Abstract
Objective
Here, we evaluate the contribution of AT-rich interaction domain-containing protein 1A (ARID1A), the most frequently mutated member of the SWItch/Sucrose Non-Fermentable (SWI/SNF) complex, in pancreatic homeostasis and pancreatic ductal adenocarcinoma (PDAC) pathogenesis using mouse models.
Design
Mice with a targeted deletion of Arid1a in the pancreas by itself and in the context of two common genetic alterations in PDAC, Kras and p53, were followed longitudinally. Pancreases were examined and analysed for proliferation, response to injury, and tumourigenesis. Cancer cell lines derived from these models were analysed for clonogenic, migratory, invasive and transcriptomic changes.
Results
Arid1a deletion in the pancreas results in progressive acinar-to-ductal metaplasia (ADM), loss of acinar mass, diminished acinar regeneration in response to injury, and ductal cell expansion. Mutant Kras cooperates with homozygous deletion of Arid1a leading to intraductal papillary mucinous neoplasm (IPMN). Arid1a loss in the context of mutant Kras and p53 leads to shorter tumour latency, with the resulting tumours being poorly differentiated. Cancer cell lines derived from Arid1a-mutant tumours are more mesenchymal, migratory, invasive and capable of anchorage-independent growth; gene expression analysis showed activation of epithelial-mesenchymal transition (EMT) and stem cell identity pathways that are partially dependent on Arid1a loss for dysregulation.
Conclusions
ARID1A plays a key role in pancreatic acinar homeostasis and response to injury. Furthermore, ARID1A restrains oncogenic KRAS-driven formation of premalignant proliferative IPMN. Arid1a-deficient PDAC are poorly differentiated and have mesenchymal features conferring migratory/invasive and stem-like properties.
Keywords: Pancreatic Ductal Adenocarcinoma, ARID1A, SWI/SNF, IPMN, EMT
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is often diagnosed at an advanced stage that precludes surgery and is largely resistant to chemotherapy.1 There is a need for improved therapies based on the molecular mechanisms underlying PDAC.2 Common genetic alterations in PDAC include KRAS, TP53, SMAD4/DPC4, and CDKN2A/p16INK4A,3-7 as well as in the chromatin remodeling SWI/SNF complex, with ARID1A being the most frequently altered.8-10 The human SWI/SNF complex exists in various combinatorial assemblies that include an ATPase, “core” elements, and other subunits including ARID1A that confer functional specificity.11 SWI/SNF is known to be involved in transcriptional regulation, DNA replication and damage repair,12-14 with ARID1A, in particular, implicated in regulation of development and cell-cycle control frequently through opposition of polycomb-group (PcG) proteins such as EZH2.15-18 Mutations in ARID1A render certain cancers susceptible to particular pharmacologic interventions including EZH2, PI3K, AKT, ATR, PARP, and HDAC6 inhibitors,14,19,20 providing potential new therapeutic options.
The SWI/SNF complex appears broadly to control malignant progression and the differentiation states of tumours. BRG1 (SMARCA4), an ATPase component of the SWI/SNF complex, is needed for terminal maturation of pancreatic ducts and its loss cooperates with oncogenic KRAS in pancreatic ductal cells to form intraductal papillary mucinous neoplasm (IPMN).21 BRG1 re-expression induces a mesenchymal phenotype in established PDAC cell lines that have developed in its absence.22 SMARCB1, via regulation of MYC, may restrain the expansion of mesenchymal subpopulations associated with de-differentiation and aggressive behaviour.23 ARID1B exhibits tumour suppressor activities when re-expressed in the human MiaPaCa2 PDAC cell line where it is repressed by promoter hypermethylation.24 In other cancers, ARID1A alters pathways related to cell-cell adhesion and invasion.13,25-27
We evaluated the impact of Arid1a loss in the pancreas using mice with conditional mutations of Arid1a, as well as the commonly occurring PDAC mutations Kras and p53. Our results show that ARID1A is critical to the maintenance of pancreatic acini and constrains proliferation of the pancreatic ductal compartment. Arid1a loss shifted the spectrum of cancer precursors from PanIN to IPMN and accelerated the onset of invasive cancer. Cell lines derived from Arid1a-mutant PDAC tumours demonstrated increased migration, invasion and anchorage-independent growth supported by an EMT and stem-like transcriptional program. Re-establishing Arid1a expression and inhibition of PDGRFb with imatinib both partially abrogate this phenotype, offering a potential path to mutation-directed therapy.
RESULTS
Loss of ARID1A leads to progressive pancreatic atrophy and cysts
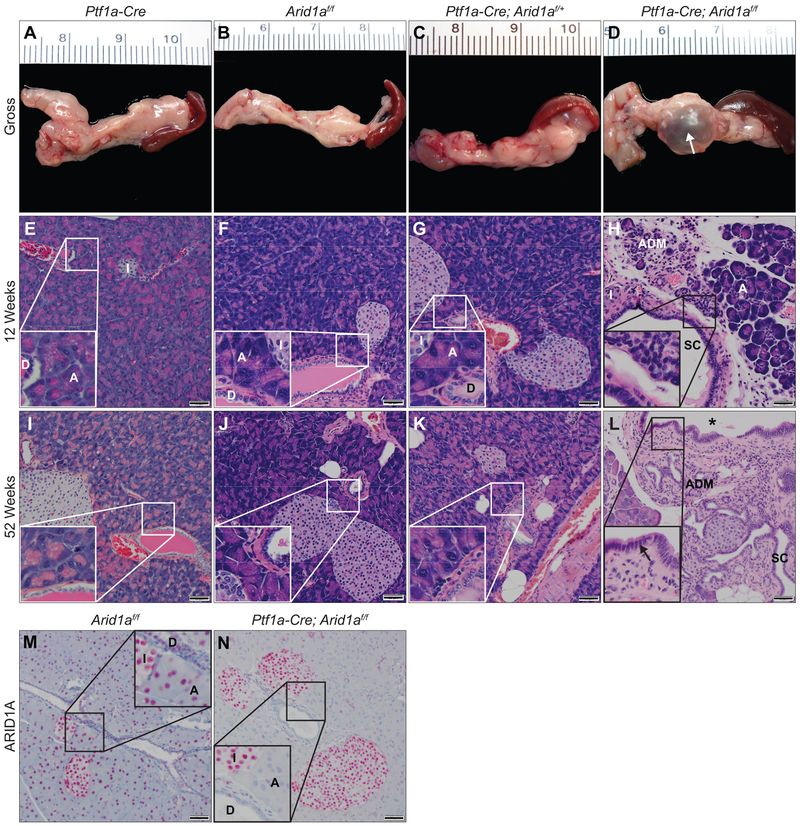
Given the importance of ARID1A in developmental processes, tissue homeostasis, and regeneration,16,17,28 we generated cohorts of mice carrying Ptf1a-Cre (C), Arid1aflox/flox(A), Ptf1a-Cre; Arid1aflox/f+ (CAf/+) or Ptf1a-Cre; Arid1aflox/flox (CAf/f) alleles (supplementary figure S1A,B). Ptf1a is expressed in multipotent progenitor cells, which give rise to the three main cellular lineages in the pancreas.29 The Arid1aflox allele enables deletion of the eighth exon of Arid1a in the presence of Cre recombinase leading to frameshift mutation and nonsense-mediated mRNA decay.17 CAf/f mice were generated at expected Mendelian ratios, developed normally, and survived through a year of age. CAf/f pancreases (figure 1D,H,L) showed progressive loss of acinar cell mass compared with all other controls (figure 1A-C,E-G,I-K), which were grossly and histologically normal. Histologic analysis at 12 weeks showed apoptosis, acinar-to-ductal metaplasia (ADM, n=12/12), focal pancreatitis (11/12), and microscopic cysts (9/12) (figure 1H and supplementary table S1) and were smaller than controls (supplementary figure S1H). With age, CAf/f demonstrated ongoing ADM, acinar loss, fat accumulation consistent with pancreatic atrophy, and dilatation of a simple columnar ductal epithelium into grossly visible fluid-filled cysts (figure 1D-white arrow,L and supplementary figure S1C-G). Immunohistochemistry (IHC) confirmed ARID1A absence in CAf/f acinar and ductal cells (figure 1M,N). Retained expression in islets suggested that islet development may depend on ARID1A and is consistent with developmental outgrowth of islets arising from non-Ptf1a-expressing progenitors.30,31 Thus, we find Arid1a mutation leads to progressive loss of the acinar cell compartment, ADM, and progression of ductal cystic lesions.
Figure 1.
ARID1A maintains pancreatic homeostasis. Gross images at ≥52 weeks of age of (A) C, (B) Af/f, (C) CAf/+, and (D) CAf/f pancreases. Hematoxylin and eosin (H&E) staining of pancreases at 12 weeks of (E) C, (F) Af/f, (G) CAf/+, (H) CAf/f and ≥52 weeks of (I) C, (J) Af/f, (K) CAf/+, (L) CAf/f showing progressive acinar attrition and ductal expansion in CAf/f compared to all controls. Immunohistochemistry (IHC) for ARID1A at 12 weeks of (M) Af/f and (N) CAf/f. Normal acinar cells (“A”), ducts (“D”), islets (“I”), acinar-to-ductal metaplasia (“ADM”), small cysts (“SC”), and large macroscopic cysts (white arrow or asterisk). Simple columnar epithelium of a large cyst denoted with black arrow. Bar=50 μm.
ARID1A is required for acinar cell recovery and suppresses ductal proliferation.
