Abstract
Sulfonylurea-receptor-1(SUR1) and its associated transient-receptor-potential cation channel subfamily-M (TRPM4) channel are key contributors to cerebral edema and intracranial hypertension in traumatic brain injury (TBI) and other neurological disorders. Channel inhibition by glyburide is clinically promising. ABCC8 (encoding SUR1) single-nucleotide polymorphisms (SNPs) are reported as predictors of raised intracranial pressure (ICP). This project evaluated whether TRPM4 SNPs predicted ICP and TBI outcome. DNA was extracted from 435 consecutively enrolled severe TBI patients. Without a priori selection, all 11 TRPM4 SNPs available on the multiplex platform (Illumina:Human-Core-Exome v1.0) were genotyped spanning the 25 exon gene. A total of 385 patients were analyzed after quality control. Outcomes included ICP and 6 month Glasgow Outcome Scale (GOS) score. Proxy SNPs, spatial modeling, and functional predictions were determined using established software programs. rs8104571 (intron-20) and rs150391806 (exon-24) were predictors of ICP. rs8104571 heterozygotes predicted higher average ICP (β = 10.3 mm Hg, p = 0.00000029), peak ICP (β = 19.6 mm Hg, p = 0.0007), and proportion ICP >25 mm Hg (β = 0.16 p = 0.004). rs150391806 heterozygotes had higher mean (β = 7.2 mm Hg, p = 0.042) and peak (β = 28.9 mm Hg, p = 0.0015) ICPs. rs8104571, rs150391806, and 34 associated proxy SNPs in linkage-disequilibrium clustered downstream. This region encodes TRPM4's channel pore and a region postulated to juxtapose SUR1 sequences encoded by an ABCC8 DNA segment containing previously identified relevant SNPs. There was an interaction effect on ICP between rs8104571 and a cluster of predictive ABCC8 SNPs (rs2237982, rs2283261, rs11024286). Although not significant in univariable or a basic multivariable model, in an expanded model additionally accounting for injury pattern, computed tomographic (CT) appearance, and intracranial hypertension, heterozygous rs8104571 was associated with favorable 6 month GOS (odds ratio [OR] = 16.7, p = 0.007951). This trend persisted in a survivor-only subcohort (OR = 20.67, p = 0.0168). In this cohort, two TRPM4 SNPs predicted increased ICP with large effect sizes. Both clustered downstream, spanning a region encoding the channel pore and interacting with SUR1. If validated, this may guide risk stratification and eventually inform treatment-responder classification for SUR1-TRPM4 inhibition in TBI. Larger studies are warranted.
Keywords: ABCC8/ SUR1, cerebral edema, SNPs, TBI, TRPM4
Introduction
Significant financial and clinical resources are dedicated to the evaluation and management of intracranial hypertension after severe traumatic brain injury (sTBI). Nonetheless, no preventative or targeted treatments are available.1 Intracranial hypertension is a complex product of many variables including injury characteristics (mechanism, severity, velocity, force, pattern, extent), patient characteristics (age, sex, race, comorbidities, intracranial compliance, autoregulation, genetics), and an expanding network of identified mechanisms underlying cerebral edema. Heterogeneity in these factors has rendered development of targeted treatments and success of clinical trials challenging; this underscores the importance of granular individualized phenotyping.
As with many diseases in this era of precision medicine, there is a growing recognition of the importance of genetic contributions to TBI variability including secondary-injury processes such as cerebral edema and intracranial hypertension.2–6 Identifying genetic variants in key molecular pathways known to contribute to underlying cerebral edema may be valuable for both predictive and prognostic enrichment. These may affect transcriptional regulation, protein expression, structure, or function, thereby directing individual differences in timing, degree, and type of cerebral edema, relative predominance of specific pathways, and response to therapy. Eventually, this may guide patient selection for clinical trials and have implications for targeted therapy.
Seminal research has identified a central pathway in cerebral edema development in many acute neurological disorders including TBI, involving the transmembrane receptor sulfonylurea-receptor 1 (SUR1) and its associated non-selective cation channel-transient receptor potential cation channel subfamily M-4 (TRPM4).7,8 Historically, SUR1 has been known for its regulation of a potassium-channel modulating insulin secretion (Kir6.2), and TRPM4 has been predominantly studied in cardiac-conductance disorders.9 The SUR1-TRPM4 channel is now emerging as an exciting potential target and biomarker in brain injury and cerebral edema; the channel is not normally expressed in the central nervous system (CNS), but upregulated after injury in multiple cell types including neurons, astrocytes, and glia.7,8,10,11 Channel opening occurs when upregulated SUR1 undergoes obligate association with TRPM4 resulting in depolarization, water influx, oncotic edema, and cell death. An existing antidiabetes medication (glibenclamide) inhibits SUR1-TRPM4 and has shown promising results against cerebral edema in recent clinical trials of stroke.12,13 This pathway and potential benefits of its inhibition have been implicated in both pre-clinical and human TBI.10,11,14–18 Regionally clustered single nucleotide polymorphisms (SNPs) in ABCC8 (the gene encoding SUR1) have been associated with intracranial pressure (ICP) in TBI.4,5 However, the impact of TRPM4 genetic variation on cerebral edema is currently unknown.
We undertook an exploratory candidate-gene association approach to examining effects of TRPM4 sequence variability on intracranial hypertension in TBI. We hypothesized that a subgroup of coding and non-coding TRPM4 SNPs would be associated with ICP and outcome. We further explored the possibility of an interaction between TRPM4 SNPs and previously reported ABCC8 SNPs associated with ICP and outcome.4,5
Methods
Study design
Subjects were prospectively enrolled through the University of Pittsburgh Brain-Trauma Research Center protocol. Informed consent was obtained from legal authorized representatives. Inclusion criteria were: being between 16 and 80 years of age, having sTBI with admission Glasgow Coma Scale (GCS) score <9 and an external ventricular drain (EVD) per standard institutional clinical protocol. Exclusion criteria were imminent brain death and pregnancy. A total of 435 subjects were consecutively enrolled between 2006 and 2014. The University of Pittsburgh Institutional Review Board approved the study.
DNA collection and genotyping
Per our previously published protocol, DNA was obtained and extracted from white blood cells by the simple-salting out method.5 Genotyping was performed using Illumina (Human-Core Exome v1.0) multiplex platform by the Center for Inherited Disease Research (CIDR) and included 11 SNPs distributed across TRPM4: rs11667393, rs3760666, rs113984787, rs146564314, rs1477363, rs10410857, rs56355369, rs909010, rs145847114, rs8104571, and rs150391806. SNPs were not identified a priori. SNP selection was unbiased and included all available TRPM4 SNPs covered by the Illumina Human-Core Exome v1.0 chip. Data cleaning and quality control included blind technical duplicates, Hardy–Weinberg Equilibrium (HWE) testing, and exclusion of SNPs that did not have a minimum 95% call rate. Principal-component analysis demonstrated that most participants clustered as Caucasian with the small numbers outside of that cluster being too few to analyze. After quality control exclusion of 50 samples, genotypes for 385 patients were included in the analysis. ABCC8 SNPs in this TBI cohort had previously been genotyped per our published protocol and evaluated for associations with ICP and outcome; significant ABCC8 SNPs were evaluated for interactions with TRPM4 SNPs in this analysis.4,5 CIDR research assistants involved in genotyping SNPs were blinded to demographic and outcome data.
Clinical data collection
Two previously established outcome measures were investigated.5,14
-
1.
Hourly ICP (continuous), subcategorized into average and peak ICP (typically 5 days of neuromonitoring at the University of Pittsburgh Medical Center), as well as proportion of ICP spikes >25 mm Hg. Each hourly measurement was a point value obtained at the end of the hour. Proportion of ICP >25 mm Hg per patient was calculated by total number of ICP measurements >25 mm Hg for the specific patient divided by their total number of ICP measurements recorded over 5 days. The threshold of 25 mm Hg was selected because it evaluates the presence of intracranial hypertension regardless of the Brain Trauma Foundation guidelines defining intracranial hypertension as either 22 (4th edition) or 20 (3rd edition) mm Hg.19
-
2.
Six-month Glasgow Outcome Scale (GOS) score (binary): favorable (GOS ≥ 4) versus unfavorable (GOS <4).
Secondary analyses censoring ICP recordings based on timing of craniectomy (if present) were also performed. Because exact matches for hourly ICP recording relative to timing of surgery were unavailable, standard censors were applied as follows. For patients who received their craniectomy on the day of presentation, ICP recordings after the first 6 h were censored, for patients who underwent craniectomy on post-TBI day 1, ICP recordings after the first 24 h were censored, for patients who underwent craniectomy on post-TBI day 2, ICP recordings after the first 48 h were censored, for patients who underwent craniectomy on post-TBI day 3, ICP recordings after the first 72 h were censored, for patients who underwent craniectomy on post-TBI day 4, ICP recordings after the first 96 h were censored, and for patients who underwent craniectomy on or after post-TBI day 5, or did not undergo craniectomy, all 120 h of ICP recordings were retained.
Spatial relationship modeling between ABCC8 and TRPM4 (gene) regions and SUR1-TRPM4 (protein) structure
Chromosomal locations were determined using the University of California, Santa Cruz (UCSC) genome browser, human genome assembly-38. Putative and canonical topology for human TRPM4 amino-acid sequence was obtained from PROTTER version-1.0, UniProt ID Q8TD43.20 Peptide sequences encoded by specific exons were identified by Ensembl (release-92) and confirmed by the UCSC genome browser.21 ProtAnnot within the Integrated-Genome Browser identified TRPM4 protein annotations in the context of TRPM4 genomic sequence.22 The established TRPM4 3-dimensional electron microscopy structure (6BQV) was obtained from the Research Collaboratory for Structural Bioinformatics Protein Data Bank.23,24 University of California, San Francisco Chimera was used to generate three-dimensional structures of TRPM4 with its regulatory associated subunit SUR1 (5WUA).25,26
Functional category of TRPM4 SNPs
Predicted functional effects for exonic-SNPs were investigated using PolyPhen, SIFT/PROVEAN, and MutPred2.27–29 RegulomeDB and Ensembl's Varient Effect Predictor were used for intronic-SNPs.30,31 Significant and proxy-SNPs were evaluated for established clinical significance via PubMed, Embase, and ClinVar searches (see Supplementary Methods at at http://www.liebertpub.com).
