Abstract
The development of new animal models, in vivo isolation approaches, and improvements in genome-wide RNA expression methods have greatly propelled molecular profiling of astrocytes and the characterization of astrocyte heterogeneity in the central nervous system (CNS). Several recent reviews have comprehensively discussed the molecular and functional diversity of mammalian astrocytes. In this brief review, we emphasize interspecies comparisons and an evolutionary perspective regarding the astro(glia) of vertebrates and invertebrates which are similar in form and function. This analysis has revealed conserved astrocyte transcriptomes in the fly, mouse and human. We also offer opinions about the pattern and origin of astrocyte heterogeneity in the CNS.
The central nervous system (CNS) is the most complex and sophisticated biological system, controlling physiology and behavior in phylogenetically diverse animal species. As a consequence of this complexity, multiple classes and unique subtypes of electrically excitable neurons cooperate with functionally unique glial cells to control CNS development, physiology and behavior. Whereas numerous studies have documented unique neuronal cell subtypes, only recently have investigators begun to do the same for glial cells of the CNS. In organisms ranging from worms and insects to rodents and humans, the glia to neuron ratio increases with nervous system complexity (Azevedo et al., 2009), suggesting that the expansion of glial cells is particularly correlated with advanced CNS functions. stroglia, in particular, are the most abundant glial cells in the mammalian CNS and they play versatile roles in regulating synaptogenesis, modulating synaptic transmission, maintaining the integrity of the blood-brain barrier, providing trophic and metabolic support, and contributing to patterns of neuronal network activity [reviewed in (Allen and Barres, 2009; Kettenmann and Ransom, 2012)]. Although astrocytes have been historically regarded as homogenous cell populations, in part due to their non-electrically excitable nature, it has become increasingly evident that astrocytes exhibit significant functional and molecular heterogeneity (Chaboub and Deneen, 2012). As distinct brain regions and neural circuits often contribute to a specific physiological or behavioral function, it is important to understand how interregional astrocyte heterogeneity might influence such functions. The morphological and functional diversity of astrocytes is thus an important topic in glial biology, and studies in this area have been greatly facilitated by the recent development of new animal models and improvements in methods for measuring genome-wide RNA expression.
This review does not intend to provide a comprehensive summary of glial biology or astrocyte heterogeneity, both of which are the subject of recent reviews (Khakh and Sofroniew, 2015; Farmer and Murai, 2017; Ben Haim and Rowitch, 2017; Losada-Perez, 2018). Instead, we will emphasize interspecies comparisons and an evolutionary perspective regarding (astro)glia diversity. We also offer opinions about the pattern and origin of astrocyte heterogeneity in the CNS.
Insects as genetic models in studying astrocyte heterogeneity
We think it is worthwhile to consider astrocyte diversity not only in mammals but also in invertebrates given the evidence for similarity of form and function in the different species, and the utility of certain insects as genetic models. The adult Drosophila brain contains multiple classes of glia with many subclasses located throughout the brain; certain classes, including astrocyte-like glia (ALG), have morphological and molecular similarities to their mammalian counterparts (Awasaki et al., 2008; Doherty et al., 2009; Hartenstein, 2011; Edwards et al., 2012; Freeman, 2015; Richier et al., 2017). Similar to mammals, fly astrocytes exhibit developmental tiling (inhabiting largely non-overlapping brain areas)(Stork et al., 2012), have processes covering a wide anatomical territory that contact many neuronal synapses, and are electrically coupled by gap junctions, but are not electrically excitable, instead employing Ca2+-dependent signaling mechanisms (MacNamee et al., 2016; Ma et al., 2016; Zhang et al., 2017).
Conserved mechanisms for astrocyte differentiation between fly and mammals
Although certain factors including glial cells missing (gcm) – which specifies the fly glial lineage [reviewed in (Granderath and Klambt, 1999;Freeman, 2010)] – do not function in mammalian glial development, we think the available evidence nonetheless supports the idea of a common evolutionary origin for vertebrate and invertebrate astroglia [reviewed in (Losada-Perez, 2018)]. This evidence includes the conservation of mechanisms regulating the differentiation and maturation of fly and mammalian glial cells. From in vitro and in vivo studies in flies and mammals, a variety of intercellular signaling pathways, including Notch, Hedgehog (Hh)/Sonic Hedgehog (Shh), Fibroblast Growth Factor (FGFs) and Bone Morphogenetic Protein (BMP) signaling have been implicated in astrocyte proliferation, migration and maturation [reviewed in (Molofsky and Deneen, 2015; Farmer and Murai, 2017; Losada-Perez, 2018)]. Hh/Shh signaling from neurons, for example, is known to regulate glial cell precursor proliferation and migration in Drosophila and mammalian eye development (Wallace and Raff, 1999; Rangarajan et al., 2001; Hummel et al., 2002) and is important for functional diversification of astrocyte subtypes in a variety of adult mouse brain regions (Garcia et al., 2010; Farmer and Murai, 2017). FGFs and BMPs have been shown to promote survival and/or maturation of immature cultured rodent astrocytes (Kang et al., 2014; Scholze et al., 2014). In vivo studies demonstrate that FGF promotes Drosophila glial cell migration/differentiation (Franzdottir et al., 2009) in the developing fly eye as well as the later morphological maturation of astrocytes in the fly visual system and central brain (Stork et al., 2014; Richier et al., 2017).
