Abstract
The chemical components of Spatholobi Caulis tannin (SCT) have a modest therapeutic effect in patients with cervical cancer. However, the active components and the mechanism of action of SCT in HeLa cervical cancer cells need to be further studied. In this paper, 3D microfluidic chip technology was applied to simulate the effects of tannins in the human body, and the appropriate dose and time of administration were calculated. The cell cycle and apoptosis experiments demonstrated that SCT inhibits proliferation and stimulated apoptosis in HeLa cells. The differentially expressed genes were screened using The Cancer Genome Atlas (TCGA) and the GEO databases to identify common differentially expressed genes. A bioinformatic analysis of relevant genes, analysis using the molecular docking technique, and survival analysis were used to predict the target genes of SCT. Circular RNAs (circRNAs) associated with the SCT target genes and the regulatory effects of SCT on these circRNAs were determined. These studies showed that SCT mediates related circRNAs in HeLa cells to inhibit proliferation and promote apoptosis in HeLa cells. Thus, SCT may be an effective strategy for treating cervical cancer.
Keywords: cervical cancer, 3D microfluidic chip, bioinformatic integration, molecular docking, circRNA
Introduction
Cervical cancer has become a global threat to the health of women, and it is common in young women who desire to maintain their fertility.1 Treatment often includes chemotherapy, and, although this treatment strategy is effective in most cases, it may cause immediate adverse effects, such as bleeding, infections, and complications.2 In some underdeveloped areas, the screening, diagnosis, and treatment of diseases are hampered by poor medical standards.3 Most patients who are diagnosed with cervical cancer are diagnosed at the advanced stage, with metastasis in some cases. Chemotherapy treatments are limited and expensive. Traditional Chinese medicine can effectively treat cancer,4 prolong the survival time of cancer patients, and prevent cancer metastasis. In addition, traditional Chinese medicine can alleviate patient pain toward the end of life. Recently, an increasing number of studies have investigated antitumor traditional Chinese medicines, which have been widely used clinically due to their high anticancer efficacy and low toxicity.5, 6, 7 Spatholobi Caulis is a traditional Chinese medicine used to improve blood and kidney circulation, relieve menstrual pain, relax the meridian, and activate collaterals. Spatholobi Caulis tannin (SCT) is extracted from Spatholobi Caulis. SCT has anticancer effects, but the anticervical cancer effects of SCT need further investigation.
Microfluidic chips have been successfully applied in laboratory and clinical settings. However, two-dimensional cell culture models fail to accurately summarize specific biological structures and functions.8, 9 For example, the functional unit structure of a target organ, the interaction among cells, and the interaction between cells and the surrounding matrix are basic components used to reconstruct physiological or pathological conditions.10, 11 Therefore, this cultivation environment cannot represent a true physiological environment. In contrast, 3D cell culture models have been demonstrated to mimic the in vivo environment of cell-cell interactions and cell-matrix interactions, resulting in higher levels of cellular differentiation and biologically relevant structural components.12, 13, 14
We built a simple and flexible 3D microfluidic chip to establish an organ chip model that can control the fluids of each channel through a sensitive gas valve system. This model allows for interactions among cancer cells in a 3D environment, and it was used to analyze drug reactions in cell clusters. Because the effect of SCT on the human body was simulated, the appropriate SCT treatment time and concentration were selected. The selected dose and time of administration were applied in the analysis of apoptosis and cell cycle. This experiment measured the effect of SCT on the apoptosis of cervical cancer cells, and it measured the drug’s effect on the cycle arrest of cervical cancer cells to provide a basis for further studies.
Gene expression profiling has been used to determine the difference between cancerous tissue and normal tissue. Often this analysis yields tens of thousands of differentially expressed genes, allowing the identification of the expression of indistinguishable genes and subtle differences in gene expression.15, 16 Gene expression profiling can elucidate the characteristic indicators of tumor transformation and progression, and it can provide a basis for future research and the determination of candidate treatment goals. In the present study, bioinformatic analyses were used to predict the potential targets of drug action, and molecular docking technology was used to predict the target proteins of SCT, thereby providing accurate target predictions for subsequent circular RNA (circRNA) assays.17, 18
circRNA was first discovered by electron microscopy in a virus study in 1976, and it has subsequently been identified in humans, mice, rats, fungi, and other organisms.19, 20, 21 circRNAs are abundant, diverse, and conserved molecules that are usually expressed in tissues at specific developmental stages.22 circRNAs can act as microRNA (miRNA) sponges to prevent the translation of miRNA and affect gene expression by regulating splicing, which affects related proteins through gene expression.23 Therefore, these molecules offer potential opportunities for therapeutic intervention and may serve as diagnostic biomarkers.24, 25 In the present study, the regulation of related circRNAs by SCT was studied to aid in the development of accurate targeted therapy for the treatment of cervical cancer.
Results
Cell Mass Growth in the 3D Microfluidic Chip
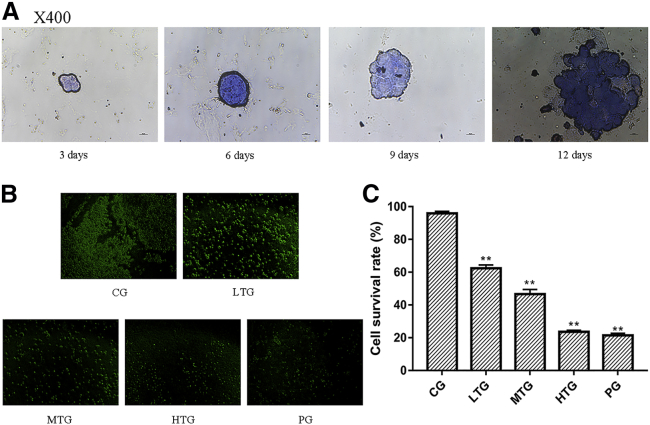
As shown in Figure 1A, the number of cells increased over time, and cell mass gradually increased. The diameter of the cultured cell mass reached 135 μm on the 12th day. Moreover, polydimethylsiloxane (PDMS) had no toxic side effects, indicating that the designed chip could be used for cell culture. The 3D microfluidic chip simulated the human environment and provided a stable environment for the growth of cell clusters.
Figure 1.
Cell Mass Growth in Chips and SCT Effects on the Cell Mass in the Chip
(A) Cell mass diameter after growth in the chip for 3, 6, 9, and 12 days. (B) Fluorescence images after cell treatment with different doses of SCT in CG, LTG, MTG, HTG, and PG. (C) All cells were counted using IPP (version 6.0) to calculate the cell survival rate (%) as follows: cell survival rate (%) = (normal cells/total number of HeLa cells) × 100% (*p < 0.05 and **p < 0.01 versus CG; mean ± SD; n = 3).
