Abstract
Chromosomal aneuploidy is a defining feature of epithelial cancers. The pattern of aneuploidies is cancer-type specific. For instance, the gain of chromosome 13 occurs almost exclusively in colorectal cancer. We used microcell-mediated chromosome transfer to generate gains of chromosome 13 in the diploid human colorectal cancer cell line DLD-1. Extra copies of chromosome 13 resulted in a significant and reproducible up-regulation of transcript levels of genes on chromosome 13 (P = .0004, FDR = 0.01) and a genome-wide transcriptional deregulation in all 8 independent clones generated. Genes contained in two clusters were particularly affected: the first cluster on cytoband 13q13 contained 7 highly up-regulated genes (NBEA, MAB21L1, DCLK1, SOHLH2, CCDC169, SPG20 and CCNA1, P = .0003) in all clones. A second cluster was located on 13q32.1 and contained five upregulated genes (ABCC4, CLDN10, DZIP1, DNAJC3 and UGGT2, P = .003). One gene, RASL11A, localized on chromosome band 13q12.2, escaped the copy number-induced overexpression and was reproducibly and significantly down-regulated on the mRNA and protein level (P = .0001, FDR = 0.002). RASL11A expression levels were also lower in primary colorectal tumors as compared to matched normal mucosa (P = .0001, FDR = 0.0001. Overexpression of RASL11A increases cell proliferation and anchorage independent growth while decreasing cell migration in +13 clones. In summary, we observed a strict correlation of genomic copy number and resident gene expression levels, and aneuploidy dependent consistent genome-wide transcriptional deregulation.
Introduction
Chromosomal aneuploidy, defined by gains or losses of chromosomes, is a defining feature of cancers of epithelial origin, i.e., carcinomas [1], [2]. Chromosomal aneuploidies occur in a tissue specific distribution: for example, essentially all cervical carcinomas carry extra copies of the long arm of chromosome 3 and the detection of 3q gain in premalignant cervical dysplasia serves as a sensitive and specific biomarker for progression to invasive disease [3]. Colorectal cancers (CRCs), on the other hand, are defined by copy number gains of chromosomes and chromosome arms 7, 8q, 13 and 20, and losses of 17p and 18q [4], [5]. Specific chromosomal aneuploidies occur at early stages of tumorigenesis, i.e., before the transition to invasive disease, and are maintained during disease progression. They persist in distant metastases, and in tumor-derived cell lines [6], [7]. We previously showed that, despite a certain degree of intratumor heterogeneity, the tumor population as a whole is defined by these aneuploidies [8]. Thus, aneuploidies can be considered drivers of carcinogenesis.
Chromosomal aneuploidies in early dysplasias facilitate the acquisition of additional genetic alterations during tumor evolution [4]. Consequently, several studies utilized yeast, mouse and human cells as model systems to examine their functional impact on the cellular transcriptome and proteome [9], [10], [11], [12]. These studies revealed that the presence of an extra chromosome is associated with elevated global transcriptional activity. Moreover, Upender and colleagues demonstrated that the insertion of exogenous chromosomes into diploid cells results in increased transcript levels of genes that reside on the trisomic chromosome; this effect is independent of the specific chromosome introduced and of the cell type [9].
To determine the consequence of aneuploidy on gene expression, we inserted chromosome 13 into the karyotypically stable, mismatch repair deficient CRC line, DLD-1. We analyzed eight independent clones to specifically address the degree of consistency in aneuploidy induced transcriptional deregulation, both locally and throughout the genome. Further elucidation of the mechanistic basis by which such initial genomic alterations confer growth advantages to pre-cancerous cells may open avenues for early targeted therapeutic interventions [13].
Materials and Methods
Cell Culture
Somatic cell hybrids were purchased from the Coriell Repository and DLD-1 cells from ATCC (American Type Culture Collection, Manassas, VA) and maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, 100 U/ml Penicillin–Streptomycin (all Invitrogen, Grand Island, NY) at 37 °C with 5% CO2.. Microcell mediated chromosome transfer (MMCT) was performed as previously described [9]. Cells that underwent MMCT were selected and maintained in 200ug/ml geneticin (G418, Invitrogen). Chromosome 13 gain was verified using fluorescence in situ hybridization (FISH) on interphase cells. Since DLD-1 cells tend to lose the extra chromosome at higher passages, some clones had fewer than 50% of cells with a gain of chromosome 13. These clones were sub-cloned to obtain a higher percentage (on average 82%) of cells with a chromosome 13 gain (Supplemental Figure 1A).
DNA and RNA Extraction
DNA, RNA and protein were extracted simultaneously from the same cell passage for each clone. DNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen, Venlo, The Netherlands); RNA was extracted using the RNeasy kit (Qiagen). DNA and RNA concentrations were quantified using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA).
Fluorescence InSitu Hybridization (FISH)
To analyze metaphase chromosomes, cells were treated with 0.02 mg/ml Colcemid (Invitrogen, Grand Island, NY) for 1 hour, suspended in hypotonic solution for 20 minutes, and then fixed using methanol/acetic acid (1:3).
To confirm the presence of an extra copy of chromosome 13, dual color FISH was performed using a contig of three overlapping bacterial artificial chromosome (BAC) clones that contained the MET gene on chromosome band 7q31 and two overlapping genes that contained the CDX2 gene on chromosome band 13q12. The MET and CDX2 contig was labeled by nick-translation with Dy505 (Dyomics, Jena, Germany) and Spectrum Orange-dUTP (Abbott Molecular, Abbott Park, IL), respectively. The FISH procedure is described at https://ccr.cancer.gov/Genetics-Branch/thomas-ried.
FISH slides were imaged using a Leica DM-RXA fluorescence microscope (Leica, Wetzlar, Germany) equipped with custom optical filters using a 40x objective.
Array CGH
Array CGH was performed with minor modifications according to the manufacturer (Agilent Technologies, Santa Clara, CA, oligonucleotide array-based CGH for genomic DNA analysis protocol (Version 6.2, October 2010)). DLD-1 clones were labeled in Cyanine 3-dUTP (Perkin Elmer, Waltham, MA) and the corresponding reference DNA (Promega, Madison, WI) was labeled with Cyanine 5-dUTP (Perkin Elmer). Data was extracted using Nexus Copy Number 6.0 Discovery Version (Biodiscovery, Hawthorne, CA).
