Abstract
Hepatocyte Nuclear Factor 4 alpha (HNF4α) is critical for hepatic differentiation. Recent studies have highlighted its role in inhibition of hepatocyte proliferation and tumor suppression. However, the role of HNF4α in liver regeneration is not known. We hypothesized that hepatocytes modulate HNF4α activity when navigating between differentiated and proliferative states during liver regeneration. Western blot analysis revealed a rapid decline in nuclear and cytoplasmic HNF4α protein levels accompanied with decreased target gene expression within 1 hour after 2/3 partial hepatectomy (post-PH) in C57BL/6J mice. HNF4α protein expression did not recover to the pre-PH levels until day 3. Hepatocyte-specific deletion of HNF4α (HNF4α-KO) in mice resulted in 100% mortality post-PH despite increased proliferative marker expression throughout regeneration. Sustained loss of HNF4α target gene expression throughout regeneration indicated HNF4α-KO mice were unable to compensate for loss of HNF4α transcriptional activity. Deletion of HNF4α resulted in sustained proliferation accompanied by c-Myc and Cyclin D1 over expression and a complete deficiency of hepatocyte function after PH. Interestingly, overexpression of degradation-resistant HNF4α in hepatocytes delayed but did not prevent initiation of regeneration after PH. Finally, AAV8-mediated reexpression of HNF4α in hepatocytes of HNF4α-KO mice post-PH restored HNF4α protein levels, induced target gene expression and improved survival of HNF4α-KO mice post-PH. In conclusion, these data indicate that HNF4α reexpression following initial decrease is critical for hepatocytes to exit from cell cycle and resume function during the termination phase of liver regeneration. These results reveal the role of HNF4α in liver regeneration and have implications for therapy of liver failure.
Introduction
HNF4α is considered the master regulator of hepatocyte differentiation because of its essential role in embryonic development (1, 2), stabilizing the hepatic transcription factor network (3) and maintaining hepatocyte function (4). HNF4α regulates genes involved in xenobiotic metabolism, carbohydrate metabolism, fatty acid metabolism, bile acid synthesis, blood coagulation, and ureagenesis (5). Expression of HNF4α induces hepatocyte-like characteristics in induced pluripotent stem cells (6) and forced expression of HNF4α induces differentiation and decrease cancer progression in hepatocellular carcinomas (HCC) (7, 8). Furthermore, decreased HNF4α expression leads to loss of hepatic function and causes decompensation in cirrhotic rats (9).
Recent studies from our laboratory and others have revealed anti-proliferative properties of HNF4α (10, 11) in the liver. Hepatocyte-specific deletion of HNF4α (HNF4α-KO) in mice results in spontaneous hepatocyte proliferation (10, 11) and promotes formation of diethylnitrosamine-induced HCCs (12).
Liver has a remarkable capacity to regenerate upon surgical resection and following viral or drug-induced liver injury. During liver regeneration after 2/3 partial hepatectomy (PH), the most widely used model to study liver regeneration, multiple redundant mechanisms regulate initiation and termination of hepatocyte regeneration (13). Understanding the mechanisms that govern adult hepatocytes to navigate between quiescent and proliferative states could result in therapeutic targets for inducing hepatocyte proliferation during impaired regeneration or inhibiting excess proliferation during carcinogenesis. Despite its role in maintaining hepatocyte differentiation and quiescence, little is known about the role of HNF4α in hepatocyte regeneration or how decreased HNF4α, a condition commonly found in diseased human livers (9, 14–16), would impact regeneration. In this study, we investigated the role of HNF4α in regulation of liver regeneration after PH using wildtype (WT) and HNF4α-KO mice. Our studies revealed that HNF4α is indispensable for survival after PH and a critical component of termination of liver regeneration.
Materials and Methods
Animal Care and Surgeries.
HNF4α-floxed mice were injected intraperitoneally with AAV8-TBG-eGFP or AAV8-TBG-CRE to generate WT and hepatocyte-specific HNF4α-KO animals, respectively. These vectors were purchased from Penn Vector Core (Philadelphia, PA) and injected as previously described (18). PH surgeries were performed on male C57BL/6J mice or HNF4α WT and KO mice and tissue samples obtained as previously described (17). Additional details are in supplementary materials.
Protein Isolation, Western blotting and Real Time PCR.
Protein isolation and Western blotting was performed as described in detailed before (20). RNA isolation, conversion to cDNA and Real time PCR analysis was performed as previously described (21).
RNA Sequencing and Ingenuity Pathway Analysis.
Equal amounts of RNA was pooled using WT and HNF4α-KO livers (n=3) from day 5 and day 7 post-PH to be used for RNA-Seq and Ingenuity Pathway Analysis (IPA). RNA sequencing and bioinformatics analysis were performed as previously described in detail (12). Additional details are in supplementary materials.
Statistical Analysis
Results are expressed as mean ± standard error. One Way ANOVA and Student’s t test was applied to all analyses with p < 0.05 being considered significant.
Detailed materials and methods are included in the supplementary materials.
Results
Decreased HNF4α Protein Expression and Transcriptional Activity During Initiation of Regeneration After Partial Hepatectomy.
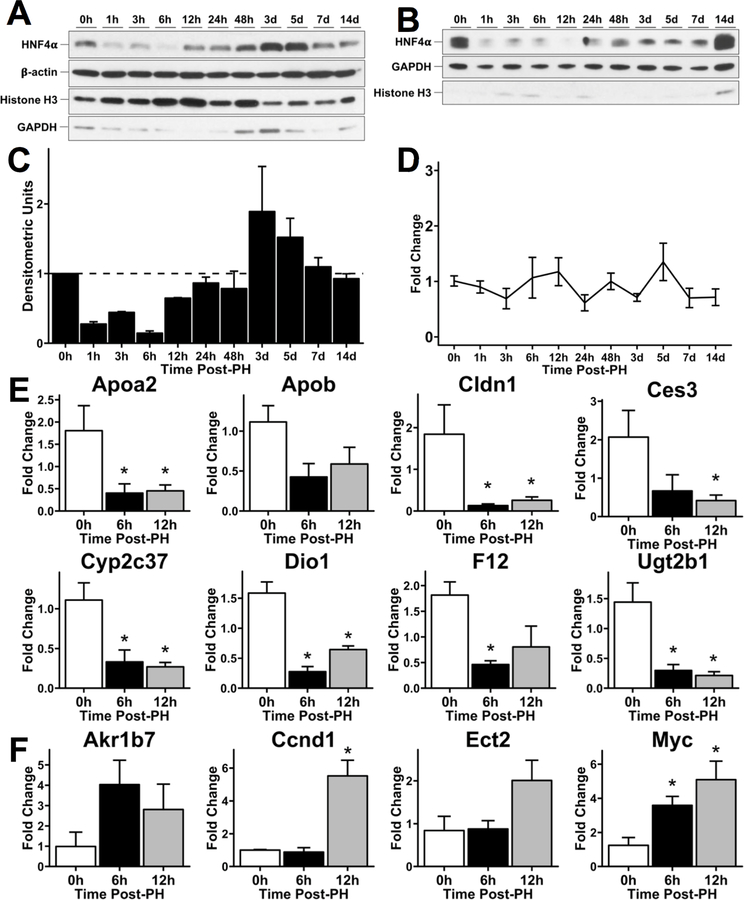
We determined changes in expression and activity of HNF4α in C57BL/6J mice over a time course after PH using Western blotting in freshly prepared nuclear (Fig. 1A, 1C) and cytoplasmic (Fig. 1B) protein extracts in. Nuclear HNF4α levels started declining at 1 (hour) hr post-PH reaching the lowest level at 6 hr post-PH. Nuclear HNF4α protein expression started increasing at 12 hr, increased higher than the 0 hr levels peaking at 3 days post PH and then declined again to reach the 0 hr levels by 7 days post-PH (Fig. 1A, 1C). Cytoplasmic HNF4α expression continually declined below 0 hr levels until 12h post-PH, then started to rise but did not return to 0 hr pre-PH levels till 14 days after PH. The expression of adult isoform of HNF4α mRNA did not change over the entire regeneration time course (Fig. 1D).