Effect of SCT on Cell Mass in Chip
As shown in Figure 1B, SCT had a significant antitumor effect in HeLa cells. The cell clusters were significantly reduced by SCT, and the intercellular force was significantly reduced. The survival rate of the SCT-treated HeLa cells was significantly lower than that of the control group, and SCT affected the survival rate in a dose-dependent manner. The cell viability of each group was as follows: 95.84% (1.32) in the control group (CG), 62.41% (2.03) in the low-dose tannin group (LTG) (p < 0.01 versus CG), 44.94% (1.51) in the medium-dose tannin group (MTG) (p < 0.01 versus CG), 23.60% (0.92) in the high-dose tannin group (HTG) (p < 0.01 versus CG), and 21.49% (1.22) in the positive drug group (PG) (p < 0.01 versus CG) (Figure 1C). Thus, increases in the SCT concentration caused significant decreases in the proportion of living cells, and the HTG was similar to the PG. These experimental results suggest that SCT has potential clinical value in the treatment of cervical cancer.
Flow Cytometry Analysis of Cell Apoptosis and Cell Cycle
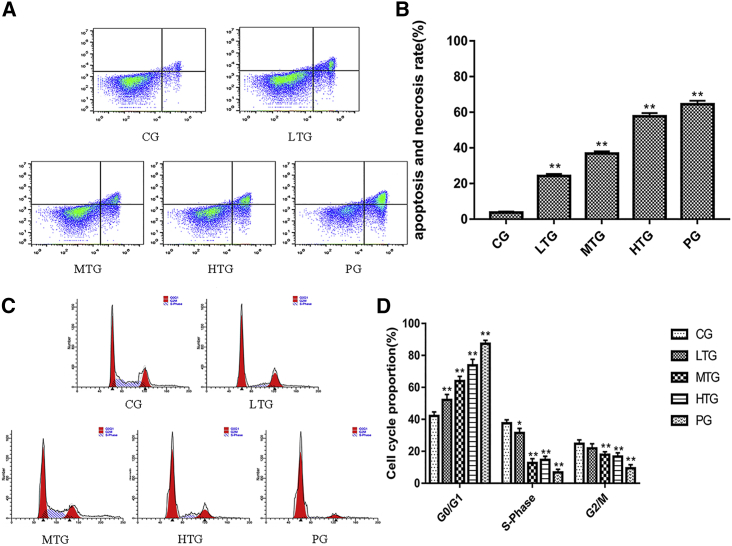
According to the analysis of the apoptosis flow cytometry data using FlowJo software, the percentage of apoptosis gradually increased as the dose increased. These results are shown in Figure 2A. The following apoptosis rates were calculated: CG, 3.73% (0.55) (p < 0.01 versus CG); LTG, 24.20% (1.16) (p < 0.01 versus CG); MTG, 36.79% (1.29) (p < 0.01 versus CG); HTG, 57.68% (1.86) (p < 0.01 versus CG); and PG, 64.46% (2.01) (p < 0.01 versus CG). Notably, the apoptosis rate in the HTG was similar to that in the PG (Figure 2B).
Figure 2.
Proapoptotic Effect of SCT in HeLa Cells and Effect of SCT on HeLa Cell Cycle
(A) Proapoptotic effect of SCT in HeLa cells (mean ± SD, n = 3) in CG, LTG, MTG, HTG, and PG. (B) Histogram of the apoptosis rate per group after treatment with different concentrations of SCT and paclitaxel for 36 h (*p < 0.05 and **p < 0.01 versus CG). (C) Effect of SCT on HeLa cell cycle (mean ± SD, n = 3) in CG, LTG, MTG, HTG, and PG. (D) Histograms of different cell cycle proportions per group after treatment with different concentrations of SCT and paclitaxel for 36 h (*p < 0.05 and **p < 0.01 versus CG).
The cell cycle flow cytometry results were analyzed using ModiFIT software (Figure 2C). The G0:G1 phase ratios among the groups were as follows: CG, 41.20% (2.50) (p < 0.01 versus CG); LTG, 52.14% (3.42) (p < 0.01 versus CG); MTG, 63.93% (2.95) (p < 0.01 versus CG); HTG, 73.79% (3.77) (p < 0.01 versus CG); and PG, 87.30% (2.19) (p < 0.01 versus CG). The G0:G1 phase ratio in the HTG was significantly higher than that in the CG, but the G2:M phase ratio and S phase proportion in the HTG were significantly lower than those in the CG (Figure 2D). These results indicate that SCT inhibits proliferation in HeLa cells by inhibiting the formation of intracellular DNA and cell mitosis.
Bioinformatic Analysis
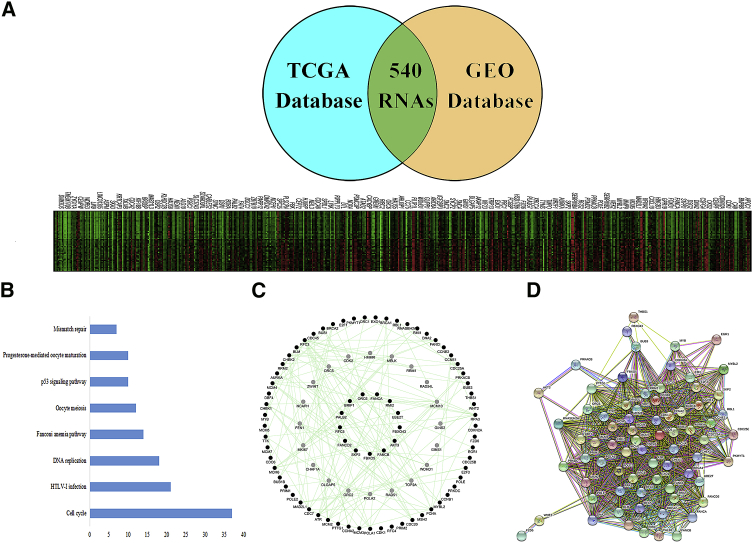
According to the analysis of the data in The Cancer Genome Atlas (TCGA) database, 5,025 genes exhibited significant differential expression between cervical cancer patients and healthy subjects, including 1,971 upregulated genes and 3,054 downregulated genes. According to the analysis of the data in the GEO database, 1,904 genes exhibited significant differential expression between cervical cancer patients and healthy subjects, including 1,699 upregulated genes and 205 downregulated genes. The differentially expressed genes shared by the integrated TCGA and GEO databases are shown in Figure 3A. In total, 540 identical genes were identified.