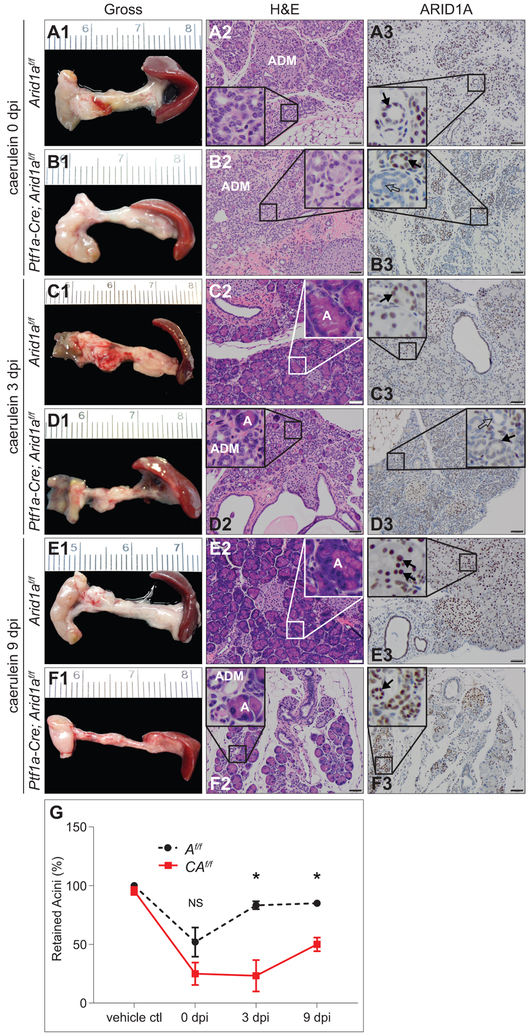
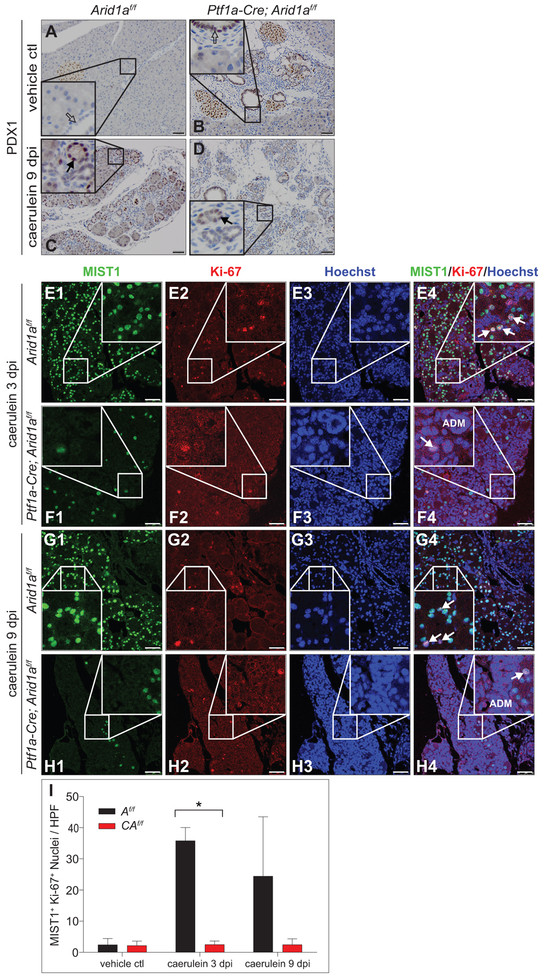
As ARID1A facilitates differentiation, maintenance of epithelial populations, and recovery after injury,16,17,28 we evaluated the effect of injury on CAf/f, C and Af/f cohorts by injection of caerulein or vehicle control at 8 weeks of age twice daily for two weeks (supplementary figure S2A).32 CAf/f, C, and Af/f were equally susceptible to injury, loss of acini, and ADM (figure 2A1&2,B1&2,G 0 days post-injection [dpi] and supplementary figure S2L,N). CAf/f pancreases were grossly smaller and failed to recover normal acinar cell mass (figure 2D1&2,F1&2,G) compared to controls at 3 and 9 dpi (figure 2C1&2,E1&2 and supplementary figure S2B-N). IHC for PDX1, critical to acinar cell development and identity,33 demonstrated robust expression in the re-emerging acinar units in caerulein-treated controls that was diminished in CAf/f, suggesting an inability to activate this program in the absence of ARID1A (figure 3A-D). Ducts in CAf/f pancreases, at baseline and after injury, stained positively for PDX1, compared with controls, consistent with previous reports that Brg1-mutant ductal structures have a de-differentiated (PDX1-positive) phenotype.22 Immunofluorescence (IF) for MIST1 (Muscle Intestine and Stomach Expression 1, a marker of terminally differentiating acinar cells),34,35 present at baseline in CAf/f and controls (figure 3E1-H1,I and supplementary figure S3A,B), and the proliferative marker Ki-67 (figure 3E2-H2) demonstrated suppression of both total MIST1-positive cells (figure 3E1-H1), and co-stained cells (proliferating acinar compartment) among CAf/f post-injury compared with controls (figure 3E-I and supplementary figure S3A-D), supporting the observed failure of CAf/f to reconstitute acinar cells. IF for the ductal marker Cytokeratin 19 (CK19) and Ki-67 similarly showed decreased proliferation in the CK19-negative (acinar) cells of caerulein-treated CAf/f (supplementary figure S3E,F,I). To exclude the possibility of Arid1a-mutant ductal structures impacting acinar recovery, we used a duct-specific, tamoxifen-inducible Cre-recombinase allele (Ck19-CreERT) to delete Arid1a in mature ducts (supplementary figure S4A-I). This had no appreciable impact on post-injury acinar recovery, MIST1 expression, or proliferation (supplementary figure S5A-I), demonstrating that impaired acinar recovery was likely due to cell autonomous effects of Arid1a mutation.
Figure 2.
ARID1A is necessary for pancreatic acinar epithelial repair following injury. (A-F) Caerulein given by intraperitoneal injection (250 μg/kg twice daily) for two weeks to 8-week-old Af/f and CAf/f mice. Gross images with corresponding H&E and ARID1A IHC of pancreases at (A1-3,B1-3) 0, (C1-3,D1-3) 3, and (E1-3,F1-3) 9 days post-injection (dpi). Both (A2) Af/f and (B2) CAf/f show loss of acini with development of ADM (enlarged in insets) at 0 dpi. At 3 and 9 dpi, (C2,E2) Af/f reconstitutes acinar epithelia and (D2,F2) CAf/f shows continued acinar depletion (enlarged in insets). ARID1A IHC shows retention of ARID1A in the epithelia of Af/f mice (A3,C3,E3) with scattered retention of ARID1A in the epithelia of CAf/f mice (B3,D3,F3). (G) Quantification of percent of retained acini in Af/f and CAf/f pancreases at the indicated time points shows earlier recovery of Af/f pancreases compared with CAf/f pancreases; n=3 per genotype per time point per treatment, mean +/− SD, *p<0.05, NS=not significant. Bar=50 μm.
Figure 3.
ARID1A is necessary for acinar cell differentiation and proliferation after injury. (A-D) PDX1 IHC of (A,C) Af/f and (B,D) CAf/f pancreases with (A,B) vehicle control or (C,D) caerulein treatments. Islets, re-emerging acini (denoted by black arrow), and progenitor-like ducts (denoted by outlined arrow) stain strongly for PDX1. Immunofluorescence (IF) of MIST1 and Ki-67 of caerulein-treated Af/f and CAf/f pancreases at (E,F) 3 and (G,H) 9 dpi separated by (E1-H1) MIST1 (green, acinar cell marker), (E2-H2) Ki-67 (red, proliferation marker), (E3-H3) Hoechst (blue, nuclear stain), and (E4-H4) overlay of all channels (white arrows denote proliferating acinar cells, ADM=acinar-to-ductal metaplasia). (I) Quantification of MIST1+ and Ki-67+ colocalised nuclei per high-power field (HPF) shows diminished MIST1 and Ki-67 co-localisation in CAf/f pancreases compared to Af/f pancreases; n=3 animals per genotype per time point with 5 HPF per animal, mean +/− SD, *p<0.05. Bar=50 μm.
ARID1A IHC showed enrichment of ARIDIA-expressing cells throughout the post-injury time course among CAf/f pancreases as compared with controls (figure 2A3-F3 and supplementary figure S2F,K), suggesting a selective advantage for ARID1A-intact cell populations (cells escaping Cre-mediated ARID1A deletion) during recovery. Consistent with an ARID1A-competent sub-population underlying this limited acinar recovery, IF for ARID1A and Ki-67 showed co-localization in many of the proliferating cells in injured CAf/f pancreases (supplementary figure S6A-D). These data further support the conclusion that ARID1A loss leads to a block of physiologic post-injury proliferation and recovery of the acinar lineage with both diminished numbers of cells directed towards an acinar fate as well as a diminished proliferative capacity. Thus, ARID1A is necessary to maintain pancreatic acinar integrity over adult life and reconstitution following insult.
