Abstract
Many indomethacin amides and esters are cyclooxygenase-2 (COX-2)–selective inhibitors, providing a framework for the design of COX-2–targeted imaging and cancer chemotherapeutic agents. Although previous studies have suggested that the amide or ester moiety of these inhibitors binds in the lobby region, a spacious alcove within the enzyme's membrane-binding domain, structural details have been lacking. Here, we present observations on the crystal complexes of COX-2 with two indomethacin-dansyl conjugates (compounds 1 and 2) at 2.22-Å resolution. Both compounds are COX-2–selective inhibitors with IC50 values of 0.76 and 0.17 μm, respectively. Our results confirmed that the dansyl moiety is localized in and establishes hydrophobic interactions and several hydrogen bonds with the lobby of the membrane-binding domain. We noted that in both crystal structures, the linker tethering indomethacin to the dansyl moiety passes through the constriction at the mouth of the COX-2 active site, resulting in displacement and disorder of Arg-120, located at the opening to the active site. Both compounds exhibited higher inhibitory potency against a COX-2 R120A variant than against the WT enzyme. Inhibition kinetics of compound 2 were similar to those of the indomethacin parent compound against WT COX-2, and the R120A substitution reduced the time dependence of COX inhibition. These results provide a structural basis for the further design and optimization of conjugated COX reagents for imaging of malignant or inflammatory tissues containing high COX-2 levels.
Keywords: cyclooxygenase (COX), enzyme structure, protein drug interaction, X-ray crystallography, enzyme kinetics, anticancer drug, inflammation, chemotherapy, prostaglandin endoperoxide synthase
Introduction
Cyclooxygenases (COX-12 and COX-2; also known as prostaglandin-endoperoxide synthases), are the first enzymes in the arachidonic acid (AA) cascade that generates prostaglandins (PGs) and thromboxane. These lipid mediators play crucial roles in a variety of biological functions, including inflammation, hemostasis, parturition, and regulation of blood pressure (1–4). The importance of the cyclooxygenase enzymes is illustrated by the fact that they are the primary targets of the isoform-nonselective or COX-2–selective nonsteroidal anti-inflammatory drugs (NSAIDs), which are widely used to treat inflammation, pain, and fever (5–8).
COX-1 and COX-2 are bifunctional enzymes that convert AA to prostaglandin G2 (PGG2) in their cyclooxygenase active site and then reduce it to prostaglandin H2 (PGH2) in their peroxidase active site. PGH2 then serves as the substrate for the remaining enzymes in the PG biosynthetic pathway. In addition to the free fatty acid, COX-2 can also oxygenate some ester and amide derivatives of AA, including the endocannabinoids 2-arachidonoylglycerol and arachidonoylethanolamide, converting them to PGH2-glyceryl ester and PGH2-ethanolamide, respectively (9, 10). Whereas COX-1 is constitutively expressed in most tissues, expression of COX-2 is strongly induced in inflammatory and malignant sites. This property of COX-2 has led to the hypothesis that it can serve as a target for molecular imaging of cancer and/or inflammation (11). Testing this hypothesis has been facilitated by the finding that many carboxylic acid–containing, isoform-nonselective NSAIDs can be converted to COX-2–selective inhibitors by esterification or amidation of the carboxyl group. In fact, a remarkably diverse assortment of chemical entities has been conjugated to indomethacin to yield potent and selective COX-2 inhibitors (12, 13). Our laboratory has exploited this approach to synthesize the fluorocoxibs, conjugates of indomethacin and carboxy-X-rhodamine that have been successfully used for optical imaging of COX-2 in inflammation and cancer in cell culture and in vivo (14–17).
Despite the availability of many crystal structures of COX enzymes complexed with various ligands and substantial literature on the potencies and mechanism of action of indomethacin esters and amides (12, 13, 18–20), the structural basis for the interaction of this class of inhibitors with the enzyme's active site has not been fully delineated. The COX enzymes are homodimeric proteins, each monomer of which consists of an epidermal growth factor (EGF) domain, membrane-binding domain (MBD), and the larger catalytic domain (Fig. 1A). The cyclooxygenase active site of the enzyme comprises a hydrophobic channel that binds AA in an L-shaped conformation, placing the catalytic tyrosyl radical (Tyr-385) in alignment to abstract the (pro)-S hydrogen atom on AA's carbon 13, thereby initiating the first step in PG biosynthesis. The opening into the active site is demarcated by a constriction formed by Arg-120, Tyr-355, and Glu-524. Beneath the constriction is a relatively spacious alcove within the MBD that is referred to as the lobby (Fig. 1B). The crystal structure of a complex of COX-2 and indomethacin reveals the inhibitor bound in the cyclooxygenase active site above the constriction (21). As the carboxylate of indomethacin interacts with Arg-120 and Tyr-355, it has been proposed that amide or ester groups attached to this carboxyl group would project through the constriction into the lobby. The relatively large size of the lobby would explain why groups as such as carboxy-X-rhodamine can be accommodated. Thus far, only one crystal structure of COX-2 in complex with an indomethacin amide has been reported (22). In this case, indomethacin was conjugated to podophyllotoxin in an effort to generate a COX-2–targeted chemotherapeutic agent. This structure revealed indomethacin bound in a pose very similar to that of the parent compound in complex with COX-2. However, the podophyllotoxin moiety, although appearing to extend into the lobby, was not visualized in the final electron density map.
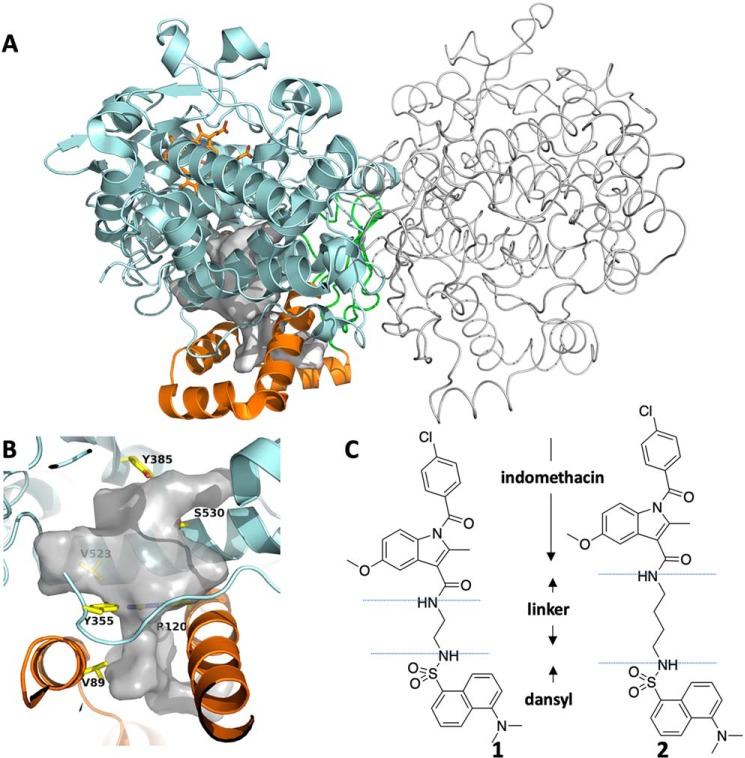
Figure 1.