Similar gene expression profiles of fly and mammalian astrocytes
The observed similarity of fly and mammalian astrocytes is also reflected in their comparable gene expression profiles (Cahoy et al., 2008; Ng et al., 2016; Zhang et al., 2016; Morel et al., 2017). For this review, we analyzed astrocyte-expressed fly, mouse and human genes (Ng et al., 2016, Zhang et al., 2016; Morel et al., 2017), revealed by Translating Ribosome Affinity Purification (TRAP) methods for mouse and fly or FACS analysis for humans. Comparison of the three species indicates that 900 of 2623 astrocyte-expressed fly genes (those with >500 reads) have mammalian orthologs that are also known to be expressed in astrocytes. Figure 1 depicts conservation of astrocyte processes and functions among flies, mice and humans, and Table 1 lists representative orthologs within several interesting functional categories that are detected (FPKM>1for mammals) in astrocytes of the three species. Notably, important transcription factors, ion channels, transporters (including mouse GLT1, GLAST and GAT-1) and potential gliotransmission regulators are found in all species. Of interest, it is known that several factors listed in Table 1 are required for normal fly behavior [Ng et al., 2016); see footnote to Table 1]. Not shown in Table 1 is a mammalian astrocyte-expressed gene (Cahoy et al., 2008) called csf4-U26 (formerly Aasdh; Drozak et al., 2014). Acsf4-U26 has a fly ortholog but it is also homologous to fly ebony. The ebony gene encodes a glial non-ribosomal peptide synthetase that conjugates β-alanine to amines; it is important for aminergic neurotransmitter recycling (Borycz et al., 2002) and circadian behavior (Suh et al., 2007). The Ebony vertebrate ortholog (ACSF4-U26) has β-alanine-activating activity, although the other substrate does not appear to be an amine (Drozak et al., 2014). Nevertheless, it may have an interesting post-translational function in mammalian astrocytes related to behavior.
Figure 1.
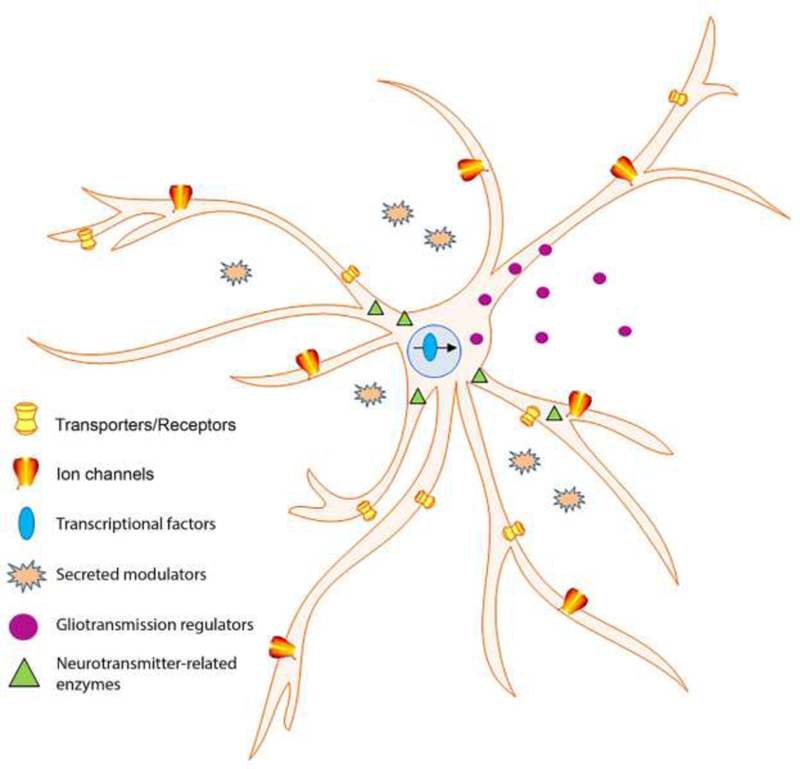
Representative conserved gene functions in fly, mouse and human adult astrocytes.
Table 1.
Representative conserved orthologs with expression in fly, mouse and human adult astrocytes.
