Abstract
Sulfiredoxin (Srx) reduces hyperoxidized 2-cysteine–containing peroxiredoxins (Prxs) and protects cells against oxidative stress. Previous studies have shown that Srx is highly expressed in primary specimens of lung cancer patients and plays a pivotal role in lung tumorigenesis and cancer progression. However, the oncogenic mechanisms of Srx in cancer are incompletely understood. In this study, we found that Srx knockdown sensitizes lung cancer cells to endoplasmic reticulum (ER) stress–induced cell death. Through MS analysis, we determined that Srx forms a complex with the ER-resident protein thioredoxin domain–containing protein 5 (TXNDC5). Using reciprocal co-immunoprecipitation, immunofluorescence imaging, subcellular fractionation, and domain-mapping assays with site-specific mutagenesis and purified recombinant proteins, we further characterized the Srx–TXNDC5 interaction. In response to ER stress but not to oxidative stress, Srx exhibits an increased association with TXNDC5, facilitating the retention of Srx in the ER. Of note, TXNDC5 knockdown in lung cancer cells inhibited cell proliferation and repressed anchorage-independent colony formation and migration, but increased cell invasion and activation of mitogen-activated protein kinases. Using immunohistochemical staining, we demonstrate that TXNDC5 is highly expressed in patient-derived lung cancer specimens. Bioinformatics analysis of publicly available data sets revealed that those with high Srx levels have significantly shorter survival and that those with high TXNDC5 levels have longer survival. We conclude that the cellular levels of Srx and TXNDC5 may be useful as biomarkers to predict the survival of individuals with lung cancer.
Keywords: antioxidant, protein-protein interaction, proteomics, enzyme mechanism, oxidative stress, protein-disulfide isomerase, pulmonary carcinoma, sulfiredoxin, thioredoxin-domain containing 5 (TXNDC5), tumorigenesis
Introduction
Lung cancer is the second most common cancer in men and women and is the leading cause of cancer death in the United States. Among major factors contributing to the high fatality rate of lung cancer are the lack of early detection methods, difficulty of treatment, tumor recurrence and metastasis, etc. In the process of lung tumorigenesis, series of genetic mutations have to occur prior to the transformation of lung normal epithelial cells, and those transformed cells may develop into benign tumors. Subsequently, additional genetic modifications may enable mutated cells to acquire invasive and metastatic traits that are characteristic features of lung cancer cells. Among all known oncogenic factors are increased levels of reactive oxygen species that lead to oxidative damage of macromolecules and also activate various antioxidant systems in the cells. Cellular peroxidases, including 2-cysteine–containing Prxs,3 are antioxidant enzymes that contribute to the development of lung cancer (1, 2). For instance, Prx1 and Prx4 have been found to enhance colony formation, cell migration in vitro, and the metastasis of lung cancer cells in vivo (3–5). In general, the cellular level of H2O2 is strictly regulated by redox enzymes, including the family of Prxs. Oxidation of Prxs can further oxidize other protein substrates, such as members of the protein-disulfide isomerase (PDI) family, to generate new disulfide bonds (6, 7). However, under oxidative stress conditions, the Cp site of Prxs can be further oxidized to generate sulfinic acid and sulfonic acid, a process known as hyperoxidation or overoxidation. As an adaptive mechanism, cells have evolved to adopt a recycling machinery that reduces hyperoxidized Cp residue of Prxs from sulfinic to sulfenic acid through the expression of Srx. Srx transfers the γ-phosphate of ATP to Cp sulfinic acid on hyperoxidized Prxs and produces sulfinic phosphoryl ester. Subsequent involvement of GSH and thioredoxin will ensure the reduction of sulfinic phosphoryl ester to sulfenic acid (8–10).
The function of Srx appears to be necessary to maintain the redox balance in cancer cells. Inhibition of Srx results in oxidative stress-induced mitochondrial damage and caspase activation, leading to the apoptosis of lung adenocarcinoma cells (11, 12). The expression level of Srx is under the coordinated control of transcription factors, including nuclear erythroid 2–related factor 2 (Nrf2) and activator protein 1 (AP-1) (13, 14). In lung normal epithelial cells, the protein level of Srx is very low and is beyond the limit of detection by regular methods such as immunostaining and Western blotting. However, an abnormally high level of Srx protein was observed in lung cancer, particularly in adenocarcinoma and squamous cell carcinoma (15, 16). Previously, we have demonstrated that the presence of Srx promotes the proliferation, colony formation, and metastasis of lung cancer cells through the activation of mitogen-activated protein kinase cascade (15). However, the oncogenic mechanisms of Srx in lung cancer have not been completely understood.
Besides antioxidant enzymes, the cellular redox state is well-maintained by many other signaling pathways, including stabilization of proteins through the generation and rearrangement of disulfide bonds. Oxidation of cysteine residue in substrate protein during disulfide bond formation is accompanied by oxidative protein folding. Electrons are thus transferred from the reduced substrate to the active sites of the PDI family members. Hence, PDIs are considered indispensable enzymes for providing maturation and structural stability for substrates (17). TXNDC5 is a member of the PDI family and plays a critical role in signal transduction and cancer development (18). However, the biochemical mechanisms of TXNDC5 have not been studied, including its role in disulfide bond exchange and redox balance in lung cancer. In this study, we found that TXNDC5 is a novel protein–protein interacting partner of Srx, and their interaction may contribute to the oncogenic mechanism of Srx in the development of lung cancer in patients.
Results
Knockdown of Srx sensitizes human lung cancer cells to ER stress–induced cell death
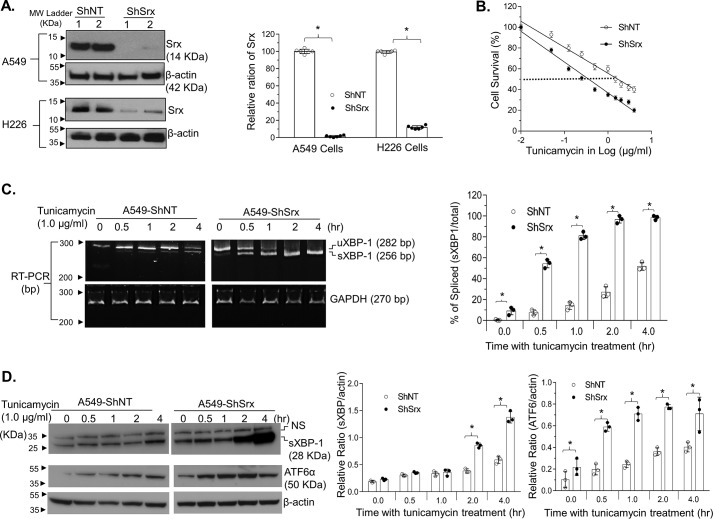
To study the role of Srx in human lung cancer cells, we used several lentiviral shRNAs targeting separate coding regions of Srx mRNA (shSrx) to knockdown the levels of endogenously expressed protein. To ensure the robustness of this study, two types of human lung cell lines, including A549 cells derived from adenocarcinoma and H226 cells derived from squamous cell carcinoma, were used in parallel in all experiments. After viral infection and antibiotic selection, cells stably expressing either of the two lentiviral shSrx constructs had significantly lower levels of Srx compared with those expressing a nontarget control shRNA (shNT) (Fig. 1A). All subsequent experiments were repeated in stable A549 or H226 cells expressing shSrx1 or shSrx2, and findings consistent in both cell types are hereby reported (unless otherwise specified).
Figure 1.
Knockdown of Srx sensitizes human lung cancer A549 cells to ER stress–induced cell death. A, representative results of Srx knockdown using two different shSrx constructs in human lung cancer cells. B, knockdown of Srx sensitizes cells to tunicamycin-induced death. Data from six replicates are presented as mean ± S.D. (error bars), and the IC50 values were calculated through log transformation and linear regression analysis. Calculated IC50 values of control cells (ShNT) and Srx knockdown cells (ShSrx) are 1.69 and 0.36 μg/ml, respectively. C, knockdown of Srx induces a rapid response of UPR, as indicated by the accelerated splicing of XBP-1 mRNA. D, expression of spliced XBP protein and activation of ATF6α in the presence of tunicamycin in A549 control and Srx knockdown cells. NS, nonspecific band. The bar graph with a dot plot on the right indicates the quantitative results (*, p < 0.05, t test).
First, we evaluated whether knockdown of Srx had any effect on cells' response to tunicamycin-induced cell death. Tunicamycin is one of the most widely used chemicals to induce ER stress in cultured cells (19). Cells in confluence were treated with increasing doses of tunicamycin for 24 h, and cell survival was evaluated by a modified XTT assay. We found that treatment of A549/H226 cells with tunicamycin resulted in a dose-dependent cell death, and Srx knockdown cells were significantly more sensitive to such treatment. For example, the IC50 of tunicamycin for A549 control cells in a treatment period of 24 h was 1.69 μg/ml, whereas the IC50 for A549-shSrx cells under the same conditions was 0.36 μg/ml, indicating a more than 4-fold increase of tunicamycin sensitivity due to the knockdown of Srx (Fig. 1B). We also did a clonogenic assay to evaluate the ability of these cells to grow into colonies under different concentrations of tunicamycin treatment for an extended period. We found that the IC50 of tunicamycin to inhibit colony formation in control cells was 101 ng/ml, whereas the IC50 in shSrx cells was 72 ng/ml (Fig. S1), indicating that Srx knockdown cells were more sensitive to the effect of tunicamycin in a clonogenic assay. Due to the known role of tunicamycin to induce ER stress, next we examined the effect of depleting Srx on ER stress response by evaluating the expression of ER stress markers, including the spliced X-box–binding protein 1 (sXBP1) and the transcription factor ATF6α (20). As representative results obtained from the treatment of A549 cells, we found that knockdown of Srx led to a significant increase of XBP1 mRNA splicing at 30 min after tunicamycin exposure, whereas equivalent levels of sXBP1 mRNA in control cell occurred at 2–4 h after treatment (Fig. 1C). Moreover, the induction of sXBP1 and ATF6α proteins also occurred much faster in Srx knockdown cells compared with those in control cells (Fig. 1D). Knockdown of Srx in H226 cells also led to a faster splicing of XBP-1 mRNA and an enhanced expression of sXBP-1 and ATF6α proteins (Fig. S2). Taken together, these data suggest that knockdown of Srx leads to a rapid activation of the unfolded protein response (UPR) and sensitizes human lung cancer cells to ER stress–induced cell death.
