Abstract
Base editing requires that the target sequence satisfy the PAM requirement of the Cas9 domain and that the target nucleotide is located within the editing window of the base editor. To increase the targeting scope of base editors, we engineered six optimized adenine base editors (ABEmax variants) that use SpCas9 variants compatible with non-NGG PAMs. To increase the range of target bases that can be modified within the protospacer, we use circularly permuted Cas9 variants to produce four cytosine and four adenine base editors with an editing window expanded from ~4–5 nucleotides to up to ~8–9 nucleotides and reduced byproduct formation. This set of base editors improves the targeting scope of cytosine and adenine base editing.
Ed sum:
Wider editing windows and different PAM requirements enable a broad set of genomic positions to be targeted with A and C base editors.
Base editing enables the programmable conversion of one base pair to another without making double-stranded DNA breaks (DSBs),1–5 and has already been widely used to install or correct point mutations in a wide range of organisms6,7. We developed two classes of base editors: cytosine base editors (CBEs) convert C•G to T•A8–10, while adenine base editors (ABEs) convert A•T to G•C11,12. Together, CBEs and ABEs enable the installation or correction of all four transition mutations, collectively accounting for ~63% of pathogenic human point mutations13,14. Because base editors do not make DSBs, they minimize the formation of editing byproducts such as insertions, deletions, translocations, or DNA rearrangements15–17.
Since base editors use a catalytically impaired Cas9 to recognize the target DNA site, they canonically require that the target site contain a PAM ~15±2 nucleotides from the target base. CBE variants that use Cas9 homologs with different PAM requirements5,10,18,19 increase the likelihood that a target site supports cytosine base editing. In contrast, far fewer ABE variants with distinct PAMs have been described20–22.
Both CBEs and ABEs mutate target base pairs within a small (typically ~4- to 5-nucleotide) window within the protospacer. The width of this editing window, together with PAM availability, define the targeting scope of base editing. In some cases, the target base is located outside the base editing window relative to an available PAM. Moreover, for applications such as mutagenizing a gene, disrupting genes by introducing premature stop codons, or abrogating splice sites or regulatory sequences, a wider editing window is desirable.
Here we introduce current-generation ABEmax23 variants optimized for mammalian cell use on target sites with non-NGG PAMs. We also report new CBEs and ABEs that use circularly permuted SpCas9 (CP-Cas9) variants to expand the base editing window from ~4–5 nucleotides to up to ~8–9 nucleotides. The resulting CP-CBEmax variants exhibit higher product purities, in addition to expanded editing windows, while CP-ABEmax variants maintain the high product purities typical of ABEs. These new CBE and ABE variants expand the targeting scope of base editing.
We and others have reported the compatibility of CBEs with CRISPR proteins that recognize PAMs other than NGG, thereby expanding their targeting scope. These variants included evolved S. pyogenes Cas9 variants SpCas9-VQR/SpCas9-VRQR (PAM: NGA)24,25, SpCas9-VRER (PAM: NGCG)24, xCas9 (PAM: NGN)19, SpCas9-NG (PAM: NG)26, S. aureus Cas9 (PAM: NNGRRT)27,28 and its modified variant KKH (PAM: NNNRRT)29, and L. bacterium Cas12a (Cpf1, PAM: TTTV where V = A, C, or G)18,30. Following the development of ABE, we hypothesized that the evolved TadA deoxyadenosine deaminase domain might be similarly compatible with other CRISPR proteins. Indeed, Yang et al. and Hua et al. recently used mouse and plant versions of VQR-ABE, VRER-ABE, SaCas9-ABE, and SaKKH-ABE to perform base editing in mouse embryos and rice, respectively20–22.