Gene Expression Microarrays
One color gene expression array was conducted according to Agilent's One-Color Microarray-Based Gene Expression Analysis protocol (Version 6.5 May 2010). Extracted RNA was labeled with Cyanine 3-CTP (Perkin Elmer) on an oligonucleotide-based whole human genome microarray. The microarray data was log2 transformed and normalized to 75 percentile according to the protocol by Agilent. PCA analysis was done with Partek Genomics Suite (Partek, St. Louis, MO).
For the current study, Gene Set Enrichment Analysis (GSEA) was used with gene sets extracted from Molecular Signature Database (MSigDB v.4–0) C2 (manually curated gene sets from online pathway databases, publications in PubMed and knowledge of domain experts). Only gene sets with more than 15 genes and less than 500 genes were included in the analysis. Analysis was based on a Fisher's exact t-test and a weighted scoring scheme with 1000 permutations.
RASL11A Plasmid Cloning, Lentivirus Production and Transduction
The RASL11A open reading frame was amplified by PCR from a template plasmid (MHS6278–213243659, Dharmacon, accession # BC136761.1) and cloned into a pDonr vector using Gateway BP Recombination. Entry clones of the expected size were Sanger sequenced and the validated insert was combinatorially subcloned by LR Gateway Recombination into a puromycin-resistant lentiviral backbone along with Tre3G promoter and IRES-eGFP. Proper recombination was determined by restriction digestion with BsrGI.
The lentiviral expression plasmid was transfected into 293FT cells along with Virapower Packaging Mix. The lentivirus was prepared per the kit manual with Lipofectamine 3000 (ThermoFisher Scientific). Virus was concentrated over Lenti-X Concentrator columns (Takara) and titer analysis was performed by qPCR.
Target cells were seeded in 12-well plates and allowed to attach for 24 hours. Lentiviruses for the RASL11A overexpression vector and the tetracycline-activated transactivator were added to a final MOI (Multiplicity of Infection) of 20 together with 8 μg/ml polybrene. Viruses were removed 24 hours and replaced by fresh medium. Selective medium (blasticidin 5 μg/ml; puromycin 3 μg/ml) was added after 48 hours and cells were selected for 21 days. RASL11A expression was induced using 0.1 ng/ml doxycycline and EGF positive cells were subsequently enriched by FACS sorting.
qRT-PCR
Total RNA was transcribed to complementary DNA using the Verso cDNA synthesis kit (Thermo Fisher Scientific). Quantitative reverse PCR (qRT-PCR) using Power Sybr Green (Applied Biosystems, Foster City, CA) was performed as previously described [14] using specific primers for RASL11A (Forward: AGCTGTATTCACGGCTGGTC Reverse: CATTTGGACAGGGAATCGAC) and the housekeeping gene YWHAZ (Forward: ACTTTTGGTACATTGTGGCTTCAA Reverse: CCGCCAGGACAAACCAGTAT) (Eurofins Genomics, Louisville, KY).
Proliferation Assay
DLD-1 and DLD-1 + 13 cells were seeded in triplicates in 96 well plates at 2000 cells per well. Cell growth was monitored using CellTiter-Blue Assay (Promega) according to the manufacturer's instructions. Briefly, cells were grown for 7 days; 20 μL of CellTiter-Blue was then added to each well and incubated at 37 °C in the dark for 90 minutes. Fluorescence generated by the conversion of the substrate by living cells was measured using a microplate reader SpectraMaxM2e (Molecular Devices, Sunnyvale, CA) at excitation 560 nm and emission 590 nm. Background fluorescence was measured in wells containing media only and subtracted from all other measured fluorescence values. Cell growth was calculated as fold over seeding: [fluorescence generated from cells at day (x) / fluorescence of cells generated just after seeding (day 0)]. Three independent experiments were performed, and the Student's t-test was used to assess statistical significance (P ≤ .05).
Soft Agar Colony Formation Assay
Soft agar assays were performed in 6well plates as described by Borowitz et al. [15]. 5000 cells/well plated in the top agar and colonies allowed to grow for 3 weeks. Cells were then labeled with nitroblue tetrazolium overnight, fixed with 4% neutral buffered formaldehyde, and plates scanned using a flatbed scanner. The number of colonies was analyzed using ImageJ 1–49, and statistical analysis performed in GraphPad Prism 7 for Mac.
Scratch Wound Migration Assay
The scratch wound migration assay was performed using the IncuCyte (Essen BioScience, Ann Arbor, MI), Briefly, 105 cells/well were plated, incubated overnight, and the cell free areas (‘scratches’) created with the WoundMaker (Essen BioScience). Plates were then placed into the IncuCyte, imaged every 4 h using for 2 days and wound closure as measured by wound density over time analyzed. To compare wound closure for multiple experiments, the area under the graph was calculated. Statistical analysis was performed in GraphPad Prism 7 for Mac.
Western Blot Analysis
For Western blot analysis, 40 μg protein was separated on a 4–12% gel using the XCell SureLock Mini-Cell (Thermo Fisher Scientific), transferred to PVDF membranes, and proteins detected using standard procedures. The following antibodies were used: RASL11A; 1:500 (NBP2–30368, Novus Biologicals, Littleton, CO), β-actin; 1:2000 (NP600-S01, Novus Biologicals, Littleton, CO), goat anti-rabbit–HRP (1:3,500, Thermo Fisher Scientific, Waltham, MA).