Identification of TXNDC5 as a component of the Srx–protein complex
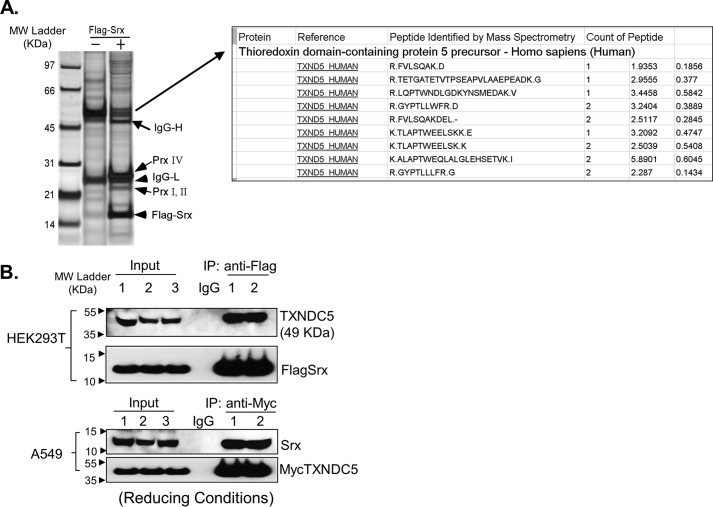
To date Srx is mainly reported as a cytosolic protein, and its role in ER homeostasis has not been revealed. We hypothesized that such a function of Srx may be mediated through a direct or indirect interaction with an ER-resident protein. To test this hypothesis, cells were transiently transfected with plasmid expressing a FLAG-Srx or an empty vector (control). The Srx–protein complex was pulled down by anti-FLAG immunoprecipitation (IP), and protein identities were determined by MS. To maximally ensure the specificity and reproducibility of protein identification, three strategies were used. First, pulldown experiments were performed in three different cell types, including HEK293T, H226, and A549 cells; second, IP eluates from either control cells or FLAG-Srx–expressing cells were concentrated and analyzed directly by reverse phase LC–MS (RPLC-MS) analysis; third, IP eluates were initially separated by SDS-PAGE and visualized by silver staining to identify differential bands, in which proteins were extracted and then analyzed by reverse-phase liquid chromatography (RPLC)-MS. Among the handful of proteins identified, Prx4 was the most abundant protein, and TXNDC5 was the second most abundant protein that was specifically pulled down by anti-FLAG IP in all cell types. For example, there were 14 counts of peptides containing nine unique sequences of TXNDC5 in the eluates of anti-FLAG IP in HEK293 cells (Fig. 2A). An example of the detailed list of proteins identified in MS is shown in Table S1. Previously, we have characterized the interaction of Srx with Prx4, and we have demonstrated its role in promoting cancer development (15). Therefore, in this study, we were focused on the characterization and understanding of the novel interaction between Srx and TXNDC5.
Figure 2.
Identification of TXNDC5 as an interacting protein of Srx. A, silver staining of protein pulldown by anti-FLAG-Srx IP in human HEK293T cells. The identity of the differential band was determined by RPLC-MS analysis. B, Srx and TXNDC5 were pulled down by reciprocal co-IP in HEK293T and A549 cells. The numbers indicate independent, experimental repeats.
To validate the association of TXNDC5 with the Srx–protein complex, reciprocal IPs were performed in HEK293 and A549 cells. Because the levels of endogenous Srx in HEK293 cells cannot be easily detected by Western blotting, FLAG-Srx was ectopically expressed, and anti-FLAG IP was performed. A549 cells do express higher levels of Srx, so Myc-TXNDC5 was thus introduced into these cells to facilitate the pulldown of TXNDC5 by anti-Myc IP. Data from these IPs indicate that TXNDC5 is a component of the Srx–protein complex (Fig. 2B).
Colocalization of Srx and TXNDC5 in the ER in cultured cells
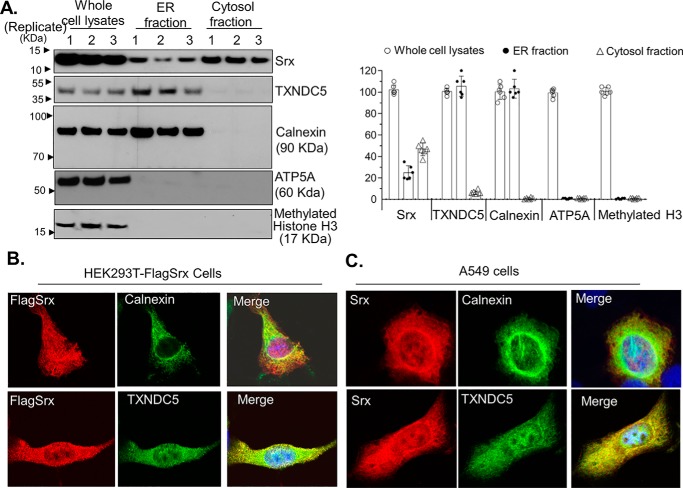
Because Srx was mainly reported as a cytosolic protein and TXNDC5 was recognized as an ER protein, one of the major concerns is that their association could result from the artifacts of ectopic overexpression, in particular, if they are not residing in the same subcellular compartment within the cell. To exclude this possibility, we investigated whether Srx is present in the ER under physiological culture conditions. A549 cells were harvested, and subcellular fractionation was obtained by using an ER enrichment kit with ultra-speed centrifugation. Collected subcellular fractions were then examined by gel electrophoresis and immunoblotting. As shown in Fig. 3A, the presence of the ER-resident protein calnexin solely in the ER and the absence of both mitochondria marker (ATP5A) and nuclear marker (methylated histone H3) confirmed that the collected ER fraction was free of cytosol, mitochondria, or nuclear contamination. After quantification, we found that the majority of Srx in the whole cell lysates was present in cytosolic fraction, and about 25% of Srx was present in the ER fraction (Fig. 3A). To further confirm the presence of Srx in the ER, immunofluorescent staining and imaging were used to visualize the colocalization of Srx and TXNDC5 in HEK293T and A549 cells. We found that co-staining of FLAG-Srx (red fluorescence) with ER protein calnexin (green fluorescence) or with TXNDC5 (green fluorescence) in HEK293T cells merged into yellow fluorescence, indicating that a portion of Srx was present in the ER and colocalized with TXNDC5 (Fig. 3B). Similarly, the yellow fluorescence in A549 cells indicates the colocalization of endogenous Srx with calnexin or TXNDC5 (Fig. 3C). Taken together, these data suggest that certain amounts of Srx do localize in the ER, which may contribute to the formation of the protein–protein complex with TXNDC5 in human lung cancer cells.
Figure 3.
Colocalization of Srx and TXNDC5 in the ER. A, subcellular fractionation and Western blotting indicate the presence of Srx and TXNDC5 in the ER. The numbers above the bands indicate independent, experimental repeats. The bar graph with a dot plot on the right indicates the quantitative results. B, immunofluorescence staining of overexpressed FLAG-Srx (red) in HEK293T cells and its colocalization with endogenous TXNDC5 (green) in the ER. The appearance of yellow in the merged image indicates possible colocalization. C, immunofluorescence staining of endogenous Srx (red) in A549 cells and its colocalization with endogenous TXNDC5 (green) in the ER. In these results, endogenously expressed calnexin, an ER-resident protein, is used as a specific marker that is only present in the ER. Error bars, S.D.
TXNDC5 directly interacts with Srx through its thioredoxin-like domains
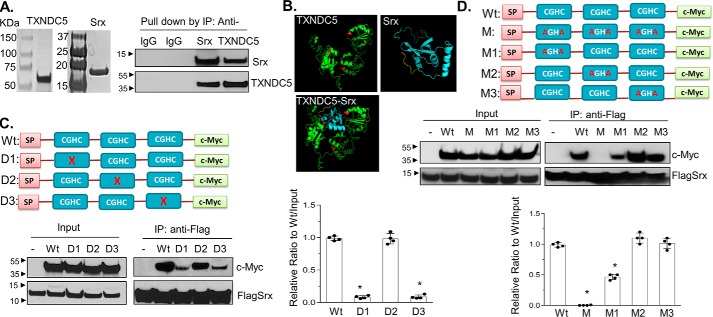
Data described above indicate that there is a protein–protein complex in the ER that contains both Srx and TXNDC5 in human lung cancer cells. However, it is not clear whether TXNDC5 directly binds to Srx because such association can also result indirectly from interacting with other components of the same complex. To determine whether Srx directly binds to TXNDC5, we constructed plasmids to express recombinant human Srx and TXNDC5 proteins in bacteria. After transformation of these plasmids into Escherichia coli, recombinant proteins were purified from bacterial lysates and used in direct binding assays in vitro. When they were mixed and present together in the solution in a test tube, we found that TXNDC5 was able to be pulled down in anti-Srx IP (but not in control IgG IP) and vice versa (Fig. 4A). Therefore, these data demonstrate that the observed association of TXNDC5 and Srx in previous experiments is most likely the result of their direct interaction with each other.
Figure 4.
Characterization and mapping of domains in TXNDC5 that directly interact with Srx. A, recombinant human Srx and TXNDC5 were purified from E. coli and visualized by Coomassie Blue staining (left). The purified proteins were mixed in the binding solution. Reciprocal IP was performed and examined by Western blotting (right). B, predicted interaction of Srx and TXNDC5 based on their structures using I-TASSER and ZDOCK software. The thioredoxin domains (with the key CGHC motif) in TXNDC5 are highlighted in red. The prediction indicates that Srx (blue) interacts with TXNDC5 (green) through the first and the third thioredoxin domain of TXNDC5. C, plasmids that express c-Myc–tagged TXNDC5 or its deletion mutants were expressed in HEK293T-FLAGSrx cells, and cell lysates were used in anti-FLAG IP and examined by Western blotting. Deletion of the first or the third CGHC motif in TXNDC5 leads to significant loss of binding to Srx. D, plasmids that express c-Myc–tagged TXNDC5 or its cysteine-specific mutants were expressed in HEK293T-FLAGSrx cells, and cell lysates were used for anti-FLAG IP and examined by Western blotting. Mutation of two cysteines in the first or the third CGHC motif in TXNDC5 leads to significant loss of binding to Srx. The bar graph with a dot plot on the right indicates the quantitative results (compared with WT; *, p < 0.05, t test). Error bars, S.D.