Figure 3.
Aberrant Gene Expression Landscape in Cervical Cancer and Genes Related to Each Other
(A) The same coding genes were integrated from the GEO and TCGA databases, and 540 common differentially expressed genes were identified. In the RNA heatmap generated by the GEO and TCGA databases, the red spots represent highly expressed RNA, and the green spots represent downregulated RNA. (B) KEGG pathway analysis. (C) Interactions among genes were analyzed using the GeneMANIA application. (D) PPI network of the host genes of the differentially expressed genes.
The Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis indicated that the differentially expressed genes were concentrated in the cell cycle, DNA replication, Fanconi anemia pathway, HTLV-I infection, mismatch repair, p53-signaling pathway, oocyte meiosis, and progesterone-mediated oocyte maturation-related pathway (p < 0.001) (Figure 3B). The gene interactions in these pathways were analyzed by the GeneMANIA application in the Cytoscape software, and the 74 genes with the highest interaction scores were identified (Figure 3C). Proteins corresponding to the selected genes were identified using a protein-protein interaction (PPI) network analysis. The network showed 73 nodes and 1,359 edges, and the 59 highest-ranking hub proteins were identified (Figure 3D). These proteins provided a basis for the SCT targets.
Molecular Docking
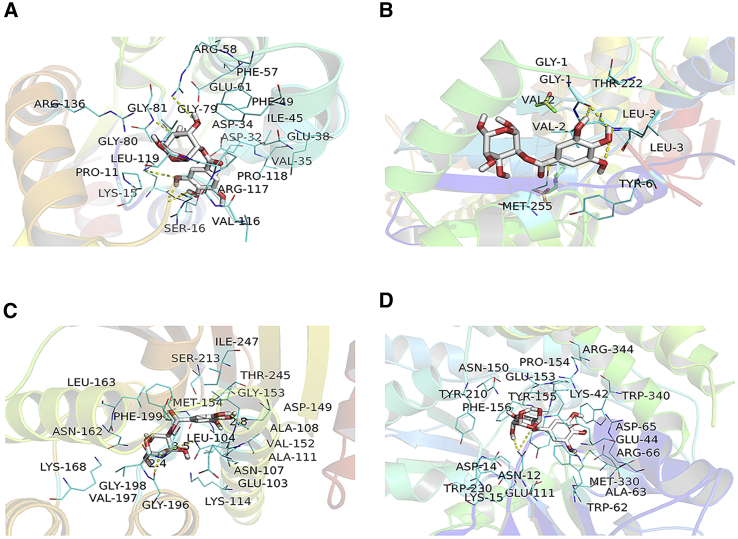
The molecular docking studies indicated that the activity of SCT on proteins included the following two factors: (1) the benzene ring in SCT forms π-π conjugates with the amino acids in the protein, and (2) the H in SCT forms hydrogen bonds with the amino acids in the protein. The docking results indicated that SCT can form the following bonds and conjugates: a hydrogen bond with ARG-58, GLY-81, PRO-11, and SER-16 in the ATR protein; a π-π conjugate with PHE-57 and PHE-49 in the ATR protein (Figure 4A); a hydrogen bond with GLY-1 in the BLM protein; a π-π conjugate with TYR-6 in the BLM protein (Figure 4B); a hydrogen bond with ASN-107, GLY-196, and LEU-104 in the E2F3 protein; a π-π conjugate with PHE-199 in the E2F3 protein (Figure 4C); a hydrogen bond with ASN-12 and TRP-62 in the FANCB protein; and a π-π conjugate with TYR-155, TYR-210, TRP-304, and TRP-62 in the FANCB protein (Figure 4D). The docking scores of SCT with ATR, BLM, E2F3, and FANCB are shown in Table 1. These results demonstrate that SCT has a good binding ability to the target proteins.
Figure 4.
Molecular Docking Results
(A) Docking results of SCT and ATR. (B) Docking results of SCT and BLM. (C) Docking results of SCT and E2F3. (D) Docking results of SCT and FANCB. Hydrogen bonds are presented as yellow dashes.
Table 1.
Molecular Docking Results for Tannin and ATR, BLM, E2F3, and FANCB
| Mode | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|---|
| ATR affinity(kcal/mol) | −7.3 | −7.2 | −7.2 | −7.1 | −7.0 | −7.0 | −7.0 | −7.0 | −6.9 |
| BLM affinity(kcal/mol) | −5.6 | −5.5 | −5.5 | −5.5 | −5.4 | −5.4 | −5.2 | −5.0 | −4.8 |
| E2F3 affinity(kcal/mol) | −7.0 | −7.0 | −6.8 | −6.7 | −6.7 | −6.6 | −6.6 | −6.6 | −6.5 |
| FANCB affinity(kcal/mol) | −7.7 | −7.7 | −7.6 | −7.3 | −7.3 | −7.2 | −7.1 | −7.1 | −7.1 |
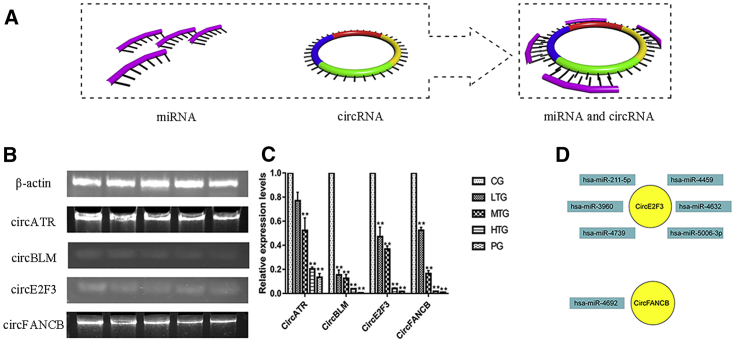
RT-PCR and miRNA Prediction of circRNAs
circRNAs constitute a class of noncoding RNA molecules that do not have a 5′ end cap or a 3′ end poly(A) tail; circRNAs form a circular structure by covalent bonds. In contrast to traditional linear RNA, circRNA molecules have a closed loop structure, are not easily degraded by exonuclease RNaseR, are more stable than linear RNA, and are highly conserved. circATR, circBLM, circE2F3, and circFANCB are under-expressed in normal cervical tissues.