Statistical analysis
Outcome variables between SNP genotypes were compared using analysis of variance (F-test, ANOVA) and Fisher's exact test. Between-group comparisons were adjusted using Bonferroni's method. Pairwise linkage-disequilibrium (LD) between SNPs was determined using LDlink for proxy-SNPs.,32 Univariable linear and logistical regression models evaluated independent relationships between TRPM4 SNPs and continuous or categorical outcomes. Basic multivariable regression models were developed with clinically relevant variables (age, gender, initial GCS score, presence of craniectomy) to control for confounders that could influence ICP and/or outcome. Expanded multivariable models also included pattern of primary injury, computed tomographic (CT) appearances (either collectively in expanded model-1, or with individual components separated out as independent predictors in expanded model-2; that is, cistern effacement, sulcal effacement, ventricular compression, midline shift, herniation), and (for clinical outcome), proportion of intracranial hypertension recordings >25 mm Hg. Multiple imputations were used for missing data; models with and without multiple imputations are presented. Odds ratios (OR) and β-coefficients were based on genotype: homozygous-wild-type (AA), heterozygous (Ab), or homozygous-variant (bb). Multiple comparisons for the regression models were adjusted using the established Benjamin–Yekutieli (B–Y) method5,33,34 yielding a significance threshold of p = 0.01656. The likelihood ratio test compared full (with SNP) versus reduced (without SNP) models to test whether the inclusion of the respective SNP significantly added to model fit. The exact test assessed HWE. Weighted and unweighted gene risk scores were calculated for TRPM4 and ABCC8 SNPs using a similar method as previously described by Lu and coworkers (see Supplementary Methods at http://www.liebertpub.com).35,36 Analyses were performed using Stata 15.0 (StataCorp, TX) and RStudio Version 1.1.453.
Results
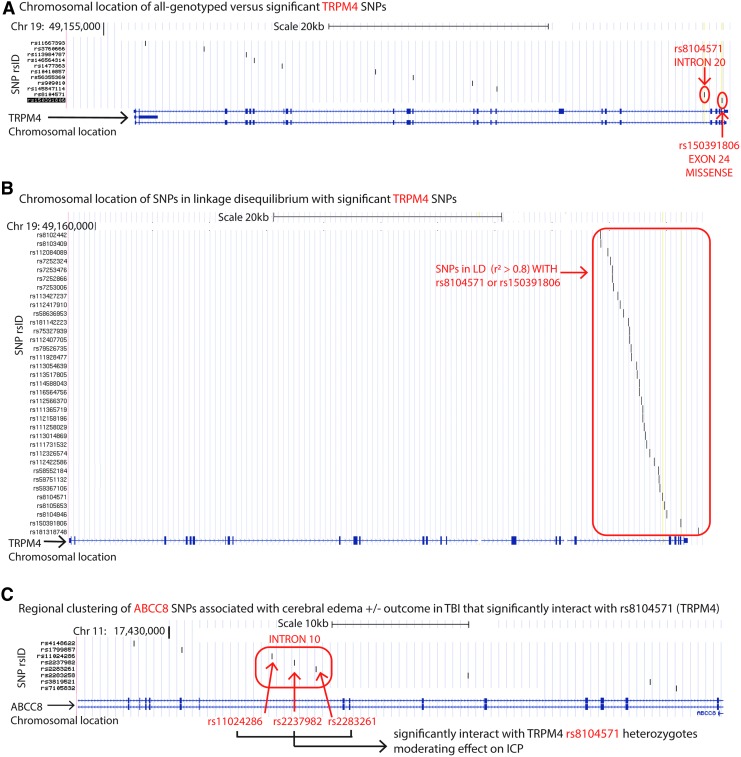
Patient characteristics are summarized in Table 1. Of 11 analyzed SNPs distributed across the length of the TRPM4 gene, two downstream SNPs, rs8104571 (intron 20) and rs150391806 (exon 24), were significantly associated with average ICP, peak ICP, and proportion of ICP spikes >25 mm Hg in univariable and multivariable single-locus analysis (Fig. 1A). Table 2 describes genotype frequencies and descriptive statistics of ICP variation and 6 month GOS between genotypes. There were no homozygous variants.
Table 1.
Patient Characteristics
| Variable (n = 385) | Mean (SD, Range) | |
|---|---|---|
| Age | 37.9 (16.8, 16–77) | |
| Initial GCS | 5.83 (1.51, 3–8) | |
| Average ICP (n = 191) | 10.4 (5.0, 4–45) | |
| Peak ICP (n = 191) | 25.9 (9–96) |
| Frequency n (%) | ||
|---|---|---|
| Gender (M) | 304 (79.0%) | |
| Proportion of ICP >25 mmHg | 5.9% | |
| Decompressive craniotomy (Y) | 128/385 (0.33) | |
| Day 0 | 87/128 (0.68) | |
| Day 1 | 18/128 (0.14) | |
| Days 2, 3, 4 or 5 | 14/128 (0.11) |
| Pattern of injury | Primary | Additional/Mixed (n = 257) |
|---|---|---|
| No radiographic lesion | 9 (2.4%) | 0 |
| Epidural | 22 (6.0%) | 6 (2.3%) |
| Subdural | 118 (32%) | 53 (20.6%) |
| Subarachnoid | 60 (16.3%) | 98 (38.1%) |
| Contusion | 107 (29%) | 74 (28.8%) |
| Intraventricular | 11 (3%) | 22 (8.6%) |
| Diffuse axonal injury | 42 (11.3%) | 4 (1.6%) |
Summarizes the patient characteristics of the severe TBI population overall (n = 385). ICP measurements were available for 191 patients.
GCS, Glasgow Coma Scale; ICP, intracranial pressure; TBI, traumatic brain injury.
FIG. 1.
Transient-receptor-potential cation channel subfamily-M (TRPM4) single nucleotide polymorphisms (SNPs) associated with intracranial pressure (ICP) in severe traumatic brain injury (sTBI). (A) Chromosomal location on chromosome 19 of all sequenced TRPM4 polymorphisms in this study plotted on the University of California, Santa Cruz (UCSC) genome browser (assembly version GRCh38/hg38, scale bar for 20 kb, shown). The schematic of the 34-exon TRPM4 gene is on the x axis where exons (separated by introns) are marked with navy blue vertical lines/blocks. Because TRPM4 is encoded on the forward strand, exon #1 is on the left extreme and exon #25 is on the right extreme of the x axis gene schematic. The vertical axis lists all SNPs by reference-SNP cluster identification (rsid) sequenced in this study. Locations of the two significant SNPs rs8104571 (intron 20) and rs150391806 (exon 24) are indicated by the red arrow. (B) Chromosomal location for all proxy SNPs in linkage-disequilibrium (LD) with either rs8104571 or rs150391806 shown on the UCSC genome browser as mentioned. The vertical axis lists all these proxy SNPs by rsid. Their corresponding locations are indicated by the red box. (C) Chromosomal location for the three ABCC8 SNPs (rs11024286, rs2237982, and rs2283261) located in ABCC8's intron 10 (on chromosome 11) that interact with rs8104571 (TRPM4) are plotted on the UCSC genome browser. The vertical axis labels rsids for all eight significant ABCC8 SNPs previously identified as important predictors of cerebral edema ± outcome after TBI. Their corresponding locations on a zoomed in version of the ABCC8 chromosome are demarcated by vertical black lines (scale bar 10 kb). The red box encircles all three significant ABCC8 SNPs that interact with rs8104571 in terms of their effects on ICP, and the red arrows label each of these SNPs individually. Chr, chromosome; hg, human genome; kb, kilo-basepair.
Table 2.
Polymorphism Distribution and Associated Characteristics in the Severe TBI Cohort
| SNP | Variable measured | Homozygous wild-type | Heterozygous | p-value (ANOVA/Fisher) |
|---|---|---|---|---|
| rs8104571 (Intron 20) wild-type C | SNP Frequency in sTBI population (%) | 377/385 (97.9%) | 8/385 (2.1%) | |
| Average ICP, mean (SE) | 10.1 ± 0.31 mmHg | 20.4 ± 6.5 mmHg | 0.0000* | |
| Peak ICP, mean (SE) | 25.4 ± 0.90 mmHg | 45.0 ± 10.7 mmHg | 0.0007* | |
| Average proportion ICP >25 mmHg (SE) | 0.055 (0.0077) | 0.213 (0.131) | 0.004* | |
| % Craniectomy Y | 32.9% | 50% | 0.45 | |
| % Favorable 6 month GOS ≥ 4, frequency (%) | 32.8% | 62.5% | 0.087 | |
| % Favorable 6 month GOS ≥4 in survivors, frequency(%) | 45.0% | 83.3% | 0.073 | |
| rs150391806 (Exon 23) Wild-type C | SNP Frequency in sTBI population (%) | 382/385 (99.2%) | 3/385 (0.8%) | |
| Average ICP, mean (SE) | 10.3 ± 0.36 mmHg | 17.5 ± 3.5 mmHg | 0.041* | |
| Peak ICP, mean (SE) | 25.6 ± 0.91 mmHg | 54.5 ± 17.5 mmHg | 0.0015* | |
| Average proportion ICP >25 mmHg (SE) | 0.058 (0.008) | 0.129 (0.103) | 0.49 | |
| % Craniectomy Y | 33.2% | 33.3% | 1.0 | |
| % Favorable 6 month GOS ≥ 4, frequency (%) | 33.5% | 33.3% | 1.0 | |
| % Favorable 6 month GOS ≥4 in survivors, frequency(%) | 45.9% | 50% | 1.0 |
Summarizes the relative frequencies of the SNP genotypes in the severe TBI population, as well as for measures of cerebral edema (average ICP, peak ICP, proportion of ICP spikes >25 mm Hg), need for craniectomy, and outcome.
Boldface, italics, and asterisks indicate significant p-value after B-Y correction for multiple comparisons.