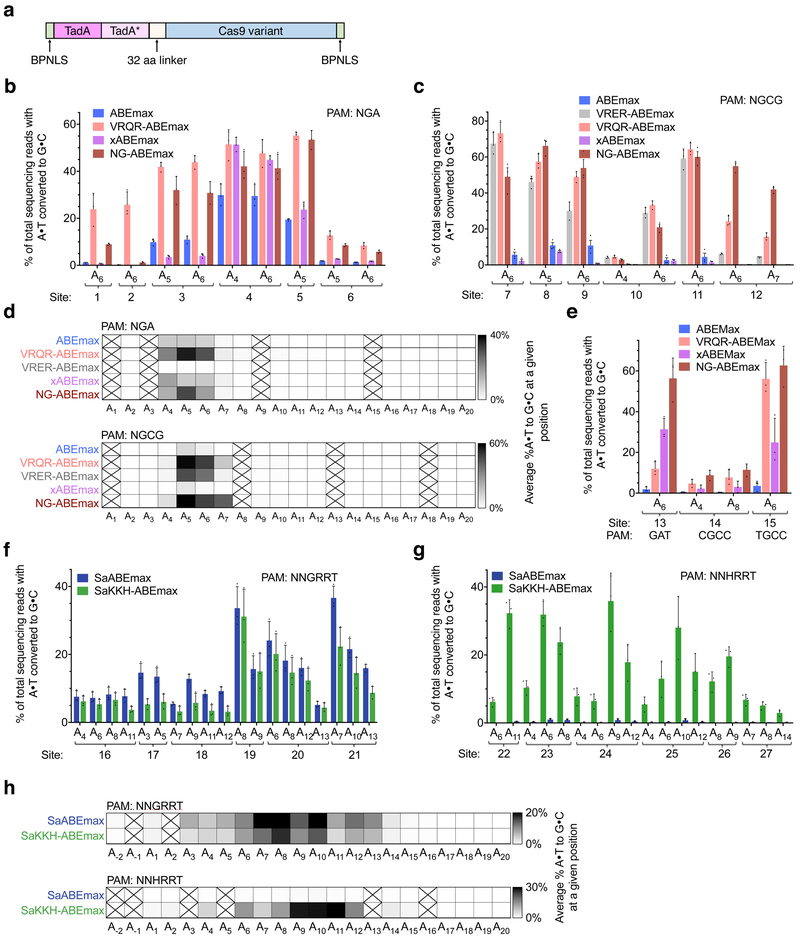
Optimization of both codon usage and nuclear localization in both cytosine and adenine base editors, resulting in BE4max (referred to hereafter as CBEmax) and ABEmax, respectively, greatly enhances base editing activity in mammalian cells23,31. We used these current-generation CBE and ABE forms for all base editors constructed in this study. First, we created ABEmax variants that replace the SpCas9 nickase component with two engineered SpCas9 variants with altered PAM specificities: VRQR-SpCas9 (PAM: NGA) and VRER-SpCas9 (PAM: NGCG) (Fig. 1a), yielding VRQR-ABEmax and VRER-ABEmax, respectively. We then evaluated base editing activity at six endogenous human genomic loci for each PAM in human HEK293T cells.
Figure 1. Cas9 PAM-variant ABE orthologues mediate A•T-to-G•C conversion in human cells.
(a) Optimized ABEmax architecture23 used for PAM variant construction. (b) Base editing in HEK293T cells by ABEmax, VRQR-ABEmax, xABEmax, and NG-ABEmax at six genomic sites containing an NGA PAM. (c) Base editing in HEK293T cells by ABEmax, VRER-ABEmax, VRQR-ABEmax, xABEmax, and NG-ABEmax at six genomic sites containing an NGCG PAM. (d) Heat maps showing average editing efficiency by SpCas9-derived ABE variants at each protospacer position across sites containing each PAM listed (n = 6). Spaces crossed out indicate a position for which no target base was present among all the genomic sites tested. (e) Base editing in HEK293T cells by ABEmax, VRQR-ABEmax, xABEmax, and NG-ABEmax at three genomic sites (PAMs: GAT, CGCC, TGCC). (f) Base editing in HEK293T cells by SaABEmax and SaKKH-ABEmax at six genomic sites containing an NNGRRT PAM. (g) Base editing in HEK293T cells by SaABEmax and SaKKH-ABEmax at six genomic sites containing an NNHRRT PAM. (h) Heat maps showing average editing efficiency by SaCas9-derived ABE variants at each protospacer position across sites containing each PAM listed (n = 6). Subscripted numbers indicate protospacer positions, counting the first base of the PAM as position 21. Values and error bars reflect the mean±s.d. of three independent biological replicates performed by different researchers on different days.
Across six endogenous NGA PAM-containing sites we observed low editing efficiency with ABEmax, averaging 11±2.1% A•T-to-G•C conversion (mean and s.d. of three biological replicates at six genomic sites, reporting the target A with the highest conversion frequency). In contrast, VRQR-ABEmax resulted in 35±4.6% A•T-to-G•C conversion across the same six genomic sites, a 3.2-fold average improvement (Fig. 1b). We also compared VRQR-ABEmax activity to that of ABEmax variants that use either xCas919 or the recently reported SpCas9-NG26, both of which are active on some NGN PAM sites (xABEmax and NG-ABEmax, respectively). The average editing activity of xABEmax was 2.7-fold lower than that of VRQR-ABEmax (Fig. 1b). NG-ABEmax exhibited comparable activity to VRQR-ABEmax at some sites, but overall lower activity than VRQR-ABEmax with an average of 24±3.9% A•T-to-G•C conversion at these six genomic sites. Thus, VRQR-SpCas9, engineered specifically to recognize NGA PAM sites28, supports more efficient editing at these sites than other evolved Cas9 variants.
