Abstract
Enzymes that act on multiple substrates are common in biology but pose unique challenges as therapeutic targets. The metalloprotease insulin-degrading enzyme (IDE) modulates blood glucose levels by cleaving insulin, a hormone that promotes glucose clearance. However, IDE also degrades glucagon, a hormone that elevates glucose levels and opposes the effect of insulin. IDE inhibitors to treat diabetes therefore should prevent IDE-mediated insulin degradation, but not glucagon degradation, in contrast with traditional modes of enzyme inhibition. Using a high-throughput screen for non-active-site ligands, we discovered potent and highly specific small-molecule inhibitors that alter IDE’s substrate selectivity. X-ray co-crystal structures, including an IDE-ligand-glucagon ternary complex, revealed substrate-dependent interactions that enable these inhibitors to potently block insulin binding while allowing glucagon cleavage, even at saturating inhibitor concentrations. These findings suggest a path for developing IDE-targeting therapeutics, and offer a blueprint for modulating other enzymes in a substrate-selective manner to unlock their therapeutic potential.
Introduction
Despite over six decades of speculation that inhibiting the degradation of insulin could offer new medicines for type-2 diabetes1–3, this concept has not yet been developed into a therapeutic strategy4,5. Insulin-degrading enzyme (IDE, Fig. 1a) is a widely expressed zinc-dependent metalloprotease that contributes to the proteolytic inactivation of insulin4–6. The precise delineation of the physiological roles of IDE on glucose regulation has been hampered by counterintuitive phenotypes observed in IDE–/– knockout studies, which may result from confounding effects on gene expression7–9 or from other intracellular roles of IDE4,10. The first examples of pharmacological inhibiton of extracellular IDE using small-molecule inhibitors11,12 suggest that IDE-targeted therapeutics have potential to improve the regulation of blood glucose levels to treat type-2 diabetes by amplifying the surge of endogenous insulin following nutrient intake, even though basal blood glucose levels are not primarily modulated by IDE4–6. Since insulin is naturally released in amounts proportional to nutrient consumption during and after meals4–6, such a strategy offers a low risk of hypoglycemia13 and may operate synergistically with current antidiabetic agents11. For example, combining drugs that boost glucose-stimulated insulin secretion, or insulin-sensitizing drugs, with extracellular IDE inhibitors might further decrease the need for regular insulin injections associated with treatment of type-2 diabetes.
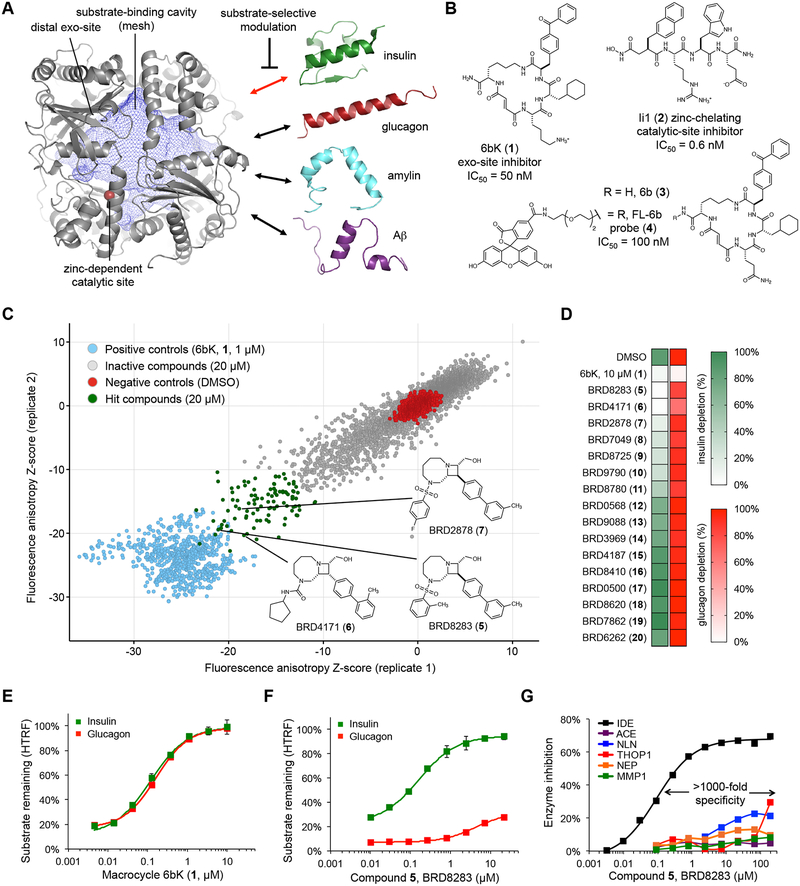
Fig. 1 |. High-throughput screen for IDE exo-site ligands and discovery of substrate-selective IDE inhibitors.
(a) Structure of the primary isoform of IDE(42–1019) comprising four homologous domains that create a large internal cavity (blue mesh)23. The therapeutic outcome of IDE inhibition arises from impeding the degradation of insulin (red double arrow), rather other in vivo IDE substrates (drawn to scale). The red sphere is the bound zinc ion in the catalytic site. (b) Macrocyclic peptide 6bK (1), zinc-chelating peptidic inhibitor Ii1 (2)23, and fluorescent high-throughput screening probe FL-6b (4) based on DNA-templated macrocycle hit 6b (3)11. (c) Small-molecule screen for displacement of FL-6b (4) from human IDE. The X and Y axes show anisotropy Z-scores from two replicates for 7,679 azetidines; see Supplementary Figures 1 and 2 for screening results on all 17,277 compounds tested. Primary assay data and counter-screening results are deposited in PubChem BioAssay databases 1259349 and 1259348, respectively (Supplementary Data Set 1). (d) IDE-mediated insulin versus glucagon depletion (green and red heatmaps, respectively), measured using HTRF with paired labeled antibodies for each substrate in the presence of hit compounds (tested at 67 μM, >10-fold EC50fluo). (e,f) Concentration-dependent profiles for 6bK (1) and BRD8283 (5) in IDE-mediated degradation assays for insulin and glucagon. See also Supplementary Figure 2, for additional substrate degradation assays using 6bK (1), BRD8283 (5), BRD4171 (6) and BRD2878 (7), respectively. (g) Fluorogenic peptide cleavage assays reveal >1,000-fold specificity of BRD8283 (5) for IDE (EC50fluo = 100 nM, IMAX = 65%) over all other metalloproteases tested: thimet oligopeptidase (THOP), neurolysin (NLN), neprilysin (NEP), matrix metalloprotease 1 (MMP1), and angiotensin converting-enzyme (ACE). See also Supplementary Figure 2 for the protease specificity profile of BRD4171 (6, >500-fold specificity). All assays include IDE alone in 2% v/v DMSO as the no-inhibitor activity reference. Points and error bars represent mean ± SEM for three technical replicates (e–f), or two technical replicates in the additional metalloprotease assays (g) and substrate depletion heatmaps (d). EC50 values are reported for endpoint degradation assays and for partial inhibitors, whereas IC50 values are calculated for kinetic assays with normal inhibitors36.
Previously we reported the discovery and optimization of the first physiologically active IDE inhibitor, 6bK (1, Fig. 1b), from a DNA-templated library of macrocycles11. Mutagenesis and X-ray crystallography studies revealed that 6bK (1) binds IDE at a novel “exo site” binding pocket that is adjacent to but non-overlapping with the catalytic site11. Since this exo site is not conserved among proteases, exo-site engagement confers the remarkable specificity of 6bK (1) for inhibiting IDE over other related metalloproteases11. We used this inhibitor to illuminate the physiological consequences of acute IDE inhibition in animal models of diabetes11. Treatment of mice with a single injection of 6bK (1) improved blood glucose clearance following oral glucose challenge, augmenting the effects of endogenous insulin release under experimental conditions that mimic a meal11. However, these and other studies revealed that IDE also degrades glucagon and amylin in vivo, in addition to insulin2,11,14, and Aβ in the brain8,15. Collectively, these observations suggest that developing a strategy for modulating IDE’s activities on a subset of its endogenous substrates is critical to exploring the potential therapeutic benefits of IDE inhibition for the treatment of type-2 diabetes8–15.
Since glucagon elevates blood glucose levels and thus opposes the action of insulin, an ideal class of IDE-targeting diabetes therapeutics should preferentially block insulin degradation without impeding glucagon degradation4,11,16. Despite the discovery of many protease inhibitors as probes and therapeutic agents17, such substrate-selective inhibition of a protease has rarely been observed18–22. Therefore, the opposing physiological effects of IDE inhibition on two key glucose-regulating hormones that exist in the same extracellular environment presents a paradox for the development of traditional small-molecule therapeutics that target IDE (Fig. 1a)4,5,11.
Two distinct classes of IDE inhibitors have emerged over the past decade.5 Traditional metalloprotease inhibitor designs use a hydroxamic acid or other zinc-chelating group to interfere directly with the biochemical mechanism of this zinc-dependent protease (e.g. Ii1, 2, Fig. 1b)23,24. Despite the observation of active-site inhibitors that block IDE more strongly in the presence of Aβ, a way to substrate-selectively inhibit IDE-mediated insulin cleavage has not been reported25–27. Recently discovered exo-site inhibitors, including 6bK (1) and others12,28,29, do not interact with the catalytic zinc ion or otherwise block the catalytic site, but instead function as competitive inhibitors by occupying part of the substrate-binding cavity11. In principle, exo-site inhibitors raise the possibility of maintaining IDE’s proteolytic activity while reshaping its substrate selectivity (Fig. 1a), but this possibility has not been realized. We hypothesized that the identification of small-molecule exo-site ligands could provide a starting point to develop substrate-selective IDE inhibitors that preferentially block insulin degradation, without susbtantially impeding glucagon degradation. To date, substrate-selective inhibition has not been systematically implemented in therapeutics discovery and development, despite the potential of this approach to modulate biology downstream of the many known poly-substrate specific targets in a sophisticated manner that cannot be achieved by blocking protein active sites. Prior to this work, only a handful of unoptimized ligands for disparate protein targets have been reported to be substrate-selective inhibitors (listed in Supplementary Table 1)18–22,30–32. Moreover, the structural basis underlying substrate-selective mode of inhibition and methods to generate such inhibitors remains largely unexplored.