The structures of cyclooxygenase-2, compound 1, and compound 2. A, the homodimer structure of COX-2 with one monomer represented in cartoon format and the other in tube format. The EGF domain is colored green, the MBD is orange, and the catalytic domain is cyan. Heme is shown in orange ball-and-stick. B, closeup of the active site of COX-2. The void space in the COX-2 active site and lobby region is colored white, and key residues are represented in yellow sticks. These include Tyr-385, site of the catalytic tyrosyl radical; Ser-530, site of inactivation by aspirin acetylation; constriction residues Tyr-355 and Arg-120; and Val-89, which plays a role in MBD structure and inhibitor binding kinetics. C, the chemical structures of compounds 1 and 2.
As part of our search to identify potential COX-2–targeted imaging agents, we synthesized compounds 1 and 2, both indomethacin-dansyl conjugates that displayed selective COX-2 inhibition (Fig. 1C). These compounds were found to be about 10-fold more potent COX-2 inhibitors (IC50 = 0.06 μm for both compounds) than the fluorocoxibs (IC50 = 0.7 μm for fluorocoxib A) in purified enzyme assays. IC50 values for COX-1 inhibition were greater than 4 μm for the dansyl derivatives and fluorocoxibs. Despite their high potency and selectivity, the spectral properties of compounds 1 and 2 (λex = 355 nm, λem = 493 nm) were suboptimal for in vivo optical imaging, limiting their potential clinical utility (14). However, due to their fairly compact amide substituents, we hypothesized that they would be suitable model compounds for elucidating the structural interactions between COX-2 and the indomethacin ester/amide class of inhibitors. Here, we report the X-ray crystallographic structures of complexes of COX-2 with compounds 1 and 2. The data confirm that the dansyl moiety occupies the lobby of the enzyme. In addition, site-directed mutagenesis studies reported here help to further illuminate structural determinants of the potency of these compounds.
Results
Characterization of compounds 1 and 2 as COX-2–selective inhibitors
We initiated our studies by reassessing the activities of compounds 1 and 2 against both WT COX isoforms using an assay in which inhibitors were preincubated with enzyme for 15 min prior to addition of AA. Consistent with the previously reported COX-2 selectivity, neither compound achieved greater than 20% inhibition of COX-1, even at concentrations of 10 μm. In contrast, compounds 1 and 2 inhibited COX-2 with IC50 values of 0.76 and 0.17 μm, respectively, values somewhat higher than those reported previously but similar to that of indomethacin (IC50 = 0.23 μm) (Table 1). Compound 1 achieved a maximum 70% inhibition of COX-2 at the concentrations tested, whereas complete inhibition was attained with compound 2. Thus, in terms of both IC50 and residual activity, compound 2 is more potent than compound 1.
Table 1.
IC50 values of compounds 1 and 2 and indomethacin (Indo) against COX-1, COX-2, and COX-2 variants
Values are mean and 95% confidence interval (in parentheses) in μm. ND, not determined.
| Inhibitor | WT COX-1 | WT COX-2 | COX-2 variants |
||||
|---|---|---|---|---|---|---|---|
| R120A | S530A | V89W | V523I | S119A | |||
| 1 | > 10 | 0.76 (0.42–1.4) | 0.28 (0.20–0.39) | 2.5 (0.9–8.6) | 1.4 (0.6–3.3) | 0.64 (0.18–2.6) | 0.34 (0.17–0.70) |
| 2 | > 10 | 0.17 (0.12–0.24) | 0.036 (0.022–0.057) | 0.21 (0.16–0.29) | 0.51 (0.18–1.4) | 0.11 (0.09–0.12) | 0.17 (0.11–0.28) |
| Indo | 0.052 (0.040–0.069) | 0.23 (0.16–0.31) | >4 | 0.22a | 0.11b | 0.45a | ND |
Having verified the selectivity of both compounds, we next hypothesized that they exhibit inhibition kinetics similar to those of their parent compound. Indomethacin is classified as a slow, tight-binding inhibitor for both COX-1 and COX-2. Its binding kinetics can be explained by a model that includes a rapid equilibrium corresponding to formation of an initial transient enzyme·inhibitor complex followed by slower formation of a more tightly bound complex (23).
| (Eq. 1) |
According to this model, incubation of enzyme with inhibitor under pseudo-first-order conditions results in an exponential loss of activity that can be measured by adding substrate at various times to the enzyme–inhibitor solution. Plotting activity versus time yields an observed first-order rate constant (kobs) for enzyme inactivation that depends on inhibitor concentration. If these constants are then plotted against the concentration of inhibitor, one obtains a hyperbola described by the following equation,
| (Eq. 2) |
where KI = k−1/k1. Graphically, the plateau of the hyperbola corresponds to k2, the y intercept to k−2, and the inhibitor concentration at which kobs reaches half of its maximal value to KI.