At the six tested endogenous genomic sites containing NGCG PAMs, we observed minimal activity from ABEmax and xABEmax in HEK293T cells (Fig. 1c). VRER-ABEmax, however, greatly improved A•T-to-G•C conversion efficiencies at all tested sites, averaging 40±3.6% conversion, a 7.0-fold improvement over ABEmax. Because the VRQR variant differs from the VQR variant (which functions on both NGA and NGCG PAMs24) only by the addition of the G1218R mutation, which is also present in VRER-SpCas9, we suspected that VRQR retains activity on NGCG PAMs. Indeed, VRQR-ABEmax exhibited a further 1.3-fold improvement in editing efficiencies (averaging 50±3.6% A•T-to-G•C conversion) at the same six NGCG PAM sites compared to VRER-ABEmax (Fig. 1c). NG-ABEmax performed equally well on the six NGCG PAM-containing genomic sites as VRQR-ABEmax, averaging 51±5.9% A•T-to-G•C conversion. VRER-ABEmax, VRQR-ABEmax, and NG-ABEmax did not exhibit significant indel formation or an apparent shift in the base editing window (Fig. 1d, Supp. Fig. 1).
Next, we evaluated the base editing activity of VRQR-ABEmax and NG-ABEmax on three genomic sites that we previously showed were edited by xABE19 containing PAMs other than NGA or NGCG. Of the three sites tested (with GAT and two NGCC PAMs), VRQR-ABEmax exhibited an average of 2.3-fold greater A•T-to-G•C conversion activity on both NGCC PAM sites, but 2.6-fold lower activity on a GAT PAM site, compared to xABEmax (Fig. 1e). NG-ABEmax exhibited a 1.5-fold greater A•T-to-G•C conversion activity on both NGCC PAM sites and a 1.7-fold greater activity on the GAT PAM site. These data indicate that the VRQR-, VRER-, and SpCas9-NG variants are compatible with the ABEmax architecture and retain base editing activity at sites containing their cognate non-NGG PAMs. In most cases, VRQR-ABEmax tends to outperform other reported SpCas9 variants on sites containing either NGCG or NGA PAMs, and at some sites with NGCC PAMs. For alternative PAMs, such as the GAT PAM site examined in this study, xABEmax or base editors derived from Cas9-NG26 are preferred. Taken together, these results indicate that targets lacking NGG PAMs can be accessed by multiple evolved or engineered Cas9 variants, and that the optimal base editor may vary on a site-by-site basis.
To further expand the targeting scope of ABE, we examined whether the Cas9 ortholog from S. aureus (SaCas9) is compatible with the ABEmax architecture. SaCas9 naturally targets NNGRRT PAMs27, and an evolved variant, SaKKH, recognizes NNNRRT PAMs29. We generated both SaCas9 and SaKKH ABEmax variants and tested them on six endogenous NNGRRT PAM sites and six endogenous NNHRRT PAM sites in HEK293T cells.
Observed A•T-to-G•C conversion activity varied substantially from site-to-site, but averaged 22±2.3% and 26±5.7% A•T-to-G•C conversion for SaABEmax on six NNGRRT PAM sites and SaKKH-ABEmax on six NNHRRT sites, respectively, with minimal indels (Fig. 1f,g, Supp. Fig. 2). SaABEmax was unable to efficiently target the six NNHRRT PAM sites (Fig. 1g). However, the editing efficiency of SaABEmax was slightly higher than that of SaKKH-ABEmax on NNGRRT sites (22±2.3% vs. 15±2.8%). The moderate editing efficiencies of SaABEmax and SaKKH-ABEmax contrast with the high activities of SaCas9-derived CBEs10, which generally edit more efficiently than the corresponding SpCas9 CBE. These results suggest further engineering or evolution may benefit targeting ABE with SaCas9 derivatives.
Consistent with our previous observations of SaCas9-derived CBEs10, SaABEmax and SaKKH-ABEmax exhibited an expanded base editing activity window from protospacer positions 4–14 (numbering the PAM as positions 21–26). Maximum editing typically occurred around positions 7–11, with the most frequent outcome being a single A•T-to-G•C edit within this window (Fig. 1h, Supp. Fig. 2). Although an expanded window increases the likelihood of bystander base editing (the editing of non-target adenines within the activity window), a larger window is useful when target adenines would otherwise be inaccessible due to the lack of a PAM, when bystander editing is not consequential, when undesired genotypes can be removed by screening, or when broad mutagenesis is desired.
