Abstract
Recombinant adeno-associated virus (rAAV) is a vector with increasing popularity in the field of gene therapy. Like other drug substances manufactured in cell lines, rAAV vectors are commonly contaminated with host cell DNA, and the levels must be carefully monitored. The current method for residual DNA quantification in rAAV was adapted from protein programs and required sample digestion by proteinase prior to qPCR analysis. While the method worked effectively, it was unclear if proteinase digestion was essential for releasing DNA from rAAV capsids and improving qPCR efficiency. In this study, we systematically investigated the role of each component and treatment with the goal to simplify and streamline the method. It was determined that the proteinase digestion step was dispensable, while the addition of Tween 20 to rAAV samples was essential for accurate quantification of residual DNA. Based on this finding, a digestion-free method has been established that requires only a one-step sample preparation—addition of Tween 20. The method has been tested extensively with an rAAV9-based drug substance and process intermediates and verified with other rAAV serotypes. This significantly simplified and faster assay can be easily automated for high-throughput applications.
Keywords: rAAV, residual host cell DNA, hAlu, qPCR
Introduction
Recombinant adeno-associated virus (rAAV) vectors are widely used for gene therapy because of several unique advantages, including long-lasting gene expression, wide tropism for mammalian cells, modest immunogenicity, non-pathogenicity, and no genome integration.1, 2 rAAV contains a single-stranded genome, which is protected by an icosahedral shell made of three different proteins: VP1, VP2, and VP3.3 Several rAAV production platforms have been established based on various host cell lines such as human HeLa and HEK293 or insect Sf9 cells.4, 5 Inside of the host cells, capsid proteins are expressed, and viral genomes are replicated and packaged into the newly assembled capsids to produce rAAV particles. However, packaging of viral DNA is not an error-proof process. It is well documented that illegitimate DNA, including genomic DNA of the host cells, could become encapsidated.4, 6 This creates significant challenges for downstream processing, as encapsidated host cell DNA cannot be removed by Benzonase treatment or through affinity purification.7 Delivery of unintended DNA sequences to patients is a major safety concern, so the level of residual host cell DNA in rAAV drug substance (DS) must be carefully monitored.
The industry standard for residual host cell DNA quantification is based on qPCR targeting repetitive DNA sequences (i.e., Alu repeats).8, 9 Because qPCR is sensitive to matrix interference, test samples are usually pretreated to remove potential PCR inhibitors. For protein-based drugs, proteinase K treatment is routinely performed for digestion of a high concentration of proteins.10, 11 This is particularly important for monoclonal antibodies, which are known to inhibit qPCR.12, 13 Some protocols also include an extra step of DNA purification.8, 14 The same method with proteinase digestion was applied to rAAV-based drugs. It is believed that digestion of the capsid protein could help release encapsidated DNA for qPCR analysis,15 although it has been reported that a brief treatment at 85°C is sufficient to break down capsids of all AAV serotypes.16 Based on this study, DNA is expected to be released from the viral particle during the denaturation step of qPCR (94°C for 10 min). Moreover, an AAV capsid contains 60 protein molecules, and for a DS with 1E+13 vg/mL of rAAV, the protein concentration is only 65 μg/mL. It is also unclear whether capsid proteins are inhibitory for qPCR analysis. Therefore, the goal of this study was to investigate if proteinase digestion is essential for residual DNA quantification in rAAV and to simplify sample preparation by eliminating unnecessary treatments.
Results
Evaluation of the Existing Method for Residual DNA Quantification in rAAV
A previously established method for residual DNA quantification includes treatment of rAAV samples by proteinase K in the presence of 0.2% SDS at 56°C for 30 min, followed by heat inactivation at 70°C for 1 h and neutralization of SDS by Tween 20 before qPCR analysis (Figure 1A). While this is much simpler than other methods that require DNA extraction, we tested if further simplification is possible by omitting proteinase digestion, assuming that the denaturation step of qPCR (94°C for 10 min) is sufficient to release encapsidated DNA for qPCR analysis. To test this possibility, rAAV9 samples (produced in HeLa cells) with or without spike of 1,000 pg/mL HeLa DNA were either digested according to Figure 1A or diluted 2.5-fold in TE buffer to adjust for sample concentration, followed by qPCR against human Alu repeats (hAlus). Assay accuracy was determined by recovery of HeLa DNA spikes. As shown in Table 1 and Figure 1B, residual DNA detected in samples that were diluted in TE buffer (9.7 pg/mL) was significantly lower than in samples that were digested (24.6 pg/mL). The difference is consistent with a decrease in assay accuracy from 106.2% to 60.1% when digestion was omitted (Figure 1B), which underscores the importance of pretreatments of rAAV samples before qPCR analysis.