To map the interacting domains between Srx and TXNDC5, we utilized the iterative threading assembly refinement (I-TASSER) software to predict the tertiary structure of TXNDC5 (21). This predicted structure was then used in protein docking (ZDOCK software version 3.0.2) to find the interface where it interacts with Srx. The structure of Srx we used in the prediction has been determined previously in a protein crystallography study (22). The model with the highest score suggests that the first (close to the N terminus) and the third (close to the C terminus) thioredoxin-like domains of TXNDC5 are most likely to be involved in direct contact with Srx (Fig. 4B). To test this prediction, we made series of expression constructs that encode WT Myc-TXNDC5 or mutants with individual thioredoxin-like domain being deleted (Fig. 4C). With the co-expression of FLAG-Srx, anti-FLAG IPs were performed in cultured human HEK293T cells. We found that deletion of the first or third thioredoxin-like domain in TXNDC5 resulted in a significant loss of its binding to Srx, whereas deletion of the second (the one in the middle) thioredoxin-like domain did not compromise its binding to Srx (Fig. 4C). To further confirm that the thioredoxin-like domains 1 and 3 were responsible for the binding to Srx, two cysteine residues within each domain were mutated to alanine by site-specific mutagenesis (Fig. 4D). After expressing these mutants and FLAG-Srx into HEK293T cells, we found that mutation of all three thioredoxin-like domains in TXNDC5 led to the complete loss of binding to Srx, and mutation of only the first or the third domain significantly reduced the binding, whereas mutation of the second domain alone had marginal effect on its binding to Srx (Fig. 4D). Taking together, these data suggest that the first and the third thioredoxin-like domains of TXNDC5 mediate its direct interaction with Srx.
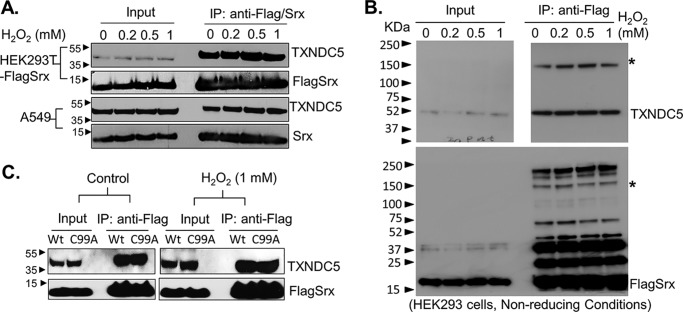
Srx and TXNDC5 form a stable complex that is not affected by the treatment of exogenous hydrogen peroxide (H2O2)
Mutation of cysteines in the thioredoxin domains of TXNDC5 leads to the loss of its binding to Srx, and cysteines are known to mediate the formation of a disulfide bond that can be affected by cellular redox. Therefore, we asked whether the association of Srx with TXNDC5 was affected by cellular oxidative stress. Cultured HEK293T-FLAGSrx and A549 cells were treated with increasing concentrations of H2O2, and cell lysates were harvested. Anti-FLAG and anti-Srx IPs were performed using lysates of HEK293T and A549, respectively. In both IPs, we found that the overall amounts of TXNDC5 associated with Srx were not affected by the exposure of cells to exogenous H2O2 as high as 1 mm (Fig. 5A).
Figure 5.
The Srx–TXNDC5 interaction is not affected by the treatment of cells with exogenous H2O2. A, HEK293-FLAGSrx or A549 cells were treated with vehicle or increasing concentrations of H2O2 for 10 min. Cell lysates were collected, and IPs were performed using anti-FLAG or anti-Srx antibodies. IP eluates were separated by SDS-PAGE under reducing conditions. Western blotting results indicate that treatment of cells with H2O2 does not affect the amount of endogenous TXNDC5 pulldown by FLAG-Srx in HEK293T or Srx in A549 cells. B, IP eluates from A were separated by SDS-PAGE under nonreducing conditions, and Western blotting indicates the position of monomer as well as possible disulfide bond formation between Srx and TXNDC5 as bands indicated by an asterisk. C, FLAG-Srx or its cysteine mutant (C99A) was expressed in HEK293T cells. Cell lysates were collected and IPs were performed using anti-FLAG antibody. IP eluates were separated by SDS-PAGE under reducing conditions. Western blotting results indicate that mutation of cysteine 99 in Srx does not affect its ability to interact with TXNDC5.
Notably, eluates from the above described IP experiments were separated using SDS-PAGE under reducing conditions. The results shown in Fig. 5A could not reveal whether Srx–TXNDC5 interaction was mediated through intermolecular disulfide bond or such a disulfide bond was affected by oxidative stress. As a member of the PDI family, TXNDC5 is known to associate with its substrates through the formation of intermolecular disulfide bond (18). To clarify whether there is intermolecular disulfide bond formation between Srx and TXNDC5, eluates from IPs of cells treated with H2O2 were separated by SDS-PAGE under nonreducing conditions. We found that around 70% of TXNDC5 in eluates was separated as monomer by SDS-PAGE, and there was also a high-molecular weight complex (about 150 kDa) containing Srx and TXNDC5 (as noted by an asterisk in Fig. 5B). This means that a portion of TXNDC5 (around 30% by quantification of band intensity) forms a complex with Srx through a possible disulfide bond. Moreover, neither the intensity of the high-molecular weight complex nor the monomer was affected by the treatment of increasing amounts of H2O2, indicating that the Srx–TXNDC5 was a relatively stable complex that was not affected by the treatment with exogenous H2O2. To further confirm this observation, we treated cells with thiol blocker N-ethylmaleimide (NEM). As shown in Fig. S3, treatment of cells with NEM also did not change the amount of Srx to form a complex with TXNDC5.
Previous studies have shown that Srx is a reducing enzyme that is important for the maintenance of cellular redox balance under oxidative stress. In this context, the Cys-99 of Srx is critical for its catalytic activity, and mutation of Cys-99 to alanine (C99A) leads to a complete loss of both enzymatic activity and its binding to substrates such as Prxs (10, 12, 23). Therefore, we expressed FLAG-tagged WT Srx or its C99A mutant into HEK293T cells and performed anti-FLAG IP. We found that equivalent amounts of TXNDC5 were pulled down by anti-FLAG IPs, regardless of the C99A mutation or the treatment by H2O2 (Fig. 5C). We then examined whether the interaction of Srx with TXNDC5 was affected by the treatment with tunicamycin. As shown in Fig. S3B, treatment of HEK293-FLAGSrx cells with tunicamycin further increased the formation of the Srx–TXNDC5 complex. Taken together, these data suggest that Srx and TXNDC5 form a relatively stable complex that can be enhanced by ER stress. However, their interaction is not easily affected by changing of cellular redox status, such as treatment of cells with exogenous H2O2.
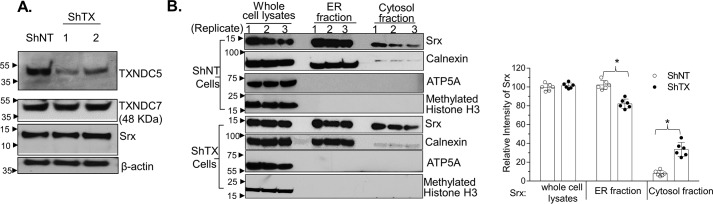
TXNDC5 facilitates the retention of Srx in the ER
To further understand the physiological role of the Srx–TXNDC5 complex, series of lentiviral shRNAs that target separate regions of TXNDC5 mRNA were used to knock down the endogenous protein expression. After screening by transient transfection and Western blotting, we found that two shRNAs targeting the coding regions of TXNDC5 had the highest efficiency to inhibit the expression of endogenous protein. These shRNAs were introduced into A549 and H226 cells by viral infection, and stable cell lines were established by puromycin resistance. In Western blot analysis, we found that cells expressing shRNA targeting TXNDC5 (shTX) had significantly down-regulated expression compared with those of control cells expressing nontarget shRNA (example data in A549 cells are shown in Fig. 6A). In addition, the knockdown was specific to TXNDC5 in that no obvious off-target effects were observed on other proteins, such as Srx and TXNDC7, a close family member of TXNDC5. Next we asked whether knockdown of TXNDC5 affected the subcellular distribution of Srx in these cells. High-speed centrifugation and fractionation were used to enrich proteins in the ER and cytosol, and the level of Srx was measured by Western blotting and quantification. We found significant decrease of Srx in the ER fraction as well as increase of Srx in the cytosolic fraction in shTX cells compared with those of control cells (Fig. 6B). Therefore, these data suggest that the presence of TXNDC5 in human lung cancer cell facilitates the retention of Srx in the ER.
Figure 6.
Knockdown of TXNDC5 in lung cancer cells leads to more localization of Srx in the cytosol. A, two shRNAs targeting different coding regions of TXNDC5 were used to establish stable knockdown in A549 cells. Knockdown of TXNDC5 does not affect the endogenous expression of Srx or TXNDC7, a close member of TXNDC5 in the PDI family. B, subcellular fractionation of A549 control (ShNT) or TXNDC5 knockdown cells for the distribution of Srx in ER and cytosol. Results from three independent replicates were shown. The bar graph with a dot plot on the right indicates the quantitative results (*, p < 0.05, ANOVA). Error bars, S.D.