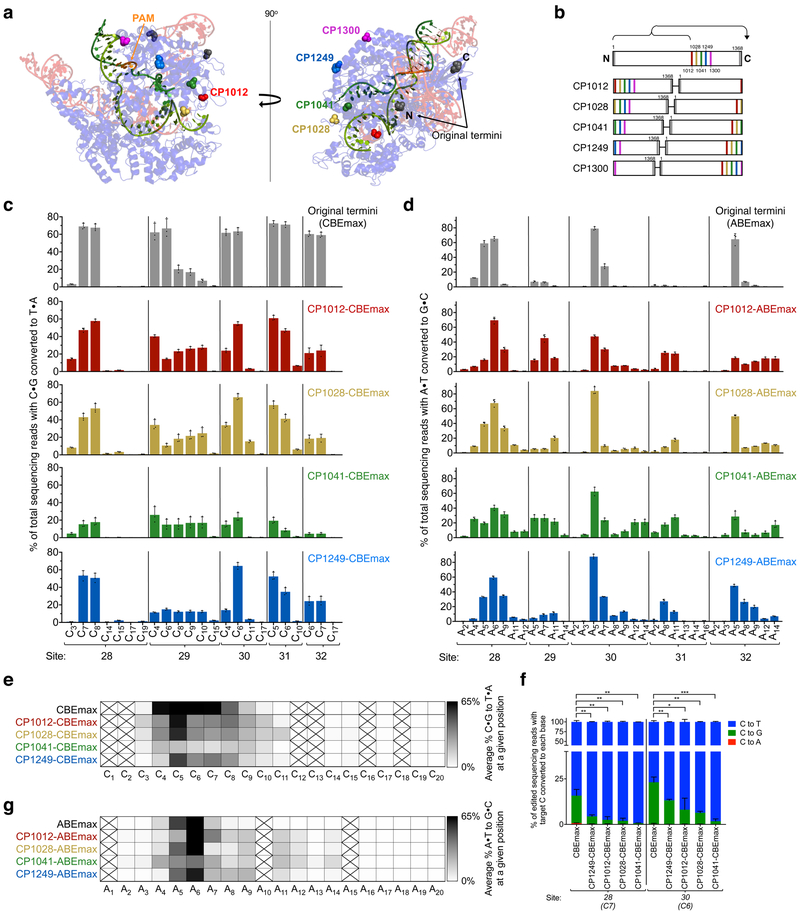
Given the potential utility of base editors with shifted or expanded activity windows32, we next sought to engineer new base editor architectures that enable editing at different protospacer positions. The activity window of base editors in mammalian cells has proven surprisingly difficult to broaden, with multimeric deaminase assembly for CBEs33 and extended guide RNAs for ABEs34 representing the only window-broadening strategies reported to date. Oakes and coworkers recently generated circularly permuted SpCas9 variants that retain both binding and DNA cleavage activity35. For several active SpCas9 circular permutants, the new termini are predicted to lie closer to the ssDNA loop that is the substrate for base editing than the original SpCas9 termini (Fig. 2a)36. We hypothesized that these circular permutants might provide the deaminase domains in CBEs and ABEs greater access to the ssDNA loop, resulting in expanded or otherwise altered activity windows.
Figure 2. Circularly permuted Cas9 variants are compatible with the cytosine and adenine base editor architectures and exhibit broadened editing windows.
(a) Two views of the SpCas9:sgRNA:DNA crystal structure (PDB: 5F9R36) showing the location of the N and C termini in the wild-type protein (black) and in circularly permuted variants tested in this work (red, yellow, green, blue, and magenta). The DNA strand paired with the guide RNA is shown in light green and the other strand, targeted for base editing and partially disordered in the structure, is shown in dark green. (b) Schematic representations of CP-Cas9 variants tested in this work, numbered to indicate the original SpCas9 amino acid that serves as the new N-terminus. (c) Base editing in HEK293T cells by CP-CBEmax variants across five genomic sites. (d) Base editing in HEK293T cells by CP-ABEmax variants across five genomic sites. For clarity, in (c) and (d), protospacer positions with editing lower than 0.5% across all editors are not shown. CP1300 data is shown in Supp. Fig. 4. (e, f) Heat maps showing average base editing efficiency at each position within the protospacer across five sites tested with (e) CP-CBEmax variants or (f) CP-ABEmax variants, normalized to the maximum observed editing within the protospacer (1.0). Boxes crossed out indicate positions for which no target base was present among all genomic sites tested. (g) The product distribution among edited DNA sequencing reads (reads in which the target C is base edited) is shown for each CP-CBEmax variant tested at two different genomic sites that are especially prone to non-C-to-T byproduct formation. Subscripted numbers indicate protospacer positions, counting the first base of the PAM as position 21. Values and error bars reflect the mean±s.d. of three biological replicates performed on different days at each site. ns, P>0.05; *P<0.05; **P<0.01; ***P<0.001, by two tailed Student’s t-test.