Figure 1.
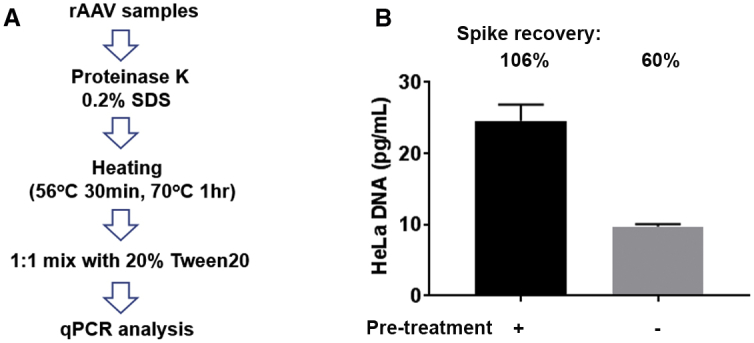
Evaluation of the Existing Residual DNA Assay with Proteinase K Digestion
(A) Key components and treatments for the method with protein K digestion. (B) rAAV samples with or without predigestion were analyzed by qPCR. Omitting digestion resulted in under-detection of residual HeLa DNA. Assay accuracy was determined by recovery of HeLa DNA spikes.
Table 1.
Pretreatment of rAAV Samples Improves Residual DNA Detection
| Replicates | With Digestion |
No Digestion |
||||
|---|---|---|---|---|---|---|
| Sample (pg/mL) | Spiked Sample (pg/mL) | Spike Measured (pg/mL) | Sample (pg/mL) | Spiked Sample (pg/mL) | Spike Measured (pg/mL) | |
| 1 | 23.6 | 1,176.2 | 1,152.6 | 10.0 | 614.9 | 605.0 |
| 2 | 27.2 | 1,058.8 | 1,031.6 | 9.8 | 569.6 | 559.8 |
| 3 | 23.0 | 1,023.8 | 1,000.8 | 9.3 | 647.5 | 638.1 |
| Average | 24.6 | 1,086.2 | 1,061.6 | 9.7 | 610.7 | 601.0 |
| Spike recovery % | 106.2 | 60.1 | ||||
rAAV9 samples with or without a spike of 1,000 pg/mL of HeLa DNA were either digested by proteinase K (Figure 1A) or directly diluted 2.5-fold in TE buffer (no digestion). The levels of HeLa DNA in samples were quantified by qPCR, and assay accuracy was determined by spike recovery.
Tween 20, Rather than Proteinase K, Is Essential for rAAV Sample Preparation
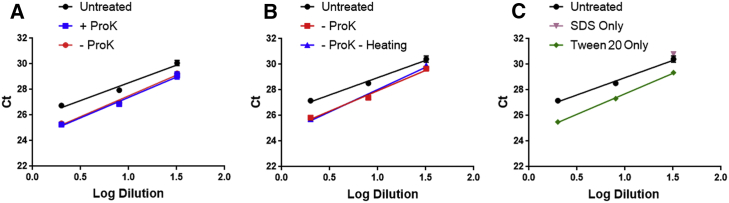
While rAAV samples require pretreatments, it was unclear if the enzymatic activity of proteinase K is essential, as the method contains other components and treatments (Figure 1A). To investigate the role of proteinase K, rAAV9 samples were treated following the method in Figure 1A (+ProK) or without proteinase K but keeping other treatments (−ProK). Samples were diluted 2-, 8-, and 32-fold in ddH2O and analyzed by qPCR against hAlu. Cycle threshold (Ct) values were plotted against the corresponding dilution factors. As shown in Figure 2A, the curves from samples + or −ProK almost completely overlap, and both are below the curve of untreated sample (direct dilution in TE). These results suggest that the enzymatic activity of proteinase K is not required for accurate determination of residual DNA levels in rAAV9 samples.
Figure 2.
Investigating Impacts of Different Steps in Residual DNA Assay with Proteinase K Digestion
(A) Proteinase K. rAAV9 samples were digested (+ProK), mock treated (−ProK), or directly diluted in TE (untreated). Samples were analyzed by qPCR against hAlu, and Ct values were plotted against the dilution factors. (B) Heat treatment. rAAV9 samples were mock treated with (−ProK) or without heating (−ProK −heating) or directly diluted in TE (untreated). Following qPCR analysis, Ct values were plotted against the dilution factors. (C) SDS and Tween 20. rAAV9 samples were treated with only SDS or Tween 20 or directly diluted in TE (untreated). Ct values were plotted against dilution factors. Values represent average of 3 replicates ± SD.