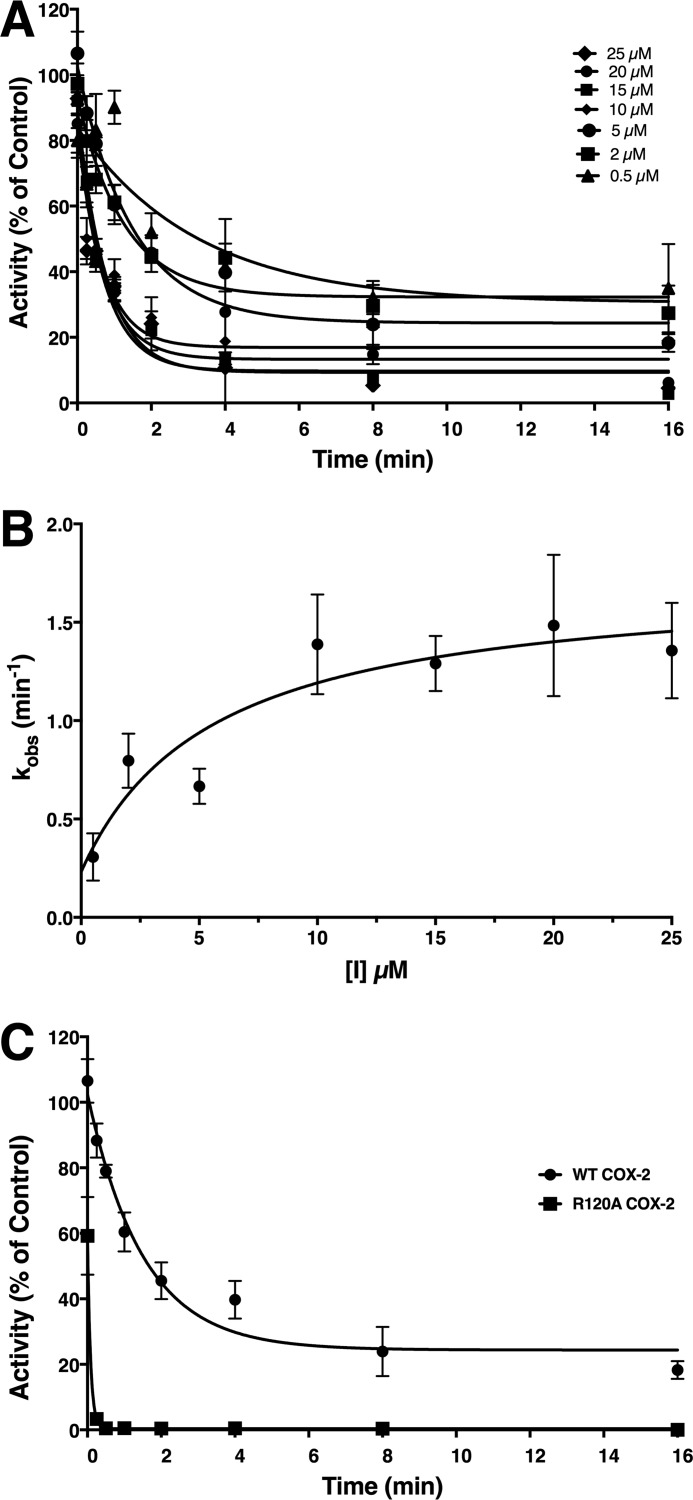
We tested our hypothesis by first confirming that both compound 1 and compound 2 exhibit time-dependent inhibition of COX-2. Then, we directly compared the COX-2 inhibitor kinetics of compound 2, the most potent of the two inhibitors, with those of indomethacin (Fig. 2). Compound 2 exhibited an affinity for initial binding (KI = 5.6 ± 4.1 μm) that was similar to that reported previously for indomethacin (KI = 7.9 ± 2.2 μm) (24). The forward rate constant for the second binding step of compound 2 to COX-2 (k2 = 0.025 ± 0.004 s−1) was lower than that of indomethacin (k2 = 0.052 ± 0.005 s−1), whereas the second dissociation constant for compound 2 (k−2 = 0.0038 s−1) was higher than that reported for indomethacin, which was too low to be determined (24). These results suggest that compound 2 interacts with the active site of COX-2 in a mode similar to that of indomethacin despite the fact that the dansyl moiety essentially eliminates COX-1 inhibitory potency.
Figure 2.
Kinetics of the time-dependent inhibition of COX-2 by compound 2. A, purified murine COX-2 was reconstituted with heme and preincubated with the designated concentrations of compound 2 at 37 °C for the indicated times prior to the addition of AA (50 μm) to assess remaining activity. Curve fitting based on first-order exponential decay yielded values for the observed rate constants (kobs) for loss of activity at each inhibitor concentration. B, a secondary plot of kobs versus compound 2 concentration was used to generate values for KI, k2, and k−2 using Equation 2. C, loss of enzymatic activity resulting from incubation of WT COX-2 or R120A COX-2 with 5 μm compound 2 for the indicated periods of time prior to addition of AA (50 μm). Data points with error bars represent the mean and standard deviation in all cases.
Crystal structure of compound 1 complexed with COX-2
The crystal structure of compound 1 complexed with murine COX-2 was obtained at 2.22-Å resolution (Protein Data Bank (PDB) code 6BL4). The data revealed an asymmetric unit of space group C2 containing four COX monomers (two homodimers) as constructed by molecular replacement and structural refinement based on the model of the structure of a COX-2·naproxen complex (PDB code 3NT1). Each of the four monomers in the completed structure comprises protein residues 33–583; the cofactor protoporphyrin IX; three posttranslational N-glycosylations at Asn-68, Asn-144, and Asn-410; and several β-octyl glucoside detergent molecules. The β-octyl glucoside molecules are located in the outer shell of the protein in the same position as reported in previously published COX-2·inhibitor crystal structures (25).
The dimers of COX-2 in the model are essentially identical to those in the complex of COX-2 and (S)-naproxen (PDB code 3NT1), with a root mean square deviation for the backbone atoms in the range of 0.11–0.25 Å, based on a comparison of all monomers from this structure with monomer A of the COX-2·naproxen complex. The following discussion of the structure is based on the model of monomer A. The residues of COX-2 are numbered based on the ovine COX-1 numbering system for ease of comparison between the two isoforms (4).
The dansyl moiety of compound 1 binds in the lobby region of the COX-2 active site
Compound 1 comprises indomethacin conjugated to a dansyl group via an ethylenediamine tether. We built the model for compound 1 in the active site and lobby region of all four monomers of COX-2 based on the well-defined electron densities that were derived from the crystallographic diffraction data (Fig. 3). The indomethacin moiety of compound 1 adopts a similar conformation to that previously reported for indomethacin bound to COX-2 (21). The chlorine atom on the phenyl ring is inserted into the hydrophobic crown made of Leu-384, Tyr-385, Trp-387, Phe-518, and Met-522, and the carbonyl oxygen between the phenyl and indole rings forms a hydrogen bond with Ser-530 at an effective distance of 3.0 Å. The methoxy group of the indomethacin moiety is directed toward the COX-2 side pocket that is exploited by the diarylheterocycle class of COX-2–selective inhibitors (3, 26, 27). The 2′-methyl group on the indole ring is adjacent to Val-523, forming hydrophobic interactions with Val-523, Val-349, and Ala-527. Insertion of this methyl group into a pocket formed by Val-349, Ala-527, Ser-530, and Leu-531 has been shown to be critical for the time dependence of COX inhibition by indomethacin (24).