Results
Generation and Characterization of DLD-1 Cells With Additional Copies of Chromosome 13
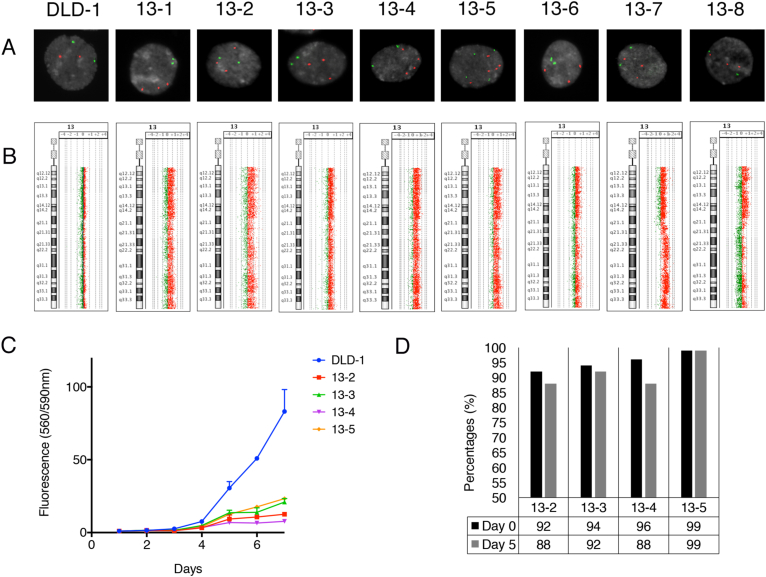
To establish models to study the consequences of chromosomal aneuploidy and the ensuing genomic imbalances on the cellular transcriptome, we used microcell mediated chromosome transfer (MMCT) to generate gains of chromosome 13 in the diploid and chromosomally stable CRC cell line DLD-1. We confirmed the gain of chromosome 13 in single-cell derived, independent clones by interphase FISH, high-resolution aCGH (Figure 1, A and B, Supplemental Figure 1, A–D), and whole chromosome paints (Supplemental Figure 1E). Clones 13–1, 13–2, 13–3 and 13–6 incorporated an intact copy of chromosome 13 (Figure 1, A and B). Approximately 50% of the population within clone 13–4 contained four copies of chromosome 13. Furthermore, clone 13–5 was tetraploid with six copies of chromosome 13 (Figure 1, A and B). Clone 13–7 had a higher copy gain extending from chromosome bands 13q21.2 to 13q31, while clone 13–8 exhibited a partial gain from the centromere to band 13q21.1 (Figure 1B, Supplemental Figure 1, B and C). The introduction of chromosome 13 did not induce any other karyotypic changes.
Figure 1.
Characterization of the eight single cell derived clones of DLD-1 + 13 cells. (A) FISH probes corresponding to the MET gene (chromosome band 7q31, green signals) and CDX2 (chromosome band 13q21, red signals) hybridized onto interphase nuclei for the parental DLD-1 WT and the eight DLD-1 + 13 clones. One hundred fifty cells were counted per clone. (B) Genomic aberration profile in the DLD-1 WT clone and all eight DLD-1 + 13 clones using aCGH. (C) Proliferation graph for the DLD-1 WT and four DLD-1 + 13 clones. (D) Percentages of cells (n = 100) with a gain of chromosome 13 as analyzed by FISH for four DLD-1 + 13 clones on day 0 and day 5 without neomycin selection. The FISH probes used were MET (7q31) and CDX2 (13q12). A gain of chromosome 13 was made if there were more copies of CDX2 as compared to MET within the same cell.
Gain of Chromosome 13 Reduces Proliferation of DLD-1 Cells
Next, we measured the consequences of chromosomal aneuploidy on cell growth. Invariably, the cell lines with an extra copy of chromosome 13 grew slower than the parental DLD-1 line (Figure 1C). Since the cell lines with extra copies of chromosome 13 were cultured in neomycin-containing medium, we asked whether the slower growth of the DLD-1 + 13 clones was attributable to the presence of neomycin. This was not the case (Supplemental Figure 1F). Over the course of 5 days, the aneuploid chromosome was maintained without selection, as confirmed by dual color interphase FISH (Figure 1D).
Aneuploidy of Chromosome 13 Alters the Cellular Transcriptome
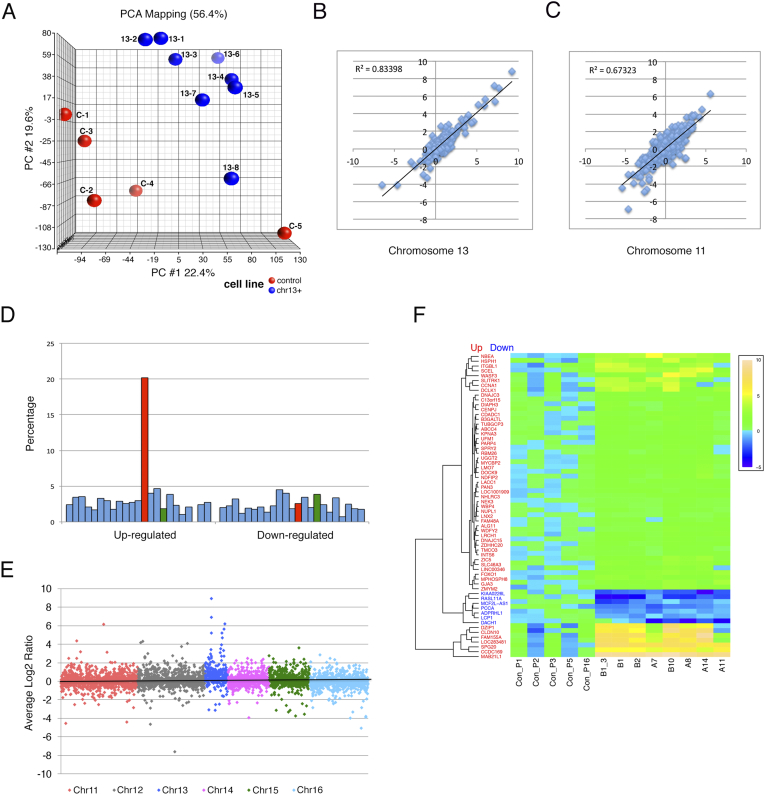
We next asked how the gain of chromosome 13 affects the cellular transcriptome by analyzing global gene expression levels using microarrays. Principal Component Analysis (PCA) (Figure 2A) showed that aneuploid clones cluster apart from the controls. An exception was clone 13–8, which has a partial chromosome 13 gain and was positioned relatively apart from to the main cluster of derived cell lines. Genome-wide, 464 genes were up-regulated, and 379 were down-regulated (fold change ≥1.5, FDR ≤0.05) in clones 13–1 to 13–8 (Supplemental Table 1, Supplemental Table 2). The reproducibility of the gene expression analyses is presented as correlation plots for chromosomes 13 (the aneuploid chromosome) and 11 (as a control) (Figure 2, B and C). The average magnitude of gene expression increase was higher for chromosome 13 (P = .0004, FDR = 0.01) compared to other chromosomes. Of the 273 genes on chromosome 13 that were present on the arrays, 20% percent showed significantly elevated expression, while 2.5% were down-regulated in cells with chromosome 13 gain (fold change >1.5, FDR ≤0.05) (Figure 2, D–F). Genes on chromosome 16 were more likely to be down-regulated in clones with gain of chromosome 13 (P = .02, FDR ≤0.05) (Figure 2, D and E, Supplemental Table 2). We did not observe other predilections for global gene expression changes on a particular chromosome after the addition of chromosome 13 (Supplemental Table 1, Supplemental Table 2). In order to explore pathways that are deregulated as a consequence of adding an additional chromosome 13, we used GSEA, which revealed a significant deregulation of genes associated with the PI3K pathway (Supplemental Figure 1G, Supplemental Table 3).