Next, we investigated the role of the heating steps that include 30-min treatment at 56°C and 60-min treatment at 70°C. It is possible that these treatments promote capsid disassembly for DNA release, or alternatively, denature capsid proteins and make them less inhibitory for qPCR. To test these possibilities, after addition of SDS, rAAV9 samples were directly mixed with 20% Tween 20 at a 1:1 ratio (−ProK −heating). As control, rAAV9 samples prepared with heating steps were included (−ProK). Samples were analyzed by qPCR, and Ct values were plotted against the corresponding dilution factors. As shown in Figure 2B, the curves from samples with or without heating are comparable, and both are below the curve of the untreated sample, which is indicative of higher qPCR efficiency. This result suggests that the heat treatment is also not necessary.
After exclusion of proteinase K and heating, the only components left were SDS and Tween 20. Therefore, rAAV9 samples were treated by simply adding SDS or Tween 20 and analyzed by qPCR, and Ct values were plotted against the corresponding dilution factors. As shown in Figure 2C, the curve of “Tween 20 only” is below the curve of “untreated,” suggesting higher qPCR efficiency. In contrast, SDS strongly inhibited qPCR, and Ct value was undetermined unless the sample was diluted by 32-fold. These results suggest that Tween 20 is the key in improving qPCR efficiency for rAAV samples.
Determining the Optimal Concentration of Tween 20
With the existing method with proteinase digestion, concentration of Tween 20 in test samples is 10%. To determine if this is optimal, Tween 20 was added to rAAV9 samples to final concentrations of 10%, 5%, 2.5%, 1.25%, 0.625%, or 0%. Samples were analyzed by qPCR, and Ct values were plotted against the corresponding Tween 20 concentrations. As shown in Figure 3, Ct values decreased as concentration of Tween 20 rose until reaching 5%. Based on this result, we concluded that Tween 20 reached its peak efficacy at 5% and proposed a “digestion-free” method by simply adding Tween 20 to rAAV samples to this concentration.
Figure 3.
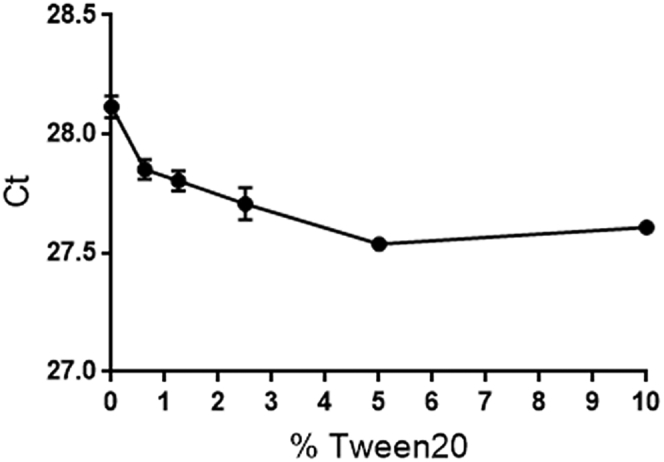
Determination of the Optimal Tween 20 Concentration for the Digestion-free Method
rAAV9 samples were prepared by adding Tween 20 to final concentrations of 10%, 5%, 2.5%, 1.25%, 0.625%, or 0%. Samples were analyzed by qPCR against hAlu. Ct values were plotted against Tween 20 concentrations. Values represent average of 3 replicates ± SD.
Qualification of the Digestion-free Method
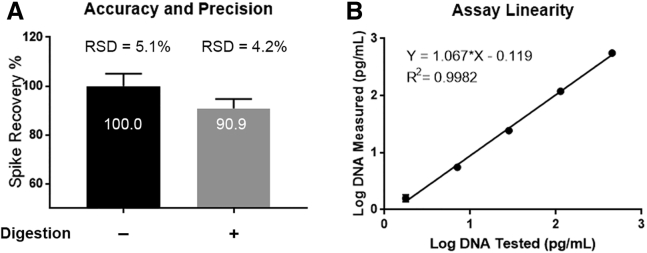
To evaluate the new “digestion-free” method, we first compared the accuracy and precision to the existing method. rAAV9 samples were first diluted to a predetermined minimum required dilution, spiked with 1,000 pg/mL HeLa DNA, and analyzed by both methods. The assay accuracy was determined by recovery of HeLa DNA spikes. As shown in Figure 4A, the digestion-free method has an accuracy of 100% and precision of 5.1% based on three independent tests, while the accuracy of the method with digestion is 90.9% with a precision of 4.2%. These results demonstrate that both methods perform well with excellent accuracy and precision.
Figure 4.
Qualifications of the Digestion-free Method
(A) Accuracy and precision were calculated based on spike recoveries in three independent experiments. Error bar represents 1 SD. (B) Assay linearity. An rAAV9 sample containing 453 pg/mL of residual HeLa DNA was 4-fold serially diluted in TE buffer with 5% Tween 20. The amounts of HeLa DNA measured were plotted against the amounts expected.