| Transcription Factors | Encoded Function(s) |
|---|---|
| Max | Myc-associated factor; cell proliferation |
| Sox5,6 | CNS development & determination of cell fate |
| Cbx1,3,5 | Histone-binding proteins |
| Jun/FosB | Major early-gene TFs |
| Crebl2 | cAMP response element (CRE)-binding protein |
| Transporters/Receptors | |
| Slc1a3 (Glast); Slc1a2 (Glt-1) | Major astrocyte glutamate transporters |
| Slc6a1 (fly Gat *) | Membrane GABA transporter |
| Gaba-a-associated | Links GABA-Areceptors to cytoskeleton |
| Gabarb1 | GABA-B receptor subunit |
| Aqp4 | Aquaporin 4 water channel |
| Npc1,2 * | Cholesterol transporters |
| ApoD | Transport of sugars and other factors |
| Egfr | Epidermal growth factor receptor |
| Ion Channels | |
| Kcnj10,16 | Potassium (K+) channels; K+ buffering |
| Grina | NMDA receptor-associated protein |
| Vdac1 | Anion channel; cell volume regulation |
| Kctd3 | Regulator of cyclic nucleotide-gated channel |
| Stim1 | Membrane Ca2+ channel; store-operated Ca2+ entry (SOCE) |
| Secreted Extracellular proteins | |
| Tgfβ2 | TGFβ receptor ligand |
| Sparcl1 | HEVIN; regulator of syaptogenesis |
| Sparc α | Antagonist of HEVIN; regulates synaptogenesis |
| Spon1 | Axon growth, guidance |
| Gliotransmission-related | |
| Snap23,25 | Regulators of neurotransmitter release |
| Synj1 | Regulates membrane phosphatidylinositol-4,5-bisphosphate |
| Syt11 | Ca2+-dependent regulation of synaptic transmission |
| Vamp2,3,4,7 | Synaptic vesicle docking and fusion |
| Stxbp1 (fly Rop *) | Regulation of neurotransmitter release |
| Vti1b | Vesicle trafficking |
| Neurotransmitter-related Enzymes | |
| Abat (fly Gabat *) | GABA transaminase |
| CSF4-U26 or Aasdh (fly Ebony *) | Aminergic neurotransmitter recycling |
| Glud1 | Glutamate/Glutamine regulation |
| Many Rab family members | GTPase; vesicle trafficking & secretion |
| Glul (fly Gs1) | Glutamine synthetase |
| GNAQ | Membrane bound GTPase linked to GPCRs |
The human gene name is shown in the first column.
deficits cause altered locomotor activity levels or circadian behavior in Drosophila.
a single Sparc gene is found in the fly genome.
Inter- and intra-regional mouse astrocyte diversity
There is ample evidence for astrocyte heterogeneity within the mammalian brain (Zhang and Barres, 2010; Chai et al., 2017; Lin et al., 2017; Morel et al., 2017; Lanjakornsiripan et al., 2018; Zeisel et al., 2018), whereas such studies have only begun in Drosophila. Mature protoplasmic astrocytes from cortex and hippocampus possess substantially more branch arborization than astrocytes from subcortical regions such as the hypothalamus (Bushong et al., 2002; Morel et al., 2014). The overall domain size of cortical/hippocampal astrocytes is also significantly greater than that of hypothalamic astrocytes (Bushong et al., 2002; Morel et al., 2014). Astroglial expression levels of the GLAST and GLT1 glutamate transporters are CNS region-dependent (Kerkerian et al., 1982; Regan et al., 2007; Danbolt, 2001). Physiological measures of astrocyte function including gap-junctional coupling (Batter et al., 1992) and Ca2+ responses (Oberheim et al., 2012; Chai et al; 2017) also differ in a region-dependent manner. Within the brain and spinal cord, astrocyte heterogeneity exhibits an interesting dorsal to ventral pattern. Dorsal Bergmann glia and ventral velate astrocytes in the cerebellum show divergent expression patterns for a number of peri-synaptic astroglial proteins, such as GLAST, Kir4.1, and GLT1. GLAST, for example, is highly expressed in dorsal Bergmann glia but is almost undetectable in velate astrocytes (Farmer et al., 2016). Based on the use of TRAP methods, it was shown that astrocytes from dorsal cortex and hippocampus have mRNA expression profiles that are quite different from those of ventral thalamic and hypothalamic astrocytes (Morel et al., 2017). Similar findings were observed for two distinct groups of telencephalon astrocytes using a single-cell RNA-seq approach (Zeisel et al, 2018). And, the profiles of mouse spinal cord astrocytes in the dorsal and ventral horns are significantly different (Molofsky et al. 2014).
In addition to interregional variation, astrocytes from a single brain region, e.g., the cortex, have been shown to display dorsal-to-ventral variations in morphology, physiology, and mRNA transcriptome. The recent study of Lanjakornsiripan et al. (2018) investigated cortical astrocyte heterogeneity and found layer-specific molecular and morphological diversity for this population of astrocytes. Based on the use of astrocyte expression reporters (Bac aldh1l1-eGFP and eaat2-tdTomato) mice, it was also recently found that three astrocyte subpopulations exist in the cortex (Morel et al., 2018). These subpopulations exhibited distinct cortical locations as well as physiological and molecular properties. Strikingly, expression of Kir4.1, an important astrocyte ion channel, is significantly underrepresented in one cortical astrocyte subpopulation that is located primarily in layers I-II. In contrast, the enpp2 and ptgds genes are uniquely and highly expressed in layer I-II astrocytes.