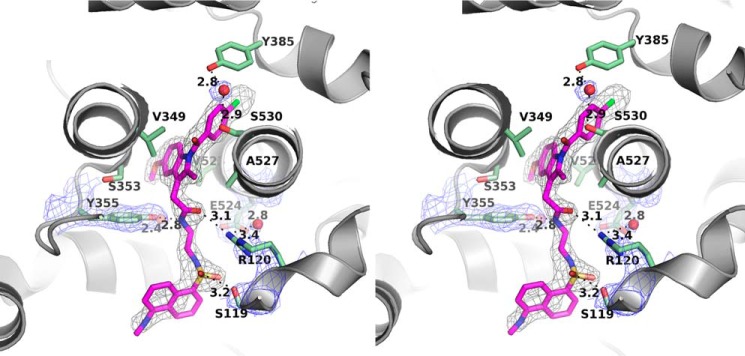
Figure 3.
Stereoview of the molecular interaction between compound 1 and COX-2. Compound 1 is colored magenta, and interacting residues of COX-2 are illustrated as green sticks. Structured water molecules are shown as red spheres. A simulated annealing composite omit map (38) around compound 1 is contoured at 3 σ in black wires. The 2Fo − Fc map around Arg-120, Glu-524, Tyr-355, and highly ordered waters is contoured at 1.5 σ in blue wires.
Consistent with the prior hypothesis, the two-carbon linker that joins the indomethacin and dansyl moieties of compound 1 passes through the constriction at the base of the active site, placing the dansyl moiety in the lobby region, surrounded by helices A–D of the MBD. To accommodate the linker, the side chain of Arg-120 is displaced from the position it occupies in the COX-2·indomethacin complex (21). Tyr-355 is hydrogen-bonded at a distance of 2.8 Å to the nitrogen atom of the amide that joins the indomethacin moiety to the linker, and the carbonyl oxygen of the amide group of compound 1 is located about 3.1 Å away from NH2 of the guanidinium group of Arg-120. Within the lobby, Val-89 and Leu-93 form hydrophobic interactions with the dansyl moiety, and the hydroxyl group of Ser-119 forms a hydrogen bond with an oxygen atom of the dansyl sulfonyl group at the effective distance of 3.2 Å (Fig. 3).
Remarkably, two highly ordered water molecules in the constriction provide additional polar binding between compound 1 and COX residues to compensate for the disorder resulting from the displacement of Arg-120. One water molecule is located immediately above the constriction, ∼2.4 Å away from the phenolic oxygen of Tyr-355. The second one is ∼3.4 Å from NH2 of the guanidinium group of Arg-120. In addition, the placement of the second water molecule in the lower part of the constriction enables it to interact with the carboxylate of Glu-524 (Fig. 3).
Crystal structure of compound 2 complexed with COX-2
We also obtained a 2.22-Å resolution crystal structure of COX-2 in complex with compound 2, a conjugate of indomethacin and a dansyl group joined by a four-carbon linker. The length of the linker, which is 1,2-diaminoethane in compound 1 and 1,4-diaminobutane in compound 2, is the only structural difference between the two inhibitors (Fig. 1C).
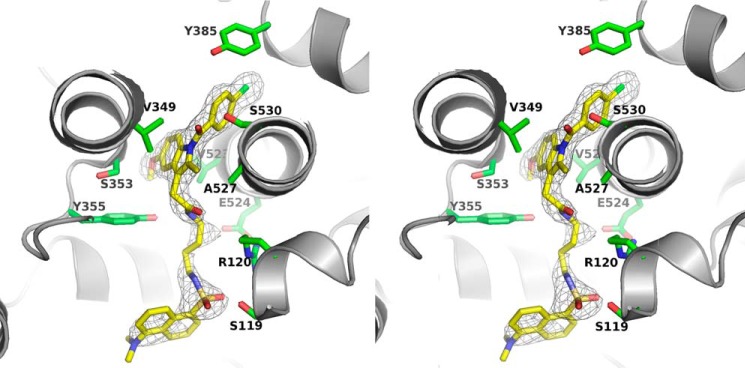
As in the case of the COX-2·compound 1 complex, the indomethacin moiety of the ligand adopts a pose very similar to that of indomethacin in complex with COX-2. Furthermore, despite the differences in the linker length between compounds 1 and 2, the dansyl moieties of the two inhibitors adopt essentially identical poses in their respective crystal structures. This suggests limited flexibility for binding of the dansyl moiety in the COX-2 lobby. To achieve correct alignment of both the indomethacin and dansyl moieties, the four-carbon linker of compound 2 must twist, thereby shortening its length to pass through the constriction, while a movement of the side chain of Ser-119 enables formation of an optimal hydrogen bond (2.7 Å) with an oxygen atom of the dansyl sulfonyl group. As in the COX-2·compound 1 complex, hydrophobic interactions between the dansyl moiety and residues Val-89 and Leu-93 are observed, and an ordered water molecule is present in the constriction where it interacts with Tyr-355 (Fig. 4).
Figure 4.
Stereoview of the molecular interaction between compound 2 and cyclooxygenase-2. A simulated annealing composite omit map (38) around compound 2 is contoured at 3 σ in black wires. Compound 2 is colored yellow, structured water molecules are shown as red spheres, and interacting residues of COX-2 are illustrated in green sticks.
Structural determinants of COX-2 inhibition of compounds 1 and 2
The crystal structure data revealed points of interaction between COX-2 and both compounds 1 and 2. To further explore the importance of these interactions, we conducted site-directed mutagenesis studies. Indomethacin forms a strong polar interaction with Arg-120 at the COX-2 constriction. In contrast, the crystal structures of compounds 1 and 2 complexed with COX-2 demonstrated no interaction between the inhibitor and Arg-120 and a displacement of Arg-120 to provide room for the linker in the constriction. Consistently, when Arg-120 was mutated to alanine, both compounds 1 and 2 displayed an increase in potency, with IC50 values of 0.28 and 0.036 μm, respectively. In contrast, indomethacin exhibited a marked loss of potency (IC50 > 4 μm) against this mutant (Table 1).
