric up to 100 μM concentrations, although there are multiple species of similar mass ranging in S values between 1.2 and 2, possibly indicating different conformations of the monomer (Figure S3B). As concentration is increased further, broadly-defined dimer- and trimer-sized species emerge (~ 40–60 kDa) with a tetramer or hexamer-sized species (100 kDa) appearing at 800 μM. A monomer-trimer-hexamer association model fits well to our experimental data, although we cannot rule a monomer-dimer-tetramer model. Together with the SEC data, these results are consistent with a model that describes continuous self-association into higher-order oligomers.
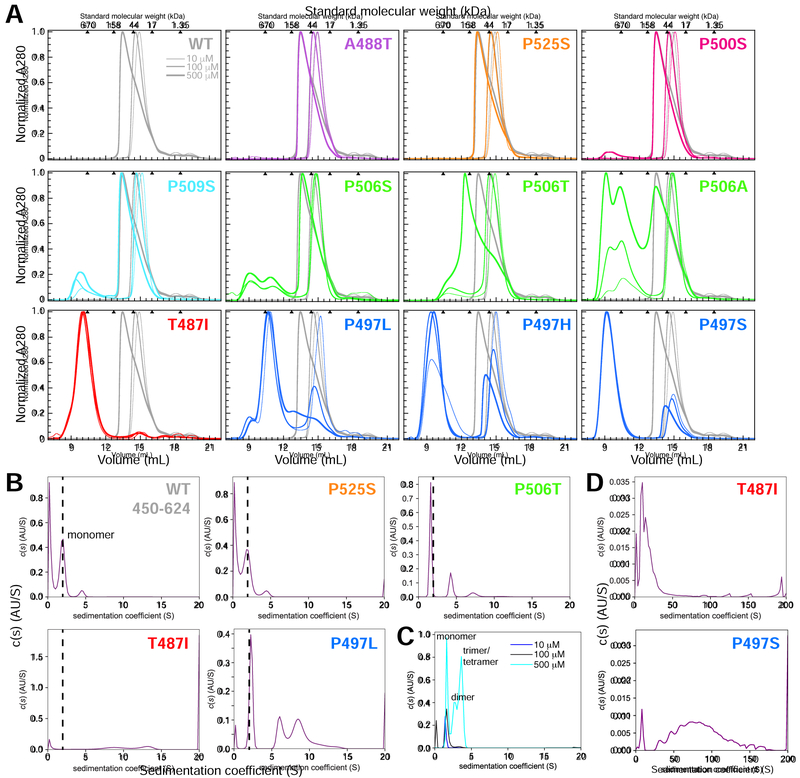
Figure 3. ALS-linked mutations increase UBQLN2 self-assembly/oligomerization.
(A) Representative SEC of UBQLN2 Pxx mutants at 10 μM (thinnest line), 100 μM (medium-thick), and 500 μM (thickest) protein concentrations using buffer containing 20 mM NaPhosphate (pH 6.8). For each mutant, WT SEC curves were plotted in gray for visual comparison. Molecular weights for globular protein standards are shown above each plot. Curves were organized and color-coded to match Figure 1. (B) Diffusion-free sedimentation coefficient distribution c(s) plots for WT and the Pxx mutants at 100 μM protein concentration from SV-AUC experiments. WT is mostly monomeric (dashed line) at this protein concentration, thus making it easier to compare higher order oligomerization states for mutant proteins. (C) Normalized c(s) curves for WT in increasing UBQLN2 protein concentration (10–500 μM). (D) c(s) plots for T487I and P497S mutants over a greater sedimentation coefficient range than used in part B.
For A488T, P500S, P509S, and P525S Pxx mutants, we observed similar SEC chromatograms compared to WT over the protein concentration ranges tested (Figure 3A). Furthermore, SV-AUC data for the P525S mutant at 100 μM concentration corroborated SEC data, as the P525S mutant exhibited a SV-AUC profile nearly identical to WT 450–624 (Figure 3B). These data support a view whereby the A488T, P500S, P509S and P525S mutations do not affect UBQLN2 oligomerization or LLPS, as spectrophotometric assay and microscopy experiments for these mutants suggest they behave similarly to WT.
By contrast, SEC profiles for the T487I, P497H, P497L, P497S, and P506A mutations were strikingly different from WT (Figure 3A). For this subclass of mutants, protein eluted as early as 9–10 mL, approaching the void volume of the SEC column. Note that relative peak volumes for these mutants vary by 10% or less depending on each protein prep, but the positions do not change. The SEC data for these mutants suggested a high degree of oligomerization, even at low 10 – 100 μM concentrations; these observations were corroborated by SV-AUC experiments (Figures 3B and 3D). The P497S and T487I mutants oligomerized extensively with species exceeding sedimentation coefficients of 20S. P497L and P506T AUC profiles were intermediate between WT and the T487I/P497S mutants. By performing a 2D size-and shape analysis for P506T, we found that the mutation promoted higher-order oligomerization states not seen for WT (Figure S3G).
Even for Pxx mutants with higher-order oligomers present, oligomerization was indeed reversible (Figure S3A). Eluted fractions for T487I or P497L were subjected to SEC and elution peaks for both monomeric and oligomeric species were observed. The relative amount of each species was not the same for the two mutants, likely due to differences in re-equilibration kinetics as well as differences in protein concentrations of the fractions. Together, these data support the interpretation that the Pxx mutations (e.g. P497 and P506 mutations) that significantly increase UBQLN2 propensity to oligomerize also lower the saturation concentration needed for UBQLN2 LLPS and/or promote amorphous and less liquid-like droplet morphology.
Pxx mutations minimally perturb UBQLN2 structure
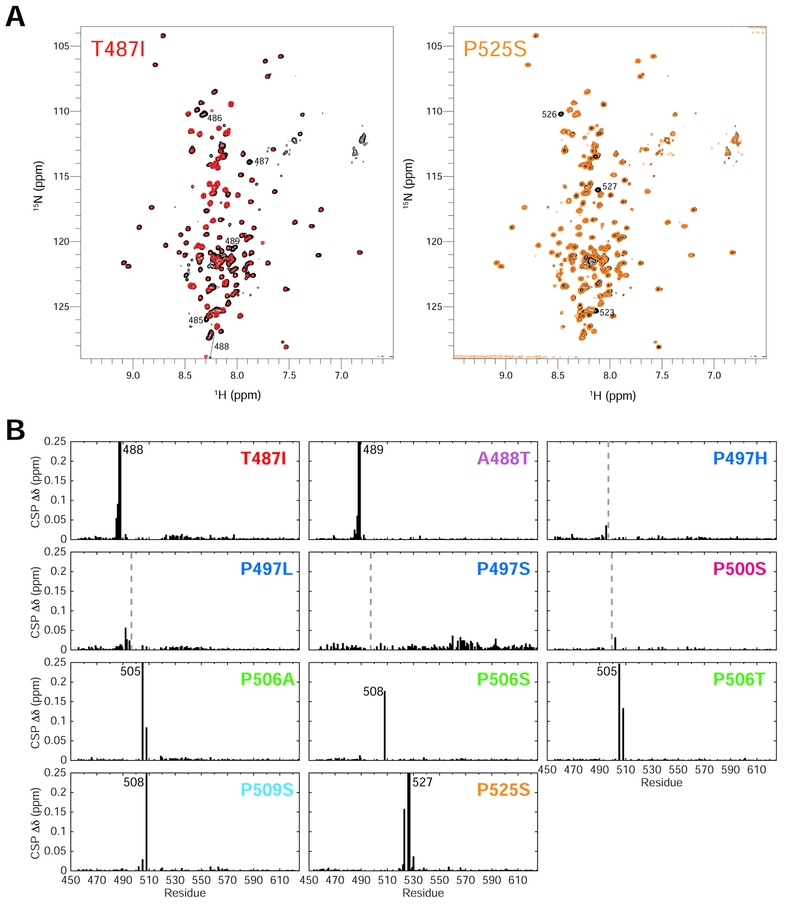
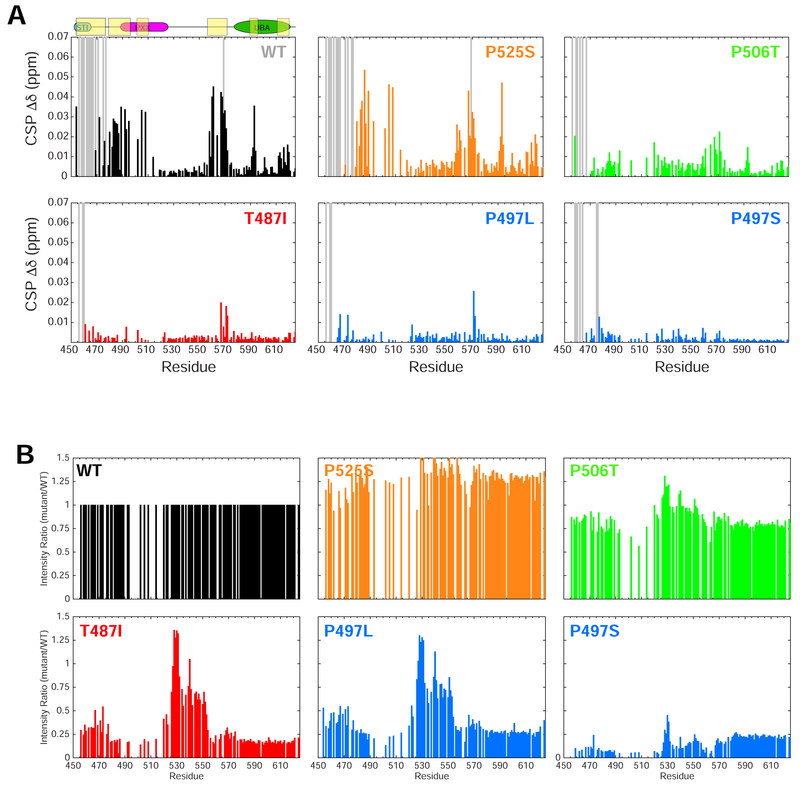
To probe how UBQLN2 Pxx mutations promote enhanced oligomerization on a residue-by-residue level, we employed NMR spectroscopy. We previously showed that residues 450–580 of the UBQLN2 450–624 are intrinsically disordered, including the Pxx region where most of the disease-linked mutations reside (Dao et al., 2018). To ascertain whether Pxx mutations disrupt UBQLN2 structure, we monitored backbone amide 1H and 15N NMR chemical shifts for all Pxx mutants and compared these to WT (at 50 μM protein concentration). As several resonances in the highly-repetitive Pxx region were difficult to assign (Dao et al., 2018), we generated numerous constructs containing point mutations or deletion of several residues to complete these assignments as best as possible (see STAR Methods). Secondary structure for the Pxx region was confirmed to be intrinsically disordered based on carbon chemical shifts (Dao et al., 2018). We compared spectra and report non-proline chemical shift perturbations (CSPs) between UBQLN2 mutants and WT in Figure 4A and Figure 4B, respectively. In all cases, NMR spectra revealed that Pxx mutations minimally perturb UBQLN2 chemical shifts. Only those residues close in primary structure to the mutation site (+/− 4–5 residues) exhibited significant CSPs (Figure 4B), suggesting that the overall structure of the protein remains unaffected.
