Abstract
Abdominal arterial aneurysm (AAA) shares many features with autoimmune diseases and appears to be a T‐cell‐mediated process. In addition, certain epigenetic changes, including DNA methylation, are associated with AAA. In this study, we investigated epigenetic modifications in regulatory T cells (Tregs) from AAA patients. We used flow cytometry to sort FOXP3+ CD4+ CD25+ Tregs from the peripheral blood of AAA patients and from healthy controls (HC), and then detected DNA methylation and histone modifications by ELISA. The DNA methylation rate of Tregs was significantly higher in AAA patients than in the HC group (0.159 ± 0.08% vs 0.098 ± 0.03%, P < 0.05), while the acetylation rates of H3 and H3K9 histones were lower in the AAA than in the HC group. We also examined the expression of mRNA encoding enzymes that catalyze making and removing epigenetic modifications by real‐time PCR: we found that mRNA levels of DNA methyltransferase (DNMT) 1 and DNMT3A were higher in the AAA than in the HC group, mRNA levels of methyl‐CpG‐binding domain protein (MBD) 2 and MBD4 were higher in the AAA than in the HC group (MBD2: 6.21 ± 2.57 vs 3.04 ± 1.45; MBD4: 7.76 ± 3.48 vs 4.97 ± 3.10; both P < 0.05), and mRNA levels of histone deacetylase (HDAC) 1 and HDAC5 were significantly up‐regulated in the AAA compared with the HC group (HDAC1: 2.17 ± 1.18 vs 1.51 ± 0.99; HDAC5: 1.35 ± 0.49 vs 0.94 ± 0.76; both P < 0.05). Together, our results reveal that rates of DNA methylation and histone modifications of Tregs are significantly altered in AAA patients.
Keywords: abdominal aortic aneurysm, DNA methylation, epigenetics, histone modifications, regulatory T cells
Abbreviations
- AAA
abdominal aortic aneurysm
- DNMT
DNA methyltransferase
- FOXP3
forkhead box P3
- HAT
histone acetyltransferase
- HC
healthy control
- HDAC
histone deacetylase
- HMT
histone methyltransferases
- MBD
methyl‐CpG‐binding domain protein
- PBMC
peripheral blood mononuclear cell
- SD
standard deviation
- Treg
regulatory T cell
Aneurysms are arterial lesions that cause a persistent extension of an artery segment; they are more common in older men. They are commonly seen in the abdominal aorta (abdominal aortic aneurysm; AAA) and the lower extremity. Such arterial lesions can be caused by multiple factors including auto‐immune disease, inflammation, arterial trauma, lipid deposits, hypertension and even smoking. Rupture of an AAA is highly dangerous 1. The incidence and the prevalence of AAA in China have been increased during the past decade 2.
AAA shares many features with autoimmune diseases, including genetic predisposition, organ specificity and chronic inflammation 3. Increasing evidence has shown that an immune response plays an important role in aneurysmal disease development, and AAA appears to be a T‐cell‐mediated process 4, 5. Vascular and molecular biology studies have revealed the role of the adaptive and innate immune systems in the initiation and propagation of the inflammatory response in aortic or aneurysmal tissue 6. Our previous studies have revealed impaired Fas‐induced apoptosis of T lymphocytes and a functional deficiency of regulatory T cells (Tregs) in AAA patients, which supports the abnormal modification of peripheral T cells in AAA tissue and implies an important role of autoimmunity in the etiology of AAA 7, 8. However, the mechanisms modulated by Tregs in AAA were still unknown.