Results
An exo site-specific screen reveals novel IDE inhibitors
Towards this goal, we designed a high-throughput screen to identify small-molecule IDE exo-site ligands by displacement of a fluorescent probe that binds away from IDE’s catalytic residues (Fig. 1b, Supplementary Table 2)33. To construct this probe, we replaced the linker attached to DNA in the original DNA-templated macrocycle library hit 6b (3) with a fluorescein group11. The resulting probe, FL-6b (4) retained strong binding activity to IDE (IC50fluo = 100 nM)11, and the interaction of FL-6b (4, 5 nM) with human IDE (0.5 μM) resulted in elevated fluorescence anisotropy. This anisotropy signal decreased upon displacement of FL-6b (4) with an excess of the non-fluorescent exo-site inhibitor 6bK (1, 1 μM). When performed in 384-well microtiter plates, this assay resulted in a robust signal suitable for high-throughput screening (Z’-factor = 0.7, Supplementary Figure 1)31,33. Importantly, the potent zinc-chelating IDE inhibitor Ii1 (2, Fig. 1b)23, which unlike 6bK (1) binds the catalytic site rather than the exo site, does not displace FL-6b (4) and did not cause any change in fluorescence anisotropy. These results establish the first high-throughput screen for discovery of novel IDE exo site-binding ligands.
Using this assay, we performed a pilot screen using a collection of 9,598 small molecules that represents the structurally diverse chemical libraries within the Broad Institute (Supplementary Figure 1)34. The pilot screen revealed a number of weakly active hits (Supplementary Data Set 1, PubChem BioAssay 1259349), primarily from azetidine-based libraries34, which feature biaryl appendages resembling the critical benzophenone group of the DNA-templated macrocyclic IDE inhibitor 6bK (1, Fig. 1b)11. Based on these results, we performed an expanded screen on the full collection of 8,959 azetidine-core compounds34 (Fig. 1c), resulting in 12 hits with average Z-scores similar to that of the positive control 6bK (1, Fig. 1c) among 100 structurally related compounds that also produced a significant decrease in the anisotropy signal. As an initial secondary screen, these compounds were individually tested for IDE inhibition using a fluorogenic decapeptide reporter substrate (Mca-RPPGFSAFK(Dnp)-OH) assay35. The top hits were all active IDE inhibitors, ranging in potency from EC50fluo = 0.1 to 5 μM (Supplementary Data Set 1, PubChem BioAssay 1259348; see Supplementary Table 2 and Supplementary Figure 1 for a summary of the secondary screening strategy).
IDE cleavage assays using the decapeptide reporter in the presence of exo-site ligands revealed partial IDE inhibition activity even at saturating inhibitor concentrations, with maximal inhibition (IMAX)36 below 100% (Supplementary Data Set 1, PubChem BioAssay 1259348). The most potent screening hits, BRD8283 (5, EC50fluo = 0.1 μM, IMAX = 67%), BRD4171 (6, EC50fluo = 0.4 μM, IMAX = 73%) and BRD2878 (7, EC50fluo = 0.34 μM, IMAX = 40%) share the same macrocycle-fused azetidine core (Fig. 1c, insets)34 and represent a structurally related collection of small-molecule exo-site ligands that could potentially alter the substrate selectivity of IDE.
IDE exo-site ligands that alter substrate selectivity
Next we prioritized identifying compound scaffolds that support the complete blockage of IDE-mediated cleavage of human insulin, but not glucagon, and considered maximizing IDE inhibition potency as a secondary goal for subsequent optimization. We tested a structurally representative group of IDE exo-site ligands, including those mentioned above, for their ability to impede IDE-catalyzed degradation of unmodified full-length hormone substrates, using homogeneous time-resolved Förster resonance energy transfer (HTRF) through paired fluorophore-conjugated monoclonal antibodies37 (Fig. 1d, Supplementary Figure 2). As expected, in these assays the competitive inhibitor 6bK (1) effectively blocked the degradation of either insulin or glucagon with similar potency (Fig. 1e) while IDE alone depleted both substrates to a similar extent. Importantly, two exo-site-binding azetidines, BRD8283 (5) and BRD4171 (6), fully prevented IDE-mediated insulin degradation preferentially over glucagon degradation (Fig. 1d). Unlike other exo-site ligands we identified, or any IDE ligand previously reported, BRD8283 (5) and BRD4171 (6) fully blocked insulin degradation in a concentration-dependent manner, while only weakly and partially inhibiting glucagon degradation (Fig. 1f, and Supplementary Figure 2). In addition to favorable discrimination of insulin versus glucagon, these compounds also exhibited modest levels of substrate-selective inhibition in assays with two other IDE substrates, amylin and Aβ40 (Supplementary Figure 2)8,15. Together, these results establish the discovery of two second-generation IDE inhibitors that selectively inhibit IDE-mediated degradation of insulin over glucagon.
To confirm that this class of inhibitors interacts with the exo site, rather than the catalytic site, we assayed their ability to inhibit IDE mutants such as A479L, in which the leucine side chain is predicted to fill the distal hydrophobic pocket of the exo site without interfering with IDE’s proteolytic activity11. These assays revealed that BRD8283 (5) and BRD4171 (6) inhibit wild-type IDE, but do not inhibit A479L exo-site variants (Supplementary Figure 2). These two inhibitors also displayed decreased affinity for IDE containing G362Q or I374Q mutations, consistent with a model in which these compounds occupy the exo-site region demarcated by these residues that is >16 Å away from the zinc-dependent catalytic site (Supplementary Figure 2)11. Assays against related and unrelated zinc-metalloproteases, including neurolysin (NLN), thimet oligopeptidase (THOP1), neprilysin (NEP), matrix metalloprotease-1 (MMP1), and angiotensin-converting enzyme (ACE) revealed that BRD8283 (5) and BRD4171 (6) (Fig. 1g, and Supplementary Figure 2) inhibit IDE with a high degree of selectivity (≥ 500-fold) over other metalloproteases, further consistent with their ability to engage the exo site, which is not conserved among proteases, rather than the catalytic site of IDE. These results collectively establish a family of small-molecule bicyclic azetidines as the first exo-site inhibitors that can alter the substrate selectivity of IDE.
Optimization of substrate-selective IDE inhibitors
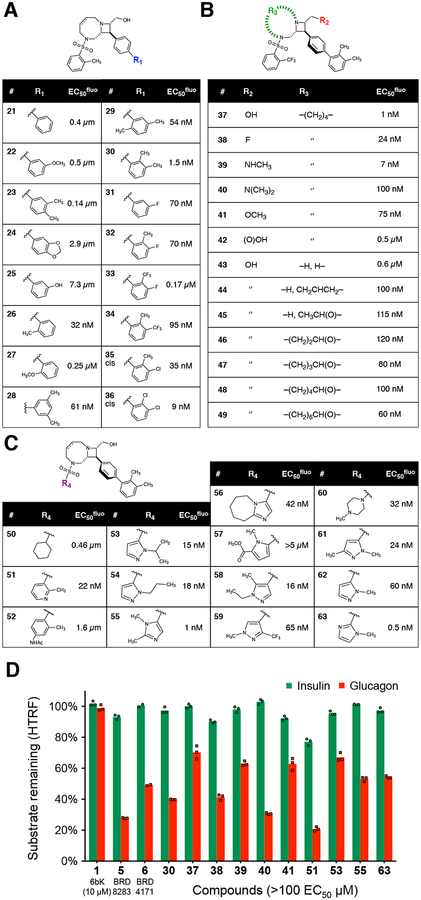
Based on the primary screen data, we chose BRD8283 (5, EC50fluo = 0.1 μM, IMAX = 67%)36 as a starting point to probe and optimize the determinants of potency and substrate selectivity (Fig. 2a–c). We began by altering the substitution pattern of the biaryl rings (analogs 20–36). Remarkably, early rounds of optimization revealed that analogs with ortho-methyl-substituted biaryl appendages displayed a >65-fold improvement in affinity, exemplified by compound 30 (EC50fluo = 1.5 nM, IMAX = 60% inhibiton of fluorogenic decapeptide proteolysis, Fig. 2a). We speculate that this substitution pattern favors a non-planar relationship between the rings of the biphenyl group. Since our previous crystal structure suggested that the aryl rings of the benzophenone group of 6b (3) must bind the exo site in a perpendicular conformation11, we hypothesize that ortho-methyl substitution of the terminal phenyl group causes these compounds to adopt a similar conformation before binding IDE, thereby reducing the entropic cost38 of adopting this conformation when bound to IDE.
Fig. 2 |. Structure-activity relationships and potency optimization of substrate-selective IDE inhibitors.
(a–c) The IDE inhibition EC50fluo values for the three families of analogs shown were determined in duplicate proteolysis assays using the fluorogenic reporter decapeptide (Mca-RPPGFSAFK(Dnp)-OH; see Supplementary Table 3, Supplementary Note and Supplementary Data Set 3). (d) Substrate-selective inhibition of IDE-mediated insulin versus glucagon degradation in the presence of selected exo-site ligand analogs (10 μM, ≥ 100× EC50fluo). Assays included two (a–c) or three technical replicates (d), and IDE alone in 2% v/v DMSO as the no-inhibitor activity reference.