Diversity of Drosophila ALG
Astrocyte heterogeneity is also observed in the nervous systems of invertebrates although it has not been explored in detail. Similar to mammals, for example, there is evidence for interregional variability in the morphology and functional diversity of fly glial cells, including astrocyte-like glia (ALG) of the fly optic lobes in the adult visual system. This is a particularly well characterized region of the fly nervous system, consisting of the lamina, medulla, lobula and lobula plate. Within the lamina, for example, ALG associate with fly photoreceptors and their targets, forming visual system columns (Edwards et al., 2012; Kremer et al., 2017). Within this region, the shape of glial cells and the number of processes associated with columns varies depending on the depth of cells within the lamina (Edwards et al., 2012), indicative of heterogeneity. ALG are also observed in the optic medulla with multiple subtypes including so-called chandelier and serpentine glia that are resident in the proximal and distal medulla, respectively. Similarly, such glia can be observed in the optic lobula and lobula plate. Although a Gal4 driver known as alrm-Gal4 (Doherty et al., 2009) is commonly employed to label or perturb fly ALG, it has been noted that the optic lobe expression pattern does not include cells of the lamina (Edwards and Meinertzhagen, 2010), again highlighting diversity among fly ALGs. The use of additional Gal4 drivers has highlighted significant interregional variability for ALG of the adult fly brain. Kremer et al., (2017) characterized thousands of different Gal4 drivers to identify those expressing in subpopulations of ALG (and other major glial cell classes) that vary in shape and position.
In the fly optic medulla, there is obvious morphological variability in cell and process alignment among ALG (Kremer et al., 2017; Richier et al., 2017), and this variability seems to depend on the depth of cells within the medulla. Based on careful morphometric analysis of medulla ALGs, Richier et al. (2017) showed that ALG of the medulla (astrocyte-like medulla neuropil glia or mng) can be categorized into at least 4 subtypes that are located in different regions. Their studies used the “Flybow” genetic system (himosako et al., 2014) to generate stochastic labeling of astrocytes with combinations of fluors, making it possible to visualize individual cells with distinct positions and morphologies. Importantly, all variants expressed GABA transaminase (GAT), the dEAAT1 glutamate transporter and glutamate synthase 2 (Gs2), indicative of astrocyte-like cell types.
Diversity of clock-containing Drosophila ALG
It is known that glial cells, particularly astrocytes, of mammals and flies are critical for the function of neuronal circuits controlling rhythmic behaviors such as sleep and circadian behavior. In mammals, astrocytes regulate sleep (homeostasis) via adenosine and an adenosine receptor-mediated mechanism (Halassa et al., 2009). In contrast, wakefulness is modulated by cholinergic stimulation of astrocytes from septal neurons, resulting in glial D-serine release and action on neuronal NMDA receptors (Papouin et al., 2017). In both mammals and flies, circadian behavior is modulated by astrocytes, which like clock neurons contain PER-based oscillators [reviewed in (Jackson et al., 2015). Perturbation of fly astrocytes results in arrhythmic behavior (Suh and Jackson, 2007) (Ng et al., 2011; Ng and Jackson, 2015; Ng et al., 2016) whereas the clock in astrocytes of the mouse suprachiasmatic nuclei (SCN) actually contributes to the determination of circadian period (Tso et al., 2017; Brancaccio et al., 2017). Thus, it is of interest that glial cells of the fly optic lobes with astrocyte-like characteristics are heterogeneous with regard to expression of PERIOD (PER), an important circadian clock protein (Gorska-Andrzejak et al., 2018). To our knowledge, glial variation in clock protein localization has not been studied in mammals. In Drosophila, however, PER expression is significantly higher in distal medulla neuropil glia (dmng) than in lamina epithelial glia (lg). In dmng cells, Glial PER oscillates in abundance, and cycling is, at least in part, regulated by release of a peptide neurotransmitter (PDF) from important circadian clock neurons. Thus, similar to mammals, heterogeneity among fly ALG can result from the action of neuronal factors that impact glial physiology.
Mechanisms regulating astrocyte diversity
The dorsal-to-ventral (DV) variation observed for mammalian astrocyte gene expression profiles provides an important clue as to how astrocyte heterogeneity might arise. Early embryonic development shows a clear DV (and posterior to anterior) axis patterning, from which neural progenitors differentiate into distinct neural cell types. Astrocyte fate-mapping experiments in spinal cord and forebrain confirmed that astrocytes are derived in a region-restricted manner from radial glia’s domains of origin, similar to neurons (Hochstim et al., 2008; Tsai et al., 2012). Therefore, astrocyte heterogeneity may arise, in part, from intrinsically defined signals that serve to regulate the early embryonic dorsal to ventral (and posterior to anterior) axis patterning. This notion is further supported by the recent observation that telencephalon and non-telencephalon astrocyte populations, with distinct molecular profiles, occupy different brain areas with very little overlap (Zeisel et al., 2018).
Given the intensive interaction between neurons and astrocytes and the temporal differentiation of these two cell types during development, it is not surprising that neuronal (and other extrinsic) signals also influence specific molecular and functional properties of astrocytes in each brain region or nucleus during the postnatal astrocyte maturation phase. It was previously demonstrated that loss of VGluT1 neuronal glutamatergic signaling selectively affects the morphological and molecular maturation of cortical, but not hypothalamic astrocytes (Morel et al., 2014). The role of neuronal signals in directing localized astrocyte molecular and morphological features is further demonstrated by the selective deletion of Dab1 in cortical neurons, which disrupts layer-specific neuronal migration during early development (Lanjakornsiripan et al., 2018). In neuronal Dab1 conditional knockout mice, the normal layer-specific branch arborization pattern of cortical astrocytes (more arborization in layers II-III relative to IV) and their molecular expression differences (lower Lef1 and Id1 expression in layers II-III) were abolished (Lanjakornsiripan et al., 2018). Additionally, there is differential expression of several transcription factors (emx2, lhx2, and hopx) in astrocytes of distinct brain regions (Morel et al., 2017). As transcription factor NF1A mediates neuronal JAG1 and DLL1-dependent activation of Notch signaling to promote gliogenesis during early development (Deneen et al., 2006; Namihira et al., 2009), it is conceivable that these differentially expressed transcription factors (emx2, lhx2, and hopx) may respond to localized and specialized neuronal signals and subsequently contribute to astrocyte heterogeneity during postnatal development.