Epigenetics is defined as the research on genome properties that are not explained by the primary DNA sequence, but are caused by modifications of the DNA and/or its associated proteins 9. Epigenetic modifications include DNA methylation, histone modifications and the role of non‐coding RNA, which can directly interact with the primary nucleotide sequence and regulate gene expression. Changes of DNA methylation have been reported to be related to AAA 5, 10, 11, 12. However, very few studies have investigated the functions of histone modifying enzymes [histone deacetylases (HDACs), histone methyltransferases (HMTs) and histone acetyltransferases (HATs)], which have been shown to be related to AAA pathology in humans 13. Thus, the epigenetic changes in the pathogenesis of AAA have been lacking. This study was designed to investigate whether DNA methylation and histone modifications were altered in Tregs of AAA patients.
Materials and methods
Patients
Thirty‐two AAA patients (the AAA group, 27 men and five women, mean age: 63.9 ± 8.1 years) at Department of Vascular Surgery, The First Hospital, China Medical University (from September 2010 to July 2013) were prospectively recruited into the study. All patients and control subjects gave written informed consent. AAA with a diameter > 50 mm was included, as measured by a computed tomography scan or magnetic resonance imaging. Patients with the following complications were excluded: atherosclerotic changes, Ehlers–Danlos syndrome, Marfan syndrome, other known vascular disorders, connective tissue disorders and medical history of other autoimmune diseases. Thirty age‐ and sex‐matched healthy controls (HCs) who received annual regular physical examinations were included as the HC group (26 men and four women, mean age: 60.5 ± 11.2 years). All the controls had a healthy peripheral vascular system, without medical history of aneurysm, type A aortic dissection, or other vascular disorders. In addition, cancer, infection, and any other immune‐mediated diseases were also excluded. The study conformed to the guidelines set by the Declaration of Helsinki. All experimental protocols were approved by the Institutional Review Board at China Medical University (Shenyang, China).
Isolation of peripheral blood mononuclear cells and Tregs
Peripheral blood was collected in 40 mL anticoagulant tubes. Thereafter, peripheral blood mononuclear cells (PBMCs) were isolated with density‐gradient centrifugation in Ficoll–sodium diatrizoate (Ficoll‐Hypaque, Haoyang TBD, Co., Tianjin, China). The cells were washed with PBS and CD4+ and CD25+ Tregs were sorted by flow cytometry. The extract of PBMCs and residual liquid Sortin were blended, and then 50 μL FITC–CD4 and 50 μL APC–CD25 fluorochrome were added into the mixture for 20 min in the dark. The dyed PBMCs were washed with 2 mL PBS once, and the supernatant was discarded. The cell pellet was diluted in 10 times, and kept at room temperature for 10 min in dark. Then, the liquid was centrifuged for 5 min, the supernatant was discarded, and washed twice with PBS. Finally, 50 μL forkhead box P3 (FOXP3) antibody was added into the cell pellet followed by culture for 30 min at dark, and then, CD4+ CD25+ Tregs were sorted by flow cytometry.
DNA methylation determination
A blood genomic DNA extraction kit (Tiangen Biochemical Technology Co., Ltd, Beijing, China) was used to extract DNA, and a centrifuge column was used to separate DNA. DNA methylation rate was detected using a DNA methylation kit (Epigentek, Group. Inc., New York, NY, USA) according to the manufacturer's instructions. 5‐Methylcytosine (5‐mc) was quantitatively analyzed using an ELISA kit. Standard absorbance was measured by a microplate reader according to the formula: 5‐mc (ng) = (sample A − negative control A)/slope × 2. The 5‐mc % = 5‐mc (ng)/amount of DNA × 100% was calculated according to manufacturer's instructions.
Histone extraction and protein quantification
Histone was extracted with a total histone protein extraction kit (Epigentek Group, Inc.) according to the manufacturer's instruction. Briefly, the cells were centrifuged at 168 g, 4 °C for 5 min and suspended in diluted pre‐lysis buffer at 107/mL. After centrifuging at 1509 g 4 °C for 5 min, the supernatant was discarded; the cells were re‐suspended in three volumes (about 107 cells/200 μL) of cell lysate and incubated on ice for 30 min. After centrifuging at 24 149 g at 4 °C for 5 min, the supernatant was moved to a new tube; 0.3 volumes of DTT and balanced salt solution at a ratio of 1 : 500 (1 μL DTT + 500 μL balanced salt solution) was added to the supernatant. Protein concentrations were determined using the enhanced BCA Protein Assay Kit (Beyotime Institute of Biotechnology, Nanjing, China). After extraction, histones were stored at −80 °C for use.