As is seen for indomethacin, the Ser-530 hydroxyl group of COX-2 forms a hydrogen bond with the carbonyl between the indole and phenyl rings of the indomethacin moiety of compounds 1 and 2 in their respective crystal complexes. Mutation of Ser-530 to alanine decreased the potency of compound 1 (IC50 = 2.5 μm) while minimally affecting the potency of compound 2 (IC50 = 0.21 μm) or indomethacin (IC50 = 0.22 μm). This result suggests that the Ser-530 hydrogen bond contributes substantially to the binding energy of compound 1 but not compound 2 or indomethacin.
Prior work had demonstrated that mutation of Val-89 to tryptophan alters the kinetics of inhibition of many weak reversible inhibitors so that they behave as time-dependent, tight-binding inhibitors (28). Crystal structure data revealed that the Trp-89 residue fills a gap between helixes B and D of the MBD, changing the opening of the lobby from a C-shaped partial ring to a fully closed donut shape. Although this mutation had minimal effect on the potency of indomethacin, we hypothesized that it might alter the potency of compounds 1 and/or 2 due to the positioning of the dansyl moiety within the lobby and formation of contacts with this residue. In support of this hypothesis, the potencies of both compounds were reduced as a result of the V89W mutation, with greater effects on compound 1 than compound 2 (IC50 values of 1.4 and 0.51 μm, respectively).
The side pocket region of COX-2 is a binding pocket that is exploited by the diarylheterocycle class of COX-2–selective inhibitors (3, 26, 27). Inhibitors access this pocket in COX-2 more easily than in COX-1 because the valine residue occupying position 523 in COX-2 is smaller than the isoleucine residue at this position in COX-1. Mutation of Val-523 in COX-2 to isoleucine leads to a loss of potency of the diarylheterocycle inhibitors. This mutation had minimal effect on the potency of indomethacin, compound 1, or compound 2, however, consistent with the structural data indicating that these inhibitors do not rely on the side pocket for binding (Table 1).
Within the COX-2 lobby, the hydroxyl group of Ser-119 forms a hydrogen bond with the dansyl moiety of compounds 1 and 2. Unexpectedly, mutation of Ser-119 to alanine increased the inhibitory potency of compound 1 (IC50 of 0.34 μm) while having essentially no effect on that of compound 2 (IC50 of 0.17 μm).
The most striking effect of all mutations evaluated was the contrast between the increased potency of compound 2 as compared with the decreased potency of indomethacin against R120A COX-2. In an attempt to elucidate the mechanism of this contrast, we evaluated the kinetics of inhibition of R120A by compound 2. Results demonstrated that the rate of inhibition of the mutant enzyme by compound 2 was substantially higher than its rate of inhibition of WT COX-2 (Fig. 2C). The inhibition rate was too rapid for accurate evaluation of the binding or kinetic constants for compound 2's interaction with R120A; however, these results suggest that the high potency of compound 2 for R120A COX-2 likely are the result of a much higher affinity for formation of the initial complex and/or a much more rapid conversion to the tightly bound complex.
Discussion
In 1996, Luong et al. (29) published the crystal structure of COX-2 complexed with an inhibitor comprising zomepirac attached to a p-iodophenyl group via an acyl sulfonamide linker. This COX-2–selective inhibitor had been created by amidation of the carboxyl group of the nonselective NSAID zomepirac in a similar fashion to our creation of COX-2–selective inhibitors by amidation or esterification of indomethacin. Although not publicly available, the crystal structure was described as showing zomepirac bound in the COX-2 active site with the linker passing through the constriction to place the p-iodophenyl ring in the lobby (29). Thus, this early finding suggested the possible use of the lobby as a binding site for functionalities attached to the carboxylates of conventional NSAIDs. Nevertheless, for many years, we have unsuccessfully attempted to obtain an X-ray crystal structure of a complex of COX-2 with an inhibitor of the indomethacin ester/amide class. In the recently reported COX-2 complex with an indomethacin-podophyllotoxin conjugate (22), we were able to visualize the indomethacin moiety and a portion of the linker but not the toxin moiety of the ligand. In that complex, the orientation of the indomethacin moiety was consistent with the hypothesis that the podophyllotoxin portion of the molecule was located in the lobby, and it was successfully placed there by computational modeling. Nevertheless, the data reported here provide the first fully observed electron density maps accounting for the placement of the amide substituent of an indomethacin amide in the lobby region of COX-2.
It is interesting to note that compound 1, with a two-carbon diamine linker, is a moderately potent COX-2–selective inhibitor, whereas compound 2, with a four-carbon diamine linker, displays much higher potency. This potency difference may be due to unfavorable tension at the constriction resulting from lack of flexibility of compound 1's short linker. Neither compound is a COX-1 inhibitor. This has been attributed to the observation that binding of carboxylate-containing substrates and inhibitors to COX-1 is much more strongly dependent on ion pairing with Arg-120 than it is in COX-2. Another contributing factor may be that the MBD of COX-1 is less flexible than that of COX-2 and may therefore be unable to accommodate the binding of a large amide or ester group in the lobby.
Compounds 1 and 2 form similar contacts with Ser-530, Ser-119, and Val-89; however, mutation studies show that compound 1 is more sensitive than compound 2 to changes that alter the nature of these contacts. A possible explanation for this difference might be that the short linker in compound 1 allows for very little flexibility in the binding conformation that would enable it to compensate for the loss of a crucial interaction. In contrast, compound 2's four-carbon linker would potentially enable it to form new contacts in the environment of a mutant enzyme.
Despite the differences in their responses to various mutations, both compounds displayed increased inhibitor potency against the R120A mutant COX-2 as compared with the WT enzyme. It is therefore notable that in both crystal structures of COX-2 complexed with the indomethacin-dansyl conjugates, the side chain of Arg-120 was difficult to fit due to relatively high B-factors, which averaged 120 and 116 Å2 for crystal complexes containing compounds 1 and 2, respectively. The side chain of Arg-120 was pushed away from the position it occupies in the crystal structure of the COX-2·indomethacin complex, presumably to accommodate the tethered linker. We examined the kinetics of inhibition of the R120A mutant COX-2 by compound 2. The results showed that the time dependence of inhibition by compound 2 was almost abolished by the R120A mutant (Fig. 2C), suggesting that Arg-120 acts as a hindrance at the constriction for both conjugates and that its displacement plays an important role in the slow rate of binding (at least in the case of compound 2) to the WT enzyme.
