Abstract
Background:
While important in diagnosis of breast cancer, the scientific assessment of the role of imaging in prognosis of outcomes and treatment planning is limited.
Purpose:
To evaluate the potential of using quantitative imaging variables for stratifying risk of distant recurrence in breast cancer patients.
Study type:
Retrospective
Population:
892 female invasive breast cancer patients.
Sequence:
Dynamic contrast-enhanced MRI with field strength 1.5T and 3T
Materials and Methods:
Computer vision algorithms were applied to extract a comprehensive set of 529 imaging features quantifying size, shape, enhancement patterns, and heterogeneity of the tumors and the surrounding tissue. Using a development set with 446 cases, we selected 20 imaging features with high prognostic value.
Statistical tests:
We evaluated the imaging features using an independent test set with 446 cases. The principal statistical measure was a concordance index between individual imaging features and patient distant recurrence-free survival.
Results:
The strongest association with distant recurrence-free survival (DRFS) that persisted after controlling for known prognostic clinical and pathology variables was found for signal enhancement ratio (SER) partial tumor volume (concordance index [C]=0.768, 95% CI: 0.679–0.856), Tumor Major Axis Length (C=0.742, 95% CI: 0.650–0.834)
Kurtosis of the SER map within tumor (C=0.640, 95% CI: 0.521–0.760), tumor cluster shade (C=0.313, 95% CI: 0.216–0.410), and wash-in rate information measure of correlation (C=0.702, 95% CI: 0.601–0.803).
Conclusions:
Quantitative assessment of breast cancer features seen in a routine breast MRI might be able to be used for assessment of risk of distant recurrence.
Keywords: breast cancer, MRI, radiomic features, metastasis distant recurrence free survival
INTRODUCTION
Outcomes of breast cancer patients vary widely based on the presentation of the disease at the time of diagnosis. Prognosis of outcomes at that time plays a crucial role in the planning of the patient’s treatment. The primary outcome of interest in non-metastatic breast cancer (the most common presentation) is recurrence-free survival. A number of factors have been shown to be associated with recurrence-free survival and are commonly used to guide decisions regarding treatment (1) including clinical variables such as the patient’s age, pathological markers such as estrogen receptor status, and more recently, genomic markers such as Oncotype DX recurrence score (2). While important in the screening and diagnosis of breast cancer(3, 4) the role of imaging in treatment planning, and particularly in prognosis of outcomes, is very limited.
Recently, an increased number studies show promise in using dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) for prognosis of outcomes. Partridge and colleagues(5) showed that tumor volume in MRI and change in tumor volume in MRI are prognostic of recurrence-free survival in breast cancer. Mazurowski et al.(6) showed the association of a specific feature that quantifies tumor enhancement kinetics with recurrence-free survival. Kim et al.(7) recently demonstrated that imaging features extracted from DCE-MRI images using standard computer-aided diagnosis software are associated with patient outcomes. However, the effect sizes found in this study were small (hazard ratios < 1.1). In another recent study, imaging features extracted manually by radiologists(8), and particularly the feature quantifying rim enhancement, showed association with outcomes. While this result is promising, manual extraction of features is highly subjective and prone to inter-reader variability. Finally, a study by Wu et al. (9) showed that algorithmically-quantified features are prognostic of outcomes. However, a set of only 60 patients with available imaging was used to evaluate these associations.
The purpose of this study was to conduct a comprehensive evaluation of a large number of algorithmically extracted image features with a large number of patients to determine whether breast MRI could be used in breast cancer prognostication.
METHODS
Patient Population
An Institutional Review Board approval was secured for this retrospective analysis. The study included 892 female invasive breast cancer cases from our institution. The details of inclusion of cases are shown in Fig. 1.
Figure 1.

Details of inclusion of patients in our study
A random split of this data was used in this study with 446 patients in the training set and 446 patients in the test set such that the training set included patients used in our earlier publications (references redacted to preserve anonymity). The training set was used for selection of a feature subset and the test set was used for evaluation of the features.
Imaging Data And Image Annotation
Axial DCE-MRIs using 1.5T or 3T scanners in the prone position were acquired. All the included studies contained a pre-contrast non-fat saturated T1-weighted sequence and a fat-saturated gradient echo T1-weighted sequence. Typically, three to four post-contrast T1-weighted sequences with fat suppression were acquired after the IV administration of contrast agent using a weight-based protocol of (0.2 mL/kg). Based on different scanners and contrast agents, the distribution of patients is shown in Table 1.
Table 1.