With regard to neuronal signals regulating heterogeneity, Kremer et al. (2017) have demonstrated variability in the thickness of fly ALG processes that is correlated with the particular neuropil they invade. Remarkably, even processes of the same cell can differ in thickness when they infiltrate adjacent neuropils, and this structural diversity is correlated with the density of invaded neuropil in different anatomical regions. This suggests that certain aspects of glial cell morphology may be determined by local synaptic density.
Concluding thoughts
While substantial progress has been made in understanding the molecular heterogeneity of astrocytes, it is important to note that most mRNA profiling studies have employed populations of astrocytes (generally >100,000 cells), isolated using a cell surface marker (i.e., by immunopanning), a genetic fluorescence reporter (fluorescent activated cell sorting) or by association with ribosomes (translating ribosome affinity purification) (Morel et al., 2017; Ng et al., 2016; Zhang et al., 2016). Therefore, these results reflect average mRNA profiles for groups of cells. In contrast, recent developments in single-cell RNA-seq methods are beginning to provide an unprecedented opportunity to examine molecular heterogeneity in individual astrocytes that reside in neighboring functional nuclei or neuronal circuits. Indeed, a recent comprehensive profiling of adult mouse central and peripheral nervous systems, using single cell RNA-seq, has classified seven distinct and regionally-restricted astrocyte subtypes (Zeisel et al., 2018). In addition to the highly specialized astrocyte subtypes of the cerebellum (Bergmann glia, ACBG), olfactory bulb (ACOB), and dorsal midbrain (Myoc-expressing, ACMB), this study characterized two astrocyte subtypes in the telencephalon (ACTE1 and ACTE2) and two subtypes in non-telencephalon brain regions (mostly diencephalon, ACNT1 and ACNT2) (Zeisel et al., 2018). The telencephalon/diencephalon differences in astrocyte expression profiles revealed by single-cell RNA-seq is consistent with previous studies using the TRAP approach with populations of astrocytes (Morel et al., 2017). These results further support the notion that astrocyte subtypes may be influenced by developmental patterning. On the other hand, the expression pattern differences between ACTE1 vs. 2 or ACNT1 vs. 2, identified using single-cell RNA-seq, also suggest the involvement of extrinsic signals, likely the local neuronal environment during postnatal development.
While such techniques have revealed a great deal about conserved genes that are expressed in astrocytes of the fly, mouse and human (Figure 1, Table 1), a large percentage of such genes remain poorly annotated and their molecular functions are unknown. Thus, it is equally important to develop new experimental reagents and approaches to explore the functions of these astrocyte-expressed genes. Such studies will ultimately provide new insights about the diverse roles of astrocytes in the CNS.
Highlights.
There are conserved mechanisms for astrocyte differentiation between fly and mammals
Similar gene expression profiles of fly and mammalian astrocytes are revealed by comparing the transcriptome of fly, mouse, and human adult astrocytes.
There is evidence of astrocyte diversity in vertebrates and invertebrates.
Inter- and intra-regional mouse astrocyte diversity follows the dorsal-to-ventral axis of the adult brain.
Acknowledgements
This work was supported by the National Institutes of Health (R01MH099554 to Y.Y. and F.R.J; R01MH106490 to Y.Y.; R21NS107804 to F.R.J.). We thank Dr. Yuqin Men for help with the Ingenuity analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare no conflict of interest.