Figure 2.
Transcriptome analysis. (A) Principal component analysis for the five DLD-1 WT biological replicates (red) and the eight DLD-1 + 13 clones (blue). (B) Correlation plots between clone 13–2 and clone 13–3 for genes (log2 ratio) on chromosome 13 (C) Correlation plots between clone 13–2 and clone 13–3 for genes (log2 ratio) on chromosome 11. (D) Chromosome-wide gene expression patterns. The ratio of significant genes on each chromosome after normalization (fold change ≥1.5, FDR ≤0.05) is shown. Each bar represents a chromosome, with chromosome 1 starting from the left. Chromosome 13 is in red and chromosome 16 is in green. (E) Genes from chromosome 11–16 were plotted according to the average log2 ratio for all clones. (F) Heat map for up-regulated (red letters) and down-regulated (blue letters) genes on chromosome 13.
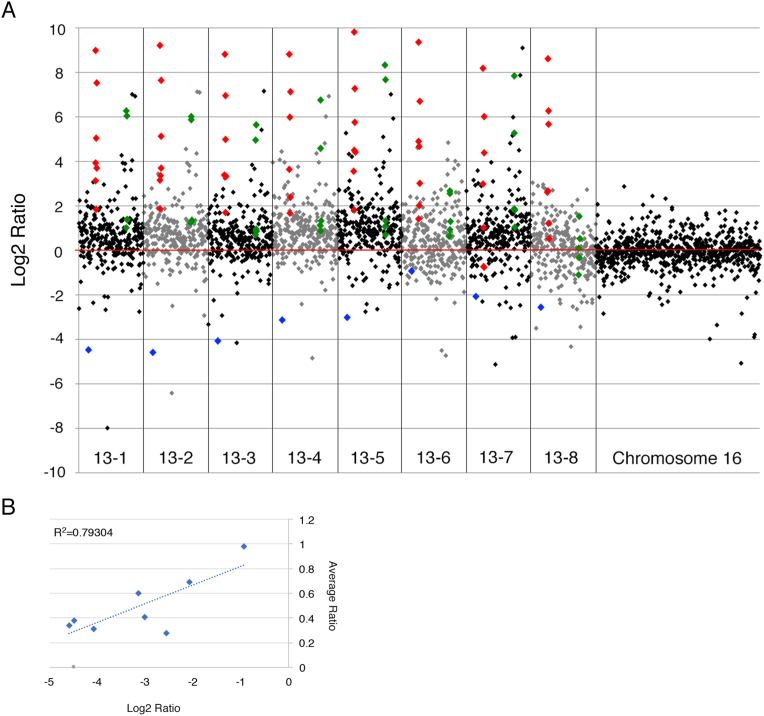
Further analysis revealed evidence of region-specific and non-random up-regulation of two discrete clusters of more than three contiguous genes, suggestive of coordinated regulation. The first cluster was located on cytoband 13q13.3 and contained seven highly upregulated genes: NBEA, MAB21L1, DCLK1, SOHLH2, CCDC169, SPG20 and CCNA1 (P = .0003) (Figure 3A, Supplemental Table 1). The second cluster was located on cytoband 13q32.1 and contained five upregulated genes (ABCC4, CLDN10, DZIP1, DNAJC3 and UGGT2) (P = .003), however, the expression was not as high as that of genes found in the first cluster (Figure 3A, Supplemental Table 1). Of note, RASL11A, which is located at cytoband 13q12.2, escaped the copy number increased up-regulation and was consistently down-regulated (P = .0001, FDR = 0.002) by, on average, 8.6-fold (Figure 3A, Supplemental Table 2).
Figure 3.
(A) All genes on chromosome 13 (log2 ratio) are plotted consecutively for each DLD-1 + 13 clone (x-axis), with each diamond representing one gene. The highest up-regulated group of genes, which belong to the contiguous locus on 13q13.3 are color-coded red. The second up-regulated group of genes on 13q32.1 is color-coded green. RASL11A, the most down-regulated gene on chromosome 13, is color-coded blue. The red line depicts the zero line. Genes on chromosome 16 are plotted at the right for comparison. (B) Correlation plot between the RASL11A gene expression levels (log2 ratio, x-axis) and the RASL11A protein expression levels (average ratio, y-axis) for all eight DLD-1 + 13 clones.
Gain of Chromosome 13 Leads to Decreased Expression of RASL11A
We next explored whether gene expression changes resulted in protein level changes of RASL11A using selected reaction monitoring (SRM) mass spectrometry to quantify protein expression levels (Supplemental Materials and Methods). Protein ratios were quantified based on a single unique peptide from duplicate analyses and the coefficient of variation (CV) determined. Indeed, RASL11A protein was down-regulated in all clones. Furthermore, we observed that RASL11A mRNA expression correlates with RASL11A protein levels (r2 = 0.79, Figure 3B), indicating that RASL11A mRNA directly determines RASL11A protein expression levels.