Fig. 1.
Decreased HNF4α protein expression and transcriptional activity during initiation of regeneration after partial hepatectomy. Western blot analysis of HNF4α adult isoform in (A) nuclear and (B) cytoplasmic lysates from mouse liver over a time course of 0 – 14 days after 2/3 PH. (C) Densitometric analysis of nuclear HNF4α blot. (D) qPCR analysis of HNF4α adult isoform mRNA over a time course of 0 – 14 days after 2/3 PH. Fold change calculated by comparison to 0 hr time point. qPCR analysis of positively regulated (E) and negatively regulated (F) HNF4α target genes at 0, 6 and 12 hr post-PH. Fold change calculated by comparison to 0 hr time point. *indicate significant difference at P ≤ 0.05.
We assessed HNF4α transcriptional activity by qPCR analysis of positive (12) and negative target genes of HNF4α (22) at the 0, 6 and 12 hr after PH. We observed decreased expression of positive target genes (APOA2, APOB, CES3, CLDN1, CYP2C37, DIO1, F12, UGT2B1) at the 6 and 12-hr time points compared to 0 hr expression levels (Fig. 1E). qPCR analysis showed induction of HNF4α negative target genes (AKR1B7, CCND1, ECT2, MYC) at the 6 and 12 hr time points (Fig. 1F). Together, these data indicate a decrease in HNF4α activity at the onset of regeneration which is consistent with the changes we observed in HNF4α protein expression (Fig. 1A-C).
Role of Src kinase in regulation of HNF4α expression after PH
Since nuclear HNF4α protein levels decreased during initiation of liver regeneration without changes to HNF4α mRNA expression, we hypothesized that the decrease in nuclear HNF4α was caused by a post-translational regulation of HNF4α protein. Src kinase has been shown to phosphorylate HNF4α leading to its cytoplasmic translocation and degradation (23). To test whether the rapid decrease in HNF4α after PH is associated with Src-mediated phosphorylation, we treated C57BL/6J mice with the Src inhibitor, PP2, before undergoing PH. Mice were sacrificed 6 hours post-PH. This timepoint was selected because this is when we observed the lowest levels of nuclear HNF4α in the WT time course. Western blot analysis showed that nuclear HNF4α protein was higher in mice treated with PP2 (Supp. Fig. 1A). Cytoplasmic HNF4α was not noticeably different in PP2 treated mice compared to Vehicle treated mice. Densitometry for these blots confirms these conclusions (Supp. Fig. 1B and 1C).
HNF4α Overexpression Delays but Does Not Prevent Hepatocyte Proliferation During Liver Regeneration
We used a Tet-On-HNF4α transgenic mouse system (24) to overexpress HNF4α in hepatocytes to test if the anti-proliferative effects of HNF4α would prevent hepatocyte proliferation during the initiation of liver regeneration (Supp. Fig. 2). Western blot analysis confirmed increased HNF4α in nuclear lysates from livers of doxycycline (Dox)-treated Tet-On-HNF4α mice at 6 hours post-PH (Supp. Fig. 3A). This resulted in fewer PCNA-positive nuclei in Dox-treated mice 6 hours post-PH and increased, although statistically similar, levels of proliferation at 48 hours post-PH. (Supp. Fig. 3B, 3C). Interestingly, Dox treatment inhibited the occurrence of transient steatosis 48 hours post-PH (Supp. Fig. 3D). qPCR analysis of HNF4α target genes demonstrated correlation between HNF4α overexpression and increased HNF4α activity. Increased expression of (Supp. Fig. 3E) HNF4α positive target genes (ALAS2, APOA2, CYP2C37, CPT1, HNF4A, UGT2B1) and decreased expression of (Supp. Fig. 2F) HNF4α negative target genes (CCND1, CDKN3, EGR1) was observed in DOX treated mice before and after PH.
Hepatocyte-Specific HNF4α-KO Mice Do Not Survive After PH.
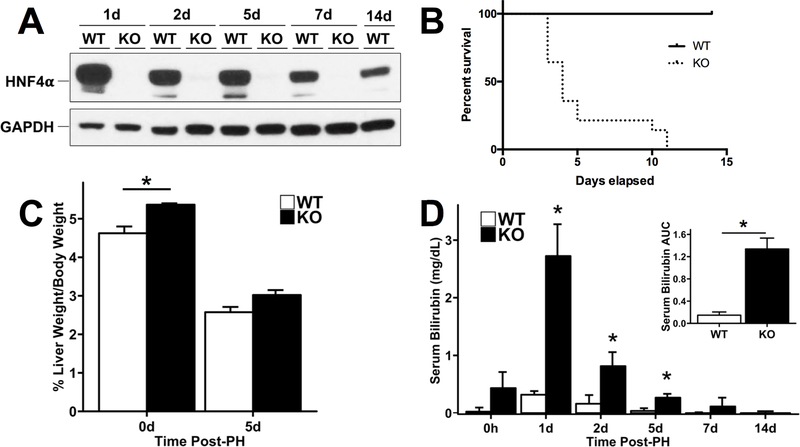
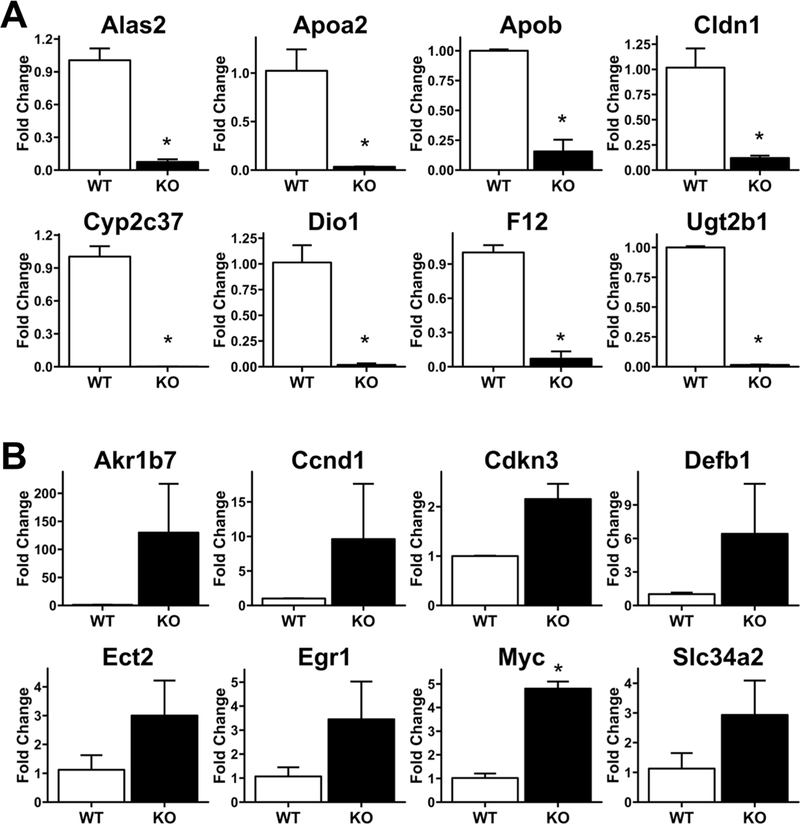
Next, we investigated the effect of hepatocyte-specific HNF4α deletion on liver regeneration after PH. WT and HNF4α-KO mice underwent PH and were euthanized at 1, 2, 5, 7 and 14 days after surgery. HNF4α protein remained undetectable in HNF4α-KO mice throughout the time course after PH (Fig 2A). We observed 100% mortality in the HNF4α-KO group by day 11 post-PH (Fig. 2B). Interestingly, while liver to body weight ratios were significantly higher in HNF4α-KO mice before PH, the recovery of liver weight was similar between WT and HNF4α-KO groups until 5 days post PH (Fig. 2C). Serum bilirubin was significantly elevated in HNF4α-KO mice at days 1, 2 and 5 post-PH (Fig. 2D). While serum bilirubin in HNF4α-KO animals did decrease over time, this assay could only be performed on surviving mice and most HNF4α-KO mice died before bilirubin could be measured at later time points. Serum bilirubin values were significantly higher in HNF4α-KO mice when summarized as the area under the curve (AUC) for the entire time course after PH. We investigated changes in HNF4α target gene expression in WT and HNF4α-KO mice at 7 days after PH, which revealed a significant decrease in expression of positive target genes including ALAS2, APOA2, APOB, CLDN1, CYP2C37, DIO1, F12 and UGT2B1 (Fig 3A). Similarly, the expression of the negative targets AKR1B7, CCND1, CDKN3, DEFB1, ECT2, EGR1, MYC and SLC34A2 was increased at 7 days post-PH (Fig. 3B).