Next, we studied the linearity of the digestion-free method. An rAAV9 sample with a previously determined amount of residual DNA was 1:4 serially diluted in Tris-EDTA (TE) buffer with 5% Tween 20. As a result, the test samples were expected to contain from 453 pg/mL to 1.8 pg/mL of residual HeLa DNA. Samples were analyzed by qPCR in triplicates, and the amounts of residual DNA measured were plotted against the amount expected. As shown in Figure 4B, the values of both slope and R2 are very close to 1, demonstrating excellent linearity for this new method.
Finally, sensitivity of the method was established by defining the lower limit of quantitation (LLOQ). HeLa DNA standard was diluted in a formulation buffer for rAAV DS to 10, 5, 2.5, and 1 pg/mL. Each sample was tested six times with acceptance criteria requiring that recoveries of at least four out of six replicates were between 70% and 130% and relative standard deviation (RSD) of acceptable results under 25%.17, 18 Based on these criteria, the LLOQ for the digestion-free method was determined to be 1 pg/mL (Table 2), a sensitivity comparable to many established residual DNA assays that require digestion.8
Table 2.
LLOQ of the Digestion-free Method
| pg/mL Tested | pg/mL Measured | Recovery % | Acceptable Runs | RSD% |
|---|---|---|---|---|
| 10 | 8.7 | 87.5 | 5 | 11.0 |
| 8.7 | 86.8 | |||
| 8.2 | 81.6 | |||
| 6.2 | 62.3 | |||
| 10.7 | 106.8 | |||
| 9.8 | 97.8 | |||
| 5 | 4.8 | 95.9 | 6 | 8.1 |
| 4.4 | 88.9 | |||
| 3.8 | 75.6 | |||
| 4.6 | 92.4 | |||
| 4.6 | 91.1 | |||
| 4.7 | 93.4 | |||
| 2.5 | 2.2 | 87.3 | 5 | 14.3 |
| 3.0 | 120.0 | |||
| 3.5 | 139.2 | |||
| 2.4 | 95.9 | |||
| 3.0 | 121.0 | |||
| 2.5 | 100.7 | |||
| 1 | 1.2 | 119.6 | 6 | 16.8 |
| 1.1 | 114.0 | |||
| 0.8 | 80.4 | |||
| 0.8 | 80.7 | |||
| 1.1 | 111.4 | |||
| 1.0 | 103.5 |
HeLa DNA standard was diluted in formulation buffer to 10, 5, 2.5, and 1 pg/mL. Samples were treated with the digestion-free method and tested six times by qPCR. Recovery was determined by the ratio between pg/mL measured and pg/mL tested. LLOQ is defined as at least four out the six tests with a recovery between 70% and 130% and RSD < 25%. Data in italic are not acceptable.
Application of the Digestion-free Method for Other Host Cell DNA and AAV Serotypes
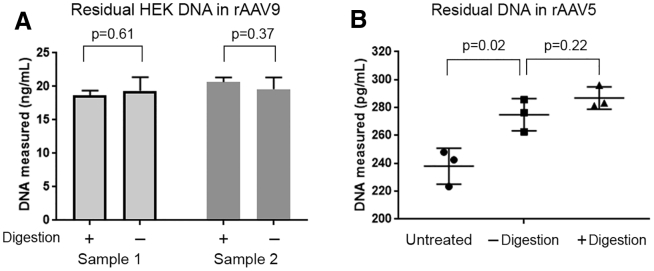
The digestion-free method was developed with rAAV9 produced in HeLa cells. Because hAlu sequences are common among cells from the human origin, this method is expected to be applicable to rAAV produced in other human cell lines, including the widely used HEK cells. To test this possibility, two rAAV9 samples produced in HEK cells were analyzed by methods with or without proteinase digestion, using HEK DNA for standard curves. As shown in Figure 5A, the levels of residual DNA determined by both methods are comparable (p > 0.05), confirming the capability for residual HEK DNA detection. To study if the digestion-free method can support samples at different processing stages, we tested an rAAV9-based DS and two intermediates from downstream purification for that DS. These samples contain significantly different amounts of viral particles and various levels of residual DNA. Testing for all samples was successful following proper dilutions, demonstrated by spike recovery within the acceptable range of 70%–130%. Overall, for residual HEK DNA quantification, the method has an accuracy of 105% and inter-assay precision of 15.7% (Table 3).
Figure 5.