Figure 4. ALS-linked mutations minimally disrupt UBQLN2 structure.
(A) Representative NMR 1H-15N spectra of T487I (red) and P525S (orange) Pxx mutants compared to WT UBQLN2 (black). Spectra were collected under identical conditions, and contour settings are the same for all spectra. Those resonances with large chemical shift changes are labeled with residue number. (B) Backbone amide chemical shift perturbations (CSPs) are calculated between resonances of Pxx mutant and WT using 50 μM protein. CSPs are calculated as described in Methods. Dotted lines denote mutation site. In some cases, no CSPs were observed due to missing backbone assignments surrounding the mutation site.
NMR data recapitulate mutant UBQLN2 self-association propensities
Using NMR spectroscopy, we previously showed that the UBQLN2 450–624 exhibited concentration-dependent CSPs, which allowed us to identify the residues involved in mediating UBQLN2 oligomerization (Dao et al., 2018). With more complete chemical shift assignments for the Pxx region (see above), we reproduced those data for WT over a protein concentration between 50 μM and 350 μM (Figure 5A). As previously identified, STI1-II residues 450–470 were significantly broadened beyond detection at 350 μM, consistent with their role in mediating UBQLN2 oligomerization (Dao et al., 2018; Kurlawala et al., 2017). Residues 470–508 exhibited small concentration-dependent CSPs > 0.02 ppm (Figure 5A), suggesting that weak interactions involving only the first half of the Pxx region (approximately residues 490–510) modulate UBQLN2 oligomerization.
Figure 5. NMR monitors protein concentration-dependent changes on a per-residue basis.
(A) Concentration-dependent CSPs for UBQLN2 (or Pxx mutant as indicated) comparing chemical shifts collected between 50 μM and 350 μM protein concentrations. Gray bars signify those resonances that broaden beyond detection at 350 μM protein concentration. (B) Comparison of amide peak intensities between Pxx mutant and WT using 50 μM protein under identical experimental conditions. All plots are organized and color-coded to match Figures 1 and 3.
Concentration-dependent CSPs for Pxx mutants revealed oligomerization propensities consistent with SEC and AUC data above (Figure 5A, Figure S4A). First, the A488T, P500S, P509S, and P525S mutants all exhibited concentration-dependent CSP profiles that mirrored WT. These CSPs likely reflect WT’s continuous self-association scheme, as the protein increases from monomeric concentrations (50 μM) to dimer/trimer species (350 μM), whose sizes are NMR-detectable (Figure S3A). By contrast, the T487I, P497L, and P497S mutants did not exhibit any notable concentration-dependent CSPs over the 50 μM – 350 μM protein concentration range. Upon close inspection, NMR peak intensities for the T487I, P497L and P497S mutants were significantly reduced compared to WT, even at 50 μM protein concentration (Figure 5B). Peak intensities were 10–20% of WT signals across all residues (except for intrinsically-disordered residues 525–555). It is well-established that NMR signals broaden with increasing protein size. Therefore, the NMR peak intensity reduction concomitant with the lack of concentration-dependent CSPs suggest that the T487I, P497L and P497S proteins exhibit higher-order oligomers even at 50 μM concentrations, in general agreement with our AUC data (Figure 3). To determine whether peak intensity reduction was a result of changes in protein dynamics between the mutant and WT, we characterized backbone 15N R1 and R2 relaxation rates for the T487I and P497L mutants at 200 μM (Figure S4B). R1 relaxation rates were higher than those of WT, while R2 relaxation rates were similar or lower than those of WT for most of the protein, except for elevated R2 rates (5 s−1 vs. 2.5 s−1) for residues 525–555 in the intrinsically-disordered region. Residues 525–555 exhibited very low hetNOE values, indicative of fast ns-ps dynamics (Figure S4B). Together, these data suggest changed dynamics for the T487I and P497L mutants compared to WT, but also suggest that the Pxx mutant NMR signals report on a species that is similar to size to WT at this concentration. As SEC and AUC experiments suggest that the T487I, P497L and P497S proteins are significantly oligomerized at 10 and 100 μM, we suspect that the NMR signals at 50 μM protein concentration are only reporting on the minor fraction of protein small enough to be NMR-visible.
Each of the mutants with increased propensity to self-associate (compared to WT) exhibited significantly reduced peak intensities (compare Figures 3 and 5B). Together, the combined biophysical characterization of the Pxx mutants by SEC, SV-AUC, NMR spectroscopy and microscopy support a model whereby only a Pxx mutation that increases UBQLN2 oligomerization propensity promotes UBQLN2 LLPS. Importantly, UBQLN2 oligomerization propensity correlates with changes to material properties of UBQLN2 liquid assemblies: those mutants that oligomerize to a significant extent (i.e. T487I, P497S) also form the most non-liquid like, amorphous UBQLN2 droplets (Figure 1D).
Ub dissolves mutant UBQLN2 aggregates and eliminates LLPS
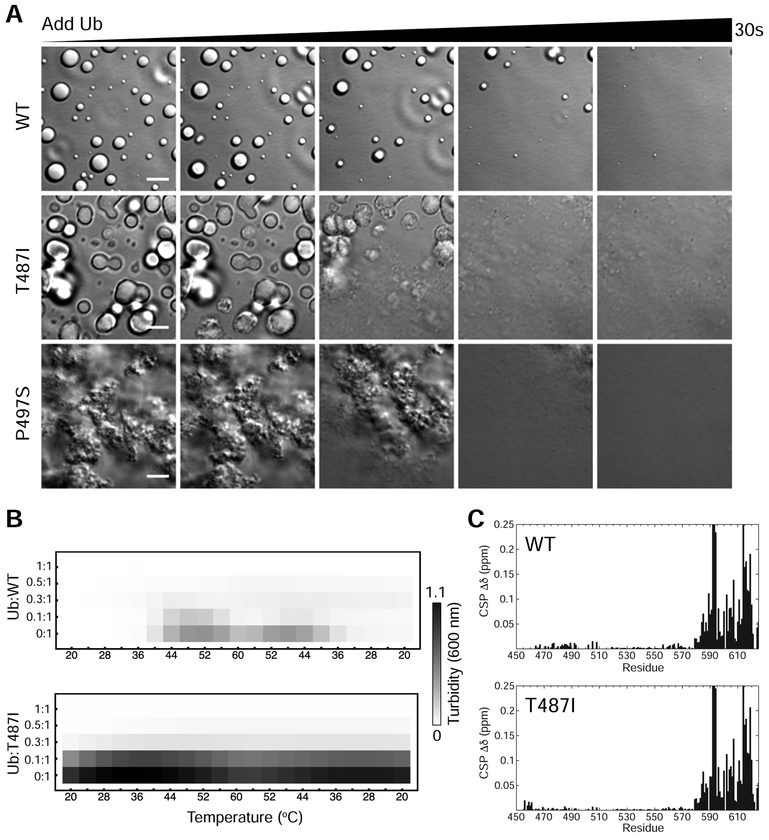
We previously showed that Ub and polyUb chains trigger a ligand-induced phase transition that eliminates UBQLN2 LLPS (Dao et al., 2018). Given the spectrum of droplet morphology and aggregates seen for UBQLN2 Pxx mutants, we investigated if Ub would affect mutant UBQLN2 LLPS. Remarkably, Ub completely cleared all mutant UBQLN2 droplet assemblies, even the aggregates visible for T487I and P497S mutants (Figure 6A). Ub cleared these mutant UBQLN2 assemblies on a timescale comparable to WT (see Movies S1–S3). We confirmed these observations with spectrophotometric assays for these mutants with Ub added up to stoichiometric amounts (Figure 6B). However, qualitative differences in Ub-mediated droplet disassembly were observed (Movies S1–S3).