RASL11A Expression is Reduced in Colorectal Tumors
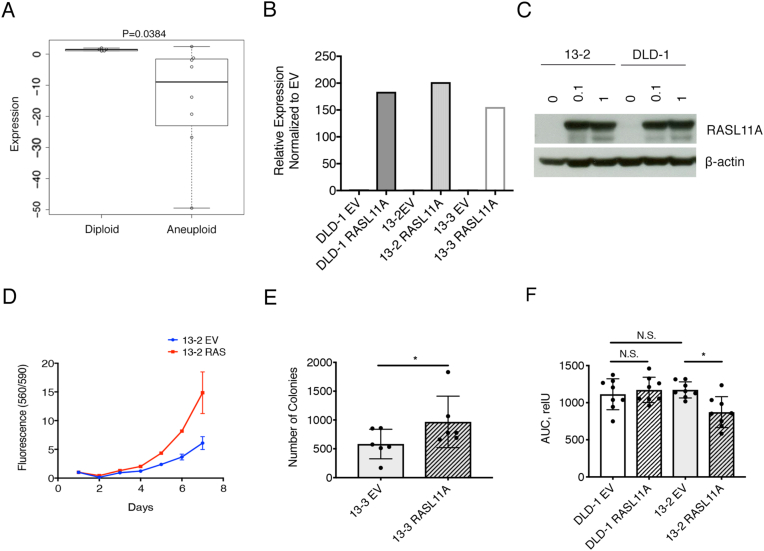
We next aimed to clarify if RASL11A expression is decreased in primary CRC in a set of 216 primary rectal cancers and matched normal mucosa [16]. We found that RASL11A mRNA levels were reduced by 1.28 fold in rectal cancers compared to matched normal mucosa (P = .0001, FDR = 0.0001) (Supplemental Table 4) [16]. When comparing 18 CRC derived cell lines that were aneuploid versus those that were diploid (i.e., deficient in mismatch repair), RASL11A had a significantly lower expression in aneuploid CRC cell lines (P = .0384) (Figure 4A). The aneuploid CRC derived cell lines all had a gain of chromosome 13 except for T84.
Figure 4.
RASL11A (A) Box plot of relative RASL11A expression levels in 8 aneuploid and 6 diploid CRC cell lines. The x-axis depicts the two groups while the y-axis depicts the relative expression level of RASL11A. (B) RASL11A relative expression in the transduced DLD-1 WT, clone 13–2 and clone 13–3 normalized to the empty vector (EV) controls. (C) Western blot for RASL11A and the loading control, β-actin for DLD-1 WT with RASL11A overexpression and DLD-1 + 13 clone 13–2 with RASL11A overexpression. RASL11A expression was induced by doxycycline in the indicated concentration. (D) Seven-day growth curve of DLD-1 + 13 clone 13–2 empty vector (EV) and RASL11A. (E) Box plot of the number of colonies (y-axis) in DLD-1 + 13 clone 13–3 EV control and RASL11A overexpression. *P ≤ .05. (F) Scratch assay between DLD-1 WT cells and DLD-1 + 13 clone 13–3 cells. Each compares EV vs RASL11A overexpression. *P ≤ .05, N.S. = P > .05.
RASL11A Overexpression Does Not Reduce InVitro Tumorigenicity of DLD-1 Cells
In order to test if RASL11A reduces tumorigenicity in vitro, we overexpressed RASL11A in DLD-1 + 13 clones (13–2, 13–3, 13–4) and DLD-1 cells (Figure 4, B and C; Supplemental Figure 2A) using a doxycycline-inducible expression system. We additionally expressed RASL11A in HT-29 cells, a aneuploid colorectal cancer cell line with a gain of chromosome 13 that showed low expression of RASL11A as compared to normal colon mucosa (Supplemental Figure 2B) [17]. Overexpression of RASL11A increased cell proliferation compared to control cells (empty vector) in the DLD-1 + 13 clones (Figure 4D). Soft agar assay showed an increased number of colonies in the DLD-1 + 13 RASL11A overexpressing cells as compared to the empty vector (Figure 4E), whereas DLD-1 cells showed no differences (Supplemental Figure 2C). Using the wound healing assay, we found that DLD-1 + 13 clones overexpressing RASL11A tended to close scratches slower than cells expressing the control vector (Figure 4F). In DLD-1 cells, there was no difference when overexpressing RASL11A. Interestingly, overexpression of RASL11A did not affect colony formation or wound closure in HT29 cells (Supplemental Figure 2, D and E). Overall, RASL11A overexpression increases cell proliferation, and anchorage independent growth while decreasing cell migration in DLD-1 + 13 cells.
In summary, we have established 8 cell lines with additional copies of chromosome 13. This aneuploidy results in a highly reproducible, consistent upregulation of genes on chromosome 13, which, in turn, affects gene expression levels throughout the genome. We conclude that aneuploidy results in a massive deregulation of the transcriptional equilibrium in cancer cells.
Discussion
Aneuploidy is a defining hallmark of carcinomas and results in a tissue specific distribution of genomic imbalances. We now systematically explored how such specific genomic imbalances affect the cellular transcriptome.
In the present study, we inserted chromosome 13 into the diploid colorectal cancer cell line DLD-1, and generated eight independent clones with additional copies of chromosome 13. We detected a consistent and highly reproducible correlation between copy number increase and gene expression levels among the biological replicates. Upender et al. [9] had determined that the correlation between genomic copy number and gene expression levels is independent of the introduced chromosome, and independent of the recipient cell line. We show that the addition of a chromosome affects a substantial number of genes residing on other chromosomes as well. This global effect was not due to genomic rearrangements as reported by Passerini et al. [18]. Of note, the transcriptional changes were highly reproducible between replicates. Our DLD-1 + 13 cell lines, not otherwise available, present clean experimental systems to study the consequences of specific aneuploidies on gene expression.
We identified two clusters on chromosome 13 that were specifically subject to gene expression increases as a consequence of the induced aneuploidy, which were not identified in our previous analysis due to the lower density of features on the arrays [9]. The overexpression of genes on the two specific loci on chromosome 13 is not known to be directly associated with colon cancer and was not found by Camps et al. to be commonly subject to focal amplifications in CRC [14]. The two identified clusters could potentially be regulating genes on other chromosomes. Nicholson and colleagues found that the overexpression of one gene, SPG20, expressed in cluster 13q13.3, promotes cytokinesis failure in the DLD-1 + 13 cells [19].
Growth impairment was previously observed in cell lines with artificially inserted chromosomes [10], [12], [20]. Consistently, all eight DLD-1 + 13 clones grew significantly slower in comparison to the parental DLD-1 cells, indicating that incorporation of extra genomic material resulted in marked growth impairment. Rutledge et al. observed higher proliferation of the DLD-1 + 13 cells cultured in serum-free conditions or with the addition on 5-FU, which could be due to a selective advantage due to phenotypic differences in the cells [20].