Quantification of Residual HEK DNA in Different rAAV Serotypes with the Digestion-free Method
(A) Levels of residual HEK DNA in two rAAV9 samples were quantified with methods with or without proteinase digestion. Results from the two methods were comparable. Values represent average of 3 replicates ± SD. (B) Level of residual HEK DNA in an rAAV5 sample was quantified with methods with or without proteinase digestion or without pretreatment. p values were calculated by two-sample t test.
Table 3.
Accuracy and Precision of the Digestion-free Method
| Sample | Dilution Factor | DNA in Unspiked Dilution (pg/mL) | DNA in Spiked Dilution (pg/mL) | DNA Spike Recovered (pg/mL) | Spike Recovery (%) |
|---|---|---|---|---|---|
| Intermediate 1 | 10 | 4,369 | 5,470 | 1,101 | 110 |
| 100 | 441 | 1,695 | 1,254 | 125 | |
| Intermediate 2 | 100 | 1,151 | 1,986 | 835 | 84 |
| 1,000 | 94 | 1,060 | 966 | 97 | |
| Drug substance | 1,000 | 376 | 1,598 | 1,222 | 122 |
| 10,000 | 31 | 963 | 932 | 93 | |
| Accuracy | 105 | ||||
| Precision | 15.7 |
An rAAV9-based drug substance and two intermediates from downstream purification were diluted in TE buffer, and HEK DNA was spiked into diluted samples to 1,000 pg/mL. Samples with or without DNA spike were analyzed with the digestion-free method. Assay accuracy and inter-assay precision were determined by the average and RSD of spike recoveries, respectively.
To study if the digestion-free method works for other AAV serotypes, we tested AAV5 whose capsid is the most stable among all serotypes.16, 19 rAAV5-GFP produced in HEK293 cells (Vigene) was analyzed by methods with or without proteinase digestion. As shown in Figure 5B, levels of residual HEK DNA measured by both methods were comparable (p = 0.22) and were significantly higher than testing conducted without any pretreatments (p = 0.02). This result is consistent with previous observations with rAAV9 (Figures 1 and 2). Moreover, we have unpublished data showing that the digestion-free method is adequate for quantification of residual DNA from a non-human primate-derived cell line in rAAV2-based DS. Because AAV2, AAV9, and AAV5 represent AAV serotypes with the least, intermediate, and the most thermal stability,16, 19 we expect that the digestion-free method is applicable to all AAV serotypes.
Discussion
In this study, we thoroughly investigated the existing method for residual DNA quantification and demonstrated that Tween 20, rather than proteinase K, is essential for rAAV sample preparation. Based on this finding, a digestion-free method has been established, which performs comparably to the existing method, but is much easier and can save hours of assay time. The digestion-free method has been extensively tested by our group with different rAAV serotypes and is in the process of being adapted for automation for high-throughput applications.
The original residual DNA assay for rAAV was adopted from those developed for protein drugs. It was assumed that AAV capsid should be digested to release encapsidated DNA and that digestion also makes capsid proteins less inhibitory for qPCR. However, these assumptions neglect three important differences between rAAV and protein-based drugs. First, concentrations of protein DSs usually reach above 50 mg/mL, which is hundreds of folds higher than the protein concentration in a typical rAAV DS (rAAV at 1E + 13 GC/mL contains less than 0.1 mg/mL of capsid protein). Second, unlike immunoglobulin G (IgG),12, 13 AAV capsid has not been reported to be a PCR inhibitor. Third, levels of residual DNA in rAAV are significantly higher than what is usually detected in protein drugs. This is largely due to the fact that the majority of residual DNA in rAAV DS is encapsiduated (>95%, unpublished data). While residual DNA is cleared during downstream purifications of protein drugs, encapsidated DNA enriches together with viral particles. The abundance of residual DNA, coupled with low levels of protein, likely explain why proteinase digestion is dispensable for rAAV samples. However, our data do highlight an essential role of Tween 20. Although the exact mechanism still waits to be discovered, we hypothesize that Tween 20 may reduce non-specific interactions between DNA and capsid proteins or prevent DNA from precipitation, thereby improving the efficiency of qPCR detection.
The AAV capsid proteins are largely conserved among different serotypes except for some small variable regions that are not involved in capsid assembly.1 The melting temperatures of all AAV serotypes are significantly lower than the denaturation temperature of qPCR, so serotype was not expected to limit the usage of the digestion-free method, as was demonstrated with AAV2, AAV9, and AAV5 in this study. Moreover, by simply switching primers and probes, this method should be applicable for quantification of other unintended DNA in rAAV, such as residual plasmid or residual Bac viral DNA depending on the production platforms. In fact, we have successfully quantified residual DNA from a non-human primate-derived cell line in rAAV2 (unpublished data). When evaluating this method for new applications, it is recommended to include spike recovery tests to monitor the assay performance and to determine the proper dilutions for different samples, as shown in Table 3. It is also recommended to compare test results with established methods. Future work will elucidate the principles of this method and reveal if it can be applied to gene therapy products based on other types of viruses.