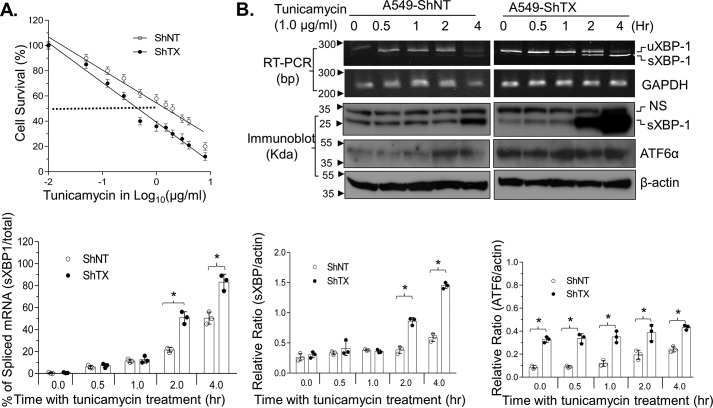
Knockdown of TXNDC5 sensitizes human lung cancer cells to ER stress–induced cell death
We found that knockdown of Srx sensitized human lung cancer cells to ER stress-induced cell death, and next we asked whether knockdown of TXNDC5 in A549 and H226 cell led to similar consequences. These cells were treated with increasing concentrations of tunicamycin for 24 h, and the rate of cell survival was evaluated by an XTT assay. Significantly more cell death was found in shTX cells treated with tunicamycin, indicating that depletion of TXNDC5 increases cells' sensitivity to the treatment by tunicamycin. For example, the IC50 of tunicamycin for A549-shTX cells was 0.45 μg/ml, which is less than one-third of the IC50 of A549 control cells (about 1.5 μg/ml in this batch of experiment) (Fig. 7A). We also examined the expression of molecular markers for stress response with tunicamycin treatment. For example, knockdown of TXNDC5 in A549 cells led to the splicing of XBP-1 mRNA at 2 h after tunicamycin exposure, whereas similar levels of splicing occurred at 4 h after treatment with the same concentration of tunicamycin (Fig. 7B, top). The induction of sXBP1 and ATF6α proteins also occurred faster in shTX cells than in control cells (Fig. 7B, bottom). Taken together, these data suggest that knockdown of TXNDC5 sensitizes human lung cancer cells to ER stress–induced cell death.
Figure 7.
Knockdown of TXNDC5 sensitizes human lung cancer cells to ER stress–induced cell death. A, knockdown of TXNDC5 (ShTX) sensitizes cells to tunicamycin-induced death. Data from six replicates are shown. B, knockdown of TXNDC5 induces a rapid response to UPR, as indicated by the accelerated splicing of XBP-1 and activation of ATF6α in the presence of tunicamycin. NS, nonspecific band. The bar graph with a dot plot indicates the quantitative results (*, p < 0.05, one-way ANOVA). Error bars, S.D.
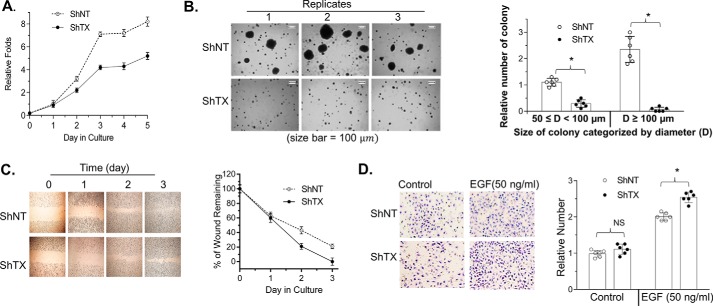
Expression of TXNDC5 stimulates anchorage-independent colony formation but inhibits cell invasion
Our previous studies demonstrate that Srx has an oncogenic function that promotes the invasion and metastasis of lung cancer cells. To further understand whether the interaction with TXNDC5 affects the oncogenic function of Srx, we examined the effect of depleting TXNDC5 on cell proliferation, migration, and invasion. For example, in A549 cells, we found that knockdown of TXNDC5 inhibited cell proliferation (Fig. 8A) and repressed anchorage colony formation in soft agar (Fig. 8B). In the wound-healing assay, we found that knockdown of TXNDC5 delayed the time for the cells to completely recover from the scratched wound (Fig. 8C). Interestingly, in the Matrigel invasion assay, we found that knockdown of TXNDC5 led to a slight increase of the cell's ability to invade through Matrigel when epidermal growth factor (EGF) was used as a chemoattractant (Fig. 8D).
Figure 8.
Knockdown of TXNDC5 in A549 cells inhibits cell proliferation but promotes cell invasion. A, representative results of cell proliferation evaluated by the modified XTT assay. B, anchorage-independent colony formation in soft agar. C and D, wound-healing assay as shown in culture (C) and quantitation (D). E and F, Matrigel invasion assay showing invaded cells (C) and quantitation (D). The bar graph with a dot plot indicates the quantitative results (*, p < 0.05; NS, no significance; one-way ANOVA). Error bars, S.D.
To further evaluate the effect of TXNDC5 on cell growth and invasion, we expressed a Myc-tagged TXNDC5 (Myc-TX) into A549 cells and established the stable cell line (Fig. S4A). We found that overexpression of TXNDC5 had a marginal effect on the growth and proliferation of A549 cells under adherent conditions (Fig. S4B), and these cells were more resistant to tunicamycin (the IC50 values for Myc-TXNDC5 cells and control vector cells were 2.6 and 1.33 μg/ml, respectively; Fig. S4C). Moreover, overexpression of TXNDC5 did not change the overall number of colonies, but the number of large colonies (those with a diameter of equal or more than 100 μm) was significantly increased (Fig. S4D). In wound-healing and Matrigel invasion assays, we found that overexpression of TXNDC5 enhanced cell migration (Fig. S4E) but inhibited Matrigel invasion induced by the addition of EGF (Fig. S4F). Therefore, these data suggest that TXNDC5 may stimulate anchorage-independent colony growth but inhibits cells' ability to invade through Matrigel in human lung cancer.
Knockdown of TXNDC5 enhances EGF-induced MAPK activation
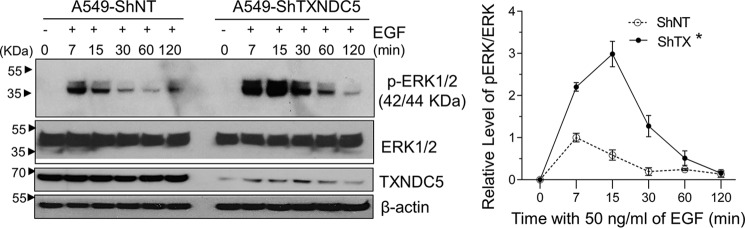
To understand why knockdown of TXNDC5 led to increased Matrigel invasion upon EGF treatment, we asked whether such an effect was due to the modulation of oncogenic signaling pathways. Control cells and shTX cells were treated with EGF and then harvested to examine the activation of MAPK in a time-dependent manner. For example, as shown in Fig. 9, treatment of control cells with EGF resulted in a temporary peak activation of ERK1/2 at 7 min and a diminishment to basal level within a course of 30 min, whereas treatment of shTX cells with the same amount of EGF led to a higher and longer period of ERK1/2 activation that lasted up to 1 h. In contrast, other signaling molecules, such as the activation of c-Jun and AKT, were not affected by the knockdown of Srx in A549 cells (Fig. S5). Taken together, these data suggest that knockdown of TXNDC5 enhances EGF-induced MAPK activation in human lung cancer cells, which may contribute to the increased invasiveness of these cells as observed above in the Matrigel invasion assay.
Figure 9.
Knockdown of TXNDC5 enhances EGF-induced MAPK activation. A549 control and TXNDC5 knockdown cells were serum-starved overnight and treated with fresh medium containing EGF for the indicated period of time. Cell lysates were harvested for Western blotting. Quantitative results of ERK1/2 activation were shown on the right. *, significant difference by comparing the area under the curve (*, p < 0.05, t test). Error bars, S.D.
Expression of TXNDC5 in human normal tissues and tumor specimens of lung cancer patients
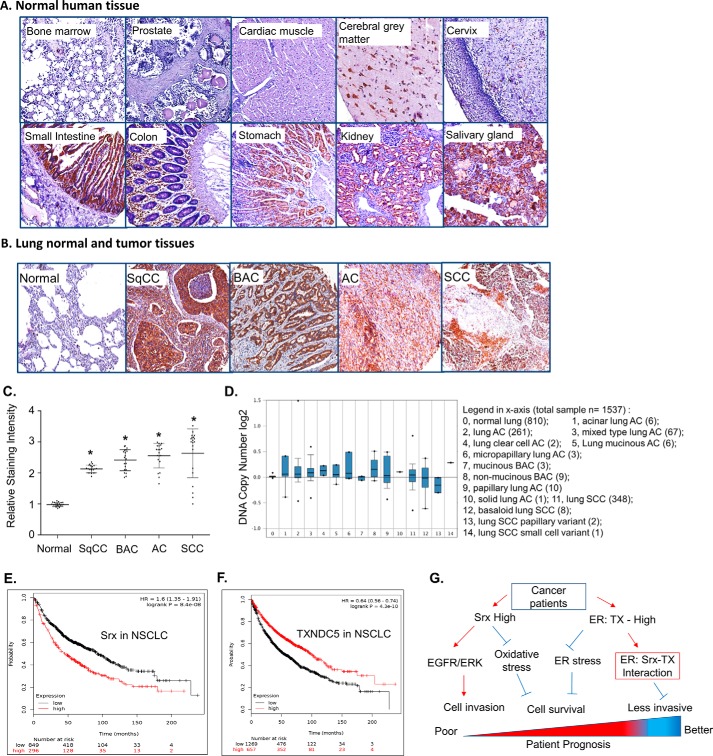
To examine the expression of TXNDC5 protein in human normal and disease tissues, we used immunohistochemical staining to visualize the endogenous levels of TXNDC5 on tissue microarray slides. These slides contain tissues prepared from a variety of human normal organs and different lung tumor specimens that were confirmed in pathology. As shown in Fig. 10A, TXNDC5 could not be detected in human normal organs, including bone marrow, prostate, cardiac muscle, and cervix. In contrast, strong positive TXNDC5 staining was consistently observed in tissues from human normal cerebral gray matter, small intestine, colon, stomach, kidney, and salivary gland. Of note, we did not find significant numbers of TXNDC5 positively stained cells in normal lung tissue. However, strongly positive staining of TXNDC5 was found in lung tumor cells, as evidenced by the dark brown color in examples of lung squamous cell carcinoma, bronchioloalveolar carcinoma, adenocarcinoma, and small cell carcinoma (Fig. 10B). Such high levels of TXNDC5 protein were commonly found in these types of human lung cancer (Fig. 10C). To our knowledge, this is the first report indicating a differential expression pattern of TXNDC5 in human lung normal and cancer specimens.
Figure 10.