The RT-PCR results showed that the expression of circATR, circBLM, circE2F3, and circFANCB in HeLa cells decreased as the drug concentration of SCT increased (Figure 5B). The RT-PCR data were analyzed using the 2−ΔΔCT method (Figure 5C). The primer sequences of the related genes are listed in Table 2. These results suggest that the expression levels of circATR, circBLM, circE2F3, and circFANCB in the drug treatment group are significantly lower than those in the CG. circRNA molecules contain a miRNA response element that binds miRNA and acts as a miRNA sponge in the cell (Figure 5A). The RegRNA analysis led to the following predictions: circE2F3 binds hsa-miR-211-5p, hsa-miR-4459, hsa-miR-3960, hsa-miR-4632, hsa-miR-4739, and hsa-miR-5006-3p and circFANCB binds hsa-miR-4692 (Figure 5D). Altogether, these results suggest that SCT inhibits growth in HeLa cells by modulating the relevant circRNAs.
Figure 5.
Expression of circRNA and the Relationship between circRNA and miRNA
(A) circRNA as a sponge model of miRNA, circRNA, and miRNA and circRNA. (B) Agarose gel electrophoresis results of related circRNAs as β-actin, circATR, circBLM, circE2F3, and circFANCB. (C) RT-PCR results. The images show that circATR, circBLM, circE2F3, and circFANCB were significantly decreased in the treatment groups (*p < 0.05 and **p < 0.01 versus CG). (D) circRNA and miRNA relationships. circE2F3 and related miRNAs and circFANCB and related miRNA are shown.
Table 2.
Primer Sequences
| Gene | Primer Sequence 5ʹ to 3ʹ |
|---|---|
| β-actin | forward: 5′-CACCCGCGAGTACAACCTTC-3′ |
| reverse: 5′-CCCATACCCACCATCACACC-3′ | |
| circATR hsa_circ_0067641 | forward: 5′-TCTGGTGTATGTCTTACTGGGT-3′ |
| reverse: 5′-ACTGGCAAGATCATGTCGAA-3′ | |
| circBLM hsa_circ_0036868 | forward: 5′-GGACATCCTGACTCAGCTGA-3′ |
| reverse: 5′-TGAGTCAGTCTTATCACCTGTCA-3′ | |
| circE2F3 hsa_circ_0075804 | forward: 5′-TGTTCCTCCTCCCTCCAAAG-3′ |
| reverse: 5′-GATGCAACGGATTGCGAGG-3′ | |
| circFANCB hsa_circ_0006971 | forward: 5′-AGCTTGGTTGTTGGAGTGAA-3′ |
| reverse: 5′-AGGCAGTCTGAGTTCAAGGA-3′ |
Discussion
Cervical cancer is considered the fourth most common malignant disease among women, and it has increasing rates of mortality and morbidity.26 The treatment of cervical cancer continues to face enormous challenges. In this paper, the application of a 3D microfluidic chip revealed that SCT can inhibit the growth of HeLa cells under 3D conditions. Cell cycle and apoptosis assays have shown that SCT can promote apoptosis and inhibit cell proliferation by cell-cycle arrest. In this study, the pathogenic genes of cervical cancer were screened by bioinformatic analysis. The targets ATR, BLM, E2F3, and FANCB of SCT were analyzed by molecular docking. These target-related genes and proteins are associated with the development of cancer. SCT was found to reduce the expression of circATR, circBLM, circE2F3, and circFANCB in the treatment of cervical cancer. This study demonstrates that SCT is valuable for the treatment of cervical cancer. SCT can be used as an alternative or complementary therapy to chemotherapy.
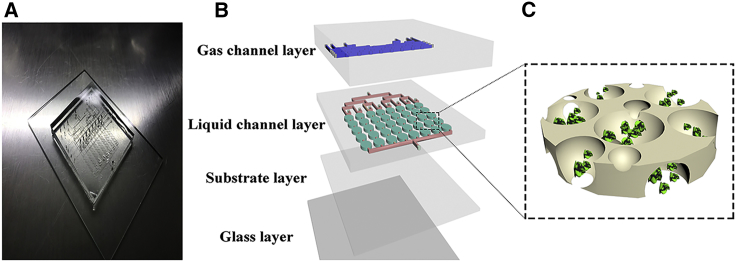
Microfluidic chip technology has great advantages and potential in research investigating traditional Chinese medicine (Figure 6A). Multilayered chips facilitate the formation and growth of cell clumps, and the growth of the cell clumps can be observed to determine the ideal time of administration through relevant staining techniques.27 Each channel has multiple culture chambers to avoid repeated operations and reduce operational errors. A sensitive valve device precisely controls the switch of the corresponding channel (Figure 6B). Simultaneously, the use of syringe pumps reduces the reagent consumption to 0.2 μL × min−1. After 36 h of treatment, the total consumption of reagents does not reach 0.5 mL, which is more economical than the traditional technique of culturing cells in 96-well plates.
Figure 6.
Design of the Microfluidic Cell Culture Chip
(A) Image of chip. (B) Schematic of the microfluidic chip with the gas channel and liquid channel layers. Blue channels indicate the gas channel layer, and red channels represent the liquid channel layer. The green ellipses indicate the cell growth chambers. (C) Cell culture chamber model.
Three-dimensional microfluidic systems provide a biomimetic experimental environment that simulates the microenvironment in the human body (Figure 6C).28 Cell viability is calculated by fluorescently stained cell smears. Compared with 2D culture, 3D culture more closely resembles the physiological environment in the human body, and 3D culture allows drugs to be transported through peristaltic pumps to the matrix around cell aggregates, indicating that 3D culture systems are more similar to human physiology. A 3D culture system was used for drug screening, which laid the foundation for the subsequent experiment. Apoptosis and cell cycle play important roles in maintaining cell viability and cell division. Drugs can participate in the regulation of apoptosis and the cell cycle.29 The present study showed that SCT promotes apoptosis in HeLa cells and arrests cells in the G0 and G1 phase, thereby inhibiting the synthesis of genes and proteins. These results indicate that SCT promotes apoptosis and inhibits cell division in HeLa cells.