Fig. 2.
Complete mortality of HNF4α-KO mice following PH. (A) Western blot of HNF4α confirming efficient KO of HNF4α in pooled liver lysates at all time points post-PH. (B) Kaplan-Meier survival analysis of WT and HNF4α-KO groups after PH. (C) Liver weight to body weight ratios and (D) serum bilirubin levels in WT and HNF4α-KO mice after PH for each time point after PH. Inset represents the area under the curve (AUC) values for bilirubin presented in the main panel. *indicate significant difference at P ≤ 0.05 between WT and HNF4α-KO.
Fig. 3.
Sustained Loss of HNF4α Transcriptional Activity In HNF4α-KO Mice 7 Days Post-PH. qPCR analysis of mRNA isolated from frozen liver in WT and HNF4α-KO mice 7 days post-PH. (A) Decreased expression of positive targets of HNF4α and (B) increased expression of negative targets of HNF4α. *indicate significant difference at P ≤ 0.05 between WT and HNF4α-KO.
Increased Hepatocyte Proliferation in HNF4α-KO Livers Throughout Regeneration.
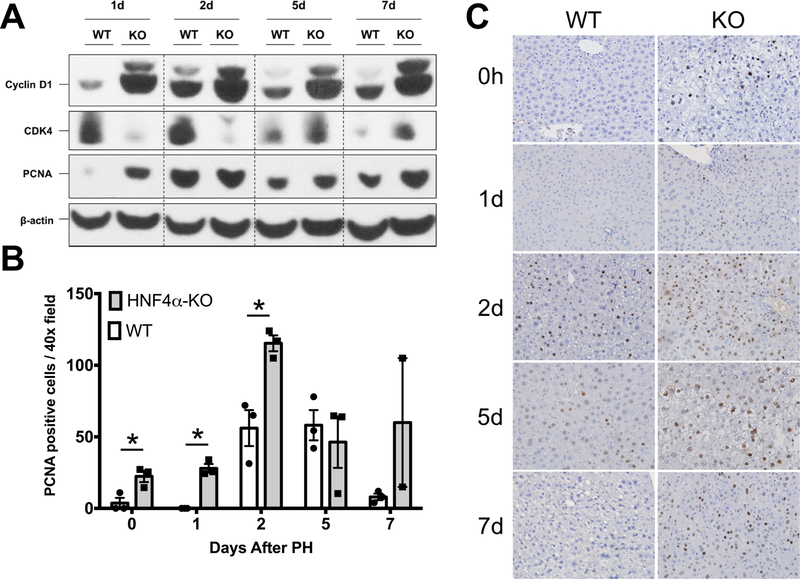
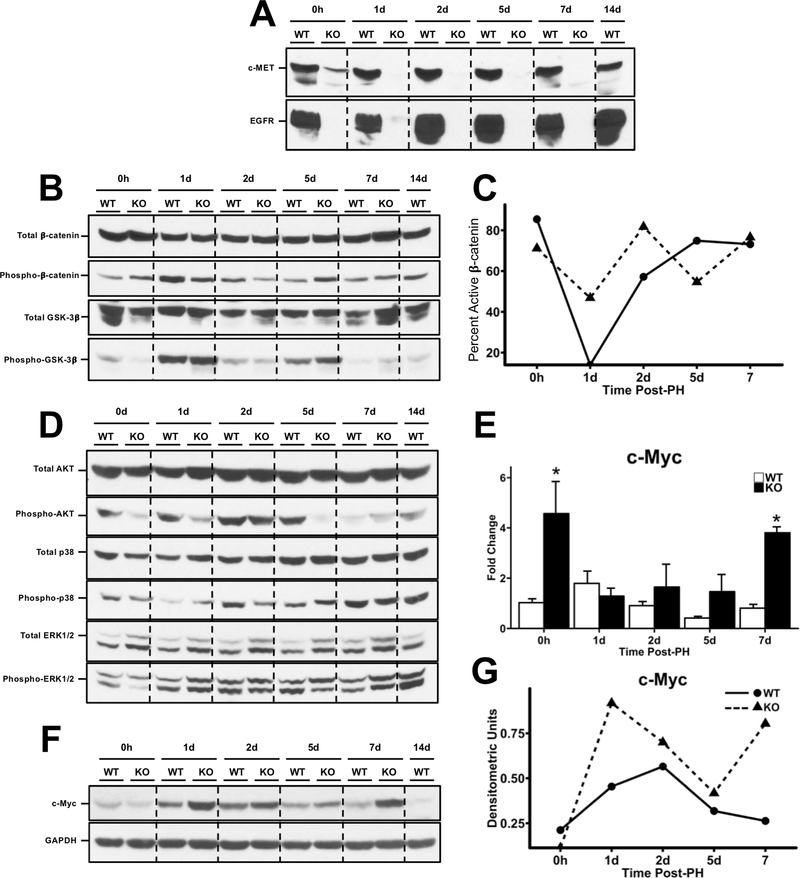
Western blot analysis revealed significant increase in Cyclin D1 protein expression in HNF4α-KO livers at all time points after PH. The expression of CDK4, the cyclin dependent kinase interacting with Cyclin D1 was lower in HNF4α-KO liver at days 1 and 2 post PH but increased to levels comparable to WT mice by days 5 post PH. Cell proliferation assessed by Western blot and immunohistochemical analysis of PCNA revealed an elevated PCNA level in HNF4α-KO mice at all time points (Fig. 4A). Immunohistochemical analysis of PCNA-positive nuclei per 40x field was significantly higher in HNF4α-KO mice throughput the 7-day time course post PH (Fig. 4B-C).
Fig. 4.