Distribution of Patients Scanner wise and Contrast Agent wise
| Characteristics | Technical Details | Manufacturer Details | Patient Count |
|---|---|---|---|
| Magnetic Field Strength | 1.5 T | Optima MR450w, GE Healthcare, Little Chalfont, UK | 96 |
| Signa HDx, GE Healthcare, Little Chalfont, UK | 51 | ||
| Signa HDxt, GE Healthcare, Little Chalfont, UK | 134 | ||
| Avanto, Siemens, Munich, Germany | 171 | ||
| 3 T | Signa Excite, GE Healthcare, Little Chalfont, UK | 8 | |
| Signa HDx, GE Healthcare, Little Chalfont, UK | 216 | ||
| Signa HDxt, GE Healthcare, Little Chalfont, UK | 106 | ||
| Skyra, Siemens Healthcare, Little Chalfont, UK | 54 | ||
| Trio, Siemens, Munich, Germany | 1 | ||
| Trio Tim, Siemens, Munich, Germany | 55 | ||
| Contrast Agent | gadobutrol | Gadavist, Bayer Healthcare, Berlin, Germany | 2 |
| gadopentetate dimeglumine | Magnevist, Bayer Healthcare, Berlin, Germany | 540 | |
| gadobenate dimeglumine | Multihance, Bracco, Milan, Italy | 254 | |
| Not known | - | 96 | |
| Repetition Time (ms) | 3.5 – 4.5 | - | 277 |
| 4.5 – 5.5 | - | 363 | |
| 5.5 – 6.5 | - | 232 | |
| 6.5 – 7.5 | - | 20 | |
|
Echo Time (ms) |
1.25–2 | - | 612 |
| 2–2.76 | - | 280 | |
| Spatial Resolution | 320 × 320 | - | 32 |
| 448 × 448 | - | 249 | |
| 512 × 512 | - | 611 |
Eight fellowship-trained readers (1– 22 years of post-fellowship experience) annotated the cases. Using an internal GUI (graphical user interface) developed in our laboratory, three sequences were displayed to the readers: (a) the pre-contrast, (b) the first post-contrast, and (c) the subtracted (obtained by subtracting the pre-contrast sequence from the first post-contrast). The reader drew a bounding rectangle on a slice and indicated the extent of a cuboid containing the abnormality by pointing out the starting and ending slices. For each tumor the box was drawn by one of the eight readers. The expected annotation time for this task is 3–5 minutes per case.
Algorithmic Image Analysis
We conducted semi-automatic analysis of the images using algorithms implemented in our laboratory. Specifically, using the boxes drawn by expert radiologists that outline the tumor as well as the breast, we used Fuzzy C-Means clustering algorithm(10) to find the exact outline of the tumor. Please note, that since the segmentation algorithm focused on bright regions, the necrotic regions were likely excluded from the tumor segmentation. We also extracted the breast masks and two fibroglandular tissue (FGT) masks (from the first post contrast and T1 non-fat saturated sequences respectively) as described in(11). The breast mask was obtained by removing the chest cavity by a parametric polynomial curve fitting technique on the T1 non-fat saturated sequence. Then, automatic global thresholding and active contour method were applied to obtain the breast mask. The breast-mask and its registered version (to the first post contrast image) were used to compute the two FGT masks using Fuzzy C-Means clustering algorithm(10).
Following segmentation of the breast into different regions, we applied a comprehensive automated analysis of the tumors and their surroundings resulting in 529 features for each patient. The complete list of features can be found in the supplementary materials of the publication(11).
The feature extraction requires the use of all of or a subset of breast masks, FGT masks, and tumor masks. Some features require the involvement of the mask only and not the intensity values of MRI sequences. This type of features are included in our set as (a) breast volume, FGT volume and density (b) tumor size and morphology related features.
We also included breast enhancement and kinetic features which use both masks and intensity values from the MRI sequence. We have included features that characterize tumor and tissue enhancement and kinetics separately. We have also included features that combine the enhancement dynamics of tumor and tissue. First, the volumetric and mean enhancement features were computed by grouping tumor/FGT voxels according to the enhancement time-point(12) and voxel-wise enhancement values to form Signal Enhancement Ratio (SER) maps, and Peak enhancement (PE) maps(13). Next, other features were computed as different measures of dispersions from these aforementioned groups and maps and fall in the distinct category of enhancement variation of tumor or FGT. Tumor enhancement variation is also captured by the features that compute different properties of the tumor uptake curves(14–16). Moreover, distinct features of tissue enhancement in the form of proportion of enhanced FGT(17) were included in our set as they offer information about the background parenchymal enhancement (BPE). Another set of features that combines both tumor and FGT enhancement dynamics(18, 19) was also included in our feature set. These features are particularly important as they account of variability in enhancement curves between patients and different MRI scanner parameters.