TBI, traumatic brain injury; SNP, single-nucleotide polymorphism; ANOVA, analysis of variance; ICP, intracranial pressure; SE, standard error of the mean; GOS, Glasgow Outcome Scale.
rs8104571 and rs150391806 are associated with ICP
Average ICP was increased by >100% in rs8104571 heterozygotes (20.4 ± 6.5 mm Hg) versus homozygous wild-type (10.1 ± 0.3 mm Hg, p = 0.0000, Table 2). Concordantly, heterozygotes had significantly higher peak ICP (45.0 ± 10.7 mm Hg vs. 25.4 ± 0.90 mm Hg, p = 0.0007) and proportion of ICP spikes >25 mm Hg (21.3 ± 13.1% vs. 5.5 ± 0.77%, p = 0.004). Univariable and multivariable regression analysis demonstrated that heterozygous rs8104571 was an independent predictor of all three measures including higher average ICP (univariable, β = 10.3 mm Hg, multivariable, β = 9.11 mm Hg [basic model], 9.55 mm Hg [expanded model 1]), peak ICP (univariable, β = 19.6 mm Hg, multivariable, β = 16.8 mm Hg [basic model], 17.6 mm Hg [expanded model 1]), and proportion of ICP spikes >25 mm Hg (univariable, β = 0.16, multivariable, β = 0.15 [basic model], 0.13 [expanded-model 1]) surviving the B–Y correction for multiple comparisons (Table 3). Effects were large. Inclusion of rs8104571 genotype in the basic multivariable regression model significantly increased the model's R2 from 0.1204 to 0.205, and further to 0.2483 in the expanded multivariable regression model that also included pattern of primary injury and acute CT findings with edema.
Table 3.
TRPM4 SNP Prediction of Intracranial Pressure and Outcome in Severe TBI
| Average ICP | Peak ICP | Proportion of ICP >25 mmHg | Favorable 6 month GOS | ||
|---|---|---|---|---|---|
| β-coefficient (95% CI, p) | β-coefficient (95% CI, p) | β-coefficient (95% CI, p) | Odds ratio (95% CI, p) | ||
| Univariable regression analyses | |||||
| rs8104571 (TC) | 10.3 mmHg (6.1–14.5, 0.0000029*) | 19.6 mmHg (8.4–30.9, 0.00071*) | 0.16 (0.05–0.26, 0.0042*) | 3.4 (0.8–14.5, 0.097) | |
| rs150391806 (TC) | 7.19 mmHg (0.28–14.1, 0.042) | 28.9 mmHg (11.2–46.6, 0.0015*) | 0.07 (−0.13–0.27, 0.5) | 0.99 (0.09–11.0, 0.99) | |
| Multivariable regression analyses | |||||
| rs8104571 (TC) | Basic model | 9.11 mmHg (5.0–13.2, 0.000021*) | 16.8 mmHg (5.6–28.1, 0.0035*) | 0.15 (0.04–0.25, 0.0095*) | 4.0 (0.74–21.6, 0.1) |
| Expanded model 1 | 9.55 mmHg (5.22–13.89, 0.000025*) | 17.6 mmHg (6.25–28.99,0.0026*) | 0.13 (0.021–0.25, 0.021) | 16.7 (2.23–165.21, 0.007951*) | |
| Expanded model 2 | 9.92 mmHg (5.52–14.31, 0.000016*) | 18.5 mmHg (6.94–30.08, 0.001912*) | 0.14 (0.022–0.26, 0.0197) | 10.97 (1.32–91.23, 0.027) | |
| Survivors only | NA | NA | NA | 20.67 (2.20–480.5, 0.016819) | |
| rs150391806 (TC) | Basic model | 5.91 mmHg (−0.77–12.6, 0.082) | 25.7 mmHg (8.2–43.3, 0.004*) | 0.05 (−0.15–0.26, 0.6) | 0.79 (0.06–10.6, 0.86) |
| Expanded model 1 | 4.34 mmHg (−2.6–11.3, 0.21) | 22.6 mmHg (5.26–39.9, 0.011*) | 0.027 (−0.18–0.24, 0.79) | 1.6e-6 (NA-1.01e61, 0.99) | |
| Expanded model 2 | 5.74 mmHg (−1.2–12.7, 0.10) | 26.6 mmHg (9.2–44.1, 0.003*) | 0.06 (−0.15–0.27, 0.59) | 1.01e-6 (NA-9.98e58, 0.99) | |
| Survivors only | NA | NA | NA | 2.9e-6 (NA-2.98e72, 0.99) | |
Provides the regression analysis results of each significant TRPM-4 single nucleotide polymorphism (SNP) effect on the outcome measures including average ICP, peak ICP, proportion of ICP spikes >25 mmHg, and odds ratio for favorable 6 month Glasgow Outcome Scale (GOS). Univariate regression results are provided first, followed by multivariate regression analysis results. For the multivariable regression analyses for ICP outcomes, basic model = SNP, sex, initial Glasgow Coma Scale (GCS) score, and presence/absence of craniectomy; expanded model 1 = SNP, age, sex, initial GCS, craniectomy, pattern of primary injury, edema on acute CT; expanded model 2 = SNP, age, sex, initial GCS, craniectomy, pattern of primary injury, sulcal effacement, cistern effacement, ventricular effacement, herniation, midline shift. For favorable 6 month GOS, basic model = multivariable model including age, sex, initial GCS; expanded model 1 = multivariable model including age, sex, initial GCS, craniectomy, pattern of primary injury, edema on acute CT, proportion of intracranial hypertension >25 mm Hg; expanded model 2 = multivariable model including age, sex, initial GCS, craniectomy, pattern of primary injury, sulcal effacement, cistern effacement, ventricular effacement, herniation, midline shift, proportion of intracranial hypertension >25 mm Hg. Survivors only subcohort = expanded model 1 in patients who were alive at discharge.
Boldface, italics, and asterisks indicate significant p-value after B-Y correction for multiple comparisons. Boldface and italics, but no asterisks, indicate p-value <0.05 standard threshold for significance but does not withstand B-Y correction for multiple comparisons.
TBI, traumatic brain injury; ICP, intracranial pressure; TC, thymine-cytosine.
rs150391806 heterozygotes had significantly higher peak ICP (54.5 ± 17.5 mm Hg vs. 25.6 ± 0.91 mm Hg, p = 0.0015) and average ICP (17.5 ± 3.5 mm Hg vs. 10.3 ± 0.36 mm Hg, p = 0.041) versus homozygous wild-type (Table 2). In univariable and multivariable analyses, heterozygous rs150391806 remained an independent predictor of peak ICP (univariable, β = 28.9 mm Hg, multivariable, β = 25.7 mm Hg, Table 3).
All results were robust to multiple imputations for missing values (Table S1) as well as expanded multivariable models that included primary injury pattern and acute CT characteristics as covariates (Table 3) (see online supplementary material at http://www.liebertpub.com). Despite exclusion of post-craniectomy ICP values in secondary sensitivity analyses resulting in an inevitable loss of data points, rs8104571 (not rs150391806) remained an independent univariable predictor of average ICP (β = 4.74, p = 0.0342), peak ICP (β = 8.89, p = 0.0256), and proportion of ICP spikes >25 mm Hg (β = 098, p = 0.0193); however, it did not withstand the B–Y correction (Table S2)(see online supplementary material at http://www.liebertpub.com). The same trend was observed in the multivariable analyses for average and peak ICP (p = 0.0625 and p = 0.0855 respectively), as well as increased proportion of ICP spikes >25 mm Hg (β = 0.095, p = 0.02391) which also did not withstand the B–Y correction.
rs8104571 and GOS
Neither rs8104571 nor rs150391806 were associated with GOS or mortality in univariable or basic multivariable models (Tables 3 and S3)(see online supplementary material at http://www.liebertpub.com). However, as proportion of intracranial hypertension and CT appearances (compression of basal cisterns, sulcal effacement, ventricular effacement, herniation, midline shift) are known to influence clinical outcome after TBI, expanded multivariable models were created to include these covariates. These expanded models suggest that variant rs8104571 but not rs150391806 may be associated with improved 6 month GOS (Table 3). Patients with variant rs8104571 in expanded model-1 had an OR of 16.7 for favorable outcome (p = 0.007951). Variant rs8104571 in expanded model-2 had a trend for favorable GOS that did not survive the B–Y correction (OR = 10.91, p = 0.027). This association with favorable GOS persisted in a subcohort of survivors-only (p = 0.0168, Table 3) and came close to surviving the B–Y correction of 0.01656.
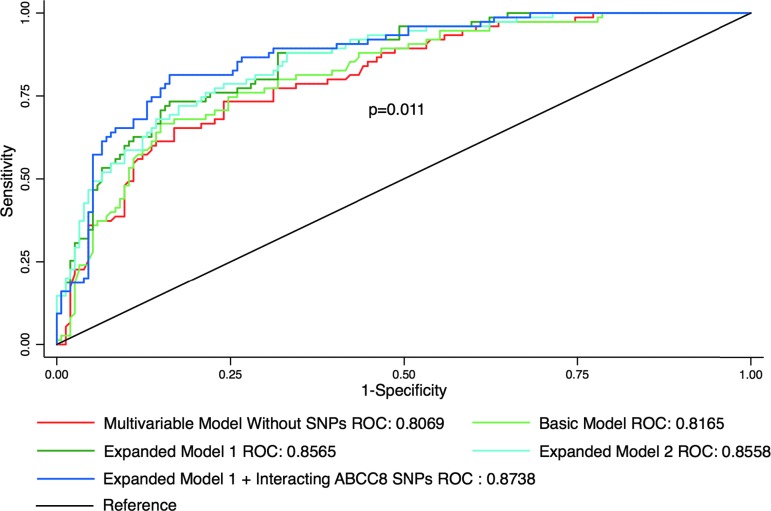
The likelihood ratio test comparing expanded model-1 versus a reduced version of this model without TRPM4 rs8104571 (but with the same covariates) was significant (p = 0.0064), suggesting that this additional predictor with genetic information added to model fit. The pseudo-R2 for the reduced model increased from 0.2946 (without TRPM4 rs8104571) to 0.3191 with inclusion of the SNP in expanded model-1 indicating that with this information, ∼31.91% of the variation in 6 month GOS could be explained by the model. Area under the receiver operating characteristic (ROC) curve (AUC) for the multivariable model without any SNP information was 0.8069, demonstrating excellent discrimination for 6 month GOS (Fig. 2). This AUC was further slightly improved by addition of rs8104571 in the basic model (0.8165), and even more so in expanded model-1 (0.8565) and expanded model-2 (0.8558). Weighted/unweighted gene risk scores in small random training and testing subsets did not significantly improve model prediction (Fig. S1) (see online supplementary material at http://www.liebertpub.com); however, the analysis was limited by the small number of subjects. Finally, including the three ABCC8 SNPs (rs2237982, rs2283261, rs11024286, see next subsection) that statistically significantly interacted with TRPM4 rs4108571 in expanded-model 1 further increased the AUC to 0.8738. These AUCs were significantly different (p = 0.011).