Determination of acetylation rate of histone H3, H3K9, H3K14 and H4
The acetylation rates of total histones H3, H3K9, H3K14 and H4 were detected by ELISA according to the manufacturer's instructions (Epigentek, Group. Inc.). The protein concentrations were adjusted to 400 ng·μL−1 and absorbance was measured at 450 nm wavelength using a microplate reader. The degree of histone acetylation rate was calculated according to the following formula: Acetylation % = A sample well−blank well)/A control well−blank well × 100%.
Real time‐polymerase chain reaction
Total RNA was extracted using an RNAiso Plus Kit (TaKaRa Biotechnology Co., Dalian, China.), and the amount and purity of the RNA were determined with spectrophotometry. High‐quality RNA samples were obtained with an A 260/A 280 ratio > 1.8. Total RNA was reverse‐transcribed with reverse transcriptase (PrimeScript RT Reagent Kit, TaKaRa) and oligo‐dT primer according to the manufacturer's instructions. The reaction conditions were: 95 °C for 30 s, then 95 °C for 5 s, and 60 °C for 30 s for 40 cycles. The expression of target genes was normalized to reference as previously described 14. The primers designed for real‐time PCR are listed in Table 1.
Table 1.
The primers for real‐time PCR.
| Gene | Primer | Sequence(5′–3′) |
|---|---|---|
| DNMT1 | Forward | CTTCGGCAACATAATGGACA |
| Reverse | CTTTGGCCAACATACAAAGCTTGA | |
| DNMT3A | Forward | CCGGAACATTGAGGACATCT |
| Reverse | CAGCAGATGGTGCAGTAGGA | |
| DNMT3B | Forward | CCCATTCGAGTCCTGTCATT |
| Reverse | GGTTCCAACAGCAATGGACT | |
| MBD1 | Forward | CACCCTCTTCGACTTCAAACAAG |
| Reverse | CAACCTGACGTTTCCGAGTCTT | |
| MBD2 | Forward | AACCCTGCTGTTTGGCTTAAC |
| Reverse | CGTACTTGCTGTACTCGCTCTTC | |
| MBD3 | Forward | CCGCTCTCCTTCAGTAAATGTAAC |
| Reverse | GGCTGGAGTTTGGTTTTCAGAA | |
| MBD4 | Forward | TGGTGGTGCATGCCTGTAAT |
| Reverse | TGAGACAGGGTCTCTCTCTGTCAT | |
| HDAC1 | Forward | AGACAGCTGTGGCCCTGGATA |
| Reverse | GGAGCTTGAAATCTGGTCCAAAGTA | |
| HDAC2 | Forward | AGTCAAGGAGGCGGCAAAA |
| Reverse | TGCGGATTCTATGAGGCTTCA | |
| HDAC4 | Forward | AAGGAGTGGACATTGCT |
| Reverse | GATTCAGCAGCTCCACT | |
| HDAC5 | Forward | TCACTGTCACCAACTCAC |
| Reverse | CAGGAATAGAGGATGTGC | |
| HDAC7 | Forward | CTTCTCCACAAGGACAAG |
| Reverse | CTCCAGGGTTCTGTAGG | |
| GAPDH | Forward | CCACCCATGGCAAATTCCCATGGCA |
| Reverse | TCTAGACGGCAGGTCAGGTCCACC |
Statistical analysis
Quantitative data are presented as mean ± standard deviation (SD). The data showing a normal distribution were analyzed by t test. Categorical variables, including gender, smoking history, hypertension history, diabetes history and coronary disease, were analyzed by the χ2 test. A value of P < 0.05 was set as the threshold for significance. The analyses were performed using spss version 16.0 software (SPSS Inc., Chicago, IL, USA).