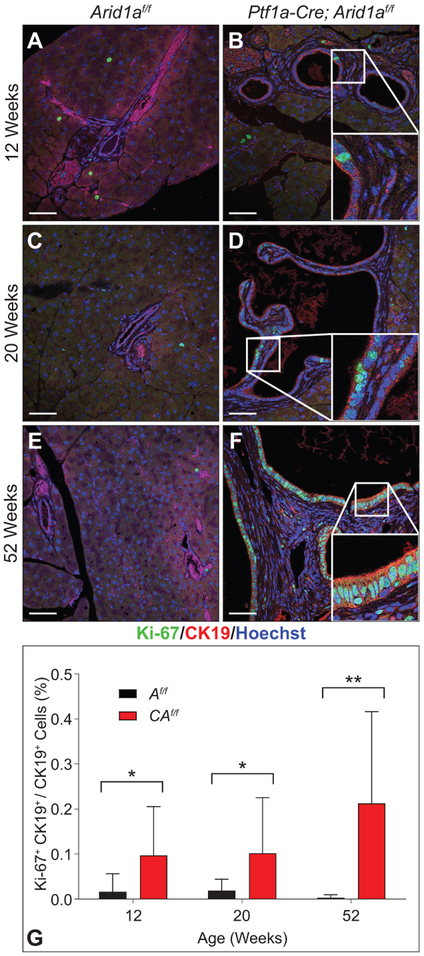
To better understand the basis for the histologic changes in CAf/f mice, we performed IHC for Ki-67 and cleaved Caspase-3 (CC3), a marker of apoptosis. At 12 weeks, CAf/f pancreases had higher rates of Ki-67 and CC3 positive cells than Af/f (supplementary figure S7A–D), signifying increased basal levels of apoptosis and proliferation. We observed that apoptosis occurred among acinar cells, consistent with progressive attrition over time, while the difference in Ki-67 staining was seen in ducts (supplementary figure S7E,F). To specifically evaluate the ductal compartment, we performed IF for CK19 and Ki-67 of CAf/f and Af/f at 12, 20, and 52 weeks. At each time point, CAf/f pancreases showed increased rates of Ki-67 in the CK19-positive compartment that increased over time (figure 4A-G and supplementary figure S3G-I), indicating that ARID1A restrains ductal proliferation.
Figure 4.
ARID1A restrains ductal proliferation. IF for CK19 (red, ductal marker), Ki-67 (green), and Hoechst (blue) of (A) 12-, (C), 20-, and (E) 52-week-old Af/f and age-matched CAf/f pancreases (B,D,F). Insets with representative duct and Ki-67+ cells. (G) Quantification of percent of Ki-67+ CK19+/CK19+ cells shows increased proliferation in CAf/f pancreases; n=3 per genotype per time point with 3-4 HPF per animal, mean +/− SD), *p<0.05, **p<0.01. Bar=50 μm.
Arid1a mutation causes IPMN in Kras and p53 mutant PDAC models
The mutational activation of KRAS is the most common genetic mutation in PDAC and is an early event in pancreatic tumourigenesis.36,37 To determine if Arid1a mutation cooperates with oncogenic KrasG12D, we utilised the Lox-stop-lox KrasG12D (LSL-KrasG12D) allele38 that undergoes Cre-mediated excision of an in-frame stop codon leading to expression of constitutively active KrasG12D under the endogenous promoter to generate Ptf1a-Cre; LSL-KrasG12D (KC), Ptf1a-Cre; LSL-KrasG12D; Arid1af/+ (KCAf/+) and Ptf1a-Cre; LSL-KrasG12D; Arid1af/f (KCAf/f) mice (supplementary figure S1A,B). KC, KCAf/+, and KCAf/f cohorts were followed longitudinally and sacrificed at signs of illness.
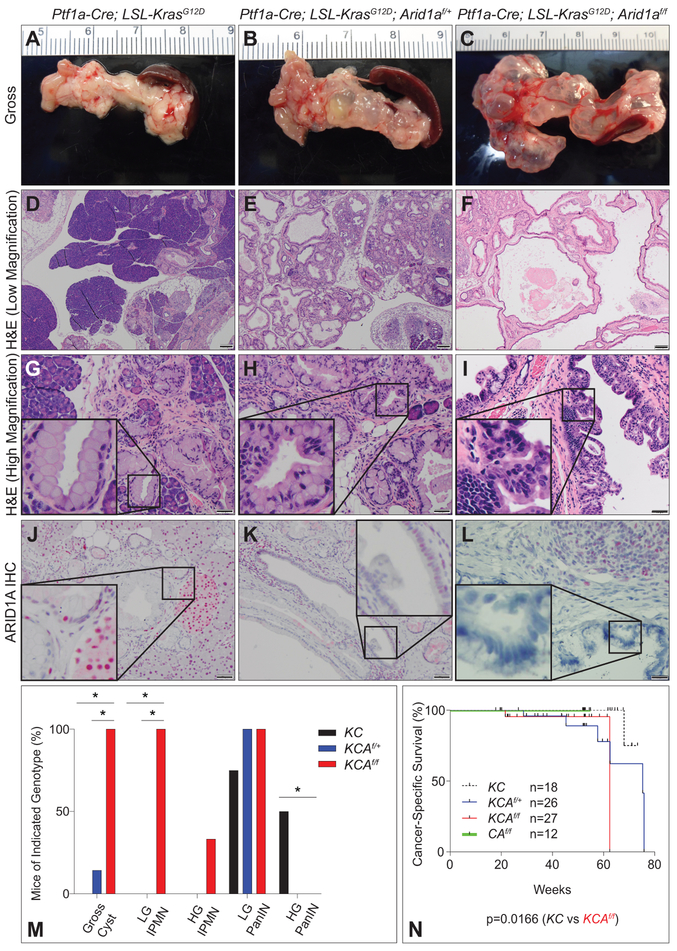
KCAf/f mice had distended abdomens beginning at ~20 weeks of age and a diminished overall survival (OS, defined by death due to any cause) compared with KC controls (supplementary figure S8A, 48.57 weeks vs. not reached, p<0.0001). At necropsy, expansive fluid-filled cystic pancreases were found in all KCAf/f mice. Analysis at 52 weeks of age demonstrated that this was not seen among KC and variably and less prominent among KCAf/+ (figure 5A-C and supplementary figure S8B,D). Widespread ductal proliferative lesions with varying degrees of cellular atypia forming large interconnecting cystic structures, morphologically consistent with IPMN, were found in KCAf/f pancreases (figure 5F,I). KCAf/f cohort harbored invasive, poorly-differentiated cancer with metastasis in 3 of 19 mice. Though most deaths were due to abdominal distention from IPMN, the KCAf/f cohort exhibited a greater incidence of invasive cancer with a diminished cancer-specific survival (CSS, defined by death with histologic evidence of cancer) compared with KC controls (62.43 weeks vs. not reached, p=0.0166, figure 5N, supplementary figure S8I-K and supplementary table S2A,C,D). Higher rates of invasive PDAC were found in the KCAf/+ cohort (n=6 of 15 [40%]) compared with KC group (n=1 of 11 [9.1%]), thus heterozygosity of Arid1a in the setting of Kras mutation may also accelerate PDAC formation (figure 5N, supplementary figure S8F-H and supplementary table S2A,B,D). Comparison of the pre-neoplastic lesions in mice sacrificed at earlier time points revealed a shift in the spectrum of precursor lesions; KCAf/f harbored more IPMN while KC harbored more high-grade, PDX1-positive PanIN (figure 5D-I,M, supplementary figure S8B-E and supplementary table S2A-D) with KCAf/+ harboring both PanIN lesions and variable cyst formation (figure 5E,H and supplementary table S2B). ARID1A IHC showed intact expression in KC (figure 5J), mosaic loss in KCAf/+ (particularly in the ducts, figure 5K), and total loss in epithelia of IPMN lesions in KCAf/f (figure 5L). These data imply that ARID1A plays a tumour suppressive role in the pancreatic epithelia in the context of oncogenic KrasG12D and is sufficient to drive expansion of IPMNs and accelerate transformation.
Figure 5.
KrasG12D and Arid1a loss cooperate to form IPMN. Cohorts of Ptf1a-Cre; KrasG12D (KC), Ptf1a-Cre; KrasG12D; Arid1af/+ (KCAf/+) and Ptf1a-Cre; KrasG12D; Arid1af/f (KCAf/f) were followed longitudinally. Gross images of (A) KC, (B) KCAf/+, and (C) KCAf/f showing large cysts in KCAf/f. H&E at low magnification of (D) KC, (E) KCAf/+, and (F) KCAf/f mice showing a shift from PanIN to IPMN with loss of Arid1a. Bar=200 μm. High magnification of (G) KC, (H) KCAf/+, and (I) KCAf/f mice demonstrates PanIN (G,H) and area of high grade dysplasia within IPMN (I). ARID1A IHC shows retained expression in PanIN in (J) KC, variable retention in (K) KCAf/+ lesions and loss of expression in (L) KCAf/f IPMN. Bar=50 μm. (M) Quantification of percent incidence of gross cysts, low-grade (LG) IPMN, high-grade (HG) IPMN, LG PanIN, and HG PanIN in mice sacrificed between the ages of 12 and 20 weeks; n=8 mice for KC, 7 mice for KCAf/+, 9 mice for KCAf/f, *p<0.05. (N) Kaplan-Meier analysis shows decreased cancer-specific survival (CSS) in KCAf/f compared to KC (p<0.0166, median CSS of KC (black, n=18) and CAf/f (green, n=12) are undefined, whereas KCAf/+ is 75.29 weeks (blue, n=26), and KCAf/f is 62.43 weeks (red, n=27)).
Kras mutation alone targeted to the murine pancreas leads to PDAC with a long latency that is accelerated with concurrent mutation in other cooperating tumour suppressor genes such as Cdkn2a or p53.3,4,7 To determine the impact of Arid1a mutation in the context of a highly penetrant and fully invasive cancer model, we utilised the p53flox/+ (p53f/+) allele that undergoes Cre-mediated excision of exons 2 through 10 rendering the gene functionally inactive.39 We generated mice carrying Ptf1a-Cre; LSL-KrasG12D; p53f/+; Arid1af/+ (KPCAf/+) as well as Ptf1a-Cre; LSL-KrasG12D; p53f/+; Arid1af/f (KPCAf/f) and compared them to littermate control Ptf1a-Cre; LSL-KrasG12D; p53f/+ (KPC) animals (supplementary figure S1A,B).