Materials and Methods
HeLa and HEK DNA Standard
Genomic DNA of HeLa cells or HEK293 cells was extracted with the DNeasy blood and tissue kit (QIAGEN, Germantown, MD, USA). DNA concentration was determined by the Picogreen dsDNA kit (Molecular Probes, Eugene, OR, USA), and DNA was stored at −70°C.
Primers and Probes
Sequences of PCR primers and probes for hAlus were previously published8 and ordered from Thermo Fisher Scientific (Carlsbad, CA, USA).
hAlu primer F, 5′-GAG GCG GGC GGA TCA-3′
hAlu primer R, 5′-CCC GGC TAA TTT TTG TAT TTT TAG TAG-3′
hAlu probe, 5′-(FAM)-CAG CCT GGC CAA CAT GGT GAA ACC-(TAMRA)-3′
rAAV Sample Preparation
The residual DNA assay with proteinase digestion was adapted from a previous publication:11 100 μL reaction mixture was prepared by adding 1 μL of 20% SDS, 5 μL of 20 mg/mL proteinase K (Thermo Fisher Scientific, Waltham, MA, USA), and 14 μL of double-distilled water (ddH2O) to 80 μL of prediluted rAAV samples. Samples were treated at 56°C for 30 min and 70°C for 1 h, followed by addition of 100 μL TE buffer with 20% Tween 20.
For the digestion-free method, rAAV samples were mixed with same volume of TE buffer with 10% Tween 20. The final concentration of Tween 20 was 5%.
qPCR and Data Analysis
qPCR reactions were prepared with TaqMan Universal MasterMix in a 96-well optical plate (Life Technologies, Carlsbad, CA, USA). Pretreated rAAV samples were mixed 1:1 with qPCR reaction mixture, and the final concentrations of primers and probe were 1 μM and 0.25 μM. Standard curves were prepared with HeLa DNA serially diluted in TE buffer with 10% Tween 20 and 0.1% SDS (method with digestion) or TE buffer with 5% Tween 20 (digestion-free method). qPCR program was as follows: 1 cycle of 50°C × 2′, 95°C × 15′; 40 cycles of 95°C × 30″, 60°C × 1′ was conducted in ViiA 7 real-time system (Life Technologies, Carlsbad, CA, USA). qPCR data were automatically analyzed by the ViiA 7 software. Ct values and DNA concentrations were reported.
Author Contributions
Y.W. designed the experiments. Y.W., R.C., and A.K. performed the experiments. S.B. and M.F. oversaw the work. Y.W. and M.F. wrote the paper.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
The authors thank Wei Zhang for critical reading of the manuscript.
Contributor Information
Yu Wang, Email: yu.wang@biogen.com.
Marina Feschenko, Email: marina.feschenko@biogen.com.
References
- 1.Samulski R.J., Muzyczka N. AAV-Mediated Gene Therapy for Research and Therapeutic Purposes. Annu. Rev. Virol. 2014;1:427–451. doi: 10.1146/annurev-virology-031413-085355. [DOI] [PubMed] [Google Scholar]; Samulski, R.J., and Muzyczka, N. (2014). AAV-Mediated Gene Therapy for Research and Therapeutic Purposes. Annu. Rev. Virol. 1, 427-451. [DOI] [PubMed]
- 2.Daya S., Berns K.I. Gene therapy using adeno-associated virus vectors. Clin. Microbiol. Rev. 2008;21:583–593. doi: 10.1128/CMR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; Daya, S., and Berns, K.I. (2008). Gene therapy using adeno-associated virus vectors. Clin. Microbiol. Rev. 21, 583-593. [DOI] [PMC free article] [PubMed]
- 3.Naso M.F., Tomkowicz B., Perry W.L., 3rd, Strohl W.R. Adeno-Associated Virus (AAV) as a Vector for Gene Therapy. BioDrugs. 2017;31:317–334. doi: 10.1007/s40259-017-0234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Naso, M.F., Tomkowicz, B., Perry, W.L., 3rd, and Strohl, W.R. (2017). Adeno-Associated Virus (AAV) as a Vector for Gene Therapy. BioDrugs 31, 317-334. [DOI] [PMC free article] [PubMed]
- 4.Penaud-Budloo M., François A., Clément N., Ayuso E. Pharmacology of Recombinant Adeno-associated Virus Production. Mol. Ther. Methods Clin. Dev. 2018;8:166–180. doi: 10.1016/j.omtm.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; Penaud-Budloo, M., François, A., Clement, N., and Ayuso, E. (2018). Pharmacology of Recombinant Adeno-associated Virus Production. Mol. Ther. Methods Clin. Dev. 8, 166-180. [DOI] [PMC free article] [PubMed]