We next probed other structural features of these compounds, including the azetidine-fused macrocycle linker, the effect of R2 group H-bond donor and charge, and the sulfonamide appendage R5 (Fig. 2b–c). The structure-activity relationships among analogs 37–63 collectively suggest a model in which the rigid bicyclic-azetidine core contributes to optimal affinity beyond acting as a scaffold for appendages, with a basic azetidine nitrogen that is a favorable but non-essential feature, and an R2 group that may be solvent-exposed but that is capable of interacting with the exo site through a hydrogen bond. Moreover, aryl-sulfonamide appendages (R5) bearing ortho-methyl groups, such as N-methyl-imidazole-2-sulfonamide, are present in several potent inhibitors. The most potent substrate-selective IDE inhibitor 63 (EC50fluo = 0.5 nM, IMAX = 60%)36 integrated all four of the favorable structural features elucidated from the structure-function analysis (Fig. 2c). Importantly, the third-generation IDE exo-site inhibitors 30, 37, and 63 maintained the ability to inhibit insulin degradation over glucagon degradation, similar to that of BRD8283 (5) throughout the stepwise potency optimization process (Fig. 2d).
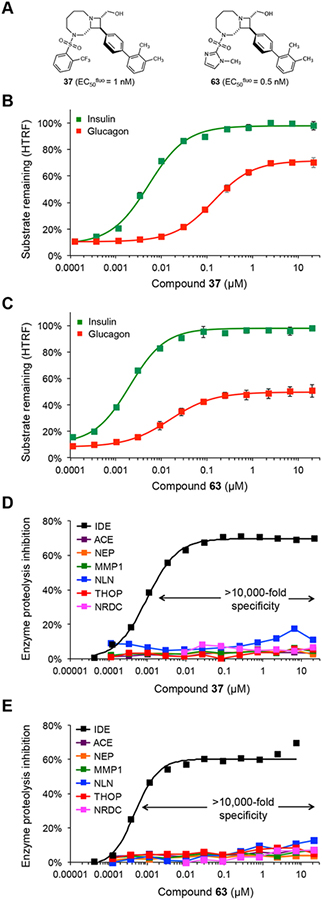
With potent substrate-selective exo-site ligands in hand, we sought to study in greater detail the concentration-dependent IDE-mediated insulin and glucagon degradation assays in the presence of analogs 37, and 63 (Fig. 3a, EC50fluo = 1 nM, and 0.5 nM, respectively, and see Supplementary Figure 2 for compound 30). The most notable difference in glucagon versus insulin degradation assays, as observed with BRD8283 (5) and BRD4171 (6), in contrast to the non-substrate-selective IDE inhibitor 6bK (1, Fig. 1e–f), was the persistent IDE proteolytic activity on glucagon spanning several orders of magnitude of concentrations of compounds 30, 37 and 63 (Fig. 3a–c, Supplementary Figure 2). Indeed, IDE-mediated processing of glucagon reached 60%, 29%, and 50% depletion over the 10-minute assays, respectively, at saturating inhibitor concentrations (Fig. 3b–c, Supplementary Figure 2; also compare with BRD8283 (5) in Fig. 1f). Moreover, the EC50 inflection points of the glucagon cleavage assays for 30, 37, and 63 were 10 to 30-fold higher than in the respective insulin degradation assays, which showed complete inhibition of insulin degradation by 30, 37, and 63 with calculated Kicomp = 6 nM, 4.3 nM, and 1.7 nM, respectively (Fig. 3b–c, Supplementary Figure 2 and Supplementary Table 4). These data demonstrate that the biochemical effects of substrate-selective inhibitors can extend beyond a substrate-dependent shift of the EC50 inflection point to preserve the processing of certain substrates even at saturating inhibitor concentrations, unlike the nearly identical glucagon and insulin degradation curves observed for the non-substrate-selective IDE inhibitor 6bK (1) (Fig. 1e). Additionally, substrate-selective inhibitors prevent IDE-mediated insulin degradation for more than one hour, comparable to 6bK (1) (Supplementary Figure 2), whereas glucagon undergoes IDE-mediated cleavage at all inhibitor concentrations, even under saturating conditions in which inhibitor concentration exceeds its EC50fluo by 10,000-fold (Fig. 3b–c)6,39. These findings indicate that ternary complexes comprising human IDE, glucagon, and a substrate-selective exo-site inhibitor are catalytically competent.
Fig. 3 |. Concentration-dependent substrate discrimination and metalloprotease specificity of potent substrate-selective IDE inhibitors.
(a) Structures of substrate-selective inhibitors 37 and 63. (b–c) Concentration-dependent profiles of IDE-mediated degradation assays for insulin and glucagon in the presence of 37 and 63 (EC50fluo = 1 nM, and 0.5 nM, respectively, in the kinetic reporter decapeptide assay; and Kicomp = 4.3 nM, and 1.7 nM, respectively, calculated from the insulin degradation assay). See Supplementary Figure 2 for expanded substrate-selectivity profiles including amylin and Aβ(1–40) (d–e) Metalloprotease specificity of 37 and 63, determined using fluorogenic peptide cleavage assays, display >10,000-fold specificity for IDE over all other metalloproteases tested: thimet oligopeptidase (THOP1), neurolysin (NLN), neprilysin (NEP), matrix metalloprotease 1 (MMP1), angiotensin converting-enzyme (ACE), and nardilysin (NRDC). Supplementary Table 6 includes additional metalloproteases assayed in the presence of 63 (1 and 10 μM) and Ii1 (2) (1 μM). See also Supplementary Figure 2 for insulin-glucagon degradation assays and the metalloprotease specificity profile for analog 30, which also displays >10,000-fold specificity. All assays include IDE alone in 2% v/v DMSO as the no-inhibitor activity reference. Values and error bars reflect mean ± SEM of three technical replicates (b–c) or two technical replicates in the additional metalloprotease assays (c–d).
To our knowledge, these compounds represent substrate-selective inhibitors with the highest potency and specificity for their target reported to date for any enzyme (Supplementary Table 1), demonstrating that substrate-selective inhibition is not exclusively a property relegated to weak ligands that can be differentially outcompeted from target binding by higher affinity substrates18–22,30–32. Importantly, the optimized analogs 30, 37, and 63 inhibit IDE with exquisite specificity (≥ 10,000-fold) over all other related and unrelated metalloproteases tested, including NLN, THOP1, NEP, MMP1, ACE, and NRDC (Fig. 2d–e, and Supplementary Figure 2). This observation is consistent with their high affinity for the IDE exo site, which is a distinctive feature not conserved among other proteases, unlike the similarity among catalytic sites of metalloproteases40. We also subjected 63 at 1 and 10 μM concentrations to assays on an unbiased panel of 18 human metalloproteases and observed minimal inhibition, in contrast to that of catalytic site zinc-chelating IDE inhibitor Ii1 (2)11,23 (Supplementary Table 6, Supplementary Data Set 2). These data further support the target selectivity advantages of inhibiting an exo site rather than the active site. Taken together, our data establish that targeting IDE’s exo site can give rise to small-molecule ligands with a superior combination of high potency, high metalloprotease specificity, and substrate-selective inhibition compared to previously described IDE inhibitors5,11,12,23,24,26–29.
Molecular basis of substrate-selective inhibition
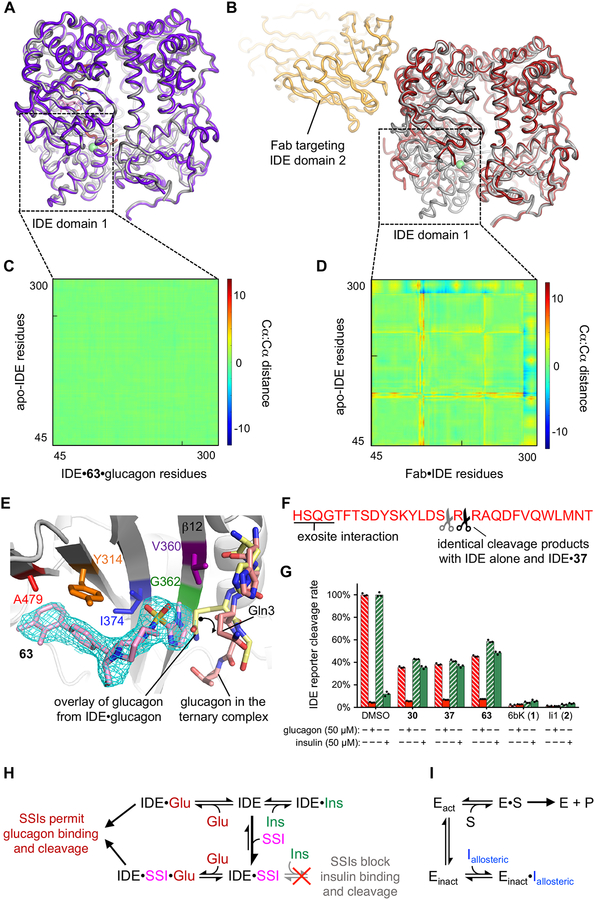
Finally, we sought to illuminate the molecular basis of substrate-selective IDE inhibition. We solved the X-ray co-crystal structure of the well-established catalytically inactive E111Q, cysteine-free form of IDE35 bound to substrate-selective inhibitors 37 and 63 (at 2.96 Å and 3.49 Å resolution, respectively), as well as the co-crystal structure of the IDE•63•glucagon ternary complex at 3.18 Å resolution (Fig. 4a, Supplementary Video, Supplementary Figures 3–4 and Supplementary Table 7). These structures revealed that substrate-selective inhibitors 37 and 63 bind to the same exo-site pocket as 6b (3), more than 16 Å away from the nearest catalytic residue (Fig. 4a–b, Supplementary Figure 3). The interactions observed between IDE and the inhibitors are consistent with the observed structure-activity relationships (Fig. 2a–c), including the conformationally locked biaryl rings of 37 and 63 filling a hydrophobic tunnel near the exo site, and the sulfonamide appendages binding a hydrophobic patch adjacent to the exo-site beta-sheet β12 (Fig. 4a, c). The protein structure of IDE bound by 37, 63, or by 63•glucagon, adopted a closed conformation nearly identical to that of wild-type IDE and IDE alone, as well as IDE•6b (3) and other previously reported IDE structures (Supplementary Figure 4)40,41. The structure of the ternary complex of IDE•63•glucagon reveals the N-terminal residues of glucagon binding to IDE’s exo-site beta-sheet β12 via backbone-backbone H-bonding interactions (Fig. 4d, Supplementary Figure 3–4) adjacent to substrate-selective inhibitor 63, precisely matching the pattern of interactions previously reported in the IDE•glucagon structure (PDB ID 2G49)40.