As noted above, we have shown previously that insertion of the 2′-methyl group on indomethacin's indole ring into a hydrophobic pocket comprising Val-349, Ala-527, Ser-530, and Leu-531 is a key determinant of the time dependence of COX-2 inhibition by indomethacin. Clearly, indomethacin's polar interaction with Arg-120 is critical to potency, as indicated by its total loss of activity against the R120A mutant, but the finding that 2′-des-methyl-indomethacin remains a rapidly reversible inhibitor of COX-2 suggests that this interaction can be established very quickly. Our data indicating the importance of Arg-120 displacement in the time dependence of compound 2 suggests a fundamental difference between this inhibitor and its parent compound with regard to their binding kinetics with the enzyme despite the similarity in their kinetic constants described above.
In summary, our data support a long-held hypothesis that indomethacin esters/amides bind to COX-2 with their indomethacin moiety in the active site above the constriction and the ester/amide functionality in the lobby. Of course, the possibility remains that some molecules of this class adopt radically different binding modes, and these data do not address all of the potential interactions that can occur between the widely varying ester/amide groups that are found among this class of inhibitors and residues in the lobby. It also does not immediately explain differences in potency between these various inhibitors, a goal that would require substantially more structural information. Regardless, these new data provide an important foundation for the future rational design and exploration of indomethacin conjugates as COX-2–targeted imaging or therapeutic agents.
Experimental procedures
Protein expression and purification
Expression and purification of murine COX-2 and its mutants were completed as described previously (25). Ovine COX-1 was purified as described previously from sheep seminal vesicles (30).
COX inhibition assay
Assay solutions contained purified enzyme (50 nm) with 2 eq of hematin per monomer of COX, 100 mm Tris-Cl, pH 8.0, 5 mm phenol, and 2% DMSO. The inhibitor was dissolved in DMSO and incubated with protein for 15 min at 37 °C prior to the addition of 5 μm AA. After 30 s, the enzymatic activity was quenched with an equal volume of ethyl acetate with 0.5% (w/w) acetic acid and 0.3 μm PGE2-d4 as internal standard for later LC-MS analysis (31). Plots of activity (percentage of uninhibited control) versus log[inhibitor] were fit to a three-parameter equation for enzyme inhibition using Prism 8 software (GraphPad).
COX time-dependent inhibition assay
Time-dependent inhibition reactions contained 50 nm purified, hematin-reconstituted native or mutant COX-2. After preincubation of enzyme with different concentrations of inhibitors (0.5, 2, 5, 10, 15, 20, and 25 μm) for the specified times (0, 0.25, 0.5, 1.0, 2, 4, 8, and 16 min), 50 μm AA was added to initiate the enzymatic reaction, which was allowed to proceed for 30 s at 37 °C. The enzymatic activity was then quenched with an equal volume of ethyl acetate with 0.5% (w/w) acetic acid and 0.3 μm PGE2-d4 as internal standard for later LC-MS analysis (31). Plots of remaining activity versus time for each inhibitor concentration were fit using nonlinear regression analysis to the equation for first-order exponential decay to obtain values for the observed rate constant (kobs) for enzyme inactivation. These values were then plotted against inhibitor concentration, and the data were fit to Equation 2 (“Results”) to obtain values for KI, k2, and k−2. All data analyses were performed using Prism 8 software.
Crystallization, X-ray data collection, structure determination, and refinement
Crystal complexes of COX-2·compound 1 and COX-2·compound 2 were obtained according to the previous publication (22). Diffraction data were collected using the synchrotron radiation X-ray source with 100 K liquid nitrogen streaming at beamline 24-ID-C in the Advance Photon Source at Argonne National Laboratory. Diffraction data were processed with XDS (32). Both complexes were determined as C2 space groups. Initial phases were determined by molecular replacement using monomer coordinates (PDB code 3NT1, chain A) with Phaser (33). A set of randomly selected data (3%) were set aside for testing and quality control. The models containing four monomers in one asymmetric unit were improved with several rounds of model building against the remaining data with a constraint of F ≥ 0 in Coot (34) and PHENIX (35). Global noncrystallographic symmetry was applied during the initial refinements and released afterward. Ligand constraints were computed using the PRODRG server (36). Water molecules were added during the last cycles of refinement, and translation, libration, and screw-rotation (TLS) refinement was applied in the last cycle of refinement (37). The potential of phase bias was excluded by simulated annealing using PHENIX (38). The values of the Ramachandran plot for the final refinement of the structure were obtained by use of the PHENIX suite (35). X-ray data collection and structural refinement statistics are reported in Table 2. Among the four monomers in each structure, no significant differences were observed, and all illustrations were prepared using the coordinates of one monomer with PyMOL (Schrödinger, LLC).
Table 2.
Statistics of X-ray data collection and structure refinement
Number of crystals for both data sets, 1. The values in parentheses are for the highest resolution shell.
| COX-2·compound 1 | COX-2·compound 2 | |
|---|---|---|
| Data collection | ||
| PDB code | 6BL4 | 6BL3 |
| Wavelength (Å) | 0.9792 | 0.9792 |
| Resolution range (Å) | 112–2.22 (2.30–2.22) | 103.3–2.22 (2.30–2.22) |
| Space group | C2 | C2 |
| Unit cell | ||
| a, b, c (Å) | 214.8, 120.4, 134.4 | 215.5, 121.8, 134.8 |
| α, β, γ (°) | 90, 123.6, 90 | 90, 123.5, 90 |
| Total reflections | 324,375 (32,417) | 480,554 (45,327) |
| Unique reflections | 138,412 (13,682) | 141,148 (13,090) |
| Multiplicity | 2.3 (2.4) | 3.4 (3.3) |
| Completeness (%) | 98.49 (97.71) | 97.83 (91.40) |
| Mean I/σ(I/σ) | 9.55 (1.58) | 5.97 (1.09) |
| Wilson B-factor (Å2) | 29.05 | 39.05 |
| Rmergea | 0.109 (0.715) | 0.1535 (1.424) |
| CC1/2b | 0.986 (0.507) | 0.989 (0.458) |
| CC*c | 0.996 (0.820) | 0.997 (0.792) |
| Refinement | ||
| Rwork/Rfreed (%) | 18.9/22.0 (28.3/33.9) | 20.5/22.9 (30.2/32.9) |
| Number of atoms | 20,177 | 19,075 |
| Protein/ligands/water | 17,896/652/1,629 | 17,932/676/467 |
| Protein residues | 2,208 | 2,236 |
| r.m.s.e (bonds/angles) | 0.01/1.22 | 0.007/1.09 |
| Ramachandran favored/outlier (%) | 97.0/0.18 | 97.0/0 |
| Average B-factor | 34.2 | 41.8 |
| Protein/ligands/solvent | 33.6/41.8/37.5 | 41.5/51.5/42.4 |
aRmerge = (ΣhklΣi|Ii(hkl) − Ii(hkl)|)/(ΣhklΣIi(hkl)) 100%; R = (Σhkl‖Fo| − |Fc‖)/(Σhkl|Fo|) 100% where Fo and Fc are the observed and calculated structure factors.
bCC1/2 = (Σ(xi − x̄)(yi − ȳ))/(Σ(xi − x)2 Σ(yi − ȳ)2) where xi and yi are the intensities of unique reflections merged across the observations randomly and evenly split into subsets X and Y and where x̄ and ȳ are the averages of the subset X,Y, respectively.
cCC* = √(2CC1/2)/(1 + CC1/2).
dRfree is the value from the test set (3.0% of all reflections).
er.m.s., root mean square.