Increased hepatocyte proliferation in HNF4α-KO livers throughout regeneration. (A) Western blot analysis of Cyclin D1, CDK4, and PCNA over a time course of 0 to 7 days post-PH. (B) Quantification of immunohistochemical analysis for PCNA positive nuclei in liver sections of WT and HNF4α-KO mice throughout time course post-PH. Bars represent means ± SEM for each group and values from individual mice indicated by black points. (C) Representative photomicrographs (400x) of PCNA-stained liver sections from WT and HNF4α-KO mice throughout time course post-PH. *indicate significant difference at P ≤ 0.05 between WT and HNF4α-KO.
Activation of Pro-Proliferative Signaling in HNF4α-KO Mice During Regeneration
We investigated the mechanism of sustained hepatocyte proliferation throughout regeneration in HNF4α-KO mice by Western blot analysis of pathways commonly involved in hepatocyte proliferation (12, 25). Most interestingly, we observed complete loss of total EGFR and total c-MET protein expression in HNF4α-KO animals at all time points throughout regeneration (Fig. 5A). Interestingly, it has been shown that signaling through these receptors is required for proliferation to occur after PH (25). Next, we examined the Wnt/β-catenin pathway (Fig. 5B). Total β-catenin levels were similar in WT and HNF4α-KO mice did not change throughout the time course. However, inactive (Thr41/Ser45- phosphorylated) β-catenin was elevated in the WT animals at days 1 and 2 post-PH. Phosphorylated β-catenin was greater in HNF4α-KO compared to WT mice 5 days post-PH. Densitometric analysis of inactive to total β-catenin ratio suggested activation of β-catenin was higher in HNF4α-KO mice at 1 and 2 days post-PH (Fig. 5C). Whereas ser-9 phosphorylated GSK-3β increased after PH, no difference was observed between WT or HNF4α-KO mice over the time course in either total of phospho-GSK3β. Next, we investigated activation of several MAPK members including AKT, p38 and ERK1/2 (Fig. 5D). No difference in total AKT and total p38 protein levels was observed between groups throughout regeneration. Phosphorylation of AKT was inhibited in the HNF4α-KO mice as compared to WT at days 0, 1 and 5 post-PH but was similar to WT at day 2 after PH. Phosphorylation (activation) of p38 was elevated in HNF4α-KO mice at days 1 and 5 post-PH. Finally, Total ERK1/2 expression was elevated in HNF4α-KO animals at all time points. Phosphorylation (activation) of ERK1 (upper band) was increased in HNF4α-KO animals compared to WT at all time points. Phosphorylation (activation) of ERK2 (lower band) was inhibited in HNF4α-KO group compared to WT at days 0 and 5 post-PH. ERK2 phosphorylation was elevated in HNF4α-KO group compared to WT at day 7 post-PH. Finally, we measured mRNA expression of known regulator of hepatocyte proliferation and negative target of HNF4α, MYC, which was elevated throughout regeneration (Fig. 5E). While c-Myc protein expression was similar between WT and HNF4α-KO animals at day 0, c-Myc expression was elevated in HNF4α-KO animals at all time points post-PH (Fig 5F, 5G).
Fig. 5.
Activation of pro-proliferative signaling in HNF4α-KO mice during regeneration. Western blot analysis of (A) total EGFR and c-Met, (B) total and phosphorylated β-catenin and total and phosphorylatedGSK3β, (C) line graphs showing densitometric signal of inactive to total β-catenin, (D) Western blots for Total AKT, Phospho-AKT, Total p38, Phospho-p38, ERK1/2, Phospho-ERK1/2, (E) qPCR analysis of c-MYC mRNA from WT and HNF4α-KO livers over time course post-PH, (F) Western blot analysis of c-Myc, (G) and densitometric analysis of c-Myc. *indicate significant difference at P ≤ 0.05between WT and HNF4α-KO.
RNAseq revealed sustained increase in pro-proliferative and anti-differentiation signaling in HNF4α-KO Mice Post-PH
To gain insights in the comprehensive signaling changes in HNF4α-KO livers after PH, we performed RNA-Seq analysis at days 2 and 5 post-PH. These time points were selected to match the peak hepatocyte proliferation (day 2 post PH) and early termination phase of regeneration (day 5 post PH) in the liver regeneration process. Overall, a similar number of genes (~1,000) increased and decreased at day 2 and day 5 after PH in HNF4α-KO mice as compared to WT mice (Supp. Table 3). An upstream regulator analysis was performed using Ingenuity Pathway Analysis (IPA) to identify activation and inhibition of key transcription factors based on gene expression changes. At 2 days post-PH (Table 1), activation of proinflammatory (EGR1, IRF3, IRF6, IRF7) and pro-proliferative transcription factors (JUN, RB1, P300, TCF3) in HNF4α-KO liver was observed. Interestingly, HNF4α-KO also showed activation of TGF-β signaling as indicated by activation of SMAD2 and SMAD4 as well as inhibition of SMAD7 (26). The transcription factors predicted to be inhibited 2 days post-PH included HNF4α, HNF1A and the HNF4α coactivator PPARGC1A (PGC1A). Furthermore, Estrogen Receptor α (Esrra), which is known to regulate gene expression in coordination with HNF4α and PGC1A (27), was also predicted to be inhibited. TAF4, a known HNF4α cofactor which is required for HNF4α transcriptional activity (28), was also inhibited. MED1, which is essential for PPARα activity and required for survival after PH (29), was inhibited in HNF4α-KO 2 days post-PH. ZBTB20, a known suppressor of hepatocyte proliferation and repressor of alpha-fetoprotein (30, 31), was predicted to be inhibited.
Table 1:
Predicted Activity of Transcription Factors in HNF4α-KO Mice Compared to WT Mice 2 Days Post-PH.
| Day 2 Post-PH | |||
|---|---|---|---|
| Activated Pathways | Activation z-score | Inhibited Pathways | Activation z-score |
| JUN | 3.31 | HNF4A | −6.498 |
| EGR1 | 2.814 | HNF1A | −4.428 |
| RB1 | 2.497 | Esrra | −3.357 |
| IRF3 | 2.42 | PPARGC1A | −3.228 |
| FOXO4 | 2.377 | NFIX | −2.433 |
| EP300 | 2.305 | ZBTB20 | −2.294 |
| SPI1 | 2.248 | MED1 | −2.272 |
| THRAP3 | 2.219 | CLOCK | −2.219 |
| IRF6 | 2.215 | NFIC | −2.2 |
| SMAD4 | 2.201 | GATA2 | −2.059 |
| MTA1 | 2.194 | NKX2–3 | −2.04 |
| Gm21596/Hmgb1 | 2.169 | E2F1 | −2.027 |
| IRF7 | 2.163 | SMAD7 | −2.027 |
| SMAD2 | 2.068 | TAF4 | −2.008 |
| TCF3 | 2.022 | ||
At day 5 post-PH (Table 2), the analysis predicted activation of transcription factor HMGB1 in HNF4α-KO mice indicating sustained inflammatory signaling in HNF4α-KO mice throughout regeneration. The TGFβ effector SMAD2 was also activated at day 5 post-PH. Proliferative marker EP300 was activated at day 5 post-PH. The HNF4α negative target gene and proliferative marker CCND1 was activated in the HNF4α-KO group at day 5 post-PH indicated sustained proliferative signaling compared to WT mice. Activation of SNAI2 was predicted in the HNF4α-KO group at 5 days post-PH. SNAI2 is repressed by HNF4α and is known to promote EMT (32). PLAG1 is a fetal gene overexpressed in hepatoblastomas (33) and was predicted to be activated in the HNF4α-KO group at day 5. Activation of TRIM24, a transcription factor with oncogenic activity (34), was predicted at day 5 post PH in the HNF4α-KO mice. Transcription factors including HNF4A, HNF1A, PPARGC1A, and Esrra were inhibited at day 2 post-PH and continued to be inhibited at day 5 post-PH. Inhibition of SIRT2 was observed at 5 days post-PH in HNF4α-KO mice consistent with its known role in sharing many of the same target genes as HNF4α(35). PPARGC1B is a known coactivator of PPARGC1A and was inhibited. NCOA2 and EBF1, known tumor suppressors(36, 37), were inhibited in HNF4α-KO mice day 5 post-PH. NFIX, a transcription factor known to inhibit development of HCC(38), was inhibited. Finally, the master regulators of sterol and lipid metabolism, SREBF1 and SREBF2, were inhibited 5 days post-PH.