Fig. 4 |. Structural basis for substrate-selective small-molecule inhibition of IDE.
(a–b) X-ray co-crystal structures of 63 and glucagon bound to IDE as a ternary complex IDE•63•glucagon (a, PDB ID 6EDS, 3.18 Å resolution) compared to the previously reported structure of insulin-bound IDE (PDB ID 2WBY, b)40. (c) View of the exo site in the IDE•63•glucagon co-crystal structure showing the space-filling model of glucagon (red). The dashed red line represents disordered residues in the central section of glucagon (see Supplementary Figures 3–4). (d–h) Matching views of the exo-site of IDE bound by 63 in which glucagon has been cloaked, and shown instead with superimposed substrates from published IDE-substrate co-crystal structures40,42. (d) Shows the superimposition with partially folded insulin bound to IDE (green, PDB ID 2WBY), (e) with unfolded insulin α-chain from cryo-EM insulin•IDE structure (green; PDB ID 6BFC), (f) with unfolded insulin β-chain from cryo-EM insulin•IDE structure (green; PDB ID 6B3Q), (g) with amylin bound to IDE (PDB ID 2G48; cyan surface), and (h) with Aβ(1–40) bound to IDE (PDB ID 2G47; purple surface)40,42. Supplementary Figure 3 includes complementary analyses for the X-ray co-crystal structures of IDE•37 and IDE•63 co-crystalized without glucagon. See the Supplementary Video associated with this figure.
Likewise, we observed additional interactions for the C-terminal residues of glucagon in the catalytic groove of IDE (Fig. 4a, Supplementary Figures 3–4)40. As previously reported in the IDE•glucagon structure40, the central section of glucagon that extends between the exo site and the catalytic site is also disordered in the IDE•63•glucagon ternary complex (Fig. 4c, dotted line represents unresolved residues); therefore, no other interactions are observed between the ligand 63 and glucagon40. In contrast, superimposition of the IDE•substrate-selective inhibitor structures with a partially folded insulin molecule from the reported IDE•insulin structure (Fig. 4d, PDB ID 2WBY)40 or with unfolded insulin molecules from IDE•insulin cryo-electron microscopy structures (Fig. 3e–f, PDB IDs 6BFC, 6B3Q)42 predict that the space occupied by the inhibitors’ aryl-sulfonamide appendages (R5) and macrocycle linker sections (R3/R4) would clash with an IDE-bound insulin molecule. Similar views from overlays of amylin•IDE and Aβ-amyloid•IDE co-crystalized substrates (Fig. 4g–h, PDB IDs 2G48, 2G47)40 suggest an intermediate extent of proteolysis in the presence of 37 and 63 (Supplementary Figure 2). These models are consistent with the above biochemistry data showing that 37 and 63 competitively block insulin binding to the IDE cavity in a mutually exclusive manner (Fig. 2b–c, and Supplementary Figure 3). These results together reveal the substrate-dependent interactions that underlie the structural basis of substrate-selective IDE inhibition.
To investigate the possibility of conformational allostery as a contributor to substrate-selective inhibition, we superimposed the IDE•63 and IDE•63•glucagon co-crystal structures with the apo-IDE structure (Fig. 5, and IDE•37 in Supplementary Figures 3–4). Strikingly, the substrate-selective inhibitors do not alter the conformation of IDE, and thus do not induce allosteric changes in the protein. Analysis of the superimposed zinc-dependent catalytic domain-1 of IDE using distance-difference matrix calculations reveals the absence of allosteric changes (Fig. 5a and c, see also Supplementary Figures 3 and 5), in contrast to the pronounced long-distance conformational changes in the catalytic site induced by an antibody-fragment (Fab) bound to IDE domain 2 (Fig. 5b and D, PDB ID 4IOF, see Supplementary Figure 5)42. These observations were corroborated by all three co-crystal structures solved in this study, demonstrating that exo-site-targeted macrocyclic inhibitors and substrate-selective IDE inhibitors act differently than allosteric inhibitors that modulate protein conformation and typically abrogate functions of the catalytic site42–46. A detailed comparison of the superimposed poses of the IDE•glucagon and the IDE•63•glucagon structures show that only a minor bond rotation of the Gln-3 side chain of glucagon has occurred to accommodate for the formation of the IDE•63•glucagon ternary complex, while preserving the cross-beta-sheet interactions between the backbone of the substrate and exo-site beta-sheet β12 (Fig. 5e)40 as well as the previously reported interactions of glucagon in the catalytic site of IDE (Supplementary Figures 4)40.
Fig. 5 |. Substrate-selective inhibitors reprogram IDE’s substrate binding without inducing conformational changes or allosteric effects on the catalytic site.
(a–b) Overlays of the structure of apo-IDE with the ternary complex IDE•63•glucagon (purple), and (b) the reported structure of an antibody fragment (Fab, yellow) targeting IDE domain 2 that generates known allosteric motions in IDE domain 1 comprising the catalytic site (brown)42. (c–d) Distance-difference matrix (DDM) heatmaps calculated for domain 1 Cα-to-Cα motions between the superimposed structures in panels A and B, which are set to the same reference scale to reveal the lack of allosteric effects of 63-bound IDE domain 1 (c), compared to the domain-1 allosteric effects previously observed with the Fab targeting IDE domain 2 (d). Supplementary Figure 5 includes complementary DDM heatmaps for the full protein domains 1–4 that shows the comparison of the relative motions of the N- and C-terminal halves for Fab-bound IDE, but not in the presence of 37 or 63. (e) Detailed view of the predicted rotation of the Gln3 side chain (curved arrow) that accommodates binding of glucagon in the IDE•63•glucagon ternary complex (red sticks), compared to the overlay of glucagon modeled from the glucagon•IDE structure (yellow sticks, PDB ID 2G49)40. Additionally, a co-crystallized polypeptide (modeled as Ala3) is observed in the cavity of the IDE•37 structure interacting with the same beta-sheet β12 of IDE’s exo site (Supplementary Figure 3). The mesh represents the composite omit electron density map of ligand 63 contoured at 1σ. The exo-site residues that were mutated to interrogate binding of the inhibitors are highlighted with matching colors to the IDE variant inhibition data shown in Supplementary Figure 2F–G. (f) Major (black scissors) and minor (grey scissors) cleavage sites of glucagon by IDE in the presence or absence of 37 (10 μM) as determined by MALDI-TOF mass spectrometry (see Supplementary Figure 6). (g) Proteolytic activity of wild-type human IDE reported by the rate of cleavage of the fluorogenic decapeptide substrate (Mca-RPPGFSAFK(Dnp)-OH) in the presence or absence of human insulin (green filled and dashed bars, respectively, 50 μM Humulin-R), or in the presence or absence of human glucagon (red filled and dashed bars, respectively, 50 μM GlucaGen) when combined with 10 μM each of substrate-selective inhibitors 30, 37, 63, with competitive inhibitor 6bK (1), or the zinc-chelating inhibitor Ii1 (2)23. A 2% v/v DMSO-alone mixture with IDE was the activity reference and negative control. Points represent three technical replicates. (h) Inhibition model of exo-site substrate-selective inhibitors (SSIs) in the presence of an allowed substrate (glucagon, left) or a blocked substrate (insulin, right), compared to (i) the inhibition model of a generic allosteric ligand driving a target enzyme (Eact) into a conformationally inactive state (Einact).
Taken together, these findings strongly support a model of substrate-selective inhibition in which glucagon avoids steric clashes with substrate-selective ligands within IDE, consistent with the efficient catalytic processing of glucagon by IDE under conditions of ligand saturation (Fig. 3a–c). The same analysis for the macrocycle 6b•IDE co-crystal structure predicts major steric clashes between 6b (3) and both insulin and glucagon (Supplementary Figure 3), explaining the inability of bulky inhibitors to substrate-selectively inhibit IDE (Fig. 1e)11. Collectively, these results provide a structural basis for the substrate-selective inhibition of IDE by third-generation exo-site ligands, and identifies key structural features of IDE inhibitors that result in this property.