FIG. 2.
Receiver operating characteristic curves for 6 month Glasgow Outcome Scale (GOS). Graph of receiver operating characteristic (ROC) curves for different multivariable models used to predict 6 month GOS score. The multivariable model without any single nucleotide polymorphism (SNP) information (red) provides excellent discrimination with an area under the curve (AUC) of 0.8069. The basic multivariable model (light green, containing TRPM4 rs8104571, age, sex, and initial Glasgow Coma Scale [GCS] score) had an AUC of 0.8165. Expanded model-1 (dark green, containing transient-receptor-potential cation channel subfamily-M [TRPM4] rs8104571, age, sex, initial GCS, pattern of primary injury, computed tomographic [CT] characteristics of edema, and proportion of intracranial hypertension >25 mm Hg) had an AUC of 0.8565. Expanded model-2 (turquoise, containing TRPM4 rs8104571, age, sex, initial GCS, pattern of primary injury, distinct CT characteristics of edema [separated into individual components of sulcal effacement, cistern effacement, ventricular compression, midline shift, and herniation], and proportion of intracranial hypertension >25 mm Hg) had an AUC of 0.8558. Addition of ABCC8 SNPs (rs2237982, rs2283261, and rs11024286 that statistically significantly interacted with TRPM4 rs8104571 in this cohort) to expanded model-1 (blue) further improved the AUC to 0.8738. These AUCs were significantly different from one another (p = 0.011).
Interaction between rs8104571 and a regional cluster of ABCC8 SNPs reported to predict ICP
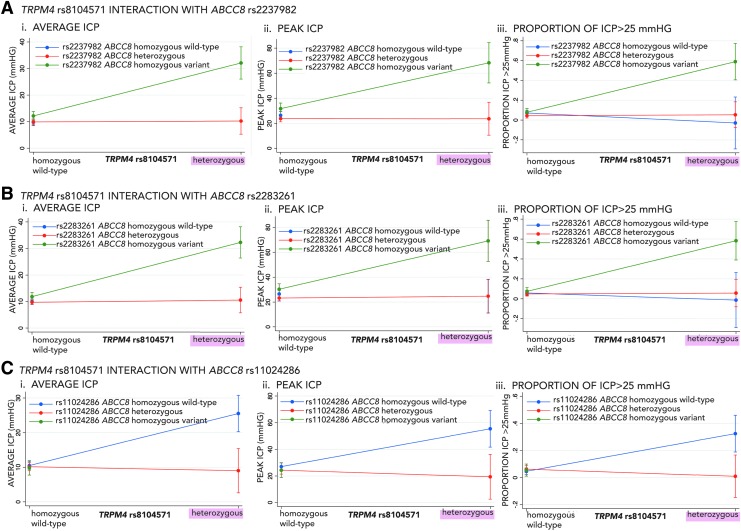
There was a significant interaction between rs8104571 genotypes and a regional cluster of ABCC8 SNPs located in intron-10 previously reported to predict ICP and/or TBI outcome (rs2237982, rs2283261, rs11024286, Fig. 1C).4,5 Effects of rs2237982, rs2283261, and rs11024286 ABCC8 genotypes on all measures of ICP (average ICP, peak ICP, and proportion of ICP spikes >25 mm Hg) were significantly different based on the TRPM4 rs8104571 genotype and vice versa (Table 4). The magnitude and direction of interactions between rs8104571 and each of the three ABCC8 SNPs on ICP is illustrated in Figure 3A–C. Patients heterozygous for rs8104571 had markedly increased average ICP, peak ICP, and proportion of ICP >25 mm Hg if they were also homozygous-variant for ABCC8 SNPs rs2237982 or rs228326, potentially indicating dose-dependent interactions (Fig. 3A and B). This maintains directional consistency with previous reports in which homozygous-variant rs2237982 and rs2283261 both independently predicted increased ICP.4,5 rs8104571 heterozygotes had markedly increased average ICP, peak ICP, and proportion of ICP >25 mm Hg if they were also homozygous wild-type for ABCC8 SNP rs1102428 (Fig. 3C), congruent with prior reports that the variant rs1102428 allele is protective.
Table 4.
TRPM4 and ABCC8 Polymorphism Interactions in Multivariable Regression Model
| ABCC8 SNP | TRM4 SNP interactions | |||||
|---|---|---|---|---|---|---|
| TRPM4 rs8104571 | TRPM4 rs150391806 | |||||
| Average ICP | Peak ICP | Proportion ICP >25 mmHg | Average ICP | Peak ICP | Proportion ICP >25 mmHg | |
| Significant interactions | Interaction p value | Non-significant interactions | ||||
| rs2237982 | 0.0000034* | 0.00079* | 0.00023* | rs2237982 | ||
| rs2283261 | 0.0000016* | 0.0004* | 0.0008* | rs2283261 | ||
| rs11024286 | 0.000021* | 0.00032* | 0.0003* | rs11024286 | ||
| Non- significant interactions | rs7105832 rs3819521 rs2283258 |
|||||
| rs7105832 | rs1799857 | |||||
| rs3819521 rs2283258 |
rs4148622 | |||||
| rs1799857 | ||||||
| rs4148622 | ||||||
Demonstrates the interactions between the two significant TRPM4 SNPs (rs8104571and rs150391806) found to predict measures of intracranial pressure (ICP) in this study, and previously reported significant ABCC8 SNPs (rs2237982, rs2283261, rs11024286, rs7105832, rs3819521, rs2283258, rs1799857, and rs4148622). There were significant interactions between rs8104571 and a cluster of significant ABCC8 SNPs located in intron-10 (rs2237982, rs2283261, and rs11024286). The direction and magnitude of these interactions on outcome measures (average, peak, and proportion of ICP spikes) is demonstrated in Figure 3.
Boldface, italics, and asterisks indicate significant p-value after B-Y correction for multiple comparisons.
FIG. 3.
Interaction effects of transient-receptor-potential cation channel subfamily-M (TRPM4) rs8104571 with previously reported significant ABCC8 single nucleotide polymorphisms (SNPs) on intracranial pressure (ICP). Graphs demonstrating the direction and magnitude of the interaction effect on ICP between TRPM4 rs8104571and each of the three ABCC8 SNPs previously implicated as significant in traumatic brain injury (TBI) organized into the following panels: (A) ABCC8- rs2237982, (B) ABCC8-rs2283261, and (C) ABCC8-rs11024286. Panel subgraphs show the interaction effects between these SNPs on various measures of ICP including (i) average-ICP, (ii) peak = ICP, and (iii) proportion of ICP spikes >25 mm Hg. The x axes for all graphs are TRPM4 rs8104571 genotypes: homozygous wild-type and heterozygous. The y axes for subgraphs A-i, B-I, and C-i are average ICP (mm Hg). The y axes for subgraphs A-ii, B-ii, and C-ii are peak ICP (mm Hg). The y axes for subgraphs A-iii, B-iii, and C-iii are proportion of ICP >25 mm Hg. Each individual graph shows the interaction between TRPM4 rs8104571 genotypes and the respective ABCC8 SNP genotypes where homozygous wild-type ABCC8 genotypes are in blue, heterozygous ABCC8 genotypes are in red, and homozygous-variant ABCC8 genotypes are in green. Error bars are 95% confidence intervals. For example, in Panel A-subgraph i, the average ICP in patients who are homozygous-wild type for TRPM4 rs8104571 is ∼10 mm Hg regardless of the ABCC8 rs2237982 genotype. However, in patients heterozygous for TRPM4 rs8104571, the average ICP is significantly higher (i.e., ∼30 mm Hg) if the patients are also homozygous-variant for ABCC8 rs2237982. In panels A-i, A-ii, B-i, and B-ii (depicting interaction effects on average and peak ICP), there are no observations for homozygous wild-type ABCC8 SNPs (blue) in any of these panels, because the interaction coefficient was omitted by the regression model as a result of collinearity between heterozygous TRPM4 rs8104571 and homozygous wild type ABCC8 rs2237982 (A-i, A-ii) as well as between TRPM4 rs8104571 and homozygous wild-type ABCC8 rs2283261 (B-i, B-ii). For Panel C (i, ii, iii) there were no patients who were both heterozygous for TRPM4 rs8104571 and homozygous variant for ABCC8 rs11024286.
There were no patients both heterozygous for TRPM4 rs8104571 and homozygous variant for ABCC8 rs11024286. For average and peak ICP, there was collinearity between heterozygous TRPM4 rs8104571 and homozygous wild-type ABCC8 rs2237982, as well as between heterozygous TRPM4 rs8104571 and homozygous wild-type ABCC8 rs2283261. There were no statistically significant interactions between rs8104571 or rs150391806 and any of the previously reported significant ABCC8 SNPs in terms of effect on GOS or mortality (Table S4)(see online supplementary material at http://www.liebertpub.com).
Predicted functional implications of significant TRPM4 and proxy-SNPs
rs8104571 and rs150391806 were not in LD with each other (r2 ≤ 0.8) thus precluding haplotype generation. Thirty-four additional TRPM4-SNPs were in LD (r2 ≥ 0.8) with either rs8104571 or rs150391806 (Fig. 1B). This region of LD extended across intron 20-exon 24 (Fig. S2). Table S5 and Figure S3 summarize association strengths, LD, and predicted functional implications of these SNPs including a missense mutation, regulatory region variants, upstream gene variants, non-coding transcript variants, and nonsense-mediated decay (see online supplementary material at http://www.liebertpub.com).