Results
DNA methylation rate and acetylation rate of histone H3 and H3K9 in Tregs were changed in AAA patients
Demographics of AAA patients and healthy controls are listed in Table 2. The sex, age, smoking history, disease history, WBC count and lymph count were comparable in the two groups. DNA methylation rate of Tregs was higher in the AAA group than in the HC group (0.159 ± 0.08% vs 0.098 ± 0.03%, P < 0.05) (Fig. 1A). The acetylation rates of histone H3 and H3K9 in peripheral blood Tregs was significantly lower in AAA patients than in HC (respectively 0.086 ± 0.06% vs 0.11 ± 0.099%, P = 0.027 for H3; 0.08 ± 0.037% vs 0.125 ± 0.09%, P = 0.016 for H3K9; Fig. 1B). By contrast, acetylation rates of H3K14 (AAA group: 0.163 ± 0.09% vs HC: 0.146 ± 0.077%, P = 0.411) and of H4 (AAA group: 0.146 ± 0.03%; HC: 0.09 ± 0.012%, P = 0.134) were not significantly different between the two groups.
Table 2.
Demographics of AAA patients and healthy controls. Comparison by Unpaired Student's t test or χ2 test. WBC (white blood cell).
| AAA group (n = 32) | Control group (n = 30) | P | |
|---|---|---|---|
| Male/female | 27/5 | 26/4 | 0.303 |
| Age (years) | 63.93 ± 8.14 | 60.45 ± 11.17 | 0.916 |
| Smoking history (n (%)) | 5 (27) | 8 (40) | 0.506 |
| Hypertension history (n (%)) | 11 (61) | 9 (45) | 0.352 |
| Diabetes history (n (%)) | 3 (16.7) | 4 (20) | 0.128 |
| Coronary disease (n (%)) | 3 (16.7) | 3 (15) | 1 |
| WBC count (×109/L) | 7.83 ± 2.56 | 6.59 ± 1.73 | 0.102 |
| Lymph count (×109/L) | 1.85 ± 0.39 | 2.17 ± 0.62 | 0.11 |
Figure 1.
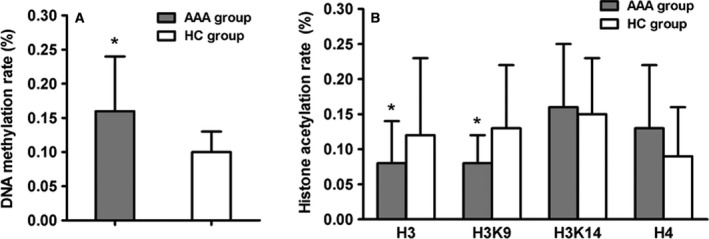
DNA methylation rate and acetylation rate of histone H3 and H3K9 in Tregs were changed in AAA patients. (A) DNA methylation rate of Tregs was up‐regulated in AAA patients; (B) Acetylation rate of histone H3 and H3K9 in Tregs was reduced in AAA patients. All data are presented as mean ± SD and analyzed by unpaired Student's t test. n = 32 in AAA group and n = 30 in HC group. *P < 0.05 compared with HC group.
mRNA expression of DNA methyltransferases 1 and 3A were up‐regulated in AAA patients
Three DNA methyltransferases (DNMTs) were detected using real time‐PCR. mRNA levels of DNMT1 and DNMT3A were significantly up‐regulated in the AAA group compared with the HC group (respectively 1.14 ± 0.77 vs 0.64 ± 0.47, P = 0.004 for DNMT1; and 1.88 ± 0.56 vs 1.01 ± 0.60, P < 0.05 for DNMT3A), which was consistent with the results of DNA hypermethylation. However, there was no differences regarding DNMT3B expression between two groups (AAA: 1.54 ± 1.25 vs HC: 1.06 ± 0.68, P = 0.298; Fig. 2).