RASL11A, a gene that escaped the genomic copy number induced overexpression, was consistently down-regulated in all DLD-1 + 13 clones. RASL11A belongs to a small family of GTPases that are highly similar to RAS proteins and are conserved in eukaryotes [21]. Functionally, little is known about RASL11A other than its role as a chromatin-associated modulator of pre-ribosomal RNA synthesis and its interaction with a putative enhancer characterized by H3K4me1 marks [22] [23]. However, it is consistently down-regulated in not only DLD-1 + 13 clones but also in primary rectal cancer samples and CRC derived cell lines. RASL11A expression is detectable in the colon, but other than the current study, there is no data regarding RASL11A expression in CRC [21]. RASL11A was found consistently down-regulated in prostate cancer, which could suggest a tumor suppressor role [21]. We found that the overexpression of RASL11A in the DLD-1 + 13 clones increases cell proliferation and colony formation in soft agar assays, while at the same time reducing cell migrations. Interestingly, these effects were limited to DLD-1 + 13 cells and were not observed in DLD-1 and HT29 cells. This could be due to genomic content since HT29 has other chromosomal aberrations as well. Orlando and colleagues reduced RASL11A levels using short interfering RNA which resulted in decreased cell viability and proliferation [23]. Our results are consistent with their findings. When we over-expressed RASL11A in HT29 cells, we found only a slight increase in cell proliferation although it was not significant. We conclude that the functions of RASL11A are not consistent with the role of a tumor suppressor protein.
In summary, our findings show that the introduction of chromosome 13 into diploid cells has a non-random effect on the expression of genes on chromosomes other than 13, and this effect can result in both down-regulation and up-regulation of gene expression. The effects were consistent among the different clones, suggesting a general, reproducible transcriptional deregulation as a consequence of chromosome-specific aneuploidy.
The following are the supplementary data related to this article.
(A) Percentage of cells (n = 100)(y-axis) with a gain of chromosome 13 using FISH probes corresponding to MET and CDX2 in all eight DLD-1+13 clones (x-axis). (B) Array-CGH depicting the gain of chromosome 13 for clone 13-7. (C) Array-CGH depicting the gain of chromosome 13 for clone 13-8. (D) Array-CGH for clone 13-4 depicting the gain of chromosome 13. The gain of chromosome 2p, 11p and loss of chromosome 6p, are known aberrations in DLD-1 WT cells. (E) Chromosome 13 paint probes (pink) were hybridized on the eight DLD-1 13 clones. (F) Proliferation graph for DLD-1, and Clone 13-2 with and without neomycin. (G) Gene set enrichment analysis for PI3K comparing DLD-1 and DLD-1+13 clones.
(A) Western blot of DLD-1 WT and clones 13-2 with RASL11A overexpression and the empty vector (EV) control with and without induction of doxycycline (0, 0.1 ng/ml and 1 ng/ml. β-actin served as the loading control. (B) RT-PCR results for HT29, DLD-1 WT and DLD-1+13 clone 13-2 overexpression of RASL11A and EV control compared to the uninduced control. (C) Box plot of the number of colonies (y-axis) between DLD-1 WT EV control and DLD-1 WT RASL11A overexpression (x-axis). N.S. = P > .05. (D) Scratch assay box plot between HT29 EV and the corresponding RASL11A overexpression. N.S. = P > .05. AUC = area under the curve. (E) Box plot of the number of colonies (y-axis) between HT29 EV and RASL11A overexpression (x-axis). N.S. = P > .05.
Up-regulated genes on all chromosomes
Down-regulated genes on all chromosomes
Pathways that are deregulated as a consequence of adding an additional chromosome 13
RASL11A mRNA expression
Supplemental Material and Methods
Acknowledgements
The study was supported by the Intramural Research Program, National Cancer Institute/NIH. RB was supported by the Deutsche Krebshilfe, GE through the Deutsche Forschungsgemeinschaft. This work was supported by the Instituto de Salud Carlos III and co-funded by the European Regional Development Fund (ERDF) [CP13/00160, CPII18/00026, PI14/00783]; the CIBEREHD program; the CERCA Program (Generalitat de Catalunya); the Agència de Gestió d'Ajuts Universitaris i de Recerca, Generalitat de Catalunya [2017 SGR 1035]. R.B. was supported by a Mildred Scheel postdoctoral scholarship of the German Cancer Aid (Deutsche Krebshilfe).
Footnotes
The authors declare no conflicts of interest.
Contributor Information
Jordi Camps, Email: jcamps@clinic.ub.es.
Thomas Ried, Email: riedt@mail.nih.gov.
References
- 1.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]; Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. (2010). The landscape of somatic copy-number alteration across human cancers Nature 463, 899-905. [DOI] [PMC free article] [PubMed]
- 2.Ried T. Homage to Theodor Boveri (1862-1915): Boveri's theory of cancer as a disease of the chromosomes, and the landscape of genomic imbalances in human carcinomas. Environ Mol Mutagen. 2009;50:593–601. doi: 10.1002/em.20526. [DOI] [PubMed] [Google Scholar]; Ried T (2009). Homage to Theodor Boveri (1862-1915): Boveri's theory of cancer as a disease of the chromosomes, and the landscape of genomic imbalances in human carcinomas Environ Mol Mutagen 50, 593-601. [DOI] [PubMed]
- 3.Heselmeyer-Haddad K, Janz V, Castle PE, Chaudhri N, White N, Wilber K, Morrison LE, Auer G, Burroughs FH, Sherman ME. Detection of genomic amplification of the human telomerase gene (TERC) in cytologic specimens as a genetic test for the diagnosis of cervical dysplasia. Am J Pathol. 2003;163:1405–1416. doi: 10.1016/S0002-9440(10)63498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; Heselmeyer-Haddad K, Janz V, Castle PE, Chaudhri N, White N, Wilber K, Morrison LE, Auer G, Burroughs FH, Sherman ME, et al. (2003). Detection of genomic amplification of the human telomerase gene (TERC) in cytologic specimens as a genetic test for the diagnosis of cervical dysplasia Am J Pathol 163, 1405-1416. [DOI] [PMC free article] [PubMed]