To investigate if the cleavage site of glucagon is altered by the presence of substrate-selective inhibitors, we evaluated IDE reactions quenched at various timepoints to capture the first cleavage products of IDE alone or in the presence of 37 (10 μM, > 10,000-fold EC50fluo), 6bK (1, 10 μM), or Ii1 (2, 1 μM) (Supplementary Figure 6). Mass spectrometry analysis using MALDI-TOF revealed a nearly identical distribution of glucagon cleavage products in the presence or the absence of saturating concentrations of 37, primarily resulting from proteolysis between Arg17-Arg18, as well as minor cleavage at Arg18-Ala19 as previously reported (labeled in Fig. 5f, full data in Supplementary Figure 6)47. Similar to the observation using antibody assays (Fig. 3b), the relative ion intensities qualitatively indicated that at ligand saturation the rate of the ternary complex of wild-type-IDE•37-mediated glucagon cleavage was approximately 50% the rate of IDE-mediated glucagon cleavage in the absence of inhibitor, whereas 6bK (1) and Ii1 (2) each fully inhibited glucagon cleavage (Supplementary Figure 6). These observations are consistent with previous studies that showed IDE substrate capture and catalysis are sequential but independent steps, in which the former is rate limiting for glucagon due to its modest binding affinity for IDE35,40.
The substrate-selective competition model (Fig. 4c versus 4d–f) predicts that IDE bound by a substrate-selective exo-site ligand should predominantly cleave smaller peptide substrates even in the presence of excess insulin, which can no longer occupy IDE’s substrate-binding cavity. To test this mechanistic prediction we used the fluorogenic decapeptide substrate as a reporter of IDE activity35, and tested the ability of substrate-selective inhibitors to redirect IDE’s activity to process this low-affinity substrate instead of insulin, the highest-affinity substrate of IDE6,39. The IDE•63•glucagon ternary structure also predicts that excess glucagon under the same set of conditions should still bind IDE and thus outcompete the fluorogenic peptide and produce no signal. We incubated IDE with excess human insulin or human glucagon (50 μM final concentration) followed by the addition of the fluorogenic decapeptide reporter substrate (5 μM), which resulted in a low fluorescence signal due to its low affinity for IDE compared to the hormone substrates (Fig. 5g)35. Indeed, IDE-mediated cleavage activity was redirected in the presence of insulin, but not glucagon, towards the small fluorogenic peptide when treated with saturating concentrations of substrate-selective inhibitors 30, 37, or 63 (Fig. 5g, cleavage rates of 40–60% compared to DMSO control, and minimally affected by the introduction of excess insulin). Importantly, these data exclude an alternative co-substrate uncompetitive mechanism stemming from an inactive ligand-enzyme-insulin ternary complex that could prevent the catalytic cycle of IDE, as observed in other systems that do not operate through bona fide substrate-selective inhibition25. Taken together, these findings reveal that the unique ability of substrate-selective inhibitors to reshape IDE’s substrate recognition mechanisms stems from substrate-specific steric clashes and competitive exclusion of insulin from the substrate-binding cavity, while allowing small and flexible substrates such as glucagon to assemble with IDE and the substrate-selective inhibitor in a partial mixed-noncompetitive mode of inhibition that allows glucagon to be processed by the nearby catalytic center.
Discussion
This study integrates insights from biochemistry, small-molecule screening, medicinal chemistry, and structural biology to develop and characterize the first series of potent and specific small drug-like molecules that substrate-selectively block IDE and redirect its proteolytic activity towards substrates other than insulin. These discoveries unlock the potential of IDE-targeting therapeutics for type-2 diabetes to avoid the paradoxical effects of blocking the degradation of glucagon11, as well as other substrates that may not yet be known to be regulated by IDE8,14. The substrate-selective inhibitors reported here represent the first pharmacological solution to mitigate IDE’s polyspecificity towards opposing glucose-modulating substrates by supporting glucagon cleavage at all tested concentrations of inhibitor, including saturating levels. Exo-site engagement by substrate-selective inhibitors enables substrate-dependent interactions that potently block insulin binding (Fig. 5h, right) while allowing the formation of a catalytically competent IDE•inhibitor•glucagon ternary complex (Fig. 5h, left). This mode-of-inhibition contrasts with traditional allostery mechanisms (Fig. 5i) in which allosteric ligand binding stabilizes a conformationally distinct enzyme state, typically abrogating catalytic site function. These results thus establish the potential of IDE exo-site substrate-selective inhibitors as alternatives to active-site or allosteric inhibitors for the treatment of post-prandial hyperglycemia in type-2 diabetes. Importantly, this new class of IDE inhibitors, by targeting an exo site unique to IDE, also avoid off-target inhibition of other zinc-dependent metalloproteases, a primary clinical challenge in this field17.
More broadly, this study may serve as a blueprint to identify substrate-selective exo-site inhibitors of other enzymes that bind their substrates through interactions not entirely tied to the catalytic site, including some kinases, phosphatases, peptidases, sheddases, metalloenzymes, ubiquitin ligases and conjugating enzymes, among others20,21,30–32,46,48–50. The development of substrate-selective inhibitors thus could enable therapeutic modulation of enzymes that operate on multiple substrates and that contain at least one exo site mediating enzyme-substrate interactions. This strategy could enable biological roles of enzymes to be tailored not by the traditional approach of abolishing catalytic activity, but instead by the more precise approach of reprogramming substrate specificity.
Online Methods
Site-directed mutagenesis, expression, and purification of human IDE
N-terminally His6-tagged human IDE(42–1019)11 was cloned into the expression plasmid pTrcHis-A (Invitrogen) using primers for uracil-specific excision reactions (USER) and Phusion U Hot-Start DNA-polymerase (ThermoFisher F555S)51. Mutant IDE constructs were generated by amplifying the complete pTrcHis-A-hIDE(42–1019) vector construct with USER cloning primers introducing a mutant overhang (Supplementary Table 8) as previously described11 and introduced by heat shock into NEB turbo E. coli cells. Transformants were selected on carbenicillin LB agar, and isolated colonies were cultured overnight in 2 mL LB media. Plasmids were extracted using a microcentrifuge membrane column kit (Miniprep, Qiagen), and the sequence of genes were confirmed by Sanger sequencing11. The plasmid constructs were transformed by heat-shock into chemically-competent expression strain Rosetta 2 (DE3) pLysS E. coli cells (EMD Millipore), and selected on carbenicillin/chloramphenicol LB agar. Cells transformed with IDE pTrcHis-A constructs were cultured overnight at 37 °C in 2×YT media (31 g in 1 L) containing 100 μg/mL ampicillin and 34 μg/mL chloramphenicol. Expression of His6-tagged IDE proteins was induced when the culture reached OD600 ~0.6 by addition of isopropyl-β-D-1-thiogalactopyranoside (IPTG) to 1 mM final concentration. The cells were incubated overnight at 37 °C, then centrifuged at 10,000 g for 30 min, 4 °C.
Recombinant His6-tagged proteins were purified by Ni(II)-affinity chromatography (IMAC sepharose beads, GE Healthcare) according to the manufacturer’s instructions. The cell pellets were resuspended in pH 8.0 buffer containing 50 mM phosphate, 300 mM NaCl, 10 mM imidazole, 1% Triton X-100 and 1 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP), and were lysed by probe sonication for 4 min at < 4 °C, followed by clearing of cell debris by centrifugation at 10,000 g for 25 min at 4 °C. The supernatant was incubated with Ni(II)-doped IMAC resin (2 mL) for 3 h at 4 °C. The resin was washed twice with the cell resuspension/lysis buffer, and three times with pH 8.0 buffer containing 50 mM phosphate, 300 mM NaCl, 50 mM imidazole and 1 mM TCEP. Elution was performed in 2 mL aliquots by raising the imidazole concentration to 250 mM and subsequently to 500 mM in the previous buffer. The fractions were combined and the buffer was exchanged to the recommended IDE buffer (R&D) using spin columns with 100 KDa molecular weight cut off membranes (Millipore). Protein yields were typically ~10 μg/L, and >90% purity based on gel electrophoresis analysis (Coomassie stained). IDE-specific protease activity was >95% as assessed by inhibition of degradation of peptide substrate Mca-RPPGFSAFK(Dnp)-OH (R&D) by 20 μM of inhibitor 6bK (1), and compared with commercially available human IDE (R&D) under the same conditions.
Fluorescence anisotropy high-throughput screening assay
Human N-His6-IDE42–1019 (E. coli expressed) was mixed with fluorescein-labeled macrocycle FL-6b (4) in 50 mM Tris buffer pH 8.0, with 1 M NaCl, at 25 °C. The optimum signal was obtained using 0.5 μM IDE (Supplementary Figure 1) and 5 to 30 nM of probe FL-6b (4) depending on the spectrophotometer used (excitation 492 nm, emission 523 nm). The pilot screen using the Broad Institute “DOS Informer Set” compound collection was transferred by pinning (100 nL/well) into 384-well plates containing the enzyme-probe mixture (50 μL/well, 0.5 μM IDE, 30 nM FL-6b, 4) using a CyBio Vario liquid handling system equipped with a pin-transfer workstation, and a Multidrop Combi-nL Reagent Dispenser (Thermo Scientific). For the second screen using azetidine-core libraries the compounds were pre-loaded in empty 384-well plates by sonication using an Echo 555 Liquid Handler (Labcyte) and the enzyme-probe mixture was added to the plate (50 μL/well, 0.5 μM IDE, 30 nM FL-6b, 4) the Multidrop Combi-nL Reagent Dispenser (Thermo Scientific). The final compound concentrations were 20 μM in both screens, and IDE inhibitor 6bK (1) was used as a positive control at 1 μM final concentration. After 30 min equilibration at room temperature, fluorescence anisotropy was recorded using an EnVision spectrophotometer (excitation 492 nm, emission 523 nm). Exclusion of compounds using auto-fluorescence measurements was not necessary in this case because the DOS compound libraries were designed to avoid fluorophores34. Primary assay data are deposited in PubChem BioAssay database 1259349 (Supplementary Data Set 1).