Expression profile of TXNDC5 in human normal tissue and lung cancer. A and B, anti-TXNDC5 staining (brown) with counterstaining by hematoxylin (blue) was performed on tissue microarray slides of human normal organs (A) and lung normal and lung tumor specimens from patients (B). C, quantitative data from tissue microarray were shown. * p < 0.05 (t test), compared with normal lung. D, an example data set from Oncomine indicates amplified DNA copy numbers in human lung cancer. E and F, Kaplan–Meier Plotter analysis of the TCGA data set in human lung cancer indicates the negative association of Srx with patient survival (E) and the positive association of TXNDC5 with patient survival (F). G, a proposed model of Srx and TXNDC5 in lung cancer and their value as potential biomarkers to predict patient survival. Error bars, S.D.
Next, bioinformatics were used to analyze existing data sets on the molecular genetics and the clinical significance of TXNDC5 in lung cancer patients. From the Oncomine database, we found that increased expression of TXNDC5 was frequently observed in several microarray data sets when the levels were compared between normal and cancer. Such an increase was mainly due to an increase in DNA copy number as shown in an example data set (Fig. 10D). Results from analyzing the Cancer Genome Atlas (TCGA) database using Kaplan–Meier Plotter indicate that higher expression of Srx in NSCLC patients is significantly associated with poor survival (Fig. 10E). In contrast, higher expression of TXNDC5 in NSCLC patients of the same data set is significantly associated with better prognosis (Fig. 10F). Taken together, we propose a model to explain why and how the expression levels of Srx and TXNDC5 in NSCLC patients may be used as potential biomarkers to predict patient survival (Fig. 10G). In this model, increased expression of Srx contributes to lung tumorigenesis through its function of reducing oxidative stress and interacting with TXNDC5 to ease ER stress; increased expression of TXNDC5 facilitates anchorage-independent colony formation and cell survival to promote tumorigenesis. The interaction of TXNDC5 with Srx also retains a portion of Srx in the ER, thus mitigating the effect of Srx to promote MAPK signaling, leading to the inhibition of cell invasion. In the future, it may be helpful to further differentiate the functional significance of Srx and TXNDC5 at different stages of lung cancer and to incorporate this knowledge into the development of precision medicine for the treatment of lung cancer patients.
Discussion
It is well-documented that Srx is a unique enzyme with the primary biochemical function of reducing the hyperoxidized Prxs. In particular, the enzymatic activity of Srx is specific to typical 2-Cys–containing Prxs, including Prx1, -2, -3, and -4 (24). Previous studies have demonstrated that Srx is overexpressed in a variety of tumors, and it may promote cancer development through Prx-dependent as well as -independent manners. In lung cancer, elevated levels of Srx promote tumorigenesis and cancer progression by enhancing intracellular phosphokinase signaling, such as the EGFR-MAPK cascade and AP-1/MMP9 signaling. Aberrantly high expression of Srx in lung squamous cell carcinoma, lung adenocarcinoma, and pancreatic cancer is correlated with poor survival of those patients (16, 25, 26). Although the importance of Srx in lung cancer is well-established, unraveling the mechanistic details of Srx function in cancer development will further help design better approaches for targeted cancer therapeutics in the era of precision medicine.
In this study, first we revealed a novel function of Srx in the ER because knockdown of Srx sensitizes human lung cancer cells to ER stress–induced cell death. The ER is an organelle that is essential for the synthesis and correct folding of nascent peptides as well as the processing of damaged proteins. When the unfolded protein load and the recovering capability of ER are not well-balanced, ER stress occurs, and the UPR pathway is activated. The activation of the UPR helps the cell restore ER homeostasis or leads to cell death if the balance is not maintained. Tunicamycin is a well-known ER stress inducer that activates the UPR pathway. Through the inhibition of the enzyme GlcNAc phosphotransferase, treatment of cancer cells with tunicamycin leads to massive accumulation of unfolded proteins and activation of the UPR and eventually induces cell death by apoptosis (27). The UPR is an adaptive response to ER stress, and its activation is characterized by the expression of major molecular markers, including the expression of sXBP-1 and ATF6α. Accumulation of unfolded proteins in the ER stimulates inositol-requiring protein-1 (IRE1α) binding and triggers its oligomerization and autophosphorylation of the cytosolic domain. The RNase activity of IRE1α cytosolic domain is thus able to cleave an intron from XBP-1 mRNA, which allows the translation and expression of the sXBP-1 protein (28). sXBP-1 functions as a transcription factor to further activate a variety of downstream target genes to restore protein homeostasis. Therefore, the splicing of XBP-1 mRNA and the expression of sXBP-1 protein reflect the activation of the UPR mediated by the IRE1α signaling pathway. Moreover, the transcription factor ATF6 is translocated to the Golgi compartment upon ER stress. It is then cleaved by the serine protease at site-1 (the luminal domain) and by the metalloprotease at site-2 in the N terminus. The cleaved N-terminal cytosolic domain of ATF6, namely ATF6α, is then translocated into the nucleus, where it binds to target genes to activate their expression (29). Therefore, XBP-1 mRNA splicing, sXBP-1, and ATF6α protein expression are recognized as molecular markers of UPR activation in ER stress (30). Our findings of increased sensitivity of Srx knockdown cells to tunicamycin indicate a disrupted ER homeostasis that results from the accumulation of unfolded proteins. Mechanistically, an accelerated activation of the UPR pathway was observed in Srx knockdown cells, which is manifested by the characteristic activation of transcription factor ATF6α and its downstream target gene sXBP-1. In other words, these data indicate that human lung cancer cells with a high level of Srx expression will be more resilient to ER stress–induced cell death.
Although Srx can be translocated into mitochondria under oxidative stress, it is mostly known as a cytosolic protein (31, 32). This study is the first demonstration of the involvement of Srx in the maintenance of ER homeostasis. To understand its molecular mechanism, we performed MS analysis and identified proteins that interact with Srx. We found that Srx forms a complex with ER-resident protein TXNDC5. This is further investigated by examining the ER colocalization of Srx with TXNDC5 using subcellular fractionation and immunofluorescent staining, demonstration of their direct interaction by IP assays using cell lysates as well as purified recombinant proteins, and characterization of their interaction by domain mapping and site-specific mutagenesis. Our data suggest that TXNDC5 directly binds with Srx through its thioredoxin-like domains, and such interaction facilitates the ER retention of Srx. Interestingly, we found that the binding of Srx with TXNDC5 involves both disulfide bond formation and disulfide-independent interaction that is presumably based on their sequence and structural features. Mutation of cysteines in the thioredoxin domain of TXNDC5 leads to the abolishment of their interaction. To further study the nature of their interaction, we used exogenous H2O2 to induce oxidative stress, and we did not find significant changes in the Srx–TXNDC5 complex formation, which suggests that their interaction is relatively stable and is not readily affected by overall redox status change. However, it will be worthwhile to investigate in the future whether a change of the local redox status within the ER may affect their association. A recent study using a diazene-based chemical probe to profile S-sulfinylated proteins seems very useful in the identification of the substrates of Srx (33). Indeed, several ER proteins (although TXNDC5 is not included) were pulled down as potential substrates of Srx. It is thus possible that Srx in the ER may be involved in the repair of damaged proteins. In the future, a similar technique can be used to further clarify the nature of the Srx–TXNDC5 interaction. In the context of ER stress induced by tunicamycin, we found that knockdown of TXNDC5 in human lung cancer cells accelerates the activation of the UPR pathway and sensitizes cells to tunicamycin-induced cell death in a similar manner as the depletion of Srx. Taken together, these findings suggest that the Srx–TXNDC5 axis contributes to the maintenance of ER homeostasis in human lung cancer cells.
Given the critical role of Srx in the promotion of cancer cell proliferation, invasion, and metastasis, we also explored how the presence of TXNDC5 contributes to the maintenance of these oncogenic phenotypes in human lung cancer cells using loss-of-function and gain-of-function strategies. Compared with control cells, we found that knockdown of TXNDC5 leads to a significant decrease of cell growth and proliferation under standard culture conditions. Such an inhibitory effect of depleting TXNDC5 on cell proliferation may likely result from the inhibition of cell cycle progression, because previously it has been shown that expression of TXNDC5 in gastric cancer cells increases the number and percentage of cells in G2/M phase, whereas knockdown of TXNDC5 leads to the accumulation of cells in G0/G1 phase (34). Moreover, we found that knockdown of TXNDC5 in human lung cancer cells also resulted in a robust inhibition of colony formation in soft agar. The ability of cells to form visible colonies in soft agar through anchorage-independent growth is the hallmark of cell transformation and is the closest correlating factor of cancer cell tumorigenicity in vivo (35–37). Due to the limit of resource and our current focus on the report of the Srx–TXNDC5 interaction, we did not carry out a xenograft experiment. Nevertheless, these data do indicate that knockdown of TXNDC5 in human lung cancer cells may inhibit their ability to form tumors in vivo. In the future, injection of these cells to immunodeficient mice and comparison of tumor xenograft growth may be carried out to further demonstrate such a role of TXNDC5 in lung cancer development. In the context of cell migration, we found that knockdown of TXNDC5 has a relatively weak effect of inhibition in a wound-healing assay. In addition, a Matrigel invasion assay was used to evaluate the effect of depleting TXNDC5 on cell invasion. We found that knockdown of TXNDC5 does not significantly affect the basal level of cell invasion but stimulates cell invasion in the presence of EGF as chemoattractant. Furthermore, the roles of TXNDC5 in ER stress, colony formation, and cell invasion were validated using cells overexpressing TXNDC5.