Gene-protein spatial model of significant TRPM4 SNPs
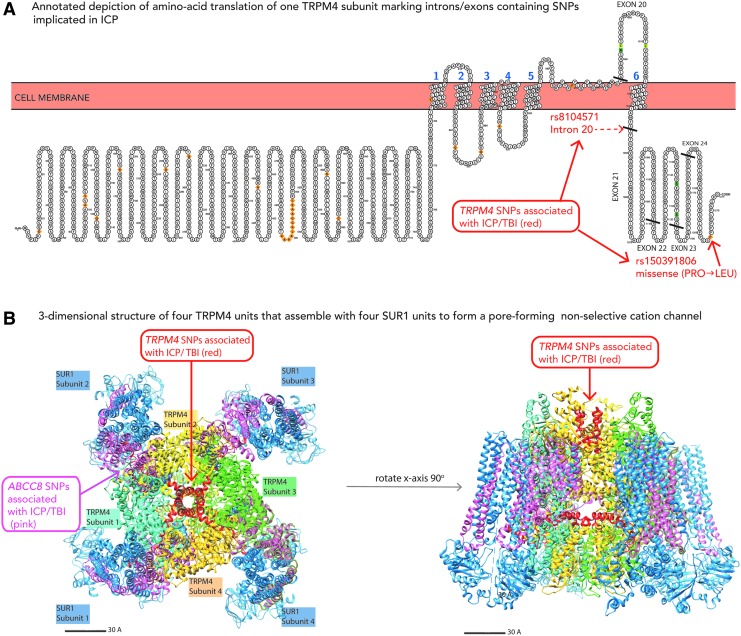
TRPM4 has 25 exons. rs8104571, rs150391806, and all 34 proxy-SNPs in this region of LD were downstream (intron 20-exon 24; Figs. 1B and S2) (see online supplementary material at http://www.liebertpub.com). Combining human TRPM4 putative and canonical amino-acid topology with TRPM4 exons, we identified protein regions encoded by exon 21 (closely flanking rs8104571 in intron 20 and other proxy SNPs in LD), and exons 23–24 (the latter containing rs150391806, Fig. 4A). A three-dimensional model demonstrates that these sequences spatially contribute to the channel-pore and a SUR1-TRPM4 binding interface (Fig. 4B). The involved binding site region juxtaposes a part of SUR1(the associated regulatory protein) encoded by a section of ABCC8 containing previously identified SNPs associated with cerebral edema and ICP after TBI.4 This may conceptually support the earlier result of interactions between genetic variations in ABCC8 and TRPM4 influencing cerebral edema and ICP.
FIG. 4.
Spatial relationships between transient-receptor-potential cation channel subfamily-M (TRPM4) and ABCC8 polymorphism regions and sulfonylurea-receptor-1 (SUR1)-TRPM4 protein structure. (A) This is an annotated depiction of amino acid translation of one TRPM4 subunit marking locations/amino acid sequences encoded by corresponding DNA regions of linkage-disequilibrium (LD) containing TRPM4 single nucleotide polymorphisms (SNPs) implicated in intracranial pressure (ICP) after traumatic brain injury (TBI). The diagrammatic representation of TRPM4 was generated using PROTTER software (UniProt ID Q8TD43). Exons 20–24 are numbered and separated by solid black lines. The specific amino acid location of the missense mutation caused by rs150391806 is indicated by a red arrow and results in substitution of leucine for proline. The dashed red arrow marks the location where intron 20 (containing rs8104571) separates exons 20 and 21. (B) The three-dimensional structure of four TRPM4 subunits (inner channel) that assemble with four regulatory SUR1 subunits to form an octameric non-selective cation channel. The TRPM4 three-dimensional structure was obtained from Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB-PDB) based on work by Autzen and coworkers23 using electron microscopy to elucidate the structure of the human TRPM4 channel in a calcium-bound state. The SUR1 three-dimensional structure was also obtained from RCSB-PDB based on work by Li and coworkers26 using cryoelectron microscopy to elucidate the structure of the pancreatic Kir6.2-SUR1 complex. University of California, San Francisco Chimera automated software was used to combine these three-dimensional structures of SUR1 and TRPM4 into the predicted octameric channel. The panel on the left is an aerial view illustrating four SUR1 subunits binding with four inner TRPM4 subunits. The SUR1 subunits are blue and pink: the pink reflecting amino acid sequences encoded by regions of DNA containing/in LD with previously reported significant ABCC8 SNPs in TBI. Each TRPM4 subunit is a different color: subunit 1 is turquoise, subunit 2 is yellow, subunit 3 is green, and subunit 4 is peach. The TRPM4 amino acid sequences in red are those encoded by regions of TRPM4 DNA in spatial proximity to rs8104571 and rs150391806. The aerial view of this octameric protein is rotated 90 degrees around the x axis to provide a coronal view. As evident from the illustration, the two protein regions (red) captured by the implicated region of TRPM4 DNA involve the channel pore, as well as interface with key regions of SUR1 (pink) associated with significant ABCC8 polymorphisms.
Discussion
In a cohort of consecutive sTBI patients, two TRPM4 SNPs spanning a region encoding the channel pore and believed to interact with SUR1, predicted increased ICP, thus adding to a growing body of research supporting the potential importance of genetic variation in this pathway and its phenotypic influence after brain injury.4,5 SUR1-TRPM4 plays a key role in cerebral edema development and intracranial hypertension in a spectrum of acute neurological disorders, the consequences of which are often life threatening.5,8,12,13,18,37 To date, no targeted preventive therapies are available. Glibenclamide inhibition of SUR1-TRPM4 in human and pre-clinical TBI has been independently linked with various favorable outcomes including reduced cerebral edema, hemorrhage progression, tissue loss, preserved white matter, and improved cognition.8,10,11,15–17,38 Results from phase I and II stroke trials of glibenclamide are encouraging, and a phase III trial is enrolling participants.12,13 Given the promising nature of this target for clinical inhibition in the context of many failed TBI trials attributed to disease heterogeneity and scarcity of biomarkers, granular phenotyping (including genetic information) could be valuable for enriching patient selection.14,39 Genetic variation may influence the cellular location, timing, and degree to which SUR1-TRPM4 contributes to edema in individual patients, and whether (or how) these patients differentially respond to targeted inhibition.
This study provides a foundation to further develop the predictive and prognostic enrichment utility of ABCC8-TRPM4 genetic variability. We report three central findings. (1) Two TRPM4 SNPs (rs8104571 and rs150391806) were independent predictors of ICP and in LD with 34 additional proxy-SNPs located between intron 20 and exon 24. (2) There was a significant interaction effect on ICP between rs8104571 and three regionally clustered ABCC8 polymorphisms (rs2237982, rs2283261, and rs11024286) previously reported as predictors of cerebral edema and ICP in TBI. (3) The implicated genetic region of TRPM4 encodes sequences contributing to the channel pore and a TRPM4-SUR1 binding interface.
TRPM4 polymorphisms and ICP
Both rs8104571 and rs150391806 were independent predictors of ICP measures in univariable and multivariable models. The predictive effect of rs8104571 was robust to multiple measures including average ICP, peak ICP, and proportion of ICP measurements >25 mm Hg. rs8105471 heterozygotes had average ICPs in the concerning range of ∼20 mm Hg (intracranial hypertension), relative to ∼10 mm Hg in homozygous wild-types. Degree of intracranial hypertension reflected by peak ICP was also significantly greater in these heterozygotes (∼ 45 mm Hg vs. 25 mm Hg). Proportion of intracranial hypertension episodes (i.e., dose) was four times greater in rs8105471 heterozygotes (21.3% vs. 5.5%). These large effects are not surprising given the low allele frequency of rs8104571 and rs150391806 variants. An inverse relationship between a variant's effect size and frequency in the population is expected/attributed to differential selection/evolutionary pressures.40 Although common variants are more frequently identified in genome-wide association studies, rare variants have a higher likelihood of informing genetic underpinnings of complex diseases, providing insight into functional consequences of the variation, and identifying causal mechanisms; all of which may eventually improve prioritization of biological therapeutic targets.40 Therefore, further study exploring the effects of rs8104571 and rs150391806 variants in biological models in vitro and in vivo may be informative.
Of the 36 genotyped and proxy SNPs, only rs8104571 and rs150391806 are reported.9,41,42 In principal-component analysis of cofactor transporters that potentially influence micronutrient requirements and individual variability in response to nutritional intervention, rs8104571 showed evidence of positive selection in an African hunter-gatherer population.42 rs150391806 causes a missense mutation (CCG→CTG) resulting in the substitution of leucine for proline. This has been associated with cardiac conduction disorders including progressive familial heart block-type-1B (ClinVar 381692) and Brugada syndrome.41 It is predicted to be “likely benign”; however, the true functional consequence is unknown.
TRPM4 polymorphisms and outcome
Consistent with the literature, in our cohort, ICP measures including average, peak, and proportion of ICP spikes >25 mm Hg predicted outcome and mortality after TBI (Table S6). rs150391806 did not predict clinical outcome. Although not statistically significant, in univariable (p = 0.097) and most multivariate models (basic model p = 0.10, expanded-model 2 p = 0.027), heterozygote rs8104571 had a trend toward favorable 6 month outcome versus homozygous wild-type. This finding was significant in expanded-model 1 (p = 0.007951) controlling for proportion of ICP >25 mm Hg and acute-CT findings in addition to the other covariates. These results are in the opposite anticipated direction of the significant and large effect on ICP in which heterozygotes had increased odds of intracranial hypertension measures. Given the small sample size and lack of consistent statistical significance surviving corrections for multiple comparisons, it is difficult to interpret the implications of this finding, which may be the result of a biological effect, consequence of sample size, or patient heterogeneity. Nonetheless, it is interesting that prior studies of ABCC8 also report different polymorphisms associated with measures of cerebral edema/ICP versus those that predict outcome. In these studies, variant alleles were also typically associated with favorable outcome; however, wild-type polymorphisms were associated with improved/reduced odds of cerebral edema measures.4,5
There are multiple speculative explanations for these findings. It is possible that to an extent, cerebral edema and intracranial hypertension may have an evolutionary function and, in patients for whom it is not fatal, it may play a role in longer-term neuroprotection. For example, astrocytic swelling may protect neurons; endothelial cell leakage may allow recruitment of important cytokines and cells involved in repair, regeneration, and neuroplasticity (akin to the dual edged sword of neuroinflammation). Intracranial hypertension may also, in part, reflect increased cerebral blood flow in attempts to improve perfusion to ischemic penumbra of the injured brain. Indeed, TRPM4 channels are thought to be important in cerebral artery myocyte depolarization, vasoconstriction, and cerebral blood flow autoregulation.43 Many prior studies have established that although intracranial hypertension is strongly associated with morbidity and mortality after severe TBI, it is not independently predictive of functional outcome in survivor-only cohorts.44–51 A recent study evaluating longitudinal ICP trajectories in severe TBI showed that groups of patients with longitudinal ICP trajectories containing multiple episodes of ICP >20 or 25 mm Hg had increased odds of favorable outcome versus other groups with either persistently low ICP trajectories or recalcitrant/severe intracranial hypertension, a finding that persisted in a survivors-only subcohort.51 The strong trend of heterozygote TRPM4 rs8104571 increasing odds of favorable GOS (OR = 20.67, p = 0.0168) in our subcohort of survivors only, although inconclusive, is therefore intriguing and potentially consistent with the hypothesis that polymorphisms increasing the risk of intracranial hypertension may also be protective in terms of clinical outcome if the patient survives.