Features that incorporate spatial relationship between voxels are categorized in either of two groups: (a) tumor enhancement texture features or (b) tumor enhancement spatial heterogeneity features. The texture features(20) are commonly used in tumor radiomics/radiogenomics to compute the spatial relationships between the enhancement patterns of the tumor. In our set we have incorporated texture features using measurements from global co-occurrence histogram(21), Discrete Fourier Transform (DFT)(22), Dynamic Local Binary Patterns (DLBP), and Dynamic Histogram of Oriented Gradients (DHOG) (23). Also, texture features were computed from first post-contrast, peak-enhancement, signal enhancement ratio and wash-in rate maps. Spatial relationships of tumor enhancement variation along the tumor margin are also included in this group(14). While the texture features that we included are indicative of spatial variations in enhancement in general, we have included four features of spatial heterogeneity of enhancement computed using global Moran’s I(24) and by clustering different types enhancement of tumor voxels. We have incorporated texture features from FGT based enhancements as well, as some studies(25–28) have reported the potential effectiveness of these tissue based features. Our set of 529 features is divided in 10 groups as shown in Fig.1 of supplemental materials.
Statistical Analysis
In order to analyze prognostic value of imaging features, we selected 20 features using the training set and then evaluated them on the test set. The details of the analysis are described below:
STEP 1: Selection of a Candidate Set of Imaging Features using the Development Set
Since our set of imaging features contains 529 variables, we identified a smaller set of candidate features with a potential association with outcomes. This step was performed using the development set of 446 patients. Specifically, we first filled in occasional missing values of the features with a mean value of a feature for the remaining patients in the training set (40 features had missing values for 1–21 patients). Then we reduced redundancy of our feature set by removing features highly correlated with other features (R function findCorrelation with cutoff correlation of 0.8). Finally, from the remaining set, we selected 20 features with the highest predictive value for the distant recurrence-free survival. Predictive value of each feature was measured using concordance index(29). This metric allows for measuring the association between the continuous imaging features and outcomes without having to decide on the cutoff threshold for the variable itself (which is the case for hazard ratio) and for time (which is the case in receiver operating characteristic).
STEP 2: Evaluation of Predictive Value of the Imaging Features using an Independent Test Set
The 20 imaging features selected in STEP 1 were evaluated using an independent test set of 446 patients. Concordance index was used as the primary metric for evaluation of each of the features. The significance of the association of each of the imaging features with distant DRFS was assessed by constructing a univariate Cox proportional hazards regression model(30). To adjust for multiple hypothesis testing, a p-value less than 0.0025 (0.05 / 20 features) was used as the significance level. Additionally, we determined whether each of the features is associated with outcomes independently of tumor volume. This allowed us to establish whether the features have additional prognostic value of outcomes other than due to expressing tumor size. To achieve this, for each of the 20 imaging features, we constructed a multivariate Cox proportional hazards regression model with the given imaging feature and tumor value as the 2 covariates. Finally, we controlled for standard clinical and pathology variables to determine whether each imaging feature is prognostic of outcomes independently of these standard factors. For this purpose, for each imaging feature, we constructed a multivariate Cox proportional hazards regression model with the imaging feature and all the standard clinical and pathology variables as covariates. Specifically, we controlled for age, staging tumor size (T staging), node involvement (N staging), Nottingham grade, estrogen receptor status, progesterone receptor status, and HER2-neu gene status.
Finally, to place the analysis in a more specific clinical context, we evaluated the prognostic value of each of the selected 20 variables for specific clinically-relevant subsets of patients. The subsets we considered were: (a) patients with ER positive tumors, (b) patients with ER negative tumors, (c) patients who received chemotherapy, (d) patients who received hormonal therapy, (e) patients who receive anti HER2-neu therapy. Concordance indices were calculated for all features for each of the subsets. Estrogen receptor status was considered as it is one of the primary factors considered when dividing patients into different treatment cohort, and (unlike e.g. HER2) it provided a relatively even split of patients. Consideration of different treatment groups provides a preliminary step toward considering imaging as a predictive biomarker of response to specific treatments.
To provide visual representation of the results, we generated a Kaplan-Meier plot for each of the features and each data subset as well as correlogram to show correlations between each pair of the 20 selected features.
RESULTS
Important patient and data characteristics are listed in Table 2. This table illustrates that the training and the independent test set have similar characteristics in terms of patient demographics and pathologic features of the tumors.
Table 2:
Patient and tumor characteristics. Median and IQR are reported for patient age, tumor major axis length and follow up time.