Figure 6. Ubiquitin binding disassembles mutant UBQLN2 droplets and aggregates.
(A) DIC microscopy of UBQLN2 droplet disassembly as ubiquitin is added. Ubiquitin was allowed to diffuse into the sample using channel slides. Ubiquitin was added in 1:1 molar stoichiometry with indicated protein at 100 μM protein in buffer containing 20 mM NaPhosphate and 200 mM NaCl (pH 6.8). Scale bar = 5 μm. (B) Spectrophotometric turbidity assay as a function of temperature for mixtures of Ub and UBQLN2 at indicated ratios. Assays used 25 μM UBQLN2 protein. (C) Chemical shift perturbations (CSPs) for residues in UBQLN2 450–624 at the titration endpoint with Ub.
To determine if Ub binding modulates mutant UBQLN2 oligomerization, we carried out SV-AUC and NMR experiments using the T487I mutant in the absence and presence of Ub under non-phase separating conditions (Figure S5). As a control, we showed that Ub and WT UBQLN2 form a complex that is consistent with 1:1 stoichiometry by AUC (Figure S5C). Using T487I, AUC experiments demonstrated that T487I oligomerization was unaffected in the presence of Ub (Figure S5D). We also monitored NMR peak intensity for the T487I mutant as Ub was titrated into a solution containing 200 μM T487I protein under non-phase separating conditions. In the absence of Ub, T487I peak intensity is significantly lower than WT (Figure S5B). Ub addition only slightly increased NMR peak intensity for T487I, far below WT peak intensity levels (Figure S5A, Figure S5B). Together, these data suggest that UBQLN2 oligomerization is not disrupted by Ub binding, at least under non-phase separating conditions. This is consistent with prior findings that UBQLN2 oligomerization is mediated by its STI1-II region (Dao et al., 2018; Kurlawala et al., 2017), which does not interact with Ub (Figure 6C).
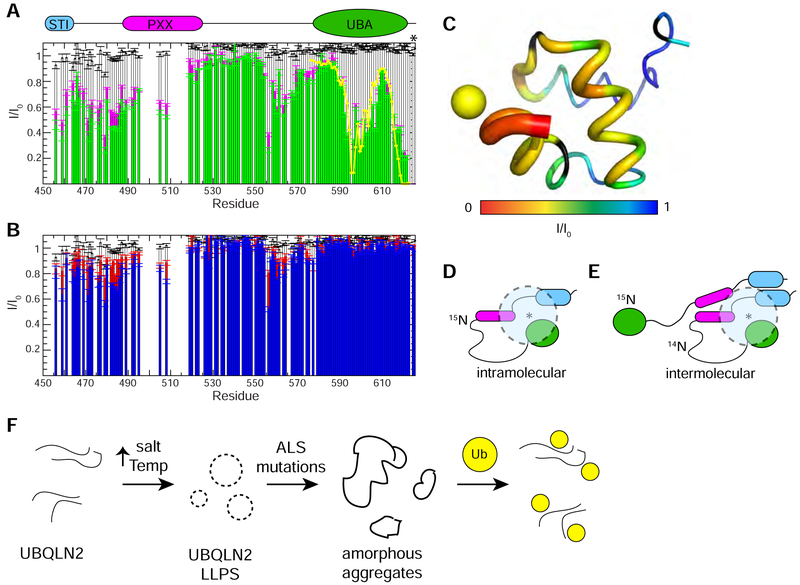
We previously determined that Ub binding to the UBA domain of UBQLN2 involves some of the same molecular contacts (residues 592–594, 616–620) that promote UBQLN2 self-association (Dao et al., 2018). Together with the CSP plot in Figure 5A, we hypothesized that the UBA domain transiently contacts the Pxx region. We tested this hypothesis using paramagnetic relaxation enhancement (PRE) experiments. We installed a paramagnetic spin label (MTSL) on residue 624 at the C-terminus of UBQLN2 to avoid perturbing UBA structure and interfering with Ub binding. Residues near the spin label experience signal attenuation. PRE experiments confirmed the position of the spin label (Figure 7A, 7C), and titration experiments confirmed that the spin label did not affect Ub binding (Figure S6). Interestingly, these PRE experiments revealed intramolecular contacts between the spin label and residues 458–495, 505–508, and 555–570 (Figure 7D). To probe intermolecular contacts between UBQLN2 molecules, we mixed equal amounts of S624C-MTSL UBQLN2 (NMR-invisible) and 15N UBQLN2 450–624 (NMR-visible). Signal attenuations were modest since the protein concentrations used were in a range where the protein was monomeric. These PRE experiments suggest that the UBA domain of one UBQLN2 molecule may contact residues 458–495, 505–508, and 555–565 but not the UBA domain of other UBQLN2 molecules (Figure 7B, 7E). Importantly, these intramolecular and intermolecular contacts are the same ones we identified in the concentration-dependent CSP screen as critical to promoting UBQLN2 self-association (Dao et al., 2018). When Ub is added, NMR signals for residues 450–490, 505–508 and 555–564 are less attenuated, but the effects are small (Figure 7A, 7B). With the important caveat that all of our biophysical experiments were conducted under non-phase separating conditions, these data indicate that the UBA domain transiently contacts the Pxx and STI1 regions. These intramolecular and intermolecular contacts may form the basis for UBQLN2 LLPS and Ub binding to the UBA domain may perturb these interactions.
Figure 7. Ubiquitin disrupts interactions between UBA and Pxx region of UBQLN2.
(A) Intramolecular paramagnetic relaxation enhancement (PRE) effects on UBQLN2 450–624 (50 μM) in the absence of Ub (green) and presence of Ub (magenta) with MTSL spin label attached to an engineered Cys at residue 624 (marked with an asterisk). Yellow line represents back-calculated PREs derived from determining the position of MTSL’s unpaired electron in the UBA structure (see part C). White bars represent PRE effects in a control 50 μM sample with unconjugated MTSL in parts A and B. (B) Intermolecular PREs on WT UBQLN2 450–624 (25 μM) in a 1:1 mixture with NMR-invisible UBQLN2 450–624 S624C (25 μM) with MTSL spin label attached. Intermolecular PREs in the absence of Ub (blue) and presence of Ub (red). Errors in I/I0 for (A) and (B) were determined using the standard error propagation formula for ratios. (C) PRE effects from MTSL are mapped onto the structure of UBQLN2 UBA via backbone thickness and color coding using PDB: 2JY6. Yellow sphere represents back-calculated position of MTSL’s unpaired electron. (D,E) Representative schematic model of results from intramolecular (D) and intermolecular (E) PRE experiments whereby spin label is represented by an asterisk, and dotted semi-transparent circle represents residues’ amide resonances that are attenuated by spin label. (F) Proposed model of how ALS-linked mutations disrupt UBQLN2 LLPS.
Discussion
An emerging feature of many ALS-linked proteins, including UBQLN2, is that they phase separate under physiological conditions and also colocalize with stress granules, liquid assemblies that likely form by LLPS (Dao et al., 2018; Taylor et al., 2016). LLPS is driven by multivalent interactions localized in “sticker” regions separated by “spacers” (Harmon et al., 2017; Rubinstein and Dobrynin, 1997). Indeed, NMR concentration-dependent chemical shifts and PRE experiments suggest that most of the disease-linked Pxx region of UBQLN2 is a “sticker” and contains some of the multivalent interactions that promote LLPS. Here, we screened eleven patient-derived Pxx mutations in UBQLN2. Using a combination of imaging and biophysical techniques, we determined that disease-linked mutations that enhanced UBQLN2 oligomerization also promoted its LLPS. Furthermore, these mutations significantly altered the material properties of UBQLN2 assemblies in vitro, forming visible but reversible aggregates in the case of P497H and P497S mutants. All mutant LLPS-induced assemblies were cleared by binding to Ub. Our findings suggest that an important function of the Pxx region in UBQLN2 is to modulate relative stabilities of the dense and dilute phases, the saturation concentrations at which UBQLN2 undergoes LLPS, and material properties of UBQLN2-containing condensates inside cells.