Table 2:
Predicted Activity of Transcription Factors in HNF4α-KO Mice Compared to WT Mice 5 Days Post-PH.
| Day 5 Post-PH | |||
|---|---|---|---|
| Activated Pathways | Activation z-score | Inhibited Pathways | Activation z-score |
| Gm21596/Hmgb1 | 2.767 | HNF4A | −6.471 |
| EP300 | 2.565 | HNF1A | −4.641 |
| TRIM24 | 2.554 | SREBF2 | −4.603 |
| SMAD2 | 2.401 | SREBF1 | −3.768 |
| CCND1 | 2.307 | PPARGC1A | −3.045 |
| SNAI2 | 2.1 | Esrra | −2.887 |
| ANKRD42 | 2 | IRF7 | −2.476 |
| PLAG1 | 2 | SIRT2 | −2.449 |
| NFIX | −2.433 | ||
| PPARGC1B | −2.424 | ||
| BCL6 | −2.229 | ||
| EBF1 | −2.177 | ||
| ZEB1 | −2.077 | ||
| NCOA2 | −2.028 | ||
IPA analysis also predicted activity of diseases and organ function based on gene expression differences between WT and HNF4α-KO mice at 2 and 5 days post-PH. The comparison at day 2 post-PH (Supp. Table 4) predicted HNF4α-KO mice would exhibit activation of pathways involving inflammation, tumorigenesis and wound healing. Inhibited functions included numerous basic liver processes such as transport, metabolism and synthesis of cholesterol, lipids, bile acids and xenobiotics. This pattern continued at day 5 post-PH (Supp. Table 5). Activated functions continued to be related to cell proliferation, tumorigenesis and inflammation. Additionally, functions related to embryonic organ tissue development were activated in the HNF4α-KO group. Functions related to transport and metabolism of lipids, cholesterol and vitamins remained inhibited in HNF4α-KO mice 5 days post-PH.
Reexpression of HNF4α Restores Hepatocyte Quiescence and Gene Expression and Extends Survival of HNF4α-KO Animals Post-PH
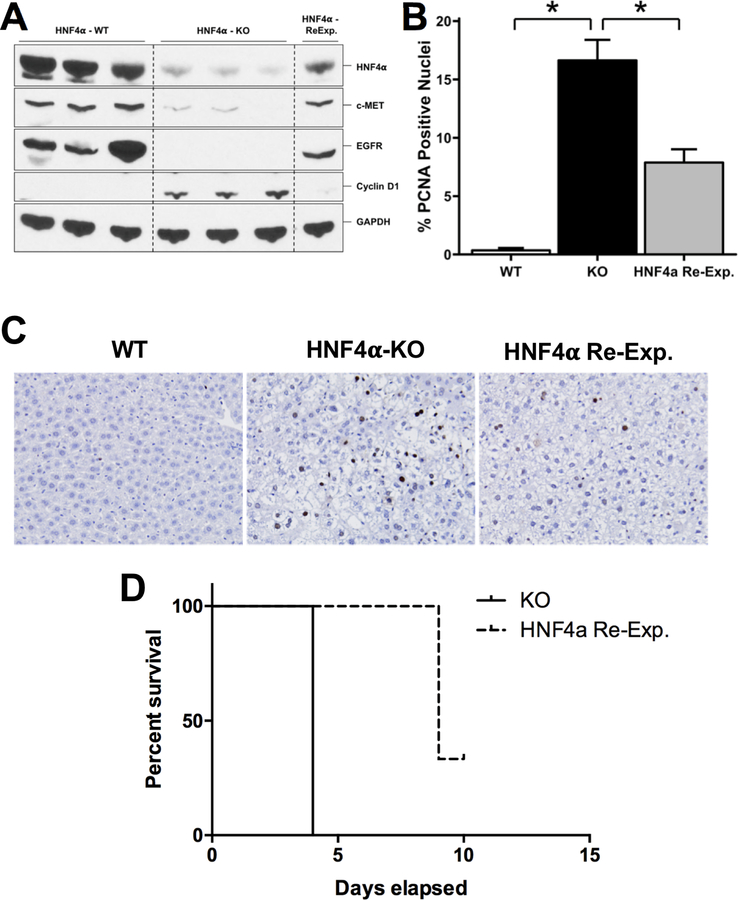
Finally, we tested if reexpression of HNF4α in hepatocytes by intravenous injection of AAV8-CMV-HNF4α could rescue HNF4α-KO mice after PH by restoring HNF4α transcriptional activity after cell division. First, we reexpressed HNF4α in HNF4α-KO mice and measured HNF4α target gene expression. HNF4α reexpression increased total c-MET and total EGFR protein expression to WT levels and reduced cyclin D1 expression to WT levels (Fig. 6A). Hepatocyte proliferation was assessed by immunohistochemical analysis of PCNA positive nuclei. High levels of hepatocyte proliferation were observed in the HNF4α-KO group. HNF4α reexpression resulted in decreased hepatocyte proliferation compared to HNF4α-KO mice (Fig. 6B,C). Finally, we reexpressed HNF4α in HNF4α-KO mice 2 days post-PH. This treatment improved survival compared to HNF4α-KO mice (Fig. 6D).
Fig. 6.
Reexpression of HNF4α restores hepatocyte quiescence and gene expression and extends survival of HNF4α-KO animals post-PH. (A) Western blot analysis of HNF4α, Cyclin D1, c-Met and EGFR in WT, HNF4α-KO and HNF4α-Reexp. mice, (B) Immunohistochemical analysis of PCNA positive nuclei in liver sections from WT, HNF4α-KO and HNF4α-Reexp. mice, (C) Representative photomicrographs (40x) of PCNA-stained liver sections, (D) Kaplan-Meier survival analysis of WT and HNF4α-Reexp mice after PH. * indicate significant difference at P ≤ 0.05.