Author contributions
S. X., J. R. K., C. A. R., and L. J. M. conceptualization; S. X., S. B., C. A. R., and L. J. M. formal analysis; S. X., M. J. U., S. B., K. D., J. M., J. R. K., and K. G. investigation; S. X. writing-original draft; M. J. U. methodology; M. J. U., S. B., J. R. K., C. A. R., and L. J. M. writing-review and editing; C. A. R. and L. J. M. supervision; C. A. R. and L. J. M. funding acquisition; C. A. R. and L. J. M. project administration.
Acknowledgments
X-ray diffraction was conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by NIGMS, National Institutes of Health Grants P30 GM124165 and S10 RR029205. We thank Life Science Collaborative Access Team beamlines at Advanced Photon Source for the earlier assistance with data collection of the COX-2·compound 2 complex. Use of the Advanced Photon Source, an Office of Science User Facility operated for the United States Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by United States DOE under Contract DE-AC02-06CH11357.
This work was supported by National Institutes of Health Grants GM15431 (to L. J. M.) and CA089450 (to L. J. M.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
The atomic coordinates and structure factors (codes 6BL4 and 6BL3) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- COX
- cyclooxygenase
- AA
- arachidonic acid
- PG
- prostaglandin
- NSAID
- nonsteroidal anti-inflammatory drug
- EGF
- epidermal growth factor
- MBD
- membrane-binding domain
- dansyl
- 5-(dimethylamino)naphthalene-1-sulfonyl.
References
- 1. Smith W. L., and Dewitt D. L. (1996) Prostaglandin endoperoxide H synthases-1 and -2. Adv. Immunol. 62, 167–215 10.1016/S0065-2776(08)60430-7 [DOI] [PubMed] [Google Scholar]
- 2. Smith W. L., DeWitt D. L., and Garavito R. M. (2000) Cyclooxygenases: structural, cellular, and molecular biology. Annu. Rev. Biochem. 69, 145–182 10.1146/annurev.biochem.69.1.145 [DOI] [PubMed] [Google Scholar]
- 3. Smith W. L., and Langenbach R. (2001) Why there are two cyclooxygenase isozymes. J. Clin. Investig. 107, 1491–1495 10.1172/JCI13271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rouzer C. A., and Marnett L. J. (2003) Mechanism of free radical oxygenation of polyunsaturated fatty acids by cyclooxygenases. Chem. Rev. 103, 2239–2304 10.1021/cr000068x [DOI] [PubMed] [Google Scholar]
- 5. Simmons D. L., Botting R. M., and Hla T. (2004) Cyclooxygenase isozymes: the biology of prostaglandin synthesis and inhibition. Pharmacol. Rev. 56, 387–437 10.1124/pr.56.3.3 [DOI] [PubMed] [Google Scholar]
- 6. Smyth E. M., Grosser T., Wang M., Yu Y., and FitzGerald G. A. (2009) Prostanoids in health and disease. J. Lipid Res. 50, (suppl.) S423–S428 10.1194/jlr.R800094-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ricciotti E., and FitzGerald G. A. (2011) Prostaglandins and inflammation. Arterioscler. Thromb. Vasc. Biol. 31, 986–1000 10.1161/ATVBAHA.110.207449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marnett L. J., and Kalgutkar A. S. (1999) Cyclooxygenase 2 inhibitors: discovery, selectivity and the future. Trends Pharmacol. Sci. 20, 465–469 10.1016/S0165-6147(99)01385-1 [DOI] [PubMed] [Google Scholar]
- 9. Rouzer C. A., and Marnett L. J. (2008) Non-redundant functions of cyclooxygenases: oxygenation of endocannabinoids. J. Biol. Chem. 283, 8065–8069 10.1074/jbc.R800005200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rouzer C. A., and Marnett L. J. (2011) Endocannabinoid oxygenation by cyclooxygenases, lipoxygenases, and cytochromes P450: cross-talk between the eicosanoid and endocannabinoid signaling pathways. Chem. Rev. 111, 5899–5921 10.1021/cr2002799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marnett L. J., Uddin M. J., and Crews B. C. (March 27, 2012) Methods and Compositions for Diagnostic and Therapeutic Targeting of COX-2. U. S. Patent 8, 143, 302
- 12. Kalgutkar A. S., Crews B. C., Rowlinson S. W., Marnett A. B., Kozak K. R., Remmel R. P., and Marnett L. J. (2000) Biochemically based design of cyclooxygenase-2 (COX-2) inhibitors: facile conversion of nonsteroidal antiinflammatory drugs to potent and highly selective COX-2 inhibitors. Proc. Natl. Acad. Sci. U.S.A. 97, 925–930 10.1073/pnas.97.2.925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kalgutkar A. S., Marnett A. B., Crews B. C., Remmel R. P., and Marnett L. J. (2000) Ester and amide derivatives of the nonsteroidal antiinflammatory drug, indomethacin, as selective cyclooxygenase-2 inhibitors. J. Med. Chem. 43, 2860–2870 10.1021/jm000004e [DOI] [PubMed] [Google Scholar]
- 14. Uddin M. J., Crews B. C., Ghebreselasie K., and Marnett L. J. (2013) Design, synthesis, and structure-activity relationship studies of fluorescent inhibitors of cycloxygenase-2 as targeted optical imaging agents. Bioconjug. Chem. 24, 712–723 10.1021/bc300693w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Uddin M. J., Crews B. C., Blobaum A. L., Kingsley P. J., Gorden D. L., McIntyre J. O., Matrisian L. M., Subbaramaiah K., Dannenberg A. J., Piston D. W., and Marnett L. J. (2010) Selective visualization of cyclooxygenase-2 in inflammation and cancer by targeted fluorescent imaging agents. Cancer Res. 70, 3618–3627 10.1158/0008-5472.CAN-09-2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cekanova M., Uddin M. J., Bartges J. W., Callens A., Legendre A. M., Rathore K., Wright L., Carter A., and Marnett L. J. (2013) Molecular imaging of cyclooxygenase-2 in canine transitional cell carcinomas in vitro and in vivo. Cancer Prev. Res. 6, 466–476 10.1158/1940-6207.CAPR-12-0358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ra H., González-González E., Uddin M. J., King B. L., Lee A., Ali-Khan I., Marnett L. J., Tang J. Y., and Contag C. H. (2015) Detection of non-melanoma skin cancer by in vivo fluorescence imaging with fluorocoxib A. Neoplasia 17, 201–207 10.1016/j.neo.2014.