The Pxx region of UBQLN2 (Figure 1A) exhibits several characteristics that likely contribute to its role in modulating UBQLN2 LLPS: (a) it is similar in sequence to elastin-like proteins known to phase separate (Quiroz and Chilkoti, 2015; Yeo et al., 2011), (b) it contributes to UBQLN2 self-association as determined from concentration-dependent NMR chemical shifts (Dao et al., 2018), and (c) it is enriched in Gly and Pro residues which exhibit high backbone pi-pi contact frequencies, a reliable predictor of LLPS in proteins (Vernon et al., 2018). While these features likely contribute to UBQLN2’s LLPS in vitro and in vivo, we note that the Pxx region can be removed from UBQLN2 and LLPS is not entirely abrogated (Dao et al., 2018). This means that the Pxx region is not the sole determinant of UBQLN2 LLPS, but rather tunes its LLPS behavior, akin to how the proline-rich region in poly-A binding protein (Pab1) modulates its LLPS (Riback et al., 2017).
Our biophysical characterization of the eleven Pxx mutants in the UBQLN2 revealed several trends. First, mutations to hydrophobic residues (e.g. T487I, P497L and P506A) lowered LCST, decreased saturation concentrations necessary for LLPS, and enhanced UBQLN2 oligomerization even in the absence of LLPS (Figure 1, Figure 3). These effects suggest that hydrophobic mutations strengthen the interactions among “stickers” and modulate UBQLN2 LLPS (Wang et al., 2018b, Yang et al., 2019). LCST behavior strongly correlates with high hydropathy index for elastin-like proteins (Quiroz and Chilkoti, 2015). Lower LCST correlates with increasing side chain hydrophobicity (I > L > A) of the Pxx mutation (Kyte and Doolittle, 1982; Riback et al., 2017; Urry et al., 1992). We note that those hydrophobic mutants with the lowest fraction of protein in the dilute phase (Figure 1C) were also those with a more viscoelastic dense phase characterized by significantly reduced droplet fusion rates (Figure 2B). Hydrophobic mutations modulate the polymer-polymer and polymer-solvent interactions to subsequently change the saturation concentrations and phase diagram of UBQLN2. Consistent with a hydrophobic-driven phase transition for UBQLN2, many polar Pxx mutations (A488T, P500S, P506S, P509S, P525S) minimally perturbed UBQLN2 LLPS, oligomerization, and material properties of the dense phase. Hydropathy plot comparisons between these mutants and WT illustrated that polar mutations at these positions minimally affected overall UBQLN2 hydrophobicity (Figure S7A). Together, these data support a model where hydrophobicity drives UBQLN2 LLPS (Dao et al., 2018).
Second, effects of Pxx mutations on UBQLN2 LLPS are dependent on sequence position. Our data suggest that P497 mutations significantly alter oligomerization and LLPS properties of UBQLN2, particularly for P497H and P497S. However, serine mutations at prolines 500, 509 and 525 confer no significant changes to UBQLN2 oligomerization or LLPS behavior. A serine mutation at position 506 increased UBQLN2 LLPS relative to WT, but is not as severe as P497S. What may be the disease mechanisms associated with UBQLN2 polar mutations that confer little or no effect on its LLPS? We noticed that polar mutations in UBQLN2 are to serine, threonine, or histidine residues, all of which can be phosphorylated. Phosphorylation disrupts LLPS of other proteins including FUS and TDP-43 (Lin et al., 2017; Monahan et al., 2017; Wang et al., 2018a; Wegmann et al., 2018). Indeed, mutating residues A488 or P509 to phosphomimetic glutamate likely increased UBQLN2 saturation concentrations as phase separation is significantly reduced at the protein concentration tested here (Figure S7B). Thus, disease mechanisms associated with polar mutations may disrupt UBQLN2’s recruitment to biomolecular condensates if LLPS was a prerequisite. Overall, our results suggest sequence-encoded phase separation properties modulated by disease-linked mutations (Weber, 2017).
Third, none of the LLPS-induced mutant UBQLN2 droplets or aggregates were irreversible. Ub clarified all mutant UBQLN2 droplets completely (Figure 6A) and 1,6-hexanediol addition cleared all mutant UBQLN2 solutions despite the appearance of visible aggregates for P497H and P497S mutants. It is important to emphasize that the underlying physical mechanisms for how Ub triggers the ligand-induced phase transitions of mutant UBQLN2 remain unknown. Wyman and Gill originally described ligand effects on shifting of phase boundaries as polyphasic linkage (Wyman and Gill, 1980). Using the principles of polyphasic linkage, a ligand (e.g. Ub) stabilizes one phase over the other. Our mutant UBQLN2 data are consistent with the interpretation that Ub preferentially binds to the diffuse state of UBQLN2, thus increasing cs values. However, to fully understand how Ub modulates mutant UBQLN2 LLPS, we need to map how Ub shifts phase boundaries (e.g. quantification of UBQLN2 cs saturation concentrations as Ub is titrated) as has been recently done for the profilin-huntingtin system (Posey et al., 2018).
In contrast to our findings, (Hjerpe et al., 2016) found that Pxx mutations did not disrupt UBQLN2 oligomerization. We reasoned that the discrepancies may be a consequence of (a) different experimental conditions (e.g. 4°C) used for their AUC and SEC experiments, and (b) different constructs using full-length UBQLN2, whereas we have used residues 450–624. We previously observed that other UBQLN2 domains, such as the UBL and the remainder of the STI1-II region, modulate UBQLN2 LLPS (Dao et al., 2018). In addition, UBQLN2’s UBL binds weakly to the UBA with a Kd of ~ 175 μM when added in trans (Nguyen et al., 2017). Therefore, we speculate that the presence of these other domains and protein-protein interactions with these domains may further modulate the effects of the Pxx mutations on LLPS.
Our work suggests that specific ALS-linked Pxx mutations promoted UBQLN2 oligomerization, reduced saturation concentrations for LLPS, increased viscoelasticity of the dense phase, and aggregated reversibly. Our in vitro results correlate with recent in vivo observations made for UBQLN2. For example, several studies suggested that ALS-linked Pxx mutations increase UBQLN2 aggregation propensity (Ceballos-Diaz et al., 2015; Kim et al., 2018; Osaka et al., 2015; Sharkey et al., 2018). Recently, (Kim et al., 2018) showed that UBQLN2 Pxx mutants are more efficiently immunoprecipitated, and pull down more Ub than WT UBQLN2 (also shown by (Chang and Monteiro, 2015; Hjerpe et al., 2016)). Using either HEK293T lysates or bacterially-expressed GST-UBQLN2, the authors showed that P497H and P506T bound more Ub than WT UBQLN2, and that P509S and P525S bound Ub similarly as WT (Kim et al., 2018). To explain this, the authors proposed that Pxx mutants promote changes in UBQLN2 folding and/or oligomerization. Indeed, our results generally correlate with their findings in that P497H and P506T mutants exhibit increased oligomerization compared to WT (perhaps binding to more Ub in pulldown assays), whereas P525S and P509S are comparable to WT (Figure 3A).
Both WT and mutant UBQLN2 form cytoplasmic puncta in cells, but puncta formation appears very sensitive to expression level (Deng et al., 2011; Hjerpe et al., 2016; Picher-Martel et al., 2015; Sharkey et al., 2018). We previously showed that UBQLN2 is diffuse in the cytoplasm, forms cytosolic puncta in cells under stress, and that puncta formation correlated with UBQLN2’s ability to phase separate (Dao et al., 2018). Drosophila studies of WT UBQLN2 and Pxx mutants (P497H and P525S) showed that WT UBQLN2 was diffuse throughout the cytoplasm of neuronal cells after one week of age. By contrast, P497H formed cytosolic puncta nearly immediately (Kim et al., 2018). P525S was intermediate between WT and P497H. Additionally, it was recently shown that P506T puncta are less liquid-like and have amorphous morphology compared to WT UBQLN2 puncta in mammalian cell culture and mouse models (Sharkey et al., 2018). These findings are generally consistent with our in vitro data, which showed P497H phase separating into aggregates and P506T droplets exhibiting more gel-like, less dynamic properties compared to WT and P525S.
UBQLN2’s role in protein quality control and colocalization with stress granules suggests that UBQLN2 may be involved in remodeling SGs or other membraneless organelles by clearing ubiquitinated proteins (Dao et al., 2018). As we demonstrated here that disease-linked Pxx mutations modulated UBQLN2 LLPS and changed material properties of UBQLN2 LLPS assemblies in vitro, we postulate that Pxx mutations disrupt how UBQLN2 regulates or localizes into biomolecular condensates in cells. Impaired UBQLN2 function may lead to disruptions in how UBQLN2 interacts with HSP70 chaperones, shuttles ubiquitinated substrates to the proteasome, or interacts with autophagic components (Chang and Monteiro, 2015; Deng et al., 2011; Hjerpe et al., 2016). ALS-linked mutations in RNA-binding proteins such as hnRNPA1, TIA-1, TDP-43, and FUS that colocalize with stress granules modify biomolecular condensates by changing their material properties and dynamics (Conicella et al., 2016; Mackenzie et al., 2017; Molliex et al., 2015; Murakami et al., 2015; Patel et al., 2015). Therefore, we surmise that mutant UBQLN2 can also alter stress granule (or other UBQLN2-containing condensate) morphology and dynamics in cells, perhaps ‘aging’ these condensates into the pathological protein-containing inclusions characteristic of ALS and other neurological disorders.