Figure 2.
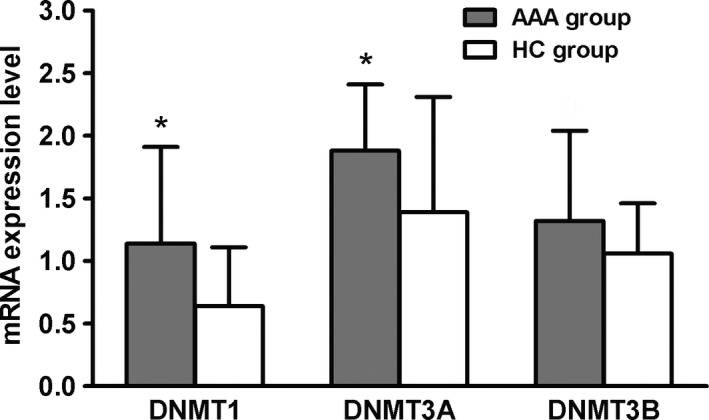
Expression of DNMT1 and DNMT3A mRNA was up‐regulated in AAA patients. All data are presented as mean ± SD and analyzed by unpaired Student's t test. n = 32 in AAA group and n = 30 in HC group. *P < 0.05 compared with HC group.
The expression of methyl‐CpG‐binding domain proteins 2 and 4 mRNA was up‐regulated in AAA patients
We examined mRNA expression of methyl‐CpG‐binding domain proteins (MBDs) in the two groups. Interestingly, mRNA expression in MBD2 and MBD4 was higher in the AAA group than in the HC group (respectively 6.21 ± 2.54 vs 3.04 ± 1.45 for MBD2; and 6.89 ± 2.24 vs 4.04 ± 1.75 for MBD4; both P < 0.05). By contrast, the mRNA expression of MBD1 and MBD3 was not different between the two groups (Fig. 3).
Figure 3.
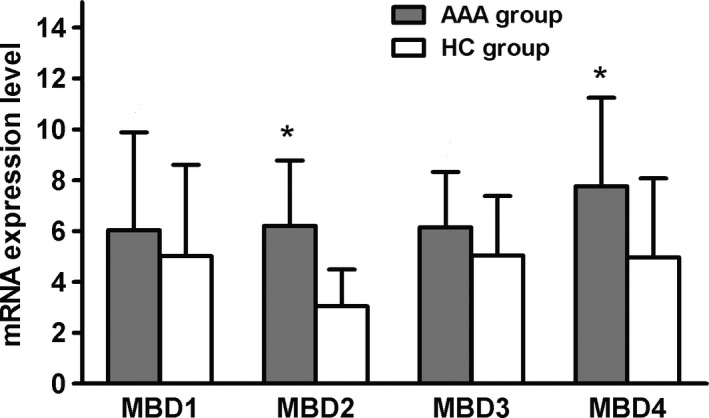
Expression of MBD2 and MBD4 mRNA was up‐regulated in AAA patients. All data are presented as mean ± SD and analyzed by unpaired Student's t test. n = 32 in AAA group and n = 30 in HC group. *P < 0.05 compared with HC group.
mRNA expression of HDAC1 and HDAC5 was increased in AAA patients
The mRNA levels in both HDAC1 and HDAC5 were higher in the AAA group than in the HC group (respectively 2.58 ± 1.66 vs 1.21 ± 0.9, P = 0.034 for HDAC1; 1.35 ± 1.01 vs 0.37 ± 0.11, P = 0.01, for HDAC5). The expression of HDAC2, HDAC4 and HDAC7 was not significantly different between the two groups (Fig. 4).
Figure 4.