Although the SUR1-TRPM4 channel is uniquely upregulated in CNS injury, ABCC8 also influences other targets in the CNS that may contribute to outcome independent of its association with TRPM4.52 TRPM4 in vivo has been associated with astrocytic swelling but not death.53 TRPM4 and ABCC8 polymorphisms may also have different local effects based on cell type, timing, and spatial distribution. It is worth emphasizing that the association between these genes and ICP is extremely complex, as is the relationship between ICP and outcome. TRPM4 genetic variation is one piece of a complex phenotype. Although inclusion of this single predictor (TRPM4 genetic variation) in a multivariable regression model of intracranial hypertension significantly increased the model's R2 by 8% (e.g., from 0.1204 to 0.205 [basic] or 0.2483 [expanded] for rs8104571) with concordantly large effect sizes, overall only 20.5–24.83% of the variation in ICP was explained by the model. Similarly although inclusion of rs8104571 in the multivariable logistic regression model for 6 month GOS resulted in an increase in pseudo R2 from 0.2946 to 0.3191, only ∼31% of the variation in GOS is explained by the model.
It is intuitive that ICP and the impact on outcome cannot solely be explained by TRPM4 genetic variation and our limited multivariable models given the underlying complexity of this process and its effect on outcome. ICP is impacted by innumerable other factors including intracranial compliance, autoregulation, and presence and degree of secondary insults such as hypoxia, as well as unmeasured genetic influences of other pathways (e.g., aquaporin-4 [AQP-4], toll-like-receptor-4 [TLR4], and matrix-metalloproteinase-9 [MMP9]).54–57 There are also potentially alternate mechanisms for neuroprotection/outcome versus edema via SUR1-TRPM4 that have been posited.4,5,8,11,58 This may relate to temporal and spatial cell-type distributions in which this pathway is upregulated after TBI (e.g., neurons vs. astrocytes vs. endothelial cells).
TRPM4–ABCC8 Interactions
Identification of ICP-predictive ABCC8 SNPs after TBI, combined with the recognized role of SUR1-TRPM4 in cerebral edema,4,5 led to the hypothesis that TRPM4 may also harbor genetic variants associated with ICP, and that these may interact with ABCC8. Analogous to ABCC8, the two significant genotyped TRPM4 SNPs predictive of ICP and their associated proxy SNPs regionally clustered in a downstream region of the gene that encoded sequences contained within the TRPM4-pore and the putative SUR1-TRPM4 binding interface. This spatial interaction supports the finding that the effect of rs8104571 on ICP was moderated by an interaction with three clustered ABCC8 SNPs: rs2237982, rs2283261, and rs11024286. The directional effect of these interactions on ICP reinforces the robustness of this result: variant rs11024286 has been reported as neuroprotective; however, variants rs2237982 and rs2283261 are associated with increased ICP.4,5 Concordantly, rs8104571 interactions with variant rs2237982 and rs2283261 but wild-type rs11024286 had dose-dependent increases in ICP measures.
If validated in larger studies, these SNPs may provide prognostic enrichment as predictors of intracranial hypertension as well as predictive enrichment for patients likely to need/benefit from targeted therapy. However, the functional consequences of these variations and the phenotypic effects of their interactions are currently unknown. Biologic causality between ABCC8 and TRPM4 genetic variation and intracranial hypertension in sTBI remains to be elucidated. Although beyond the scope of this study, this is an important avenue for future research, as it may partly explain the pathophysiology underlying differential cerebral edema development among individuals and potentially identify treatment responders.
Additional limitations
This study has several limitations. ICP measurements were point values recorded at the end of each hour and, therefore, do not integrate to provide a true proportion of time spent in intracranial hypertension. The complexity and heterogeneity of all possible variables influencing ICP and outcome after TBI could not be captured by our models. Further, although the SUR1-TRPM4 pathway is a key contributor to edema and intracranial hypertension, it is one of several known (and unknown) mechanisms. Investigation of potentially relevant genes either related to this pathway (e.g., TNFA, KCJN11, MMP-9 and CASP3) or other known mechanisms of cerebral edema (e.g. AQP-4, TLR4) may be valuable.57 Dedicated tag-SNP sequencing may provide better coverage across coding and non-coding regions of the TRPM4 gene. Another approach that would capture genetic variations in established and unknown contributors to cerebral edema is a genome-wide association study. This would require an extremely large sample size and multi-center collaborations from initiatives such as Transforming Research and Clinical Knowledge in TBI (TRACK-TBI) or Collaborative European NeuroTrauma Effectiveness Research in TBI (CENTER-TBI). To place our cohort in context, after 4 years with 11 enrolling sites, recent genetic-association studies published in the TRACK-TBI cohort have included between 93 and 220 subjects, and most have had mild rather than sTBI.2,3,59,60 There are currently ∼230 sTBI patients enrolled in TRACK-TBI; although this number is expected to grow over several years, data such as ours may importantly support development and funding of these vital multi-center resources.
Our work needs to be validated with other cohorts. Although this report represents a large polymorphism study in sTBI, it is small for a genetics study.5,60–62 Nonetheless, it is a candidate-gene targeted approach, reducing sample-size requirements. Our findings had large effect sizes, retained significance in multivariable analyses after correcting for multiple comparisons, and had a strong pathophysiological basis.34
Results from our single-center cohort are prone to selection bias. Variables such as “therapeutic intensity level” or secondary insults that could influence ICP while potentially useful, were unavailable. Historically at the University of Pittsburgh Medical Center during the study time period, treatment thresholds of 20 mm Hg have been used to target therapy, with sustained values >25 mm Hg triggering a neurosurgical (re)assessment. Our ICP calculations included patients after craniectomy. Importantly, our models withstood controlling for surgical treatment (craniectomy), which has one of the most powerful therapeutic influences on ICP; however, we recognize that ICP values in this context may no longer reflect biology that can be modulated by genetic variance. Although secondary analyses censoring post-craniectomy ICP values suggested similar trends, these models did not withstand the B–Y correction, likely because of imperfect censoring and the inevitable loss of data points. Subgroup analyses by injury pattern may also have been informative; however, meaningful statistical comparisons were precluded by insufficient sample sizes within each subgroup.
Conclusions
The impact of genetic polymorphisms on guiding precision medicine has been demonstrated across various medical subspecialties, and is increasingly informing clinical practice.63–67 We report two TRPM4 polymorphisms, rs8104571 and rs150391806, associated with intracranial hypertension after TBI. These polymorphisms and associated proxy SNPs spatially clustered around critical regions of DNA encoding sequences in the channel pore and the SUR1-TRPM4 binding interface. rs8104571's effect on ICP was moderated by three previously reported ABCC8 SNPs clustered in intron-10, further adding credence to the hypothesis that there may be important interactions between the two genes (ABCC8, TRPM4) and resultant associated proteins (SUR1, TRPM4). Our findings are consistent with SUR1-TRPM4's pathophysiological basis in cerebral edema development. Although the functional consequences of these SNPs are unknown and need exploration in biological models, our results support the hypothesis that genetic variability in this pathway may play a role in phenotyping intracranial hypertension after TBI. Identifying effects of ABCC8 and TRPM4 genetic variability on intracranial hypertension is a first step toward understanding their functional significance and causal mechanisms. In addition to clinical prognostication and risk stratification, this may help characterize treatment responders and inform patient selection for precision medicine based trials. This is particularly important in the context of optimally evaluating glibenclamide, a targeted therapy inhibiting SUR1-TRPM4 that has shown promise in early clinical and pre-clinical TBI studies but is yet to have proven benefit in large randomized trials.
Supplementary Material
Acknowledgments
The authors are grateful to funding from following grants: National Institutes of Health (NIH)/National Institute of Neurological Disorders and Stroke (NINDS) K23NS101036 (R.M.J.), NIH/NINR R00 NR013176 (A.M.P.), NIH P50 NS30318 (D.O.O.), NIH/NINR T32NR007969 (T.A.K.), NIH/NINDS 1R01NS087978-01 (P.M.K.), NIH/NINR R01NR013342 (Y.P.C.), University of Pittsburgh Physicians Foundation Award (R.M.J.), and University of Pittsburgh School of Medicine Dean's Faculty Advancement Award (R.M.J.).
Author Disclosure Statement
RMJ has provided consulting services for Biogen.