- 5.Clément N., Grieger J.C. Manufacturing of recombinant adeno-associated viral vectors for clinical trials. Mol. Ther. Methods Clin. Dev. 2016;3:16002. doi: 10.1038/mtm.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]; Clement, N., and Grieger, J.C. (2016). Manufacturing of recombinant adeno-associated viral vectors for clinical trials. Mol. Ther. Methods Clin. Dev. 3, 16002. [DOI] [PMC free article] [PubMed]
- 6.Wright J.F. Product-Related Impurities in Clinical-Grade Recombinant AAV Vectors: Characterization and Risk Assessment. Biomedicines. 2014;2:80–97. doi: 10.3390/biomedicines2010080. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wright, J.F. (2014). Product-Related Impurities in Clinical-Grade Recombinant AAV Vectors: Characterization and Risk Assessment. Biomedicines 2, 80-97. [DOI] [PMC free article] [PubMed]
- 7.Schnödt M., Büning H. Improving the Quality of Adeno-Associated Viral Vector Preparations: The Challenge of Product-Related Impurities. Hum. Gene Ther. Methods. 2017;28:101–108. doi: 10.1089/hgtb.2016.188. [DOI] [PubMed] [Google Scholar]; Schnodt, M., and Buning, H. (2017). Improving the Quality of Adeno-Associated Viral Vector Preparations: The Challenge of Product-Related Impurities. Hum. Gene Ther. Methods 28, 101-108. [DOI] [PubMed]
- 8.Zhang W., Wu M., Menesale E., Lu T., Magliola A., Bergelson S. Development and qualification of a high sensitivity, high throughput Q-PCR assay for quantitation of residual host cell DNA in purification process intermediate and drug substance samples. J. Pharm. Biomed. Anal. 2014;100:145–149. doi: 10.1016/j.jpba.2014.07.037. [DOI] [PubMed] [Google Scholar]; Zhang, W., Wu, M., Menesale, E., Lu, T., Magliola, A., and Bergelson, S. (2014). Development and qualification of a high sensitivity, high throughput Q-PCR assay for quantitation of residual host cell DNA in purification process intermediate and drug substance samples. J. Pharm. Biomed. Anal. 100, 145-149. [DOI] [PubMed]
- 9.Wang X., Morgan D.M., Wang G., Mozier N.M. Residual DNA analysis in biologics development: review of measurement and quantitation technologies and future directions. Biotechnol. Bioeng. 2012;109:307–317. doi: 10.1002/bit.23343. [DOI] [PubMed] [Google Scholar]; Wang, X., Morgan, D.M., Wang, G., and Mozier, N.M. (2012). Residual DNA analysis in biologics development: review of measurement and quantitation technologies and future directions. Biotechnol. Bioeng. 109, 307-317. [DOI] [PubMed]
- 10.Hussain M. A direct qPCR method for residual DNA quantification in monoclonal antibody drugs produced in CHO cells. J. Pharm. Biomed. Anal. 2015;115:603–606. doi: 10.1016/j.jpba.2015.03.005. [DOI] [PubMed] [Google Scholar]; Hussain, M. (2015). A direct qPCR method for residual DNA quantification in monoclonal antibody drugs produced in CHO cells. J. Pharm. Biomed. Anal. 115, 603-606. [DOI] [PubMed]
- 11.Peper G., Fankhauser A., Merlin T., Roscic A., Hofmann M., Obrdlik P. Direct real-time quantitative PCR for measurement of host-cell residual DNA in therapeutic proteins. J. Pharm. Biomed. Anal. 2014;100:123–130. doi: 10.1016/j.jpba.2014.07.032. [DOI] [PubMed] [Google Scholar]; Peper, G., Fankhauser, A., Merlin, T., Roscic, A., Hofmann, M., and Obrdlik, P. (2014). Direct real-time quantitative PCR for measurement of host-cell residual DNA in therapeutic proteins. J. Pharm. Biomed. Anal. 100, 123-130. [DOI] [PubMed]
- 12.Al-Soud W.A., Jönsson L.J., Râdström P. Identification and characterization of immunoglobulin G in blood as a major inhibitor of diagnostic PCR. J. Clin. Microbiol. 2000;38:345–350. doi: 10.1128/jcm.38.1.345-350.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]; Al-Soud, W.A., Jonsson, L.J., and Radstrom, P. (2000). Identification and characterization of immunoglobulin G in blood as a major inhibitor of diagnostic PCR. J. Clin. Microbiol. 38, 345-350. [DOI] [PMC free article] [PubMed]