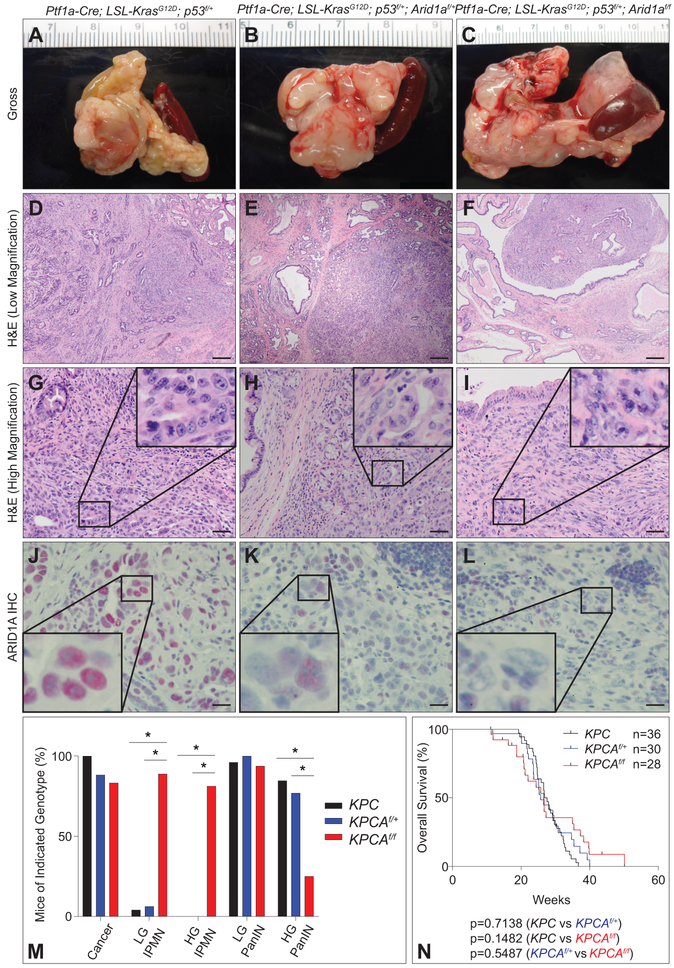
Survival analyses did not reveal any differences in OS between KPC, KPCAf/+, and KPCAf/f cohorts (figure 6N). KPCAf/f mice had large, cystic pancreases (figure 6C), similar to KCAf/f, often with focal nodularity harboring invasive cancer, while KPC and KPCAf/+ harbored firm, diseased pancreases with invasive cancer (figure 6A,B) absent of extensive cystic changes. KPCAf/f mice had inferior CSS compared with controls (26.71 vs. 28.00 weeks, p=0.048, supplementary figure S9A) and Arid1a loss shifted the histologic spectrum of precursors from low- and high-grade PanINs, the dominant lesions found in KPC, to predominantly low- and high-grade IPMNs in KPCAf/f cohorts (figure 6D,F,M and supplementary figure S9B-E), with both PanINs and IPMNs in KPCAf/+ (figure 6E,M). PDAC in KPCAf/f was associated with large IPMN-like cystic lesions (similar to KCAf/f) with varying grades of dysplasia and cancer (figure 6F,I and supplementary figure S9D). ARID1A IHC confirmed retained expression in KPC and KPCAf/+ but was lost in KPCAf/f (figure 6J-L). The majority of KPCAf/f PDAC (n=12/13 [92.3%]) were poorly differentiated compared to KPC (n=18/30 [60.0%]) or KPCAf/+ PDAC (n=6/13 [46.2%]) (figure 6G-I and supplementary table S3A-D), but with similarly aggressive metastatic rates (supplementary figure S9F-K and supplementary table S3D). These results indicate that in the setting of Kras and p53 mutation, ARID1A loss leads to IPMN and the development of fully-invasive poorly-differentiated adenocarcinoma with a small decrease in CSS.
Figure 6.
Arid1a mutation in KrasG12D; p53 PDAC model. (A-C) Gross images at necropsy of (A) Ptf1a-Cre; KrasG12D; p53f/+ (KPC), (B) Ptf1a-Cre; KrasG12D; p53f/+; Arid1af/+ (KPCAf/+), and (C) Ptf1a-Cre; KrasG12D; p53f/+; Arid1af/f (KPCAf/f). (D-F) Low magnification H&E of (D) KPC PDAC with associated PanIN, (E) KPCAf/+ PDAC with mix of precursors, and (F) KPCAf/f PDAC with associated IPMN. Bar=200 μm. (G-I) High magnification H&E reveals (I and inset) high grade KPCAf/f PDAC compared to (G and inset) KPC and (H and inset) KPCAf/+. Bar=50 μm. (J-L) ARID1A IHC demonstrates intact ARID1A staining in (J) KPC PDAC and (K) KPCAf/+ PDAC and loss of ARID1A staining in (L) KPCAf/f PDAC. Bar=20 μm. (M) Quantification of percent incidence of cancer, LG IPMN, HG IPMN, LG PanIN, and HG PanIN in mice sacrificed at signs of distress; n=36 mice for KPC, 30 mice for KPCAf/+, and 26 mice for KPCAf/f, *p<0.05. (N) Kaplan-Meier plot of overall survival (OS) for KPC (black, n=36, 27.22 weeks), KPCAf/+ (blue, n=30, 25.71 weeks), and KPCAf/f (red, n=28, 26.57 weeks) shows no significant differences in survival.
As compared with human pathology, KCAf/f and KPCAf/f IPMN lesions were found to most closely resemble the gastric subtype, predominantly lacking expression of MUC1 and MUC2 (supplementary figure S10A-C, I-K), with discrete areas of high-grade dysplasia within IPMNs demonstrating a transition resembling either intestinal-type IPMN (MUC2 positive, supplementary figure S10F,G,L-N,R) or pancreatobiliary-type IPMN (MUC1 positive, supplementary figure S10E,O-R), which is consistent with progression of human IPMNs.40,41 Furthermore, the epithelia expressed PDX1 (marker of ducts with a progenitor-like phenotype, supplementary figure S8E,S9E) and did not show nuclear expression of ERα in the stroma around the ducts (supplementary figure S10D,H), indicating that these lesions are not consistent with mucinous cystic neoplasms (MCNs).
Arid1a-mutant PDAC cell lines are more mesenchymal and invasive
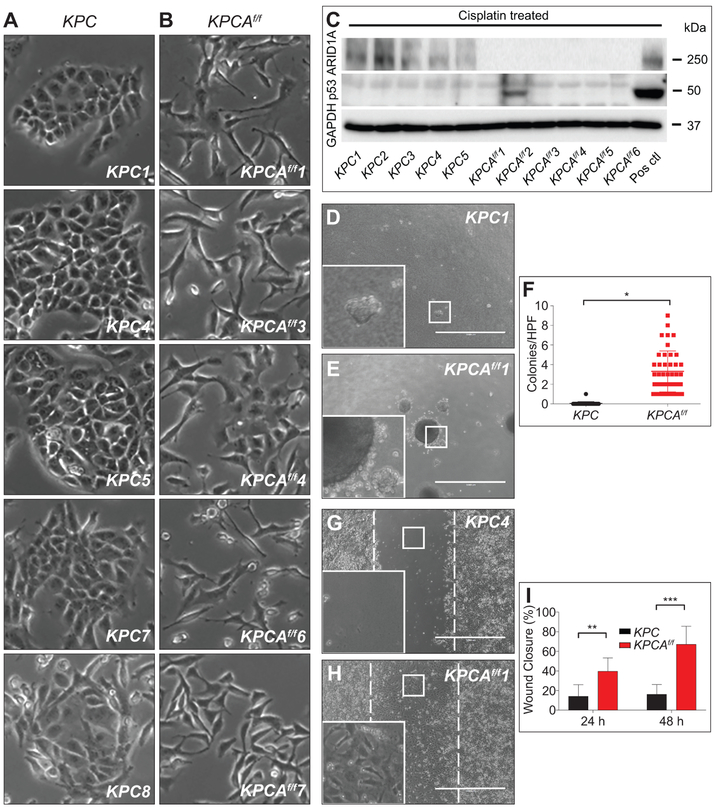
Primary cell lines derived from KPCAf/f tumours using established methods21 were morphologically more mesenchymal compared to KPC lines, which retained an epithelial appearance, corresponding to histologic analysis of the original tumours (figure 7A,B and supplementary figure S11A,B). Somatic mutation of Arid1a in KPCAf/f cancer lines was shown by PCR (supplementary figure S11C) and confirmed by western blot analysis (figure 7C). Cisplatin failed to induce p53 in all KPC and all but one KPCAf/f lines, demonstrating an impaired p53 axis (figure 7C), presumably due to spontaneous loss of heterozygosity (supplementary figure S11C). Comparison of KPCAf/f with KPC lines (n=2 per genotype) showed that Arid1a-mutant lines exhibited enhanced anchorage-independent growth in a clonogenic assay (figure 7D-F and supplementary figure S11E,F) and faster closure in a wound healing assay (figure 7G-I and supplementary figure S11G-L), consistent with increased migratory potential, while KPC lines generally had higher proliferation rates (supplementary figure S11D). Evaluation of the boundaries of colonies revealed extension and invasion of KPCAf/f cells into the agar, a feature notably absent among KPC lines (figure 7D,E insets). Mirroring the more aggressive development of cancer in KPCAf/f mice, these results were suggestive of ARID1A’s role in suppressing cell migration and anchorage-independent growth.
Figure 7.