In our previous studies, we have demonstrated that one of the major molecular mechanisms for Srx to promote cancer cell invasion and metastasis is through enhancing the activation of the EGFR-MAPK cascade (15). This function of Srx presumably occurs in the cytosol. As we identified that the interaction of TXNDC5 with Srx contributes to the ER retention of Srx, we hypothesized that the presence of TXNDC5 may thus have a negative effect on the activation of the MAPK cascade due to the reduced distribution of Srx to the cytosol. By comparing the levels of phosphorylated AP-1, ERK1/2, and AKT using phospho-specific antibodies, we found that knockdown of TXNDC5 in human lung cancer cells indeed enhances the activation of MAPK induced by EGF without significantly affecting the activation of AP-1 and AKT. The increased activation of MAPK in TXNDC5 knockdown cells may thus lead to an increased number of cells invading through Matrigel in a cell invasion assay. Although such an effect of TXNDC5 on cell invasion is statistically significant in a cell culture model, we did not investigate whether knockdown of TXNDC5 is sufficient to affect the tumorigenesis and metastasis of human lung cancer cells in vivo. In the future, a combination of mouse models, including a xenograft experiment through subcutaneous injection and orthotopic implantation through intrathoracic injection, will be valuable to determine the role of TXNDC5 in lung cancer growth and metastasis. In fact, such experiments have been performed in other types of human cancer cells. For example, TXNDC5 has been found to be up-regulated in prostate cancer, and its presence stimulates tumor xenograft growth in nude mice in vivo (38).
We also examined the expression of TXNDC5 in a variety of human normal organs and tumor specimens from lung cancer patients. We found a tissue-specific expression pattern of TXNDC5 in human organs, including strong positive expression in human normal cerebral gray matter, small intestine, colon, stomach, kidney, and salivary gland, whereas there was weak or no expression in other examined organs. The potential role of TXNDC5 expression in these normal organs is not clear. A recent study indicates that genomic loss of TXNDC5 does not generate any developmental defects in mice but protects them against cardiac fibrosis and contractile dysfunction (39). Compared with its nondetectable expression in human lung normal tissue, we found that TXNDC5 is highly expressed in cancer specimens, including lung squamous cell carcinoma, bronchioloalveolar carcinoma, adenocarcinoma, and small cell carcinoma. Such significantly high expression of TXNDC5 in human lung cancer may indicate its potential use as a therapeutic target or molecular diagnostic indicator for human lung cancer pathogenesis with or without the combination of Srx. In addition, previous studies indicate that nearly 85% of tumor specimens from human colon cancer have an increased level of TXNDC5 (40); similar findings were also found in the study of breast invasive ductal carcinoma, cervical squamous cell carcinoma, esophageal squamous cell carcinoma, gastric carcinoma, hepatocellular carcinoma, ovarian papillary serous carcinoma, prostate cancer, and adrenal cell carcinoma. To further evaluate the value of Srx and TXNDC5 as biomarker, we performed bioinformatics analysis using the existing data sets. Interestingly, we found that a combination of Srx and TXNDC5 expression pattern can be used to predict the survival probability of lung cancer patients.
In summary, our data indicate that Srx and TXNDC5 may play a complex role in lung tumorigenesis and cancer progression. In the future, it will be helpful to further clarify the functional significance of Srx and TXNDC5 in lung tumorigenesis: for example, whether and how their interaction in the ER contributes to the invasion and metastasis of lung cancer and whether targeting Srx alone or in combination with the inhibition of TXNDC5 can be used as a potential therapeutic strategy for the treatment of lung cancer in patients.
Experimental procedures
Cell lines and chemicals
Human embryonic kidney cells (HEK293T) and human lung cancer cell lines (A549 and H226) were purchased from ATCC. Dulbecco's modified Eagle's medium (Lonza Bio Whittaker) supplemented with 10% fetal bovine serum (Atlanta Biologicals) was used to culture HEK293T cells. RPMI 1640 medium (HyClone) supplemented with 10% fetal bovine serum was used to culture cancer cell lines. Penicillin-streptomycin solution (Thermo Scientific) (where penicillin is 100 units/ml and streptomycin 100 μg/ml) and 5 μg/ml gentamycin (Gibco, Life Technologies) were added to the medium before use. Puromycin (Gibco, Life Technologies), 1 μg/ml, was added to establish stable cell lines infected with lentiviral shRNAs. Cells were cultured in a 100% humidified atmosphere with 5% CO2 at 37 °C. Reagents including tunicamycin, H2O2, NEM, EGF, etc., were purchased primarily from Sigma-Aldrich.
Plasmid constructs, lentiviral shRNA, and stable knockdown cells
To make pcDNA 3.1/TXNDC5-Myc expression construct, total RNA was extracted from HEK293T cells following the procedure of the RNeasy Mini Kit (Qiagen). First-strand cDNA of TXNDC5 was synthesized by the ProtoScript II Reverse Transcriptase kit (New England Biolabs). The PCR was used to amplify the coding region of TXNDC5, and the PCR product was cloned into the XhoI/XbaI sites of pcDNA3.1-Myc plasmid. The viral expression vector for TXNDC5 was made by subcloning into the pLVX-IRES-Puro expression vector. All cloning sequences were verified by DNA sequencing, and protein expression was confirmed by Western blotting using specific antibody. To make stable knockdown cells, MISSION pLKO.1-puro control vector (vector control), MISSION Non-Target shRNA (shNT), and shRNAs specifically targeting TXNDC5 (shTX) were commercially obtained, and the lentiviral particles were generated following instructions in the manual (Sigma-Aldrich). Briefly, lentiviral particles were produced in HEK293T cells using the provider's plasmid packaging system and PolyJet transfection reagent (3:1). Stable cells were established and maintained in puromycin-containing medium.
RPLC-MS
Cells were transiently transfected with plasmid expressing FLAG-Srx or empty vector as control. Cells were harvested in radioimmunoprecipitation assay buffer, and anti-FLAG IPs were performed. The IP eluates were concentrated by lyophilization and then subjected to trypsin digestion and used directly for MS analysis or separated by SDS-PAGE and visualized by silver staining (Invitrogen). The differential bands in FLAG-Srx IP on silver staining were excavated and destained in the solution (50% acetonitrile, 25 mm NH4HCO3, pH 8.4). For trypsin digestion, samples were lyophilized and digested overnight at 37 °C in the digestion buffer (25 mm NH4HCO3, pH 8.4) containing 20 ng/μl trypsin. The reaction was then treated with the extraction buffer (70% acetonitrile and 5% formic acid). The extracted peptide solution was lyophilized and reconstituted in 0.1% formic acid before being subjected to the RPLC using the C-18 nano-RPLC column and the Agilent 1100 nano-LC system, which is coupled to a linear ion-trap mass spectrometer (LTQ). Peptides were eluted using a stepwise gradient of mobile phase A (0.1% formic acid in water) and B (0.1% formic acid in acetonitrile). The tandem mass spectra were searched against the UniProt human proteomic database using SEQUEST software.
Site-directed mutagenesis and RT-PCR
QuikChange II kit (Agilent Technologies) was used for site-specific mutagenesis. Primers used to make internal deletion mutants of TXNDC5 include the 5′ forward primer ATCCAGAGCGCCGCGCACAATGACCTGGGAGAC and the 3′ reverse primer GTCTCCCAGGTCATTGTGCGCGGCGCTCTGGAT to generate the N terminus thioredoxin-like domain deletion (Del1); the 5′ forward primer GTTGCACAAGGCGACCACGAGCAGCTGGCTCTGGGCCTTG and the 3′ reverse primer CAAGGCCCAGAGCCAGCTGCTCGTGGTCGCCTTGTGCAAC to generate the second thioredoxin-like domain deletion (Del2); and the 5′ forward primer CGATGACACCATTGCAGAAGGAATAACCGAGGAACTCTCTAAAAAGG and the 3′ reverse primer CCTTTTTAGAGAGTTCCTCGGTTATTCCTTCTGCAATGGTGTCATC to generate the C terminus thioredoxin-like domain deletion (Del3). All targeted mutations were verified by DNA sequencing. For RT-PCR, cells were harvested by scraping, and total RNA was extracted using the RNeasy kit (Qiagen). First-strand cDNA was synthesized using the Protoscript II Reverse Transcriptase kit (New England Biolabs). Primers for PCR included the 5′ forward primer TTACGAGAGAAAACTCATGGCC and the 3′ reverse primer GGGTCCAAGTTGTCCAGAATGC to amplify XBP-1 and the 5′ forward primer CAACGAATTTGGCTACAGCA and the 3′ reverse primer AGGGGTCTACATGGCAACTG to amplify GAPDH. PCR products were separated in 3% agarose gel by electrophoresis, stained with SYBR Green, and visualized using a UV-imaging station.
Recombinant protein purification
Srx and TXNDC5 coding regions were subcloned into pRSET(B) plasmids, which were then transformed into BL21 E. coli. Target protein expression was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside, and nickel-affinity chromatography was performed following the protocol in the protein purification kit (Qiagen). Target proteins were eluted with 250 mm imidazole buffer and dialyzed against 1× PBS buffer using the Pur-A-Lyzer Midi Dialysis Kit (Sigma). Protein samples were aliquoted in the presence of 10% glycerol and stored in −80 °C before being used.
IP, subcellular fractionation, and Western blotting
Cells were cultured in a 100-mm dish and harvested in radioimmunoprecipitation assay buffer containing 50 mm Tris at pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 1 mm EDTA, 0.25% sodium deoxycholate, 0.6 mm phenylmethylsulfonyl fluoride, and a mixture of 1% protease mixture inhibitors (Santa Cruz Biotechnology, Inc., Dallas, TX). Protein A or G magnetic beads (New England Biolabs) or anti-FLAG M2 magnetic beads (Sigma-Aldrich) were used in IP. A magnetic separator was used to collect the beads and washed in cold IP buffer five times before being resuspended in Laemmli buffer with β-mercaptoethanol. For UPR assays, cells were cultured in 35-mm dishes and treated with 1.0 μg/ml tunicamycin diluted in culture medium for 4, 6, 8, 12, or 24 h before being harvested. Total cell lysates and cytosolic and nuclear fraction were collected using a commercial kit (Thermo Scientific) following the manufacturer's suggested protocol. For ER fractionation, cells were lysed in chilled homogenization buffer (0.25 m sucrose, 25 mm KCl, 10 mm HEPES) with a freshly added mixture of protease inhibitors. The lysates were homogenized using glass beads and the MiniBeadBeater-16 (BioSpec, model 607). Samples were centrifuged at 15,000 rpm at 4 °C for 15 min to remove mitochondrial, nuclear, and cytoskeletal fractions. The supernatant was centrifuged at 35,000 rpm for 70 min, and the microsomal fractions were washed with 1× PBS by centrifugation at 35,000 rpm for 10 min. The sediments were lysed in Laemmli sample buffer with β-mercaptoethanol.