| Characteristics | Training Set (N=446) | Test Set (N=446) |
|---|---|---|
| Median Age(years) | 52.68 | 51.62 |
| Age Range(years) | 21.75–79.91 | 23.98–89.49 |
| Race/Ethnicity | ||
| White | 316 | 312 |
| Black | 104 | 96 |
| Asian | 6 | 8 |
| Native | 3 | 0 |
| Hispanic | 4 | 13 |
| Multi | 2 | 6 |
| Hawaiian | 1 | 0 |
| American Indian | 1 | 2 |
| Not Available | 9 | 9 |
| Menopausal Status- | ||
| Pre | 186 | 205 |
| Post | 256 | 231 |
| Not Available | 4 | 10 |
| Estrogen Receptor Status | ||
| Positive | 330 | 335 |
| Negative | 116 | 111 |
| Tumor Staging (Size) | ||
| T1 | 198 | 206 |
| T2 | 191 | 187 |
| T3 | 48 | 35 |
| T4 | 7 | 14 |
| Not Available | 2 | 4 |
| Node Involvement | ||
| N0 | 257 | 272 |
| N1 | 130 | 125 |
| N2 | 33 | 24 |
| N3 | 21 | 17 |
| Not available | 5 | 8 |
| Nottingham grade | ||
| Low | 88 | 72 |
| Intermediate | 223 | 219 |
| High | 129 | 147 |
| Not Available | 6 | 8 |
| Median Follow up(years) | 4.08 | 3.48 |
| Number of distant Recurrences | 41 | 32 |
The results of our primary analysis are presented in Table 3. Twenty features were selected for further evaluation using the training set. We found that five of these features remained significantly associated with DRFS in the test set in the univariate analysis. All these features were extracted from the tumor and hence the tumor mask was employed in the calculation procedure for all of these features. These features were the following: (a) signal enhancement ratio (SER) partial tumor volume (p<0.0001): volume of the voxels in tumor where the ratio of the early enhancement (subtraction of the pre-contrast from the first post-contrast) to late enhancement (subtraction of the pre-contrast from the late post-contrast) exceeds 0.9, (b) maximum axial extent of the tumor (p<0.0001): obtained as the maximum major axis length (in mm) of the tumor over the slices, (c) tumor cluster shade (p=0.0003): calculated from the first post-contrast image and the tumor mask to capture the asymmetry in the intensity of the tumor in the first post-contrast image., (d) wash-in rate information measure of correlation 1 (p=0.0009): to calculate this, first, the time-point at which the mean percentage enhancement of the tumor is maximum is obtained. Thereafter, the rate of enhancement is found at the same time-point to obtain the wash-in rate map. Texture analysis is then conducted on this map to obtain the value this feature., and (e) kurtosis of the SER map (p<0.0001): the ratio of the early enhancement (subtraction of the pre-contrast from the first post-contrast) to late enhancement (subtraction of the pre-contrast from the late post-contrast) to obtain the SER map of the tumor. Kurtosis of this tumor SER map is the feature. This list of features indicates that the tumor size, as well as textural and volumetric measures related to the enhancement, play the dominant role in prediction of patient outcomes. Both early and late post-contrast sequences play important role in the computation of these features.
Table 3:
Prognostic value of individual features measured by the concordance index and evaluated on the independent test set. P-values are provided for the univariate analysis, for the multivariate analysis where we control for clinical and pathology variables and for multivariate analysis where we control for MR-based tumor volume.
| Name of the feature | C-index (95% CI) | p-value | p-value adjusted for clinical variables |
p-value adjusted for tumor volume |
|---|---|---|---|---|
| SER_Partial_tumor_vol_cu_mm | 0.768 (0.679–0.856) | <0.0001 | <0.0001 | 0.0016 |
| Variance_of_RGH_values_tumor | 0.304 (0.211–0.397) | 0.0380 | 0.2471 | 0.0714 |
| WashinRate_map_difference_entropy_tumor | 0.325 (0.237–0.413) | 0.0017 | 0.1479 | 0.0757 |
| SER_map_Cluster_Prominence_tumor | 0.303 (0.193–0.413) | 0.1912 | 0.5704 | 0.2952 |
| TumorMajorAxisLength | 0.742 (0.650–0.834) | <0.0001 | <0.0001 | <0.0001 |
| Mean_norm_DHOG_max_timepoint_binsize_6_with_filling_tumor | 0.528 (0.449–0.607) | 0.9882 | 0.9220 | 0.9853 |
| Uptake_rate_from_char_curve_tumor | 0.591 (0.483–0.699) | 0.3606 | 0.7106 | 0.2998 |
| Cluster_Shade_tumor | 0.313 (0.216–0.410) | 0.0003 | 0.0021 | 0.0047 |
| PE_map_Cluster_Shade_tumor | 0.366 (0.285–0.447) | 0.4487 | 0.6798 | 0.5351 |
| PE_map_Cluster_Prominence_tumor | 0.307 (0.202–0.412 | 0.0384 | 0.1204 | 0.0695 |
| BEDR1_tumor | 0.470 (0.377–0.564) | 0.6905 | 0.9335 | 0.5152 |
| F1_T1(T11=0.02,T12=0.5) | 0.488 (0.388–0.588) | 0.5966 | 0.5001 | 0.6397 |
| WashinRate_map_Correlation2_tumor | 0.480 (0.385–0.575) | 0.4522 | 0.2172 | 0.1202 |
| Max_Enhancement_from_char_curve_tumor | 0.583 (0.478–0.688) | 0.3619 | 0.9434 | 0.3165 |
| SER_map_Correlation2_tissue_PostCon | 0.509 (0.417–0.602) | 0.4454 | 0.3530 | 0.5375 |
| BEDR2_tumor | 0.473 (0.383–0.564) | 0.8719 | 0.9582 | 0.7871 |
| WashinRate_map_information_measure_correlation1_tumor | 0.702 (0.601–0.803) | 0.0009 | 0.0091 | 0.0015 |
| SER_map_kurtosis_tumor | 0.640 (0.521–0.760) | <0.0001 | <0.0001 | <0.0001 |
| F1_T1(T11=0.05,T12=0.1) | 0.527 (0.422–0.633) | 0.3919 | 0.8115 | 0.3895 |
| WashinRate_map_Autocorrelation_tissue_PostCon | 0.453 (0.344–0.561) | 0.5160 | 0.6676 | 0.7128 |
In the multivariate analysis, all features described above remain a significant predictor of DRFS after controlling for clinical variables, except for wash-in rate map information measure of correlation 1. Similarly, all features described above remained a significant predictor of DRFS after controlling for tumor volume, except for tumor cluster shade. Thus, the tumor enhancement features remained prognostic after controlling for clinical variables and tumor volume. This demonstrates a potential for prognostic utility of these imaging features in addition to the standard, currently used, clinical and pathologic prognostic variables.