Conclusions
Disease-linked mutations in the Pxx region of UBQLN2 alter phase separation and material properties of UBQLN2 LLPS-induced assemblies in a sequence-dependent and site-specific manner. Extensive biophysical characterization of eleven Pxx mutants revealed that several disease-linked mutations increased UBQLN2 oligomerization. However, Pxx disease mutations promote a reversible aggregated state that is disassembled by Ub. These studies suggest that disease mutations change saturation concentrations for UBQLN2 LLPS, and alter material properties of UBQLN2-containing condensates resulting in impaired protein quality control mechanisms normally mediated by UBQLN2.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Carlos Castañeda (cacastan@syr.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial Culture
Unlabeled proteins were expressed in either NiCo21 (DE3) or Rosetta 2 (DE3) pLysS cells in Luria-Bertani (LB) broth. Uniformly 15N or 15N/13C labeled proteins were expressed in M9 minimal media with 15N ammonium chloride and 13C-glucose as the sole nitrogen and carbon sources, respectively. Cells were induced with IPTG and harvested after 12–16 hours at 37°C. Cell pellets were frozen, lysed and cleared by centrifugation at 22,000 × g for 20 min at 4°C.
METHOD DETAILS
Subcloning, Protein Expression, and Purification
Ubiquitin was expressed and purified as detailed elsewhere (Beal et al., 1996). UBQLN2 Pxx mutants were generated from UBQLN2 450–624 using Phusion Site-Directed Mutagenesis Kit (Thermo Scientific). A tryptophan (W) codon was added to the end of all constructs to facilitate determination of protein concentration. UBQLN2 450–624 and Pxx mutants were expressed and purified as described in (Dao et al., 2018). Briefly, the constructs were expressed in Escherichia coli Rosetta 2 (DE3) pLysS cells in Luria-Bertani (LB) broth at 37°C overnight. Bacteria were pelleted, frozen, lysed, then purified via a “salting out” process. Briefly, NaCl was added to the cleared lysate to the final concentration of 0.5 M-1 M. UBQLN2 droplets were pelleted and then resuspended in 20 mM NaPhosphate, 0.5 mM EDTA (pH 6.8). For P497H mutant, pellet was redissolved in a solution containing the above buffer and 6 M urea. Leftover NaCl was removed through HiTrap desalting column (GE Healthcare). For P497H, urea was removed by several exchanges with the HiTrap desalting column using no-salt buffer. Protein samples for NMR spectroscopy were produced in M9 minimum media supplemented with 15N ammonium chloride and 13C glucose as appropriate for the experiment. Purified proteins were frozen at −80°C.
Fluorescent Labeling
UBQLN2 450–624 constructs were fluorescently labelled with Dylight-488 or DyLight-650 NHS Ester (Thermo Scientific), according to the manufacturer’s instructions. Mole dye per mole protein ratio for all samples was determined to be around 0.5–0.9.
Spectrophotometric Absorbance/Turbidity Measurements
Protein samples (300 μL) were prepared by adding protein (from stock to a final concentration of 25 μM unless otherwise noted) to cold sodium phosphate buffer (pH 6.8, 20 mM NaPhosphate, 0.5 mM EDTA) containing 200 mM NaCl and were kept on ice for at least 10 minutes before the assay. We chose 25 μM so that we could capture and compare the phase behaviors of the mutants as clearly as possible. Absorbance at 600 nm as a function of temperature was recorded by a Beckman DU-640 UV/Vis spectrophotometer using a temperature ramp rate of 2°C/min increasing from 20°C to 60°C (or 44°C) and then decreasing back to 20°C. Net absorbance values were recorded after subtracting the absorbance value of a buffer control.
DIC/Fluorescence Imaging of Phase Separation
UBQLN2 450–624 constructs were prepared to contain 100 μM protein (or spiked with fluorophore-labeled UBQLN2 (Dylight-488 or Dylight-650), 1:1000 molar ratio) in 20 mM NaPhosphate, 200 mM NaCl, and 0.5 mM EDTA (pH 6.8). Samples were added to MatTek glass bottom dishes that had been incubated with 3% BSA to reduce rapid coating of protein droplets onto the glass surface (Lin et al., 2015) and incubated at 30 °C. Phase separation was imaged on a Leica DiM8 STP800 (Leica, Bannockburn, IL) equipped with a Lumencor SPECTRA X (Lumencor, Beaverton, Or), and Hamamatsu ORCAflash 4.0 V2 CMOS C11440–22CU camera using a 100×/1.4 N.A. HC Pl Apo objective to visualize the formation and fusion of UBQLN2 droplets over time. Images were taken every 0.5 seconds for up to 3 minutes to visualize these events. Exposures were 50 ms each on all channels. To observe the effect of Ub on mutant UBQLN2 assemblies, we began a time-lapse of a phase-separated UBQLN2 sample before gently adding a small volume of highly concentrated ubiquitin (to a final molar ratio of 1:1) to an area removed from where the camera was imaging and observed the droplets disappear as Ub diffused into the sample.
Measurements of Dilute Phase Concentrations
Due to the low extinction coefficient for the UBQLN2 450–624 construct (5500 M−1 cm−1), we quantified the fraction in the dilute phase using SDS-PAGE gels. To determine the concentration of the dilute phase, UBQLN2 450–624 constructs were prepared to contain 100 μM protein in 20 mM NaPhosphate, 200 mM NaCl, and 0.5 mM EDTA (pH 6.8), and stored in 50 L aliquots on ice. After the microcentrifuge was incubated for at least 30 minutes at certain temperature (every 5 °C from 5 °C to 60 °C), samples were incubated for 10 minutes, then centrifuged for 2 minutes at 21000 × g. 10 μL of the supernatant of each samples were immediately mixed with 10 μL of 2x SDS-PAGE dye. 4 μL of each samples were load onto Mini-PROTEAN TGX Stain-Free Precast Gels, imaged using the BioRad Gel Doc, and band volumes were determined with BioRad Image Lab software. A 50 μM standard was also loaded, analyzed and used to calculate the concentration of the samples. The dilute phase measurements were carried out in 3 separate trials. In agreement with our turbidity assays, the fraction of mutant protein in the dilute phase was lowest under conditions when the solution was most turbid.
Fluorescence Recovery After Photobleaching (FRAP)
WT and T487I UBQLN2 450–624 were prepared to contain 80 μM protein spiked with Dylight-650-labeled UBQLN2, 1:1000 molar ratio) in 20 mM NaPhosphate, 200 mM NaCl, and 0.5 mM EDTA (pH 6.8) and incubated for 10 min at 30°C. FRAP experiments were performed using a Leica SP5 laser scanning confocal microscope (Leica, Bannockburn, IL) with an HCX Plan Apochromat 63 ×/1.40–0.06 N.A. OIL objective. The LAS-X (Leica application suite advanced fluorescence) software (Leica) FRAP wizard was used to acquire images every 220 milliseconds.
Analytical Size Exclusion Chromatography
Purified UBQLN2 constructs at different concentrations were subjected to chromatography over a ENrich™ SEC 650 10 × 300 column (Biorad) to analyze concentration-dependent activity. Experiments were conducted at ambient temperatures at 1 mL/min in pH 6.8 buffer containing 20 mM NaPhosphate, 0.5 mM EDTA, with no added NaCl. Standard molecular weights were determined by subjecting a sample of Gel Filtration Standard (BioRad #1511901) over the column at the same conditions. Wild-type UBQLN2 450–624 and P497S and T487I mutant SEC experiments were similar or identical in the absence or presence of urea.
Sedimentation Velocity Analytical Ultracentrifugation
Purified wild type and mutant UBQLN2 proteins with and without added equimolar amounts of ubiquitin were loaded into 3- or 12-mm two-sector charcoal-filled Epon centerpieces with sapphire windows. All experiments were carried out using a Beckman Coulter ProteomeLab XLA analytical ultracentrifuge equipped with absorbance optics and a 4-hole An-60 Ti rotor at 20,000 or 60,000 rpm that was pre-equilibrated to 25°C prior to running the experiment. The samples were scanned with a zero second time interval between scans for 50–300 scans and analyzed by the continuous distribution (c(s)) method in the program SEDFIT (Schuck, 2000). Two-dimensional size and shape distributions (c(s,fr)) were calculated using SEDFIT and plotted as sedimentation coefficient-molar mass distributions (c(s,M)) using GUSSI (Brautigam, 2015). Integration of individual peaks in c(s,fr) plots gave an estimate of the molecular weight of each species in each distribution. The program SEDNTERP (Laue et al., 1992) was used to calculate the buffer density (1.00 g/mL), viscosity (0.009 P), and partial specific volume of UBQLN2 ( = 0.74), which was based on the amino acid sequence. The concentration series of WT UBQLN2 was used to construct a signal-weighted isotherm that was then fit to a monomer-dimer-tetramer or monomer-trimer-hexamer model.