Arid1a loss increases anchorage-independent growth and migration. (A) KPC tumour cell lines (n=5) demonstrating epithelial morphology compared to (B) KPCAf/f tumour cell lines (n=5) showing mesenchymal morphology. (C) Western blot of protein lysates from KPC and KPCAf/f cell lines shows absence of ARID1A as expected and absence of P53 induction following cisplatin treatment in all but one KPCAf/f cell lines. GAPDH served as a loading control. Positive control is an Arid1a/p53 intact PDAC line (Pos ctl). (D-F) Clonogenic assay and quantification showing (E,F) KPCAf/f cell lines (n=2) form greater numbers of colonies in soft agar compared to (D,F) KPC (n=2), *p=8.5×10−13. Colonies ≥125 μm were counted. (G-I) Wound healing assay and quantification demonstrate that (H,I) KPCAf/f cell lines (n=2) have increased ability to fill the wound area compared to (G,I) KPC (n=2) at (I) 24 and 48 hours, **p=6.45×10−8, ***p=4.81×10−15. Analysis by TScratch. Bar=1000 μm for (D,E,G,H).
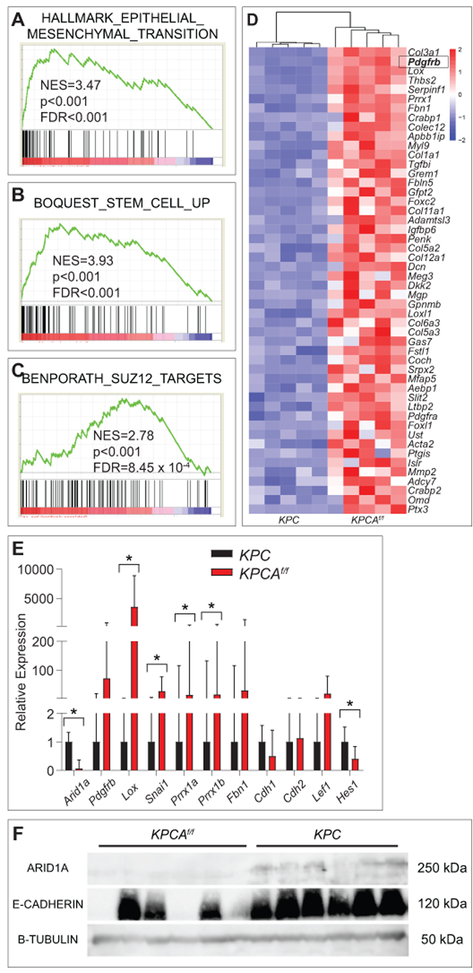
Global transcriptome analyses comparing KPC and KPCAf/f cell lines (n=5 independent lines for each genotype, supplementary table S4) identified 1109 differentially expressed genes (846 upregulated genes and 263 downregulated genes) with p-adj <0.05 (supplementary figure S12A,B). Sample-to-sample distance plot and principal components analysis demonstrated clustering of KPC lines separate from KPCAf/f lines (supplementary figure S12C,D). Gene Set Enrichment Analysis (GSEA) identified “SUZ12 targets” as a significantly enriched pathway (figure 8C) and analyses on the Enrichr platform identified both SUZ12 and EZH2 as transcriptional regulators of the differentially expressed genes (supplementary figure S12E), consistent with other studies that show SWI/SNF complexes oppose epigenetic silencing by polycomb repressive complex 2 (PRC2).18 Additionally, gene signatures for EMT and stem cells were enriched in KPCAf/f lines (figure 8A,B,D), consistent with above phenotypic observations. Other gene signatures associated with KPCAf/f cell lines include invasiveness, alterations to cell-matrix adhesions, and differentiation (supplementary table S5).
Figure 8.
EMT and stem cell transcriptional programs are activated in Arid1a-mutant PDAC. (A) Gene set enrichment analysis of differentially expressed genes from RNA-Seq of KPC vs KPCAf/f using the “Hallmark” gene sets demonstrates EMT signature is the most enriched signature in KPCAf/f lines. NES=normalised enrichment score; FDR=false discovery rate. (B,C) Gene set enrichment analysis using the “Curated chemical and genetic perturbations” gene sets demonstrates that stem cell and SUZ12 targets are highly enriched in KPCAf/f lines. (D) Expression data of the 50 genes with the highest log2FC between KPCAf/f and KPC cell lines from the gene sets in (A-C) with Pdgfrb highlighted in box. (E) qPCR validation of indicated genes in KPCAf/f (n=8) compared to KPC (n=6) cell lines, *p<0.05. (F) Western blot for ARID1A, E-Cadherin, and B-Tubulin in KPCAf/f and KPC cell lines (n=6 each) shows loss of expression of E-Cadherin in 3 of 6 KPCAf/f cell lines.
Differential expression of genes affecting EMT and metastasis including Lox (log2FC 8.33, p-adj 2.55 × 10−38), Prrx1 (log2FC 7.43, p-adj 3.96 × 10−38), and Fbn1 (log2FC 7.18, p-adj 8.30 × 10−25) was validated using quantitative polymerase chain reaction (qPCR) in three each of the original cell lines used in RNA-Seq and additionally derived cell lines from KPC (n=3 new) and KPCAf/f (n=5 new) tumours (figure 8E and supplementary table S4). Pdgfrb (log2FC 8.63, p-adj 4.00 × 10−31) and Fbn1 (log2FC 7.18, p-adj 8.30 × 10−25) did not demonstrate a significant expression difference between KPC and KPCAf/f, likely due to heterogeneity within the group, although the majority of lines were consistent with the RNA-seq results (supplementary figure S13A,B). Western blot analysis for E-Cadherin showed loss of expression in 3 of 6 KPCAf/f cell lines as compared to intact expression in all KPC cell lines (figure 8F), correlating with a more invasive phenotype.42 These results suggest that KPCAf/f cell lines are less differentiated with increased migratory and invasive potential compared to KPC cell lines.
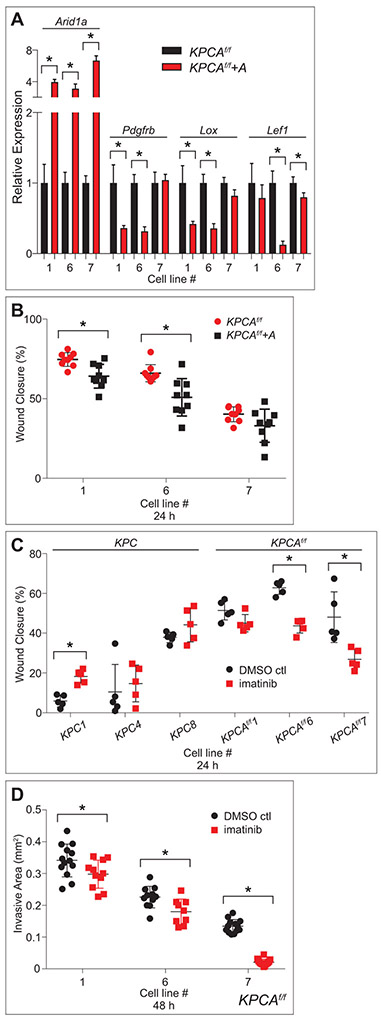
To assess whether ARID1A directly regulates the migration of derivative cancer lines as well as the genes identified by RNA-Seq, Arid1a was re-expressed in three of the Arid1a-mutant derived cancer lines described above (KPCAf/f lines 1,6,7) using a Sleeping Beauty transposon system.43 ARID1A re-expression was confirmed by qPCR and Western blot (figure 9A and supplementary figure S14B). Additionally, qPCR confirmed that re-expression decreased mRNA levels of Pdgfrb, Lox, and Lef1 in two of three cell lines tested for each gene though not of all genes tested (figure 9A and supplementary figure S14A), suggesting a direct role of ARID1A in persistent regulation of some of these metastasis-associated genes. Furthermore, ARID1A reexpression led to diminished migration using the wound healing assay, demonstrating that its mutation is important to maintaining this migratory phenotype (figure 9B).
Figure 9.
Re-expression of Arid1a and inhibition of PDGFRb partially abrogate the phenotypes of KPCAf/f cell lines. (A) qPCR data showing re-expression (KPCAf/f+A) and controls (KPCAf/f) in three KPCAf/f cell lines demonstrate significantly increased expression of Arid1a mRNA in all three lines, and significantly decreased expression of Pdgfrb, Lox, and Lef1 in two of three lines each. (B) Quantification of wound healing assay in three KPCAf/f cell lines with (KPCAf/f+A) and without (KPCAf/f) Arid1a re-expression demonstrates that KPCAf/f+A have decreased ability to fill the wound area compared to controls in two of three cell lines tested. (C) Quantification of wound healing assay in three KPC and KPCAf/f cell lines treated with imatinib (1 μM) and DMSO control demonstrates that imatinib has no effect on most KPC cell lines tested while imatinib-treated KPCAf/f cell lines have decreased ability to fill the wound area compared to controls in two of three lines tested. (B,C) All measured at 24 hours, *p<0.05, analysis by TScratch. (D) Quantification of cancer cell spheroid invasion assay in three KPCAf/f cell lines treated with imatinib (1 μM) and DMSO control reveals decreased total area invaded by imatinib-treated KPCAf/f cells leaving the spheroid in all three cell lines tested. All measured at 48 hours, *p<0.05, analysis by ImageJ.