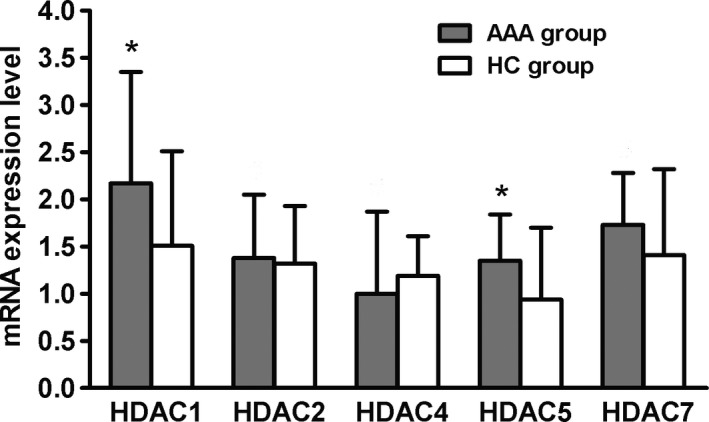
HDAC1 and HDAC5 mRNA expression was increased in AAA patients. All data are presented as mean ± SD and analyzed by unpaired Student's t test. n = 32 in AAA group and n = 30 in HC group. *P < 0.05 compared with HC group.
Discussion
Abdominal arterial aneurysm is a complex disease that develops due to the interaction of environmental risk factors and genetic predisposition 15. DNA methylation and histone modification influence disease onset and progression as effectors of the environment, such as lifestyle or diet, and have been studied extensively in many diseases 16. In this study, we revealed that DNA methylation and histone modifications in FOXP3+ CD4+ CD25+ Tregs were different in AAA patients.
DNA methylation occurs naturally as a result of aging and cell differentiation but is also recognized as an important modifier of disease risk. DNMTs induce and maintains DNA methylation. There are two types: DNMT1 (a maintenance enzymes) and DNMT3 (a de novo enzyme) 17. DNMT1 recognizes hemi‐methylated sites opposite newly synthesized DNA strands in mitosis. Recognizing the role of epigenetic modifications in autoimmune diseases is crucial for a full understanding of autoimmunity. In this study, the level of DNA methylation in Tregs was significantly higher in AAA patients than in healthy subjects, indicating that Tregs were in the state of transcriptional repression, affecting the cellular function. The results were consistent with a previous study 11. On the contrary, another study pointed out that DNA methylation in peripheral T cells was significantly lower in an AAA group than a HC group 5. Thus, Toghill et al. 12 considered that no direct evidence yet confirms the association between DNA methylation and AAA. DNMT3 enzymes are divided into two groups: DNMT3A and DNMT3B. DNMT3A is thought to have a preference for unmethylated DNA, but DNMT3B is equally efficient at methylating unmethylated and hemimethylated CpG sites 46 17. Our results showed that DNMT3A was significantly higher in the AAA group than in the HC group, while DNMT3B was not different between the two groups, indicating that DNMT3A and 3B perform different functions in the two groups.
Previous studies identified disease‐associated changes in the expression of DNMTs and MBDs in some autoimmune diseases such as systemic lupus erythematosus and subacute cutaneous lupus erythematosus. Increased expression of DNMT1 would maintain the hypermethylation of Tregs and cause a decrease in the transcriptional activity of Tregs, resulting in insufficient numbers of cells and affecting the normal cell function. Abnormal expression of DNMT3A reveals that Treg cells in some AAA patients have aberrant methylation. The increased expression of MBD2 and MBD4 in the AAA group was also consistent with the increased rate of DNA methylation, further enhancing the main function of regulatory T cells to inhibit autoimmune diseases.