The subgroup analysis by the estrogen receptor status is shown in Fig. 2. Among the strongly prognostic variables, the concordance index remained similar for the ER positive and ER negative patients. This remained true in general when the prognostic value of the features was analyzed for subgroups defined by the systemic therapy received by patients (shown in Fig. 3), although some variability was observed. Of interest, the prognostic value of SER partial tumor volume increased to an even higher value of concordance index, equal to 0.843 (95% CI: 0.753–0.932), when patients with endocrine therapy were considered as the subgroup. This increase demonstrated the promise of using imaging variables as a part of a biomarker to predict tumor response to individual systemic therapies.
Figure 2.
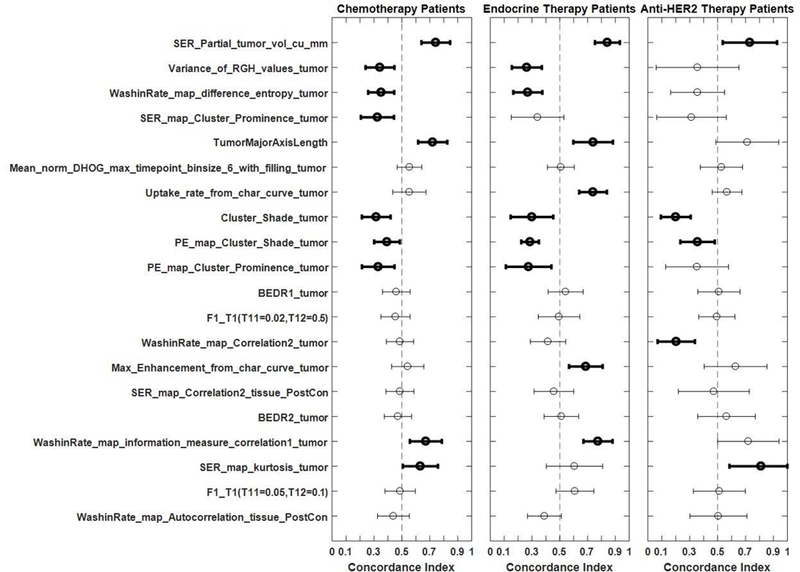
Concordance indices with confidence intervals for the entire cohort as well as for ER positive and ER negative patient subgroups. Bold markers and lines signify that the confidence interval for the concordance index does not overlap with 0.5.
Figure 3.
Concordance indices with confidence intervals for the subgroups of patients receiving different types of systemic therapies. Bold markers and lines signify that the confidence interval for the concordance index does not overlap with 0.5.
DISCUSSION
In this study, we analyzed a cohort of 892 patients to identify potential features that were prognostic of distant progression-free survival. We first identified a candidate set of 20 features of interest using a training set and then evaluated them on the independent test set. We concluded that 5 of those features, reflecting tumor size, enhancement dynamics, and heterogeneity, remained significant predictors of outcomes in the test cohort.
Breast MRI is commonly acquired in the clinical setting for a large portion of breast cancer patients. Therefore, identifying prognostic variables in MRI scans could valuably guide treatment planning by more accurately stratifying patients to appropriate treatment regimens at no additional cost to patients. Our study demonstrates the promise of this approach.