Differential gene expression between cervical cancer patients and healthy subjects was analyzed using the GEO and TCGA databases,15 and 540 shared differentially expressed genes were identified. By comparing the differentially expressed gene sets from these two databases, false positives were reduced. The GEO and TCGA data provided mutual verifiability. These differentially expressed genes may be involved in the pathogenesis of cervical cancer. The KEGG analysis identified that the differentially expressed genes were enriched in the following pathways: cell cycle, DNA replication, p53-signaling pathway, and other pathways. These signaling pathways are important in multicellular eukaryotes, and they are not only involved in the processes of cell proliferation, differentiation, and apoptosis but also in the regulation of gynecological diseases, viral infections, and other physiological processes.30
RNA from these pathways was screened and prepared for a gene analysis to identify gene-gene interactions. The interactive network of genes allowed the identification of genes with close interactions, providing a powerful screening range and basis for targeted therapy using SCT. PPI was used to analyze the target proteins of SCT.31 This study used molecular docking to establish links between target disease-associated prion proteins and SCT. According to the molecular docking analysis, SCT had a higher docking score for the following pathogenic proteins: ATR, BLM, E2F3, and FANCB. ATR can inhibit the development of cervical cancer by regulating the ATR/Chk1-signaling pathway and thereby promoting nuclear contraction and apoptosis.32 In addition, inhibition of BLM expression can increase the rate of cancer development.33 The E2F3-mediated pathway participates in the regulation of cervical cancer.34 Although FANCB has not been found to be involved in the regulation of cervical cancer, it has a regulatory role in sporadic head and neck squamous cell carcinomas.35 The results of this study suggest that cervical cancer can be treated by drug interference with FANCB expression. These results indicate that SCT can effectively act on these pathogenic proteins, demonstrating that SCT can treat cervical cancer by interfering with genes associated with these proteins.
In contrast to traditional linear RNA, circRNA is not easily decomposed by enzymes and can be stably expressed in corresponding cells. The SCT-induced inhibition of pathogenic circRNA expression in HeLa cells indicated that SCT has a certain therapeutic effect on cervical cancer. SCT downregulated the expression levels of pathogenic circATR, circBLM, circE3F2, and circFANCB. There are currently no studies on these circRNAs. Through database and molecular docking analyses, the genes and proteins associated with these circRNAs were shown to be involved in the regulation of cervical cancer. The experimental results demonstrate that decreases in the expression of circATR, circBLM, circE3F2, and circFANCB inhibit the proliferation of HeLa cells. Thus, these findings demonstrate that SCT can treat cervical cancer by acting on these circRNAs. The binding of circRNA to related miRNAs can alter the expression of the circRNA. Regulating the expression of miRNAs can directly control the expression of circRNA. The experimental results show that multiple miRNAs can regulate circE3F3 and that circE2F3 can be used as a main therapeutic target for cervical cancer. Therefore, miRNAs that interact with circRNA were screened, providing a basis for further research investigating the targeting of SCT.
The three-dimensional microfluidic chip assays and analysis of the HeLa cell cycle and apoptosis showed that SCT inhibits HeLa cell proliferation but promotes HeLa cell apoptosis. The pathogenic genes involved in cervical cancer were analyzed using relevant databases. The main pathogenic targets were analyzed by KEGG, GeneMANIA, and PPI analyses. Proteins with higher scores in terms of the interaction with SCT were analyzed by molecular docking techniques. SCT was found to modulate the identified pathogenic circRNAs, demonstrating that SCT can treat diseases by modulating circRNAs. The present study also predicted the miRNAs that interact with circRNA, thereby laying the foundation for further explorations of the deeper mechanism of action.
Conclusions
Our findings suggest that SCT can be used to treat cervical cancer via precise therapeutic effects on related circRNA.
Materials and Methods
Extract Preparation
For the sample preparation, the samples were ground and sieved through a 40-mesh screen. The sample powder (50 g) was accurately weighed in a round-bottom flask and extracted twice with 10 times the volume of 60% ethanol solution at reflux (1 h each). Then, the filtrates were combined, and the final concentration of the drug was adjusted to 0.20 g × mL−1 (m/v). The sample was extracted twice with an equal volume of n-butanol solution, and then the n-butanol was evaporated.36, 37
Cell Culture
The human cervical carcinoma cell line HeLa was obtained from the Chinese Academy of Sciences (Shanghai, China). DMEM (RPMI DMEM, Gibco, Grand Island, NY, USA) containing 10% fetal calf serum (FCS, Gibco) was used to culture the cells at 5% CO2, 100% humidity, and 37°C (Nuaire, USA).38
Microfluidic Chip Fabrication
The 3D microfluidic chip designed in this experiment contained three layers of PDMS (Dow Corning, Midland, MI, USA) and a layer of glass. The chip contained 10 gas valves and 10 liquid valves to control the flow of liquid in each channel. The layers were designed as follows: the first layer was the gas channel layer, the second layer was the liquid channel layer, the third layer was the base layer, and the fourth layer was glass. The layers were bonded together by oxygen plasma to form an irreversible chemical bond.
The elliptical gas structure in the gas passage layer had dimensions of approximately 1.00 mm (length) by 0.50 mm (width) by 0.1 mm (height). The cell culture module in the fluid channel layer was approximately 1.50 mm (length) by 1.00 mm (width) by 0.20 mm (height). The device was manufactured by a classical soft lithography process, and the acid-treated silicon wafer and glass plate were cleaned. The surface moisture was drained with nitrogen and dried. SU8-2075 negative photoresist (MicroChem, Newton, MA, USA) was spin coated, lithographically patterned, and baked to obtain the chip model. The layers of the chips were cast and baked using a curing agent and PDMS (Sylgard 184, Dow Corning, Midland, MI, USA), trimmed, and bonded to form the 3D microfluidic chip.39, 40
Cell Growth Activity in the Chip
A cell suspension containing 1% sodium alginate and a solution containing 1% calcium chloride (Sigma-Aldrich, USA) were prepared and injected into the chip for culture using a peristaltic syringe pump (Longer Pump, LSP04-1A, China).41 The cell morphology was observed after 3, 6, 9, and 12 days of culture. The colloid was dissolved in a solution containing 1% sodium citrate, and the cells were collected to prepare a cell smear. The state of the cell pellet was observed by Wright-Giemsa (Solarbio, USA) reagent staining, which stained the nuclei pink and the cytoplasm blue.