References
- 1. Shah S., and Kimberly W.T. (2016). Today's approach to treating brain swelling in the neuro intensive care unit. Semin. Neurol.36, 502–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yue J.K., Winkler E.A., Rick J.W., Burke J.F., McAllister T.W., Oh S.S., Burchard E.G., Hu D., Rosand J., Temkin N.R., Korley F.K., Sorani M.D., Ferguson A.R., Lingsma H.F., Sharma S., Robinson C.K., Yuh E.L., Tarapore P.E., Wang K.K.W., Puccio A.M., Mukherjee P., Diaz-Arrastia R., Gordon W.A., Valadka A.B., Okonkwo D.O., Manley G.T., and TRACK-TBI Investigators. (2017). DRD2 C957T polymorphism is associated with improved 6-month verbal learning following traumatic brain injury. Neurogenetics 18, 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yue J.K., Pronger A.M., Ferguson A.R., Temkin N.R., Sharma S., Rosand J., Sorani M.D., McAllister T.W., Barber J., Winkler E.A., Burchard E.G., Hu D., Lingsma H.F., Cooper S.R., Puccio A.M., Okonkwo D.O., Diaz-Arrastia R., Manley G.T., COBRIT Investigators, and TRACK-TBI Investigators. (2015). Association of a common genetic variant within ANKK1 with six-month cognitive performance after traumatic brain injury. Neurogenetics 16, 169–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jha R.M., Koleck T.A., Puccio A.M., Okonkwo D.O., Park S.-Y., Zusman B.E., Clark R.S.B., Shutter L.A., Wallisch J.S., Empey P.E., Kochanek P.M., and Conley Y.P. (2018). Regionally clustered ABCC8 polymorphisms in a prospective cohort predict cerebral oedema and outcome in severe traumatic brain injury. J. Neurol. Neurosurg. Psychiatry 89, 1152–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jha R.M., Puccio A.M., Okonkwo D.O., Zusman B.E., Park S.-Y., Wallisch J., Empey P.E., Shutter L.A., Clark R.S.B., Kochanek P.M., and Conley Y.P. (2017). ABCC8 single nucleotide polymorphisms are associated with cerebral edema in severe TBI. Neurocrit. Care 26, 213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jha R.M., and Kochanek P.M. (2017). Adding insight to injury: a new era in neurotrauma. Lancet Neurol. 16, 578–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Woo S.K., Kwon M.S., Ivanov A., Gerzanich V., and Simard J.M. (2013). The sulfonylurea receptor 1 (Sur1)-transient receptor potential melastatin 4 (Trpm4) channel. J. Biol. Chem. 288, 3655–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Simard J.M., Woo S.K., Schwartzbauer G.T., and Gerzanich V. (2012). Sulfonylurea receptor 1 in central nervous system injury: a focused review. J. Cereb. Blood Flow Metab. 32, 1699–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yue Z., Xie J., Yu A.S., Stock J., Du J., and Yue L. (2015). Role of TRP channels in the cardiovascular system. Am. J. Physiol. Heart Circ. Physiol. 308, H157-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Simard J.M., Kilbourne M., Tsymbalyuk O., Tosun C., Caridi J., Ivanova S., Keledjian K., Bochicchio G., and Gerzanich V. (2009). Key role of sulfonylurea receptor 1 in progressive secondary hemorrhage after brain contusion. J. Neurotrauma 26, 2257–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Patel A.D., Gerzanich V., Geng Z., and Simard J.M. (2010). Glibenclamide reduces hippocampal injury and preserves rapid spatial learning in a model of traumatic brain injury. J. Neuropathol. Exp. Neurol. 69, 1177–1190 [DOI] [PubMed] [Google Scholar]
- 12. Kimberly W.T., Battey T.W.K., Pham L., Wu O., Yoo A.J., Furie K.L., Singhal A.B., Elm J.J., Stern B.J., and Sheth K.N. (2014). Glyburide is associated with attenuated vasogenic edema in stroke patients. Neurocrit. Care 20, 193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sheth K.N., Elm J.J., Molyneaux B.J., Hinson H., Beslow L.A., Sze G.K., Ostwaldt A.-C., Del Zoppo G.J., Simard J.M., Jacobson S., and Kimberly W.T. (2016). Safety and efficacy of intravenous glyburide on brain swelling after large hemispheric infarction (GAMES-RP): a randomised, double-blind, placebo-controlled phase 2 trial. Lancet Neurol. 15, 1160–1169 [DOI] [PubMed] [Google Scholar]
- 14. Jha R.M., Puccio A.M., Chou S.H.-Y., Chang C.-C.H., Wallisch J.S., Molyneaux B.J., Zusman B.E., Shutter L.A., Poloyac S.M., Janesko-Feldman K.L., Okonkwo D.O., and Kochanek P.M. (2017). Sulfonylurea receptor-1: a novel biomarker for cerebral edema in severe traumatic brain injury. Crit. Care Med. 45, e255–e264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jha R.M., Molyneaux B.J., Jackson T.C., Wallisch J.S., Park S.-Y., Poloyac S., Vagni V.A., Janesko-Feldman K.L., Hoshitsuki K., Minnigh M.B., and Kochanek P.M. (2018). Glibenclamide produces region-dependent effects on cerebral edema in a combined injury model of traumatic brain injury and hemorrhagic shock in mice. J. Neurotrauma 35, 2125–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zweckberger K., Hackenberg K., Jung C.S., Hertle D.N., Kiening K.L., Unterberg A.W., and Sakowitz O.W. (2014). Glibenclamide reduces secondary brain damage after experimental traumatic brain injury. Neuroscience 272, 199–206 [DOI] [PubMed] [Google Scholar]
- 17. Xu Z.-M., Yuan F., Liu Y.-L., Ding J., and Tian H.-L. (2016). Glibenclamide attenuates blood-brain barrier disruption in adult mice after traumatic brain injury. J. Neurotrauma 34, 925–933 [DOI] [PubMed] [Google Scholar]
- 18. Martínez-Valverde T., Vidal-Jorge M., Martínez-Saez E., Castro L., Arikan F., Cordero E., Rădoi A., Poca M.-A., Simard J.M., and Sahuquillo J. (2015). Sulfonylurea receptor 1 in humans with post-traumatic brain contusions. J. Neurotrauma 32, 1478–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carney N., Totten A.M., O'Reilly C., Ullman J.S., Hawryluk G.W.J., Bell M.J., Bratton S.L., Chesnut R., Harris O.A., Kissoon N., Rubiano A.M., Shutter L., Tasker R.C., Vavilala M.S., Wilberger J., Wright D.W., and Ghajar J. (2017). Guidelines for the management of severe traumatic brain injury, fourth edition. Neurosurgery 80, 6–15 [DOI] [PubMed] [Google Scholar]
- 20. Omasits U., Ahrens C.H., Müller S., and Wollscheid B. (2014). Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30, 884–886 [DOI] [PubMed] [Google Scholar]
- 21. Yates A., Akanni W., Amode M.R., Barrell D., Billis K., Carvalho-Silva D., Cummins C., Clapham P., Fitzgerald S., Gil L., Girón C.G., Gordon L., Hourlier T., Hunt S.E., Janacek S.H., Johnson N., Juettemann T., Keenan S., Lavidas I., Martin F.J., Maurel T., McLaren W., Murphy D.N., Nag R., Nuhn M., Parker A., Patricio M., Pignatelli M., Rahtz M., Riat H.S., Sheppard D., Taylor K., Thormann A., Vullo A., Wilder S.P., Zadissa A., Birney E., Harrow J., Muffato M., Perry E., Ruffier M., Spudich G., Trevanion S.J., Cunningham F., Aken B.L., Zerbino D.R., and Flicek P. (2016). Ensembl 2016. Nucleic Acids Res. 44, D710–D716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mall T., Eckstein J., Norris D., Vora H., Freese N.H., and Loraine A.E. (2016). ProtAnnot: an app for integrated genome browser to display how alternative splicing and transcription affect proteins. Bioinformatics 32, 2499–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Autzen H.E., Myasnikov A.G., Campbell M.G., Asarnow D., Julius D., and Cheng Y. (2018). Structure of the human TRPM4 ion channel in a lipid nanodisc. Science 359, 228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., and Bourne P.E. (2000). The protein data bank. Nucleic Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., and Ferrin T.E. (2004). UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 26. Li N., Wu J.-X., Ding D., Cheng J., Gao N., and Chen L. (2017). Structure of a pancreatic ATP-sensitive potassium channel. Cell 168, 101–110.e10. [DOI] [PubMed] [Google Scholar]
- 27. Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., and Sunyaev S.R. (2010). A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Choi Y., Sims G.E., Murphy S., Miller J.R., and Chan A.P. (2012). Predicting the functional effect of amino acid substitutions and indels. PLoS One 7, e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stenson P.D., Mort M., Ball E.V., Evans K., Hayden M., Heywood S., Hussain M., Phillips A.D., and Cooper D.N. (2017). The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet. 136, 665–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., Flicek P., and Cunningham F. (2016). The ensembl variant effect predictor. Genome Biol. 17, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S., Cherry J.M., and Snyder M. (2012). Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22, 1790–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Machiela M.J., and Chanock S.J. (2015). LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31, 3555–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Narum S.R. (2006). Beyond Bonferroni: less conservative analyses for conservation genetics. Conserv. Genet. 7, 783–787 [Google Scholar]
- 34. Balding D.J. (2006). A tutorial on statistical methods for population association studies. Nat. Rev. Genet. 7, 781–791 [DOI] [PubMed] [Google Scholar]
- 35. Lu Y., Feskens E.J.M., Boer J.M.A., Imholz S., Verschuren W.M.M., Wijmenga C., Vaarhorst A., Slagboom E., Müller M., and Dollé M.E.T. (2010). Exploring genetic determinants of plasma total cholesterol levels and their predictive value in a longitudinal study. Atherosclerosis 213, 200–205 [DOI] [PubMed] [Google Scholar]
- 36. Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.-C., and Müller M. (2011). pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lin Z., Huang H., Gu Y., Huang K., Hu Y., Ji Z., Wu Y., Wang S., Yang T., and Pan S. (2017). Glibenclamide ameliorates cerebral edema and improves outcomes in a rat model of status epilepticus. Neuropharmacology 121, 1–11 [DOI] [PubMed] [Google Scholar]