Transmission Electron Microscopy
Samples of wild-type UBQLN2 and P497S constructs were prepared under phase separating conditions to contain 60 μM protein, 200 mM NaCl in 20 mM Tris, pH 7.4, incubated for 10 minutes, then diluted 1 to 20 into 20 mM Tris, pH 7.4 buffer and immediately used to prepare for transmission electron microscopy (TEM). Protein solutions were spread on glow discharged carbon coated copper grids for one minute, and the grids were washed with water, stained with 2% uranyl acetate for one minute, and excess stain was removed by blotting with filter paper. The stained samples were observed in a JEOL JEM-2100F electron microscope operating at 200 kV and electron micrographs were recorded on a 4k × 4k CMOS camera (Gatan OneView).
NMR Experiments
NMR experiments were performed at 10°C, 25°C, 40°C, or 55°C on a Bruker Avance III 800 MHz spectrometer equipped with TCI cryoprobe. Proteins were prepared in 20 mM NaPhosphate buffer (pH 6.8), 0.5 mM EDTA, 0.02% NaN3, and 5% D2O. All NMR data were processed using NMRPipe (Delaglio et al., 1995) and analyzed using CCPNMR 2.4.2 (Vranken et al., 2005).
NMR Assignments
We previously obtained 85% of backbone N, HN, Ca, Cβ and C=O chemical shifts for WT UBQLN2 450–624 (Dao et al., 2018) with lowest coverage in part of the repetitive Pxx region (488–519). Using Pxx deletion constructs (e.g. Δ496–502 and Δ510–518) or single point Pxx mutants (e.g. G499S, G514S), we collected additional triple resonance experiments (HNCACB, CBCA(CO)NH, and HNN), and 13C-detect experiments ((HACA)N(CA)CON and (HACA)N(CA)NCO). With these judiciously chosen UBQLN2 constructs, we increased overall backbone N, HN, Ca, Cβ, and C=O chemical shift coverage to >94%. Only a handful of amide resonances in the Pxx region could not be unambiguously assigned. For UBQLN2 Pxx mutants, backbone amide signals in 1H-15N HSQC spectra were assigned based on visual comparison with WT UBQLN2 spectra using identical protein concentrations and experimental conditions.
NMR Spectra
1H-15N TROSY-HSQC (or CP-HISQC as noted) experiments were acquired using spectral widths of 15 and 26 ppm in the direct 1H and indirect 15N dimensions, and corresponding acquisition times of 200 ms and 47 ms. Centers of frequency axes were 4.7 and 116 ppm for 1H and 15N dimensions, respectively. 1H-15N TROSY spectra were processed and apodized using a Lorentz-to-Gauss window function with 15 Hz line sharpening and 20 Hz line broadening in the 1H dimension, while 15N dimension was processed using a cosine squared bell function. Where applicable, chemical shift perturbations (CSPs) were quantified as follows: Δδ = [(ΔδH)2 + (ΔδN/5)2]1/2 where ΔδH and ΔδN are the differences in 1H and 15N chemical shifts, respectively, for the same residue in UBQLN2 at different protein concentrations, or in the absence and presence of ubiquitin. For assignments, 15N-13CO spectra were collected using the (HACA)CON 13C-detect pulse program (Bastidas et al., 2015). Spectra were acquired with 16 or 32 transients using spectral widths of 40 and 36 ppm in the 13CO and 15N dimensions, respectively, with corresponding acquisition times of 64 ms and 47 ms. Centers of 15N and 13CO frequence axes were 122 ppm and 175 ppm, respectively.
15N Relaxation Experiments
Longitudinal (R1) and transverse (R2) 15N relaxation rates, and {1H}−15N steady-state heteronuclear Overhauser enhancement (hetNOE) were measured for T487I or P497L UBQLN2 samples (200 μM) using established interleaved relaxation experiments and protocols (Castañeda et al., 2016; Hall and Fushman, 2003). Relaxation inversion recovery periods for R1 experiments were 4 ms (x 2), 500 ms (x 2), and 840 ms (x 2), using an interscan delay of 2.5 s. Total spin-echo durations for R2 experiments were 8 ms, 32 ms, 64 ms, 88 ms, 112 ms, and 200 ms using an interscan delay of 2.5 s. Heteronuclear NOE experiments were acquired with an interscan delay of 4.5 s. All relaxation experiments were acquired using spectral widths of 12 and 24 ppm in the 1H and 15N dimensions, respectively, with corresponding acquisition times of 110 ms and 31 ms. Spectra were processed using squared cosine bell apodization in both 1H and 15N dimensions. Relaxation rates were derived by fitting peak heights to a mono-exponential decay using RELAXFIT (Fushman et al., 1997). Relaxation rates for wild-type 15N UBQLN2 were previously reported in (Dao et al., 2018).
NMR Binding Experiments
Unlabeled ligand (Ub) was titrated into 200 M samples of 15N UBQLN2 (or T487I) at fixed concentrations (see Figure 6 and Figure S6), and binding was monitored by recording 1H-15N TROSY-HSQC spectra as a function of ligand concentration.
Paramagnetic Relaxation Enhancement (PRE) Experiments
The paramagnetic spin label, 1-oxyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl methanesulfonate (MTSL, Toronto Research Chemicals), was attached to an Cys at position 624, introduced via site-directed mutagenesis, as described (Varadan et al., 2005a). The PRE effects were quantitated as the ratio (I/I0) of the signal intensities in the HSQC spectra recorded with MTSL in the oxidized and reduced states. Using the distance constraints imparted by the spin label, the location of the spin label was determined using SLFIT (Ryabov and Fushman, 2006). Unconjugated MTSL control experiment was carried out by mixing 50 μM spin label with 50 μM UBQLN2. For a structure of the UBQLN2 UBA:Ub complex, we used the UBQLN1 UBA:Ub NMR-derived structure from PDB 2JY6 (Zhang et al., 2008). The two UBA sequences are 98% identical.
Hydropathy Plots
UBQLN2 hydropathy plots in Figure S7 were calculated using the Kyte and Doolittle hydrophobicity scale for amino acids (Kyte and Doolittle, 1982) with Protscale (Gasteiger et al., 2005).
QUANTIFICATION AND STATISTICAL ANALYSIS
Turbidity Assays and Phase Diagrams
Data were collected using proteins from two separate preps and three trials for each (total n = 6) at the same condition for both WT and mutant UBQLN2 450–624. For Ub assays, n = 3 trials were conducted. For LLPS reversibility assays, samples at the end of spectrophotometric measurements were kept cold on ice for one hour and were then subjected to the same assay again. Results were averaged over n=6 trials (two protein preps and three trials for each) for WT and mutant UBQLN2. For the low-concentration arm of phase diagrams, dilute phase measurements were carried out in 3 separate trials.
Droplet Fusion Rates
The characteristic relaxation times for wild-type and mutant droplets (except for P497S and P497H) were calculated as described (Brangwynne et al., 2011). Briefly, we fit a single exponential function to the aspect ratio of two fusing droplets vs. fusing time to obtain the relaxation times. Error bars represent the standard deviation of 9 separate fusion events for each protein. See legend for Figure 2.
FRAP Experiments
The ImageJ FRAP profiler plug-in was used to generate normalized FRAP curves to calculate the half-life and immobile fraction values. Graphs were then produced in Prism GraphPad software. An average FRAP curve was generated by averaging FRAP data from 10 separate droplets of similar size. FRAP curve error bars represent the standard deviation of the FRAP data. Fluorescence recovery times were derived by fitting a single exponential fit to the data. The student’s unpaired T-test was used to determine statistical significance between the mobile fractions for WT and mutant UBQLN2 (T487I) as described in legend for Figure 2.
NMR Relaxation
As described above, relaxation rates were derived by fitting peak heights to a mono-exponential decay using RELAXFIT (Fushman et al., 1997). Errors in 15N R1 and R2 relaxation rates were determined using 500 Monte Carlo trials in RELAXFIT. Errors in hetNOE measurements were determined using the standard error propagation formula.
Paramagnetic Relaxation Enhancement (PRE) Experiments
Errors in I/I0 were determined using the standard error propagation formula for ratios. Noise was estimated using the median intensity of 5000 points in NMR spectra.
DATA AND SOFTWARE AVAILABILITY
Microscopy imaging data is available on Mendeley (http://dx.doi.org/10.17632/9cr27cbmkz.1).
Supplementary Material
Movie S1. Ubiquitin eliminates UBQLN2 phase separation. Ubiquitin was added to one part of sample well, and was allowed to diffuse into a different area of the well where UBQLN2 450–624 droplets were imaged. Related to Figure 6.
Movie S2. Ubiquitin eliminates UBQLN2 T487I droplets. Ubiquitin was added to one part of sample well, and was allowed to diffuse into a different area of the well where UBQLN2 T487I droplets were imaged. Related to Figure 6.
Movie S3. Ubiquitin eliminates UBQLN2 P497S aggregates. Ubiquitin was added to one part of sample well, and was allowed to diffuse into a different area of the well where UBQLN2 P497S aggregates were imaged. Related to Figure 6.
All movies are approximately 40 seconds of real time. Final concentrations of Ub and UBQLN2 were 100 μM each in pH 6.8 buffer containing 200 mM NaCl. The temperature used for all experiments was 30°C. Scale bar = 5 μm.
Highlights.
ALS-linked mutations in proline-rich region disrupt UBQLN2 phase separation (LLPS).
ALS-linked mutations promote UBQLN2 oligomerization and self-association.