Pdgfrb, among the top genes found differentially expressed between KPC and KPCAf/f cell lines (figure 8D), is a known driver of invasion, migration, and metastasis.44 To assess whether pharmacologic inhibition of this actionable target could impact these behaviors, we treated cell 3 KPC and 3 KPCAf/f lines with inhibitors of PDGFRb (imatinib). Imatinib (1 μM) significantly inhibited the migratory (2 of 3 lines, figure 9C and supplementary figure S15F-M) and invasive (3 of 3 lines, figure 9D and supplementary figure S16M-R) abilities of KPCAf/f lines, while having no impact on proliferative (supplementary figure S15A), migratory (figure 9C and supplementary figure S15B-E), clonogenic or invasive characteristics of KPC lines compared to KPCAf/f (supplementary figure S16A-R). These data suggest that dysregulation of PDGFRb downstream of Arid1a loss is partly responsible for the invasive characteristics of KPCAf/f cell lines and potentially targetable with existing drugs. Interestingly, the cell line whose migratory capacity was unaffected by Arid1a re-expression was the same cell line in which Arid1a re-expression did not alter Pdgfrb expression yet was still sensitive to imatinib treatment (cell line 7, figure 9A-C). In summary, KPCAf/f cell lines which are migratory and enriched in gene signatures associated with PRC2, EMT, and stem-like phenotypes can be rendered less migratory with Arid1a re-expression or with targeting of the PDGFRb axis with imatinib.
DISCUSSION
We report here the characterisation of a pancreas-specific conditional knockout of Arid1a by itself and in the context of other known driver mutations, Kras and p53. Arid1a is critical to maintain acinar cell mass through the adult life of the organism and for the normal function of an acinar re-differentiation program following injury, while serving in the ductal compartment to restrain proliferation. In pancreatic cancer models, mutation of Arid1a leads to IPMN precursors and the acceleration of cancer formation with a more migratory and mesenchymal phenotype supported by transcriptomic programs consistent with unopposed PRC2 function as well as activated EMT and stem cell pathways. These phenotypes and select downstream targets including PDGFRb are dependent on ARID1A loss for de-regulation and are potentially targetable.
Pancreatic acinar cells are highly differentiated and specialised, functioning to produce digestive enzymes important for the breakdown of nutrients in the gut. ARID1A has been found to be important for achieving or maintaining states of differentiation in hepatocytes and cardiomyocytes16,17,28 and similarly we find here that ARID1A is necessary for supporting acinar cell function and repopulation following injury with altered expression of key transcription factors required for acinar fate specification, PDX1 and MIST1.33,34 The importance of ARID1A in maintaining enhancer accessibility13 and the recent identification of a transcriptional enhancer critical to Mist1 expression35 raises the possibility of Arid1a mutation leading to dysfunction at this critical locus. In contrast to the phenotype in acinar cells, we observed expansion and proliferation of a PDX1-expressing ductal compartment that is amplified with progressive age and consistent with a progenitor-like state within the ducts. This proliferative phenotype may be caused by dysregulation of cell-cycle checkpoints,15 failure to achieve or maintain terminal differentiation, as seen with loss of BRG121 or suppression of senescence.45-47
The shift of pancreatic precursors, from low- through high-grade PanINs to IPMNs and low-grade PanINs with homozygous deletion of Arid1a in the context of KrasG12D with or without hemizygous deletion of p53 raises the possibility that Arid1a is required for the transition to high-grade PanIN. Consistent with a recent report,48 we show that CAf/fmice develop macroscopic proliferative cysts, wherein addition of oncogenic KRAS gives rise to IPMNs, suggesting ARID1A is a barrier to ductal expansion. Studies showing that acinar cells give rise to PanIN5,49,50 while ductal cells give rise to IPMN using compartment-specific Cre recombinases are in keeping with this.21 However, PanINs have been reported to develop in KCAf/f mouse pancreases,48 and indeed we did see PanINs in KCAf/f, but no examples of high-grade PanINs, whereas similarly-aged KC mice did show these lesions, suggesting the possibility of bystander PanINs in cells escaping the effect of Cre51 or different biological outcomes of Arid1a deletion in embryologic as opposed to adult pancreases. Thus, ARID1A may play a dichotomous role in each epithelial compartment, inhibiting IPMN progression while potentially being necessary for the progression to high grade PanIN, consistent with data on the effect of Brg1 and other chromatin modulators in the pancreas21,22,52 and pleomorphic phenotypic outcomes observed in the liver.43
Concomitant with the shift to IPMN, we observed a decreased latency in cancer-related death, demonstrating that Arid1a plays a potent tumour suppressive role in the development of PDAC from IPMNs consistent with its observed loss in human disease. The IPMNs in KCAf/f and KPCAf/f cohorts mostly resembled the gastric subtype, but some areas of higher-grade pathology had characteristics resembling intestinal and pancreatobiliary subtypes, consistent with transitions in phenotype seen in human disease.40,41,48 Taken together, our data suggest that in the presence of oncogenic KRAS, biallelic loss of Arid1a drives the expansion and transformation of the ductal compartment.
Arid1a-deficient PDAC are poorly differentiated and derivative cell lines have a greater capacity for migration, invasion, anchorage-independent growth, and an EMT as well as stem-like transcriptomic profile. Poorly differentiated tumours have been found to be associated with overexpression of genes enriched in embryonic stem cells53 and other models of cancer with loss of function of SWI/SNF components exhibit similar characteristics.13 Beyond genetic lesions, down-regulated expression of SWI/SNF components are known to underlie states of mesenchymal differentiation and reprogramming.23 Loss of these complexes can promote differentiation in breast and ovarian cancers;27,54 however, consistent with our data, other gastrointestinal cancers become more mesenchymal and/or have poorer prognosis in the absence of ARID1A.25,26,43,55 ARID1A has been shown to target the SWI/SNF complex to enhancers in a colon model13 and SMARCB1 loss has been shown to alter binding to typical enhancers with redistribution of SWI/SNF complexes that may impair SMARCB1-deficient cells to differentiate while maintaining other features that promote tumourigenesis, such as self-renewal, survival and proliferation.56
Signaling through PDGFRb contributes to multiple tumour-associated processes, including proliferation, transvascular transport of chemotherapeutic agents, cell invasion and metastasis via autocrine, paracrine and angiogenic pathways44 and may be a targetable downstream pathway in pancreatic cancer.57,58 Accordingly, we present evidence for the role of PDGFRb as one possible targeted therapeutic for ARID1A-deficient tumours as it was affected by Arid1a re-expression, and its inhibition by imatinib dampened the invasive phenotypes observed in KPCAf/f cell lines. Additional studies would be helpful to explore the mechanism by which ARID1A regulates PDGFRb, such as utilizing histone markers of activation and repression via ChIP-Seq or chromatin availability with ATAC-Seq.59 Additional studies to correlate these features of ARID1A loss as a prognostic or therapeutic opportunity using human PDAC samples will also be important.
Our mouse models, in vitro, and transcriptomic data show that ARID1A: 1) maintains the acinar epithelia while constraining the proliferation of ductal epithelia, 2) cooperates with KRAS to accelerate the expansion of IPMN precursors and cancer formation, and 3) in the setting of Kras and p53 mutations, leads to the formation of more poorly differentiated cancer associated with EMT and stem-like properties, which can be partially abrogated by Arid1a re-expression or PDGFRb inhibition. Our findings demonstrate the importance of ARID1A in normal pancreatic function and suppression of the development of premalignant ductal pathologies, as well as establish its role in restraining mesenchymal transition of invasive cancers.
MATERIALS AND METHODS
A detailed description of materials and methods can be found in Supplementary Materials and Methods.
Mouse lines
All animal studies were conducted in accordance with the AAALAC accredited University Committee on Animal Resources (UCAR). Experimental animals were generated using the alleles described in supplementary figure S1 and Supplementary Materials and Methods. Caerulein (Sigma) was administered by intraperitoneal injection at a dose of 250 μg/kg twice daily for two weeks. Tumour-derived cell lines were generated as previously described21 and genotyped as detailed in Supplementary Materials and Methods. Arid1a was re-expressed using the Sleeping Beauty transposon system as previously described.43
Immunohistochemistry and immunofluorescence
Mouse tissue was fixed in 10% neutral buffered formalin, embedded in paraffin, and cut to 4 μm sections that were processed for staining as described in Supplementary Materials and Methods.
Quantitative PCR (qPCR) analysis
Total RNA was purified, cDNA was synthesised, and qPCR was carried out as described in Supplementary Materials and Methods.
Western blotting
SDS-PAGE was performed on whole-cell extracts and protein was detected as described in Supplementary Materials and Methods.
RNA sequencing and data analysis
RNA was isolated, sequenced, and analysed as detailed in Supplementary Materials and Methods.
Wound healing assay
Cell migration post-wounding was assessed as described in Supplementary Materials and Methods.
Clonogenic assay in soft agar
Anchorage-independent growth was evaluated as described in Supplementary Materials and Methods.
Cancer cell spheroid invasion assay60
Spheroids were generated, and invasion was measured as described in Supplementary Materials and Methods.
Statistical analysis
Results were presented as mean ± SD and p-values were calculated using the two-tailed Student’s t test using GraphPad Prism or Excel software. Survival was determined using the Kaplan-Meier method and comparisons between genotypes were determined using the log-rank test. Animals that were euthanised for signs of illness or were found dead were included as events with the overall survival curve. Animals that were found to have histologic evidence of cancer were included as events while those that died for reasons other than cancer were censored with the cancer-specific survival curve.
Supplementary Material
Significance of this study.
What is already known about this subject?
Mutations in the SWI/SNF complex are found in approximately a third of PDAC, implicating deregulation of epigenetic processes in the development and progression of this disease.
ARID1A is the most frequently mutated member of the SWI/SNF complex.
In other cancers, ARID1A loss establishes synthetic vulnerabilities in critical cellular processes including DNA damage repair, proliferation/survival signaling, and other epigenetic regulatory pathways.
What are the new findings?