Metalloprotease cleavage activity assays using fluorogenic peptide substrates
Recombinant human IDE42–1019 (R&D, #2496-ZN, or variants of His6-IDE42–1019 expressed in house from E. coli), neprilysin (NEP, R&D, #1182-ZNC), and angiotensin-converting enzyme (ACE, R&D, #929-ZN) were assayed using the fluorophore/quencher-tagged decapeptide substrate V, Mca-RPPGFSAFK(Dnp)-OH (R&D, #ES005) according to the manufacturer’s instructions and recommended buffers (Supplementary Table 5). For IDE assays the recommended buffer is 50 mM Tris pH 7.5, 1 M NaCl. The enzyme mixtures (48 μL) were transferred to a 96-well plate and combined with 2 μL of inhibitor in DMSO stock solutions, prepared in 3-fold dilution series. The mixtures were allowed to equilibrate for 10 min and the enzymatic reaction was started by addition of substrate peptide in assay buffer (50 μL, 20 μM), mixed, and monitored on a SpectraMAX fluorescence plate reader in kinetic mode for 5 min (excitation 320 nm, emission 405 nm). Similarly, thimet oligopeptidase (THOP, R&D, #3439-ZN) and neurolysin (NLN, R&D, #3814-ZN) were assayed using substrate Mca-PLGPK(Dnp)-OH (Bachem, #M-2710) according to the manufacturer’s instructions and recommended buffers (Supplementary Table 5). Matrix metalloproteinase-1 (MMP-1, R&D, #901-MP) was activated and assayed according to the manufacturer’s instructions with substrate Mca-KPLGL-Dpa-AR-NH2 (R&D, #ES010). For the insulin and glucagon competition experiments, an IDE enzyme mixture (R&D #2496-ZN, 48 μL, 85 ng/mL final dilution) in assay buffer (50 mM Tris pH 7.5, 1 M NaCl) was combined with freshly prepared Humulin-R (Eli Lilly) in the same buffer (25 μL, 50 μM final dilution) or GlucaGen (Novo Nordisk, 25 μL, 50 μM final concentration), respectively, and inhibitors in DMSO stock solutions (2 μL). The reaction was started by the addition of fluorogenic decapeptide substrate Mca-RPPGFSAFK(Dnp)-OH (R&D, #ES005) in the same assay buffer (25 μL, 10 μM final dilution), mixed and immediately measured on a SpectraMAX fluorescence plate reader in kinetic mode for 5 min (excitation 320 nm, emission 405 nm). Counter-screen data are deposited in PubChem BioAssay database 1259348 (Supplementary Data Set 1).
Dual-antibody HTRF assay for IDE-mediated degradation of insulin
A solution of 0.8 μg/mL recombinant human IDE (R&D, #2496-ZN) in pH 7.5 buffer containing 20 mM HEPES, 135 mM NaCl (24 μL) was transferred to a 200-μL tube strip, and combined with 1 μL of each inhibitor in DMSO stock solutions, or prepared as 3-fold dilution series. A solution of insulin in assay buffer Diluent (25 μL, CisBio 62INSPEB) was added to a final concentration of 20 ng/mL, and incubated at 30 °C for 15 min. This procedure was optimized to result in ~85% degradation of insulin in no-inhibitor DMSO control reactions. All reactions were terminated at the same time by adding 25 μL of inhibitor Ii1 (2)23 in the same buffer (200 nM) and chilled on ice. The remaining insulin was quantified using 10 μL of the quenched enzymatic reaction using the sensitive-range protocol Homogeneous Time-Resolved FRET Insulin assay (CisBio 62INSPEB) in 20 μL total volume according to the manufacturer’s instructions (384 well-plate Greiner 784904 non-binding). Fluorescence was measured using a Tecan M1000Pro plate reader (excitation 320 nm, emission 665 and 620 nm, lag time 60 μs) according the assay manufacturer’s recommendations. Blank wells and an insulin standard curve were included in the assay.
Dual-antibody HTRF assay for IDE-mediated degradation of glucagon
A solution of 0.05 μg/mL recombinant human IDE (R&D, #2496-ZN) in assay buffer Diluent #5 (1×, 24 μL, CisBio 62GLCPEF) was transferred to a 200 μL tube strip, and combined with 1 μL of each inhibitor in DMSO stock solutions, or prepared as 3-fold dilution series. A solution of glucagon in the same buffer (25 μL) was added to a final concentration of 4 ng/mL, and incubated at 25 °C for 10 min. This procedure was optimized to result in ~85% degradation of glucagon in no-inhibitor DMSO control reactions. All reactions were terminated at the same time by the addition of 1 μL of inhibitor Ii1 (2) (5 μM)23 and chilled on ice. The remaining glucagon in each reaction was quantified using 10 μL of the quenched enzymatic reaction using the Homogeneous Time-Resolved FRET Glucagon assay (CisBio 62GLCPEF) in 20 μL total volume according to the manufacturer’s instructions (384 well-plate Greiner 784904 non-binding). Fluorescence was measured using a Tecan M1000Pro plate reader (excitation 340 nm, emission 665 and 620 nm, lag time 60 μs) according the assay manufacturer’s recommendations. Blank wells and a glucagon standard curve were included in the assay. Manufacturer reported specificity validation: Glucagon, 100%; Glucagon fragment 1–18, <1.81%, Glucagon fragment 19–29, <0.03%, Oxyntomodulin, <0.07%; Glicentin, <0.07%; GLP-1 (7–36) amide, <0.06%, GLP-1 (7–37); <0.11%, GLP-2, <0.3%; GRPP (Glicentin-Related Pancreatic Peptide) <0.01%.
ELISA-based assay for measurement of IDE-mediated degradation of amylin
A solution of 0.75 μg/mL recombinant human IDE (R&D, #2496-ZN) in pH 7.5 buffer containing 20 mM HEPES, 135 mM NaCl (24 μL) was transferred to a 200-μL tube strip, and combined with 1 μL of each inhibitor in DMSO stock solutions, or prepared as 3-fold dilution series. A solution of amylin (25 μL, Millipore #E8051-K) was added to a final concentration of 175 pM (685 pg/mL), and incubated at 25 °C for 15 min. This procedure was optimized to result in ~85% degradation of amylin in no-inhibitor DMSO control reactions. All reactions were terminated at 15 min by the addition of 50 μL of a solution of ELISA Assay Buffer containing inhibitor Ii1 (2)23 (200 nM) and chilled on ice. The remaining amylin in each reaction was quantified by transferring 100 μL of the quenched enzymatic reaction to the ELISA plate (Millipore, #EZHA-52K), and incubated for 1 hour at room temperature as indicated in the ELISA manufacturer’s instructions. Following the washing steps and antibody detection procedure using the alkaline phosphatase substrate 4-methylumbelliferyl phosphate (MUP), fluorescence was measured using a Tecan M1000Pro plate reader (excitation 355 nm, emission 460 nm) according the assay manufacturer’s recommendations. Blank wells and an amylin standard curve in the same DMSO-treated reaction buffer were included in the assay. Manufacturer reported specificity: the capture antibody recognizes the N-terminus of human amylin (disulfide bridge Cys2-Cys7), but not the reduced form of amylin; and the detection antibody recognizes the C-terminus of human amylin, but not a 1–20 fragment of amylin.
Dual-antibody HTRF assay for IDE-mediated degradation of Aβ(40)
A solution of 2.9 μg/mL recombinant human IDE (R&D, #2496-ZN) in pH 7.5 buffer containing 20 mM HEPES, 135 mM NaCl (24 μL) was transferred to a 200-μL tube strip, and combined with 1 μL of each inhibitor in DMSO stock solutions prepared as 3-fold dilution series. A solution of Aβ(40) in assay buffer Diluent (25 μL, CisBio 62INSPEB) was added to a final concentration of 1600 pg/mL, and incubated at 30 °C for 35 min. This procedure was optimized to result in ~85% degradation of Aβ(40) in no-inhibitor DMSO control reactions. All reactions were terminated at the same time by adding 25 μL of inhibitor Ii1 (2)23 in the same buffer (200 nM) and chilled on ice. The remaining Aβ(40) was quantified using 10 μL of the quenched enzymatic reaction in Homogeneous Time-Resolved FRET Aβ(40) assay (CisBio 62B40PEG) in 20 μL total volume according to the manufacturer’s instructions (384 well-plate Greiner 784904 non-binding). Fluorescence was measured using a Tecan M1000Pro plate reader (excitation 320 nm, emission 665 and 620 nm, lag time 60 μs) according the assay manufacturer’s recommendations. Blank wells and an Aβ(40) standard curve were included in the assay. Manufacturer reported specificity: No cross-reaction with Amyloid β1–42 and β1–43, limit of detection: 13.64 pg/mL, assay range: 25 to 1600 pg/mL.
Mass spectrometry IDE-mediated glucagon cleavage analysis using MALDI-TOF
A solution of 0.5 μg/mL recombinant human IDE (R&D, #2496-ZN) in pH 7.5 buffer containing 20 mM Tris, 100 mM NaCl (200 μL) was transferred to a 1.5-mL LoBind Eppendorf tube, and combined with 1 μL of each inhibitor in DMSO stock solutions. A solution of human glucagon (Eli Lilly) in the same buffer (200 μL) was added to a final concentration of 10 ng/mL, and incubated at room temperature. At the indicated timepoints a 25-μL aliquot of each reaction was quenched by transferring into a new Eppendorf tube preloaded with 5 μL of 1% TFA solution, and chilled on ice. For MALDI-TOF analysis, an aliquot of quenched reaction (10 μL) was combined with an equal volume (10 μL) of a freshly prepared saturated solution of alpha-cyano-4-hydroxycinnamic acid in 1:1 (v/v) acetonitrile:water containing 0.1% TFA, spotted on the MALDI plate target and allowed to air-dry slowly while covered. Spectra were recorded on a Bruker Ultraflextreme MALDI-TOF/TOF mass spectrometer and FlexControl software, calibrated using ProteoMass peptide calibration kit (Sigma, #MSCAL2).