Hydrophobic mutations decrease saturation concentration necessary for UBQLN2 LLPS.
Ubiquitin disassembles mutant UBQLN2 liquid and solid-like aggregates.
Acknowledgements
This work was supported by ALS Association grants (17-IIP-369 and 18-IIP-400) and the National Science Foundation (CAREER award 1750462) to C.A.C.. H.H. was supported by NIH R00 GM107355, DOD grant PC160083, and the Carol Baldwin Foundation for Central New York. M.S.C. was supported by NIH 2R01CA140522. Data collected on a Bruker 800 MHz NMR magnet was supported by NIH shared instrumentation grant 1S10OD012254. We thank SUNY-Upstate for access to the Leica laser-scanning confocal microscope. We acknowledge Stephan Wilkens for expertise and assistance with TEM experiments. TEM data were collected on a JEOL electron microscope obtained by NSF MRI grant 1531757. We thank Ananya Majumdar, Andrea Sorrano, and J. Paul Taylor for useful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Alberti S, and Carra S (2018). Quality Control of Membraneless Organelles. J. Mol. Biol 430, 4711–4729. [DOI] [PubMed] [Google Scholar]
- Bastidas M, Gibbs EB, Sahu D, and Showalter SA (2015). A primer for carbon-detected NMR applications to intrinsically disordered proteins in solution. Concepts Magn. Reson. Part A 44, 54–66. [Google Scholar]
- Beal R, Deveraux Q, Xia G, Rechsteiner M, and Pickart C (1996). Surface hydrophobic residues of multiubiquitin chains essential for proteolytic targeting. Proc. Natl. Acad. Sci. U. S. A 93, 861–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard JJ, Otero JH, Scott DC, Szulc E, Martin EW, Sabri N, Granata D, Marzahn MR, Lindorff-Larsen K, Salvatella X, et al. (2018). Cancer Mutations of the Tumor Suppressor SPOP Disrupt the Formation of Active, Phase-Separated Compartments. Mol. Cell 72, 19–36.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Mitchison TJ, and Hyman AA (2011). Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci 108, 4334–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautigam CA (2015). Chapter Five - Calculations and Publication-Quality Illustrations for Analytical Ultracentrifugation Data In Methods in Enzymology, Cole JL, ed. (Academic Press; ), pp. 109–133. [DOI] [PubMed] [Google Scholar]
- Brown PH, and Schuck P (2006). Macromolecular Size-and-Shape Distributions by Sedimentation Velocity Analytical Ultracentrifugation. Biophys. J 90, 4651–4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Kolaitis R-M, Taylor JP, and Parker R (2013). Eukaryotic Stress Granules Are Cleared by Autophagy and Cdc48/VCP Function. Cell 153, 1461–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañeda CA, Dixon EK, Walker O, Chaturvedi A, Nakasone MA, Curtis JE, Reed MR, Krueger S, Cropp TA, and Fushman D (2016). Linkage via K27 Bestows Ubiquitin Chains with Unique Properties among Polyubiquitins. Structure 24, 423–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos-Diaz C, Rosario AM, Park H-J, Chakrabarty P, Sacino A, Cruz PE, Siemienski Z, Lara N, Moran C, Ravelo N, et al. (2015). Viral expression of ALS-linked ubiquilin-2 mutants causes inclusion pathology and behavioral deficits in mice. Mol. Neurodegener 10, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, and Monteiro MJ (2015). Defective Proteasome Delivery of Polyubiquitinated Proteins by Ubiquilin-2 Proteins Containing ALS Mutations. PLoS ONE 10, e0130162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conicella AE, Zerze GH, Mittal J, and Fawzi NL (2016). ALS Mutations Disrupt Phase Separation Mediated by α-Helical Structure in the TDP-43 Low-Complexity C-Terminal Domain. Structure 24, 1537–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao TP, Kolaitis R-M, Kim HJ, O’Donovan K, Martyniak B, Colicino E, Hehnly H, Taylor JP, and Castañeda CA (2018). Ubiquitin Modulates Liquid-Liquid Phase Separation of UBQLN2 via Disruption of Multivalent Interactions. Mol. Cell 69, 965–978.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, and Bax A (1995). NMRPIPE - A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Deng H-X, Chen W, Hong S-T, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H, et al. (2011). Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 477, 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahed AC, McDonough B, Gouvion CM, Newell KL, Dure LS, Bebin M, Bick AG, Seidman JG, Harter DH, and Seidman CE (2014). UBQLN2 mutation causing heterogeneous X-linked dominant neurodegeneration. Ann. Neurol 75, 793–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushman D, Cahill S, and Cowburn D (1997). The main-chain dynamics of the dynamin pleckstrin homology (PH) domain in solution: analysis of 15N relaxation with monomer/dimer equilibration. J. Mol. Biol 266, 173–194. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, and Bairoch A (2005). Protein Identification and Analysis Tools on the ExPASy Server In The Proteomics Protocols Handbook, (Humana Press; ), pp. 571–607. [Google Scholar]
- Gellera C, Tiloca C, Del Bo R, Corrado L, Pensato V, Agostini J, Cereda C, Ratti A, Castellotti B, Corti S, et al. (2013). Ubiquilin 2 mutations in Italian patients with amyotrophic lateral sclerosis and frontotemporal dementia. J. Neurol. Neurosurg. Psychiatry 84, 183–187. [DOI] [PubMed] [Google Scholar]
- Hall JB, and Fushman D (2003). Characterization of the overall and local dynamics of a protein with intermediate rotational anisotropy: Differentiating between conformational exchange and anisotropic diffusion in the B3 domain of protein G. J. Biomol. NMR 27, 261–275. [DOI] [PubMed] [Google Scholar]
- Harmon TS, Holehouse AS, Rosen MK, and Pappu RV (2017). Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. ELife 6, e30294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjerpe R, Bett JS, Keuss MJ, Solovyova A, McWilliams TG, Johnson C, Sahu I, Varghese J, Wood N, Wightman M, et al. (2016). UBQLN2 Mediates Autophagy-Independent Protein Aggregate Clearance by the Proteasome. Cell 166, 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, and Parker R (2016). ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell 164, 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Stiles SG, Feichtmeier JM, Ramesh N, Zhan L, Scalf MA, Smith LM, Bhan Pandey U, and Tibbetts RS (2018). Mutation-dependent aggregation and toxicity in a Drosophila model for UBQLN2-associated ALS. Hum. Mol. Genet 27, 322–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S, Maharana S, Mateju D, Malinovska L, Nüske E, Poser I, Richter D, and Alberti S (2015). Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. ELife 4, e06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S, Maharana S, and Simon A (2017). Hexanediol: a chemical probe to investigate the material properties of membrane-less compartments. Matters 3, e201702000010. [Google Scholar]
- Kurlawala Z, Shah PP, Shah C, and Beverly LJ (2017). The STI and UBA Domains of UBQLN1 are Critical Determinants of Substrate Interaction and Proteostasis. J. Cell. Biochem 118, 2261–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S, Zhang Y, and Matthias P (2007). The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 21, 3381–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J, and Doolittle RF (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Laue TM, Shah BD, Ridgeway TM, and Pelletier SL (1992). Computer-aided interpretation of analytical sedimentation data for proteins In Analytical Ultracentrifugation in Biochemistry and Polymer Science, (Cambridge, UK: The Royal Society of Chemistry; ), pp. 90–125. [Google Scholar]
- Le NTT, Chang L, Kovlyagina I, Georgiou P, Safren N, Braunstein KE, Kvarta MD, Dyke AMV, LeGates TA, Philips T, et al. (2016). Motor neuron disease, TDP-43 pathology, and memory deficits in mice expressing ALS–FTD-linked UBQLN2 mutations. Proc. Natl. Acad. Sci 113, E7580–E7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DSW, Rosen MK, and Parker R (2015). Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol. Cell 60, 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Mori E, Kato M, Xiang S, Wu L, Kwon I, and McKnight SL (2016). Toxic PR Poly-Dipeptides Encoded by the C9orf72 Repeat Expansion Target LC Domain Polymers. Cell 167, 789–802.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Currie SL, and Rosen MK (2017). Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J. Biol. Chem 292, 19110–19120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Nicholson AM, Sarkar M, Messing J, Purice MD, Pottier C, Annu K, Baker M, Perkerson RB, Kurti A, et al. (2017). TIA1 Mutations in Amyotrophic Lateral Sclerosis and Frontotemporal Dementia Promote Phase Separation and Alter Stress Granule Dynamics. Neuron 95, 808–816.