While previous studies(5, 7–9) showed the potential of using MR imaging as a prognostic tool in breast cancer, those studies suffered limitations including a small sample size, manual extraction of few features, greatly limited set of CAD-based features, or lack of adjustment for clinical variables or MR tumor volume. Our study builds on the previous evidence by addressing the limitations of these studies. Additionally, the promise of using the MR imaging features can be found in numerous recent radiogenomic studies(12, 18, 19, 27, 28, 31–35) showing the association between imaging features and genomic markers, including intrinsic molecular subtype or Oncotype DX recurrence score. While these studies are of high importance, they only suggest imaging features might be associated with outcomes; analysis involving actual outcomes data is needed to evaluate this hypothesis. Our study related the imaging features to distant recurrence-free survival to directly address the question of whether imaging features are prognostic of outcomes in breast cancer.
An important aspect of evaluating the prognostic value of imaging features is placing it in the proper context of how they would be clinically used. If an imaging feature merely duplicates the prognostic value of a variable already used to stratify patients, it is of limited use. To determine the independent prognostic value of each of the imaging variables, a multivariate analysis including standard prognostic variables is needed. Our analysis showed most of the variables that were identified as strongly prognostic of outcomes in the univariate analysis were also independently prognostic of outcomes after controlling for clinical and pathologic variables. Moreover, while principally evaluated here as general prognostic features, we also observed that the MRI features could potentially be used for prognostication within specific treatment groups and thus increasing their applicability in clinical settings. The further research could evaluate whether the features could be used for an improved selection of treatment by evaluating the interaction between treatment and feature values. Of interest here is the added value of imaging as compared to tumor size based on the TNM staging. We found that the major axis length in imaging, as well as other imaging features, was prognostic of outcomes independent of T staging demonstrating the orthogonal value of imaging features as compared to clinical and pathology variables.
Some of the imaging features, while intended to evaluate various aspects of tumor radiological appearance, might be correlated with tumor size as seen in imaging. Therefore, we additionally evaluated the prognostic value of the selected features against tumor value as measured in MRI. We found that most of the identified features were prognostic of outcomes independently of tumor volume. However, it is important to note that MRI tumor size, consistent with previous studies, was one of the most prognostic features.
A strength of this study is that the features were calculated using deterministic computer algorithms which means that, given a location of the tumor indicated by a radiologist’s box, the value of the feature will be the same every time, making it fully reproducible. However, some variability has been observed among radiologists when annotating breast tumors in MRI(36) which might result in some feature differences due to this variability. A fully automatic algorithm relying on automatic segmentation of breast tumors (which is currently an unsolved problem) could resolve this issue.
A limitation of this study was its retrospective nature, which meant that we were unable to control scanning parameters such as equipment manufacturer, magnet strength, scanning resolution, and contrast protocol. A previous study(11) showed some of these parameters could bias some of the imaging features. It is encouraging to see strong associations of the imaging features with outcomes, despite these limitations, demonstrating the robustness of selected features for this purpose. A follow-up study with a larger cohort could evaluate the prognostic value of imaging features in more uniform cohorts.
Another limitation is related to inter-reader variability in the annotation of the lesions by radiologists. Each of the lesions analyzed in this study was drawn by one radiologist (see details in (6)and (37)). In a prior study (36), we observed that there is a substantial inter-reader variability between readers when annotating breast lesions in MRI although the impact of the variability on feature extraction is moderate. Future studies should evaluate whether this variability between readers has an impact on an overall accuracy of a prognostic model such as the one presented in this study. Furthermore, automatic segmentation algorithms such as the one in (38) will allow for eliminating this effect and allow for reproducible prediction of outcomes.
Please note that the distant-recurrence rates in our population (8.2% with the medial follow up time around 4 years) was slightly higher than that reported Nguyen et al.(39) report 6.3% at 5 years with 95% CI of 4.7% to 8.3%). It was however at the same level as that in Solin et al.(40) who reported 9% (95% CI: 5% to 14%) at 5 years for invasive cancer and DCIS patients with MRI and breast conservation surgery. Solin et al. also showed a slightly lower (not statistically significant) 5-year distant recurrence rate for patients that did not have an MRI of 6% (95% CI: 4% - 9%) indicating that the recurrence rate might be higher in patients that had and MRI. The fairly high rate of recurrence in our study is likely associated with the fact that due to the nature of this study only patient with MRI and invasive cancer are included. Given this population the observed distant recurrence rate can be considered within expectation.
In conclusion, we provide strong evidence of the prognostic value of imaging independent of standard prognostic variables. Future studies will validate our conclusions in other data and propose how to incorporate these imaging variables in the decision-making process in managing early-stage breast cancer.
Supplementary Material
Acknowledgments
Grant Support
The authors would like to acknowledge funding from the North Carolina Biotechnology Center and the National Institutes of Health.
Abbreviations:
- (FGT)
Fibroglandular tissue
- (SER)
Signal enhancement ratio
- (DCE)
Dynamic contrast enhanced
- (DRFS)
Distant recurrence-free survival
Footnotes
CONFLICTS OF INTEREST STATEMENT
The authors have no conflict of interests to disclose.