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Harman C. A., Turman M. V., Kozak K. R., Marnett L. J., Smith W. L., and Garavito R. M. (2007) Structural basis of enantioselective inhibition of cyclooxygenase-1 by S-α-substituted indomethacin ethanolamides. J. Biol. Chem. 282, 28096–28105 10.1074/jbc.M701335200 [DOI] [PubMed] [Google Scholar]
- 19. Konkle M. E., Blobaum A. L., Moth C. W., Prusakiewicz J. J., Xu S., Ghebreselasie K., Akingbade D., Jacobs A. T., Rouzer C. A., Lybrand T. P., and Marnett L. J. (2016) Conservative secondary shell substitution in cyclooxygenase-2 reduces inhibition by indomethacin amides and esters via altered enzyme dynamics. Biochemistry 55, 348–359 10.1021/acs.biochem.5b01222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Timofeevski S. L., Prusakiewicz J. J., Rouzer C. A., and Marnett L. J. (2002) Isoform-selective interaction of cyclooxygenase-2 with indomethacin amides studied by real-time fluorescence, inhibition kinetics, and site-directed mutagenesis. Biochemistry 41, 9654–9662 10.1021/bi0203637 [DOI] [PubMed] [Google Scholar]
- 21. Kurumbail R. G., Stevens A. M., Gierse J. K., McDonald J. J., Stegeman R. A., Pak J. Y., Gildehaus D., Miyashiro J. M., Penning T. D., Seibert K., Isakson P. C., and Stallings W. C. (1996) Structural basis for selective inhibition of cyclooxygenase-2 by anti-inflammatory agents. Nature 384, 644–648 10.1038/384644a0 [DOI] [PubMed] [Google Scholar]
- 22. Uddin M. J., Crews B. C., Xu S., Ghebreselasie K., Daniel C. K., Kingsley P. J., Banerjee S., and Marnett L. J. (2016) Antitumor activity of cytotoxic cyclooxygenase-2 inhibitors. ACS Chem Biol 11, 3052–3060 10.1021/acschembio.6b00560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rome L. H., and Lands W. E. (1975) Structural requirements for time-dependent inhibition of prostaglandin biosynthesis by anti-inflammatory drugs. Proc. Natl. Acad. Sci. U.S.A. 72, 4863–4865 10.1073/pnas.72.12.4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Prusakiewicz J. J., Felts A. S., Mackenzie B. S., and Marnett L. J. (2004) Molecular basis of the time-dependent inhibition of cyclooxygenases by indomethacin. Biochemistry 43, 15439–15445 10.1021/bi048534q [DOI] [PubMed] [Google Scholar]
- 25. Xu S., Hermanson D. J., Banerjee S., Ghebreselasie K., Clayton G. M., Garavito R. M., and Marnett L. J. (2014) Oxicams bind in a novel mode to the cyclooxygenase active site via a two-water-mediated H-bonding network. J. Biol. Chem. 289, 6799–6808 10.1074/jbc.M113.517987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blobaum A. L., and Marnett L. J. (2007) Structural and functional basis of cyclooxygenase inhibition. J. Med. Chem. 50, 1425–1441 10.1021/jm0613166 [DOI] [PubMed] [Google Scholar]
- 27. Garavito R. M., and Mulichak A. M. (2003) The structure of mammalian cyclooxygenases. Annu. Rev. Biophys. Biomol. Struct. 32, 183–206 10.1146/annurev.biophys.32.110601.141906 [DOI] [PubMed] [Google Scholar]
- 28. Blobaum A. L., Xu S., Rowlinson S. W., Duggan K. C., Banerjee S., Kudalkar S. N., Birmingham W. R., Ghebreselasie K., and Marnett L. J. (2015) Action at a distance: mutations of peripheral residues transform rapid reversible inhibitors to slow, tight binders of cyclooxygenase-2. J. Biol. Chem. 290, 12793–12803 10.1074/jbc.M114.635987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Luong C., Miller A., Barnett J., Chow J., Ramesha C., and Browner M. F. (1996) Flexibility of the NSAID binding site in the structure of human cyclooxygenase-2. Nat. Struct. Biol. 3, 927–933 10.1038/nsb1196-927 [DOI] [PubMed] [Google Scholar]
- 30. Odenwaller R., Chen Y. N., and Marnett L. J. (1990) Preparation and proteolytic cleavage of apoprostaglandin endoperoxide synthase. Methods Enzymol. 187, 479–485 10.1016/0076-6879(90)87054-7 [DOI] [PubMed] [Google Scholar]
- 31. Kingsley P. J., Rouzer C. A., Saleh S., and Marnett L. J. (2005) Simultaneous analysis of prostaglandin glyceryl esters and prostaglandins by electrospray tandem mass spectrometry. Anal. Biochem. 343, 203–211 10.1016/j.ab.2005.05.005 [DOI] [PubMed] [Google Scholar]
- 32. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McCoy A. J. (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 10.1107/S0907444906045975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schüttelkopf A. W., and van Aalten D. M. (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 10.1107/S0907444904011679 [DOI] [PubMed] [Google Scholar]
- 37. Winn M. D., Isupov M. N., and Murshudov G. N. (2001) Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 10.1107/S0907444900014736 [DOI] [PubMed] [Google Scholar]
- 38. Terwilliger T. C., Grosse-Kunstleve R. W., Afonine P. V., Moriarty N. W., Zwart P. H., Hung L. W., Read R. J., and Adams P. D. (2008) Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 10.1107/S090744490705024X [DOI] [PMC free article] [PubMed] [Google Scholar]