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzahn MR, Marada S, Lee J, Nourse A, Kenrick S, Zhao H, Ben-Nissan G, Kolaitis R-M, Peters JL, Pounds S, et al. (2016). Higher-order oligomerization promotes localization of SPOP to liquid nuclear speckles. EMBO J. 35, 1254–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateju D, Franzmann TM, Patel A, Kopach A, Boczek EE, Maharana S, Lee HO, Carra S, Hyman AA, and Alberti S (2017). An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J. e201695957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrea DM, Cika JA, Stanley CB, Nourse A, Onuchic PL, Banerjee PR, Phillips AH, Park C-G, Deniz AA, and Kriwacki RW (2018). Self-interaction of NPM1 modulates multiple mechanisms of liquid–liquid phase separation. Nat. Commun 9, 842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moelbert S, and De Los Rios P (2003). Hydrophobic Interaction Model for Upper and Lower Critical Solution Temperatures. Macromolecules 36, 5845–5853. [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, and Taylor JP (2015). Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization. Cell 163, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan Z, Ryan VH, Janke AM, Burke KA, Rhoads SN, Zerze GH, O’Meally R, Dignon GL, Conicella AE, Zheng W, et al. (2017). Phosphorylation of the FUS low complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 36, 2951–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Qamar S, Lin JQ, Schierle GSK, Rees E, Miyashita A, Costa AR, Dodd RB, Chan FTS, Michel CH, et al. (2015). ALS/FTD Mutation-Induced Phase Transition of FUS Liquid Droplets and Reversible Hydrogels into Irreversible Hydrogels Impairs RNP Granule Function. Neuron 88, 678–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen K, Puthenveetil R, and Vinogradova O (2017). Investigation of the adaptor protein PLIC-2 in multiple pathways. Biochem. Biophys. Rep 9, 341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka M, Ito D, Yagi T, Nihei Y, and Suzuki N (2015). Evidence of a link between ubiquilin 2 and optineurin in amyotrophic lateral sclerosis. Hum. Mol. Genet 24, 1617–1629. [DOI] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. (2015). A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 162, 1066–1077. [DOI] [PubMed] [Google Scholar]
- Patel SS, Belmont BJ, Sante JM, and Rexach MF (2007). Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell 129, 83–96. [DOI] [PubMed] [Google Scholar]
- Picher-Martel V, Dutta K, Phaneuf D, Sobue G, and Julien J-P (2015). Ubiquilin-2 drives NF-κB activity and cytosolic TDP-43 aggregation in neuronal cells. Mol. Brain 8, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posey AE, Ruff KM, Harmon TS, Crick SL, Li A, Diamond MI, and Pappu RV (2018). Profilin reduces aggregation and phase separation of huntingtin N-terminal fragments by preferentially binding to soluble monomers and oligomers. J. Biol. Chem 293, 3734–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroz FG, and Chilkoti A (2015). Sequence heuristics to encode phase behaviour in intrinsically disordered protein polymers. Nat. Mater 14, 1164–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riback JA, Katanski CD, Kear-Scott JL, Pilipenko EV, Rojek AE, Sosnick TR, and Drummond DA (2017). Stress-Triggered Phase Separation Is an Adaptive, Evolutionarily Tuned Response. Cell 168, 1028–1040.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein M, and Dobrynin AV (1997). Solutions of Associative Polymers. Trends Polym. Sci 6, 181–186. [Google Scholar]
- Ruff KM, Roberts S, Chilkoti A, and Pappu RV (2018). Advances in Understanding Stimulus-Responsive Phase Behavior of Intrinsically Disordered Protein Polymers. J. Mol. Biol 430, 4619–4635. [DOI] [PubMed] [Google Scholar]
- Ryan VH, Dignon GL, Zerze GH, Chabata CV, Silva R, Conicella AE, Amaya J, Burke KA, Mittal J, and Fawzi NL (2018). Mechanistic View of hnRNPA2 Low-Complexity Domain Structure, Interactions, and Phase Separation Altered by Mutation and Arginine Methylation. Mol. Cell 69, 465–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P (2000). Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J 78, 1606–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey LM, Safren N, Pithadia AS, Gerson JE, Dulchavsky M, Fischer S, Patel R, Lantis G, Ashraf N, Kim JH, et al. (2018). Mutant UBQLN2 promotes toxicity by modulating intrinsic self-assembly. Proc. Natl. Acad. Sci 115, E10495–E10504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JP, Brown RH Jr, and Cleveland DW (2016). Decoding ALS: from genes to mechanism. Nature 539, 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teyssou E, Chartier L, Amador M-D-M, Lam R, Lautrette G, Nicol M, Machat S, Da Barroca S, Moigneu C, Mairey M, et al. (2017). Novel UBQLN2 mutations linked to Amyotrophic Lateral Sclerosis and atypical Hereditary Spastic Paraplegia phenotype through defective HSP70-mediated proteolysis. Neurobiol. Aging 58, 239.e11–239.e20. [DOI] [PubMed] [Google Scholar]
- Turakhiya A, Meyer SR, Marincola G, Böhm S, Vanselow JT, Schlosser A, Hofmann K, and Buchberger A (2018). ZFAND1 Recruits p97 and the 26S Proteasome to Promote the Clearance of Arsenite-Induced Stress Granules. Mol. Cell 70, 906–919.e7. [DOI] [PubMed] [Google Scholar]
- Urry DW, Gowda DC, Parker TM, Luan C-H, Reid MC, Harris CM, Pattanaik A, and Harris RD (1992). Hydrophobicity scale for proteins based on inverse temperature transitions. Biopolymers 32, 1243–1250. [DOI] [PubMed] [Google Scholar]
- Vengoechea J, David MP, Yaghi SR, Carpenter L, and Rudnicki SA (2013). Clinical variability and female penetrance in X-linked familial FTD/ALS caused by a P506S mutation in UBQLN2. Amyotroph. Lateral Scler. Front. Degener 14, 615–619. [DOI] [PubMed] [Google Scholar]
- Vernon RM, Chong PA, Tsang B, Kim TH, Bah A, Farber P, Lin H, and Forman-Kay JD (2018). Pi-Pi contacts are an overlooked protein feature relevant to phase separation. ELife 7, e31486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, and Laue ED (2005). The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins Struct. Funct. Bioinforma 59, 687–696. [DOI] [PubMed] [Google Scholar]
- Walters RW, and Parker R (2015). Coupling of Ribostasis and Proteostasis: Hsp70 Proteins in mRNA Metabolism. Trends Biochem. Sci 40, 552–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Conicella AE, Schmidt HB, Martin EW, Rhoads SN, Reeb AN, Nourse A, Montero DR, Ryan VH, Rohatgi R, et al. (2018a). A single N terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. EMBO J. 37, e97452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Choi J-M, Holehouse AS, Lee HO, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, et al. (2018b). A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 174, 688–699.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber SC (2017). Sequence-encoded material properties dictate the structure and function of nuclear bodies. Curr. Opin. Cell Biol 46, 62–71. [DOI] [PubMed] [Google Scholar]
- Wegmann S, Eftekharzadeh B, Tepper K, Zoltowska KM, Bennett RE, Dujardin S, Laskowski PR, MacKenzie D, Kamath T, Commins C, et al. (2018). Tau protein liquid–liquid phase separation can initiate tau aggregation. EMBO J. 37, e98049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M-T, Elbaum-Garfinkle S, Holehouse AS, Chen CC-H, Feric M, Arnold CB, Priestley RD, Pappu RV, and Brangwynne CP (2017). Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nat. Chem 9, 1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KL, Warraich ST, Yang S, Solski JA, Fernando R, Rouleau GA, Nicholson GA, and Blair IP (2012). UBQLN2/ubiquilin 2 mutation and pathology in familial amyotrophic lateral sclerosis. Neurobiol. Aging 33, 2527.e3–2527.e10. [DOI] [PubMed] [Google Scholar]
- Wyman J, and Gill SJ (1980). Ligand-Linked Phase Changes in a Biological System: Applications to Sickle Cell Hemoglobin. Proc. Natl. Acad. Sci. U. S. A 77, 5239–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Jones HB, Dao TP, Castañeda CA Single amino acid substitutions in stickers, but not spacers, substantially alter UBQLN2 phase transitions and dense phase material properties. J. Phys. Chem. B 2019; 10.1021/acs.jpcb.9b01024 [DOI] [PubMed] [Google Scholar]
- Yeo GC, Keeley FW, and Weiss AS (2011). Coacervation of tropoelastin. Adv. Colloid Interface Sci 167, 94–103. [DOI] [PubMed] [Google Scholar]
- Zhang D, Raasi S, and Fushman D (2008). Affinity makes the difference: nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J. Mol. Biol 377, 162–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Ubiquitin eliminates UBQLN2 phase separation. Ubiquitin was added to one part of sample well, and was allowed to diffuse into a different area of the well where UBQLN2 450–624 droplets were imaged. Related to Figure 6.
Movie S2. Ubiquitin eliminates UBQLN2 T487I droplets. Ubiquitin was added to one part of sample well, and was allowed to diffuse into a different area of the well where UBQLN2 T487I droplets were imaged. Related to Figure 6.
Movie S3. Ubiquitin eliminates UBQLN2 P497S aggregates. Ubiquitin was added to one part of sample well, and was allowed to diffuse into a different area of the well where UBQLN2 P497S aggregates were imaged. Related to Figure 6.
All movies are approximately 40 seconds of real time. Final concentrations of Ub and UBQLN2 were 100 μM each in pH 6.8 buffer containing 200 mM NaCl. The temperature used for all experiments was 30°C. Scale bar = 5 μm.
Data Availability Statement
Microscopy imaging data is available on Mendeley (http://dx.doi.org/10.17632/9cr27cbmkz.1).