Discussion
The role of HNF4α in hepatocyte differentiation is well known (3–5) and recent studies have revealed its anti-proliferative effects in hepatocytes (12, 15, 16) but its role in liver regeneration is not known. We investigated the role of HNF4α in liver regeneration after PH, where adult hepatocytes exit quiescence, enter cell cycle, and proliferate before returning to differentiated, quiescent state upon completion of regeneration. Based on its anti-proliferative effects, we hypothesized HNF4α expression and function would decrease during the initiation of regeneration. Indeed, we observed decreased nuclear and cytoplasmic levels of HNF4α occurring within hours after surgery. This decreased protein expression was not due to changes in transcription of the HNF4α gene because there were no changes in HNF4α mRNA post-PH. We also observed decreased HNF4α activity as measured by its target gene expression(12). Our observations independently reproduce a microarray study describing decreased HNF4α target gene expression 4 hours post-PH (43). This highlights functional differences between quiescent and proliferating hepatocytes and suggests restoring HNF4α in hepatocytes responding to chronic injury could successfully restore hepatic function (9, 44). These studies are consistent with previous observation of decreased periportal HNF4α staining after PH (39). The mechanism behind this rapid decrease in HNF4α expression remains to be studied. Posttranslational modifications (PTMs) of HNF4α which result in decreased nuclear localization are known (40, 41). Phosphorylation of HNF4α by PKC or Src can lead to proteasomal degradation of HNF4α (23, 42). We investigated the role of Src Kinase in regulation of HNF4α protein expression after PH. Interestingly, pretreatment of C57BL6/J male mice with a Src inhibitor PP2, resulted in higher levels of nuclear HNF4α at 6 hr post-PH, providing evidence that phosphorylation of HNF4α by Src could be one of the mechanisms through which HNF4α exits the nucleus after PH. Future studies will determine changes in individual PTMs on HNF4α and mechanisms associated with altering those PTMs after PH.
Next, we hypothesized that overexpressing HNF4α would prevent hepatocytes from proliferating and halt or delay initiation of liver regeneration. Interestingly, our experiments with Tet-On driven HNF4α expression demonstrated significantly decreased hepatocyte proliferation 6 hours post-PH and an almost statistically significant increase in proliferation 48 hours post-PH. Decreased proliferation 6 hours post-PH was likely driven by decreased expression of pro-proliferative HNF4α negative target genes CCND1 and CDKN3. These data suggest rapid downregulation of HNF4α after PH may accelerate hepatocyte cell cycle entry, but is not necessary for initiation of cell cycle progression after PH. It also suggests activation of compensatory proliferative pathways which can activate liver regeneration despite increased HNF4α expression, highlighting the redundant nature of pro-regeneration pathways. Another possibility is HNF4α overexpression resulted in delayed initiation because the mechanisms responsible for HNF4α degradation, like Src, were still active, and more time was needed to degrade the overexpressed protein. Variable overexpression of HNF4α between hepatocytes is another potential cause of the increased proliferation observed in DOX treated mice at 48 hours. Additional studies will determine if an inverse correlation exists between hepatocytes overexpressing HNF4α and hepatocytes expressing PCNA. Interestingly, lack of steatosis was observed in HNF4α overexpressing mice, which is known to occur after PH (45). This might be caused by increased expression of apolipoproteins, like APOA2, which are well established HNF4α target genes. These data indicate that the initial loss of HNF4α could be mechanistically involved in transient steatosis observed following PH and those mechanisms may be independent of initiation of cell proliferation.
The most striking observation from our studies is the 100% mortality in HNF4α-KO mice following PH within 11 days secondary to loss of hepatic function. Ureagenesis requires HNF4α and liver-specific HNF4α-KO mice have increased serum ammonia and decreased serum urea (46). Several HNF4α-KO mice exhibited symptoms of hepatic encephalopathy such as loss of righting reflex before death (data not shown). Decreased hepatocyte differentiation in HNF4α-KO mice post-PH significantly diminished liver function, as shown by elevated bilirubin. Conversely, significantly increased nuclear HNF4α expression occurred in control mice (Fig. 1) at days 3 and 5 post-PH, when cell proliferation decreases and redifferentiation starts. The fact that HNF4α-KO hepatocytes were not able to compensate for loss of HNF4α function further supports the role of HNF4α as a master regulator of hepatic differentiation. Together, these data demonstrate that HNF4α-mediated differentiation of the newly divided hepatocytes is an essential part of the termination of regeneration and failure to redifferentiate can lead to mortality.
Another important observation in our studies was the complete loss of c-MET and EGFR expression in HNF4α-KO mice which could contribute to the death observed in HNF4α-KO mice after PH. Recent studies have shown that c-MET deletion combined with EGFR inhibition results in death of mice (47) due to decreased serum albumin, blood glucose and expression of genes involved in urea synthesis, lipid metabolism and carbohydrate metabolism. Microarray analysis of MET-EGFR inhibited mice revealed significant HNF4α inhibition. This study shows that baseline signaling through c-MET and EGFR is required for hepatocyte function in the quiescent liver and combined with our data is evidence of a positive regulation feedback loop between EGFR, c-MET and HNF4α in normal liver. Combined loss of EGFR, c-MET and HNF4α, observed in the HNF4α-KO mice, could have contributed to death after PH by removing several redundant pro-differentiation pathways.
This observation also leads to an interesting conundrum. It is known that HGF and EGF family members (EGF, TGFα etc.) signaling via their cognate receptors c-MET and EGFR is essential for liver regeneration after PH (25). Mice with deletion of c-MET and inhibition of EGFR do not exhibit any hepatocyte proliferation after PH (25). However, HNF4α-KO mice seem to be an exception to this rule. Despite the loss of c-MET and EGFR expression in HNF4α-KO mice, hepatocyte proliferation occurred at high levels before PH and a surge of proliferation occured at the same time as peak proliferation in WT mice before returning to baseline levels, although remaining higher than WT levels. Two important conclusions can be made from this observation. First, increased proliferation in HNF4α-KO mice before and after PH is consistent with previous findings that HNF4α is anti-proliferative in hepatocytes (10). Second, although proliferation is higher in HNF4α-KO mice before PH, the surge of additional proliferation that occurs in response to PH in the HNF4α-KO mice suggests activation of EGFR and c-MET independent pro-proliferative pathways. Furthermore, the regenerative response in HNF4α-KO mice, which do not express EGFR or c-MET, must be caused by activation of a pro-proliferative pathway normally repressed by HNF4α. Our studies revealed significant activation of c-Myc in HNF4α-KO mice consistent with studies showing activation of c-Myc in the absence of HNF4α (12). The role of c-Myc in cancer is well documented (48), and c-Myc competes with HNF4α to repress the anti-proliferative p21 gene (49). Our studies show HNF4α-KO hepatocytes lose two of the primary mitogenic pathways, c-MET and EGFR, but proliferation continues mainly via c-Myc activation.
Deletion of hepatic HNF4α increases liver to body weight ratios before PH, consistent with other observations in HNF4α-KO mice (11, 50). However, despite exhibiting increased hepatocyte proliferation throughout the time after PH, there were no differences in recovery of liver weight between WT and HNF4α-KO mice. Liver mass is a product of cell number and cell size. Increased proliferation would contribute to increased cell number, but restoration of cell size after PH involves the nutrient, protein and water content of hepatocytes to return to normal levels. Considering the importance of HNF4α activity in hepatocyte function, processes required to restore cell size would likeley take more time in HNF4α-KO mice, but HNF4α-KO mice die before this occurs. Thus, we cannot determine if regeneration in HNF4α-KO mice would overshoot target liver mass due to excess proliferation.