Expression and purification of recombinant cysteine-free hIDE (CF-IDE-E111Q)
Cysteine-free, catalytically inactive human IDE (CF-IDE-E111Q) was expressed in BL21-CodonPlus(DE3)-RIL E. coli cells and purified using Ni-affinity and anion exchange chromatography as previously described11. Generating the IDE•37, IDE•63 co-crystal complexes as well as the IDE•63•glucagon ternary complex required three consecutive cycles of purification using a HiLoadTM 16/16 Superdex S200 size exclusion column (GE LifeScience) in 20 mM Tris pH 8.0, 50 mM NaCl, 0.1 mM PMSF. An additional 20 mM EDTA pH 8.0 was added to all the buffers associated with IDE•63•glucagon purification to inhibit the catalytic degradation of glucagon by IDE. After purifying CF-IDE-E111Q by size exclusion chromatography once, CF-IDE-E111Q eluent fractions were pooled and mixed with a two-fold molar excess of 37 or 63. The protein-drug mixture was incubated for a minimum of 30 min on ice to facilitate complex formation. The protein-inhibitor complexes were then subjected to two further rounds of size exclusion chromatography with the addition of a two-fold molar excess of inhibitors 37 or 63 and/or glucagon (0 or two-fold molar excess), respectively, to the eluent after each round of purification. After a final 30 min incubation step, IDE•37, IDE•63, or the IDE•63•glucagon complex was concentrated to 15 – 20 mg/mL and used immediately for crystallization.
Co-crystallization and X-ray diffraction of IDE•37 and IDE•63•Glucagon
The complexes IDE•37, IDE•63, and IDE•63•glucagon were crystallized by hanging drop vapor diffusion. Specifically, 1 μL of mother liquor (0.1 M HEPES pH 6.8, 12% Tacsimate pH 7, 20% PEGMME-5000, 10% 1,4-dioxane) was mixed with 1 μL of 20 mg/mL IDE•37 and incubated at room temperature. For the IDE•63 and IDE•63•glucagon complexes, 1 μL of mother liquor (0.1 M HEPES pH 7.0, 5% Tacsimate pH 7, 13% PEGMME-5000, 10% 1,4-dioxane) was mixed with 1 μL of 15 mg/mL IDE•63 or IDE•63•glucagon and incubated at room temperature. Crystals for all the CF-IDE-E111Q protein complexes were visible after 3 days and displayed an urchin-like morphology. When harvesting crystals for X-ray diffraction, the crystals were first separated into individual needles. The largest needles were soaked in a cryoprotective crystallography buffer composed of mother liquor containing 30% glycerol, and then snap-frozen in liquid nitrogen. X-ray diffraction data for IDE•37 was obtained from a single crystal at a temperature of 100 Kelvin (K) and wavelength of 1.116 Å using the SIBYLS Beamline located at the Advanced Light Source (ALS), operated by Lawrence Berkeley National Laboratory. Diffraction data for the IDE•63 and IDE•63•glucagon complexes were obtained from a single crystal at 100 K using the AMX and FMX beamlines respectively at the National Synchrotron Light Source II operated by Brookhaven National Laboratory. Data for IDE•63 was collected at a wavelength of 0.978 Å, and data for IDE•63•glucagon was collected at 0.979 Å.
IDE•37, IDE•63 and IDE•63•glucagon structure refinement
Diffraction data for IDE•37 were indexed, integrated and scaled using X-ray Detector Software (XDS), and data for IDE•63 and IDE•63•glucagon were indexed, integrated and scaled using XDS via autoPROC52. All crystals were established in space group P65 (Supplementary Table 7)52. We phased the data via molecular replacement in Phaser53, using our previously solved structure of human CF-IDE-E111Q complexed with the macrocyclic inhibitor 6b (3) (PDB ID 4LTE),11 as our search model. Coot54 was used to build the structural model for both IDE•37, IDE•63 and IDE•63•glucagon. All refinements to the model were performed in PHENIX55, using NCS (torsion-angle) and TLS (9 groups per chain). The finished model for IDE•17 had an Rwork of 0.161 and an Rfree of 0.203 with 0.0% of residues forming Ramachandran outliers, and 97.5% of residues falling within Ramachandran favored regions. The finished model for IDE•63 had an Rwork of 0.162 and an Rfree of 0.211 with 0.1% of residues forming Ramachandran outliers, and 95.7% of residues falling within Ramachandran favored regions. Our model for IDE•63•glucagon had an Rwork of 0.177 and an Rfree of 0.222 with 0.1% of residues forming Ramachandran outliers, and 96.2% of residues falling within Ramachandran favored regions. Additional data collection and crystal refinement statistics are listed in Supplementary Table 7).
Structure visualization, superimposition, and distance difference matrix plots
We used PHENIX55 to compute the distance difference matrix plots between apo-IDE (PDB ID 2JG4), Fab•IDE (PDB ID 4IOF)42, IDE•37 (PDB ID 6BYZ), IDE•63 (PDB ID 6MQ3), and IDE•63•glucagon (PDB ID 6EDS). PyMOL was used to generate the graphics for the structural models as well as the overlays of previously solved structures with IDE bound to insulin (PDB ID 2WBY), glucagon (PDB ID 2G49), amylin (PDB ID 2G48), and amyloid-β (1–40) (PDB ID 2G47)40.
X-Ray Crystallography for synthetic intermediate 66
A crystal mounted on a diffractometer was collected data at 100 K. The intensities of the reflections were collected by means of a Bruker APEX II CCD diffractometer (MoKα radiation, λ=0.71073 Å), and equipped with an Oxford Cryosystems nitrogen flow apparatus. The collection method involved 0.5° scans in ω at 28° in 2θ. Data integration down to 0.78 Å resolution was carried out using SAINT V8.34 C56, with reflection spot size optimization. Absorption corrections were made with the program SADABS57. The structure was solved by the Intrinsic Phasing methods and refined by least-squares methods again F2 using SHELXT-201458, and SHELXL-201459, with OLEX 2 interface60. Non-hydrogen atoms were refined anisotropically, and hydrogen atoms were allowed to ride on the respective atoms. Crystal data as well as details of data collection and refinement are summarized in Supplementary Table 9. The Ortep plot were produced with SHELXL-2014 program59.
Data availability statement
Complete results of the IDE high-throughput screens and the IDE inhibition counter-screen are available in the PubChem BioAssay database (1259348, 1259349), the IDE•37, IDE•63, and the IDE•63•glucagon X-ray structures are available in the Protein Data Bank (PDB IDs 6BYZ, 6MQ3, and 6EDS, respectively).
Supplementary Material
Acknowledgements
We thank A. Saghatelian, S. Schreiber, and M. Morningstar for helpful discussions. We are grateful to S. Trauger and J. Wang for mass spectrometry assistance. We thank J. Bittker, M. Wawer, and V. Dancik for assistance with library management and analysis. We thank Z. Foda and A. Lyczek for ligand docking studies, and D. Dobrovolsky for assistance with assays. We thank S.-L. Zheng for small-molecule structural determination. IDE X-ray diffraction data were collected at ALS, operated by LBNL on behalf of DOE, and is supported by DOE Office of Biological and Environmental Research and NIH (R01GM105404 and S10OD018483). This research was supported by the NIH R35 GM118062 (D.R.L.), R01 EB022376 (D.R.L.), R35 GM119437 (M.A.S.), R56 DK106200 (M.A.S.), and the Howard Hughes Medical Institute (D.R.L.). The Fonds de Recherche en Santé du Québec and Alfred Bader Fund provided fellowship support to J.P.M.
Footnotes
Competing interests
J.P.M. and D.R.L. are co-inventors on patents and patent applications based on this work, and are co-founders of Exo Therapeutics, a small-molecule drug discovery company. The authors declare no competing non-financial interests.