Arid1a deletion in the pancreas results in progressive acinar-to-ductal metaplasia with evolving pancreatitis, loss of acinar mass with pancreatic atrophy and diminished ability to regenerate acinar tissue mass in response to injury, and ductal expansion into macroscopic cystic structures.
Mutant Kras cooperates with concurrent homozygous deletion of Arid1a leading to intraductal papillary mucinous neoplasm (IPMN), a clinically recognised PDAC precursor, instead of the predominant pancreatic intraepithelial neoplasia (PanIN) precursors seen in mutant Kras alone; however, these IPMNs rarely progress to fully invasive PDAC.
Arid1a loss in the context of mutant Kras and hemizygous p53 leads to diminished cancer-specific survival, with the resulting tumour being poorly differentiated. Cancer cell lines derived from these Kras; p53; Arid1a mutant models are more mesenchymal, migratory, invasive and capable of anchorage-independent growth as compared with Arid1a-proficient PDAC cells. Gene expression analysis of these cell lines identified enriched gene sets that suggest activation of pathways including EMT and gain of stem cell identity, including Pdgfrb.
How might it impact on clinical practice in the foreseeable future?
This study sheds light on the role of ARID1A in establishing IPMN precursors in the context of two common genetic alterations in PDAC, Kras and p53.
Gene expression analysis identifies a relationship between ARID1A loss with EMT and stem cell pathways, offering actionable targets for the development of mutation-directed therapy, such as imatinib.
Acknowledgements
We thank Zhong Wang from the Department of Cardiac Surgery at the University of Michigan for providing the Arid1aflox mouse allele. We thank Hao Zhu from the Children’s Medical Center Research Institute at UTSW for providing the Sleeping Beauty transposon system. We greatly appreciate the help from Loralee McMahon and Mary Georger with embedding, sectioning, and histologic staining of experimental tissue.
Funding AFH is supported by The John and Ethel Heselden Professorship, Pancreatic Cancer Association of Western New York, Michael Contestable Golf Tournament, and NCI R01 CA172302. WW and BG are supported by internal funding through the Wilmot Cancer Research Fellowship Program.
ABBREVIATIONS
- ARID1A
AT-rich interaction domain-containing protein 1A
- SWI/SNF
SWItch/Sucrose Non-Fermentable
- PDAC
Pancreatic ductal adenocarcinoma
- ADM
acinar-to-ductal metaplasia
- IPMN
intraductal papillary mucinous neoplasm
- EMT
epithelial-mesenchymal transition
- PanIN
pancreatic intraepithelial neoplasia
- PDGFRb
platelet-derived growth factor receptor beta
- PI3K
phosphoinositide 3-kinase
- ATR
ataxia telangiectasia and Rad3 related
- PARP
poly(ADP-ribose) polymerase
- HDAC6
histone deacetylase 6
- ARID1B
AT-rich interaction domain-containing protein 1B
- Ptf1a
pancreas-specific transcription factor 1a
- IHC
immunohistochemistry
- dpi
days post-injection
- PDX1
pancreas/duodenum homeobox protein 1
- IF
immunofluorescence
- MIST1
muscle intestine and stomach expression 1
- ERα
estrogen receptor α
- MCN
mucinous cystic neoplasms
- GSEA
Gene Set Enrichment Analysis
- PRC2
polycomb repressive complex 2
- qPCR
quantitative polymerase chain reaction
- RNA-Seq
RNA sequencing
- ATAC-Seq
Assay for Transposase-Accessible Chromatin using sequencing
Footnotes
Competing interests None declared.
Provenance and peer review Not commissioned; externally peer reviewed.
Data sharing statement We are willing to share the data related to any of the work reported.
REFERENCES
- 1.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med 2014;371 (11): 1039–49. doi: 10.1056/NEJMra1404198 [published Online First: 2014/09/11] [DOI] [PubMed] [Google Scholar]
- 2.Ying H, Dey P, Yao W, et al. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev 2016;30(4):355–85. doi: 10.1101/gad.275776.115 [published Online First: 2016/02/18] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hingorani SR, Petricoin EF, Maitra A, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 2003;4(6):437–50. [published Online First: 2004/01/07] [DOI] [PubMed] [Google Scholar]
- 4.Hingorani SR, Wang L, Multani AS, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 2005;7(5):469–83. doi: 10.1016/j.ccr.2005.04.023 [published Online First: 2005/05/17] [DOI] [PubMed] [Google Scholar]
- 5.Bailey JM, Hendley AM, Lafaro KJ, et al. p53 mutations cooperate with oncogenic Kras to promote adenocarcinoma from pancreatic ductal cells. Oncogene 2016;35(32):4282–8. doi: 10.1038/onc.2015.441 [published Online First: 2015/11/26] [DOI] [PubMed] [Google Scholar]
- 6.Izeradjene K, Combs C, Best M, et al. Kras(G12D) and Smad4/Dpc4 haploinsufficiency cooperate to induce mucinous cystic neoplasms and invasive adenocarcinoma of the pancreas. Cancer Cell 2007;11(3):229–43. doi: 10.1016/j.ccr.2007.01.017 [published Online First: 2007/03/14] [DOI] [PubMed] [Google Scholar]
- 7.Aguirre AJ, Bardeesy N, Sinha M, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev 2003; 17(24):3112–26 doi: 10.1101/gad.1158703 [published Online First: 2003/12/19] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biankin AV, Waddell N, Kassahn KS, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012;491(7424):399–405. doi: 10.1038/nature11547 [published Online First: 2012/10/30] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shain AH, Giacomini CP, Matsukuma K, et al. Convergent structural alterations define SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proc Natl Acad Sci U S A 2012;109(5):E252–9. doi: 10.1073/pnas.1114817109 [published Online First: 2012/01/12] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Witkiewicz AK, McMillan EA, Balaji U, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun 2015;6:6744. doi: 10.1038/ncomms7744 [published Online First: 2015/04/10] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandler RL, Brennan J, Schisler JC, et al. ARID1a-DNA interactions are required for promoter occupancy by SWI/SNF. Mol Cell Biol 2013;33(2):265–80. doi: 10.1128/MCB.01008-12 [published Online First: 2012/11/07] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Euskirchen G, Auerbach RK, Snyder M. SWI/SNF chromatin-remodeling factors: multiscale analyses and diverse functions. J Biol Chem 2012;287(37):30897–905. doi: 10.1074/jbc.R111.309302 [published Online First: 2012/09/07] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathur R, Alver BH, San Roman AK, et al. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat Genet 2017;49(2):296–302. doi: 10.1038/ng.3744 [published Online First: 2016/12/13] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen J, Peng Y, Wei L, et al. ARID1A Deficiency Impairs the DNA Damage Checkpoint and Sensitizes Cells to PARP Inhibitors. Cancer Discov 2015;5(7):752–67. doi: 10.1158/2159-8290.CD-14-0849 [published Online First: 2015/06/13] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flores-Alcantar A, Gonzalez-Sandoval A, Escalante-Alcalde D, et al. Dynamics of expression of ARID1A and ARID1B subunits in mouse embryos and in cells during the cell cycle. Cell Tissue Res 2011;345(1):137–48. doi: 10.1007/s00441-011-1182-x [published Online First: 2011/06/08] [DOI] [PubMed] [Google Scholar]
- 16.Lei I, Gao X, Sham MH, et al. SWI/SNF protein component BAF250a regulates cardiac progenitor cell differentiation by modulating chromatin accessibility during second heart field development. J Biol Chem 2012;287(29):24255–62. doi: 10.1074/jbc.M112.365080 [published Online First: 2012/05/25] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao X, Tate P, Hu P, et al. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci U S A 2008; 105(18):6656–61. doi: 10.1073/pnas.0801802105 [published Online First: 2008/05/02] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson BG, Wang X, Shen X, et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 2010;18(4):316–28. doi: 10.1016/j.ccr.2010.09.006 [published Online First: 2010/10/19] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bitler BG, Fatkhutdinov N, Zhang R. Potential therapeutic targets in ARID1A-mutated cancers. Expert Opin Ther Targets 2015; 19(11): 1419–22. doi: 10.1517/14728222.2015.1062879 [published Online First: 2015/07/01] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bitler BG, Wu S, Park PH, et al. ARID1A-mutated ovarian cancers depend on HDAC6 activity. Nat Cell Biol 2017; 19(8):962–73. doi: 10.1038/ncb3582 [published Online First: 2017/07/25] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Figura G, Fukuda A, Roy N, et al. The chromatin regulator Brg1 suppresses formation of intraductal papillary mucinous neoplasm and pancreatic ductal adenocarcinoma. Nat Cell Biol 2014;16(3):255–67. doi: 10.1038/ncb2916 [published Online First: 2014/02/25] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roy N, Malik S, Villanueva KE, et al. Brg1 promotes both tumor-suppressive and oncogenic activities at distinct stages of pancreatic cancer formation. Genes Dev 2015;29(6):658–71. doi: 10.1101/gad.256628.114 [published Online First: 2015/03/21] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Genovese G, Carugo A, Tepper J, et al. Synthetic vulnerabilities of mesenchymal subpopulations in pancreatic cancer. Nature 2017;542(7641):362–66. doi: 10.1038/nature21064 [published Online First: 2017/02/09] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khursheed M, Kolla JN, Kotapalli V, et al. ARID1B, a member of the human SWI/SNF chromatin remodeling complex, exhibits tumour-suppressor activities in pancreatic cancer cell lines. Br J Cancer 2013; 108(10):2056–62. doi: 10.1038/bjc.2013.200 [published Online First: 2013/05/11] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan HB, Wang XF, Zhang Q, et al. Reduced expression of the chromatin remodeling gene ARID1A enhances gastric cancer cell migration and invasion via downregulation of Ecadherin transcription. Carcinogenesis 2014;35(4):867–76. doi: 10.1093/carcin/bgt398 [published Online First: 2013/12/03] [DOI] [PubMed] [Google Scholar]