For Western blotting, proteins were separated in SDS-polyacrylamide gel by electrophoresis and transferred to polyvinylidene difluoride membrane. Membranes were blocked for 1 h in 5% nonfat dry milk in Tris buffer before the overnight incubation with diluted primary antibody. The membrane was then washed with TBST and incubated for 1 h with the horseradish peroxidase–conjugated secondary antibody. After multiple washing steps, the signal was detected using Western Dura chemiluminescence substrate (Pierce), and bands were visualized onto X-ray film. Primary antibodies used include anti-Srx (Proteintech, Chicago, IL) anti-TXNDC5, anti-calnexin, anti-GAPDH, anti-TXNDC7, anti-XBP-1, anti-ATP5A, anti-methylated histone H3 (Santa Cruz Biotechnology), anti-ATF6α (Abcam), and anti-FLAG, anti-β-actin (Sigma-Aldrich). Antibodies for phosphorylated c-Jun, total c-Jun, phosphorylated ERK1/2, total ERK1/2, phosphorylated AKT, and total AKT were from Cell Signaling (Billerica, MA).
Prediction of interacting domains by molecular docking
Molecular docking was used to predict protein–protein interaction according to previously published methods (41–43). Briefly, protein sequences of Srx and TXNDC5 were uploaded to the program I-TASSER. The structure with the highest score was used in the molecular docking and imported to the ZDOC server. Molecular visualization of the results was prepared using PyMOL software (Schroedinger, LLC). The model with the highest score was used in the experiments to predict the domains involved in the Srx–TXNDC5 complex formation.
XTT assay, clonogenic assay, anchorage-independent colony formation, wound-healing assay, and Matrigel invasion assay
The XTT assay was performed using Trevigen's TACS XTT cell proliferation and viability assay kit. Absorbance was measured at 490 nm with 600 nm as reference. In the cell proliferation assay, the values are plotted as -fold increase in growth in each day compared with day 0 (mean ± S.D.). In the cell viability assay, confluent cells were treated with different concentrations of tunicamycin (μg/ml) for 24 h before the absorbance was measured. For the clonogenic assay, cells were seeded onto 6-well plates (1000 cell/well), and tunicamycin treatment was started the next day and maintained. Culture medium was changed every 3 days with fresh tunicamycin. Cells were cultured for 10 days before being stained with crystal violet. Colonies with more than 50 cells were counted using the ImageJ software. For the anchorage-independent colony formation assay, cells suspended in 0.3% agar were seeded on top of a layer of 0.6% agar. Cells were cultured for 30 days at 37 °C with medium changed every 4 days. Colonies were fixed by methanol and stained with 0.25% Crystal Violet. Colonies were examined under a ×4 microscope, and those with a diameter > 50 μm were counted and recorded for data analysis. For the wound-healing assay, cells were cultured until confluence, and the wound was made using a 200-μl tip, perpendicular to the surface of the cells. Floating cells were removed by extensive washing. Images of wound healing were taken each 24 h, and data were measured through imaging analysis. Transwell Matrigel invasion assays using a BD invasion chamber were performed following the manufacturer's suggested protocol with or without 50 ng/ml EGF as chemo-attractant. Invaded cells were stained by Diff-QuikTM staining, and images were taken under a microscope. Numbers of invaded cells were counted using the ImageJ software.
Immunofluorescence imaging and immunohistochemistry (IHC) staining
For immunofluorescence imaging, cells were cultured in a chamber slide and then fixed with chilled methanol for 10 min. After PBS washing, cells were incubated in 5% goat serum blocking buffer and then incubated with primary and fluorescence-labeled secondary antibodies. A drop of 4′,6-diamidino-2-phenylindole–containing ProLong antifade solution was added to the slide and covered with a coverslip, which was then examined by using the confocal microscope (Nikon). For IHC staining, human lung normal and cancer tissue microarray slides were commercially obtained (Biomax US). Slides of a multiple-organ normal tissue array and lung disease spectrum tissue microarray were used. The lung cancer slides contained at least 20 cases each for tumor-adjacent normal lung tissue, squamous cell carcinoma, bronchioloalveolar carcinoma and adenocarcinoma and 15 cases of small cell carcinoma. Tissue slides were rehydrated in xylene, and antigen retrieval was processed in the buffer containing 1× target retrieval solution (Dako), 90% glycerol, and 1 mm EDTA. Primary anti-TXNDC5 antibody was used in a dilution of 1:50, and the IHC staining was carried out using the Dako LSAB2 System–horseradish peroxidase kit following the manufacturer's suggested protocol. After counterstaining with hematoxylin, positive anti-TXNDC5 staining appeared brown in color (3,3′-diaminobenzidine was used as the chromatogen). The intensity of the positive staining was analyzed by using Image Scope software (version 11.2.0; Aperio Technologies).
Statistical and bioinformatics analysis
Quantitative data were obtained from the measurement of band intensity or cell or colony number using the ImageJ software with a combination of plug-ins. All data are presented as mean ± S.D. (x̄ ± S.D.). Data were analyzed, and graphs of original data sets were made using the software GraphPad Prism (version 6.01). Statistical analyses were carried out in GraphPad Prism using data transformation, linear regression, Student's t test, paired t test, and one-way or two-way analysis of variance (ANOVA) as specified in each result. In all analysis, the p value was calculated using a two-tailed 95% confidence interval, and the p value of ≤0.05 was considered statistically significant. Bioinformatics analysis was carried using the open source tools, including Oncomine, cBioportal, and Kaplan–Meier Plotter to process currently available cancer genomic data sets from microarrays and TCGA. Example results from these analysis are shown in this study.
Author contributions
H. A. C., H. J., Q. Y., N. D., P. T., and Q. W. data curation; H. A. C., H. J., and Q. W. formal analysis; H. A. C., H. J., and Q. W. investigation; H. A. C., H. J., Q. Y., and Q. W. methodology; H. A. C., H. J., and Q. W. writing-original draft; H. J., Q. Y., N. D., P. T., and Q. W. validation; H. J. and Q. W. project administration; Q. W. conceptualization; Q. W. resources; Q. W. supervision; Q. W. funding acquisition; Q. W. visualization; Q. W. writing-review and editing.
Supplementary Material
Acknowledgments
We thank Dr. Jianhang Jia and Garretson Epperly (Markey Cancer Center) for assistance with the confocal imaging.
This work was supported in part by NCI, National Institutes of Health, Grant R01CA222596; Department of Defense Grant W81XWH-16-1-0203; American Cancer Society Grant RSG-16-213-01-TBE; and Kentucky Lung Cancer Research Program Grant KLCRP2016. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or other funding agencies.
This article contains Table S1 and Figs. S1–S5.
- Prx
- peroxiredoxin
- Srx
- sulfiredoxin
- TXNDC5
- thioredoxin domain–containing protein 5
- UPR
- unfolded protein response
- PDI
- protein-disulfide isomerase
- XTT
- sodium(2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide
- IP
- immunoprecipitation
- RPLC
- reverse-phase liquid chromatography
- AP-1
- activator protein-1
- ATF6α
- activating transcription factor 6α
- XBP-1
- X-box–binding protein 1
- MAPK
- mitogen-activated protein kinase
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- ER
- endoplasmic reticulum
- NEM
- N-ethylmaleimide
- EGF
- epidermal growth factor
- EGFR
- EGF receptor
- IHC
- immunohistochemistry
- ANOVA
- analysis of variance
- TCGA
- the Cancer Genome Atlas.