- 4.Ried T, Heselmeyer-Haddad K, Blegen H, Schrock E, Auer G. Genomic changes defining the genesis, progression, and malignancy potential in solid human tumors: a phenotype/genotype correlation. Genes Chromosomes Cancer. 1999;25:195–204. doi: 10.1002/(sici)1098-2264(199907)25:3<195::aid-gcc1>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]; Ried T, Heselmeyer-Haddad K, Blegen H, Schrock E, Auer G (1999). Genomic changes defining the genesis, progression, and malignancy potential in solid human tumors: a phenotype/genotype correlation Genes Chromosomes Cancer 25, 195-204. [DOI] [PubMed]
- 5.Ried T, Hu Y, Difilippantonio MJ, Ghadimi BM, Grade M, Camps J. The consequences of chromosomal aneuploidy on the transcriptome of cancer cells. Biochim Biophys Acta. 2012;1819:784–793. doi: 10.1016/j.bbagrm.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ried T, Hu Y, Difilippantonio MJ, Ghadimi BM, Grade M, Camps J (2012). The consequences of chromosomal aneuploidy on the transcriptome of cancer cells Biochim Biophys Acta 1819, 784-793. [DOI] [PMC free article] [PubMed]
- 6.Al-Mulla F, Keith WN, Pickford IR, Going JJ, Birnie GD. Comparative genomic hybridization analysis of primary colorectal carcinomas and their synchronous metastases. Genes Chromosomes Cancer. 1999;24:306–314. doi: 10.1002/(sici)1098-2264(199904)24:4<306::aid-gcc3>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]; Al-Mulla F, Keith WN, Pickford IR, Going JJ, Birnie GD (1999). Comparative genomic hybridization analysis of primary colorectal carcinomas and their synchronous metastases Genes Chromosomes Cancer 24, 306-314. [DOI] [PubMed]
- 7.Sylvester BE, Vakiani E. Tumor evolution and intratumor heterogeneity in colorectal carcinoma: insights from comparative genomic profiling of primary tumors and matched metastases. J Gastrointest Oncol. 2015;6:668–675. doi: 10.3978/j.issn.2078-6891.2015.083. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sylvester BE, Vakiani E (2015). Tumor evolution and intratumor heterogeneity in colorectal carcinoma: insights from comparative genomic profiling of primary tumors and matched metastases J Gastrointest Oncol 6, 668-675. [DOI] [PMC free article] [PubMed]
- 8.Heselmeyer-Haddad K, Berroa Garcia LY, Bradley A, Ortiz-Melendez C, Lee WJ, Christensen R, Prindiville SA, Calzone KA, Soballe PW, Hu Y. Single-cell genetic analysis of ductal carcinoma in situ and invasive breast cancer reveals enormous tumor heterogeneity yet conserved genomic imbalances and gain of MYC during progression. Am J Pathol. 2012;181:1807–1822. doi: 10.1016/j.ajpath.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]; Heselmeyer-Haddad K, Berroa Garcia LY, Bradley A, Ortiz-Melendez C, Lee WJ, Christensen R, Prindiville SA, Calzone KA, Soballe PW, Hu Y, et al. (2012). Single-cell genetic analysis of ductal carcinoma in situ and invasive breast cancer reveals enormous tumor heterogeneity yet conserved genomic imbalances and gain of MYC during progression Am J Pathol 181, 1807-1822. [DOI] [PMC free article] [PubMed]
- 9.Upender MB, Habermann JK, McShane LM, Korn EL, Barrett JC, Difilippantonio MJ, Ried T. Chromosome transfer induced aneuploidy results in complex dysregulation of the cellular transcriptome in immortalized and cancer cells. Cancer Res. 2004;64:6941–6949. doi: 10.1158/0008-5472.CAN-04-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]; Upender MB, Habermann JK, McShane LM, Korn EL, Barrett JC, Difilippantonio MJ, Ried T (2004). Chromosome transfer induced aneuploidy results in complex dysregulation of the cellular transcriptome in immortalized and cancer cells Cancer Res 64, 6941-6949. [DOI] [PMC free article] [PubMed]
- 10.Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]; Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A (2007). Effects of aneuploidy on cellular physiology and cell division in haploid yeast Science 317, 916-924. [DOI] [PubMed]
- 11.Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]; Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A (2008). Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells Science 322, 703-709. [DOI] [PMC free article] [PubMed]
- 12.Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, Storchova Z. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol Syst Biol. 2012;8:608. doi: 10.1038/msb.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, Storchova Z (2012). Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells Mol Syst Biol 8, 608. [DOI] [PMC free article] [PubMed]
- 13.Auslander N H-HK, Patkar S, Hirsch D, Camps J, Brown M, Bronder D, Chen WD, Lokanga R, Wangsa D, Wangsa D, Hu Y, Lischka A, Braun R, Emons G, Ghadimi BM, Gaedcke J, Grade M, Montagna C, Lazebnik Y, Difilippantonio M, Habermann JK, Auer G, Ruppin E, Ried T (2019). Cancer-type specific aneuploidies hard-wire chromosome-wide gene expression patterns of their tissue of origin.
- 14.Camps J, Pitt JJ, Emons G, Hummon AB, Case CM, Grade M, Jones TL, Nguyen QT, Ghadimi BM, Beissbarth T. Genetic amplification of the NOTCH modulator LNX2 upregulates the WNT/beta-catenin pathway in colorectal cancer. Cancer Res. 2013;73:2003–2013. doi: 10.1158/0008-5472.CAN-12-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]; Camps J, Pitt JJ, Emons G, Hummon AB, Case CM, Grade M, Jones TL, Nguyen QT, Ghadimi BM, Beissbarth T, et al. (2013). Genetic amplification of the NOTCH modulator LNX2 upregulates the WNT/beta-catenin pathway in colorectal cancer Cancer Res 73, 2003-2013. [DOI] [PMC free article] [PubMed]
- 15.Borowicz S, Van Scoyk M, Avasarala S, Karuppusamy Rathinam MK, Tauler J, Bikkavilli RK, Winn RA. e51998. 2014. The soft agar colony formation assay J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]; Borowicz S, Van Scoyk M, Avasarala S, Karuppusamy Rathinam MK, Tauler J, Bikkavilli RK, Winn RA (2014). The soft agar colony formation assay J Vis Exp, e51998. [DOI] [PMC free article] [PubMed]