We chose five SpCas9 circular permutants (CP1012, CP1028, CP1041, CP1249, and CP1300, in which the number identifies the amino acid that serves as the new N-terminus) based on both retention of DNA binding activity and predicted proximity to the ssDNA loop35. We generated five CP-CBEmax and five CP-ABEmax variants by fusing the circularly permuted Cas9 nickase variants in bis-bpNLS and codon-optimized forms (Fig. 2a,b).
The resulting CP-CBEmax and CP-ABEmax variants were transfected into HEK293T cells and tested for base editing activity at five endogenous genomic sites containing adenines and cytosines throughout the target 20-nt protospacer (Fig. 2c-f). We chose genomic loci for which target bases were mostly located outside of the canonical editing window of positions 4–8 to more precisely define the editing window of these new base editors. Four of the five CP-CBE variants were capable of base editing at all five sites without substantial indel formation (generally < 2%) (Fig. 2c, Supp. Fig. 3), while CP1300-CBEmax demonstrated highly site-dependent base editing activity (Supp. Fig. 4). Three of the remaining four CP-CBEmax variants exhibited efficient editing activity, averaging 47±3.6%, 46±4.9%, 18±5.6% and 42±4.6% C•G-to-T•A conversion for CP1012-CBEmax, CP1028-CBEmax, CP1041-CBEmax, and CP1249-CBEmax, respectively, compared to 66±5.9% C•G-to-T•A conversion for CBEmax at the same genomic sites. Two of the variants, CP1012-CBEmax and CP1028-CBEmax, showed broadening of the editing window from the canonical positions 4–8 to positions 4–11 of the protospacer, averaging 12±1.6% and 15±3.9% C•G-to-T•A conversion at positions 9–11, respectively, compared to 5.8±2.2% for CBEmax at these positions (Fig. 2c,e). These results together establish that circularly permuted CBEmax variants indeed exhibit broadened editing windows.
Surprisingly, at three of the five genomic sites tested with CP-CBEmax variants, CP1012-CBEmax, CP1028-CBEmax, and CP1041-CBEmax also edited bases upstream of the protospacer on both the target strand (the strand normally targeted for nucleobase deamination) and the non-target strand. This out-of-protospacer editing was particularly evident for CP1012-CBEmax, with editing observed as far upstream as the −13 position of the target strand (Supp. Fig. 5). These upstream editing events may arise from extended R-loop formation by some circular permutants at sites prone to unwinding, creating larger accessible ssDNA regions.
As we previously reported1,8, CBEs can generate both desired C-to-T edits and unanticipated C-to-G and C-to-A mutations resulting from error-prone base excision repair of the uracil intermediate. Among the five genomic sites tested, three sites when treated with CBEmax resulted in < 1% non-C-to-T byproducts, but two sites unusually prone to unanticipated editing byproducts showed an average of 19±3.3% non-C-to-T byproducts among CBEmax-edited products. Surprisingly, CP1012-CBEmax, CP1028-CBEmax, CP1041-CBEmax, and CP1249-CBEmax demonstrated greatly reduced (2.1- to 19-fold lower than CBEmax) byproduct formation at these two problematic sites (Fig. 2g, Supp. Table 1). The improved product purity of CP-CBEmax variants might result from the newly positioned termini of Cas9 allowing the C-terminal UGI (or a UGI•UNG complex) to better impede uracil excision by UNG. Consistent with this model, the minimum linear distance between the predicted location of the C-termini and the ssDNA target in two different SpCas9 crystal structures36,37 is inversely related to observed product purity (decreasing distance and increasing product purities: CBEmax, CP1249-CBEmax ~ CP1012-CBEmax, CP1028-CBEmax, CP1041-CBEmax) (Supp. Fig. 6).