- 26.Ozawa Y, Nakamura Y, Fujishima F, et al. Decreased expression of ARID1A contributes to infiltrative growth of esophageal squamous cell carcinoma. Tohoku J Exp Med 2015;235(3):185–91. doi: 10.1620/tjem.235.185 [published Online First: 2015/03/12] [DOI] [PubMed] [Google Scholar]
- 27.Zhai Y, Kuick R, Tipton C, et al. Arid1a inactivation in an Apc- and Pten-defective mouse ovarian cancer model enhances epithelial differentiation and prolongs survival. J Pathol 2016;238(1):21–30. doi: 10.1002/path.4599 [published Online First: 2015/08/19] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun X, Chuang JC, Kanchwala M, et al. Suppression of the SWI/SNF Component Arid1a Promotes Mammalian Regeneration. Cell Stem Cell 2016;18(4):456–66. doi: 10.1016/j.stem.2016.03.001 [published Online First: 2016/04/06] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawaguchi Y, Cooper B, Gannon M, et al. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet 2002;32(1):128–34. doi: 10.1038/ng959 [published Online First: 2002/08/20] [DOI] [PubMed] [Google Scholar]
- 30.Krapp A, Knofler M, Ledermann B, et al. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev 1998;12(23):3752–63. [published Online First: 1998/12/16] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burlison JS, Long Q, Fujitani Y, et al. Pdx-1 and Ptf1a concurrently determine fate specification of pancreatic multipotent progenitor cells. Dev Biol 2008;316(1):74–86. doi: 10.1016/j.ydbio.2008.01.011 [published Online First: 2008/02/26] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ardito CM, Gruner BM, Takeuchi KK, et al. EGF receptor is required for KRAS-induced pancreatic tumorigenesis. Cancer Cell 2012;22(3):304–17. doi: 10.1016/j.ccr.2012.07.024 [published Online First: 2012/09/15] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roy N, Takeuchi KK, Ruggeri JM, et al. PDX1 dynamically regulates pancreatic ductal adenocarcinoma initiation and maintenance. Genes Dev 2016;30(24):2669–83. doi: 10.1101/gad.291021.116 [published Online First: 2017/01/15] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lo HG, Jin RU, Sibbel G, et al. A single transcription factor is sufficient to induce and maintain secretory cell architecture. Genes Dev 2017;31(2):154–71. doi: 10.1101/gad.285684.116 [published Online First: 2017/02/09] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang M, Azevedo-Pouly A, Deering TG, et al. MIST1 and PTF1 Collaborate in Feed-forward Regulatory Loops that Maintain the Pancreatic Acinar Phenotype in Adult Mice. Mol Cell Biol 2016. doi: 10.1128/MCB.00370-16 [published Online First: 2016/09/21] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanda M, Matthaei H, Wu J, et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology 2012;142(4):730–33 e9. doi: 10.1053/j.gastro.2011.12.042 [published Online First: 2012/01/10] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amato E, Molin MD, Mafficini A, et al. Targeted next-generation sequencing of cancer genes dissects the molecular profiles of intraductal papillary neoplasms of the pancreas. J Pathol 2014;233(3):217–27. doi: 10.1002/path.4344 [published Online First: 2014/03/08] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson EL, Willis N, Mercer K, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 2001;15(24):3243–8. doi: 10.1101/gad.943001 [published Online First: 2001/12/26] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jonkers J, Meuwissen R, van der Gulden H, et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet 2001;29(4):418–25. doi: 10.1038/ng747 [published Online First: 2001/11/06] [DOI] [PubMed] [Google Scholar]
- 40.Furukawa T, Kloppel G, Volkan Adsay N, et al. Classification of types of intraductal papillary-mucinous neoplasm of the pancreas: a consensus study. Virchows Arch 2005;447(5):794–9. doi: 10.1007/s00428-005-0039-7 [published Online First: 2005/08/10] [DOI] [PubMed] [Google Scholar]
- 41.Adsay V, Mino-Kenudson M, Furukawa T, et al. Pathologic Evaluation and Reporting of Intraductal Papillary Mucinous Neoplasms of the Pancreas and Other Tumoral Intraepithelial Neoplasms of Pancreatobiliary Tract: Recommendations of Verona Consensus Meeting. Ann Surg 2016;263(1):162–77. doi: 10.1097/SLA.0000000000001173 [published Online First: 2015/03/17] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vleminckx K, Vakaet L Jr., Mareel M, et al. Genetic manipulation of E-cadherin expression by epithelial tumor cells reveals an invasion suppressor role. Cell 1991;66(1): 107–19. [published Online First: 1991/07/12] [DOI] [PubMed] [Google Scholar]
- 43.Sun X, Wang SC, Wei Y, et al. Arid1a Has Context-Dependent Oncogenic and Tumor Suppressor Functions in Liver Cancer. Cancer Ce11 2017;32(5):574–89 e6. doi: 10.1016/j.ccell.2017.10.007 [published Online First: 2017/11/15] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pietras K, Sjoblom T, Rubin K, et al. PDGF receptors as cancer drug targets. Cancer Cell 2003;3(5):439–43. [published Online First: 2003/06/05] [DOI] [PubMed] [Google Scholar]
- 45.Tu Z, Zhuang X, Yao YG, et al. BRG1 is required for formation of senescence-associated heterochromatin foci induced by oncogenic RAS or BRCA1 loss. Mol Cell Biol 2013;33(9):1819–29. doi: 10.1128/MCB.01744-12 [published Online First: 2013/02/27] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He L, Chen Y, Feng J, et al. Cellular senescence regulated by SWI/SNF complex subunits through p53/p21 and p16/pRB pathway. Int J Biochem Cell Biol 2017;90:29–37. doi: 10.1016/j.biocel.2017.07.007 [published Online First: 2017/07/19] [DOI] [PubMed] [Google Scholar]
- 47.Li ZY, Zhu SS, Chen XJ, et al. ARID1A suppresses malignant transformation of human pancreatic cells via mediating senescence-associated miR-503/CDKN2A regulatory axis. Biochem Biophys Res Commun 2017. doi: 10.1016/j.bbrc.2017.09.099 [published Online First: 2017/09/25] [DOI] [PubMed] [Google Scholar]
- 48.Kimura Y, Fukuda A, Ogawa S, et al. ARID1A Maintains Differentiation of Pancreatic Ductal Cells and Inhibits Development of Pancreatic Ductal Adenocarcinoma in Mice. Gastroenterology 2018. doi: 10.1053/j.gastro.2018.03.039 [published Online First: 2018/04/01] [DOI] [PubMed] [Google Scholar]
- 49.De La OJ, Emerson LL, Goodman JL, et al. Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc Natl Acad Sci U S A 2008; 105(48):18907–12. doi: 10.1073/pnas.0810111105 [published Online First: 2008/11/26] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Habbe N, Shi G, Meguid RA, et al. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc Natl Acad Sci U S A 2008; 105(48):18913–8. doi: 10.1073/pnas.0810097105 [published Online First: 2008/11/26] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferreira RMM, Sancho R, Messal HA, et al. Duct- and Acinar-Derived Pancreatic Ductal Adenocarcinomas Show Distinct Tumor Progression and Marker Expression. Cell Rep 2017;21(4):966–78. doi: 10.1016/j.celrep.2017.09.093 [published Online First: 2017/10/27] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mallen-St Clair J, Soydaner-Azeloglu R, Lee KE, et al. EZH2 couples pancreatic regeneration to neoplastic progression. Genes Dev 2012;26(5):439–44. doi: 10.1101/gad.181800.111 [published Online First: 2012/03/07] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ben-Porath I, Thomson MW, Carey VJ, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 2008;40(5):499–507. doi: 10.1038/ng.127 [published Online First: 2008/04/30] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jordan NV, Prat A, Abell AN, et al. SWI/SNF chromatin-remodeling factor Smarcd3/Baf60c controls epithelial-mesenchymal transition by inducing Wnt5a signaling. Mol Cell Biol 2013;33(15):3011–25. doi: 10.1128/MCB.01443-12 [published Online First: 2013/05/30] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simbolo M, Vicentini C, Ruzzenente A, et al. Genetic alterations analysis in prognostic stratified groups identified TP53 and ARID1A as poor clinical performance markers in intrahepatic cholangiocarcinoma. Sci Rep 2018;8(1):7119. doi: 10.1038/s41598-018-25669-1 [published Online First: 2018/05/10] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang X, Lee RS, Alver BH, et al. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat Genet 2017;49(2):289–95. doi: 10.1038/ng.3746 [published Online First: 2016/12/13] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hwang RF, Yokoi K, Bucana CD, et al. Inhibition of platelet-derived growth factor receptor phosphorylation by STI571 (Gleevec) reduces growth and metastasis of human pancreatic carcinoma in an orthotopic nude mouse model. Clin Cancer Res 2003;9(17):6534–44. [published Online First: 2003/12/26] [PubMed] [Google Scholar]
- 58.Weissmueller S, Manchado E, Saborowski M, et al. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor beta signaling. Cell 2014; 157(2):382–94. doi: 10.1016/j.cell.2014.01.066 [published Online First: 2014/04/15] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buenrostro JD, Giresi PG, Zaba LC, et al. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 2013;10(12):1213–8. doi: 10.1038/nmeth.2688 [published Online First: 2013/10/08] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berens EB, Holy JM, Riegel AT, et al. A Cancer Cell Spheroid Assay to Assess Invasion in a 3D Setting. J Vis Exp 2015(105) doi: 10.3791/53409 [published Online First: 2015/12/10] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.