SCT Efficacy Studies in the Chip
After the cells were cultured for 12 days in the chip, different doses of SCT (0.5 mg × mL−1 in LTG, 1.0 mg × mL−1 in MTG, and 2.0 mg × mL−1 in HTG) and positive control drugs (6 μg·mL−1 paclitaxel; Sichuan, China) were injected into the chip. After 36 h, the treatments were dissolved in a solution containing 1% sodium citrate. The cells were collected to generate cell smears. The living and dead cells were detected using two dyes, namely, calcein acetoxymethyl ester (AM) and propidium iodide (PI). For the staining, 5 μL 8 mM PI stock solution and 5 μL 4 mM calcein AM stock solution were formulated into working solutions for the fluorescent staining of the cell smears under an inverted fluorescence microscope (Nikon ECLIPSE TI, Nikon, Japan). The cell smears were observed, and the cell viability (%) was calculated using Image-Pro Plus (IPP) software.42
Flow Cytometry Analysis of Apoptosis and the Cell Cycle
The HeLa cells (2 × 105 cells/well) were cultured in a 6-well plate in an incubator. Once the cells reached 80% confluency, different doses of SCT and positive drugs were added (6 μg × mL−1 paclitaxel; Sichuan, China). After 36 h the cells were collected, and apoptosis detection was performed using a cell cycle assay kit and a flow cytometer (BD Accuri C6, USA). After treatment, the cells were collected, fixed in ethanol for 24 h, and subjected to a cycle analysis by flow cytometry using a cell cycle detection kit (KeyGEN BioTECH, Nanjing, China) (BD Accuri C6, USA).43
Bioinformatic Analysis
The cervical cancer gene expression levels in TCGA database (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga), which contains 312 subjects (309 cervical cancer patients and 3 healthy subjects), were analyzed with Tpm correction and p value adjustments, using the Perl and edgeR software packages (Bioconductor), to describe the matrix of the differentially expressed genes (fold change R2 and p < 0.05).44 The cervical cancer gene expression levels were investigated in the GEO database (https://www.ncbi.nlm.nih.gov/geo/; the GEO: GSE63514 dataset containing 28 cervical cancer patients and 24 healthy subjects) using the LIMMA software package (Bioconductor) to describe the matrix of differentially expressed genes (fold change R2 and p < 0.05).45
The differentially expressed genes shared in both databases were screened and subjected to a KEGG analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/).31 Screening conditions for defining KEGG-related pathways were applied using the R language (gene = gene, organization = human, qvalueCutoff = 0.05, and readable = TRUE; pathways of p < 0.001 were extracted from the screening results). The gene interactions in the relevant pathways were analyzed by the GeneMANIA application in Cytoscape software, and the interactions among related proteins were analyzed by STRING (https://string-db.org/) to provide a basis for the precise targeting of SCT.
Molecular Docking
Integrated proteins that were the precise targets of SCT were identified by molecular docking studies. Proteins containing ligands similar to SCT structures were screened in the PDB database (http://www.rcsb.org/) and then dehydrated and energized by Pymol and Autodock-Vina software.46 The basic structure of SCT was docked with the corresponding protein using the following parameters: the grid spacing was set to 0.375; the grid spacing was centered to the binding site; and a 22 × 22 × 22 search box was used in the x, y, and z dimensions. Then, the molecular docking results were analyzed.
RT-PCR Analysis of circRNAs and miRNA Target Analysis
The total RNA was extracted from the cells using TransZol Up (TransGen Biotech, China). cDNA was synthesized using the TransScript One-Step gDNA Removal and cDNA synthesis SuperMix kit (TransGen Biotech, China). The designed back-to-back primers were combined with cDNA using a Piko Thermal Cycler 96-well system and TransStart Top Green qPCR SuperMix (TransGen Biotech, China) to amplify the circRNA. RegRNA (http://regrna2.mbc.nctu.edu.tw/detection.html) was used to analyze the miRNAs that target the circRNAs.
Statistical Analysis
One-way ANOVA was used to test for statistical significance in all in vitro experiments using SPSS software (SPSS, Chicago, IL, USA). *p < 0.05, **p < 0.01, and ***p < 0.001 versus CG were considered statistically significant. The data are presented as the mean ± SD. The datasets supporting the conclusions of this article are included within the article and additional files.
Author Contributions
N.W. collected the data and drafted the manuscript. J.W. and X.M. obtained funding. Y.B., S.W., and T.L. revised the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This research was successfully completed with assistance from Jiaxin Fan, Yibo Zheng, Yan Wang, and Linna Guo. This study was supported by grants from the National Natural Science Foundation of China (81874342 and 81241111), the Project of Institutions of Higher Learning Talents to Support in Liaoning Province (LR2013044), the Education General Project of Liaoning Provincial Department (L201605 and L201608), and the National Key R&D Project (2018YFC1704805).
Contributor Information
Jiayi Wang, Email: 414566224@qq.com.
Xiansheng Meng, Email: mxsvvv@163.com.
References
- 1.Martel-Billard C., Faller E., Delaine M., Boisramé T., Baldauf J.J., Akladios C.Y. Type B Laparoscopic Radical Trachelectomy With Pelvic Lymphadenectomy for Early Cervical Cancer. J. Minim. Invasive Gynecol. 2017;24:14–15. doi: 10.1016/j.jmig.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Yadav R., Jee B., Rao K.S. How homeopathic medicine works in cancer treatment: deep insight from clinical to experimental studies. J. Exp. Ther. Oncol. 2019;13:71–76. [PubMed] [Google Scholar]
- 3.Ferris D.G., Shapiro J., Fowler C., Cutler C., Waller J., Guevara Condorhuaman W.S. The Impact of Accessible Cervical Cancer Screening in Peru-The Día del Mercado Project. J. Low. Genit. Tract Dis. 2015;19:229–233. doi: 10.1097/LGT.0000000000000112. [DOI] [PubMed] [Google Scholar]