References
- 1. Kinnula V. L., Pääkkö P., and Soini Y. (2004) Antioxidant enzymes and redox regulating thiol proteins in malignancies of human lung. FEBS Lett. 569, 1–6 10.1016/j.febslet.2004.05.045 [DOI] [PubMed] [Google Scholar]
- 2. Hampton M. B., Vick K. A., Skoko J. J., and Neumann C. A. (2018) Peroxiredoxin Involvement in the Initiation and Progression of Human Cancer. Antioxid. Redox Signal. 28, 591–608 10.1089/ars.2017.7422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jiang H., Wu L., Mishra M., Chawsheen H. A., and Wei Q. (2014) Expression of peroxiredoxin 1 and 4 promotes human lung cancer malignancy. Am. J. Cancer Res. 4, 445–460 [PMC free article] [PubMed] [Google Scholar]
- 4. Kim J. H., Bogner P. N., Baek S. H., Ramnath N., Liang P., Kim H. R., Andrews C., and Park Y. M. (2008) Up-regulation of peroxiredoxin 1 in lung cancer and its implication as a prognostic and therapeutic target. Clin. Cancer Res. 14, 2326–2333 10.1158/1078-0432.CCR-07-4457 [DOI] [PubMed] [Google Scholar]
- 5. Hwang J. A., Song J. S., Yu D. Y., Kim H. R., Park H. J., Park Y. S., Kim W. S., and Choi C. M. (2015) Peroxiredoxin 4 as an independent prognostic marker for survival in patients with early-stage lung squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 8, 6627–6635 [PMC free article] [PubMed] [Google Scholar]
- 6. Ramming T., and Appenzeller-Herzog C. (2013) Destroy and exploit: catalyzed removal of hydroperoxides from the endoplasmic reticulum. Int. J. Cell Biol. 2013, 180906 10.1155/2013/180906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kakihana T., Nagata K., and Sitia R. (2012) Peroxides and peroxidases in the endoplasmic reticulum: integrating redox homeostasis and oxidative folding. Antioxid. Redox Signal. 16, 763–771 10.1089/ars.2011.4238 [DOI] [PubMed] [Google Scholar]
- 8. Biteau B., Labarre J., and Toledano M. B. (2003) ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature 425, 980–984 10.1038/nature02075 [DOI] [PubMed] [Google Scholar]
- 9. Woo H. A., Chae H. Z., Hwang S. C., Yang K.-S., Kang S. W., Kim K., and Rhee S. G. (2003) Reversing the inactivation of peroxiredoxins caused by cysteine sulfinic acid formation. Science 300, 653–656 10.1126/science.1080273 [DOI] [PubMed] [Google Scholar]
- 10. Chang T. S., Jeong W., Woo H. A., Lee S. M., Park S., and Rhee S. G. (2004) Characterization of mammalian sulfiredoxin and its reactivation of hyperoxidized peroxiredoxin through reduction of cysteine sulfinic acid in the active site to cysteine. J. Biol. Chem. 279, 50994–51001 10.1074/jbc.M409482200 [DOI] [PubMed] [Google Scholar]
- 11. Kim J., Lee G. R., Kim H., Jo Y. J., Hong S. E., Lee J., Lee H. I., Jang Y. S., Oh S. H., Lee H. J., Lee J. S., and Jeong W. (2016) Effective killing of cancer cells and regression of tumor growth by K27 targeting sulfiredoxin. Free Radic. Biol. Med. 101, 384–392 10.1016/j.freeradbiomed.2016.11.001 [DOI] [PubMed] [Google Scholar]
- 12. Baek J. Y., Han S. H., Sung S. H., Lee H. E., Kim Y. M., Noh Y. H., Bae S. H., Rhee S. G., and Chang T. S. (2012) Sulfiredoxin protein is critical for redox balance and survival of cells exposed to low steady-state levels of H2O2. J. Biol. Chem. 287, 81–89 10.1074/jbc.M111.316711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Soriano F. X., Léveillé F., Papadia S., Higgins L. G., Varley J., Baxter P., Hayes J. D., and Hardingham G. E. (2008) Induction of sulfiredoxin expression and reduction of peroxiredoxin hyperoxidation by the neuroprotective Nrf2 activator 3H-1,2-dithiole-3-thione. J. Neurochem. 107, 533–543 10.1111/j.1471-4159.2008.05648.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wei Q., Jiang H., Matthews C. P., and Colburn N. H. (2008) Sulfiredoxin is an AP-1 target gene that is required for transformation and shows elevated expression in human skin malignancies. Proc. Natl. Acad. Sci. U.S.A. 105, 19738–19743 10.1073/pnas.0810676105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wei Q., Jiang H., Xiao Z., Baker A., Young M. R., Veenstra T. D., and Colburn N. H. (2011) Sulfiredoxin-Peroxiredoxin IV axis promotes human lung cancer progression through modulation of specific phosphokinase signaling. Proc. Natl. Acad. Sci. U.S.A. 108, 7004–7009 10.1073/pnas.1013012108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Merikallio H., Pääkkö P., Kinnula V. L., Harju T., and Soini Y. (2012) Nuclear factor erythroid-derived 2-like 2 (Nrf2) and DJ1 are prognostic factors in lung cancer. Hum. Pathol. 43, 577–584 10.1016/j.humpath.2011.05.024 [DOI] [PubMed] [Google Scholar]
- 17. Lu J., and Holmgren A. (2014) The thioredoxin superfamily in oxidative protein folding. Antioxid. Redox Signal. 21, 457–470 10.1089/ars.2014.5849 [DOI] [PubMed] [Google Scholar]
- 18. Chawsheen H. A., Ying Q., Jiang H., and Wei Q. (2018) A critical role of the thioredoxin domain containing protein 5 (TXNDC5) in redox homeostasis and cancer development. Genes Dis. 5, 312–322 10.1016/j.gendis.2018.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oslowski C. M., and Urano F. (2011) Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods Enzymol. 490, 71–92 10.1016/B978-0-12-385114-7.00004-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kennedy D., Samali A., and Jäger R. (2015) Methods for studying ER stress and UPR markers in human cells. Methods Mol. Biol. 1292, 3–18 10.1007/978-1-4939-2522-3_1 [DOI] [PubMed] [Google Scholar]
- 21. Yang J., and Zhang Y. (2015) Protein structure and function prediction using I-TASSER. Curr. Protoc. Bioinformatics 52, 5.8.1–15 10.1002/0471250953.bi0508s52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jönsson T. J., Johnson L. C., and Lowther W. T. (2008) Structure of the sulphiredoxin-peroxiredoxin complex reveals an essential repair embrace. Nature 451, 98–101 10.1038/nature06415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jeong W., Park S. J., Chang T. S., Lee D. Y., and Rhee S. G. (2006) Molecular mechanism of the reduction of cysteine sulfinic acid of peroxiredoxin to cysteine by mammalian sulfiredoxin. J. Biol. Chem. 281, 14400–14407 10.1074/jbc.M511082200 [DOI] [PubMed] [Google Scholar]
- 24. Woo H. A., Jeong W., Chang T. S., Park K. J., Park S. J., Yang J. S., and Rhee S. G. (2005) Reduction of cysteine sulfinic acid by sulfiredoxin is specific to 2-cys peroxiredoxins. J. Biol. Chem. 280, 3125–3128 10.1074/jbc.C400496200 [DOI] [PubMed] [Google Scholar]
- 25. Kim Y. S., Lee H. L., Lee K. B., Park J. H., Chung W. Y., Lee K. S., Sheen S. S., Park K. J., and Hwang S. C. (2011) Nuclear factor E2-related factor 2 dependent overexpression of sulfiredoxin and peroxiredoxin III in human lung cancer. Korean J. Intern. Med. 26, 304–313 10.3904/kjim.2011.26.3.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Soini Y., Eskelinen M., Juvonen P., Karja V., Haapasaari K. M., Saarela A., and Karihtala P. (2014) Nuclear Nrf2 expression is related to a poor survival in pancreatic adenocarcinoma. Pathol. Res. Pract. 210, 35–39 10.1016/j.prp.2013.10.001 [DOI] [PubMed] [Google Scholar]
- 27. Noda I., Fujieda S., Seki M., Tanaka N., Sunaga H., Ohtsubo T., Tsuzuki H., Fan G. K., and Saito H. (1999) Inhibition of N-linked glycosylation by tunicamycin enhances sensitivity to cisplatin in human head-and-neck carcinoma cells. Int. J. Cancer. J. Int. Cancer 80, 279–284 [DOI] [PubMed] [Google Scholar]
- 28. Hassler J., Cao S. S., and Kaufman R. J. (2012) IRE1, a double-edged sword in pre-miRNA slicing and cell death. Dev. Cell 23, 921–923 10.1016/j.devcel.2012.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bailey D., and O'Hare P. (2007) Transmembrane bZIP transcription factors in ER stress signaling and the unfolded protein response. Antioxid. Redox Signal. 9, 2305–2321 10.1089/ars.2007.1796 [DOI] [PubMed] [Google Scholar]
- 30. Han J., and Kaufman R. J. (2017) Physiological/pathological ramifications of transcription factors in the unfolded protein response. Genes Dev. 31, 1417–1438 10.1101/gad.297374.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Noh Y. H., Baek J. Y., Jeong W., Rhee S. G., and Chang T. S. (2009) Sulfiredoxin translocation into mitochondria plays a crucial role in reducing hyperoxidized peroxiredoxin III. J. Biol. Chem. 284, 8470–8477 10.1074/jbc.M808981200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kil I. S., Ryu K. W., Lee S. K., Kim J. Y., Chu S. Y., Kim J. H., Park S., and Rhee S. G. (2015) Circadian oscillation of sulfiredoxin in the mitochondria. Mol. Cell 59, 651–663 10.1016/j.molcel.2015.06.031 [DOI] [PubMed] [Google Scholar]
- 33. Akter S., Fu L., Jung Y., Conte M. L., Lawson J. R., Lowther W. T., Sun R., Liu K., Yang J., and Carroll K. S. (2018) Chemical proteomics reveals new targets of cysteine sulfinic acid reductase. Nat. Chem. Biol. 14, 995–1004 10.1038/s41589-018-0116-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang L., Hou Y., Li N., Wu K., and Zhai J. (2010) The influence of TXNDC5 gene on gastric cancer cell. J. Cancer Res. Clin. Oncol. 136, 1497–1505 10.1007/s00432-010-0807-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Colburn N. H., Bruegge W. F., Bates J. R., Gray R. H., Rossen J. D., Kelsey W. H., and Shimada T. (1978) Correlation of anchorage-independent growth with tumorigenicity of chemically transformed mouse epidermal cells. Cancer Res. 38, 624–634 [PubMed] [Google Scholar]
- 36. Freedman V. H., and Shin S. I. (1974) Cellular tumorigenicity in nude mice: correlation with cell growth in semi-solid medium. Cell 3, 355–359 10.1016/0092-8674(74)90050-6 [DOI] [PubMed] [Google Scholar]
- 37. Shin S. I., Freedman V. H., Risser R., and Pollack R. (1975) Tumorigenicity of virus-transformed cells in nude mice is correlated specifically with anchorage independent growth in vitro. Proc. Natl. Acad. Sci. U.S.A. 72, 4435–4439 10.1073/pnas.72.11.4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang L., Song G., Chang X., Tan W., Pan J., Zhu X., Liu Z., Qi M., Yu J., and Han B. (2015) The role of TXNDC5 in castration-resistant prostate cancer-involvement of androgen receptor signaling pathway. Oncogene 34, 4735–4745 10.1038/onc.2014.401 [DOI] [PubMed] [Google Scholar]
- 39. Shih Y. C., Chen C. L., Zhang Y., Mellor R. L., Kanter E. M., Fang Y., Wang H. C., Hung C. T., Nong J. Y., Chen H. J., Lee T. H., Tseng Y. S., Chen C. N., Wu C. C., Lin S. L., et al. (2018) Endoplasmic reticulum protein TXNDC5 augments myocardial fibrosis by facilitating extracellular matrix protein folding and redox-sensitive cardiac fibroblast activation. Circ. Res. 122, 1052–1068 10.1161/CIRCRESAHA.117.312130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang Y., Ma Y., Lu B., Xu E., Huang Q., and Lai M. (2007) Differential expression of mimecan and thioredoxin domain-containing protein 5 in colorectal adenoma and cancer: a proteomic study. Exp. Biol. Med. (Maywood) 232, 1152–1159 10.3181/0701-RM-8 [DOI] [PubMed] [Google Scholar]
- 41. Roy A., Kucukural A., and Zhang Y. (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang J., Yan R., Roy A., Xu D., Poisson J., and Zhang Y. (2015) The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8 10.1038/nmeth.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pierce B. G., Wiehe K., Hwang H., Kim B.-H., Vreven T., and Weng Z. (2014) ZDOCK server: interactive docking prediction of protein–protein complexes and symmetric multimers. Bioinformatics 30, 1771–1773 10.1093/bioinformatics/btu097 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.