REFERENCES
- 1.Page DL: Prognosis and breast cancer: recognition of lethal and favorable prognostic types. Am J Surg Pathol 1991; 15:334–349. [DOI] [PubMed] [Google Scholar]
- 2.Paik S, Shak S, Tang G, et al. : A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 2004; 351:2817–2826. [DOI] [PubMed] [Google Scholar]
- 3.Myers ER, Moorman P, Gierisch JM, et al. : Benefits and harms of breast cancer screening: a systematic review. Jama 2015; 314:1615–1634. [DOI] [PubMed] [Google Scholar]
- 4.Peters NHGM, Borel Rinkes IHM, Zuithoff NPA, Mali WPTM, Moons KGM, Peeters PHM: Meta-analysis of MR imaging in the diagnosis of breast lesions. Radiology 2008; 246:116–124. [DOI] [PubMed] [Google Scholar]
- 5.Partridge SC, Gibbs JE, Lu Y, et al. : MRI measurements of breast tumor volume predict response to neoadjuvant chemotherapy and recurrence-free survival. Am J Roentgenol 2005; 184:1774–1781. [DOI] [PubMed] [Google Scholar]
- 6.Mazurowski MA, Grimm LJ, Zhang J, et al. : Recurrence-free survival in breast cancer is associated with MRI tumor enhancement dynamics quantified using computer algorithms. Eur J Radiol 2015; 84:2117–2122. [DOI] [PubMed] [Google Scholar]
- 7.Kim JJ, Kim JY, Kang HJ, et al. : Computer-aided Diagnosis–generated Kinetic Features of Breast Cancer at Preoperative MR Imaging: Association with Disease-free Survival of Patients with Primary Operable Invasive Breast Cancer. Radiology 2017; 284:45–54. [DOI] [PubMed] [Google Scholar]
- 8.Song SE, Shin SU, Moon H-G, Ryu HS, Kim K, Moon WK: MR imaging features associated with distant metastasis-free survival of patients with invasive breast cancer: a case–control study. Breast Cancer Res Treat 2017; 162:559–569. [DOI] [PubMed] [Google Scholar]
- 9.Wu J, Cui Y, Sun X, et al. : Unsupervised clustering of quantitative image phenotypes reveals breast cancer subtypes with distinct prognoses and molecular pathways. Clin Cancer Res 2017; 23:3334–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bezdek JC: Pattern Recognition with Fuzzy Objective Function Algorithms. Norwell, MA, USA: Kluwer Academic Publishers; 1981. [Google Scholar]
- 11.Saha A, Yu X, Sahoo D, Mazurowski MA: Effects of MRI scanner parameters on breast cancer radiomics. Expert Syst Appl 2017; 87:384–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashraf AB, Daye D, Gavenonis S, et al. : Identification of intrinsic imaging phenotypes for breast cancer tumors: Preliminary associations with gene expression profiles. Radiology 2014; 272:374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arasu VA, Chen RCY, Newitt DN, et al. : Can signal enhancement ratio (SER) reduce the number of recommended biopsies without affecting cancer yield in occult MRI-detected lesions? Acad Radiol 2011; 18:716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilhuijs KGA, Giger ML, Bick U: Computerized analysis of breast lesions in three dimensions using dynamic magnetic‐resonance imaging. Med Phys 1998; 25:1647–1654. [DOI] [PubMed] [Google Scholar]
- 15.Chen W, Giger ML, Bick U, Newstead GM: Automatic identification and classification of characteristic kinetic curves of breast lesions on DCE‐MRI. Med Phys 2006; 33:2878–2887. [DOI] [PubMed] [Google Scholar]
- 16.Chen W, Giger ML, Lan L, Bick U: Computerized interpretation of breast MRI: investigation of enhancement-variance dynamics. Med Phys 2004; 31:1076–1082. [DOI] [PubMed] [Google Scholar]
- 17.Wu S, Weinstein SP, DeLeo MJ, et al. : Quantitative assessment of background parenchymal enhancement in breast MRI predicts response to risk-reducing salpingo-oophorectomy: preliminary evaluation in a cohort of BRCA1/2 mutation carriers. Breast Cancer Res 2015; 17:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazurowski MA, Zhang J, Grimm LJ, Yoon SC, Silber JI: Radiogenomic analysis of breast cancer: Luminal B molecular subtype is associated with enhancement dynamics at MR imaging. Radiology 2014; 273:365–372. [DOI] [PubMed] [Google Scholar]
- 19.Grimm LJ, Zhang J, Mazurowski MA: Computational Approach to Radiogenomics of Breast Cancer: Luminal A and Luminal B Molecular Subtypes Are Associated With Imaging Features on Routine Breast MRI Extracted Using Computer Vision Algorithms. J Magn Reson Imaging 2015. [DOI] [PubMed] [Google Scholar]
- 20.Bhooshan N, Giger ML, Jansen SA, Li H, Lan L, Newstead GM: Cancerous breast lesions on dynamic contrast-enhanced MR images: computerized characterization for image-based prognostic markers. Radiology 2010; 254:680–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haralick RM, Shanmugam K, Dinstein I: Textural Features for Image Classification. IEEE Trans Syst Man Cybern 1973; 3:610–621. [Google Scholar]