References
- 1.Mirsky IA & Broh-Kahn RH The inactivation of insulin by tissue extracts; the distribution and properties of insulin inactivating extracts. Arch Biochem 20, 1–9, (1949). [PubMed] [Google Scholar]
- 2.Duckworth WC & Kitabchi AE Insulin and glucagon degradation by the same enzyme. Diabetes 23, 536–543, (1974). [DOI] [PubMed] [Google Scholar]
- 3.Roglic G & World Health Organization. Global report on diabetes. (World Health Organization, 2016). [Google Scholar]
- 4.Costes S & Butler PC Insulin-degrading enzyme inhibition, a novel therapy for type 2 diabetes? Cell Metab 20, 201–203, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang WJ Targeting Insulin-Degrading Enzyme to Treat Type 2 Diabetes Mellitus. Trends in endocrinology and metabolism: TEM 27, 24–34, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duckworth WC, Bennett RG & Hamel FG Insulin degradation: progress and potential. Endocr Rev 19, 608–624, (1998). [DOI] [PubMed] [Google Scholar]
- 7.Abdul-Hay SO et al. Deletion of insulin-degrading enzyme elicits antipodal, age-dependent effects on glucose and insulin tolerance. PLoS One 6, e20818, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farris W et al. Insulin-degrading enzyme regulates the levels of insulin, amyloid beta-protein, and the beta-amyloid precursor protein intracellular domain in vivo. Proc Natl Acad Sci U S A 100, 4162–4167, (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villa-Perez P et al. Liver-specific ablation of insulin-degrading enzyme causes hepatic insulin resistance and glucose intolerance, without affecting insulin clearance in mice. Metabolism: clinical and experimental 88, 1–11, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steneberg P et al. The type 2 diabetes-associated gene ide is required for insulin secretion and suppression of alpha-synuclein levels in beta-cells. Diabetes 62, 2004–2014, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maianti JP et al. Anti-diabetic activity of insulin-degrading enzyme inhibitors mediated by multiple hormones. Nature 511, 94–98, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durham TB et al. Dual Exosite-binding Inhibitors of Insulin-degrading Enzyme Challenge Its Role as the Primary Mediator of Insulin Clearance in Vivo. J Biol Chem 290, 20044–20059, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahren B Avoiding hypoglycemia: a key to success for glucose-lowering therapy in type 2 diabetes. Vascular health and risk management 9, 155–163, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bennett RG, Duckworth WC & Hamel FG Degradation of amylin by insulin-degrading enzyme. J Biol Chem 275, 36621–36625, (2000). [DOI] [PubMed] [Google Scholar]
- 15.Malito E, Hulse RE & Tang WJ Amyloid beta-degrading cryptidases: insulin degrading enzyme, presequence peptidase, and neprilysin. Cellular and molecular life sciences: CMLS 65, 2574–2585, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Unger RH & Cherrington AD Glucagonocentric restructuring of diabetes: a pathophysiologic and therapeutic makeover. The Journal of clinical investigation 122, 4–12, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drag M & Salvesen GS Emerging principles in protease-based drug discovery. Nat Rev Drug Discov 9, 690–701, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berg DT, Wiley MR & Grinnell BW Enhanced protein C activation and inhibition of fibrinogen cleavage by a thrombin modulator. Science 273, 1389–1391, (1996). [DOI] [PubMed] [Google Scholar]
- 19.Xu X, Chen Z, Wang Y, Bonewald L & Steffensen B Inhibition of MMP-2 gelatinolysis by targeting exodomain-substrate interactions. Biochem J 406, 147–155, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knapinska AM et al. SAR Studies of Exosite-Binding Substrate-Selective Inhibitors of A Disintegrin And Metalloprotease 17 (ADAM17) and Application as Selective in Vitro Probes. J Med Chem 58, 5808–5824, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madoux F et al. Discovery of an enzyme and substrate selective inhibitor of ADAM10 using an exosite-binding glycosylated substrate. Scientific reports 6, 11, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panwar P et al. Tanshinones that selectively block the collagenase activity of cathepsin K provide a novel class of ectosteric antiresorptive agents for bone. British journal of pharmacology 175, 902–923, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leissring MA et al. Designed inhibitors of insulin-degrading enzyme regulate the catabolism and activity of insulin. PLoS One 5, e10504, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deprez-Poulain R et al. Catalytic site inhibition of insulin-degrading enzyme by a small molecule induces glucose intolerance in mice. Nature communications 6, 8250, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hendriks BS, Seidl KM & Chabot JR Two additive mechanisms impair the differentiation of ‘substrate-selective’ p38 inhibitors from classical p38 inhibitors in vitro. BMC Syst Biol 4, 23, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abdul-Hay SO et al. Optimization of peptide hydroxamate inhibitors of insulin-degrading enzyme reveals marked substrate-selectivity. J Med Chem 56, 2246–2255, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charton J et al. Imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, substrate-dependent modulators of insulin-degrading enzyme in amyloid-beta hydrolysis. European journal of medicinal chemistry 79, 184–193, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abdul-Hay SO et al. Selective Targeting of Extracellular Insulin-Degrading Enzyme by Quasi-Irreversible Thiol-Modifying Inhibitors. ACS chemical biology 10, 2716–2724, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Charton J et al. Structure-activity relationships of imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, dual binders of human insulin-degrading enzyme. European journal of medicinal chemistry 90, 547–567, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Busschots K et al. Substrate-selective inhibition of protein kinase PDK1 by small compounds that bind to the PIF-pocket allosteric docking site. Chem Biol 19, 1152–1163, (2012). [DOI] [PubMed] [Google Scholar]
- 31.Rettenmaier TJ et al. A small-molecule mimic of a peptide docking motif inhibits the protein kinase PDK1. Proc Natl Acad Sci U S A 111, 18590–18595, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shah NG et al. Novel Noncatalytic Substrate-Selective p38alpha-Specific MAPK Inhibitors with Endothelial-Stabilizing and Anti-Inflammatory Activity. J Immunol 198, 3296–3306, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hall MD et al. Fluorescence polarization assays in high-throughput screening and drug discovery: a review. Methods Appl Fluoresc 4, 022001, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowe JT et al. Synthesis and profiling of a diverse collection of azetidine-based scaffolds for the development of CNS-focused lead-like libraries. The Journal of organic chemistry 77, 7187–7211, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malito E et al. Molecular bases for the recognition of short peptide substrates and cysteine-directed modifications of human insulin-degrading enzyme. Biochemistry 47, 12822–12834, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sebaugh JL Guidelines for accurate EC50/IC50 estimation. Pharm Stat 10, 128–134, (2011). [DOI] [PubMed] [Google Scholar]
- 37.Degorce F et al. HTRF: A technology tailored for drug discovery - a review of theoretical aspects and recent applications. Current chemical genomics 3, 22–32, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leung CS, Leung SS, Tirado-Rives J & Jorgensen WL Methyl effects on protein-ligand binding. J Med Chem 55, 4489–4500, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shroyer LA & Varandani PT Purification and characterization of a rat liver cytosol neutral thiol peptidase that degrades glucagon, insulin, and isolated insulin A and B chains. Archives of biochemistry and biophysics 236, 205–219, (1985). [DOI] [PubMed] [Google Scholar]
- 40.Shen Y, Joachimiak A, Rosner MR & Tang WJ Structures of human insulin-degrading enzyme reveal a new substrate recognition mechanism. Nature 443, 870–874, (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leissring MA & Selkoe DJ Structural biology: enzyme target to latch on to. Nature 443, 761–762, (2006). [DOI] [PubMed] [Google Scholar]
- 42.McCord LA et al. Conformational states and recognition of amyloidogenic peptides of human insulin-degrading enzyme. Proc Natl Acad Sci U S A 110, 13827–13832, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song ES, Juliano MA, Juliano L & Hersh LB Substrate activation of insulin-degrading enzyme (insulysin). A potential target for drug development. J Biol Chem 278, 49789–49794, (2003). [DOI] [PubMed] [Google Scholar]
- 44.Im H et al. Structure of substrate-free human insulin-degrading enzyme (IDE) and biophysical analysis of ATP-induced conformational switch of IDE. J Biol Chem 282, 25453–25463, (2007). [DOI] [PubMed] [Google Scholar]
- 45.Song ES, Rodgers DW & Hersh LB A monomeric variant of insulin degrading enzyme (IDE) loses its regulatory properties. PLoS One 5, e9719, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duggan KC et al. (R)-Profens are substrate-selective inhibitors of endocannabinoid oxygenation by COX-2. Nat Chem Biol 7, 803–809, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rose K et al. Insulin proteinase liberates from glucagon a fragment known to have enhanced activity against Ca2+ + Mg2+-dependent ATPase. Biochem J 256, 847–851, (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vandenbroucke RE & Libert C Is there new hope for therapeutic matrix metalloproteinase inhibition? Nat Rev Drug Discov 13, 904–927, (2014). [DOI] [PubMed] [Google Scholar]
- 49.McMurray JJ Neprilysin inhibition to treat heart failure: a tale of science, serendipity, and second chances. Eur J Heart Fail 17, 242–247, (2015). [DOI] [PubMed] [Google Scholar]
- 50.Zeke A et al. Systematic discovery of linear binding motifs targeting an ancient protein interaction surface on MAP kinases. Mol Syst Biol 11, 837, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Online Methods References
- 51.Geu-Flores F, Nour-Eldin HH, Nielsen MT & Halkier BA USER fusion: a rapid and efficient method for simultaneous fusion and cloning of multiple PCR products. Nucleic acids research 35, e55, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vonrhein C et al. Data processing and analysis with the autoPROC toolbox. Acta crystallographica. Section D, Biological crystallography 67, 293–302, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCoy AJ et al. Phaser crystallographic software. Journal of applied crystallography 40, 658–674, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta crystallographica. Section D, Biological crystallography 60, 2126–2132, (2004). [DOI] [PubMed] [Google Scholar]
- 55.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D 66, 213–221, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.APEX2 v. 2014.11–0 (Bruker AXS, Madison, Wisconsin, USA, 2014).
- 57.Krause L, Herbst-Irmer R, Sheldrick GM & Stalke D Comparison of silver and molybdenum microfocus X-ray sources for single-crystal structure determination. Journal of applied crystallography 48, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sheldrick GM SHELXT - Integrated space-group and crystal-structure determination. Acta crystallographica. Section A, Foundations and advances 71, 3–8, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sheldrick GM Crystal structure refinement with SHELXL. Acta Crystallographica Section C Structural Chemistry 71, 3–8, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dolomanov OV, Bourhis LJ, Gildea RJ, Howard JAK & Puschmann H OLEX2: a complete structure solution, refinement and analysis program. Journal of applied crystallography 42, 339–341, (2009). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Complete results of the IDE high-throughput screens and the IDE inhibition counter-screen are available in the PubChem BioAssay database (1259348, 1259349), the IDE•37, IDE•63, and the IDE•63•glucagon X-ray structures are available in the Protein Data Bank (PDB IDs 6BYZ, 6MQ3, and 6EDS, respectively).