Further insights into the mechanisms driving liver regeneration in HNF4α-KO mice came from the RNAseq analysis. HNF4α-KO livers exhibited a proinflammatory and profibrotic transcriptional profile after PH. Elevated inflammation in HNF4α-KO mice was not caused by hepatocellular injury, as there was no difference in ALT levels or histopathological changes between groups (data not shown). The increase in inflammatory signaling is consistent with the observation that HNF4α inhibits pro-inflammatory genes such as EGR1. IPA predicted consistent inhibition of several prominent hepatocyte functions in HNF4α-KO supporting our findings of dedifferentiation and loss of hepatocyte function in HNF4α-KO mice. However, we observed a shift in the characteristics of activated functions between the two time points. Most of the activated functions in HNF4α-KO mice at day 2 included those involved in inflammation. However, the activated functions in HNF4α-KO mice at day 5 included many more functions involved in proliferation, tumorigenesis and a developmentally immature phenotype in addition to inflammation. Furthermore, HNF4α-KO mice at day 5 were predicted to express higher levels of known oncogenes (TRIM24, SNAI2, PLAG1) which were not activated at day 2. This transformation from an abnormal regenerative response to the beginnings of a pathological condition suggests a role of HNF4α in regulation of regenerative response following chronic or intermittent low-level liver injury and indicate that loss of HNF4α in chronic liver diseases may trigger oncogenic growth leading to HCC.
HNF4α reexpression in HNF4α-KO mice resulted in restoration of quiescence and expression of c-MET and EGFR. However, when the HNF4α construct was introduced after PH, we observed only partial prevention of death. This may be due to the timing of HNF4α reexpression, the possibility that dividing cells may have decreased sensitivity to AAV8 or dividing cells may reject AAV8-mediated introduction of exogenous material. Nevertheless, the significantly higher survival of HNF4α-KO mice after PH after HNF4α reexpression and the delayed mortality due to partial restoration of hepatic function through HNF4α expression supports hepatic failure being the cause of death in these animals.
In summary, our studies are the first to examine and manipulate HNF4α expression over multiple time points throughout the initiation, progression and termination of liver regeneration after partial hepatectomy. Our findings suggest downregulation of HNF4α may contribute to hepatocyte cell cycle entry during initiation of regeneration. More importantly, reestablishment of HNF4α activity during termination of regeneration is absolutely required for termination of regeneration and resumption of hepatocyte function. This study uncovers new evidence of HNF4α mediated expression of EGFR and c-MET and demonstrate that HNF4α-KO mice are capable of mounting a regenerative response despite lacking these receptors. Furthermore, regeneration in the absence of HNF4α results in a dedifferentiated, pro-carcinogenic hepatocyte phenotype. These results confirm the role of HNF4α as a major player in hepatocyte proliferation and differentiation during liver regeneration.
Supplementary Material
Acknowledgments
Financial Support: These studies were supported by NIH-COBRE (P20 RR021940–03, P30 GM118247), NIEHS Toxicology Training Grant (T32ES007079–34) NIH R01DK 0198414 and NIH RDK112768A
References
- 1.Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G, Kaestner KH, et al. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet 2003;34:292–296. [DOI] [PubMed] [Google Scholar]
- 2.Duncan SA, Manova K, Chen WS, Hoodless P, Weinstein DC, Bachvarova RF, Darnell JE Jr. Expression of transcription factor HNF-4 in the extraembryonic endoderm, gut, and nephrogenic tissue of the developing mouse embryo: HNF-4 is a marker for primary endoderm in the implanting blastocyst. Proc Natl Acad Sci U S A 1994;91:7598–7602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kyrmizi I, Hatzis P, Katrakili N, Tronche F, Gonzalez FJ, Talianidis I. Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes Dev 2006;20:2293–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morimoto A, Kannari M, Tsuchida Y, Sasaki S, Saito C, Matsuta T, Maeda T, et al. An HNF4alpha-microRNA-194/192 signaling axis maintains hepatic cell function. J Biol Chem 2017;292:10574–10585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez FJ. Regulation of hepatocyte nuclear factor 4 alpha-mediated transcription. Drug Metab Pharmacokinet 2008;23:2–7. [DOI] [PubMed] [Google Scholar]
- 6.Hu X, Xie P, Li W, Li Z, Shan H. Direct induction of hepatocyte-like cells from immortalized human bone marrow mesenchymal stem cells by overexpression of HNF4alpha. Biochem Biophys Res Commun 2016;478:791–797. [DOI] [PubMed] [Google Scholar]
- 7.Wei L, Dai Y, Zhou Y, He Z, Yao J, Zhao L, Guo Q, et al. Oroxylin A activates PKM1/HNF4 alpha to induce hepatoma differentiation and block cancer progression. Cell Death Dis 2017;8:e2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin C, Lin Y, Zhang X, Chen YX, Zeng X, Yue HY, Hou JL, et al. Differentiation therapy of hepatocellular carcinoma in mice with recombinant adenovirus carrying hepatocyte nuclear factor-4alpha gene. Hepatology 2008;48:1528–1539. [DOI] [PubMed] [Google Scholar]
- 9.Nishikawa T, Bell A, Brooks JM, Setoyama K, Melis M, Han B, Fukumitsu K, et al. Resetting the transcription factor network reverses terminal chronic hepatic failure. J Clin Invest 2015;125:1533–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walesky C, Gunewardena S, Terwilliger EF, Edwards G, Borude P, Apte U. Hepatocyte-specific deletion of hepatocyte nuclear factor-4alpha in adult mice results in increased hepatocyte proliferation. Am J Physiol Gastrointest Liver Physiol 2013;304:G26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonzo JA, Ferry CH, Matsubara T, Kim JH, Gonzalez FJ. Suppression of hepatocyte proliferation by hepatocyte nuclear factor 4alpha in adult mice. J Biol Chem 2012;287:7345–7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walesky C, Edwards G, Borude P, Gunewardena S, O’Neil M, Yoo B, Apte U. Hepatocyte nuclear factor 4 alpha deletion promotes diethylnitrosamine-induced hepatocellular carcinoma in rodents. Hepatology 2013;57:2480–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michalopoulos GK. Hepatostat: Liver regeneration and normal liver tissue maintenance. Hepatology 2017;65:1384–1392. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka T, Jiang S, Hotta H, Takano K, Iwanari H, Sumi K, Daigo K, et al. Dysregulated expression of P1 and P2 promoter-driven hepatocyte nuclear factor-4alpha in the pathogenesis of human cancer. J Pathol 2006;208:662–672. [DOI] [PubMed] [Google Scholar]
- 15.Hatziapostolou M, Polytarchou C, Aggelidou E, Drakaki A, Poultsides GA, Jaeger SA, Ogata H, et al. An HNF4alpha-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell 2011;147:1233–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai WY, Lin LY, Hao H, Zhang SM, Ma F, Hong XX, Zhang H, et al. Yes-associated protein/TEA domain family member and hepatocyte nuclear factor 4-alpha (HNF4alpha) repress reciprocally to regulate hepatocarcinogenesis in rats and mice. Hepatology 2017;65:1206–1221. [DOI] [PubMed] [Google Scholar]