Histone modification occurs mainly during cell cycling, cell development and differentiation. The most common modification is acetylation 18. In this study, the acetylation rates of H3 and H3K9 were lower in AAA patients than in HC, which reduced the transcriptional activity and might contribute to the decrease of Tregs. These processes involve selected lysine residues in the tails of nucleosomal histones, which are induced by HAT and HDAC 9. Histone modifying enzymes, including HAT, HMT and HDACs, have been implicated in cardiovascular disease, cancer and Alzheimer's disease 19, 20. The latest study has reported that HDAC can regulate non‐histone acetylation 21; 1750 proteins were proposed as substrates of HDAC, including p53, GATA1–3, signal transducer and activator of transcription (STAT) 3, STAT5, FOXP3, estrogen and estrogen receptor, HSP90, and α‐tubulin 22. Acetylation of these non‐histone proteins has pleiotropic effects, such as increased expression of FOXP3 22, decreased expression of DNMT1, interference between proteins, and increased p53 transcriptional activity 23. Our results showed that the decreased acetylation rates of H3 and H3K9 might be affected by increased HDAC1 and HDAC5, which inhibits the transcriptional activity of regulatory T cells and reduces the number of Tregs. However, the contrary result has been reported that H3K9 was increased in AAA patients 13.
Conclusions
The rates of DNA methylation and histone modifications of Tregs in peripheral blood were altered in AAA patients compared with healthy subjects. However, there is still controversy regarding the increase or decrease of H3K9 in AAA patients. Thus, further studies are needed to settle this discrepancy.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
QX contributed significantly to analysis and manuscript preparation. JZ contributed to the conception of the study. YH performed the data collection and collation. XZ performed the data analyses. HJ, YL, XW, QG, ZL, DB and ZD participated in some experiments. SX wrote the manuscript.
Acknowledgements
This study is funded by National Nature Science Foundation of China (Grant No. 304717076) and Educational Department of Liaoning Province, China (Key Laboratory Research Project, No. LS 2010172).
References
- 1. Kent KC (2014) Clinical practice. Abdominal aortic aneurysms. New Engl J Med 371, 2101–2108. [DOI] [PubMed] [Google Scholar]
- 2. Song JQ, Zhang J, Yin MD, Wu B, Shan SY, Duan ZQ and Xin SJ (2008) Clinico‐epidemiological features of infrarenal abdominal aortic aneurysm and relevant prognostic factors. Zhonghua yi xue za zhi 88, 1613–1617. [PubMed] [Google Scholar]
- 3. Jagadesham VP, Scott DJ and Carding SR (2008) Abdominal aortic aneurysms: an autoimmune disease? Trends Mol Med 14, 522–529. [DOI] [PubMed] [Google Scholar]
- 4. Eagleton MJ (2012) Inflammation in abdominal aortic aneurysms: cellular infiltrate and cytokine profiles. Vascular 20, 278–283. [DOI] [PubMed] [Google Scholar]
- 5. Jiang H, Xia Q, Xin S, Lun Y, Song J, Tang D, Liu X, Ren J, Duan Z and Zhang J (2015) Abnormal epigenetic modifications in peripheral T cells from patients with abdominal aortic aneurysm are correlated with disease development. J Vasc Res 52, 404–413. [DOI] [PubMed] [Google Scholar]
- 6. Doderer SA, Gabel G, Kokje VBC, Northoff BH, Holdt LM, Hamming JF and Lindeman JHN (2018) Adventitial adipogenic degeneration is an unidentified contributor to aortic wall weakening in the abdominal aortic aneurysm. J Vasc Surg 67, 1891–1900. [DOI] [PubMed] [Google Scholar]
- 7. Zhang J, Bockler D, Ryschich E, Klemm K, Schumacher H, Schmidt J and Allenberg JR (2007) Impaired Fas‐induced apoptosis of T lymphocytes in patients with abdominal aortic aneurysms. J Vasc Surg 45, 1039–1046. [DOI] [PubMed] [Google Scholar]