- 38. Khalili H., Derakhshan N., Niakan A., Ghaffarpasand F., Salehi M., Eshraghian H., Shakibafard A., and Zahabi B. (2017). Effects of oral glibenclamide on brain contusion volume and functional outcome of patients with moderate and severe traumatic brain injuries: a randomized double-blind placebo-controlled clinical trial. World Neurosurg. 101, 130–136 [DOI] [PubMed] [Google Scholar]
- 39. Schwamm L.H. (2014). Progesterone for traumatic brain injury–resisting the sirens' song. N. Engl. J. Med. 371, 2522–2523 [DOI] [PubMed] [Google Scholar]
- 40. Bomba L., Walter K., and Soranzo N. (2017). The impact of rare and low-frequency genetic variants in common disease. Genome Biol. 18, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stallmeyer B., Zumhagen S., Denjoy I., Duthoit G., Hébert J.-L., Ferrer X., Maugenre S., Schmitz W., Kirchhefer U., Schulze-Bahr E., Guicheney P., and Schulze-Bahr E. (2012). Mutational spectrum in the Ca(2+)-activated cation channel gene TRPM4 in patients with cardiac conductance disturbances. Hum. Mutat. 33, 109–117 [DOI] [PubMed] [Google Scholar]
- 42. Parolo S., Lacroix S., Kaput J., and Scott-Boyer M.-P. (2017). Ancestors' dietary patterns and environments could drive positive selection in genes involved in micronutrient metabolism-the case of cofactor transporters. Genes Nutr. 12, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Earley S. (2013). TRPM4 channels in smooth muscle function. Pflugers Arch. 465, 1223–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Badri S., Chen J., Barber J., Temkin N.R., Dikmen S.S., Chesnut R.M., Deem S., Yanez N.D., and Treggiari M.M. (2012). Mortality and long-term functional outcome associated with intracranial pressure after traumatic brain injury. Intensive Care Med. 38, 1800–1809 [DOI] [PubMed] [Google Scholar]
- 45. Balestreri M., Czosnyka M., Hutchinson P., Steiner L.A., Hiler M., Smielewski P., and Pickard J.D. (2006). Impact of intracranial pressure and cerebral perfusion pressure on severe disability and mortality after head injury. Neurocrit. Care 4, 8–13 [DOI] [PubMed] [Google Scholar]
- 46. Lannoo E., Van Rietvelde F., Colardyn F., Lemmerling M., Vandekerckhove T., Jannes C., and De Soete G. (2000). Early predictors of mortality and morbidity after severe closed head injury. J. Neurotrauma 17, 403–414 [DOI] [PubMed] [Google Scholar]
- 47. Czosnyka M., Hutchinson P.J., Balestreri M., Hiler M., Smielewski P., and Pickard J.D. (2006). Monitoring and interpretation of intracranial pressure after head injury. Acta Neurochir. Suppl. 96, 114–118 [DOI] [PubMed] [Google Scholar]
- 48. Struchen M.A., Hannay H.J., Contant C.F., and Robertson C.S. (2001). The relation between acute physiological variables and outcome on the Glasgow Outcome Scale and Disability Rating Scale following severe traumatic brain injury. J. Neurotrauma 18, 115–125 [DOI] [PubMed] [Google Scholar]
- 49. Levin H.S., Eisenberg H.M., Gary H.E., Marmarou A., Foulkes M.A., Jane J.A., Marshall L.F., and Portman S.M. (1991). Intracranial hypertension in relation to memory functioning during the first year after severe head injury. Neurosurgery 28, 196–200 [DOI] [PubMed] [Google Scholar]
- 50. Chesnut R., Videtta W., Vespa P., Le Roux P., and Participants in the International Multidisciplinary Consensus Conference on MultimodalityMonitoring. (2014). Intracranial pressure monitoring: fundamental considerations and rationale for monitoring. Neurocrit. Care 21, Suppl. 2, S64–84 [DOI] [PubMed] [Google Scholar]
- 51. Jha R.M., Elmer J., Zusman B.E., Desai S., Puccio A.M., Okonkwo D.O., Park S.Y., Shutter L.A., Wallisch J.S., Conley Y.P., and Kochanek P.M. (2018). Intracranial pressure trajectories: a novel approach to informing severe traumatic brain injury phenotypes. Crit. Care Med. 46, 1792–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Castro L., Noelia M., Vidal-Jorge M., Sanchez-Ortiz D., Gándara D., Martínez-Saez E., Cicuendez M., Poca M.A., Simard J.M., and Sahuquillo J. (2018). Kir6.2, the pore-forming subunit of ATP-sensitive K+ channels, is overexpressed in human post-traumatic brain contusions.J. Neurotrauma [Epub ahead of print; doi: 10.1089/neu.2017.5619.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gorse K.M., Lantzy M.K., Lee E.D., and Lafrenaye A.D. (2018). transient receptor potential melastatin 4 induces astrocyte swelling but not death after diffuse traumatic brain injury. J. Neurotrauma 35, 1694–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liang F., Luo C., Xu G., Su F., He X., Long S., Ren H., Liu Y., Feng Y., and Pei Z. (2015). Deletion of aquaporin-4 is neuroprotective during the acute stage of micro traumatic brain injury in mice. Neurosci. Lett. 598, 29–35 [DOI] [PubMed] [Google Scholar]
- 55. Laird M.D., Shields J.S., Sukumari-Ramesh S., Kimbler D.E., Fessler R.D., Shakir B., Youssef P., Yanasak N., Vender J.R., and Dhandapani K.M. (2014). High mobility group box protein-1 promotes cerebral edema after traumatic brain injury via activation of toll-like receptor 4. Glia 62, 26–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tisherman S.A., Schmicker R.H., Brasel K.J., Bulger E.M., Kerby J.D., Minei J.P., Powell J.L., Reiff D.A., Rizoli S.B., and Schreiber M.A. (2015). Detailed description of all deaths in both the shock and traumatic brain injury hypertonic saline trials of the Resuscitation Outcomes Consortium. Ann. Surg. 261, 586–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stokum J.A., Gerzanich V., and Simard J.M. (2016). Molecular pathophysiology of cerebral edema. J. Cereb. Blood Flow Metab. 36, 513–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jha R., Yan H., Dixon C.E., Poloyac S., Jackson T., Hoshitsuki K., Ma X., Henchir J., Janesko-Feldman K., and Kochanek P. (2015). Evaluation of glibenclamide in the Pittsburgh Controlled Cortical Impact Model of Traumatic Brain Injury: an OBTT Consortium Study. J. Neurotrauma 32, 119 [Google Scholar]
- 59. Winkler E.A., Yue J.K., Ferguson A.R., Temkin N.R., Stein M.B., Barber J., Yuh E.L., Sharma S., Satris G.G., McAllister T.W., Rosand J., Sorani M.D., Lingsma H.F., Tarapore P.E., Burchard E.G., Hu D., Eng C., Wang K.K.W., Mukherjee P., Okonkwo D.O., Diaz-Arrastia R., Manley G.T., and TRACK-TBI Investigators. (2017). COMT Val158Met polymorphism is associated with post-traumatic stress disorder and functional outcome following mild traumatic brain injury. J. Clin. Neurosci. 35, 109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Winkler E.A., Yue J.K., McAllister T.W., Temkin N.R., Oh S.S., Burchard E.G., Hu D., Ferguson A.R., Lingsma H.F., Burke J.F., Sorani M.D., Rosand J., Yuh E.L., Barber J., Tarapore P.E., Gardner R.C., Sharma S., Satris G.G., Eng C., Puccio A.M., Wang K.K.W., Mukherjee P., Valadka A.B., Okonkwo D.O., Diaz-Arrastia R., Manley G.T., and TRACK-TBI Investigators. (2016). COMT Val 158 Met polymorphism is associated with nonverbal cognition following mild traumatic brain injury. Neurogenetics 17, 31–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Osier N.D., Bales J.W., Pugh B., Shin S., Wyrobek J., Puccio A.M., Okonkwo D.O., Ren D., Alexander S., Conley Y.P., and Dixon C.E. (2017). Variation in PPP3CC genotype is associated with long-term recovery after severe brain injury. J. Neurotrauma 34, 86–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dardiotis E., Paterakis K., Tsivgoulis G., Tsintou M., Hadjigeorgiou G.F., Dardioti M., Grigoriadis S., Simeonidou C., Komnos A., Kapsalaki E., Fountas K., and Hadjigeorgiou G.M. (2014). AQP4 tag single nucleotide polymorphisms in patients with traumatic brain injury. J. Neurotrauma 31, 1920–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shuldiner A.R., O'Connell J.R., Bliden K.P., Gandhi A., Ryan K., Horenstein R.B., Damcott C.M., Pakyz R., Tantry U.S., Gibson Q., Pollin T.I., Post W., Parsa A., Mitchell B.D., Faraday N., Herzog W., and Gurbel P.A. (2009). Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA 302, 849–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. SEARCH Collaborative Group, Link E., Parish S., Armitage J., Bowman L., Heath S., Matsuda F., Gut I., Lathrop M., and Collins R. (2008). SLCO1B1 variants and statin-induced myopathy–a genomewide study. N. Engl. J. Med. 359, 789–799 [DOI] [PubMed] [Google Scholar]
- 65. Mega J.L., Close S.L., Wiviott S.D., Shen L., Hockett R.D., Brandt J.T., Walker J.R., Antman E.M., Macias W., Braunwald E., and Sabatine M.S. (2009). Cytochrome p-450 polymorphisms and response to clopidogrel. N. Engl. J. Med. 360, 354–362 [DOI] [PubMed] [Google Scholar]
- 66. Nishimura J., Yamamoto M., Hayashi S., Ohyashiki K., Ando K., Brodsky A.L., Noji H., Kitamura K., Eto T., Takahashi T., Masuko M., Matsumoto T., Wano Y., Shichishima T., Shibayama H., Hase M., Li L., Johnson K., Lazarowski A., Tamburini P., Inazawa J., Kinoshita T., and Kanakura Y. (2014). Genetic variants in C5 and poor response to eculizumab. N. Engl. J. Med. 370, 632–639 [DOI] [PubMed] [Google Scholar]
- 67. Nissen S.E., Pillai S.G., Nicholls S.J., Wolski K., Riesmeyer J.S., Weerakkody G.J., Foster W.M., McErlean E., Li L., Bhatnagar P., Ruotolo G., and Lincoff A.M. (2018). ADCY9 Genetic variants and cardiovascular outcomes with evacetrapib in patients with high-risk vascular disease: a nested case-control study. JAMA Cardiol. 3, 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.