To probe the relationship between product purity and UGI positioning, we generated CP-CBEmax base editors without UGI, denoted CP-CBEmax-B variants. At one of the genomic sites prone to product mixtures, CP-CBEmax-B variants no longer showed a correlation between linear distance and product purity (Supp. Fig. 7, Supp. Table 1). This correlation was still evident, however, at the other site, suggesting that factors other than UGI positioning can also govern product purity12 on a site-dependent basis. Together, these results reveal that circularly permuted CBEmax variants generate undesired byproducts less frequently than CBEmax, possibly by enhancing access of the UGI domain to the edited site.
Most CP-ABEmax variants similarly exhibited a broadening of the editing window (Fig. 2d). CP-ABEmax variants retained efficient editing activity similar to that of ABEmax, averaging 45±3.5%, 52±3.9%, 40±5.4%, and 51±2.2% A•T-to-G•C conversion for CP1012-ABEmax, CP1028-ABEmax, CP1041-ABEmax, and CP1249-ABEmax, respectively compared to an average of 54±4.3% A•T-to-G•C conversion for ABEmax at the same four genomic sites. Both ABEmax and the circularly permuted variants generated minimal indels (generally < 2%) (Supp. Fig. 3). While no out-of-protospacer editing was observed among circularly permutated ABEmax variants, the window-broadening effect was pronounced, generally resulting in an expansion from the canonical window of protospacer positions 4–7 for ABEmax to a window spanning positions 4–12 (Fig. 2f). Base editing efficiencies at protospacer positions 8–12 averaged 20±1.9%, 18±2.2%, 24±4.0%, and 23±2.3% A•T-to-G•C conversion for CP1012-ABEmax, CP1028-ABEmax, CP1041-ABEmax, and CP1249-ABEmax, respectively, a 6.4- to 8.6-fold increase over that of ABEmax at these distal protospacer positions. Intriguingly, CP1012-ABEmax also exhibited a shifted (rather than broadened) base editing window, with maximal editing taking place at A6 or A7, compared to A5 or A6 for ABEmax (Fig. 2f). CP1041-ABEmax showed the broadest editing window, with up to 11±3.3% average editing at position 14 of the protospacer (Fig. 2f). CP-ABEmax variants thus are able to edit target As that lie outside the canonical ABE editing window.
To assess possible effects of circular permutation on off-target base editing, we measured off-target editing of all ten CP-CBEmax and CP-ABEmax variants at nine genomic off-target sites previously identified by GUIDE-Seq as the most highly edited off-target substrates of SpCas9 nuclease for three target loci38. Off-target base editing efficiency of circularly permuted base editors was similar to or less than that of CBEmax or ABEmax for C or A nucleotides within the canonical editing window. As expected, for C or A nucleotides outside the canonical editing window, the expanded editing windows of circularly permuted base editors in some cases allowed higher off-target editing than CBEmax or ABEmax (Supp. Tables 2-7).
Together, these results demonstrate that circularly permuting the Cas9 nickase domain of base editors results in CBEmax and ABEmax variants with broadened or shifted editing windows. These altered targeting properties enable efforts to perform base editing at currently inaccessible target nucleotides, and can also substantially improve product purity. Indeed, an analysis of human pathogenic SNPs in ClinVar17,18 reflects a substantial improvement in the fraction of targetable SNPs when considering the expanded CP-CBEmax or CP-ABEmax editing windows (51% of SNPs correctable by A•T-to-G•C conversion or 51% of SNPs correctable by C•G-to-T•A conversion, respectively) compared to their unpermuted CBEmax and ABEmax counterparts (27% and 31%, respectively) (Supp. Fig. 8). Alternative-PAM ABEmax variants and circularly permuted CBEmax and ABEmax variants thus expand the capabilities of base editors, now widely used in the biomedical research community5.
Methods
General methods
PCR was performed using Phusion U Green Multiplex PCR Master Mix (ThermoFisher Scientific). All plasmids were assembled by the USER cloning method as previously described2. The amino acid sequences for maxed base editor variants are listed in Supplementary Sequences 1. Guide RNA plasmids for SpCas9, SaCas9, and all engineered variants were assembled as previously described10. Sequences of protospacers used in this study are listed in Supplementary Table 8. Plasmids for mammalian cell transfections were prepared using the ZymoPURE Plasmid Midiprep kit (Zymo Research Corporation).
Cell culture
HEK293T cells (ATCC CRL-3216) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Corning) supplemented with 10% fetal bovine serum (FBS, ThermoFisher Scientific) and maintained at 37 °C with 5% CO2.