- 8. Yin M, Zhang J, Wang Y, Wang S, Bockler D, Duan Z and Xin S (2010) Deficient CD4+CD25+ T regulatory cell function in patients with abdominal aortic aneurysms. Arterioscl Throm Vas 30, 1825–1831. [DOI] [PubMed] [Google Scholar]
- 9. Meda F, Folci M, Baccarelli A and Selmi C (2011) The epigenetics of autoimmunity. Cell Mol Immunol 8, 226–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Krishna SM, Dear A, Craig JM, Norman PE and Golledge J (2013) The potential role of homocysteine mediated DNA methylation and associated epigenetic changes in abdominal aortic aneurysm formation. Atherosclerosis 228, 295–305. [DOI] [PubMed] [Google Scholar]
- 11. Ryer EJ, Ronning KE, Erdman R, Schworer CM, Elmore JR, Peeler TC, Nevius CD, Lillvis JH, Garvin RP, Franklin DP et al (2015) The potential role of DNA methylation in abdominal aortic aneurysms. Int J Mol Sci 16, 11259–11275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Toghill BJ, Saratzis A, Harrison SC, Verissimo AR, Mallon EB and Bown MJ (2015) The potential role of DNA methylation in the pathogenesis of abdominal aortic aneurysm. Atherosclerosis 241, 121–129. [DOI] [PubMed] [Google Scholar]
- 13. Han Y, Tanios F, Reeps C, Zhang J, Schwamborn K, Eckstein HH, Zernecke A and Pelisek J (2016) Histone acetylation and histone acetyltransferases show significant alterations in human abdominal aortic aneurysm. Clin Epigenetics 8, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhu G, Li J, He L, Wang X and Hong X (2015) MPTP‐induced changes in hippocampal synaptic plasticity and memory are prevented by memantine through the BDNF‐TrkB pathway. Bri J Pharmacol 172, 2354–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van de Luijtgaarden KM, Heijsman D, Maugeri A, Weiss MM, Verhagen HJ, IJpma A, Brüggenwirth HT and Majoor‐Krakauer D (2015) First genetic analysis of aneurysm genes in familial and sporadic abdominal aortic aneurysm. Hum Genet 134, 881–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Robertson KD (2005) DNA methylation and human disease. Nat Rev Genet 6, 597–610. [DOI] [PubMed] [Google Scholar]
- 17. Robertson AK, Geiman TM, Sankpal UT, Hager GL and Robertson KD (2004) Effects of chromatin structure on the enzymatic and DNA binding functions of DNA methyltransferases DNMT1 and Dnmt3a in vitro. Biochem Bioph Res Co 322, 110–118. [DOI] [PubMed] [Google Scholar]
- 18. Jenuwein T and Allis CD (2001) Translating the histone code. Science 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- 19. Bauer AJ and Martin KA (2017) Coordinating regulation of gene expression in cardiovascular disease: interactions between chromatin modifiers and transcription factors. Frontiers Cardiovasc Med 4, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tran TQ, Lowman XH and Kong M (2017) Molecular pathways: metabolic control of histone methylation and gene expression in cancer. Clin Cancer Res 23, 4004–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zheng C, Zhong M, Qi Z, Shen F, Zhao Q, Wu L, Huang Y, Tsang SY and Yao X (2017) Histone deacetylase (HDAC) inhibitors relax mouse aorta partly by their inhibitory action on L‐type Ca2+ channels. J Pharmacol Exp Ther 363, 211–220. [DOI] [PubMed] [Google Scholar]
- 22. Olzscha H, Bekheet ME, Sheikh S and La Thangue NB (2016) HDAC inhibitors. Methods Mol Bio 1436, 281–303. [DOI] [PubMed] [Google Scholar]
- 23. Li B, Samanta A, Song X, Iacono KT, Bembas K, Tao R, Basu S, Riley JL, Hancock WW, Shen Y et al (2007) FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression. Proc Natl Acad Sci USA 104, 4571–4576. [DOI] [PMC free article] [PubMed] [Google Scholar]