- 22.Zheng Y, Englander S, Baloch S, et al. : STEP: spatiotemporal enhancement pattern for MR-based breast tumor diagnosis. Med Phys 2009; 36:3192–3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wan T, Bloch BN, Plecha D, et al. : A radio-genomics approach for identifying high risk estrogen receptor-positive breast cancers on DCE-MRI: preliminary results in predicting OncotypeDX risk scores. Sci Rep 2016; 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song C, Smith M, Huang Y, Jeraj R, Fain S: Heterogeneity of vascular permeability in breast lesions with dynamic contrast enhanced MRI. In Proc 17th Int Symp Magn Reson Med; 2009. [Google Scholar]
- 25.King V, Brooks JD, Bernstein JL, Reiner AS, Pike MC, Morris EA: Background parenchymal enhancement at breast MR imaging and breast cancer risk. Radiology 2011; 260:50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahn HS, Kim SM, Jang M, Yun B La: Quantitative analysis of breast parenchymal background enhancement (BPE) on magnetic resonance (MR) imaging: Association with mammographic breast density and aggressiveness of the primary cancer in postmenopausal women. Volume 31 26_suppl; :38. [Google Scholar]
- 27.Uematsu T, Kasami M, Yuen S: Triple-negative breast cancer: correlation between MR imaging and pathologic findings. Radiology 2009; 250:638–647. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Kato F, Oyama-Manabe N, et al. : Identifying Triple-Negative Breast Cancer Using Background Parenchymal Enhancement Heterogeneity on Dynamic Contrast-Enhanced MRI: A Pilot Radiomics Study. PLoS One 2015; 10:e0143308–e0143308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pencina MJ, D’Agostino RB: Overall C as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation. Stat Med 2004; 23:2109–2123. [DOI] [PubMed] [Google Scholar]
- 30.Cox DR: Regression Models and Life-Tables In Break Stat. Springer; 1992:527–541. [Google Scholar]
- 31.Li H, Zhu Y, Burnside ES, et al. : MR Imaging Radiomics Signatures for Predicting the Risk of Breast Cancer Recurrence as Given by Research Versions of MammaPrint, Oncotype DX, and PAM50 Gene Assays. Radiology 2016; 281:382–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Zhu Y, Burnside ES, et al. : Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. npj Breast Cancer 2016; 2:16012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutton EJ, Dashevsky BZ, Oh JH, et al. : Breast cancer molecular subtype classifier that incorporates MRI features. J Magn Reson Imaging 2016:n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agner SC, Rosen MA, Englander S, et al. : Computerized image analysis for identifying triple-negative breast cancers and differentiating them from other molecular subtypes of breast cancer on dynamic contrast-enhanced MR images: a feasibility study. Radiology 2014; 272:91–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blaschke E, Abe H: MRI phenotype of breast cancer: Kinetic assessment for molecular subtypes. J Magn Reson Imaging 2015; 42:920–924. [DOI] [PubMed] [Google Scholar]
- 36.Saha A, Grimm LJLJ, Harowicz M, et al. : Interobserver variability in identification of breast tumors in MRI and its implications for prognostic biomarkers and radiogenomics. Med Phys 2016; 43:4558–4564. [DOI] [PubMed] [Google Scholar]
- 37.Saha A, Harowicz MR, Grimm LJ, et al. : A machine learning approach to radiogenomics of breast cancer: a study of 922 subjects and 529 DCE-MRI features. Br J Cancer 2018; 119:508–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, Saha A, Zhu Z, Mazurowski MA: Hierarchical Convolutional Neural Networks for Segmentation of Breast Tumors in MRI with Application to Radiogenomics. IEEE Trans Med Imaging - online before print 2018. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen PL, Taghian AG, Katz MS, et al. : Breast cancer subtype approximated by estrogen receptor, progesterone receptor, and HER-2 is associated with local and distant recurrence after breast-conserving therapy. J Clin Oncol 2008; 26:2373–2378. [DOI] [PubMed] [Google Scholar]
- 40.Solin LJ, Orel SG, Hwang W-T, Harris EE, Schnall MD: Relationship of breast magnetic resonance imaging to outcome after breast-conservation treatment with radiation for women with early-stage invasive breast carcinoma or ductal carcinoma in situ. J Clin Oncol 2008; 26:386–391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