- 4.Qi F., Zhao L., Zhou A., Zhang B., Li A., Wang Z., Han J. The advantages of using traditional Chinese medicine as an adjunctive therapy in the whole course of cancer treatment instead of only terminal stage of cancer. Biosci. Trends. 2015;9:16–34. doi: 10.5582/bst.2015.01019. [DOI] [PubMed] [Google Scholar]
- 5.Chai F.N., Ma W.Y., Zhang J., Xu H.S., Li Y.F., Zhou Q.D., Li X.G., Ye X.L. Coptisine from Rhizoma coptidis exerts an anti-cancer effect on hepatocellular carcinoma by up-regulating miR-122. Biomed. Pharmacother. 2018;103:1002–1011. doi: 10.1016/j.biopha.2018.04.052. [DOI] [PubMed] [Google Scholar]
- 6.Zhai Q.L., Hu X.D., Xiao J., Yu D.Q. [Astragalus polysaccharide may increase sensitivity of cervical cancer HeLa cells to cisplatin by regulating cell autophagy] Zhongguo Zhongyao Zazhi. 2018;43:805–812. doi: 10.19540/j.cnki.cjcmm.20171113.018. [DOI] [PubMed] [Google Scholar]
- 7.Shi X., Yang J., Wei G. Ginsenoside 20(S)-Rh2 exerts anti-cancer activity through the Akt/GSK3β signaling pathway in human cervical cancer cells. Mol. Med. Rep. 2018;17:4811–4816. doi: 10.3892/mmr.2018.8454. [DOI] [PubMed] [Google Scholar]
- 8.Nasseri B., Soleimani N., Rabiee N., Kalbasi A., Karimi M., Hamblin M.R. Point-of-care microfluidic devices for pathogen detection. Biosens. Bioelectron. 2018;117:112–128. doi: 10.1016/j.bios.2018.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vriend J., Nieskens T.T.G., Vormann M.K., van den Berge B.T., van den Heuvel A., Russel F.G.M., Suter-Dick L., Lanz H.L., Vulto P., Masereeuw R., Wilmer M.J. Screening of Drug-Transporter Interactions in a 3D Microfluidic Renal Proximal Tubule on a Chip. AAPS J. 2018;20:87. doi: 10.1208/s12248-018-0247-0. [DOI] [PubMed] [Google Scholar]
- 10.Hübner J., Raschke M., Rütschle I., Gräßle S., Hasenberg T., Schirrmann K., Lorenz A., Schnurre S., Lauster R., Maschmeyer I. Simultaneous evaluation of anti-EGFR-induced tumour and adverse skin effects in a microfluidic human 3D co-culture model. Sci. Rep. 2018;8:15010. doi: 10.1038/s41598-018-33462-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan L., Skoko J., Yu J., Ozdoganlar O.B., LeDuc P.R., Neumann C.A. Mimicking Embedded Vasculature Structure for 3D Cancer on a Chip Approaches through Micromilling. Sci. Rep. 2017;7:16724. doi: 10.1038/s41598-017-16458-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frick C., Dettinger P., Renkawitz J., Jauch A., Berger C.T., Recher M., Schroeder T., Mehling M. Nano-scale microfluidics to study 3D chemotaxis at the single cell level. PLoS ONE. 2018;13:e0198330. doi: 10.1371/journal.pone.0198330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han S., Kim J., Li R., Ma A., Kwan V., Luong K., Sohn L.L. Hydrophobic Patterning-Based 3D Microfluidic Cell Culture Assay. Adv. Healthc. Mater. 2018;7:e1800122. doi: 10.1002/adhm.201800122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen C., Townsend A.D., Hayter E.A., Birk H.M., Sell S.A., Martin R.S. Insert-based microfluidics for 3D cell culture with analysis. Anal. Bioanal. Chem. 2018;410:3025–3035. doi: 10.1007/s00216-018-0985-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jing H., Qu X., Liu L., Xia H. A Novel Long Noncoding RNA (lncRNA), LL22NC03-N64E9.1, Promotes the Proliferation of Lung Cancer Cells and is a Potential Prognostic Molecular Biomarker for Lung Cancer. Med. Sci. Monit. 2018;24:4317–4323. doi: 10.12659/MSM.908359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li T., Gao X., Han L., Yu J., Li H. Identification of hub genes with prognostic values in gastric cancer by bioinformatics analysis. World J. Surg. Oncol. 2018;16:114. doi: 10.1186/s12957-018-1409-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saeki K., Hayakawa S., Nakano S., Ito S., Oishi Y., Suzuki Y., Isemura M. In Vitro and In Silico Studies of the Molecular Interactions of Epigallocatechin-3-O-gallate (EGCG) with Proteins That Explain the Health Benefits of Green Tea. Molecules. 2018;23:E1295. doi: 10.3390/molecules23061295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yadav P., Bandyopadhyay A., Chakraborty A., Sarkar K. Enhancement of anticancer activity and drug delivery of chitosan-curcumin nanoparticle via molecular docking and simulation analysis. Carbohydr. Polym. 2018;182:188–198. doi: 10.1016/j.carbpol.2017.10.102. [DOI] [PubMed] [Google Scholar]
- 19.Wu Y., Zhang Y., Niu M., Shi Y., Liu H., Yang D., Li F., Lu Y., Bo Y., Zhang R. Whole-Transcriptome Analysis of CD133+CD144+ Cancer Stem Cells Derived from Human Laryngeal Squamous Cell Carcinoma Cells. Cell. Physiol. Biochem. 2018;47:1696–1710. doi: 10.1159/000490992. [DOI] [PubMed] [Google Scholar]
- 20.Wang H.X., Huang Q.L., Shen J.Y., Xu T., Hong F., Gong Z.Y., Li F., Yan Y., Chen J.X. Expression profile of circular RNAs in IDH-wild type glioblastoma tissues. Clin. Neurol. Neurosurg. 2018;171:168–173. doi: 10.1016/j.clineuro.2018.06.020. [DOI] [PubMed] [Google Scholar]
- 21.Zhang J., Jiang J., Huang R., Wang Y., Nie X., Gui R. Circular RNA expression profiles are significantly altered in mice bone marrow stromal cells after total body irradiation. Leuk. Res. 2018;70:67–73. doi: 10.1016/j.leukres.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 22.Li X.M., Ge H.M., Yao J., Zhou Y.F., Yao M.D., Liu C., Hu H.T., Zhu Y.X., Shan K., Yan B., Jiang Q. Genome-Wide Identification of Circular RNAs as a Novel Class of Putative Biomarkers for an Ocular Surface Disease. Cell. Physiol. Biochem. 2018;47:1630–1642. doi: 10.1159/000490982. [DOI] [PubMed] [Google Scholar]
- 23.Dragomir M., Calin G.A. Circular RNAs in Cancer - Lessons Learned From microRNAs. Front. Oncol. 2018;8:179. doi: 10.3389/fonc.2018.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye P., Shi Y., An N., Zhou Q., Guo J., Long X. miR-145 overexpression triggers alteration of the whole transcriptome and inhibits breast cancer development. Biomed. Pharmacother. 2018;100:72–82. doi: 10.1016/j.biopha.2018.01.167. [DOI] [PubMed] [Google Scholar]