- 16.Hu Y, Gaedcke J, Emons G, Beissbarth T, Grade M, Jo P, Yeager M, Chanock SJ, Wolff H, Camps J. Colorectal cancer susceptibility loci as predictive markers of rectal cancer prognosis after surgery. Genes Chromosomes Cancer. 2018;57:140–149. doi: 10.1002/gcc.22512. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hu Y, Gaedcke J, Emons G, Beissbarth T, Grade M, Jo P, Yeager M, Chanock SJ, Wolff H, Camps J, et al. (2018). Colorectal cancer susceptibility loci as predictive markers of rectal cancer prognosis after surgery Genes Chromosomes Cancer 57, 140-149. [DOI] [PMC free article] [PubMed]
- 17.Grade M, Hummon AB, Camps J, Emons G, Spitzner M, Gaedcke J, Hoermann P, Ebner R, Becker H, Difilippantonio MJ. A genomic strategy for the functional validation of colorectal cancer genes identifies potential therapeutic targets. Int J Cancer. 2011;128:1069–1079. doi: 10.1002/ijc.25453. [DOI] [PMC free article] [PubMed] [Google Scholar]; Grade M, Hummon AB, Camps J, Emons G, Spitzner M, Gaedcke J, Hoermann P, Ebner R, Becker H, Difilippantonio MJ, et al. (2011). A genomic strategy for the functional validation of colorectal cancer genes identifies potential therapeutic targets Int J Cancer 128, 1069-1079. [DOI] [PMC free article] [PubMed]
- 18.Passerini V, Ozeri-Galai E, de Pagter MS, Donnelly N, Schmalbrock S, Kloosterman WP, Kerem B, Storchova Z. The presence of extra chromosomes leads to genomic instability. Nat Commun. 2016;7 doi: 10.1038/ncomms10754. [DOI] [PMC free article] [PubMed] [Google Scholar]; Passerini V, Ozeri-Galai E, de Pagter MS, Donnelly N, Schmalbrock S, Kloosterman WP, Kerem B, Storchova Z (2016). The presence of extra chromosomes leads to genomic instability Nat Commun 7, 10754. [DOI] [PMC free article] [PubMed]
- 19.Nicholson JM, Macedo JC, Mattingly AJ, Wangsa D, Camps J, Lima V, Gomes AM, Doria S, Ried T, Logarinho E. Chromosome mis-segregation and cytokinesis failure in trisomic human cells. Elife. 2015;4 doi: 10.7554/eLife.05068. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nicholson JM, Macedo JC, Mattingly AJ, Wangsa D, Camps J, Lima V, Gomes AM, Doria S, Ried T, Logarinho E, et al. (2015). Chromosome mis-segregation and cytokinesis failure in trisomic human cells Elife 4. [DOI] [PMC free article] [PubMed]
- 20.Rutledge SD, Douglas TA, Nicholson JM, Vila-Casadesus M, Kantzler CL, Wangsa D, Barroso-Vilares M, Kale SD, Logarinho E, Cimini D. Selective advantage of trisomic human cells cultured in non-standard conditions. Sci Rep. 2016;6 doi: 10.1038/srep22828. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rutledge SD, Douglas TA, Nicholson JM, Vila-Casadesus M, Kantzler CL, Wangsa D, Barroso-Vilares M, Kale SD, Logarinho E, Cimini D (2016). Selective advantage of trisomic human cells cultured in non-standard conditions Sci Rep 6, 22828. [DOI] [PMC free article] [PubMed]
- 21.Louro R, Nakaya HI, Paquola AC, Martins EA, da Silva AM, Verjovski-Almeida S, Reis EM. RASL11A, member of a novel small monomeric GTPase gene family, is down-regulated in prostate tumors. Biochem Biophys Res Commun. 2004;316:618–627. doi: 10.1016/j.bbrc.2004.02.091. [DOI] [PubMed] [Google Scholar]; Louro R, Nakaya HI, Paquola AC, Martins EA, da Silva AM, Verjovski-Almeida S, Reis EM (2004). RASL11A, member of a novel small monomeric GTPase gene family, is down-regulated in prostate tumors Biochem Biophys Res Commun 316, 618-627. [DOI] [PubMed]
- 22.Pistoni M, Verrecchia A, Doni M, Guccione E, Amati B. Chromatin association and regulation of rDNA transcription by the Ras-family protein RasL11a. EMBO J. 2010;29:1215–1224. doi: 10.1038/emboj.2010.16. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pistoni M, Verrecchia A, Doni M, Guccione E, Amati B (2010). Chromatin association and regulation of rDNA transcription by the Ras-family protein RasL11a EMBO J 29, 1215-1224. [DOI] [PMC free article] [PubMed]
- 23.Orlando G, Law PJ, Cornish AJ, Dobbins SE, Chubb D, Broderick P, Litchfield K, Hariri F, Pastinen T, Osborne CS. Promoter capture Hi-C-based identification of recurrent noncoding mutations in colorectal cancer. Nat Genet. 2018;50:1375–1380. doi: 10.1038/s41588-018-0211-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; Orlando G, Law PJ, Cornish AJ, Dobbins SE, Chubb D, Broderick P, Litchfield K, Hariri F, Pastinen T, Osborne CS, et al. (2018). Promoter capture Hi-C-based identification of recurrent noncoding mutations in colorectal cancer Nat Genet 50, 1375-1380. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Percentage of cells (n = 100)(y-axis) with a gain of chromosome 13 using FISH probes corresponding to MET and CDX2 in all eight DLD-1+13 clones (x-axis). (B) Array-CGH depicting the gain of chromosome 13 for clone 13-7. (C) Array-CGH depicting the gain of chromosome 13 for clone 13-8. (D) Array-CGH for clone 13-4 depicting the gain of chromosome 13. The gain of chromosome 2p, 11p and loss of chromosome 6p, are known aberrations in DLD-1 WT cells. (E) Chromosome 13 paint probes (pink) were hybridized on the eight DLD-1 13 clones. (F) Proliferation graph for DLD-1, and Clone 13-2 with and without neomycin. (G) Gene set enrichment analysis for PI3K comparing DLD-1 and DLD-1+13 clones.
(A) Western blot of DLD-1 WT and clones 13-2 with RASL11A overexpression and the empty vector (EV) control with and without induction of doxycycline (0, 0.1 ng/ml and 1 ng/ml. β-actin served as the loading control. (B) RT-PCR results for HT29, DLD-1 WT and DLD-1+13 clone 13-2 overexpression of RASL11A and EV control compared to the uninduced control. (C) Box plot of the number of colonies (y-axis) between DLD-1 WT EV control and DLD-1 WT RASL11A overexpression (x-axis). N.S. = P > .05. (D) Scratch assay box plot between HT29 EV and the corresponding RASL11A overexpression. N.S. = P > .05. AUC = area under the curve. (E) Box plot of the number of colonies (y-axis) between HT29 EV and RASL11A overexpression (x-axis). N.S. = P > .05.
Up-regulated genes on all chromosomes
Down-regulated genes on all chromosomes
Pathways that are deregulated as a consequence of adding an additional chromosome 13
RASL11A mRNA expression
Supplemental Material and Methods