Reference List
- Allen NJ, Barres BA (2009) Neuroscience: Glia - more than just brain glue. Nature 457:675–677. [DOI] [PubMed] [Google Scholar]
- Azevedo FA, Carvalho LR, Grinberg LT, Farfel JM, Ferretti RE, Leite RE, Jacob Filho W, Lent R, Herculano-Houzel S (2009) Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J Comp Neurol 513: 532–541. [DOI] [PubMed] [Google Scholar]
- Awasaki T, Lai SL, Ito K, Lee T (2008) Organization and postembryonic development of glial cells in the adult central brain of Drosophila. J Neurosci 28:13742–13753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batter DK, Corpina RA, Roy C, Spray DC, Hertzberg EL, Kessler JA (1992) Heterogeneity in gap junction expression in astrocytes cultured from different brain regions. Glia 6:213–221. [DOI] [PubMed] [Google Scholar]
- Ben Haim L and Rowitch DH (2017). Functional diversity of astrocytes in neural circuit regulation. Nat.Rev.Neurosci 18:31–41. [DOI] [PubMed] [Google Scholar]
- Borycz J, Borycz JA, Loubani M, Meinertzhagen IA (2002) tan and ebony genes regulate a novel pathway for transmitter metabolism at fly photoreceptor terminals. J Neurosci 22:10549–10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancaccio M, Patton AP, Chesham JE, Maywood ES, Hastings MH (2017) Astrocytes Control Circadian Timekeeping in the Suprachiasmatic Nucleus via Glutamatergic Signaling. Neuron 93:1420–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushong EA, Martone ME, Jones YZ, Ellisman MH (2002) Protoplasmic astrocytes in CA1 stratum radiatum occupy separate anatomical domains. J Neurosci 22: 183–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA (2008) A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci 28: 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]; Annotation: One of the first studies to investigate and compare the cell-type specific transcriptome of major CNS cell types
- Chaboub LS, Deneen B (2012) Developmental origins of astrocyte heterogeneity: the final frontier of CNS development. Dev Neurosci 34: 379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai H, Diaz-Castro B, Shigetomi E, Monte E, Octeau JC, Yu X, Cohn W, Rajendran PS, Vondriska TM, Whitelegge JP, Coppola G, Khakh BS (2017) Neural Circuit-Specialized Astrocytes: Transcriptomic, Proteomic, Morphological, and Functional Evidence. Neuron 95:531–549. [DOI] [PMC free article] [PubMed] [Google Scholar]; Annotation: A comprehensive analysis of the transcriptomic, proteomic, morphological, and functional properties of hippocampal and striatal astrocytes in the mouse NS
- Danbolt ONC (2001), Glutamate uptake. Prog.Neurobiol 65:1–105. [DOI] [PubMed] [Google Scholar]
- Deneen B, Ho R, Lukaszewicz A, Hochstim CJ, Gronostajski RM, Anderson DJ (2006) The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron 52: 953–68. [DOI] [PubMed] [Google Scholar]
- Doherty J, Logan MA, Tasdemir OE, Freeman MR (2009) Ensheathing glia function as phagocytes in the adult Drosophila brain. J Neurosci 29:4768–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozak J, Veiga-da-Cunha M, Kadziolka B, Van SE (2014) Vertebrate Acyl CoA synthetase family member 4 (ACSF4-U26) is a beta-alanine-activating enzyme homologous to bacterial non-ribosomal peptide synthetase. FEBS J 281:1585–1597. [DOI] [PubMed] [Google Scholar]
- Edwards TN, Meinertzhagen IA (2010) The functional organization of glia in the adult brain of Drosophila and other insects. Prog Neurobiol 90:471–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TN, Nuschke AC, Nern A, Meinertzhagen IA (2012) Organization and metamorphosis of glia in the Drosophila visual system. J Comp Neurol 520:2067–2085. [DOI] [PubMed] [Google Scholar]
- Farmer OWT, Abrahamsson T, Chierzi S, Lui C, Zaelzer C, Jones EV, Bally BP, Chen GG, Theroux JF, Peng J, Bourque CW, Charron F, Ernst C, Sjostrom PJ, Murai KK 2016) Neurons diversify astrocytes in the adult brain through sonic hedgehog signaling. Science 351: 849–854. [DOI] [PubMed] [Google Scholar]; Annotation: Demonstrated that neuron-derived sonic hedgehog signaling is able to differentially affect dorsal Bergmann glia and ventral velate astrocytes in the cerebellum
- Farmer WT, Murai K (2017) Resolving Astrocyte Heterogeneity in the CNS. Front Cell Neurosci 11:300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzdottir SR, Engelen D, Yuva-Aydemir Y, Schmidt I, Aho A, Klambt C(2009) Switch in FGF signalling initiates glial differentiation in the Drosophila eye. Nature 460:758–761. [DOI] [PubMed] [Google Scholar]
- Freeman MR (2010) Specification and morphogenesis of astrocytes. Science 330:774–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman MR (2015) Drosophila Central Nervous System Glia. Cold Spring Harb Perspect Biol 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia AD, Petrova R, Eng L, Joyner AL (2010) Sonic hedgehog regulates discrete populations of astrocytes in the adult mouse forebrain. J Neurosci 30:13597–13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorska-Andrzejak J, Chwastek EM, Walkowicz L, Witek K (2018) On Variations in the Level of PER in Glial Clocks of Drosophila Optic Lobe and Its Negative Regulation by PDF Signaling. Front Physiol 9:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granderath S, Klambt C (1999) Glia development in the embryonic CNS of Drosophila. Curr Opin Neurobiol 9:531–536. [DOI] [PubMed] [Google Scholar]
- Halassa MM, Florian C, Fellin T, Munoz JR, Lee SY, Abel T, Haydon PG, Frank MG (2009) Astrocytic modulation of sleep homeostasis and cognitive consequences of sleep loss. Neuron 61:213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V (2011) Morphological diversity and development of glia in Drosophila. Glia [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstim C, Deneen B, Lukaszewicz A, Zhou Q, Anderson DJ (2008) Identification of positionally distinct astrocyte subtypes whose identities are specified by a homeodomain code. Cell 133:510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel T, Attix S, Gunning D, Zipursky SL (2002) Temporal control of glial cell migration in the Drosophila eye requires gilgamesh, hedgehog, and eye specification genes. Neuron 33:193–203. [DOI] [PubMed] [Google Scholar]