- 25.Chen S., Zhao Y. Circular RNAs: Characteristics, function, and role in human cancer. Histol. Histopathol. 2018;33:887–893. doi: 10.14670/HH-11-969. [DOI] [PubMed] [Google Scholar]
- 26.Small W., Jr., Bacon M.A., Bajaj A., Chuang L.T., Fisher B.J., Harkenrider M.M., Jhingran A., Kitchener H.C., Mileshkin L.R., Viswanathan A.N., Gaffney D.K. Cervical cancer: A global health crisis. Cancer. 2017;123:2404–2412. doi: 10.1002/cncr.30667. [DOI] [PubMed] [Google Scholar]
- 27.Yang C., Zhang N., Wang S., Shi D., Zhang C., Liu K., Xiong B. Wedge-shaped microfluidic chip for circulating tumor cells isolation and its clinical significance in gastric cancer. J. Transl. Med. 2018;16:139. doi: 10.1186/s12967-018-1521-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sano E., Mori C., Nashimoto Y., Yokokawa R., Kotera H., Torisawa Y.S. Engineering of vascularized 3D cell constructs to model cellular interactions through a vascular network. Biomicrofluidics. 2018;12:042204. doi: 10.1063/1.5027183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao X.B., Qin Y., Niu Y.L., Yang J. Matrine inhibits hypoxia/reoxygenation-induced apoptosis of cardiac microvascular endothelial cells in rats via the JAK2/STAT3 signaling pathway. Biomed. Pharmacother. 2018;106:117–124. [Google Scholar]
- 30.Liu D., Xu X., Wen J., Xie L., Zhang J., Shen Y., Jiang G., Chen J., Fan M. Integrated Genome-Wide Analysis of Gene Expression and DNA Copy Number Variations Highlights Stem Cell-Related Pathways in Small Cell Esophageal Carcinoma. Stem Cells Int. 2018;2018:3481783. doi: 10.1155/2018/3481783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu K., Yi Y., Liu F., Wu W., Chen Y., Zhang W. Identification of key pathways and genes in the progression of cervical cancer using bioinformatics analysis. Oncol. Lett. 2018;16:1003–1009. doi: 10.3892/ol.2018.8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W.Y., Cao Y.X., Zhou X., Wei B., Zhan L., Fu L.T. HMGA2 gene silencing reduces epithelial-mesenchymal transition and lymph node metastasis in cervical cancer through inhibiting the ATR/Chk1 signaling pathway. Am. J. Transl. Res. 2018;10:3036–3052. [PMC free article] [PubMed] [Google Scholar]
- 33.Srivas R., Shen J.P., Yang C.C., Sun S.M., Li J., Gross A.M., Jensen J., Licon K., Bojorquez-Gomez A., Klepper K. A Network of Conserved Synthetic Lethal Interactions for Exploration of Precision Cancer Therapy. Mol. Cell. 2016;63:514–525. doi: 10.1016/j.molcel.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yao T., Lu R., Zhang J., Fang X., Fan L., Huang C., Lin R., Lin Z. Growth arrest-specific 5 attenuates cisplatin-induced apoptosis in cervical cancer by regulating STAT3 signaling via miR-21. J. Cell. Physiol. 2019;234:9605–9615. doi: 10.1002/jcp.27647. [DOI] [PubMed] [Google Scholar]
- 35.Richardson T.E., Sathe A.A., Kanchwala M., Jia G., Habib A.A., Xiao G., Snuderl M., Xing C., Hatanpaa K.J. Genetic and Epigenetic Features of Rapidly Progressing IDH-Mutant Astrocytomas. J. Neuropathol. Exp. Neurol. 2018;77:542–548. doi: 10.1093/jnen/nly026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan H.J., Li W., Jin J.M., Jia X.B., Zhou T., Wang H., Sun E., Ni H.Y. [Characterization of sediment of water extract of Guizhi decoction and its effect on relevant compound (fractions) in decoction] Zhongguo Zhongyao Zazhi. 2018;43:1633–1641. doi: 10.19540/j.cnki.cjcmm.20180117.004. [DOI] [PubMed] [Google Scholar]
- 37.Huo X.K., Liu J., Yu Z.L., Wang Y.F., Wang C., Tian X.G., Ning J., Feng L., Sun C.P., Zhang B.J., Ma X.C. Alisma orientale extract exerts the reversing cholestasis effect by activation of farnesoid X receptor. Phytomedicine. 2018;42:34–42. doi: 10.1016/j.phymed.2018.03.017. [DOI] [PubMed] [Google Scholar]
- 38.Plotkin B.J., Sigar I.M., Swartzendruber J.A., Kaminski A., Davis J. Differential expression of cytokines and receptor expression during anoxic growth. BMC Res. Notes. 2018;11:406. doi: 10.1186/s13104-018-3520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christoffersson J., Bergström G., Schwanke K., Kempf H., Zweigerdt R., Mandenius C.F. A Microfluidic Bioreactor for Toxicity Testing of Stem Cell Derived 3D Cardiac Bodies. Methods Mol. Biol. 2016;1502:159–168. doi: 10.1007/7651_2016_340. [DOI] [PubMed] [Google Scholar]
- 40.Daniel G.S.T., Thiruppathy M., Aswath N., Narayanan S.R. Lab on a Chip: Conquer Disease at the Earliest. J. Pharm. Bioallied Sci. 2018;10:106–108. doi: 10.4103/JPBS.JPBS_210_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adutler-Lieber S., Friedman N., Geiger B. Expansion and Antitumor Cytotoxicity of T-Cells Are Augmented by Substrate-Bound CCL21 and Intercellular Adhesion Molecule 1. Front. Immunol. 2018;9:1303. doi: 10.3389/fimmu.2018.01303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu Z., Zhu J., Wang X., Yao J., Wu Y., Wang D. [Apoptosis and Ca∼(2+) balance disorder of BV-2 cells induced by polychlorinated biphenyl] Wei Sheng Yan Jiu. 2018;47:108–112. [PubMed] [Google Scholar]
- 43.Czarnomysy R., Surażyński A., Muszynska A., Gornowicz A., Bielawska A., Bielawski K. A novel series of pyrazole-platinum(II) complexes as potential anti-cancer agents that induce cell cycle arrest and apoptosis in breast cancer cells. J. Enzyme Inhib. Med. Chem. 2018;33:1006–1023. doi: 10.1080/14756366.2018.1471687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han J., Chen M., Wang Y., Gong B., Zhuang T., Liang L., Qiao H. Identification of Biomarkers Based on Differentially Expressed Genes in Papillary Thyroid Carcinoma. Sci. Rep. 2018;8:9912. doi: 10.1038/s41598-018-28299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sang L., Wang X.M., Xu D.Y., Zhao W.J. Bioinformatics analysis of aberrantly methylated-differentially expressed genes and pathways in hepatocellular carcinoma. World J. Gastroenterol. 2018;24:2605–2616. doi: 10.3748/wjg.v24.i24.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Masuda T. Molecular dynamics simulation of a myosin subfragment-1 docking with an actin filament. Biosystems. 2013;113:144–148. doi: 10.1016/j.biosystems.2013.06.001. [DOI] [PubMed] [Google Scholar]