Transfections
HEK293T cells were seeded on 48-well Poly-D-Lysine plates (Corning) in the same culture medium. Cells were transfected 12–16 hours after plating with 1.5 µL Lipofectamine 2000 (ThermoFisher Scientific) using 750 ng of base editor plasmid, 250 ng of guide RNA plasmid, and 10 ng of green fluorescent protein as a transfection control. Cells were cultured for 3 d with media exchanged following the first day, then washed with 1x phosphate-buffered saline (PBS, ThermoFisher Scientific), followed by genomic DNA extraction by addition of 100 µL of freshly prepared lysis buffer (10 mM Tris-HCl, pH 7.5, 0.05 % SDS, 25 µg/mL proteinase K (ThermoFisher Scientific)) directly into each transfected well. The mixture was incubated at 37 ºC for 1 h then heat inactivated at 80 ºC for 30 min. Genomic DNA lysate was subsequently used immediately for HTS sequencing.
HTS of genomic DNA samples
HTS of genomic DNA from HEK293T cells was performed as previously described2. Primers for PCR 1 of target genomic site amplification are listed in Supplementary Table 9. Following Illumina barcoding, PCR products were pooled and purified by electrophoresis with a 2% agarose gel using a Monarch DNA Gel Extraction Kit (NEB), eluting with 30 μL H2O. DNA concentration was quantified with Qubit dsDNA High Sensitivity Assay Kit (ThermoFisher Scientific) and sequenced on an Illumina MiSeq instrument (paired-end read – R1: 250–280 cycles, R2: 0 cycles) according to the manufacturer’s protocols.
HTS data analysis
Sequencing reads were demultiplexed using the MiSeq Reporter (Illumina) and fastq files were analyzed using Crispresso239. Representative analysis input and usage is described in Supplementary Note 1. Base editing values are representative of n= 3 independent biological replicates collected over different days by different researchers with the mean ± s.d. shown. Base editing values are reported as a percentage of the number of reads with cytosine or adenine mutagenesis over the total aligned reads.
Statistics and reproducibility
All statistical analyses were performed on n = 3 biologically independent experiments using the unpaired two tailed Student’s t-test. Biologically independent experiments reported here were performed by different researchers using independent splits of the mammalian cell type used. Test values can be found in Supplementary Table 1.
Life sciences reporting summary
Further information on experimental design is available in the Life Sciences Reporting Summary linked to this article.
Code availability
Access to and usage for the custom Crispresso2 script can be found in Supplementary Note 139.
ClinVar analysis of pathogenic human SNPs targetable by the base editors described in this study was executed using a custom Matlab script described previously1,2.
Data availability
Plasmids encoding modified PAM adenine base editors and circularly permuted cytidine and adenine base editors have been deposited to Addgene. High-throughput sequencing data are deposited in the NCBI Sequence Read Archive (PRJNA498804).
Supplementary Material
Acknowledgements
This work was supported by US NIH U01 AI142756, RM1 HG009490, R01 EB022376, R35 GM118062, St. Jude Research Consortium, the Ono Pharma Foundation, DARPA HR0011–17-2–0049, and HHMI. T.P.H. and K.T.Z. were supported by the Harvard Chemical Biology Program NIH Training Grant T32 GM095450. S.M.M. was supported by an NSF graduate fellowship. B.L.O. was supported by the Innovative Genomic Institute Entrepreneurial Fellowship Program. C.F. is supported by US NIH K99 GM118909 and R00 GM118909. D.F.S. was supported by US NIH DP2 EB018658.
Footnotes
Competing Financial Interests
The authors declare competing financial interests: D.R.L. is a consultant and co-founder of Editas Medicine, Pairwise Plants, and Beam Therapeutics, companies that use genome editing. The authors have filed patent applications on base editors with altered targeting properties. The authors declare no competing non-financial interests.