- 17.Borude P, Edwards G, Walesky C, Li F, Ma X, Kong B, Guo GL, et al. Hepatocyte-specific deletion of farnesoid X receptor delays but does not inhibit liver regeneration after partial hepatectomy in mice. Hepatology 2012;56:2344–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yanger K, Zong Y, Maggs LR, Shapira SN, Maddipati R, Aiello NM, Thung SN, et al. Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev 2013;27:719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 2010;328:1036–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolfe A, Thomas A, Edwards G, Jaseja R, Guo GL, Apte U. Increased activation of the Wnt/beta-catenin pathway in spontaneous hepatocellular carcinoma observed in farnesoid X receptor knockout mice. J Pharmacol Exp Ther 2011;338:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Apte U, Singh S, Zeng G, Cieply B, Virji MA, Wu T, Monga SP. Beta-catenin activation promotes liver regeneration after acetaminophen-induced injury. Am J Pathol 2009;175:1056–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffman BG, Robertson G, Zavaglia B, Beach M, Cullum R, Lee S, Soukhatcheva G, et al. Locus co-occupancy, nucleosome positioning, and H3K4me1 regulate the functionality of FOXA2-, HNF4A-, and PDX1-bound loci in islets and liver. Genome Res 2010;20:1037–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chellappa K, Jankova L, Schnabl JM, Pan S, Brelivet Y, Fung CL, Chan C, et al. Src tyrosine kinase phosphorylation of nuclear receptor HNF4alpha correlates with isoform-specific loss of HNF4alpha in human colon cancer. Proc Natl Acad Sci U S A 2012;109:2302–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das AT, Tenenbaum L, Berkhout B. Tet-On Systems For Doxycycline-inducible Gene Expression. Curr Gene Ther 2016;16:156–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paranjpe S, Bowen WC, Mars WM, Orr A, Haynes MM, DeFrances MC, Liu S, et al. Combined systemic elimination of MET and epidermal growth factor receptor signaling completely abolishes liver regeneration and leads to liver decompensation. Hepatology 2016;64:1711–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fabregat I, Moreno-Caceres J, Sanchez A, Dooley S, Dewidar B, Giannelli G, Ten Dijke P, et al. TGF-beta signalling and liver disease. FEBS J 2016;283:2219–2232. [DOI] [PubMed] [Google Scholar]
- 27.Charos AE, Reed BD, Raha D, Szekely AM, Weissman SM, Snyder M. A highly integrated and complex PPARGC1A transcription factor binding network in HepG2 cells. Genome Res 2012;22:1668–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alpern D, Langer D, Ballester B, Le Gras S, Romier C, Mengus G, Davidson I. TAF4, a subunit of transcription factor II D, directs promoter occupancy of nuclear receptor HNF4A during post-natal hepatocyte differentiation. Elife 2014;3:e03613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia Y, Viswakarma N, Reddy JK. Med1 subunit of the mediator complex in nuclear receptor-regulated energy metabolism, liver regeneration, and hepatocarcinogenesis. Gene Expr 2014;16:63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weng M-Z, Zhuang P-Y, Hei Z-Y, Lin P-Y, Chen Z-S, Liu Y-B, Quan Z-W, et al. ZBTB20 is involved in liver regeneration after partial hepatectomy in mouse. Hepatobiliary & Pancreatic Diseases International 2014;13:48–54. [DOI] [PubMed] [Google Scholar]
- 31.Xie Z, Zhang H, Tsai W, Zhang Y, Du Y, Zhong J, Szpirer C, et al. Zinc finger protein ZBTB20 is a key repressor of alpha-fetoprotein gene transcription in liver. Proc Natl Acad Sci U S A 2008;105:10859–10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santangelo L, Marchetti A, Cicchini C, Conigliaro A, Conti B, Mancone C, Bonzo JA, et al. The stable repression of mesenchymal program is required for hepatocyte identity: a novel role for hepatocyte nuclear factor 4alpha. Hepatology 2011;53:2063–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Juma AR, Damdimopoulou PE, Grommen SV, Van de Ven WJ, De Groef B. Emerging role of PLAG1 as a regulator of growth and reproduction. J Endocrinol 2016;228:R45–56. [DOI] [PubMed] [Google Scholar]
- 34.Appikonda S, Thakkar KN, Barton MC. Regulation of gene expression in human cancers by TRIM24. Drug Discov Today Technol 2016;19:57–63. [DOI] [PubMed] [Google Scholar]
- 35.Palu RA, Thummel CS. Sir2 Acts through Hepatocyte Nuclear Factor 4 to maintain insulin Signaling and Metabolic Homeostasis in Drosophila. PLoS Genet 2016;12:e1005978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Donnell KA, Keng VW, York B, Reineke EL, Seo D, Fan D, Silverstein KA, et al. A Sleeping Beauty mutagenesis screen reveals a tumor suppressor role for Ncoa2/Src-2 in liver cancer. Proc Natl Acad Sci U S A 2012;109:E1377–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Armartmuntree N, Murata M, Techasen A, Yongvanit P, Loilome W, Namwat N, Pairojkul C, et al. Prolonged oxidative stress down-regulates Early B cell factor 1 with inhibition of its tumor suppressive function against cholangiocarcinoma genesis. Redox Biol 2018;14:637–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu Y, Guo X, Wang J, Liu Y, Gao H, Fan H, Nong X, et al. A novel microRNA identified in hepatocellular carcinomas is responsive to LEF1 and facilitates proliferation and epithelial-mesenchymal transition via targeting of NFIX. Oncogenesis 2018;7:22. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Fukuda T, Fukuchi T, Yagi S, Shiojiri N. Immunohistochemical analyses of cell cycle progression and gene expression of biliary epithelial cells during liver regeneration after partial hepatectomy of the mouse. Exp Anim 2016;65:135–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yokoyama A, Katsura S, Ito R, Hashiba W, Sekine H, Fujiki R, Kato S. Multiple post-translational modifications in hepatocyte nuclear factor 4alpha. Biochem Biophys Res Commun 2011;410:749–753. [DOI] [PubMed] [Google Scholar]
- 41.Wang Z, Salih E, Burke PA. Quantitative analysis of cytokine-induced hepatocyte nuclear factor-4alpha phosphorylation by mass spectrometry. Biochemistry 2011;50:5292–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun K, Montana V, Chellappa K, Brelivet Y, Moras D, Maeda Y, Parpura V, et al. Phosphorylation of a conserved serine in the deoxyribonucleic acid binding domain of nuclear receptors alters intracellular localization. Mol Endocrinol 2007;21:1297–1311. [DOI] [PubMed] [Google Scholar]
- 43.Jiao H, Zhu Y, Lu S, Zheng Y, Chen H. An Integrated Approach for the Identification of HNF4alpha-Centered Transcriptional Regulatory Networks During Early Liver Regeneration. Cell Physiol Biochem 2015;36:2317–2326. [DOI] [PubMed] [Google Scholar]
- 44.Yue HY, Yin C, Hou JL, Zeng X, Chen YX, Zhong W, Hu PF, et al. Hepatocyte nuclear factor 4alpha attenuates hepatic fibrosis in rats. Gut 2010;59:236–246. [DOI] [PubMed] [Google Scholar]
- 45.Shteyer E, Liao Y, Muglia LJ, Hruz PW, Rudnick DA. Disruption of hepatic adipogenesis is associated with impaired liver regeneration in mice. Hepatology 2004;40:1322–1332. [DOI] [PubMed] [Google Scholar]
- 46.Inoue Y, Hayhurst GP, Inoue J, Mori M, Gonzalez FJ. Defective ureagenesis in mice carrying a liver-specific disruption of hepatocyte nuclear factor 4alpha (HNF4alpha ). HNF4alpha regulates ornithine transcarbamylase in vivo. J Biol Chem 2002;277:25257–25265. [DOI] [PubMed] [Google Scholar]
- 47.Tsagianni A, Mars WM, Bhushan B, Bowen WC, Orr A, Stoops J, Paranjpe S, et al. Combined Systemic Disruption of MET and Epidermal Growth Factor Receptor Signaling Causes Liver Failure in Normal Mice. Am J Pathol 2018. [DOI] [PMC free article] [PubMed]
- 48.Gabay M, Li Y, Felsher DW. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med 2014;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hwang-Verslues WW, Sladek FM. Nuclear receptor hepatocyte nuclear factor 4alpha1 competes with oncoprotein c-Myc for control of the p21/WAF1 promoter. Mol Endocrinol 2008;22:78–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol 2001;21:1393–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.