- Jackson FR, Ng FS, Sengupta S, You S, Huang Y (2015) Glial cell regulation of rhythmic behavior. Methods Enzymol 552:45–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang W, Balordi F, Su N, Chen L, Fishell G, Hebert JM (2014) Astrocyte activation is suppressed in both normal and injured brain by FGF signaling. Proc Natl Acad Sci U SA 111:E2987–E2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkerian L, Nieoullon A, Dusticier N (1982) Brain glutamate uptake: Regional distribution study from sensorimotor areas in the cat. Neurochem Int 4:275–281. [DOI] [PubMed] [Google Scholar]
- Kettenmann H, Ransom BR, eds. Neuroglia, 3rd edition, Oxford niversity Press, 2012. [Google Scholar]
- Khakh BS, Sofroniew VM (2015) Diversity of astrocyte functions and phenotypes in neural circuits. Nat Neurosci 18:942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer MC, Jung C, Batelli S, Rubin GM, Gaul (2017) The glia of the adult Drosophila nervous system. Glia 65:606–638. [DOI] [PMC free article] [PubMed] [Google Scholar]; Annotation: Analyzed thousands of rosophila Gal4 reporter strains to identify diverse glial cell subtypes, including distinct types of fly astrocytes
- Lanjakornsiripan D, Pior BJ, Kawaguchi D, Furutachi S, Tahara T, Katsuyama Y, Suzuki Y, Fukazawa Y, Gotoh Y (2018) Layer-specific morphological and molecular differences in neocortical astrocytes and their dependence on neuronal layers. Nature Communications 9:1623. [DOI] [PMC free article] [PubMed] [Google Scholar]; Annotation: Characterized layer-specific differences in astrocyte morphology and gene expression in adult cortex
- Lin JCC, Yu K, Hatcher A, Huang TW, Lee HK, Carlson J, Weston MC, Chen F, Zhang Y, Zhu W, Mohila CA, Ahmed N, atel AJ, Arenkiel BR, Noebels JL, Creighton CJ, Deneen B (2017) Identification of diverse astrocyte populations and their malignant analogs. Nat Neurosci 20:396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]; Annotation: Identified five molecularly and functionally distinct subpopulations of Aldh1l1– GFP-expressing mouse astrocytes in normal brain tissue and malignant glioma
- Losada-Perez M (2018) Glia: from ‘just glue’ to essential players in complex nervous systems: a comparative view from flies to mammals. J Neurogenet 32:78–91. [DOI] [PubMed] [Google Scholar]
- Ma Z, Stork T, Bergles DE, Freeman MR (2016) Neuromodulators signal through astrocytes to alter neural circuit activity and behaviour. Nature 539:428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]; Annotation: Identified neuromodulators that act on fly astrocytes to regulate behavior via astrocytic Ca2+ signaling mechanisms
- MacNamee SE, Liu KE, Gerhard S, Tran CT, Fetter RD, Cardona A, Tolbert LP, Oland LA (2016) Astrocytic glutamate transport regulates a Drosophila CNS synapse that lacks astrocyte ensheathment. J Comp Neurol 524:1979–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Deneen B (2015) Astrocyte development: A Guide for the Perplexed. Glia 63:1320–1329. [DOI] [PubMed] [Google Scholar]
- Morel L, Higashimori H, Tolman M, Yang Y (2014) VGluT1+ neuronal glutamatergic signaling regulates postnatal developmental maturation of cortical protoplasmic astroglia. J Neurosci 34: 10950–10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel L, Chiang MSR, Higashimori H, Shoneye T, Iyer LK, Yelick J, Tai A, Yang Y (2017) Molecular and Functional Properties of Regional Astrocytes in the Adult Brain. J Neurosci 37:8706–8717. [DOI] [PMC free article] [PubMed] [Google Scholar]; Annotation: Demonstrated regional heterogeneity among astrocytes of different mouse brain regions using TRAP-seq analysis, and identified a dorsal-to-ventral pattern of astroglial gene expression
- Morel L, Men Y, Chiang MSR, Tian Y, Jin S, Yelick J, Higashimori H, Yang Y (2018) ntracortical astrocyte subpopulations defined by astrocyte reporter mice in adult brain. Glia, in press. [DOI] [PubMed] [Google Scholar]; Annotation: Defined heterogeneous astrocyte subtypes within the mouse cortex using TRAP/RNA-seq methods
- Ng FS, Jackson FR (2015) The ROP vesicle release factor is required in adult Drosophila glia for normal circadian behavior. Front Cell Neurosci 9:256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng FS, Sengupta S, Huang Y, Yu AM, You S, Roberts MA, Iyer LK, Yang Y, Jackson FR (2016) TRAP-seq Profiling and RNAi-Based Genetic Screens Identify Conserved Glial Genes Required for Adult Drosophila Behavior. Front Mol Neurosci 9:146. [DOI] [PMC free article] [PubMed] [Google Scholar]; Annotation: Described the genome-wide Drosophila astrocyte gene-expression profile and identified astrocyte-expressed genes required for behavior
- Ng FS, Tangredi MM, Jackson FR (2011) Glial cells physiologically modulate clock neurons and circadian behavior in a calcium-dependent manner. Curr Biol 21:625–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberheim NA, Goldman SA, Nedergaard M(2012) Heterogeneity of astrocytic form and function. Methods Mol Biol 814:23–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papouin T, Dunphy JM, Tolman M, ineley KT, Haydon PG (2017) Septal Cholinergic Neuromodulation Tunes the Astrocyte-Dependent Gating of Hippocampal NMDA Receptors to Wakefulness. Neuron 94:840–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan R, Courvoisier H, Gaul U (2001) Dpp and Hedgehog mediate neuron-glia interactions in Drosophila eye development by promoting the proliferation and motility of subretinal glia. Mech Dev 108:93–103. [DOI] [PubMed] [Google Scholar]
- Regan MR, Huang YH, Kim YS, Dykes-Hoberg MI, Jin L, Watkins AM, Bergles DE, Rothstein JD (2007) Variations in promoter activity reveal a differential expression and physiology of glutamate transporters by glia in the developing and mature CNS. J Neurosci 27:6607–6619. [DOI] [PMC free article] [PubMed] [Google Scholar]; Annotation: Developed Bac transgenic mice for GLT1 and GLAST used to show that GLT1 and GLAST promoters are diversely expressed in particular CNS regions.