- 13.Queipo-Ortuño M.I., De Dios Colmenero J., Macias M., Bravo M.J., Morata P. Preparation of bacterial DNA template by boiling and effect of immunoglobulin G as an inhibitor in real-time PCR for serum samples from patients with brucellosis. Clin. Vaccine Immunol. 2008;15:293–296. doi: 10.1128/CVI.00270-07. [DOI] [PMC free article] [PubMed] [Google Scholar]; Queipo-Ortuño, M.I., De Dios Colmenero, J., Macias, M., Bravo, M.J., and Morata, P. (2008). Preparation of bacterial DNA template by boiling and effect of immunoglobulin G as an inhibitor in real-time PCR for serum samples from patients with brucellosis. Clin. Vaccine Immunol. 15, 293-296. [DOI] [PMC free article] [PubMed]
- 14.Cai H., Gu X., Scanlan M.S., Lively C.R. Development of a quantitative PCR assay for residual mouse DNA and comparison of four sample purification methods for DNA isolation. J. Pharm. Biomed. Anal. 2011;55:71–77. doi: 10.1016/j.jpba.2011.01.010. [DOI] [PubMed] [Google Scholar]; Cai, H., Gu, X., Scanlan, M.S., and Lively, C.R. (2011). Development of a quantitative PCR assay for residual mouse DNA and comparison of four sample purification methods for DNA isolation. J. Pharm. Biomed. Anal. 55, 71-77. [DOI] [PubMed]
- 15.Werling N.J., Satkunanathan S., Thorpe R., Zhao Y. Systematic Comparison and Validation of Quantitative Real-Time PCR Methods for the Quantitation of Adeno-Associated Viral Products. Hum. Gene Ther. Methods. 2015;26:82–92. doi: 10.1089/hgtb.2015.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; Werling, N.J., Satkunanathan, S., Thorpe, R., and Zhao, Y. (2015). Systematic Comparison and Validation of Quantitative Real-Time PCR Methods for the Quantitation of Adeno-Associated Viral Products. Hum. Gene Ther. Methods 26, 82-92. [DOI] [PMC free article] [PubMed]
- 16.Rayaprolu V., Kruse S., Kant R., Venkatakrishnan B., Movahed N., Brooke D., Lins B., Bennett A., Potter T., McKenna R. Comparative analysis of adeno-associated virus capsid stability and dynamics. J. Virol. 2013;87:13150–13160. doi: 10.1128/JVI.01415-13. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rayaprolu, V., Kruse, S., Kant, R., Venkatakrishnan, B., Movahed, N., Brooke, D., Lins, B., Bennett, A., Potter, T., McKenna, R., et al. (2013). Comparative analysis of adeno-associated virus capsid stability and dynamics. J. Virol. 87, 13150-13160. [DOI] [PMC free article] [PubMed]
- 17.Wang Y., Bergelson S., Feschenko M. Determination of Lentiviral Infectious Titer by a Novel Droplet Digital PCR Method. Hum. Gene Ther. Methods. 2018;29:96–103. doi: 10.1089/hgtb.2017.198. [DOI] [PubMed] [Google Scholar]; Wang, Y., Bergelson, S., and Feschenko, M. (2018). Determination of Lentiviral Infectious Titer by a Novel Droplet Digital PCR Method. Hum. Gene Ther. Methods 29, 96-103. [DOI] [PubMed]
- 18.Wang Y., Cooper R., Bergelson S., Feschenko M. Quantification of residual BHK DNA by a novel droplet digital PCR technology. J. Pharm. Biomed. Anal. 2018;159:477–482. doi: 10.1016/j.jpba.2018.07.022. [DOI] [PubMed] [Google Scholar]; Wang, Y., Cooper, R., Bergelson, S., and Feschenko, M. (2018). Quantification of residual BHK DNA by a novel droplet digital PCR technology. J. Pharm. Biomed. Anal. 159, 477-482. [DOI] [PubMed]
- 19.Bennett A., Patel S., Mietzsch M., Jose A., Lins-Austin B., Yu J.C., Bothner B., McKenna R., Agbandje-McKenna M. Thermal Stability as a Determinant of AAV Serotype Identity. Mol. Ther. Methods Clin. Dev. 2017;6:171–182. doi: 10.1016/j.omtm.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bennett, A., Patel, S., Mietzsch, M., Jose, A., Lins-Austin, B., Yu, J.C., Bothner, B., McKenna, R., and Agbandje-McKenna, M. (2017). Thermal Stability as a Determinant of AAV Serotype Identity. Mol. Ther. Methods Clin. Dev. 6, 171-182. [DOI] [PMC free article] [PubMed]