References Cited
- 1.Komor AC, Kim YB, Packer MS, Zuris JA & Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424, doi: 10.1038/nature17946 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaudelli NM et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551, 464–471, doi: 10.1038/nature24644 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zong Y et al. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol 35, 438–440, doi: 10.1038/nbt.3811 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Zeng Y et al. Correction of the Marfan Syndrome Pathogenic FBN1 Mutation by Base Editing in Human Cells and Heterozygous Embryos. Mol Ther, doi: 10.1016/j.ymthe.2018.08.007 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rees HA & Liu DR Base Editing: Precision Chemistry on the Genome and Transcriptome of Living Cells. Nature Reviews Genetics (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chadwick AC, Wang X & Musunuru K In Vivo Base Editing of PCSK9 (Proprotein Convertase Subtilisin/Kexin Type 9) as a Therapeutic Alternative to Genome Editing. Arterioscler Thromb Vasc Biol 37, 1741–1747, doi: 10.1161/ATVBAHA.117.309881 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Villiger L et al. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat Med 24, 1519–1525, doi: 10.1038/s41591-018-0209-1 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Komor AC et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci Adv 3, eaao4774, doi: 10.1126/sciadv.aao4774 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rees HA et al. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat Commun 8, 15790, doi: 10.1038/ncomms15790 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim YB et al. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol 35, 371–376, doi: 10.1038/nbt.3803 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan F et al. Highly Efficient A.T to G.C Base Editing by Cas9n-Guided tRNA Adenosine Deaminase in Rice. Mol Plant 11, 631–634, doi: 10.1016/j.molp.2018.02.008 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Kang BC et al. Precision genome engineering through adenine base editing in plants. Nat Plants 4, 427–431, doi: 10.1038/s41477-018-0178-x (2018). [DOI] [PubMed] [Google Scholar]
- 13.Landrum MJ et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res 44, D862–868, doi: 10.1093/nar/gkv1222 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landrum MJ et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 42, D980–985, doi: 10.1093/nar/gkt1113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosicki M, Tomberg K & Bradley A Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol 36, 765–771, doi: 10.1038/nbt.4192 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haapaniemi E, Botla S, Persson J, Schmierer B & Taipale J CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med 24, 927–930, doi: 10.1038/s41591-018-0049-z (2018). [DOI] [PubMed] [Google Scholar]
- 17.Ihry RJ et al. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat Med 24, 939–946, doi: 10.1038/s41591-018-0050-6 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Li X et al. Base editing with a Cpf1-cytidine deaminase fusion. Nat Biotechnol 36, 324–327, doi: 10.1038/nbt.4102 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Hu JH et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63, doi: 10.1038/nature26155 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hua K, Tao X, Yuan F, Wang D & Zhu JK Precise A.T to G.C Base Editing in the Rice Genome. Mol Plant 11, 627–630, doi: 10.1016/j.molp.2018.02.007 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Hua K, Tao X & Zhu JK Expanding the base editing scope in rice by using Cas9 variants. Plant Biotechnol J, doi: 10.1111/pbi.12993 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang L et al. Increasing targeting scope of adenosine base editors in mouse and rat embryos through fusion of TadA deaminase with Cas9 variants. Protein Cell 9, 814–819, doi: 10.1007/s13238-018-0568-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koblan LW et al. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat Biotechnol 36, 843–846, doi: 10.1038/nbt.4172 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleinstiver BP et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523, 481–485, doi: 10.1038/nature14592 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleinstiver BP et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495, doi: 10.1038/nature16526 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishimasu H et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 361, 1259–1262, doi: 10.1126/science.aas9129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ran FA et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191, doi: 10.1038/nature14299 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu T et al. Highly efficient base editing in Staphylococcus aureus using an engineered CRISPR RNA-guided cytidine deaminase. Chem Sci 9, 3248–3253, doi: 10.1039/c8sc00637g (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleinstiver BP et al. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol 33, 1293–1298, doi: 10.1038/nbt.3404 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zetsche B et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771, doi: 10.1016/j.cell.2015.09.038 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zafra MP et al. Optimized base editors enable efficient editing in cells, organoids and mice. Nat Biotechnol 36, 888–893, doi: 10.1038/nbt.4194 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zong Y et al. Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat Biotechnol, doi: 10.1038/nbt.4261 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Jiang W et al. BE-PLUS: a new base editing tool with broadened editing window and enhanced fidelity. Cell Res, doi: 10.1038/s41422-018-0052-4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryu SM et al. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat Biotechnol 36, 536–539, doi: 10.1038/nbt.4148 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Oakes BL FC, Rishi H, Taylor KL, Ren SM, Nadler D, Yokoo R, Arkin A, Doudna JA, Savage DF. CRISPR-Cas9 circular permutants as programmable scaffolds for genome modification. Cell (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang F et al. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 351, 867–871, doi: 10.1126/science.aad8282 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huai C et al. Structural insights into DNA cleavage activation of CRISPR-Cas9 system. Nat Commun 8, 1375, doi: 10.1038/s41467-017-01496-2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai SQ et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 33, 187–197, doi: 10.1038/nbt.3117 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clement K et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol 37, 224–226, doi: 10.1038/s41587-019-0032-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Plasmids encoding modified PAM adenine base editors and circularly permuted cytidine and adenine base editors have been deposited to Addgene. High-throughput sequencing data are deposited in the NCBI Sequence Read Archive (PRJNA498804).