- Richier B, Vijandi CM, Mackensen S, Salecker I (2017) Lapsyn controls branch extension and positioning of astrocyte-like glia in the Drosophila optic lobe. Nat Commun 8:317. [DOI] [PMC free article] [PubMed] [Google Scholar]; Annotation: Described astrocytes with distinct morphologies, positions and orientations within the optic medulla of the Drosophila visual system
- Scholze AR, Foo LC, Mulinyawe S, Barres BA (2014) BMP signaling in astrocytes downregulates EGFR to modulate survival and maturation. PLoS One 9:e110668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimosako N, Hadjieconomou D, Salecker I (2014) Flybow to dissect circuit assembly in the Drosophila brain. Methods Mol Biol 1082:57–69. [DOI] [PubMed] [Google Scholar]
- Stork T, Bernardos R, Freeman MR (2012) Analysis of glial cell development and function in Drosophila. Cold Spring Harb Protoc 2012:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork T, Sheehan A, Tasdemir-Yilmaz OE, Freeman MR (2014) Neuron-Glia Interactions through the Heartless FGF Receptor Signaling Pathway Mediate Morphogenesis of Drosophila Astrocytes. Neuron 83:388–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh J, Jackson FR (2007) Drosophila ebony activity is required in glia for the circadian regulation of locomotor activity. Neuron 55:435–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HH, Li H, Fuentealba LC, Molofsky AV, Taveira-Marques R, Zhuang H, Tenney A, Murnen AT, Fancy SP, Merkle F, Kessaris N, Alvarez-Buylla A, Richardson WD, Rowitch DH (2012) Regional astrocyte allocation regulates CNS synaptogenesis and repair. Science 337:358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]; Annotation: Demonstrated the heterogeneous regulatory role of astrocytes on neuronal synaptogenesis in the spinal cord
- Tso CF, Simon T, Greenlaw AC, Puri T, Mieda M, Herzog ED (2017) Astrocytes Regulate Daily Rhythms in the Suprachiasmatic Nucleus and Behavior. Curr Biol 27:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace VA, Raff MC (1999) A role for Sonic hedgehog in axon-to-astrocyte signalling in the rodent optic nerve. Development 126:2901–2909. [DOI] [PubMed] [Google Scholar]
- Zeisel A, Hochgerner H, Lonnerberg P, Johnsson A, Memic F, van der Zwan J, Haring M, Braun E, Borm LE, La MG, Codeluppi S, Furlan A, Lee K, Skene N, Harris KD, Hjerling-Leffler J, Arenas E, Ernfors P, Marklund U, Linnarsson S (2018) Molecular Architecture of the Mouse Nervous System. Cell 174:999–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]; Annotation: Defined subtypes of major CNS cell types, based on single-cell, genome-wide sequence analysis, in the mouse central and peripheral nervous system
- Zhang Y, Barres BA (2010) Astrocyte heterogeneity: an underappreciated topic in neurobiology. Curr Opin Neurobiol 20:588–594. [DOI] [PubMed] [Google Scholar]
- Zhang OY, Sloan SA, Clarke L ., Caneda C, Plaza CA, Blumenthal PD, Vogel H, Steinberg GK, Edwards MS, Li G, Duncan JA III, Cheshier SH, Shuer LM, Chang EF, Grant G ., Gephart MG, Barres BA (2016) Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron 89:37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]; Annotation: A comprehensive study to compare the transcriptome of different CNS cell types in mouse and human
- Zhang YV, Ormerod KG, Littleton JT (2017) Astrocyte Ca2+ Influx Negatively Regulates Neuronal Activity. eNeuro 4. [DOI] [PMC free article] [PubMed] [Google Scholar]; Annotation: Demonstrates the importance of astrocytic Ca2+ in the regulation of Drosophila neuronal activity


